Abstract
The human general transcription factor TFIID is composed of the TATA-binding protein (TBP) and 13 TBP-associated factors (TAFs). In eukaryotic cells, TFIID is thought to nucleate RNA polymerase II (Pol II) preinitiation complex formation on all protein coding gene promoters and thus, be crucial for Pol II transcription. In a child with intellectual disability, mild microcephaly, corpus callosum agenesis and poor growth, we identified a homozygous splice-site mutation in TAF8 (NM_138572.2: c.781–1G > A). Our data indicate that the patient’s mutation generates a frame shift and an unstable TAF8 mutant protein with an unrelated C-terminus. The mutant TAF8 protein could not be detected in extracts from the patient’s fibroblasts, indicating a loss of TAF8 function and that the mutation is most likely causative. Moreover, our immunoprecipitation and proteomic analyses show that in patient cells only partial TAF complexes exist and that the formation of the canonical TFIID is impaired. In contrast, loss of TAF8 in mouse embryonic stem cells and blastocysts leads to cell death and to a global decrease in Pol II transcription. Astonishingly however, in human TAF8 patient cells, we could not detect any cellular phenotype, significant changes in genome-wide Pol II occupancy and pre-mRNA transcription. Thus, the disorganization of the essential holo-TFIID complex did not affect global Pol II transcription in the patient’s fibroblasts. Our observations further suggest that partial TAF complexes, and/or an altered TFIID containing a mutated TAF8, could support human development and thus, the absence of holo-TFIID is less deleterious for transcription than originally predicted.
Introduction
In order for a gene to be transcribed, a transcriptional preinitiation complex (PIC) needs to assemble at its promoter. The first complex to bind the promoter is the general transcription factor, TFIID, consisting of the TATA-binding protein (TBP) and 13 different TBP-associated factors (TAFs) (1,2). Following TFIID binding, other general transcription factors (TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) and RNA polymerase II (Pol II) are recruited to the promoter (3). Nine of the TAFs contain a histone fold domain (HFD), and form five histone fold pairs. TAF8 is an HFD-containing TAF which forms a histone fold pair with TAF10 in TFIID (4,5). Biochemical studies revealed that the TFIID complex is assembled in a step-wise manner, first forming a stable core complex consisting of TAF5, TAF6-TAF9 and TAF4-TAF12. This core is then bound by TAF2 associated with the TAF8-TAF10 heterodimer, forming the 8TAF complex. Next, TBP and the remaining TAFs join the 8TAF complex, to form holo-TFIID (6,7).
Studies performed in yeast show comprehensively that TFIID is recruited to almost all promoters and is required for the transcription of all Pol II transcribed genes (8). In agreement, individual yeast TAFs are essential for viability. Similarly, germ line knock-out of several TFIID subunits in mouse (Taf7, Taf8, Taf10 and Tbp) results in early embryonic lethality around E4.0 (9–12), suggesting that these mammalian TFIID subunits are absolutely essential for blastocyst viability and early development. Furthermore, conditional deletion of Taf4 or Taf10, in embryonic keratinocytes (13,14), deletion of Taf10 in embryonic liver (15), or ablation of Taf7 in CD4–CD8– thymocytes (9) compromises the viability of the mutant cells, suggesting that these TFIID subunits play an essential role in transcription in these cell types.
Curiously, however, deletion of Taf10 in postnatal liver resulted in a less dramatic phenotype compared with the lack of Taf10 during embryogenesis, with mild effect on transcription of liver genes (15). Similarly, in adult mouse keratinocytes, deletion of Taf10 had no effect on epidermal function and no apparent effect on transcription (13). Also, mouse CD4+CD8+ T-cells lacking Taf7 appear to be viable with no apparent effect on transcription (9). Together these animal studies demonstrate that many cell types absolutely require the canonical TFIID complex for transcription and survival; however, certain cell types are seemingly unaffected by loss of TFIID subunits. How and why these transcription regulatory differences occur amongst different cell types of embryonic and/or adult origin is not understood.
Several different human TFIID subunit coding genes (TBP, MIM: 600075; TAF1, MIM: 313650; TAF2, MIM: 604912; TAF6, MIM: 602955; and TAF13, MIM: 600774) have been implicated in human diseases including many with neurological outcomes and intellectual disability (16–28). A polyglutamine expansion in TBP has a well-characterized gain of function effect resulting in a neurodegenerative disorder, spinocerebellar ataxia-17 (17,19,22,24,26,27,28). In contrast, most of the other reported mutations in genes encoding TFIID subunits are recessive missense mutations in different TAFs found in patients with intellectual disability and various degree of developmental delay and microcephaly (16,18,21,23,25,28). The outcome of these mutations on TFIID structure and function was not assessed in detail, but it can be expected that they cause, to a variable degree, a partial loss of function of the corresponding protein and potentially of TFIID.
In the present study, we identified a homozygous splice site mutation in TAF8 in a patient presenting with intellectual disability, developmental delay and mild microcephaly. The mutation resulted in a frame shift changing the C-terminus of TAF8. The mutant TAF8 protein was undetectable by either western blot analysis or mass spectrometry in cell extracts as well as in TAF-containing complexes purified from the patient fibroblasts. We show that TFIID assembly is impaired in the patient fibroblasts, but astonishingly, there was no significant reduction of the genome-wide RNA Pol II occupancy, elongation rates and pre-mRNA synthesis.
Results
Intellectually disabled child identified with a TAF8: c.781–1G > A mutation
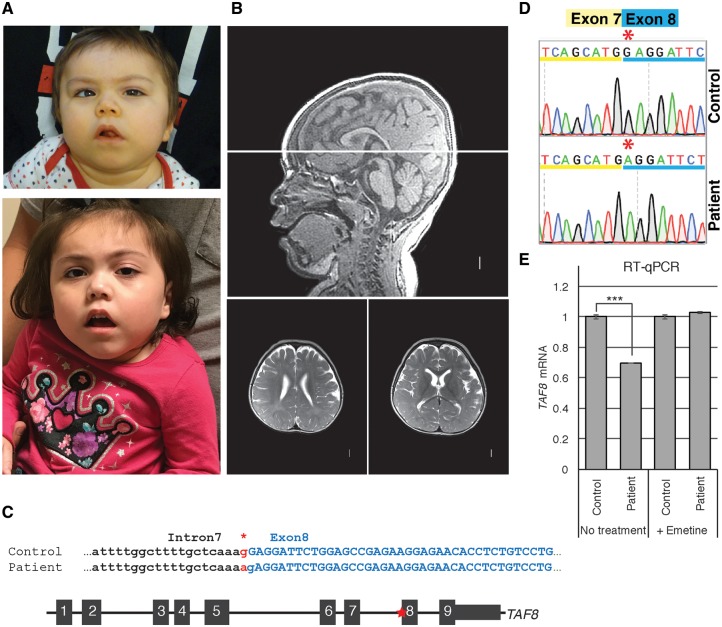
A girl, first seen at 15 months of age, presented with severe global developmental delay and intellectual disability. She was born to non-consanguineous 15- and 17-year-old Hispanic parents after an uncomplicated term pregnancy. Growth parameters have been at –4–5 SD for height and weight and –2 SD for head circumference. Her development has been severely delayed with smiling by 3–4 months and rolling at a year. At age 4.5 years she cannot sit or speak. She has developed progressive spasticity and equinovarus foot deformities. However, she does not have seizures and her general health is reasonably good without intercurrent hospitalizations. Her craniofacial findings include brachycephaly, a mildly short forehead, a tented mouth, mild left sided ptosis and a long philtrum (Fig. 1A). Her hands and feet are unremarkable except for the progressive deformities of her feet.
Figure 1.
Developmentally delayed child identified with c.781–1G > A mutation in TAF8 gene. (A) Photographs of patient at 3 years of age (top) and 4 years of age (bottom). (B) Brain MRI acquired at 2 years and 7 months of age shows (sagittal on top and two transversal levels on bottom) mildly prominent extra-axial spaces, diffuse thinning of white matter with delayed myelinization, borderline enlarged lateral ventricles, short corpus callosum with narrow posterior body and absent splenium, normal brainstem, borderline small cerebellar vermis, and mildly small posterior fossa size. (C) (Top) Patient and control genomic sequences at the intron 7–exon 8 boundary of interest. Asterisk highlights the G > A splice site mutation (in red). Capital letters show exon 8 coding nucleotides. (Bottom) Schematic representation of the TAF8 gene (not to scale) with the position of the mutation indicated by the red asterisk. (D) Sequencing chromatogram highlighting the G nucleotide missing from the beginning of exon 8 in the patient cDNA. (E) RT-qPCR performed with primer annealing to the boundaries between exons 2 and 3, showing abundance of TAF8 mRNA in control and patient cells treated with and without emetine. Error bars represent SEM of three technical replicates. ***P < 0.001.
Extensive genetic evaluation revealed a normal chromosomal SNP microarray without excessive homozygosity, normal metabolic studies including glycosylated transferrin, lactate, acylcarnitine profile, very long chain fatty acids and urine organic and plasma amino acids. Two MRI’s have revealed mild microcephaly, reduced myelination, a small cerebellar vermis and partial absence of the corpus callosum (Fig. 1B).
Exome sequencing revealed a homozygous splice site variant in TAF8 [NC_000006.12: g.42077099G > A (NM_138572.2: c.781–1G > A)] (Fig. 1C), whereas no other mutation or variant in genes possibly associated with the phenotype could be identified. Both parents were found to be heterozygous for the same variant. This change was observed in the Genome Aggregation Database (gnomAD) in the heterozygous state in ten individuals, including five of Hispanic origin, out of 277264 alleles.
TAF8: c.781–1G > A variant generates a cryptic splice site resulting in a frame shift in the TAF8 mRNA
As the TAF8: c.781–1G > A mutation in the 3′ end of intron 7 of the TAF8 gene occurred in a predicted splice acceptor site (Fig. 1C), we analyzed the consequences of this mutation on TAF8 mRNA and protein. To determine the outcome of the mutation, reverse transcriptase-polymerase chain reaction (RT-PCR) analyses were performed on RNA extracted from control and patient fibroblasts using a combination of primers (Supplementary Material, Table S1). These analyses excluded skipping of exon 8 or intron 7 retention (Supplementary Material, Fig. S1A and B). Sequencing of a cDNA fragment generated by RT-PCR using primers annealing in exons 7 and 8 revealed that a cryptic splice acceptor site, one base pair downstream of the original one, was utilized instead of the original splice acceptor site, in the patient fibroblasts. As a result, exon 8 was missing the first G nucleotide (Fig. 1C and D), causing a frame shift. Identical results were observed when the samples were treated with emetine, a non-sense mediated decay inhibitor (29), confirming that this cryptic acceptor site is exclusively used in the mutant fibroblasts. Quantitative PCR (qPCR) was carried out on mutant and control cDNA, using primers amplifying the junction between exon 2 and 3, which revealed that the abundance of the mutant TAF8 mRNA was mildly reduced by about 30% in the patient cells. This mild reduction is likely caused by non-sense mediated decay, because treatment with emetine restored normal levels of TAF8 mRNA (Fig. 1E).
The TAF8: c.781–1G > A gene encodes a highly unstable protein lacking the C-terminal nuclear localization signal
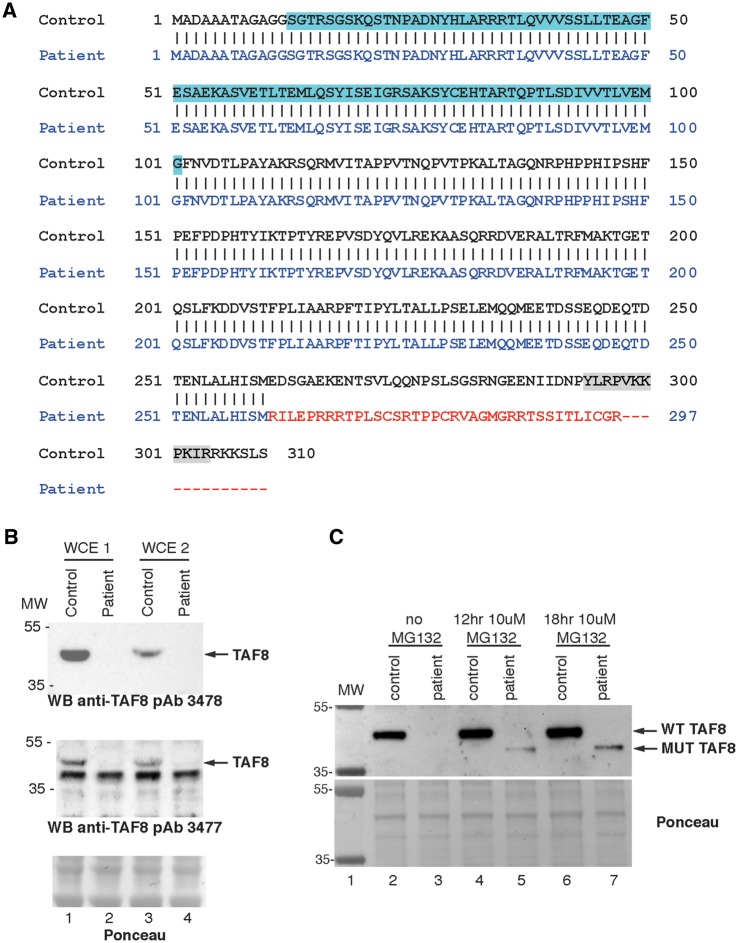
The amino acid sequence produced from the mutant TAF8 cDNA was predicted using an in silico translation tool. Loss of the first G nucleotide in exon 8, results in a frame shift, affecting the final 50 amino acids of the ‘wild-type’ (WT) TAF8 protein, creating a different C-terminus in the mutant TAF8 protein and a premature stop codon encoded in the 8th exon (Fig. 2A). In the WT TAF8 sequence, the stop codon is in the last 9th exon (Fig. 1C). The region encompassing the TAF8 HFD, which is required for its pairing with TAF10, is upstream of the frame-shift and is therefore unaffected in the mutant TAF8 protein. The C-terminal region of the WT TAF8 contains a nuclear localization signal (NLS) (30), which is lost in the predicted mutant TAF8 protein (Fig. 2A). WT TAF8 protein has a predicted molecular weight of 34.3 kDa, but migrates around the 43 kDa position (5). To detect the endogenous mutant TAF8 protein, whole cell extracts (WCEs) were prepared from control and mutant fibroblasts using two different extraction methods (see Materials and Methods) and were subjected to western blot analysis with two different polyclonal anti-TAF8 antibodies (pAb 3477 and 3478), which were generated against the entire TAF8 protein. Unexpectedly, mutant TAF8 protein was not detected in patient fibroblasts (Fig. 2B and C lanes 2 and 3), irrespective of mutant TAF8 mRNA levels (Fig. 1E). To exclude the possibility that the anti-TAF8 pAbs are unable to detect the mutant TAF8 protein because of its unique carboxyl terminus, we generated recombinant baculoviruses expressing the mutant TAF8 protein fused to an N-terminal FLAG tag. The anti-TAF8 pAb readily detected the mutant FLAG-TAF8 fusion protein when highly over-expressed in Sf9 insect cells (Fig. 3A).
Figure 2.
The TAF8: c.781–1G > A mutant protein lacks an NLS and is degraded by the proteasome. (A) Amino acid sequence of WT TAF8 protein aligned to the in silico predicated amino acid sequence generated by the TAF8: c.781-1G > A mutation, which possesses a unique carboxy-terminus (depicted in red), and lacks an NLS (depicted in grey highlight in the WT sequence). Histone fold domain in the WT sequence is highlighted in blue. (B) Two different protein WCE (WCE1 and WCE2) preparations (see Materials and Methods) were analyzed by western blot. Blots were probed with two different polyclonal anti-TAF8 antibodies (affinity purified pAb 3478 and crude pAb 3477 serum) targeting the entire TAF8 protein, as indicated. (C) Patient and control fibroblasts were treated with MG132 for 12 or 18 h, WCEs were prepared and probed with an anti-TAF8 antibody (affinity purified pAb 3477). The presence of a smaller protein, likely corresponding to the mutant TAF8 protein was detected in the presence of MG132.
Figure 3.
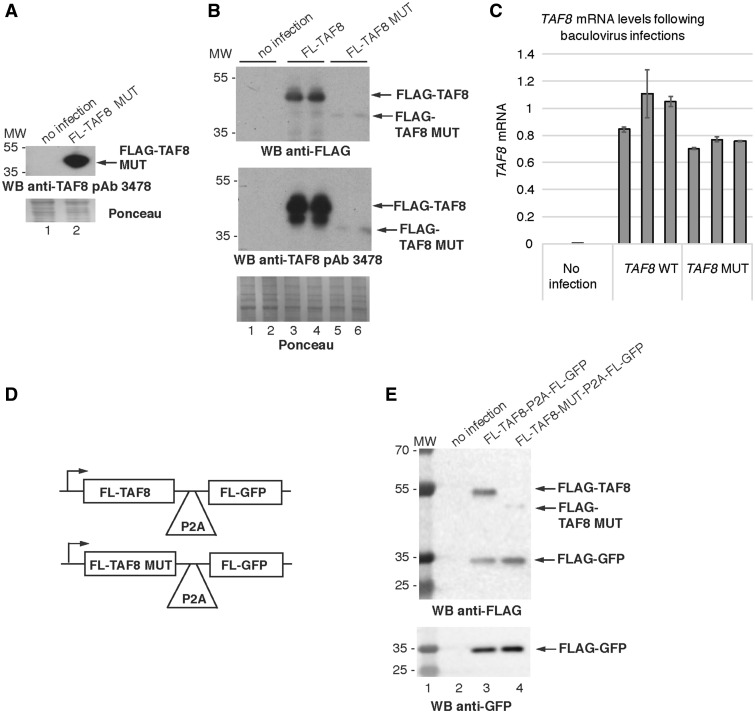
Exogenously expressed TAF8 mutant protein is unstable. (A) WCEs from insect Sf9 cells were prepared with either no infection, or with infection of a recombinant baculovirus encoding the TAF8: c.781-1G > A mutant protein fused to an FLAG tag and western blot assays were carried out using purified anti-TAF8 pAb 3478. (B) Western blot analyses of WCEs generated from Sf9 cells either with no infection, infection with recombinant baculovirus encoding FLAG tagged WT TAF8, or FLAG-tagged mutant TAF8 (FL-TAF8 MUT) protein, probed either with anti-FLAG antibody (upper blot), or affinity purified anti-TAF8 pAb 3478 (middle blot). Each lane represents an independent infection. (C) RT-qPCR performed with primers annealing to the boundary between exons 2 and 3 of TAF8 mRNA normalized to Sf9 housekeeping genes 28S, GAPDH and L35, performed on cDNA generated from Sf9 cells with either no infection, or infected with recombinant baculovirus encoding WT TAF8 protein or TAF8: c.781-1G > A mutant protein. All values were normalized to the average of the WT TAF8 infection. Error bars represent SEM of three technical replicates. See Supplementary Material, Table S1 for primer sequences. (D) Schematic representations of the cassettes expressing a single mRNA encoding either WT FLAG-TAF8 or mutant FLAG-TAF8 together with FLAG-GFP separated by the self-cleavage P2A sequence. (E) Western blot analyses of WCEs generated from Sf9 cells either with no infection, infection with recombinant baculovirus encoding FLAG tagged WT TAF8-P2A-FL-GFP, or FLAG-tagged mutant TAF8 (FL-TAF8 MUT- P2A-FL-GFP) protein, probed either with anti-FLAG antibody (upper blot), anti-GFP (lower blot). In A and B, for Ponceau staining 5× the amount of protein was loaded as compared with the western blots. In A, B and E, the molecular weight (MW) markers are indicated in kDa.
These results suggest that the mutant TAF8 protein, as compared with WT, may be less stable and prone to degradation in patient cells. To verify the stability of the mutant TAF8 in a different cellular system, first insect Sf9 cells were infected with baculoviruses expressing either FLAG-tagged WT, or mutant TAF8. When identical total protein amounts from both WT and mutant TAF8 expressing cells were analyzed by western blot, the mutant TAF8 protein was hardly detectable, although high levels of WT TAF8 were detected (Fig. 3B, compare lanes 3 and 4 with 5 and 6). Importantly, however, both WT and mutant TAF8 expressing Sf9 cells contained comparable amounts of TAF8 mRNAs (Fig. 3C), suggesting that the altered final 50 C-terminal amino acids lead to instability of the mutant TAF8 protein. Secondly, to determine whether the mutant TAF8 mRNA is translated into protein at a similar rate as the WT TAF8 mRNA, we generated a single cassette system which co-expresses FLAG-tagged WT, or mutated TAF8 proteins, together with FLAG-green fluorescent protein (GFP), separated by the P2A ‘self-cleaving’ 22 amino acid peptide. Importantly, the P2A sequence mediates ribosome-skipping, enabling the generation of equal amounts of two separate peptide products from a single mRNA (31,32). When the WT FLAG-TAF8 was expressed with FLAG-GFP from the same cassette (Fig. 3D), using a baculovirus expression system in insect cells, approximately equal amount of the two proteins were detected (Fig. 3E, lane 3). However, in good agreement with our earlier results, when the mutated FLAG-TAF8 was expressed with FLAG-GFP from the same cassette (Fig. 3D), the mutated FLAG-TAF8 was hardly detectable, although the FLAG-GFP, produced from the same mRNA, was readily detected (Fig. 3E, lane 4). These results unequivocally demonstrate that the mutated TAF8, containing the unrelated 50 C-terminal amino acids, is intrinsically unstable.
Degradation of the mutant TAF8 protein could be mediated by the proteasome. In order to test this hypothesis, patient and control fibroblasts were treated with the proteasome inhibitor MG132 for 12 or 18 h, and then western blot analysis was conducted. In untreated samples, the WT TAF8 protein was readily detected, although the mutant TAF8 protein was not. However, in the patient fibroblasts treated with MG132, a protein, smaller than the WT TAF8 protein was detected, likely corresponding to the mutant TAF8 protein (Fig. 2C). These results suggest that degradation of the mutant TAF8 protein is likely carried out, at least partially, by the proteasome.
Together these results show that the TAF8: c.781–1G > A mutation generates a protein with a unique C-terminus that is prone to degradation, causing an apparently complete loss of function of TAF8, further indicating that TAF8: c.781–1G > A, p.Glu261Argfs*38 is most likely the causative mutation in this patient.
Ablation of mouse TAF8 protein in embryonic stem cells results in cell death caused by transcriptional defects
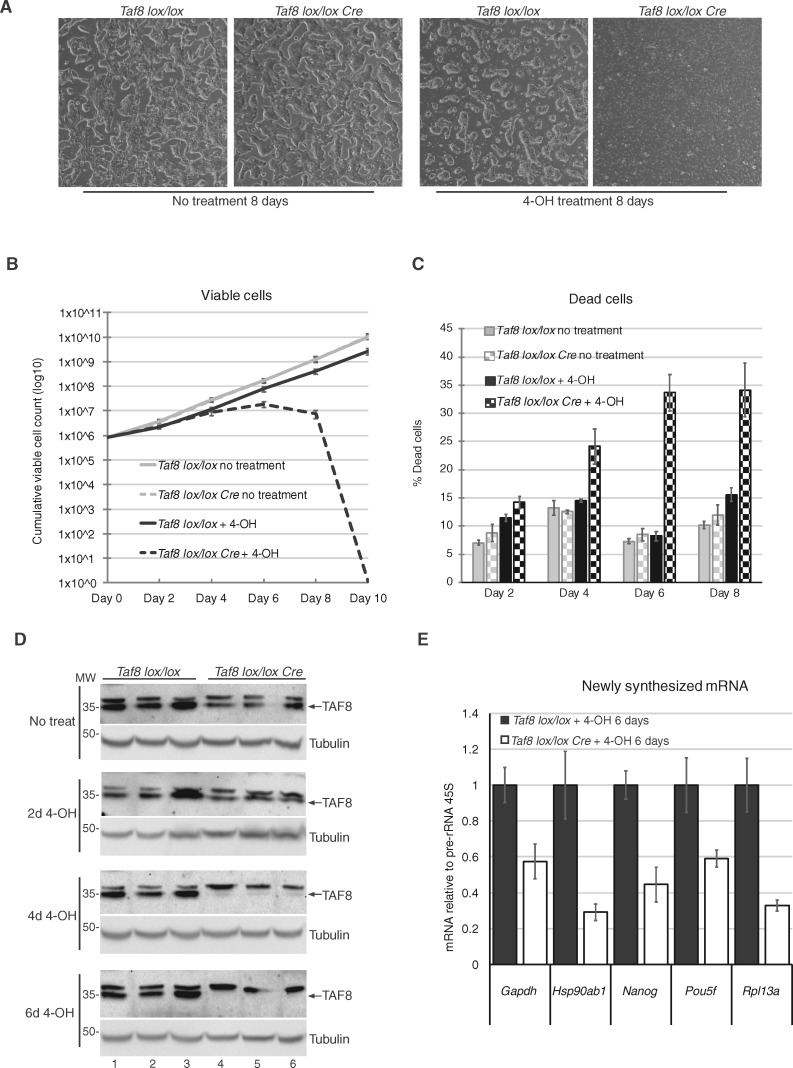
The human patient fibroblasts lacking any detectable TAF8 protein were apparently healthy. Thus, we decided to investigate mouse cells in which TAF8 protein elimination can be induced by Taf8 knock-out. Mouse blastocysts lacking Taf8 die very early during development, at around embryonic day 4.0 (E4.0), because of apoptosis in the inner cell mass (ICM) (12). To be able to monitor the direct consequences of inducible TAF8 protein ablation, we used Taf8lox/lox: Rosa26-CreERT2 mouse embryonic stem cells (mESCs), in which Taf8 deletion can be induced by the addition of 4-hydroxy Tamoxifen (4-OH) (Supplementary Material, Fig. S2A). Cells were seeded at a density of 8 × 105 cells per well and every second day, live and dead cells were counted, then 8 × 105 cells were reseeded. After 8 days of treatment, there was a massive reduction in the number of live cells, and an increase in dead cells, in the Taf8lox/lox: Rosa26-CreERT2 cultures treated with 4-OH tamoxifen, as compared with control cultures (Fig. 4A–C). The cell death observed in the tamoxifen treated Taf8lox/lox: Rosa26-CreERT2 mESCs correlated with a gradual increase in the deletion of the Taf8 allele (Supplementary Material, Fig. S2B) and a gradual depletion of TAF8 protein, with no TAF8 protein detected after day 6 (Fig. 4D).
Figure 4.
mESCs induced to delete Taf8 undergo cell death potentially as a result of transcriptional defects. (A) Representative images of Taf8lox/lox or Taf8lox/lox-Rosa26-CreERT2 mESCs after 8 days of culture with or without 0.5 µM 4-hydroxy tamoxifen treatment (4-OH). See also Supplementary Material, Figure S2A. (B) Curve showing the cumulative number of viable mESCs over the course of the treatment, cell counts were plotted taking into account the dilution factor at each passage. (C) Cell death was measured by trypan blue exclusion over the course of the treatment (as indicated). (D) Western blot analyses to detect TAF8 protein with the affinity purified 3478 pAb during several days (d) of 4-hydroxy tamoxifen (4-OH) treatment. Equal loading was measured by using an anti-γ-tubulin antibody. MW markers are indicated in kDa. (E) RT-qPCR of randomly selected, highly expressed genes, measuring newly synthesized mRNA relative to Pol I transcribed pre-rRNA 45S. The list of the primers is indicated in Supplementary Material, Table S1. For all experiments three biological replicates were carried out per genotype. Error bars represent SEM.
Next, we wanted to determine if TAF8 depletion causes global transcriptional defects. As steady-state mRNA levels can be buffered by modulation of mRNA decay, especially in situations where Pol II transcription is globally impaired [(8,33,34) and references therein], we measured newly synthesized pre-mRNA using primers that amplify an intron–exon boundary in unspliced pre-mRNA. Pre-mRNA transcript abundance was normalized to the RNA Pol I transcribed 45S pre-rRNA, which is not affected by loss of Taf8. For all tested genes, the abundance of pre-mRNAs was reduced by about 2-fold upon TAF8 depletion, suggesting a global defect in Pol II transcription in the absence of TAF8. Together, these experiments show that mESCs die in the absence of mouse TAF8, similarly to the in vivo Taf8–/– blastocyst phenotype reported by Voss et al. [12], and our preliminary transcriptional analysis suggests that a global decrease in PIC formation correlates with cell death.
The lack of detectable human TAF8 protein dramatically alters the assembly of the TFIID complex in patient fibroblasts
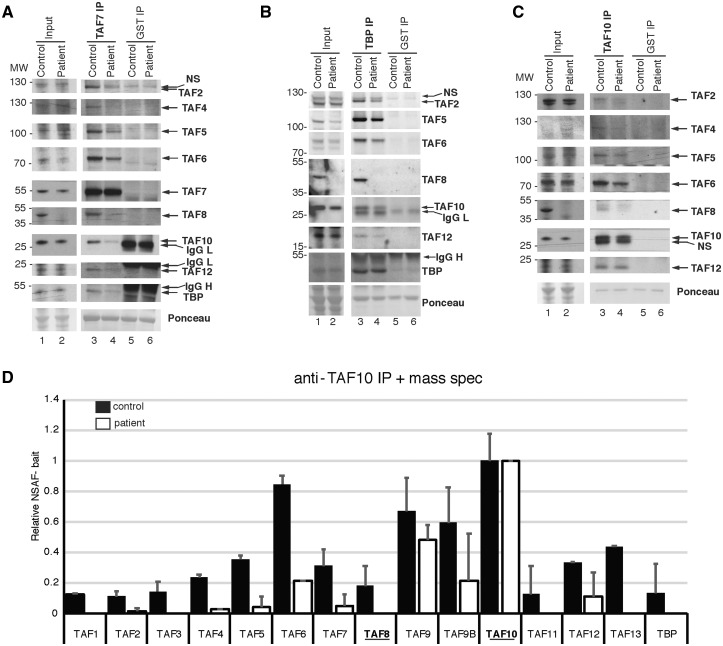
As the patient fibroblasts did not present any obvious phonotype when compared with control fibroblasts, we next assessed if TFIID integrity was affected in the cells harboring the TAF8: c.781–1G > A mutation. To this end, WCEs were prepared from control and TAF8 mutant fibroblasts and subjected to immunoprecipitations (IPs) using anti-TAF7, anti-TBP or anti-TAF10 antibodies to capture the TAF7-, TBP- or TAF10-containing complexes. The abundance of the TFIID subunits tested was similar in the WCEs from control and patient cells (Fig. 5A–C, lanes 1 and 2). Likewise, the amount of bait protein captured by the corresponding antibody was similar (Fig. 5A–C, lanes 3 and 4). However, in complexes purified using either anti-TAF7, anti-TBP or anti-TAF10 antibodies, the levels of several of the tested TFIID subunits was markedly reduced in the patient fibroblasts when compared with intact TFIID complexes purified from control cells (Fig. 5A–C, compare lanes 3 and 4).
Figure 5.
TFIID assembly is defective in fibroblasts harboring the TAF8: c.781–1G > A mutation. (A–C) Coimmunoprecipitation performed on WCEs prepared from control, or TAF8: c.781-1G > A patient fibroblasts, using either anti-TAF7 and anti-GST (negative control) antibodies (A) or anti-TBP and anti-GST (B) or anti-TAF10 and anti-GST (C). Input extracts or immunopurified complexes were separated by SDS-PAGE and western blot analyses were carried out by probing the blots with antibodies against the indicated TFIID subunits. The anti-GST antibody heavy (IgG H) and light chains (IgG L) are indicated. Ponceau-stained membranes in A–C show equal loading. In A–C the MW markers are indicated in kDa. (D) Mass spectroscopy performed on proteins co-IP-ed with anti-TAF10 antibody, normalized to TAF10 bait protein. Relative NSAFbait was calculated as described previously (52). Error bars represent SEM of two technical replicates. See Supplementary Material, Table S2 for raw mass spectrometry peptide counts.
To detect potential very low levels of mutant TAF8 protein and additional TFIID subunits with a higher sensitivity and in a more quantitative way, anti-TAF10 immunopurified complexes were analyzed by matrix assisted laser desorption ionisation-time of flight mass spectrometry. To be able to compare the composition of complexes purified from control and Taf8 mutant cells, normalized spectral abundance factor (NSAF) values were calculated (35). Although all the known TFIID subunits were identified by mass spectrometry analysis of TAF10-containing complexes isolated from control fibroblasts, we did not detect any TAF8 peptides from the patient cells. We also did not detect TAF1, TAF3, TAF11, TAF13 and TBP, which are supposed to be present in a functional module of TFIID, which does not contain the TAF8/10 histone fold pair (6) (Fig. 5D, Supplementary Material, Table S2). Moreover, the bait-normalized anti-TAF10 immunopurification performed on patient cells identified eight TFIID subunits (TAF2, TAF4, TAF5, TAF6, TAF7, TAF9/9b and TAF12) with significantly less peptide counts than detected in the control sample (Fig. 5D). It should be noted that the overall peptide counts were rather low, likely owing to the overall low level of TFIID subunits in fibroblasts relative to other cell types (Supplementary Material, Fig. S3). Therefore, we cannot exclude the possibility that low levels of mutant TAF8 protein are present in the TFIID complexes purified with anti-TAF10 antibodies, but the levels were below the detection limit.
Taken together, these results indicate that in the fibroblasts harboring the TAF8: c.781–1G > A mutation, the lack of detectable mutant TAF8 results in TFIID disassembly to partial TAF-containing complexes, in spite of the fact that the abundance of TFIID subunits (with the exception of TAF8 protein) in the input extracts seem to be unaffected (Fig. 5A–C). Because of the crucial role of TFIID in transcription initiation and during early embryonic mouse development, these observations together offer a likely causative explanation for the homozygous TAF8: c.781–1G > A mutation in the patient phenotype.
A defective TFIID complex does not cause a global reduction of the genome-wide RNA Pol II occupancy and elongation rates
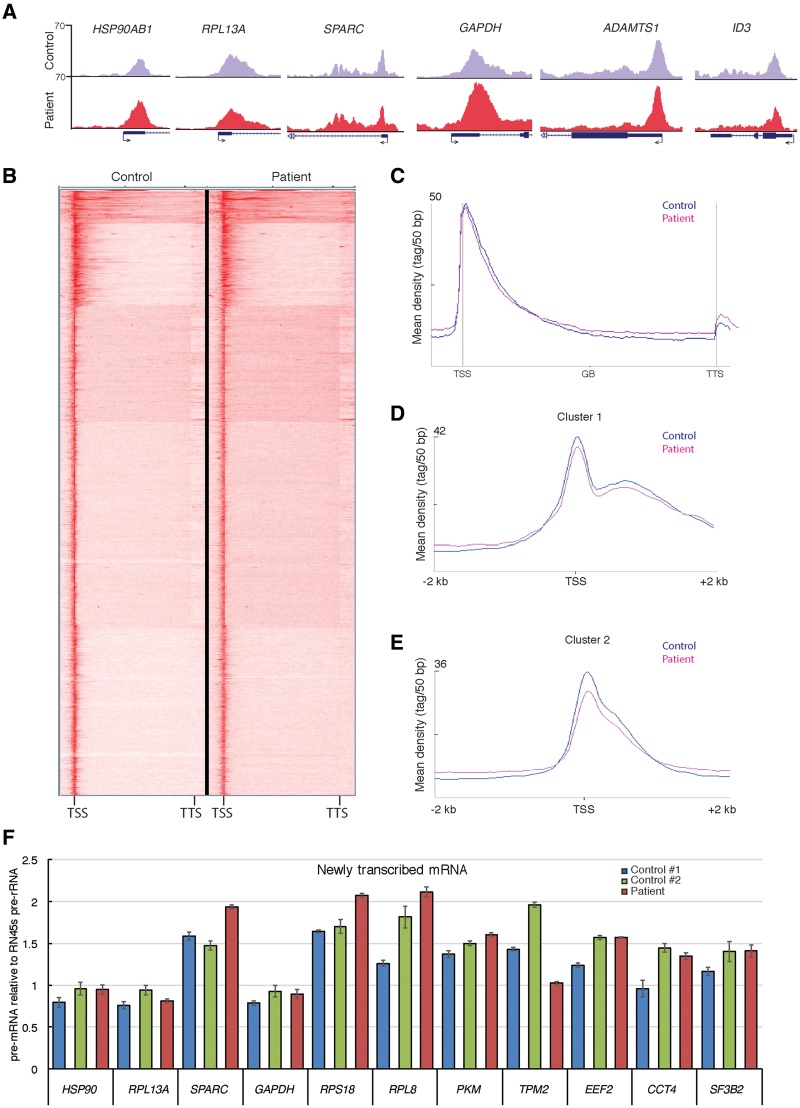
Because TFIID was disassembled in fibroblasts harboring the TAF8: c.781–1G > A mutation (Fig. 5), we set out to verify whether the global recruitment of RNA polymerase II to active promoters may also be compromised. To test this possibility, control and patient fibroblasts were subjected to chromatin immunoprecipitation (ChIP) coupled to high throughput sequencing (ChIP-seq), using a mouse monoclonal antibody recognizing the C-terminal domain (CTD) of the largest subunit of Pol II (RPB1, hereafter called anti-Pol II ChIP-seq). Anticipating that subtle difference may be present between the Pol II occupancies at promoters in the control and patient fibroblasts, which may be difficult to quantify, we added a consistent trace amount of Drosophila chromatin to the human fibroblast chromatin at the beginning of the ChIP procedure. As the antibody used in this experiment recognizes the CTD of both human and Drosophila RPB1, the two distinct human chromatin preparations were normalized to the Drosophila ‘spike-in.’ Surprisingly, Pol II occupancy at selected representative Pol II transcribed genes (Fig. 6A), and genome-wide, analyzed by either K-means clustering (Fig. 6B) or by metagene plots (Fig. 6C), did not change significantly when compared with control cells. Selecting a few representative Pol II-bound promoters (Fig. 6A), the ChIP-seq results were validated by ChIP-qPCR. This validation further confirmed the surprising observation that in patient fibroblasts the Pol II occupancy at the selected promoters was not significantly altered (Supplementary Material, Fig. S4A). Thus, in spite of the complete disorganization of holo-TFIID, Pol II occupancy across all transcribed genes did not change dramatically in patient fibroblasts harboring the TAF8: c.781–1G > A mutation (Fig. 6A–C).
Figure 6.
Genome wide RNA polymerase II occupancy and transcription is not significantly effected in fibroblasts harboring the TAF8: c.781–1G > A mutation. (A) UCSC genome browser snapshots of randomly selected representative genes showing Pol II occupancy in control fibroblasts (HFF-1, purple) and TAF8: c.781-1G > A patient fibroblasts (red). Arrows at TSSs show the direction of transcription. (B) Heatmap of anti-Pol II ChIP-seq generated by K-means clustering in a window showing TSS to transcription termination site (TTS) of all genes in control and patient fibroblasts. (C) Metagene plots of all expressed Pol II transcribed genes, generated from spike-in normalized anti-Pol II ChIP-seq, on control fibroblasts and TAF8 mutant fibroblasts. Average Pol II profiles were calculated for every expressed refseq gene from –5 kb region upstream of every TSS, including the gene bodies (GBs), until +5 kb downstream from the TTS. The Y-axis shows mean tag/50 bp densities. (D and E) On the basis of the clustering of Pol II occupancy in the gene bodies (Supplementary Material, Fig. S3B) Cluster 1 (D), containing 580 genes (Supplementary Material, Table S3), and Cluster 2 (E), containing 1102 genes (Supplementary Material, Table S4), profiles from –2 to +2 kb relative to TSS are shown. From these profiles, the corresponding Pol II traveling ratios were calculated and are depicted in the Supplementary Material, Figure S3C and D. In C–E, control graphs are in blue and patient graphs in pink. (F) RT-qPCR of newly transcribed mRNA in control fibroblasts and TAF8: c.781-1G > A patient fibroblasts of 11 representative genes normalized to the Pol I transcribed 45S pre-rRNA. Primers for each gene amplify fragments either only in introns or overlapping intron–exon boundaries from the indicated pre-mRNA. See Supplementary Material, Table S1 for primer sequences.
Next, we wanted to determine if Pol II pausing at promoters and/or elongation rates in gene bodies was altered in the patient cells compared with control. To this end we measured the Pol II travelling ratio, which is defined as the ratio of the Pol II occupancy signal at the promoter [from –100 to +300 bp around the transcription start site (TSS)] relative to the Pol II signal in the gene body (from +300 to +2 kb) (36,37). Because the Pol II ChIP-seq signal in the gene body can be close to background, we clustered the genes on the basis of Pol II signal in their gene body (Supplementary Material, Fig. S4B), and calculated Pol II travelling ratio [(37) and see Materials and Methods] for two gene clusters having significant gene body signal. In the first cluster, consisting of 580 genes, the Pol II promoter pausing and gene body signals as well as Pol II travelling ratios were similar between patient and control fibroblasts (Fig. 6D and Supplementary Material, Fig. S4C). In cluster 2, consisting of 1102 genes (Supplementary Material, Fig. S4B), Pol II promoter pausing was slightly decreased and the Pol II signal in the gene body was somewhat higher in patient’s fibroblasts compared with control fibroblasts (Fig. 5E). As a consequence, the Pol II travelling ratio was shifted to the left in patient fibroblasts compared with control (Supplementary Material, Fig. S4D), suggesting that in cluster 2 genes, Pol II was slightly redistributed from promoters to genes bodies in the mutant fibroblasts.
Loss of TAF8 and holo-TFIID complex do not alter mRNA synthesis from selected genes
The lack of obvious changes in the overall distribution of RNA Pol II despite the undetectable mutant TAF8 protein and holo-TFIID (Figs 5 and 6), suggested that transcription may not be dramatically affected in the patient fibroblasts. To verify this hypothesis, the level of pre-mRNA transcription of randomly selected genes was measured by RT-qPCR. Because steady-state levels of mRNA, determined by assessing total mRNA abundance, are modulated by mRNA decay rates [(8,33,34) and references therein], we measured the abundance of newly synthesized unspliced pre-mRNA relative to the abundance of the newly synthesized RNA polymerase I transcribed 45S pre-rRNA (Fig. 6F). Consistent with the lack of significant Pol II occupancy changes (Fig. 6A–E), we found that the level of transcription of all the tested genes was not affected in the patient fibroblasts as compared with two control fibroblast cell lines (Fig. 6F). In addition, we tested the abundance of some Pol II transcribed histone mRNAs, snRNA and snoRNAs in patient fibroblasts compared with control fibroblasts (Supplementary Material, Fig. S4E) and found no significant changes. Together, these results indicate that in the fibroblasts harboring the TAF8: c.781–1G > A mutation, on average, genome-wide RNA pol II occupancy, Pol II elongation rates and pre-mRNA transcription are not dramatically impaired compared with control fibroblasts.
Discussion
In this study, we identified a patient with severe intellectual disability and developmental delay, who inherited a homozygous splice site mutation in TAF8 (c.781–1G > A), encoding a subunit of the general transcription factor TFIID. In the patient fibroblasts, TAF8 protein was undetectable by western blotting and/or mass spectrometry analyses in WCEs as well as in immunopurified TAF-containing complexes (Figs 2B and C and 5). The much lower levels of mutant TAF8 compared with WT TAF8 protein in the different recombinant expression systems used, clearly shows that the mutant protein is intrinsically unstable (Fig. 3). The identified mutation leads to a seemingly complete loss of TAF8 function, and subsequent disassembly of the TFIID complex. These observations are highly unexpected given the key role attributed to TFIID in transcription. Indeed, in the mouse, germline deletion of Taf8 results in embryonic lethality at E4.0, because of apoptosis of the pluripotent ICM cells of the blastocyst (12). This early lethality demonstrates that in the absence of TAF8 protein mouse blastocysts cannot undergo gastrulation, organogenesis and further embryonic development. Moreover, we also demonstrate here that the inducible deletion of Taf8 in mESCs leads to cell death and globally affects Pol II transcription (Fig. 4). Mouse and human TAF8 proteins are 95.5% identical, and the TFIID complex as a whole is highly conserved between mouse and human. Although suffering from severe neurological deficits, the child identified in this study is physically mostly healthy and has developed well beyond the stage that one would anticipate if this mutation resulted in complete elimination of the mutated TAF8 protein in all cell types during the early stages of development.
We speculate that the TAF8 (c.781–1G > A) mutation did not cause a complete loss of TAF8 function during early embryogenesis and that residual levels of mutated TAF8 could have been sufficient to maintain blastocyst survival and embryo development. In support of this hypothesis, we observed that mESCs with low residual levels of TAF8 protein can survive and divide similarly to controls (see day 4 of 4-OH treatment in Fig. 4B and D); however, when TAF8 protein is completely lost, mESCs rapidly undergo massive apoptosis. Thus, in the patient cells, low residual TAF8 protein incorporated in limiting amounts of holo-TFIID, and partial TAF-containing complexes lacking TAF8 (Fig. 5) are sufficient to maintain Pol II transcription (Fig. 7). Alternatively, in the patient the mutant TAF8 protein might have been present in higher quantities during embryo development, and/or maybe are higher in different cell types of the patient, to which we had no access. Indeed, it has been recognized that proteins can have different stabilities in different cell types (38,39), likely owing to the interaction with different factors in the varying cellular environments. Nonetheless, from the experiments carried out on the human mutated TAF8 patient fibroblasts and by comparison with the effect of a complete loss of TAF8 function in mice, we conclude that the homozygous TAF8: c.781–1G > A mutation is the disease causing mutation.
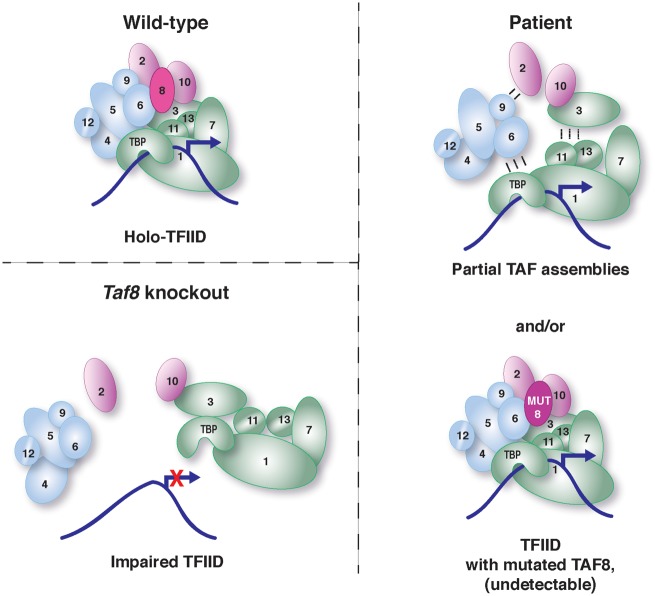
Figure 7.
Schematic representation of the TFIID composition within WT, Taf8 KO and patient cells. In WT cells holo-TFIID, containing TBP and 13 TAFs, including WT TAF8, is abundant in cells and drives faithful transcription. In Taf8 knockout cells, such as the mESCs described in Figure 4 of this study, loss of TAF8 results in impaired TFIID and no transcription. In contrast, in the patient fibroblasts either only partial TAF assemblies form, or the tiny (undetectable) amount of mutant TAF8 protein integrates into a TFIID-like complex. Either, or both of these complexes could mediate Pol II transcription initiation in patinet fibroblasts.
In the patient fibroblasts described in this study, TAF8 protein is not detectable, and TFIID was disassembled. However, recruitment of RNA polymerase II to promoters was not significantly affected on a genome-wide scale. This result together with previous knowledge (9,13,15) suggests that the PIC may exhibit more diversity than commonly thought and warrants further mechanistic investigations to determine the heterogeneity of PIC/TFIID function in different cell types.
Patient fibroblasts were used in this study, and although they were useful for performing biochemical analyses, they fail to explain the patient’s neurological phenotype. Our analyses did not reveal a global defect in Pol II occupancy/transcription, which could have been expected considering the extent of TAF8 depletion and TFIID disassembly. However, it is likely that Pol II transcription is mildly impaired as a result of the TAF8 mutation (see Fig. 7) leading to subtle changes in gene expression, which may explain some aspects of the patient phenotype when occurring during neurogenesis and brain development. This is supported by our analyses suggesting putative minor changes in Pol II elongation rates (see for example cluster 2 in Fig. 6 and Supplementary Material, Fig. S4) which should be confirmed by ChIP-seq using antibodies against phosphorylated serine 5 and serine 2 Pol II CTD.
Although many links have been made between mutations in genes encoding TFIID subunits and intellectual disabilities, the outcome of the identified mutations appears different from that of the TAF8: c.781–1G > A mutation identified here. This present mutation appears to cause a nearly complete loss of TAF8 protein, whereas all other mutations identified in other TAF genes were missense mutations expected to result in only partial loss of function of the corresponding gene product. Homozygous missense mutation and compound heterozygous mutations in TAF2 were identified in two independent consanguineous families presenting with autosomal recessive intellectual disability associated with microcephaly and thin corpus callosum (18,40). The impact of the TAF2 mutations on TFIID function was not explored experimentally, but TAF2 is known to interact with TAF8 (7), which could explain the apparent similarities between the TAF2 and TAF8 patients. Recently, homozygous TAF13 missense mutations, identified in two families with mild intellectual disability, growth retardation and microcephaly, were also suggested to affect TFIID structure (25). Missense mutations in TAF6 were also found associated with intellectual disability (16) or an autism spectrum disorder (41). Because TAF6 forms part of the TFIID core (6), it is plausible that small alterations in its structure could impact TFIID assembly. Of note, homozygous missense mutations in TAF6 were found to be associated with a Cornelia de Lange like syndrome and were reported to affect TFIID assembly (42).
Hemizygous missense mutations in the X-linked TAF1 gene were identified in male patients with a global developmental delay, intellectual disability, facial dysmorphic features, microcephaly and hypoplasia of the corpus callosum (23). The authors predicted that the TAF1 mutations likely disrupted the TAF1–TAF7 interaction, or the TAF1 bromodomain, likely affecting the structure and/or function of TFIID (23). As noted earlier, the clinical phenotypes observed in patients with mutations in several TAFs partially overlap, in particular sharing common features of variable intellectual disability and microcephaly. These similarities suggest that a common molecular mechanism, namely TFIID disruption, results in a secondary variable effect on Pol II transcription in certain, but not all cell types.
In conclusion, our detailed TAF8 patient study, including biochemical characterization of general transcription complexes by mass spectrometry and genome-wide high throughput ChIP analyses, together with previous less mechanistic reports of patients with mutations in TAF genes, demonstrates that perturbed TFIID function is less deleterious for Pol II transcription initiation than originally predicted. Additional studies using animal model systems are needed to further delineate the exact mechanism(s) responsible for the developmental, growth and neurological abnormalities seen in individuals with TAF mutations and to understand how cells can survive without holo-TFIID.
Materials and Methods
Subject
Written parents’ consent was obtained for conducting the study and showing MRIs and photographs of their affected child.
Exome sequencing
Exome sequencing performed by GeneDx revealed a homozygous splice site variant in TAF8 [NC_000006.12: g.42077099G > A (NM_138572.2: c.781–1G > A)] (Fig. 1C), whereas no other mutation or variant in genes possibly associated with the phenotype could be identified. Both parents were found to be heterozygous for the same variant. This change was observed in the gnomAD in the heterozygous state in 10 individuals, including 5 individuals of Hispanic origin. This variant was observed once out of 12 065 alleles in the NHBLI exome sequencing project.
Plasmids
The baculovirus expression vector for FLAG-tagged human TAF8 WT was PCR amplified from the pPBAC MultiBac vector, expressing TAF8 and TAF10 together, described in (7), and the cDNA was inserted in the pVL1393 baculovirus expression vector digested with Bam HI and Eco RI together with a primer coding for a FLAG-tag on the 5′ end of the hTAF8 cDNA. The mutated TAF8 (FLAG-TAF8 MUT) was generated by site-directed mutagenesis using pVL1393-FLAG-TAF8 WT vector as a template to delete the G residue at the border of exon 7 and 8 (see Fig. 1D). The WT FLAG-TAF8 and the mutated FLAG-TAF8 cassettes encoding the P2A self-cleaving peptide [GSGATNFSLLKQAGDVEENPGP (31,32)] and the FLAG-GFP sequences were inserted the pVL1393 baculovirus expression vector driven by the polyhedrin promoter. The constructs were verified by sequencing.
Isolation and culturing of human fibroblasts and Drosophila S2 cells
A biopsy was taken from the patient which was used to establish a fibroblast cell line. Control fibroblasts were either HFF1 fibroblasts (ATCC) or control FD137 (male 3 years old). Fibroblasts were cultured on 15 cm2 plates in standard fibroblast medium (DMEM/F12 with GLUTAMAX, 10% fetal calf serum (FCS), 15 mM HEPES and pen/strep) at 37°C. Drosophila S2 cells, used for ChIP spike-in were cultured in standard medium (Schneider, 10% heat inactivated FCS and 0.5% pen/strep) at 28°C with no CO2.
Generation of conditional Taf8 mutant mice and isolation of mESCs
A targeting construct was produced using a 17.6-kb fragment from a C57BL/6-derived BAC (RP23–182C6) containing the entire Taf8 gene. A loxP site was introduced 2.5 kb 5′ of exon 1 and a neomycin phosphotransferase expression cassette flanked by FRT sites also containing a single loxP site at the 3′ end was introduced between exons 8 and 9 using the recombineering method (43). This construct was electroporated into C57BL/6 ESCs, and correctly targeted cell lines were confirmed by Southern blot analysis. PCR genotyping using three oligonucleotides allowed the identification of the Taf8 exons 1 to 8-deleted allele (Taf8–), the undeleted allele (Taf8lox) and the WT allele (Taf8+) simultaneously. Amplification of the WT and undeleted alleles using primers (see Supplementary Material, Table S1) resulted in products of 222 and 345 bps, respectively. Amplification of the deleted allele resulted in a product of 429 bps.
Mice carrying the Taf8lox allele were bred to mice carrying the Rosa26Cre-ERT2 allele (44) to produce Taf8lox/lox; Rosa26-CreERT2 mice. mESCs were isolated from E3.5 blastocysts of Taf8lox/lox × Taf8lox/lox; Rosa26-CreERT2 matings, essentially as described previously (45) with the modification that 2i medium (46) and no feeder cells were used. Mouse ESCs were cultured in medium containing two inhibitors (2i; CHIR99021 and PD0325901, Axon MedChem), leukemia inhibitory factor, and 1% FCS, on gelatin-coated plates. To induce deletion of Taf8, cells were treated with 0.5 µM 4-OH tamoxifen (Sigma). Cell death was measured by trypan blue exclusion.
WCE preparations
Two methods were used for WCE isolation. In the first method (used for generating the WCE used in the co-IPs, and in the first lane of Fig. 2B), pelleted cells were resuspended in 1× volume of buffer containing 400 mM KCl, 20 mM Tris–HCl pH 7.5, 2 mM dithiothreitol (DTT), 20% glycerol and 1× complete protease inhibitors. The cells were frozen and thawed in liquid N2 four times before being centrifuged at 13 000 RPM for 10 min to pellet debris and isolate WCE supernatant. In the second method, pelleted cells were first resuspended in 1× volume of 600 mM KCl, 50 mM Tris–HCl pH 7.9, 25% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 5 mM MgCl2, 0.5% Nonidet P-40 and 1× complete protease inhibitors, then incubated on ice for 10 min, then 3× volume of another buffer containing 25 mM Tris–HCl pH 7.9, 5% glycerol, 0.5 mM DTT, 5 mM MgCl2, 0.1% Nonidet P-40, and 1× protease inhibitor cocktail was added and incubated on ice for a further 10 min, followed by centrifugation at 13 000 RPM for 10 min to pellet debris and isolate WCE supernatant.
Recombinant protein production
Recombinant baculoviruses were generated as described (47) and used for protein complex production in Sf9 insect cell culture. Infected insect cells were harvested 48 h after cell infection by centrifugation and stored at –80°C until further use.
Pellets of baculovirus-infected Sf9 insect cells expressing TAF8 constructs were resuspended in lysis buffer (400 mM KCl, 50 mM Tris–HCl pH 7.9, 10% glycerol, 0.2 mM EDTA, 0.5 mM DTT, containing 1× protease inhibitor cocktail) and crude extracts were prepared by three round freeze thawing cleared by centrifugation.
Coimmunoprecipitation experiments
Confluent plates of TAF8 mutant and control fibroblasts were harvested and lysed in WCE buffer no.1 (described earlier), and 3 mg of WCE from each genotype was used for each coimmunoprecipitation. Protein G or A magnetic Dynabeads (Invitrogen 10002D or 10004D) were incubated with 6 μl of anti-TAF10 antibody (1H8), anti-TAF7 antibody (3475) or anti-GST antibody (15TF21D10) for 2 h, then unbound antibody was removed with two washes in IP100 buffer (25 mM Tris–HCl pH 7.5, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 100 mM KCl, 2 mM DTT and 1× complete protease inhibitors). Fibroblast WCE was made up to 600 μl in IP100 buffer, precleared then added to Dynabeads coupled to antibody, and incubated overnight at 4°C with gentle agitation. The following day the magnetic beads were isolated with a magnetic rack, the unbound material was collected, and the beads were subjected to two rounds of washing for 10 min each with IP500 buffer (25 mM Tris–HCl pH 7.5, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 500 mM KCl, 2 mM DTT and 1× complete protease inhibitors), followed by IP100 buffer. For the anti-TAF10 coimmunopurification, proteins bound to the beads were eluted by adding 60 μl of 2 mg/ml competing TAF10 peptide for 4 h, then again for 2 h (48). For the anti-TAF7 co-IP, proteins were eluted with 0.1 M pH 2.8 glycine, then neutralized with 2 M Tris pH 8.8. Co-IP-ed proteins were separated on an 4–12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, along with the input extract and flow through samples, and were probed with antibodies against TFIID subunits.
Antibodies
Mouse monoclonal (mAb) and rabbit polyclonal (pAb) antibodies raised against the following proteins have been described previously, or were purchased commercially: anti-TBP (mAb 3G3) (49), anti-TAF2 (pAb 3083) (7), anti-TAF4 (mAb 32TA2B9) (11), anti-TAF5 (mAb 1TA1C2) (50), anti-TAF6 (mAb 25TA2G7) (50), anti-TAF7 (mAb 19TA) (51) and (pAb 3475) (52), anti-TAF8 (pAb 3478 and 3477) (52), anti-TAF10 (mAb 6TA2B11) (48) and TAF12 (22TA) (53), anti-GST antibody (mAb 15TF21D10) (54), anti-GFP (Abcam Ab290), anti-γ-Tubulin (Sigma Aldrich T6557), and anti-FLAG (Sigma Aldrich M2).
Mass spectrometry
IP-ed protein samples were analyzed using an Ultimate 3000 nano-RSLC (Thermo Scientific, San Jose, California) coupled in line with a linear trap Quadrupole (LTQ)-Orbitrap ELITE mass spectrometer via a nano-electrospray ionization source (Thermo Scientific). Data were analyzed by calculation of the NSAFbait (52,55). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (56,57) with the dataset identifier PXD008084.
ChIP-seq and ChIP-qPCR
For each ChIP experiment, a minimum of 20 × 15 cm2 plates were used per genotype. Approximately 2 × 106 fibroblast cells were seeded onto each 15 cm2 plate, then the following day the fibroblasts were fixed in 1% PFA for 10 min at room temperature, then the PFA was quenched with glycine at a final concentration of 125 mM for 5 min at room temperature. After two washes in cold 1× PBS, fibroblasts were scraped, and pelleted. Nuclei were isolated by incubating cells with nuclear isolation buffer (50 mM Tris–HCl pH 8.0, 2 mM EDTA pH 8.0, 0.5% Nonidet P-40, 10% glycerol and 1× protease inhibitors) for 10 min at 4°C with gentle agitation, followed by centrifugation at 13 000 RPM to pellet the nuclei. Nuclei were resuspended in sonication buffer (0.1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.0 and 1× protease inhibitors) then chromatin was sheared with the Covaris E220 sonicator and chromatin concentration was measured with the Qubit 3.0 (ThermoFischer). A minimum of 50 µg of chromatin was used for each IP, which was diluted in ChIP dilution buffer (0.5% Nonidet P-40, 16.7 mM Tris–HCl pH 8.0, 1.2 mM EDTA, 167 mM NaCl and 1× protease inhibitor) and each sample was spiked with 1/10 amount of Drosophila sheared chromatin for normalization. Antibodies used for the ChIP included anti-RPB1 CTD (1PB 7G5) (58), and mouse IgG (Jackson Laboratories) which were incubated with the chromatin overnight with gentle agitation at 4°C. The following day, magnetic beads were isolated and washed for 5 min at 4°C, once with low salt wash buffer (0.1% SDS, 0.5% Nonidet P-40, 2 mM EDTA, 150 mM NaCl, 20 mM and Tris–HCl pH 8.0), high salt wash buffer (0.1% SDS, 0.5% Nonidet P-40, 2 mM EDTA, 500 mM NaCl, 20 mM and Tris–HCl pH 8.0), and LiCl wash buffer (0.2 M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH8.0), then twice with TE buffer, then the beads were incubated in freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C with shaking to elute complexes. Crosslinks were reversed with by adding NaCl at a final concentration of 0.2 M overnight as well as 50 μg/ml RNase A at 65°C and the following day the samples were treated with 20 µg Protinase K, 26.6 of 1 M Tris–HCl pH 7.9, and 13.3 µl of 0.5 M EDTA, and DNA was isolated with phenol chloroform. The precipitated DNA was used to generate libraries with the Diagenode MicroPlex Library Preparation kit v2 for ChIP-seq according to the manufacturer’s instructions, with read lengths of 1 × 50 bp, then the samples were sequenced on Hiseq 4000, reads were mapped to the human hg38 and Drosophila dm3 assemblies. Samples were normalized on the basis of Drosophila spike-in as described in (59) then peak calling was performed using the MACS2 software (60). Heatmaps and Metagene plots were generated by the seqMINER software (61). The ChIP-seq data were deposited on the Gene Expression Omnibus (GEO) database with the accession number GSE106252. ChIP-qPCR primers were designed to span a region directly under the RNA Pol II peaks identified by the Pol II ChIP-seq, for randomly selected genes including GAPDH, HSP90AB1, SPARC, RPL13a, ADAMST1 and ID3 as well as a negative region at chromosome 9. Primer sequences are detailed in Supplementary Material, Table S1. The clustering of the genes according their Pol II density in the –2 to +2 kb regions relative to the TSS was carried out by the seqMINER software (61). Pol II traveling ratio was calculated as described in (37), except that the TSS tag density values in –100 to +300 bp regions for each gene in clusters 1 and 2 were divided by the corresponding gene body tag density values in +300 to 2 kb regions.
Newly synthesized mRNA analysis
Cells were harvested, and RNA was extracted followed by DNase I treatment, according to the manufacturer’s instructions (Macherey-Nagel). cDNA was generated with Super Script II (Thermo Fischer Scientific) using random hexamer primers, according to the manufacturer’s instructions. To measure genomic DNA contamination in the cDNA preparations, cDNA synthesis reactions were conducted without reverse transcriptase enzyme (–RT). The CP values generated from RT-qPCR with the –RT samples were always >35 cycles, indicating that genomic DNA contamination contribution was negligible. RT-qPCR primers were designed to amplify intron-exon boundaries (Supplementary Material, Table S1).
Accession numbers
The ChIP-seq data were deposited on the GEO database with the accession number GSE106252. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (56,57) with the dataset identifier PXD008084.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
Without the consent and cooperation of the family in this study, this work would not have been possible. We are grateful to the IGBMC cell culture facility for assistance in the expansion of the patient primary fibroblast cells. The two control cell lines used in this study, HFF-1 (ATCC) and FD137, were gifts from E. Weiss and H. Puccio, respectively. We thank J.-L. Mandel for helpful discussions, J. Chelly for critically reading the manuscript, F. Mattioli for advice with the emetine experiment and S. Bour for help with Figure 7. We are grateful to the IGBMC proteomics facility for performing the mass spectrometry, and we thank the IGBMC GenomEast platform [a member of the ‘France Génomique’ consortium (ANR-10-INBS-0009)] for preparing libraries and sequencing the ChIP material.
Conflict of Interest statement. None declared.
Funding
This work was supported by funds from Agence Nationale de la Recherche (ANR-13-BSV8–0021-03 DiscoverIID to LT and IB) and the European Research Council Advanced grant (ERC-2013–340551, Birtoaction to L.T.). This work was also supported by funds from CNRS, INSERM, Strasbourg University and Investissements d'Avenir grants (ANR-10-IDEX-0002–02 and ANR-10-LABX-0030-INRT) to D.D. and L.T., by the Australian National Health and Medical Research Council (NHMRC) though grants and research fellowships (575512, 1003435, 1084504, 1081421) and the Independent Research Institutes Infrastructure Support (IRIIS) Scheme and by the Victorian State Government OIS (Operational Infrastructure Support) grant to A.K.V. and T.T. I.B. is recipient of a senior investigator award from the Wellcome Trust (106115/Z/14/Z). Funding to pay the Open Access publication charges for this article was provided by the European Research Council Advanced grant (ERC-2013-340551, Birtoaction) to L.T.
References
- 1. Muller F., Zaucker A., Tora L. (2010) Developmental regulation of transcription initiation: more than just changing the actors. Curr. Opin. Genet. Dev., 20, 533–540. [DOI] [PubMed] [Google Scholar]
- 2. Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]
- 3. Roeder R.G. (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci., 21, 327–335. [PubMed] [Google Scholar]
- 4. Gangloff Y.G., Sanders S.L., Romier C., Kirschner D., Weil P.A., Tora L., Davidson I. (2001) Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol., 21, 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guermah M., Ge K., Chiang C.M., Roeder R.G. (2003) The TBN protein, which is essential for early embryonic mouse development, is an inducible TAFII implicated in adipogenesis. Mol. Cell, 12, 991–1001. [DOI] [PubMed] [Google Scholar]
- 6. Bieniossek C., Papai G., Schaffitzel C., Garzoni F., Chaillet M., Scheer E., Papadopoulos P., Tora L., Schultz P., Berger I. (2013) The architecture of human general transcription factor TFIID core complex. Nature, 493, 699–702. [DOI] [PubMed] [Google Scholar]
- 7. Trowitzsch S., Viola C., Scheer E., Conic S., Chavant V., Fournier M., Papai G., Ebong I.O., Schaffitzel C., Zou J.. et al. (2015) Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat. Commun., 6, 6011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warfield L., Ramachandran S., Baptista T., Devys D., Tora L., Hahn S. (2017) Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol. Cell, 68, 118–129 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gegonne A., Tai X., Zhang J., Wu G., Zhu J., Yoshimoto A., Hanson J., Cultraro C., Chen Q.R., Guinter T.. et al. (2012) The general transcription factor TAF7 is essential for embryonic development but not essential for the survival or differentiation of mature T cells. Mol. Cell. Biol., 32, 1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martianov I., Viville S., Davidson I. (2002) RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science, 298, 1036–1039. [DOI] [PubMed] [Google Scholar]
- 11. Mohan W.S. Jr., Scheer E., Wendling O., Metzger D., Tora L. (2003) TAF10 (TAF(II)30) is necessary for TFIID stability and early embryogenesis in mice. Mol. Cell. Biol., 23, 4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Voss A.K., Thomas T., Petrou P., Anastassiadis K., Scholer H., Gruss P. (2000) Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos. Development, 127, 5449–5461. [DOI] [PubMed] [Google Scholar]
- 13. Indra A.K., Mohan W.S. 2nd, Frontini M., Scheer E., Messaddeq N., Metzger D., Tora L. (2005) TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Dev. Biol., 285, 28–37. [DOI] [PubMed] [Google Scholar]
- 14. Fadloun A., Kobi D., Pointud J.C., Indra A.K., Teletin M., Bole-Feysot C., Testoni B., Mantovani R., Metzger D., Mengus G.. et al. (2007) The TFIID subunit TAF4 regulates keratinocyte proliferation and has cell-autonomous and non-cell-autonomous tumour suppressor activity in mouse epidermis. Development, 134, 2947–2958. [DOI] [PubMed] [Google Scholar]
- 15. Tatarakis A., Margaritis T., Martinez-Jimenez C.P., Kouskouti A., Mohan W.S. 2nd, Haroniti A., Kafetzopoulos D., Tora L., Talianidis I. (2008) Dominant and redundant functions of TFIID involved in the regulation of hepatic genes. Mol. Cell, 31, 531–543. [DOI] [PubMed] [Google Scholar]
- 16. Alazami A.M., Patel N., Shamseldin H.E., Anazi S., Al-Dosari M.S., Alzahrani F., Hijazi H., Alshammari M., Aldahmesh M.A., Salih M.A.. et al. (2015) Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep., 10, 148–161. [DOI] [PubMed] [Google Scholar]
- 17. Bauer P., Laccone F., Rolfs A., Wullner U., Bosch S., Peters H., Liebscher S., Scheible M., Epplen J.T., Weber B.H.. et al. (2004) Trinucleotide repeat expansion in SCA17/TBP in white patients with Huntington's disease-like phenotype. J. Med. Genet., 41, 230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hellman-Aharony S., Smirin-Yosef P., Halevy A., Pasmanik-Chor M., Yeheskel A., Har-Zahav A., Maya I., Straussberg R., Dahary D., Haviv A.. et al. (2013) Microcephaly thin corpus callosum intellectual disability syndrome caused by mutated TAF2. Pediatr. Neurol., 49, 411–416 e411. [DOI] [PubMed] [Google Scholar]
- 19. Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. (1999) A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum. Mol. Genet., 8, 2047–2053. [DOI] [PubMed] [Google Scholar]
- 20. Makino S., Kaji R., Ando S., Tomizawa M., Yasuno K., Goto S., Matsumoto S., Tabuena M.D., Maranon E., Dantes M.. et al. (2007) Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am. J. Hum. Genet., 80, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P.. et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature, 478, 57–63. [DOI] [PubMed] [Google Scholar]
- 22. Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. (2001) SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet., 10, 1441–1448. [DOI] [PubMed] [Google Scholar]
- 23. O’Rawe J.A., Wu Y., Dörfel M.J., Rope A.F., Au P.Y., Parboosingh J.S., Moon S., Kousi M., Kosma K., Smith C.S.. et al. (2015) TAF1 variants are associated with dysmorphic features, intellectual disability, and neurological manifestations. Am. J. Hum. Genet., 97, 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stevanin G., Fujigasaki H., Lebre A.S., Camuzat A., Jeannequin C., Dode C., Takahashi J., San C., Bellance R., Brice A.. et al. (2003) Huntington's disease-like phenotype due to trinucleotide repeat expansions in the TBP and JPH3 genes. Brain, 126, 1599–1603. [DOI] [PubMed] [Google Scholar]
- 25. Tawamie H., Martianov I., Wohlfahrt N., Buchert R., Mengus G., Uebe S., Janiri L., Hirsch F.W., Schumacher J., Ferrazzi F.. et al. (2017) Hypomorphic pathogenic variants in TAF13 are associated with autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet., 100, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toyoshima Y., Yamada M., Onodera O., Shimohata M., Inenaga C., Fujita N., Morita M., Tsuji S., Takahashi H. (2004) SCA17 homozygote showing Huntington's disease-like phenotype. Ann. Neurol., 55, 281–286. [DOI] [PubMed] [Google Scholar]
- 27. Wu Y.R., Fung H.C., Lee-Chen G.J., Gwinn-Hardy K., Ro L.S., Chen S.T., Hsieh-Li H.M., Lin H.Y., Lin C.Y., Li S.N.. et al. (2005) Analysis of polyglutamine-coding repeats in the TATA-binding protein in different neurodegenerative diseases. J. Neural. Transm. (Vienna), 112, 539–546. [DOI] [PubMed] [Google Scholar]
- 28. Zuhlke C., Hellenbroich Y., Dalski A., Kononowa N., Hagenah J., Vieregge P., Riess O., Klein C., Schwinger E. (2001) Different types of repeat expansion in the TATA-binding protein gene are associated with a new form of inherited ataxia. Eur. J. Hum. Genet., 9, 160–164. [DOI] [PubMed] [Google Scholar]
- 29. Noensie E.N., Dietz H.C. (2001) A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol., 19, 434–439. [DOI] [PubMed] [Google Scholar]
- 30. Soutoglou E., Demeny M.A., Scheer E., Fienga G., Sassone-Corsi P., Tora L. (2005) The nuclear import of TAF10 is regulated by one of its three histone fold domain-containing interaction partners. Mol. Cell. Biol., 25, 4092–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., Choi S.Y. (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One, 6, e18556.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y., Wang F., Wang R., Zhao P., Xia Q. (2015) 2A self-cleaving peptide-based multi-gene expression system in the silkworm Bombyx mori. Sci. Rep., 5, 16273.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baptista T., Grunberg S., Minoungou N., Koster M.J.E., Timmers H.T.M., Hahn S., Devys D., Tora L. (2017) SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell, 68, 130–143 e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonnet J., Wang C.Y., Baptista T., Vincent S.D., Hsiao W.C., Stierle M., Kao C.F., Tora L., Devys D. (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev., 28, 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res., 5, 2339–2347. [DOI] [PubMed] [Google Scholar]
- 36. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. (2010) c-Myc regulates transcriptional pause release. Cell, 141, 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen F.X., Woodfin A.R., Gardini A., Rickels R.A., Marshall S.A., Smith E.R., Shiekhattar R., Shilatifard A. (2015) PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell, 162, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Danielsson J., Mu X., Lang L., Wang H., Binolfi A., Theillet F.X., Bekei B., Logan D.T., Selenko P., Wennerstrom H.. et al. (2015) Thermodynamics of protein destabilization in live cells. Proc. Natl. Acad. Sci. U. S. A., 112, 12402–12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Offord E.A., Chappuis P.O., Beard P. (1993) Different stability of AP1 proteins in human keratinocyte and fibroblast cells: possible role in the cell-type specific expression of human papillomavirus type 18 genes. Carcinogenesis, 14, 2447–2455. [DOI] [PubMed] [Google Scholar]
- 40. Halevy A., Basel-Vanagaite L., Shuper A., Helman S., Har-Zahav A., Birk E., Maya I., Kornreich L., Inbar D., Nurnberg G.. et al. (2012) Microcephaly-thin corpus callosum syndrome maps to 8q23.2-q24.12. Pediatr. Neurol., 46, 363–368. [DOI] [PubMed] [Google Scholar]
- 41. C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z.. et al. (2017) Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci., 20, 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yuan B., Pehlivan D., Karaca E., Patel N., Charng W.L., Gambin T., Gonzaga-Jauregui C., Sutton V.R., Yesil G., Bozdogan S.T.. et al. (2015) Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Invest., 125, 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu P., Jenkins N.A., Copeland N.G. (2003) A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res., 13, 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Soriano P. (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet., 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 45. Voss A.K., Thomas T., Gruss P. (1997) Germ line chimeras from female ES cells. Exp. Cell Res., 230, 45–49. [DOI] [PubMed] [Google Scholar]
- 46. Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature, 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trowitzsch S., Bieniossek C., Nie Y., Garzoni F., Berger I. (2010) New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol., 172, 45–54. [DOI] [PubMed] [Google Scholar]
- 48. Wieczorek E., Brand M., Jacq X., Tora L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- 49. Brou C., Chaudhary S., Davidson I., Lutz Y., Wu J., Egly J.M., Tora L., Chambon P. (1993) Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J., 12, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dantonel J.C., Murthy K.G., Manley J.L., Tora L. (1997) Transcription factor TFIID recruits factor CPSF for formation of 3' end of mRNA. Nature, 389, 399–402. [DOI] [PubMed] [Google Scholar]
- 51. Lavigne A.C., Mengus G., May M., Dubrovskaya V., Tora L., Chambon P., Davidson I. (1996) Multiple interactions between hTAFII55 and other TFIID subunits. Requirements for the formation of stable ternary complexes between hTAFII55 and the TATA-binding protein. J. Biol. Chem., 271, 19774–19780. [DOI] [PubMed] [Google Scholar]
- 52. Bardot P., Vincent S.D., Fournier M., Hubaud A., Joint M., Tora L., Pourquie O. (2017) The TAF10-containing TFIID and SAGA transcriptional complexes are dispensable for early somitogenesis in the mouse embryo. Development, 144, 3808–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brand M., Moggs J.G., Oulad-Abdelghani M., Lejeune F., Dilworth F.J., Stevenin J., Almouzni G., Tora L. (2001) UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J., 20, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nagy Z., Riss A., Fujiyama S., Krebs A., Orpinell M., Jansen P., Cohen A., Stunnenberg H.G., Kato S., Tora L. (2010) The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci., 67, 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fournier M., Orpinell M., Grauffel C., Scheer E., Garnier J.M., Ye T., Chavant V., Joint M., Esashi F., Dejaegere A.. et al. (2016) KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun., 7, 13227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S.. et al. (2017) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res., 45, D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vizcaino J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T.. et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res., 44, 11033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lebedeva L.A., Nabirochkina E.N., Kurshakova M.M., Robert F., Krasnov A.N., Evgen'ev M.B., Kadonaga J.T., Georgieva S.G., Tora L. (2005) Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc. Natl. Acad. Sci. U. S. A., 102, 18087–18092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Orlando D.A., Chen M.W., Brown V.E., Solanki S., Choi Y.J., Olson E.R., Fritz C.C., Bradner J.E., Guenther M.G. (2014) Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep., 9, 1163–1170. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ye T., Ravens S., Krebs A.R., Tora L. (2014) Interpreting and visualizing ChIP-seq data with the seqMINER software. Methods Mol. Biol., 1150, 141–152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.