Abstract
Powdery mildew caused by the obligate biotrophic fungus Podosphaera xanthii poses a serious threat to melon (Cucumis melo L.) production worldwide. Frequent occurrences of the disease in different regions of South Korea hints at the potential existence of several races which need to be identified. The races of five isolates collected from different powdery mildew affected regions were identified based on the pathogenicity tests of these isolates on eight known differential melon cultigens namely, SCNU1154, PMR 45, WMR 29, PMR 5, MR-1, PI124112, Edisto 47 and PI414723. None of the isolates have shown same disease responses to those of the known races tested in this study and in previous reports on these identical differential melon cultigens. This indicates that the tested uncharacterized isolates are new races. Among the isolates, the isolates from Hadong, Buyeo, Yeongam and Gokseong have shown same pathogenicity indicating the possibility of these isolates being one new race, for which we propose the name ‘race KN1’. The isolate of Janghueng have also shown unique disease response in the tested differential melon cultigens and hence, we identified it as another new race with a proposed name ‘race KN2’. Report of these new races will be helpful in taking effective control measures in prevalent regions and for future breeding programs aimed at developing varieties that are resistant to these race(s).
Keywords: melon, pathogenicity, Podosphaera xanthii, powdery mildew, race
Cucurbits greatly suffer from the chronic powdery mildew disease. Multiple species of the order Erysiphales can cause this disease on cucurbits (McGrath, 1997). On melon (Cucumis melo L.), the disease has been reported to be caused by two Erysiphaceae fungi namely, Podosphaera xanthii (formerly Sphaerotheca fuliginea) and Golovinomyces cichoracearum (formerly Erysiphe cichoracearum) with the former being most prevalent and devastating particularly in Asia (Cohen et al., 2004; Jahn et al., 2002; Kasiamdari et al., 2016; Kuzuya et al., 2006). The disease is easily distinguishable by the characteristic white flour-like fungal structures on leaves and stems (Pérez-García et al., 2009). In the severely affected plants, leaves become chlorotic and wrinkled, canopy decreases and quality and yield of fruits are reduced (Keinath and Dubose, 2004; McGrath and Thomas, 1996). Infection at initial stages is detrimental to plant itself or can drastically reduce fruit yield whereas infection at later stages generally reduces fruit quality. The disease thrives in moderate temperature and high humidity which usually make greenhouse atmosphere more prone to the disease (Huang et al., 2000). The fact that climate is being changed to more subtropical nature and the greenhouses are being used increasingly, melon cultivation in South Korea is more threatened by increasing outbreak of the disease (Lee et al., 2010).
Since its appearance on melon in 1925, the commercial cultivation of melon is in a biological “arms race” against powdery mildew with the reports of occurrence of this disease in several countries including Czechoslovakia (Lebeda, 1983; Lebeda et al., 2004), Israel (Cohen et al., 1984), United States (McCreight and Coffey, 2011; McCreight et al., 1987; Sowell, 1982), France (Epinat et al., 1992), Sudan (Mohamed et al., 1995), Japan (Hosoya et al., 1999), Czech Republic (Křístková et al., 2004) and Spain (Pérez-García et al., 2009; Del Pino et al., 2002) etc. Several races of the fungus were subsequently identified based on their pathogenicity or virulence on known resistant cultigens. A total of 28 different races of P. xanthii was reported to cause powdery mildew worldwide (McCreight, 2006). The strain that failed to overcome the resistance of PMR45, the most resistant melon cultivar of that time (Jagger et al., 1938), was named as ‘race 1’ while the strain that overcame the resistance of PMR45 was named as ‘race 2’. The name ‘race 3’ was given to the strain that broke the resistance of another resistant variety, PMR5 (Hosoya et al., 1999; Pryor et al., 1946). Similarly, race 4 & 5 were named for strains that broke the resistance of other resistant varieties in France (Bardin et al., 1997). These races were reported in several countries including race 1 and 2 in Spain and Israel (Cohen and Eyal, 1988; Kenigsbuch and Cohen, 1987), Israel (Cohen et al., 1984), Greece (Vakalounakis and Klironomou, 1995), United States (McCreight and Coffey, 2011; McCreight et al., 1987; Sowell, 1982), and race 3 in Israel (Cohen et al., 1996). on watermelon, two pathogenically distinct races namely, 1W and 2W were reported in the United States (Davis et al., 2001, 2007; Tetteh et al., 2010). In Japan, race 1, N1, N2 and 5 were first reported by Hosoya et al. (1999) and after that Kuzuya et al. (2004, 2006) identified seven races namely, race 1 through race 7 against eight selected melon lines (PMR45, Fuyu3, MR29, PI414723, PMR5, Edisto47, PI124112 and MR-1). Later, Izumikawa et al. (2008) reported three additional races namely, race A, S and O which were identified using a modified version of the method described by Bardin et al (1999b). In addition, few sub races namely, race 2US, race 3.5, and race 4.5 were identified in the US (McCreight and Coffey, 2011).
Despite the continual efforts by breeders, recurrent outbreak of the disease is reported in major melon producing countries of the world which is mainly manifested by the breakdown of the existing resistance by the appearance of new physiological races of the fungus. The classical example of the resistance breakdown is observed in US where only two races of P. xanthii were known before 2003 when a new race S were identified. Since then, these three races occurred in different regions in different years (CucCAP, 2017). Also in South Korea, increased occurrence of the disease was observed and more and more melon growing areas are being affected by the disease every year. Besides, breakdown of resistance of existing commercial resistant varieties were observed which raised the speculation of the appearance of new powdery mildew races. Such widespread occurrence of this disease in different regions indicates the existence of several races or pathotypes in South Korea which are yet to be thoroughly characterized. Recently, Kim et al. (2016a) reported the appearance of three potential new races of powdery mildew fungus on melon which necessitated the constant monitoring and characterization of the newly appeared races. Safeguarding the melon production requires to tackle the disease either by chemical control measure or by developing resistant cultivars with the latter being the most sustainable, inexpensive and environmentally friendly approach. Identification of the newly appeared races are thus important so that race specific, preferably multi-race resistant varieties can be developed for different regions based on the pathogen profile of that regions. This study reports the identification of two uncharacterized races from major melon growing areas based on the pathogenicity of the races in standard resistant cultivars collected from all around the world.
Materials and Methods
Plant materials and growing conditions
For identification of the unknown races of P. xanthii, the disease response of a set of eight differential standard melon cultigens namely, SCNU1154, PMR 45, WMR 29, PMR 5, MR-1, PI124112, Edisto 47 and PI414723 were used (Table 1). These cultigens were collected from various sources as indicated in Kim et al (2016b). Seeds were germinated in plastic pots containing garden soil in a culture room at a controlled day/night temperature of 25/20°C, 16 h day length, 80–120 μmol.m−2.s−1 light intensity and 65% relative humidity without any agro-chemical based protection.
Table 1.
In vitro disease response (leaf disc bioassay) of eight differential melon genotypes against different races of powdery mildew fungus Podosphaera xanthii. S, susceptible; R, Resistance; I, Intermediate
| Races | KN1 | KN2 | N1 | N1 | N1 | N1 | N1 | N1 | 1 | 1 | 1 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locations | Hadong | Buyeo | Yeongam | Gokseong | Janghueng | Namwon | Damyang | Yeoju | Hadong | Changnyeong | Suncheon | Changnyeong | Hadong | Ansung | |
| Cultigens | SCNU1154 | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| PMR 45 | S | S | S | S | S | R | R | R | R | R | R | R | R | R | |
| WMR 29 | S | S | S | S | S | R | R | R | R | R | R | R | R | R | |
| PMR 5 | S | S | S | S | R | R | R | R | R | R | R | R | R | R | |
| MR-1 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| PI124112 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| Edisto 47 | S | S | S | S | R | R | R | R | R | R | R | R | R | R | |
| PI414723 | S | S | S | S | S | S | S | S | S | S | S | R | R | R | |
Collection of isolates
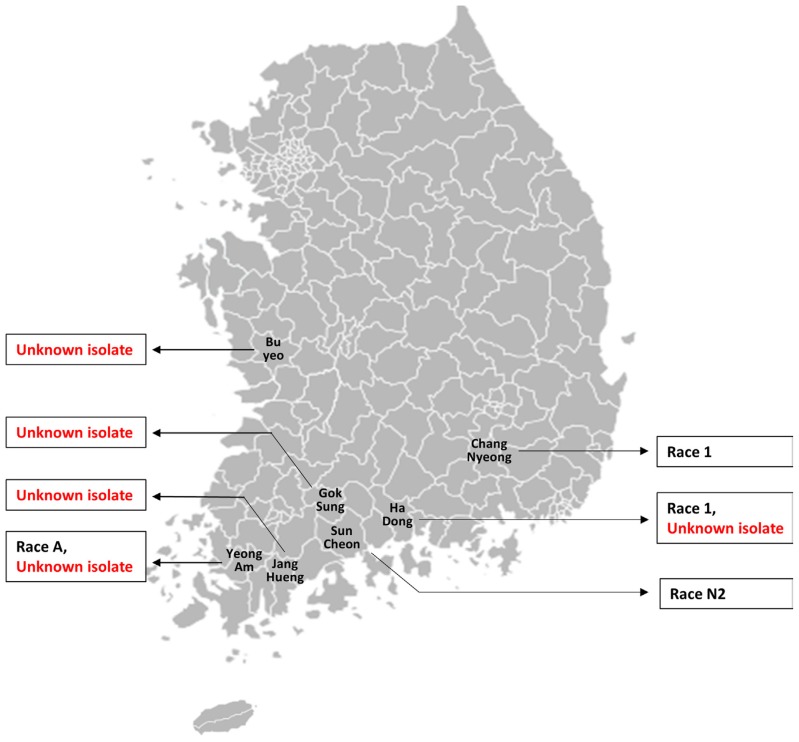
Several uncharacterized isolates of the powdery mildew fungus were collected from the affected melon farms of different regions of South Korea namely, Hadong, Buyeo, Yeongam, Gokseong and Janghueng (Fig. 1). Besides, few known races were also isolated from various regions such as Namwon, Damyang, Yeoju, Hadong, Changnyeong, Suncheon and Ansung to identify if the same race is available (along with those uncharacterized isolates) in the affected region (Table 1).
Fig. 1.
Regional distribution of known races and isolates of Podosphaera xanthii that were collected from powdery mildew infested melon farms for testing against standard differential cultigens. Known race 1, race N2 and race A were reported in the year 2013 by Kim et al (2016a). The isolates that were characterized in this study were shown in red colored text.
Incubation and inoculation
The pathogens were preliminary identified as P. xanthii based on their morphological characters under light microscopic examination (data not shown). The biotropic fungal isolates were maintained in vitro on cut leaves of melon placed on water agar medium in square petridishes as per the method described by Bertrand (2002). The fungus was sub-cultured seven times from the areas of cut leaves showing typical powdery mildew symptoms to obtain pure isolates. The culture was maintained in growth room with controlled day/night temperature (25/20°C), relative humidity (65%) and light intensity (80–120 μmol.m−2.s−1) at a photoperiod of 16 h.
For the pathogenicity test, the upper surface of the leaf discs (2 × 2 cm) of differential melon cultigens were inoculated by tapping and dusting (surface contact) of the sporulating mycelia (2–3 weeks old), originating from cotyledons of C. sativus cv. Marketer, with a tree stick as a modified version of the techniques described by Bardin et al (1999a). For each cultigens five leaf discs were inoculated which were then incubated in a growth chamber as described above.
Assessment of pathogenicity
The disease response on the melon lines were characterized visually as resistant, susceptible and intermediate based on the severity of the spread of fungal spores on the leaf discs as estimated by the percentage of infected area of leaf discs (0%–10%, 11%–30%, and 31%–100%, respectively) after two weeks of inoculation. The experiment was repeated five times.
Results
Disease response against two known races, race N1 and race 1
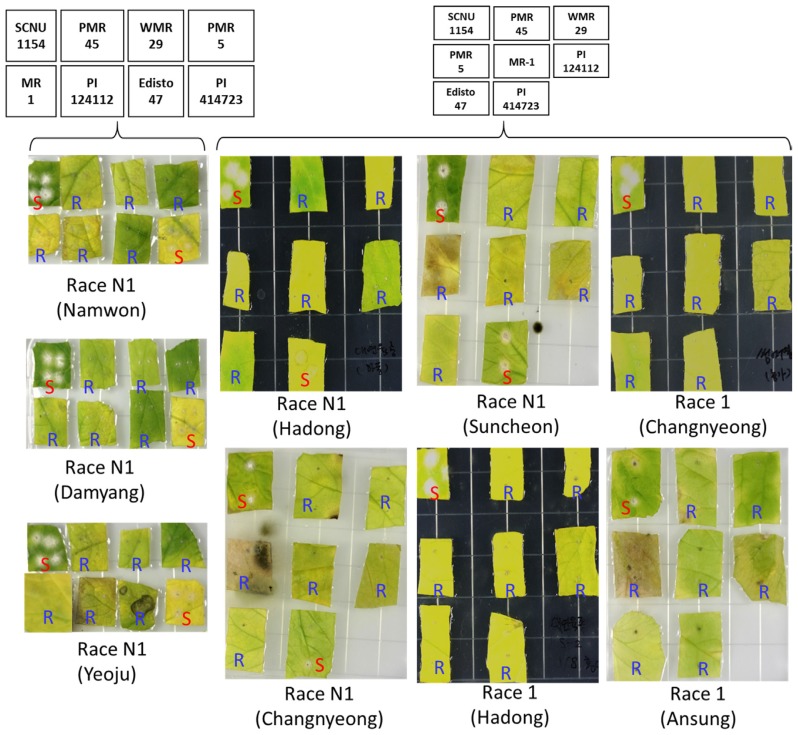
The pathogenicity of the collected known races (race N1 from Namwon, Damyang, Yeoju, Hadong, Changnyeong and Suncheon regions and race 1 from Changnyeong, Hadong and Ansung regions) was tested against eight well characterized differential cultigens in in vitro condition using cut leaf disc experiment and the appearance of characteristic powdery mildew symptom was visually assessed after two weeks of inoculation. Despite being collected from different regions, each of the races showed consistent pathogenicity on the tested genotypes (Fig. 2, Table 1). Race N1 (of all six regions) caused disease only on SCNU1154 and PI414723 while the remaining six cultigens were resistant against this race. Race 1 (of all three regions), on the other hand were only able to cause disease on the cut leaves of SCNU1154 with the other 7 cultigens being resistant against this race (Fig. 2, Table 1). This consistency in the pathogenicity of a particular race indicates its uniqueness in the ability to cause disease on a set of differential cultivars despite being collected from different regions. This indicates towards the fact that any unknown race can be identified based on their pathogenicity on these differential cultivars.
Fig. 2.
Disease response of eight well characterized differential melon genotypes against two known races of powdery mildew fungus Podosphaera xanthii namely, Race N1 and Race 1 collected from six and three different locations, respectively as determined by invitro bioassay using leaf discs after 2 weeks of inoculation. Conidial growth was observed in susceptible cultigens and are indicated by red ‘S’ letter. The layout of leaf discs samples in petri-dishes are shown in right. S, susceptible; R, Resistance. The experiment were repeated five times and similar responses were observed every time.
It is noteworthy that the pathogenicity of the known races, race N1 and race 1 studied in the current work were compared with that of their previous reports on same differential cultivars to reconfirm if the races show same pathogenicity. This is necessary as the consistency of the races in terms of pathogenicity should be checked prior to determine any new race(s) based on this standard disease causing ability against known differential cultigens. It was observed that the race N1 and race 1 (of all regions) showed same pathogenicity to that of their previous reports (Hosoya et al., 1999; Izumikawa et al., 2009; Kim et al., 2016b; McCreight, 2006). This confirmed the consistency of the pathogenicity of the races in differential cultigens to which the uncharacterized races can now be confidently compared.
Disease response against the collected isolates
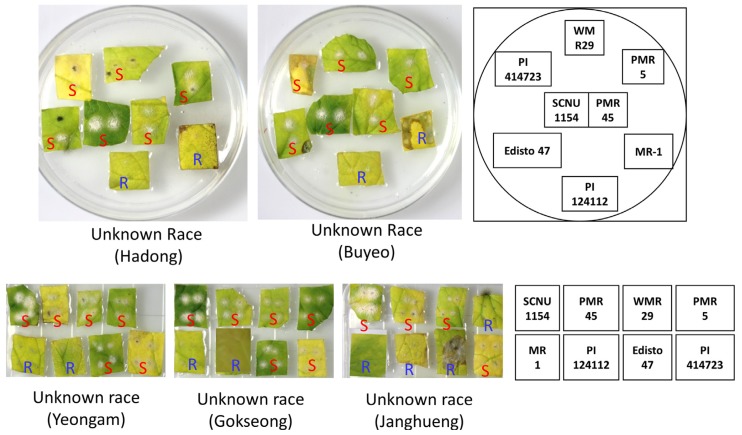
In the same experimental setting, the pathogenicity of the collected isolates were tested against the same set of differential melon cultigens. It was observed that the newly collected isolates of Hadong, Buyeo, Yeongam and Gokseong regions showed same patterns of pathogenicity in the differential melon cultigens (Fig. 3, Table 1). Only the cultigens, MR-1 and PI124112 have shown resistance against these isolates of Hadong, Buyeo, Yeongam and Gokseong regions while the other six cultigens namely, SCNU1154, PMR 45, WMR 29, PMR 5, Edisto 47 and PI414723 were found to be susceptible as evident by the appearance of characteristic powdery mildew symptoms after 14 days of pathogen inoculation (Fig. 3). This same pattern of pathogenicity indicates that the isolates of these four regions are essentially the same pathogenic strain. Unfortunately, the other unknown race collected from Icheon region were not able to be tested for pathogenicity as the culture of the race failed in growth room.
Fig. 3.
Disease response of eight well characterized differential melon genotypes against unknown races of powdery mildew fungus Podosphaera xanthii collected from five different regions of Korea as determined by in-vitro bioassay using leaf discs after 2 weeks of inoculation. Conidia growth was observed in susceptible cultigens and are indicated by red ‘S’ letter. The layout of leaf discs samples in petri-dishes are shown in right. S, susceptible; R, Resistance. The experiment were repeated five times and same responses were observed every time.
When the disease response of eight standard differential cultigens against the collected isolates of four different regions namely, Hadong, Buyeo, Yeongam and Gokseong were compared with that of known races tested in this study and in previous reports (Hosoya et al., 1999; Izumikawa et al., 2008; Kim et al., 2016b; McCreight, 2006), it became obvious that none of the isolates showed same pattern of pathogenicity to any of the known races (Table 1). This indicates towards the possibility of these four isolates to be a new race in South Korea and we propose the name ‘race KN1’ for this race as indicated in Table 1.
The isolate of Jang-Hueng, on the other hand, was able to cause disease only in four cultigens such as SCNU1154, PMR 45, WMR 29and PI414723. The cultigens PMR 5 and Edisto 47 which were susceptible against the isolates of Hadong, Buyeo, Yeongam and Gokseong (now named as ‘race KN1’), have shown resistance against the isolate collected from Janghueng region (Fig. 3, Table 1). This pathogenicity profile of the isolate of Janghueng region is also unique as it didn’t match with that of any of the known races in these differential cultigens (Table 1). We thus conclude this isolate of Janghueng region as a new race with a new proposed name ‘race KN2’.
Discussion
Increased occurrence of powdery mildew and occasional breakdown of existing resistance indicates the existence of different races and appearance of new (unknown) races of Podosphaera xanthii in South Korea which need to be identified. In this study, few isolates of powdery mildew fungus were collected from affected melon farms of different region of South Korea (Fig. 1) and were tested for their pathogenicity against the standard differential cultigens whose resistance status is known against the identified races (Table 1). Idea behind this technique is that the isolates that show same pathogenic effect on a set of cultivars should essentially belong to same race and vice-versa. Bardin et al. (1999a, 1999b) identified two races of E. cichoracearum and 7 races of S. fuliginea using 10 differential melon genotypes namely, PI124112, PI414723, PMR45, PMR 5, MR-1, WMR29, Edisto 47, Nantais oblong, Védrantais and Iran. Kim et al. (2016b) identified a new race, race N5 of powdery mildew of melon based on its unique disease response in ten standard differential cultivars compared to that of against seven known races of P. xanthii (race 1, N1, N2, 5, A, S, O). In the current study, we used eight differential cultigens namely, SCNU1154, PMR 45, WMR 29, PMR 5, MR-1, PI124112, Edisto 47 and PI414723 based on their prior reports of being used in pathogenicity test to identify newly appeared races.
Our results identified the isolates of Hadong, Buyeo, Yeongam and Gokseong region as one single race based on their similarity in pathogenicity in the tested differential cultigens. Again, the uniqueness of the pathogenicity of these four isolates compared to that of two known races (tested in this study) and to that of the previous reports against 16 known races (Hosoya et al., 1999; Izumikawa et al., 2009; Kim et al., 2016b; McCreight, 2006) indicated it as a new race in Korea; for which the name ‘race KN1’ is proposed. Similarly, the uniqueness of the fungal isolate of Janghueng region indicated it as another new race with a new proposed name, ‘race KN2’.
These differential responses of the cultigens against different races of Podosphaera xanthii may be due to the inherent genetic makeup of the cultigen itself and/or due to unique interaction between virulent genes of pathogen and resistant/susceptibility genes of host plants. For example, melon line PI124112 and its derivative line MR-1 were resistant against two powdery mildew species namely, Erysiphe cichoracearum (formerly Golovinomyces cichoracearum) and Sphaerotheca fuliginea (Alvarez et al., 2000; Bardin et al., 1999b; Lebeda, 1991; Pitrat, 2002; Pitrat et al., 1996, 1998). The resistant line PI124112 was suggested to carry four different closely linked resistance genes namely, Pm-4, Pm-5, Pm-F and Pm-G where the former- and the latter- two were shown to confer resistance against powdery mildew fungus S. fuliginea and E. cichoracearum, respectively (Bardin et al., 1999a). Moreover, Pm-3 gene of PI124111 was responsible for resistance to S. fuliginea race 1 and Pm-x gene of PI 414723 to were responsible for resistance to S. fuliginea (Epinat et al., 1992; Harwood and Markarian, 1968a, 1968b; Pitrat, 1996; Pitrat and Besombes, 2008; Pitrat et al., 1998). The pattern of inheritance of the powdery mildew genes was reported as dominant inheritance such as resistant Pm-1 gene against race 1 fungus (Thomas and Webb, 1982); Pm-W gene of resistant line WMR29 and PI414723 against race 2 and Pm-X gene against race 1 (Pitrat, 1991). In addition, QTLs for powdery mildew resistance were mapped and molecular markers were developed using differential segregating populations of melon (Fukino et al., 2008; Kim et al., 2015; Perin et al., 2002).
Thus race determination based on these differential cultivars offers a quick, effective and reasonably robust approach as the resistance against a particular race are determined by the genetic resistance of the host plants. However, the results should be viewed with caution as the differential lines may possess several resistant genes, most of which are yet to be fully characterized (Bardin et al., 1999a). Moreover, the cultigens of one country may show differential levels of resistance in another country due to variation in environmental conditions s etc. which is evident from the case of the virulence shift of S. fuliginea races across different growing seasons (Hosoya et al., 1999) and across geographical and seasonal variations (CucCAP, 2017). Furthermore, the disease response of differential cultivars may also be manifested by the variation in the virulence and aggressiveness of the races of different locations and their interactions with host plant’s resistance mechanism.
In our case however, we tested the pathogenicity of the two known races in the standard differential cultivars and observed exactly same pathogenicity to that of previous reports indicating the robustness of the comparison between our tests to that of previous reports. Nevertheless, besides this pathogenicity test based identification of new races, further molecular validation using sequence based technologies or simply by using race specific molecular markers may confirm the accurate determination of the races.
The fact that race diversity can vary in different years and in different regions along with the appearance of new races as evidently shown in Table 2, the monitoring of the race diversity and identification of the new races is necessary. Our results provide a preliminary assessment of the unknown races in this regard which will be helpful in designing any future breeding program aiming at the development of multi-race resistant melon cultivars depending on the race diversity of different melon growing regions of South Korea.
Table 2.
Year-wise reports of appearances of melon powdery mildew races in South Korea
| Region | Year-wise reports of appeared races* | ||
|---|---|---|---|
|
| |||
| 2013 (Kim et al., 2016a) | 2014 (This Study) | 2015 (This Study) | |
| Changnyeong | Race 1 | - | Race 1, N1 |
| Hadong | - | Race KN1 | Race 1, N1 |
| Suncheon | Race N2 | - | Race N1 |
| Ansung | - | Race 1 | |
| Yeongam | Race A | Race KN1 | - |
| Icheon | Race A | - | - |
| Buyeo | - | Race KN1 | - |
| Gokseong | - | Race KN1 | - |
| Namwon | - | Race N1 | - |
| Damyang | - | Race N1 | - |
| Yeoju | - | Race N1 | - |
| Janghueng | - | Race KN2 | - |
Races can appear year wise and a particular location can have multiple races in a particular Year. Race KN1 and KN2 (shown in red text) were identified by this study based on the pathogenicity test against standard differential cultigens.
Increased occurrence of powdery mildew in South Korea calls for characterization of the prevalent races of different regions. This study identified the races of five isolates collected from five different powdery mildew affected regions based on pathogenicity test on known differential cultigens via comparing with previously reported pathogenicity of other known races. The suspected Podosphaera xanthii isolates were identified as two new races and were named as ‘race KN1’ and ‘race KN2’. Identification of these newly appeared races will be helpful for any future breeding programs aiming at developing resistant and high yielding varieties and will open up avenues for new race characterization research using sequence based genomic and molecular techniques.
Acknowledgments
This study was supported by the Golden Seed Project (Center for Horticultural Seed Development, Grant No. 213007-05-2-CG100 and Melon Project, Grant No. 312065-05-5-HD030) of the Ministry of Agriculture, Food and Rural affairs in the Republic of Korea (MAFRA).
References
- Alvarez JM, Gómez-Guillamon ML, Torés NA, Cánovas I, Floris E. Virulence differences between two Spanish isolates of Sphaerotheca fuliginea race 2 on melon. Acta Hortic. 2000;510:67–69. doi: 10.17660/ActaHortic.2000.510.10. [DOI] [Google Scholar]
- Bardin M, Dogimont C, Nicot P, Pitrat M. Genetic analysis of resistance of melon line PI 124112 to Sphaerotheca fuliginea and erysiphe cichoracearum studied in recombinant inbred lines. Acta Hortic. 1999a;492:163–168. doi: 10.17660/ActaHortic.1999.492.19. [DOI] [Google Scholar]
- Bardin M, Dogimont C, Pitrat M, Nicot PC. Virulence of Sphaerotheca fuliginea and Erysiphe cichoracearum on melon and genetic analysis of resistance of melon genotypes “PI 124112” and “PI 414723”. Proceedings of the First International Powdery Mildew Conference; August 29–September 2, 1999; Avignon, France. 1999b. pp. 85–86. [Google Scholar]
- Bardin M, Nicot PC, Normand P, Lemaire JM. Virulence variation and DNA polymorphism in Sphaerotheca fuliginea, causal agent of powdery mildew of cucurbits. Eur J Plant Pathol. 1997;103:545–554. doi: 10.1023/A:1008608413984. [DOI] [Google Scholar]
- Bertrand F. AR Hale’s Best Jumbo, a new differential melon variety for Sphaerotheca fuliginea races in leaf disk test. Proc Cucurbitaceae 2002; Naples, Florida. 2002. pp. 234–237. [Google Scholar]
- Cohen Y, Eyal H, Thomas CE. Stabilizing resistance in Cucumis melo against downy and powdery mildews in Israel and the USA. Phytopathology. 1984;74:829. (Abstract) [Google Scholar]
- Cohen Y, Eyal H. Reaction of muskmelon genotypes to races 1 and 2 of Sphaerotheca fuliginea in Israel. Cucurbit Genet Coop Rep. 1988;11:47–49. [Google Scholar]
- Cohen R, Burger Y, Katzir N. Monitoring physiological races of Podosphaera xanthii (syn. Sphaerotheca fuliginea), the causal agent of powdery mildew in cucurbits: factors affecting race identification and the importance for research and commerce. Phytoparasitica. 2004;32:174–183. doi: 10.1007/BF02979784. [DOI] [Google Scholar]
- Cohen R, Katzir N, Schreiber S, Greenberg R, Yarden O. Occurrence of Sphaerotheca fuliginea race 3 on cucurbits in Israel. Plant Dis. 1996;80:344. doi: 10.1094/PD-80-0344D. [DOI] [Google Scholar]
- Davis AR, Levi A, Tetteh A, Wehner T, Russo V, Pitrat M. Evaluation of watermelon and related species for resistance to race 1W powdery mildew. J Am Soc Hortic Sci. 2007;132:790–795. [Google Scholar]
- Davis AR, Bruton BD, Pair SD, Thomas CE. Powdery mildew: an emerging disease of watermelon in the United States. Rep Cucurbit Genet Coop. 2001;24:42–48. [Google Scholar]
- Del Pino D, Olalla L, Pérez-García A, Rivera ME, García S, Moreno R, Torés JA. Occurrence of races and pathotypes of cucurbit powdery mildew in southeastern Spain. Phytoparasitica. 2002;30:459–466. doi: 10.1007/BF02979750. [DOI] [Google Scholar]
- Epinat C, Pitrat M, Bertrand F. Genetic analysis of resistance of five melon lines to powdery mildews. Euphytica. 1992;65:135–144. doi: 10.1007/BF00022575. [DOI] [Google Scholar]
- Fukino N, Ohara T, Monforte AJ, Sugiyama M, Sakata Y, Kunihisa M, Matsumoto S. Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.) Theor Appl Genet. 2008;118:165–175. doi: 10.1007/s00122-008-0885-1. [DOI] [PubMed] [Google Scholar]
- Harwood RR, Markarian D. Agenetic survey of resistance to powdery mildew in muskmelon. J Hered. 1968a;59:213–217. doi: 10.1093/oxfordjournals.jhered.a107695. [DOI] [Google Scholar]
- Harwood RR, Markarian D. The inheritance of resistance to powdery mildew in the cantaloupe variety Seminole. J Hered. 1968b;59:126–130. doi: 10.1093/oxfordjournals.jhered.a107663. [DOI] [Google Scholar]
- Hosoya K, Narisawa K, Pitrat M, Ezura H. Race identification in powdery mildew (Sphaerotheca fuliginea) on melon (Cucumis melo L.) in Japan. Plant Breed. 1999;118:259–262. doi: 10.1046/j.1439-0523.1999.118003259.x. [DOI] [Google Scholar]
- Huang XQ, Hsam SLK, Zeller FJ, Wenzel G, Mohler V. Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet. 2000;101:407–414. doi: 10.1007/s001220051497. [DOI] [Google Scholar]
- Izumikawa Y, Fukino N, Kuzuya M, Miyagi M. Phenotypic analysis of powdery mildew resistant gene originated from commercial cultivar of melon. 1. Resistant gene linked with SSR marker, CMBR8. Hort Res (Japan) 2009;8:173. [Google Scholar]
- Izumikawa Y, Kuzuya M, Takazusu Y, Miyagi M. Occurrence of several pathogenic strains of melon powdery mildew with different host-specificity and search for melon breeding materials resistant to these strains. The 114th Meeting of the Japanese Society of Breeding. 2008;10(10):196. [Google Scholar]
- Jagger IC, Whitaker TW, Porter DR. A new biologic form of powdery mildew on Muskmelons in the Imperial Valley of California. Plant Dis Rep. 1938;22:275–276. [Google Scholar]
- Kasiamdari RS, Riefani MK, Daryono BS. AIP conference proceedings. 1. Vol. 1744. AIP Publishing; 2016. The occurrence and identification of powdery mildew on melon in Java, Indonesia; p. 020050. [DOI] [Google Scholar]
- Keinath AP, Dubose VB. Evaluation of fungicides for prevention and management of powdery mildew on watermelon. Crop Prot. 2004;23:35–42. doi: 10.1016/S0261-2194(03)00165-0. [DOI] [Google Scholar]
- Kenigsbuch D, Cohen Y. Inheritance of resistance to powdery mildew race 0 and race 1 in muskmelon. Phytopathology. 1987;77:1724. doi: 10.1094/PHYTO-02-15-0048-R. (Abstract) [DOI] [PubMed] [Google Scholar]
- Kim HT, Park JI, Ishikawa T, Kuzuya M, Horii M, Yashiro K, Nou IS. Development of molecular marker to select resistant lines and to differentiate the races related to powdery mildew in melon (Cucumis melo L.) J Plant Biotechnol. 2015;42:284–289. doi: 10.5010/JPB.2015.42.4.284. [DOI] [Google Scholar]
- Kim HT, Park JI, Nou IS. Identification of fungal races that cause powdery mildew in melon (Cucumis melo L.) and selection of resistant commercial melon cultivars against the identified races in Korea. J Plant Biotechnol. 2016a;43:58–65. doi: 10.5010/JPB.2016.43.1.58. [DOI] [Google Scholar]
- Kim HT, Park JI, Robin AHK, Ishikawa T, Kuzuya M, Horii M, Nou IS. Identification of a new race and development of DNA markers associated with powdery mildew in melon. Plant Breed Biotechnol. 2016b;4:225–233. doi: 10.9787/PBB.2016.4.2.225. [DOI] [Google Scholar]
- Křístková E, Lebeda A, Sedláková B. Virulence of Czech cucurbit powdery mildew isolates on Cucumis melo genotypes MR-1 and PI 124112. Sci Hortic. 2004;99:257–265. doi: 10.1016/S0304-4238(03)00105-5. [DOI] [Google Scholar]
- Kuzuya M, Yashiro K, Tomita K. Melon breeding for resistance to powdery mildew in respect to its races. Proc Vege Tea Sci. 2004;1:39–43. [Google Scholar]
- Kuzuya M, Yashiro K, Tomita K, Ezura H. Powdery mildew (Podosphaera xanthii) resistance in melon is categorized into two types based on inhibition of the infection processes. J Exp Bot. 2006;57:2093–2100. doi: 10.1093/jxb/erj166. [DOI] [PubMed] [Google Scholar]
- Lebeda A. The genera and species spectrum of cucumber powdery mildew in Czechoslovakia. J of Phytopathology. 1983;108:71–77. doi: 10.1111/j.1439-0434.1983.tb00565.x. [DOI] [Google Scholar]
- Lebeda A. Resistance in muskmelons to Czechoslovak isolates of Pseudoperonospora cubensis from cucumbers. Sci Hortic. 1991;45:255–260. doi: 10.1016/0304-4238(91)90071-6. [DOI] [Google Scholar]
- Lebeda A, Sedláková B, Křístková E. Distribution, harmfulness and pathogenic variability of cucurbit powdery mildew in the Czech Republic. Acta Fytotechnica et Zootechnica. 2003;7:174–176. [Google Scholar]
- Lee WM, Son SH, Park DK, Lee HJ, Huh YC, Cho IH. Selection of watermelon germplasm with disease resistance to powdery mildew (Sphaerotheca fusca) Kor J Hort Sci Technol. 2010;28(Suppl II):73. [Google Scholar]
- Jahn M, Munger HM, McCreight JD. Breeding cucurbit crops for powdery mildew resistance. The powdery mildews: a comprehensive treatise. 2002:239–248. [Google Scholar]
- McCreight JD, Pitrat M, Thomas CE, Kishaba AN, Bohn GW. Powdery mildew resistance genes in muskmelon. J Am Soc Hortic Sci. 1987;112:156–160. [Google Scholar]
- McCreight JD, Coffey MD. Inheritance of resistance in melon PI 313970 to cucurbit powdery mildew incited by Podosphaera xanthii race S. HortScience. 2011;46:838–840. [Google Scholar]
- McCreight JD. Melon-powdery mildew interactions reveal variation in melon cultigens and Podosphaera xanthii races 1 and 2. J Amer Soc Hort Sci. 2006;131:59–65. [Google Scholar]
- McGrath MT, Thomas CE. Powdery mildew. In: Zitter TA, Hopkins DL, Thomas CE, editors. Compendium of cucurbit disease. The American Phytopathological Society; MN, USA: 1996. pp. 28–30. [Google Scholar]
- McGrath MT. Powdery mildew of cucurbits. 1997 URL http://vegetablemdonline.ppath.cornell.edu/factsheets/Cucurbits_PM.htm/
- CucCAP. Melon Powdery - Mildew Database at UC Riverside. 2017 URL https://cuccap.org/2017/07/06/powdery-mildew-melon-powdery-mildew-database-at-uc-riverside/
- Mohamed YF, Bardin M, Nicot PC, Pitrat M. Causal agents of powdery mildew of cucurbits in Sudan. Plant Dis. 1995;79:634–636. doi: 10.1094/PD-79-0634. [DOI] [Google Scholar]
- Pérez-García A, Romero D, Fernández-Ortuño D, López-Ruiz F, De Vicente A, Torés JA. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol Plant Pathol. 2009;10:153–160. doi: 10.1111/j.1364-3703.2008.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périn C, Hagen L, De Conto V, Katzir N, Danin-Poleg Y, Portnoy V, Pitrat M. A reference map of Cucumis melo based on two recombinant inbred line populations. Theor Appl Genet. 2002;104:1017–1034. doi: 10.1007/s00122-002-0864-x. [DOI] [PubMed] [Google Scholar]
- Pitrat M. Linkage groups in Cucumis melo L. J Hered. 1991;82:406–411. doi: 10.1093/oxfordjournals.jhered.a111112. [DOI] [Google Scholar]
- Pitrat M. 2002 gene list for melon. Rep Cucurbit Genet Coop. 2002;25:76–93. [Google Scholar]
- Pitrat M, Besombes D. Inheritance of Podosphaera xanthii resistance in melon line ‘90625’. In: Pitrat M, editor. Cucurbitaceae 2008, IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae. INRA; Avignon, France: 2008. pp. 135–142. [Google Scholar]
- Pitrat M, Dogimont C, Bardin M. Resistance to fungal diseases of foliage in melon. In: McCreight JD, editor. Cucurbitaceae 98, evaluation and Enhancement of cucurbit germplasm. ASHS Press; Alexandria, VA, USA: 1998. pp. 167–173. [Google Scholar]
- Pitrat M, Risser G, Bertrand F, Blancard D, Lecoq H. In: Gómez-Guillamón ML, Soria C, Cuartero J, Torés JA, Fernández-Muòoz R, editors. Evaluation of a melon collection for disease resistance; Cucurbits towards 2000, proceedings of the VIth eucarpia meeting on cucurbit genetics and breeding; May 28–30, 1996; Málaga, Spain: 1996. pp. 49–58. [Google Scholar]
- Pryor DE, Whitaker TW, Davis GN. The development of powdery mildew resistant cantaloupes. J Am Soc Hortic Sci. 1946;47:347–356. [Google Scholar]
- Sowell G., Jr Population shift of Sphaerotheca fuliginea on musk melon. J Am Soc Hortic Sci. 1982;112:156–160. [Google Scholar]
- Tetteh AY, Wehner TC, Davis AR. Identifying resistance to powdery mildew race 2W in the USDA-ARS watermelon germplasm collection. Crop Sci. 2010;50:933–939. doi: 10.2135/cropsci2009.03.0135. [DOI] [Google Scholar]
- Thomas CE, Nugent PE. Resistance to powdery mildew incited by race-3 of Sphaerotheca-fuliginea in the united-states plant introduction collection of Cucumis melo. Hortscience. 1983;18:172. [Google Scholar]
- Vakalounakis DJ, Klironomou E. Race and mating type identification of powdery mildew on cucurbits in Greece. Plant Pathol. 1995;44:1033–1038. doi: 10.1111/j.1365-3059.1995.tb02662.x. [DOI] [Google Scholar]