Abstract
Fragile X syndrome (FXS) is a monogenic form of intellectual disability and autism spectrum disorder caused by the absence of the fragile X mental retardation protein (FMRP). In biological models for the disease, this leads to upregulated mRNA translation and as a consequence, deficits in synaptic architecture and plasticity. Preclinical studies revealed that pharmacological interventions restore those deficits, which are thought to mediate the FXS cognitive and behavioral symptoms. Here, we characterized the de novo rate of protein synthesis in patients with FXS and their relationship with clinical severity. We measured the rate of protein synthesis in fibroblasts derived from 32 individuals with FXS and from 17 controls as well as in fibroblasts and primary neurons of 27 Fmr1 KO mice and 20 controls. Here, we show that levels of protein synthesis are increased in fibroblasts of individuals with FXS and Fmr1 KO mice. However, this cellular phenotype displays a broad distribution and a proportion of fragile X individuals and Fmr1 KO mice do not show increased levels of protein synthesis, having measures in the normal range. Because the same Fmr1 KO animal measures in fibroblasts predict those in neurons we suggest the validity of this peripheral biomarker. Our study offers a potential explanation for the comprehensive drug development program undertaken thus far yielding negative results and suggests that a significant proportion, but not all individuals with FXS, may benefit from the reduction of excessive levels of protein synthesis.
Introduction
Adaptations of synaptic strength to neuronal activity are thought to be cardinal in learning and memory (1). Synaptic integrity is compromised in many neurodevelopmental disorders including broad clinical categories such as intellectual disabilities (IDs), autism and schizophrenia as well as specific monogenic or monolocus forms of IDs and autism such as tuberous sclerosis, 16p11.2 deletion and fragile X syndrome (FXS) (2–4). The latter is caused by the loss of function of FMR1 and subsequent lack of the resulting protein FMRP (5). One of the molecular mechanisms regulating spine shaping is local dendritic protein synthesis that affords spatial and temporal regulation of gene expression enabling synapses to autonomously alter their structure and function (6–8). FMRP is crucial in regulating this process and partial or complete lack of FMRP leads to an increase in protein translation at synapses (9–12).
The metabotropic glutamate receptor subtype 5 (mGluR5) theory of FXS posits that imbalance of mechanisms involved in protein translation and synaptic shaping is driving many of the symptoms observed in patients with FXS (13). Compelling data shows that altered mechanisms regulating levels of protein synthesis, as well as cognitive and behavioral symptoms, can be restored by reducing mGluR5 signaling genetically or with pharmacological treatments in mouse and fly models of FXS (Fmr1 KO) (14–18). Furthermore, pharmacological or peptide-based interventions can partially or fully restore appropriate rates of de novo protein synthesis as well as synaptic architecture and plasticity. These include several mGluR5 antagonists, gamma-butyric acid (GABAB) agonists, statins, lithium and ribosomal protein tyrosine kinase S6 (S6K) inhibitors. Genetic interventions [including mGluR5 reduction by haplo-insufficiency, striatal enriched tyrosine phosphatase (STEP) signaling reduction, MMP9 reduction and S6K signaling reduction] can also restore these molecular and cellular phenotypes (14,19–34). Recently it has been shown that mGluR-mediated increase of protein synthesis is sustained by the excessive production of soluble amyloid beta precursor protein α (sAPPα) due to the impaired processing of amyloid beta precursor protein (APP) during a critical developmental window (35). More importantly, treatment of FXS mice with a cell permeable peptide able to modulate ADAM metallopeptidase domain 10 (ADAM10) activity, and therefore APP processing, restores protein synthesis to wild type (WT) levels and rescues behavioral deficits that constitute a hallmark of the disease (35).
The above mentioned preclinical data led to the development of one of the most comprehensive drug development programs undertaken thus far for a genetically defined group of neurodevelopmental disorders. It was conducted in parallel by several pharmaceutical companies, and academic research centers assessing the effect of mGluR5 antagonists, GABAA and GABAB agonists in children, adults and adolescents with FXS. Unfortunately, what appeared to be an optimal translational scenario in FXS has not led to the expected results (36,37), and none of the human studies have demonstrated yet efficacy in children, adolescents or adults with FXS on the primary outcome measures which were mainly behavioral questionnaires (37–39). These sobering results have not deterred the community and despite these setbacks, molecular mechanisms controlling protein synthesis continue to be the prime targets in FXS and other neurodevelopmental disorders. This is illustrated by several large and innovative ongoing clinical trials targeting these mechanisms (www.clinicaltrial.gov). These ‘second generation’ trials attempt to avoid pitfalls potentially related to the aforementioned negative results by enrolling younger patients, using objective cognitive measures and biomarkers including EEG and eye tracking (40,41). Dysregulated protein synthesis has been observed in the animal models of FXS (42,43) and suggested to be pathogenic in FXS. Altered protein synthesis in human patients with FXS has only been investigated in a few studies. In a study of immortalized lymphoblastoid cell lines derived from one patient with FXS and one healthy control that used a fluorescent metabolic labeling method to measure nascent protein synthesis, higher rates of synthesis were observed in patient-derived cells compared to control (44). In a L-[1–11C]leucine positron emission tomography study, rates of cerebral protein synthesis (rCPS) were found to be lower in 15 FXS individuals compared to 12 age-matched controls (45). This unexpected finding, discordant with what is seen in the Fmr1 KO mice was interpreted as a side effect of propofol sedation used in the study and subsequently shown to decrease rates of protein synthesis in Fmr1 KO mice (45,46). A study based on human fibroblasts derived from FXS patients focused on demonstrating the relevance of fibroblasts as in-vitro model and in particular, whether these cells can be used to evaluate drugs prior to their use in clinical trials. Basal [3H]leucine incorporation rates in fibroblast lines derived from eight individuals with FXS showed elevated rates of protein synthesis compared to nine controls along with increased levels of phosphorylated mechanistic target of rapamycin (p-mTOR). A similar trend was observed in peripheral blood mononuclear cells (PBMCs) derived from patients with FXS for phosphorylated extracellular signal regulated kinase 1/2 (p-ERK 1/2) and phosphorylated p70 ribosomal S6 kinase 1 (p-S6K1) (46,47). Treatment with small molecules that inhibit S6K1, and a known FMRP target, phosphoinositide 3-kinase (PI3K) catalytic subunit p110β, lowered the rates of protein synthesis in both control and FXS fibroblasts (44). The small sample size, and the lack of clinical data prevented from investigating the distribution and variance of this cellular trait as well as its relationship with clinical manifestations in humans. To which extent excessive protein synthesis is associated with cognitive and behavioral impairments remains unknown. The aim of the current study was to validate in humans the protein synthesis findings obtained in the Fmr1 KO mouse. We characterized the distribution of de novo rates of protein synthesis in patients with FXS using fibroblast cells. To establish cross-tissue correlations, we performed in parallel, measures on fibroblasts and primary neurons from the same Fmr1 KO mice. In addition, we investigated the relationship between basal rates of protein synthesis and the severity of the clinical symptoms in individuals with FXS.
Results
Fibroblasts from patients with FXS show variable levels of protein synthesis
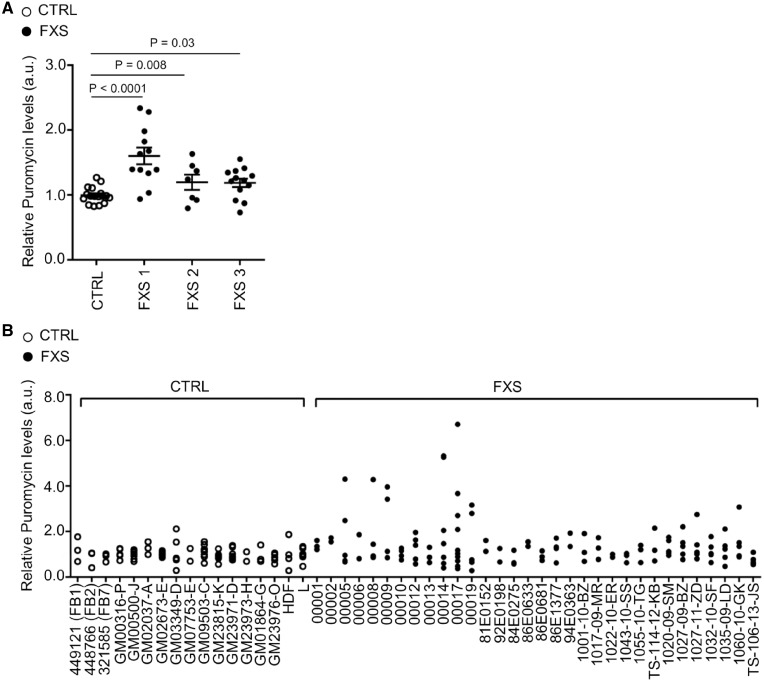
We characterized de novo protein synthesis in primary fibroblasts from 32 individuals with FXS (age range 6–69) in three independent cohort datasets and from 17 controls (age range from 10 to 50) described in the Supplementary Material, Table S1. We have chosen the SUnSET assay based on puromycin incorporation, as it has been shown to be a robust and accurate method for measuring relative rates of protein synthesis in cell culture (48). Fibroblasts derived from patients with FXS were established at the University Hospital in Lausanne (FXS1), at the Erasmus Medical Center in Rotterdam (FXS2) and at the MIND Institute in Sacramento (FXS3). While within the control group each individual presented a comparable level of protein synthesis and very low technical and biological variability, some patients with FXS presented very high rates of protein synthesis (+200% of the normal levels) and increased technical and biological variability (Fig. 1A and B; and Supplementary Material, Fig. S1A). We observed a 34% increase (corresponding to a d-Cohen effect size of 0.48, non-parametric ANOVA P = 0.0008) in the total level of de novo protein synthesis for the FXS group (FXS 1 + 2 + 3, n = 32) compared to controls (n = 17). Of note, a third of FXS patients showed rates within the normal range (Fig. 1B). In particular, 21.9% of the FXS individuals displayed levels below the 50th percentile while 9.4% showed levels between the 50th and the 90th percentile of the control group. This variability was not explained by participant’s age, gender or the number of cell passages. In particular, we conducted replicates to investigate the effect of multiple cell passages on the level of protein synthesis, three measures were performed for each cell line at the first, second and third passage (Supplementary Material, Fig. S2). A linear regression excluded any correlation between the number of cell passages and measures of de novo protein synthesis in the control group (P = 0.40, non-parametric ANOVA) as well as in the FXS group (P = 0.31, non-parametric ANOVA).
Figure 1.
Global protein synthesis in human fibroblasts. (A) Quantification of puromycin incorporation normalized to Coomassie and Vinculin in control and FXS fibroblasts (FXS1, FXS2 and FXS3 cohort, respectively). Each dot represents the average of at least two independent experiments performed on successive cell passages per individual. The bars represent the SEM; non-parametric ANOVA (n = 17 controls, n = 12 FXS1, n = 7 FXS2, n = 13 FXS3). (B) The panel shows multiple data points per individual representing at least two independent experiments performed on successive cell passages (n = 17 controls, n = 32 FXS).
Levels of protein synthesis are not correlated to measures of FMR1 expression
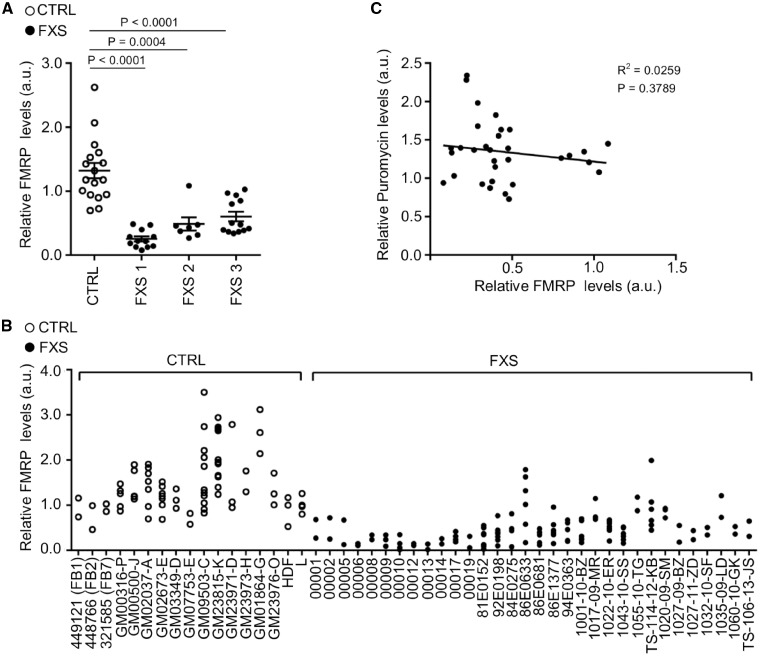
It has been largely shown that FMRP regulates the rate of protein synthesis by inhibiting translation, so the most straightforward hypothesis to explain the variability of protein synthesis levels was differences in levels of FMRP (33,49,50). Indeed, individuals with fragile X syndrome present different levels of FMR1 methylation and mosaicism resulting in variable FMRP expression (Fig. 2A and B;Supplementary Material, Table S2 and Fig. S1B). As expected, we observed a higher FMRP levels (1.86 fold-increase) in the mosaic participants (P = 0.002) compared to individuals with a full mutation without mosaicism. However, FMRP levels in the FXS group did not show any correlation with de novo protein synthesis (Pearson correlation coefficient −0.16, P = 0.3789, Spearman correlation coefficient −0.18, P = 0.3052) (Fig. 2C). All the parameters relative to FMR1 gene expression were correlated among themselves, specifically: the status of FMR1 promoter methylation in blood, FMR1 mRNA expression in blood and fibroblasts, FMRP staining in lymphocytes (Table 1). The lack of correlation between lymphocyte and fibroblast FMRP-related measures may be partially explained by the use of two different techniques for the evaluation of FMRP level, a semi-quantitative western blot for fibroblasts and a less accurate and qualitative method based on the immunostaining for lymphocyte (percentage of lymphocytes stained by FMRP antibodies). Furthermore, this inconsistency may potentially be affected by the level of intra tissue mosaicism. Indeed, Pretto and colleagues described differences in methylation and size mosaicism occurring between blood and fibroblasts in a cohort of 18 FXS individuals (51).
Figure 2.
FMRP levels in human fibroblasts. (A) FMRP levels normalized to Coomassie and Vinculin in control and FXS fibroblast groups. Each dot represents the average of at least two independent determinations per individual. The bars represent the SEM; non-parametric ANOVA (n = 17 controls, n = 12 FXS1, n = 7 FXS2, n = 13 FXS3). (B) In the panel each dot represents a single technical replicate (n = 17 controls, n = 32 FXS). (C) Correlation between FMRP levels and puromycin signals measured by the SUnSET assay in fibroblast cultures from individuals with FXS.
Table 1.
Correlation between FMR1 gene expression and levels of protein synthesisa
|
FMR1 mRNA |
FMRP |
Protein Synthesis |
|||
|---|---|---|---|---|---|
| Blood | Fibroblasts | Lymphocytes IHC | Fibroblasts W Blot | Fibroblasts | |
| N = 20 | N = 30 | N = 20 | N = 32 | N = 32 | |
| FMR1 Methylation, blood n=20 |
|
NS |
|
NS | NS |
| FMR1 mRNA, blood | NS |
|
NS | NS | |
| FMR1 mRNA, Fibroblasts | NS |
|
NS | ||
| FMRP Lymphocytes IHC | NS | NS | |||
| FMRP Fibroblasts W Blot | NS | ||||
Correlation analysis between protein synthesis measured in fibroblasts and different measures of FMR1 expression in blood, and fibroblasts from patients with FXS (Pearson correlation test). Correlation analysis indicates that the same measures in blood do not correlate with those performed in fibroblasts. Measures of protein synthesis in fibroblasts do not correlate with any of the other measures of FMR1 expression. Spearman correlation analysis (data not shown) does not change any of the results reported in the Table 1. (NS= Pearson and Spearman tests not significant).
We did not observe any correlation between the levels of de novo protein synthesis, FMR1 mRNA and FMRP protein levels in blood as well as in fibroblasts. Furthermore, amongst the different interactors of FMRP (52) we analyzed the expression level of CYFIP1 (Cytoplasmic FMR1-interacting Protein1), a specific translational regulator (10,53) and of eIF4E, a general regulator of protein translation. We did not observe any change between FXS and controls and no correlation was observed with the level of protein synthesis (data not shown). This suggests that additional factors, independent from FMR1 gene expression, contribute to the variability of the rate of de novo protein synthesis in individuals with fragile X syndrome.
Fmr1 KO neurons and fibroblasts show an upregulation of de novo protein synthesis
To further explore the validity of our observations in human fibroblasts and address the contribution of heterogeneous genetic background and individual variability, we analyzed the level of protein synthesis in 30 Fmr1 KO mice (27 MEFs and 30 neuronal primary cultures), in two different isogenic backgrounds (C57BL/6J and FVB.129P2) and with no expression of functional FMRP (54).
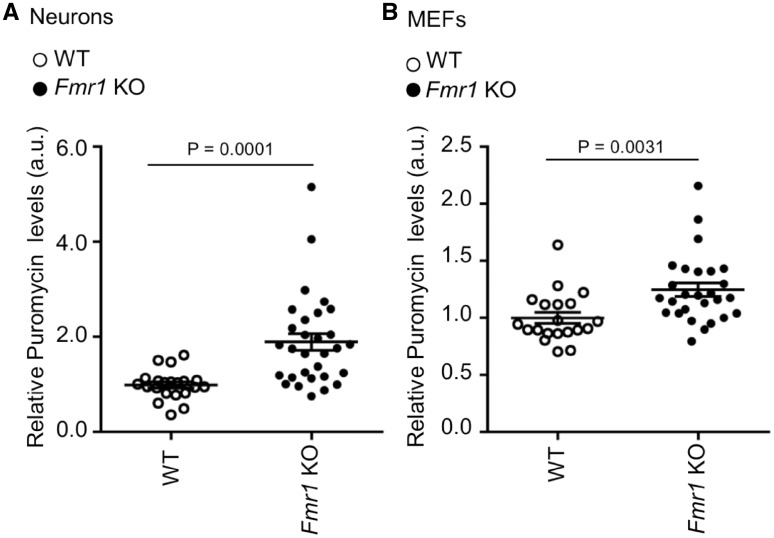
We monitored protein synthesis by the SUnSET analysis in primary cortical neurons and mouse embryonic fibroblasts (MEFs) of Fmr1 KO mice (Fig. 3;Supplementary Material, Fig. S1C). As opposed to FXS in humans which is caused by a CGG expansion, leading to variable epigenetic silencing and low FMRP expression in mosaic cases, the knockout mouse completely lacks FMRP expression (54). Levels of puromycin incorporation were measured in independent neuronal cultures from Fmr1 KO (n = 30) and WT mice (n = 21). In neurons, protein synthesis levels were significantly increased (non-parametric ANOVA, P = 0.0001) compared to WT (Fig. 3A), consistent with previous findings (35). Of note, levels of puromycin incorporation measured in MEF from Fmr1 KO (n = 27) and WT (n = 20) mice were increased as well (Fig. 3B) although the effect size was smaller (non-parametric ANOVA, P = 0.0031). Interestingly, in the FXS mouse model in which the expression of FMRP is absent, measures in 17% of the neuronal lines and 19% of the MEFs were within the normal range and showed a broad distribution. This suggests again that variance in the dysregulation of de novo protein synthesis is determined by factors likely independent of FMRP.
Figure 3.
Levels of protein synthesis in mouse primary neurons and embryonic fibroblasts. (A) Puromycin incorporation in primary neurons from WT and Fmr1 KO mice normalized to Coomassie and Vinculin. Each dot represents the average of at least two technical replicates per animal. The bars represent the SEM; non-parametric ANOVA (WT n = 21; Fmr1 KO n = 30). (B) Puromycin incorporation in MEF from WT and Fmr1 KO mice, normalized to Coomassie and Vinculin. Each dot represents the average of at least two technical replicates per animal. The bars represent the SEM; non-parametric ANOVA (WT n = 20, Fmr1 KO n = 27).
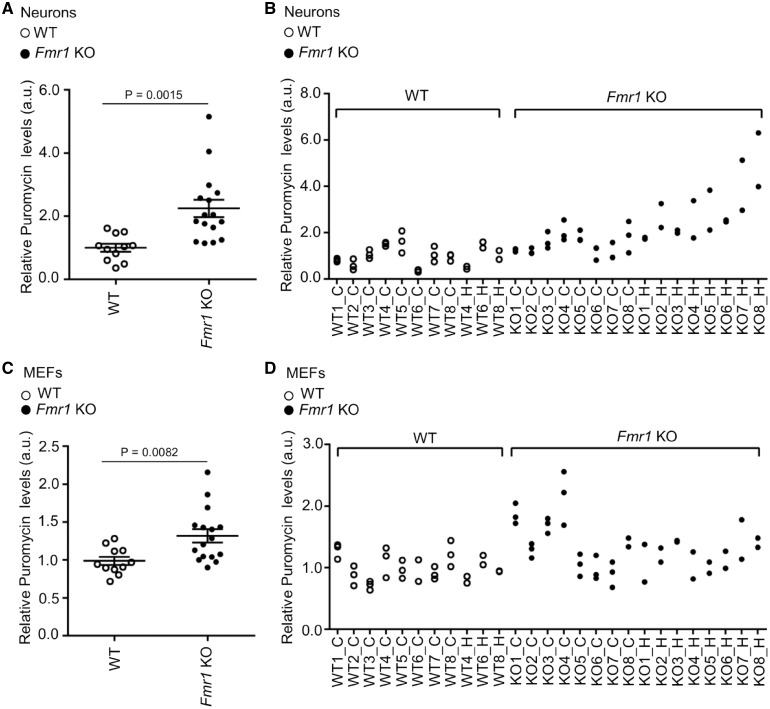
To establish whether levels of de novo protein synthesis in neurons could be predicted by those measured in fibroblasts, we established a method for preparing primary neurons and MEFs from the same animal. Puromycin incorporation was measured in Fmr1 KO (n = 16) and WT (n = 11) mice (Fig. 4A–D;Supplementary Material, Fig. S1C) showing a significant correlation between basal measures performed in neurons and in fibroblasts (Spearman correlation coefficient 0.54, P = 0.004). This relationship was not linear and log-transformation of puromycin levels in primary neurons was required to observe the same level of correlation using Pearson’s method (0.46, P = 0.02).
Figure 4.
Levels of protein synthesis in matched mouse primary neurons and embryonic fibroblasts. Puromycin incorporation in neurons (A and B) and MEF (C and D) derived from the same animal. In A and C, each dot represents the average of at least two technical replicates per animal. Bars represent the SEM; non-parametric ANOVA (WT n = 11; Fmr1 KO n = 16). B and D show multiple data points per each animal (independent replicates).
As an exploratory aim, we investigated whether molecular measures (including protein synthesis, FMRP levels and methylation status) may explain some of the variance in several clinical measures including the Vineland subscales, the Vineland composite score and the presence of an autism diagnosis. This analysis was performed in the fragile X group only, since clinical data was not available in the control group (Supplementary Material, Table S3). The Vineland rather than IQ was used because IQ measures show important floor effects on standardized cognitive tests in low functioning individuals that are normed primarily for the typically developing population, and are thus unable to capture the variance in the severity among individuals with FXS (55). The Vineland adaptive behavioral scale (VABS) and the diagnosis of Autism were performed in 21 FXS individuals from the Sacramento and Lausanne cohorts (Table 2). This represented 32 tests and results were Bonferroni corrected. None of the clinical measures showed significant correlation with one of the four molecular measures (Table 2).
Table 2.
Correlation between the Vineland adaptive behavior scale, protein synthesis and FMRP related biomarkersa
| Variable 1 | Variable 2 | Pearsons correlation | Pp | Spearman correlation | Ps |
|---|---|---|---|---|---|
| Protein synthesis in patients with FXS (n = 21) | Vineland_Co | 0.41 | 0.0618 | 0.51 | 0.0171 |
| Vineland_DL | 0.59 | 0.0046 | 0.68 | 0.0007 | |
| Vineland_S | 0.44 | 0.0437 | 0.53 | 0.0128 | |
| Vineland_AB | 0.28 | 0.2178 | 0.05 | 0.8360 | |
| FMRP levels in patients with FXS (n = 21) | Vineland_Co | 0.01 | 0.9740 | −0.02 | 0.9220 |
| Vineland_DL | −0.26 | 0.2545 | −0.26 | 0.2465 | |
| Vineland_S | −0.06 | 0.7841 | −0.05 | 0.8270 | |
| Vineland_AB | 0.43 | 0.0543 | 0.44 | 0.0453 | |
| FMR1 mRNA levels (n = 25) | Vineland_Co | 0.06 | 0.8173 | −0.23 | 0.3468 |
| Vineland_DL | −0.28 | 0.2526 | −0.52 | 0.0220 | |
| Vineland_S | −0.15 | 0.5402 | −0.33 | 0.1683 | |
| Vineland_AB | 0.36 | 0.1251 | 0.34 | 0.1535 | |
| FMR1 methylation levels (n = 15) | Vineland_Co | 0.06 | 0.8553 | 0.33 | 0.2887 |
| Vineland_DL | 0.42 | 0.1761 | 0.59 | 0.0442 | |
| Vineland_S | 0.22 | 0.4937 | 0.50 | 0.0971 | |
| Vineland_AB | −0.71 | 0.0099 | −0.64 | 0.0255 |
Vineland Co (Communication), DL (Daily Living), S (Socialization), AB (Adaptive Behavior). Data on motor skills was not recorded for all cohorts. Spearman correlation scores are similar across subscales except for abnormal behavior. The regression analysis did not show any main effect of age or cohort. The threshold for significance is calculated at 0.0015 on the basis of 32 tests. No clear correlations are identified.
Discussion
Despite intense efforts, there are still no therapies targeting the core mechanisms of FXS. Significant studies have focused on rescuing protein synthesis-dependent synaptic plasticity in mouse models of FXS by manipulating receptors that regulate FMRP signaling and activity. Drugs targeting group 1 metabotropic glutamate receptors (mGluR1, 5) and GABA receptors have shown very promising results in preclinical studies but a series of large human trials have failed to demonstrate any benefits of these drugs (36). Like for other neurological and psychiatric disorders, clinical trials in FXS and neurodevelopmental disorders have been hindered by two related issues: a partial understanding of the mechanisms and a lack of biomarkers measuring these mechanisms (41). In this study, we investigated the relevance of basal levels of de novo protein synthesis measured in peripheral tissue as a biomarker of translational control, one of the most extensively studied functions of FMRP.
Our observations show significantly increased levels of protein synthesis in FXS human fibroblasts and suggest that the non-radioactive SUnSET assay provides a robust readout to study molecular and clinical correlates in patients with FXS. Importantly, protein synthesis levels are not increased in all individuals with fragile X suggesting that compounds restoring translational regulation may not be helpful in a substantial proportion of patients who show normal levels of basal protein synthesis. We also show that de novo protein synthesis measured in mouse fibroblasts are reasonably correlated with levels measured in neurons of the same animal. This suggests that peripheral measures of protein synthesis may serve as a relevant biomarker of the mechanisms underlying this condition as well as response to treatments aiming at restoring translational control in FXS and in other other neurodevelopmental disorders.
Although, several studies demonstrated a correlation between FMRP levels and clinical phenotypes, these correlations were mainly driven by groups of patients with detectable levels of FMRP such as mosaic males with unmethylated full mutation (56). In our samples, FMRP did not correlate to VABS likely do to the fact that patients presented a classic form of fragile X with little or no expression of FMRP. In those cases, the observed clinical variability was most likely due to additional factors.
Mounting evidence suggests that aberrant synaptic protein synthesis may be a major contributing mechanism underlying many forms of neurodevelopmental disorders including those caused by mutations in MECP2, TSC1, TSC2, CYFIP1, SYNGAP1 as well as deletions of the 16p11.2 locus (57–61). Mechanisms regulating protein synthesis will therefore continue to be important targets in future preclinical and clinical studies. However, all of these studies have been conducted in animal models and the contribution of aberrant synaptic protein synthesis to the nature and severity of symptoms in patients remains unknown. We provide the first evidence on a cohort of 32 patients that measures of basal protein synthesis in fibroblast cell cultures are relevant markers of this mechanism and that protein synthesis may explain some of the variance in symptoms observed in patients.
Our most puzzling observation is that some individuals with FXS and Fmr1 KO mice have normal levels of basal de novo protein synthesis in fibroblasts as well as in primary neurons. Of note cell division is not a contributing factor to the observed variability since primary neurons are non-dividing cells. This suggests, therefore, that this alteration may not be responsible of all symptoms observed in patients and that individuals with normal levels of protein synthesis would not benefit from drugs that further decrease these levels. We recently observed in the FXS murine model (Fmr1 KO) that the absence of FMRP leads to a dysregulation in the anabolic pathway of the amyloid precursor protein (APP). The exaggerated production of the APP processing metabolite sAPPα contributes to the three major hallmarks of FXS: increased protein synthesis, aberrant spine morphology and altered synaptic function and behavior (35).
While this study analyzed a large collection of primary cells from patients and controls, the potential correlations between increased levels of protein synthesis and clinical measures is limited by the sample size. As an example, previous studies have shown correlations between FMRP levels and cognitive traits in 144 families with FXS, (56) as well as FMRP and autistic traits in 83 FXS individuals (62). It is likely that the net increase of protein synthesis in fragile X syndrome is not capturing the complexity of translational dysregulation caused by the lack of FMRP. In particular, it is unknown to which extent FXS may be due to broad translational dysregulation as opposed to abnormal regulation of a few transcripts.
In conclusion, protein synthesis remains a primary mechanism for neurodevelopmental disorders and FMRP clearly modulates this cellular phenotype but it is likely that many other molecular factors independent of FMRP (environmental and genetic) contribute to the modulation of homeostasis of molecules involved in synaptic plasticity. Our findings demonstrate that only a subgroup of individuals with FXS shows a net increase in protein synthesis and this may serve to stratify future clinical trials aiming at restoring translational regulation.
Materials and Methods
Participants
The participants were enrolled in three independent hospitals/centers [Department of Clinical Genetics, University Hospital of Lausanne, Switzerland, Medical Investigation of Neurodevelopmental Disorders (MIND) Institute, University of California, Davis Medical Center, Sacramento, US and Department of Clinical Genetics, Erasmus Medical Center, the Netherlands]. Demographics are summarized in Supplementary Material, Table S1.
Lausanne cohort (FXS 1)
Inclusion criteria: Subjects were male, aged 12–45 years, with a confirmed diagnosis of FXS based on genetic testing (full mutation, >200 CGG repeats). They were required to have a Clinical Global Impressions of Severity (CGI-S) score of ≥4 (moderately ill) and a score of ≥20 on the ABC-C scale (at screening, Supplementary Material, Table S1). Exclusion criteria: Clinically significant systemic illness or symptoms that might deteriorate or affect their safety or ability to cooperate during the study. Subjects were required to list all concomitant medications taken prior to the start of the study. The study protocol and all amendments were reviewed by the Independent Ethics Committee of the University hospital of Lausanne. Informed written consent was obtained from each parent/legal guardian as well as the participant.
Rotterdam cohort (FXS 2)
The study protocol and all amendments were reviewed by the Independent Ethics Committee of the Erasmus Medical Center, Rotterdam, The Netherlands.
Sacramento cohort (FXS 3)
All participants signed one or more consents for the collection of a blood sample, skin biopsy and fibroblast culture. They were enrolled in clinical studies assessing the phenotype and genotype correlations. Inclusion criteria: Confirmed diagnosis of FXS with a FMR1 full mutation (>200 CGG repeats) with or without a mosaic status (Supplementary Material, Table S1). Subjects were male, aged 12–69 years (inclusive). At screening, the medical history was reviewed and the study eligibility according to the inclusion and exclusion criteria was assessed according to which study they were enrolled. Biological specimens were collected at day 1 and the FXS phenotype was assessed either at Day 1 or at Day 2, if the subject could not complete all the tests in one day. The study protocol and all amendments for each study were reviewed by the Institutional Review Board of the University of California Davis Medical Center. Informed written consent was obtained from each parent/legal guardian as well as the participant assent.
Clinical assessments
Clinical assessments were available for the Sacramento and the Lausanne cohort. No phenotypic data were available for the Rotterdam samples (FXS2). Controls (cell lines from the biobanks of Coriell Institute for Medical Research, CliniSciences, Lonza) were not clinically assessed. Clinical assessments are summarized in Supplementary Material, Tables S1 and S3. All participants from the Sacramento cohort had IQ testing performed using the Stanford-Binet Intelligence Scales, Fifth Edition (SB-5), (63) the Wechsler Preschool and Primary Scale of Intelligence, third edition (WPPSI 3) (64) or the Wechsler Adult Intelligence Scale-Third Edition (WAIS 3) (65). For the participants from the Lausanne cohort a mental age was calculated based on the scores from the Wechsler nonverbal scale or WPPSI, Wechsler preschool and primary scale of intelligence. Adaptive functioning was measured using the Vineland Adaptive Behavior Scales, Second Edition (66) A diagnosis of Autism Spectrum Disorder was established by the Autism Diagnostic Observation Schedule (ADOS) Module 3 or Module 4 (67,68).
Primary human fibroblasts
The cell lines analyzed in this study are listed in Supplementary Material, Table S2. The control fibroblasts were derived from individuals who were free of any neurological disease (n = 17, age range 10–50 years inclusive) and were purchased from the Coriell Cell Repositories (Camden, NJ), LONZA and Cliniscience supplier and provided by the UZ Leuven biobank (n = 3). The human male FXS fibroblasts (n = 32, age range 6–69 years inclusive) were derived at the Cytogenetics Department, University Hospital of Lausanne (FXS1), MIND Institute Fragile X Research and Treatment Center, UC DAVIS (FXS3), Department of Neurosciences, Erasmus MC, Rotterdam (FXS2).
Fibroblasts were prepared from dermal biopsies obtained from individuals with a confirmed diagnosis of FXS based on clinical features of the disease, expression of FRAXA (fragile site, X chromosome, A site), genetic testing (full mutation, >200 CGG repeats). 17 subjects had a hypermethylated FMR1 full mutation while 15 subjects showed methylation or size mosaicism. Cells were maintained in DMEM F12 medium supplemented with 10% Fetal Bovine Serum, 1X GlutaMaxTM, 1X Penicillin-Streptomycin (Life Technologies) and Mycozap reagent (LONZA). FXS (n = 32) and controls (n = 17) fibroblast cell lines were analyzed for expression levels of FMR1 mRNA and FMRP at University of Rome Tor Vergata (Supplementary Material, Table S2). A subset of these samples was also analyzed in parallel at UC Davis.
Methylation status
Genomic DNA from primary fibroblast cell lines was isolated using Gentra Puregene Blood Kit (Qiagen). CGG repeat allele size and methylation status were obtained using a combination of PCR and Southern Blot analysis as previously described (69–71).
FMR1 mRNA levels in fibroblasts
Measurements of FMR1 mRNA expression levels were performed by quantitative RT-PCR upon total RNA extraction from control and FXS cells. Total RNA was isolated from the fibroblasts using TrizolTM reagent (Life Technologies) according to manufacturer’s instructions. For the cDNA synthesis 500 ng of total RNA was used as input into a 20 μl reaction using p(dN)6 and 200 U/μl M-MLV RTase (Invitrogen). FMR1 and hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNAs were quantified by real-time PCR using SYBR® Green Master Mix (Bio-Rad) on StepOnePlusTM Real-Time PCR machine (Life Technologies) according to the manufacturer’s instructions. FMR1 mRNAs levels were expressed in relative abundance compared to HPRT1 gene. Specific primers were used to amplify FMR1 and HPRT1 mRNAs:
FMR1: (5′-TGT CAG ATT CCC ACC TCT TG-3′; 5′-TAA CCA CCA ACA GCA AGG CT- 3′)
HPRT1: (5′-TGC TGA GGA TTT GGA AAG GGT-3′; 5′-TGC AGC AAG ACG TTC AGT CC-3′).
Obtained values were reported in Supplementary Material, Table S2 using the hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA as reference gene. Normalized relative quantities (RQ) were calculated according to the method described by Hellemans et al. (72). The arithmetic mean of the quantification cycles (Cq) of all healthy normal volunteer samples was used as a reference Cq for the calculation of RQ, FMR1 mRNA varied between 0 and 2.88 (mean 0.64).
FMR1 mRNA levels in blood samples
FMR1 mRNA expression levels in blood (cohort FXS1) were measured by qRT-PCR. Complementary desoxyribonucleic acid (cDNA) synthesis was performed using a high capacity cDNA reverse transcription kit with RNAse Inhibitor (Applied Biosystems, CA, USA). For the cDNA synthesis, 500 ng of RNA was used as input into a 20 μl cDNA synthesis reaction to generate 25 ng/μl cDNA (total RNA equivalents). RT-PCR was performed using the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystem). The following TaqMan assays obtained from Applied Biosystems were used: FMR1: Hs00924544_m1; actin B (ACTB): Hs99999903_m1; beta-glucuronidase (GUSB): Hs99999908_m1. All samples were processed in triplicate with a 25 ng cDNA (total RNA equivalent) for FMR1 and 10 ng cDNA (total RNA equivalent) for reference gene assays (ACTB, GUSB). The RT-PCR consisted of one step at 50°C for 2 min, one denaturing step at 95°C for 10 min followed by 40 cycles of melting (15 s at 95°C) and annealing/extension (1 min at 60°C). Normalized relative quantities (RQ) were calculated according to the method described by Hellemans et al. (72). The arithmetic mean of the quantification cycles (Cq) of all healthy normal volunteer samples was used as a reference Cq for the calculation of RQ. RQ values were normalized using the geometric mean of the RQs of the two reference genes, ACTB and GUSB.
FMRP levels in different cell types
All fibroblast cell lines were analyzed by Western blotting. FMRP levels in human fibroblasts were measured on total cell lysates by Western blotting using a specific antibody against FMRP (73). FMRP expression in blood was determined by immunostaining of lymphocytes in blood smear using anti-FMRP antibody. Briefly, blood smear slides were air-dried and fixed in acetone/methanol (1:1) for 20 minutes at room temperature. After washing with phosphate buffered saline (PBS) solution followed by reaction buffer (Tris-based buffer solution at pH 7.6 ± 0.2, Ventana Medical Systems, AZ, USA), the slides were loaded onto the Ventana Discovery XT (Res IHC Omni-UltraMap HRP XT); Ventana Medical System for anti-FMRP immunostaining, following the auto-staining procedure. The blood smear slides were fixed with acetone/methanol (1:1) and then treated with the cell conditioning solutions, CC1 and CC2 (Ventana Medical System) to allow the anti-FMRP monoclonal antibody (Millipore, Billerica, MA, USA) to penetrate the lymphocyte cell membrane and bind to FMRP. After washing the slides with reaction buffer (Ventana Medical System), the anti-mouse secondary antibody Omni-Map™ HRP (Ventana Medical System), the ChromoMap DAB Hematoxylin II substrate (Ventana Medical System) and the Blue reagent (Ventana) were added sequentially to visualize the FMRP expression density and distribution. Quantification was done measuring the % of cells expressing FMRP.
Animal experiments
All animal procedures were conducted conforming to institutional guidelines in compliance with international laws and policies (European Community Guidelines for Animal Care, DL 116/92, application of the European Communities Council Directive, 86/609/EEC). Studies were approved by the Institutional Ethical Board at the University of Rome Tor Vergata, according to the Guideline of the Italian Institute of Health (protocol n. 1088/2016-PR). C57BL/6J and FVB.129P2 WT and Fmr1 KO male mice (54) were used in this study.
Mouse neuronal cell cultures
Mouse primary cortical neurons were prepared as previously described (74) from embryonic day (E) E15 mice. The brains were removed, neocortices were freed of meninges, treated with 0.025% trypsin, minced and plated on poly-l-lysine (Sigma-Aldrich) wells in Neurobasal medium supplemented with 2% B27, 0.5 mM glutamine and Penicillin-Streptomycin (GIBCO BRL); cells were maintained at 37°C and 5% CO2. For the SUnSET assay cells were seeded on 24 well plates and treated with puromycin at DIV14. In a subset of experiments from the same embryo, cortices were used for primary neurons and the rest for the production of MEFs.
Mouse embryo fibroblasts (MEFs) cultures
MEF cultures were prepared as previously described (75) from embryonic day (E) E14–16 mice, by dissociation of the whole embryo after removal of internal organs and heads. Embryos were treated with 0.25% trypsin at 4°C overnight, tissues minced by pipetting up and down. Cells were plated on 6 well plate, 24 h later the medium was replaced, cells were washed by shaking vigorously twice to remove debris, detached cell clumps and dead cells. Fresh media were added and cells were maintained at 37°C and 5% CO2 to reach confluence (2–3 days). At this stage cells were frozen and used for later procedures. For the SUnSET assay each culture was divided into 2 or 3 wells (cells were plated in duplicate or triplicate in 12w-plate) and treated with puromycin as described in the text.
SUnSET assay
A protein synthesis assay was performed as previously described using the SUnSET method (48). Cells (80 000/well) were seeded on 12-multiwell plate wells, deprived of serum for 16 h and after 4 h of recovery in complete medium supplemented with 10% FBS, were treated with puromycin 5 μg/ml for 30 min. To ascertain the specificity of the assay cells were pre-incubated in the absence or presence of cycloheximide 60 µM for 15 min. After puromycin pulse, cells were chased with fresh complete medium for 15 min then washed with ice-cold PBS and lysed directly in Laemmly buffer. Samples were analysed by WB and puromycin incorporation was detected using the mouse monoclonal antibody PMY-2A4. Coomassie staining of total proteins and immunolabelling of housekeeping proteins as GAPDH and Vinculin was used as loading control.
To assess the stability of measures within subjects, in addition to replicates performed by means of split passages performed for all fibroblasts, independent SUnSET assays were performed over a period of 1 year, on a subset of 10 cell lines frozen, re-thawed and cultured again.
Western blotting
Standard methodologies were used. Protein extracts were separated by SDS–PAGE electrophoresis, and transferred to a PVDF membrane (GE Healthcare). Membranes were incubated using specific antibodies including mouse anti-puromycin PMY-2A4 (1:500 DSHB), mouse anti-GAPDH (1:2000, Thermo Fisher), mouse anti-Vinculin (1:2000, Sigma) and rabbit anti-FMRP (1:1000) (73).
Secondary HRP-conjugated anti-rabbit, anti-mouse antibodies (1:10 000) were purchased from Promega or Chemicon. Proteins were revealed using an enhanced chemiluminescence kit (Bio-Rad) and the imaging system LAS-4000 mini (GE). Quantification was performed using the IQ ImageQuant TL software (GE Healthcare). Coomassie staining of the membranes, GAPDH and Vinculin signals were used as normalizers.
Statistical analysis
All statistical analyses were conducted using SAS. 9.3. A linear regression was used to evaluate correlation between the number of cell passages and measures. A non-parametric analysis of variance (ANOVA) was used to compare the differences of protein synthesis between groups. Correlation between molecular levels and clinical parameters was accessed by Pearson and Spearman correlation analysis.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to all patients and caregivers who participated in the clinical studies. We would like to thank Veronica Nobile and Eleonora Rosina for sharing preliminary data and Tilmann Achsel for discussions and critical reading of the manuscript.
Conflict of Interest statement: S.J. has served on the Novartis Fragile X Advisory Board, has received funding from Novartis to conduct clinical trials and has consulted for Novartis and WG Pharma. A.E.J. participated in clinical trials led by Novartis Pharma and has no further competing interests to declare. F.G. is an employee and a shareholder of Novartis. B.G.M., I.R. and Y.H. are employees of Novartis Institutes for Biomedical Research. R.H. has received funding from Novartis, Roche/Genetech, Alcobra, Marinus and Neuren for clinical trials in fragile X syndrome and has consulted with Roche/Genentech, Zynerba and Novartis regarding fragile X treatment and clinical trial design. F.T. has received funding from Asuragen, Inc. The authors C.B., L.P., G.C., L.D.A., G.P., R.W. and M.E. report no biomedical financial interests or potential conflicts of interest.
Funding
This work was supported by Telethon (GGP15257 to C.B.); Jerome Lejeune Foundation (to C.B.); Associazione Italiana Sindrome X Fragile (FWO G.0532.12N to C.B.); Fondazione Cariplo (to C.B.); Etat de Vaud (to C.B.). NCCR Synapsy (51NF40-158776) to C.B. Part of this work was supported by Novartis Pharma AG, Basel, Switzerland (B.G.M.); National Institute of Child Health and Human Development (HD036071 to F.T. and R.H., GM113929 to F.T.); the MIND Institute Intellectual and Developmental Disability Research Center (U54HD07912 to F.T. and R.H.); S.J. is recipient of a Canada research Chair, the Jeanne et Jean-Louis Lévesque Chair and the Swiss National Science Foundation Professorship. A.E.J. is recipient of the Odense University Hospital Free Research Fund (15-A857), the Region of Zealand and Region of Southern of Denmark joint research fund (14–001308 to A.E.J.); and a Ph.D. scholarship from the Region of Southern of Denmark and the FAculty of Health sciences, University of Southern of Denmark. Funding to pay the Open Access publication charges for this article was provided by Etat de Vaud, University of Lausanne (to C.B.).
References
- 1. Sah P., Westbrook R.F. (2008) Behavioural neuroscience: the circuit of fear. Nature, 454, 589–590. [DOI] [PubMed] [Google Scholar]
- 2. Fiala J.C., Spacek J., Harris K.M. (2002) Dendritic spine pathology: cause or consequence of neurological disorders?. Brain Res. Brain Res. Rev., 39, 29–54. [DOI] [PubMed] [Google Scholar]
- 3. Penzes P., Cahill M.E., Jones K.A., VanLeeuwen J.E., Woolfrey K.M. (2011) Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci., 14, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valnegri P., Sala C., Passafaro M. (2012) Synaptic dysfunction and intellectual disability. Adv. Exp. Med. Biol., 970, 433–449. [DOI] [PubMed] [Google Scholar]
- 5. Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. (1992) DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet., 1, 397–400. [DOI] [PubMed] [Google Scholar]
- 6. Jung H., Yoon B.C., Holt C.E. (2012) Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat. Rev. Neurosci., 13, 308–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin K.C., Ephrussi A. (2009) mRNA localization: gene expression in the spatial dimension. Cell, 136, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steward O., Schuman E.M. (2001) Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci., 24, 299–325. [DOI] [PubMed] [Google Scholar]
- 9. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Napoli I., Mercaldo V., Boyl P.P., Eleuteri B., Zalfa F., De Rubeis S., Di Marino D., Mohr E., Massimi M., Falconi M.. et al. (2008) The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell, 134, 1042–1054. [DOI] [PubMed] [Google Scholar]
- 11. Narayanan U., Nalavadi V., Nakamoto M., Pallas D.C., Ceman S., Bassell G.J., Warren S.T. (2007) FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J. Neurosci., 27, 14349–14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zalfa F., Giorgi M., Primerano B., Moro A., Di Penta A., Reis S., Oostra B., Bagni C. (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell, 112, 317–327. [DOI] [PubMed] [Google Scholar]
- 13. Huber K.M., Gallagher S.M., Warren S.T., Bear M.F. (2002) Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. U. S. A., 99, 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gross C., Hoffmann A., Bassell G.J., Berry-Kravis E.M. (2015) Therapeutic strategies in fragile X syndrome: from bench to bedside and back. Neurotherapeutics, 12, 584–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacquemont S., Berry-Kravis E., Hagerman R., von Raison F., Gasparini F., Apostol G., Ufer M., Des Portes V., Gomez-Mancilla B. (2014) The challenges of clinical trials in fragile X syndrome. Psychopharmacology (Berl.), 231, 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krueger D.D., Bear M.F. (2011) Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu. Rev. Med., 62, 411–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lozano R., Hare E.B., Hagerman R.J. (2014) Modulation of the GABAergic pathway for the treatment of fragile X syndrome. Neuropsychiatr. Dis. Treat., 10, 1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pop A.S., Gomez-Mancilla B., Neri G., Willemsen R., Gasparini F. (2014) Fragile X syndrome: a preclinical review on metabotropic glutamate receptor 5 (mGluR5) antagonists and drug development. Psychopharmacology (Berl.), 231, 1217–1226. [DOI] [PubMed] [Google Scholar]
- 19. Bhattacharya A., Kaphzan H., Alvarez-Dieppa A.C., Murphy J.P., Pierre P., Klann E. (2012) Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron, 76, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bilousova T.V., Dansie L., Ngo M., Aye J., Charles J.R., Ethell D.W., Ethell I.M. (2009) Minocycline promotes dendritic spine maturation and improves behavioural performance in the fragile X mouse model. J. Med. Genet., 46, 94–102. [DOI] [PubMed] [Google Scholar]
- 21. Boda B., Mendez P., Boury-Jamot B., Magara F., Muller D. (2014) Reversal of activity-mediated spine dynamics and learning impairment in a mouse model of Fragile X syndrome. Eur. J. Neurosci., 39, 1130–1137. [DOI] [PubMed] [Google Scholar]
- 22. Busquets-Garcia A., Gomis-Gonzalez M., Guegan T., Agustin-Pavon C., Pastor A., Mato S., Perez-Samartin A., Matute C., de la Torre R., Dierssen M.. et al. (2013) Targeting the endocannabinoid system in the treatment of fragile X syndrome. Nat. Med., 19, 603–607. [DOI] [PubMed] [Google Scholar]
- 23. Dolan B.M., Duron S.G., Campbell D.A., Vollrath B., Shankaranarayana Rao B.S., Ko H.Y., Lin G.G., Govindarajan A., Choi S.Y., Tonegawa S. (2013) Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc. Natl. Acad. Sci. U. S. A., 110, 5671–5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dölen G., Osterweil E., Rao B.S.S., Smith G.B., Auerbach B.D., Chattarji S., Bear M.F. (2007) Correction of fragile X syndrome in mice. Neuron, 56, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gross C., Nakamoto M., Yao X., Chan C.-B., Yim S.Y., Ye K., Warren S.T., Bassell G.J. (2010) Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J. Neurosci., 30, 10624–10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hayashi M.L., Rao B.S., Seo J.S., Choi H.S., Dolan B.M., Choi S.Y., Chattarji S., Tonegawa S. (2007) Inhibition of p21-activated kinase rescues symptoms of fragile X syndrome in mice. Proc. Natl. Acad. Sci. U. S. A., 104, 11489–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hebert B., Pietropaolo S., Meme S., Laudier B., Laugeray A., Doisne N., Quartier A., Lefeuvre S., Got L., Cahard D.. et al. (2014) Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by a BKCa channel opener molecule. Orphanet. J. Rare Dis., 9, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michalon A., Sidorov M., Ballard T.M., Ozmen L., Spooren W., Wettstein J.G., Jaeschke G., Bear M.F., Lindemann L. (2012) Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron, 74, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas A.M., Bui N., Graham D., Perkins J.R., Yuva-Paylor L.A., Paylor R. (2011) Genetic reduction of group 1 metabotropic glutamate receptors alters select behaviors in a mouse model for fragile X syndrome. Behav. Brain Res., 223, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tian M., Zeng Y., Hu Y., Yuan X., Liu S., Li J., Lu P., Sun Y., Gao L., Fu D.. et al. (2015) 7, 8-Dihydroxyflavone induces synapse expression of AMPA GluA1 and ameliorates cognitive and spine abnormalities in a mouse model of fragile X syndrome. Neuropharmacology, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 31. Udagawa T., Farny N.G., Jakovcevski M., Kaphzan H., Alarcon J.M., Anilkumar S., Ivshina M., Hurt J.A., Nagaoka K., Nalavadi V.C.. et al. (2013) Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat. Med., 19, 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Osterweil E.K., Krueger D.D., Reinhold K., Bear M.F. (2010) Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J. Neurosci., 30, 15616–15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richter J.D., Bassell G.J., Klann E. (2015) Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci., 16, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sawicka K., Pyronneau A., Chao M., Bennett M.V., Zukin R.S. (2016) Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in fragile X mice. Proc. Natl. Acad. Sci. U. S. A., 113, E6290–e6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasciuto E., Ahmed T., Wahle T., Gardoni F., D’Andrea L., Pacini L., Jacquemont S., Tassone F., Balschun D., Dotti C.G.. et al. (2015) Dysregulated ADAM10-mediated processing of APP during a critical time window leads to synaptic deficits in fragile X syndrome. Neuron, 87, 382–398. [DOI] [PubMed] [Google Scholar]
- 36. Berry-Kravis E., Des Portes V., Hagerman R., Jacquemont S., Charles P., Visootsak J., Brinkman M., Rerat K., Koumaras B., Zhu L.. et al. (2016) Mavoglurant in fragile X syndrome: results of two randomized, double-blind, placebo-controlled trials. Sci. Transl. Med., 8, 321ra5.. [DOI] [PubMed] [Google Scholar]
- 37. Jacquemont S., Curie A., des Portes V., Torrioli M.G., Berry-Kravis E., Hagerman R.J., Ramos F.J., Cornish K., He Y., Paulding C.. et al. (2011) Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci. Transl. Med., 3, 64ra1.. [DOI] [PubMed] [Google Scholar]
- 38. Berry-Kravis E.M., Hessl D., Rathmell B., Zarevics P., Cherubini M., Walton-Bowen K., Mu Y., Nguyen D.V., Gonzalez-Heydrich J., Wang P.P.. et al. (2012) Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Sci. Transl. Med., 4, 152ra127.. [DOI] [PubMed] [Google Scholar]
- 39. Berry-Kravis E., Hagerman R., Visootsak J., Budimirovic D., Kaufmann W.E., Cherubini M., Zarevics P., Walton-Bowen K., Wang P., Bear M.F.. et al. (2017) Arbaclofen in fragile X syndrome: results of phase 3 trials. J. Neurodev. Disord., 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leigh M.J., Nguyen D.V., Mu Y., Winarni T.I., Schneider A., Chechi T., Polussa J., Doucet P., Tassone F., Rivera S.M.. et al. (2013) A randomized double-blind, placebo-controlled trial of minocycline in children and adolescents with fragile x syndrome. J. Dev. Behav. Pediatr., 34, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berry-Kravis E.M., Lindemann L., Jønch A.E., Apostol G., Bear M.F., Carpenter R.L., Crawley J.N., Curie A., Des Portes V., Hossain F., Gasparini F.. et al. (2017) Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat. Rev. Drug Discov. doi: 10.1038/nrd.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolduc F.V., Bell K., Cox H., Broadie K.S., Tully T. (2008) Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci., 11, 1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qin M., Kang J., Burlin T.V., Jiang C., Smith C.B. (2005) Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J. Neurosci., 25, 5087–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gross C., Bassell G.J. (2012) Excess protein synthesis in FXS patient lymphoblastoid cells can be rescued with a p110beta-selective inhibitor. Mol. Med., 18, 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qin M., Schmidt K.C., Zametkin A.J., Bishu S., Horowitz L.M., Burlin T.V., Xia Z., Huang T., Quezado Z.M., Smith C.B. (2013) Altered cerebral protein synthesis in fragile X syndrome: studies in human subjects and knockout mice. J. Cereb. Blood Flow Metab., 33, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumari D., Bhattacharya A., Nadel J., Moulton K., Zeak N.M., Glicksman A., Dobkin C., Brick D.J., Schwartz P.H., Smith C.B.. et al. (2014) Identification of fragile X syndrome specific molecular markers in human fibroblasts: a useful model to test the efficacy of therapeutic drugs. Hum. Mutat., 35, 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoeffer C.A., Sanchez E., Hagerman R.J., Mu Y., Nguyen D.V., Wong H., Whelan A.M., Zukin R.S., Klann E., Tassone F. (2012) Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav., 11, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt E.K., Clavarino G., Ceppi M., Pierre P. (2009) SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods, 6, 275–277. [DOI] [PubMed] [Google Scholar]
- 49. Bagni C., Tassone F., Neri G., Hagerman R. (2012) Fragile X syndrome: causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest., 122, 4314–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huber K.M., Klann E., Costa-Mattioli M., Zukin R.S. (2015) Dysregulation of mammalian target of rapamycin signaling in mouse models of autism. J. Neurosci., 35, 13836–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pretto D., Yrigollen C.M., Tang H.T., Williamson J., Espinal G., Iwahashi C.K., Durbin-Johnson B., Hagerman R.J., Hagerman P.J., Tassone F. (2014) Clinical and molecular implications of mosaicism in FMR1 full mutations. Front. Genet., 5, 318.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pasciuto E., Bagni C. (2014) SnapShot: fMRP interacting proteins. Cell, 159, 218–e211. [DOI] [PubMed] [Google Scholar]
- 53. Panja D., Kenney J.W., D’Andrea L., Zalfa F., Vedeler A., Wibrand K., Fukunaga R., Bagni C., Proud C.G., Bramham C.R. (2014) Two-stage translational control of dentate gyrus LTP consolidation is mediated by sustained BDNF-TrkB signaling to MNK. Cell Rep., 9, 1430–1445. [DOI] [PubMed] [Google Scholar]
- 54.No Author (1994) Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell, 78, 23–33. [PubMed] [Google Scholar]
- 55. Hessl D., Nguyen D.V., Green C., Chavez A., Tassone F., Hagerman R.J., Senturk D., Schneider A., Lightbody A., Reiss A.L.. et al. (2009) A solution to limitations of cognitive testing in children with intellectual disabilities: the case of fragile X syndrome. J. Neurodev. Disord, 1, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loesch D.Z., Huggins R.M., Hagerman R.J. (2004) Phenotypic variation and FMRP levels in fragile X. Ment. Retard. Dev. Disabil. Res. Rev., 10, 31–41. [DOI] [PubMed] [Google Scholar]
- 57. Auerbach B.D., Osterweil E.K., Bear M.F. (2011) Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature, 480, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barnes S.A., Wijetunge L.S., Jackson A.D., Katsanevaki D., Osterweil E.K., Komiyama N.H., Grant S.G.N., Bear M.F., Nägerl U.V., Kind P.C.. et al. (2015) Convergence of hippocampal pathophysiology in Syngap+/− and Fmr1-/y mice. J. Neurosci., 35, 15073–15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bozdagi O., Sakurai T., Dorr N., Pilorge M., Takahashi N., Buxbaum J.D. (2012) Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One, 7, e42422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tao J., Wu H., Coronado A.A., de Laittre E., Osterweil E.K., Zhang Y., Bear M.F. (2016) Negative allosteric modulation of mGluR5 partially corrects pathophysiology in a mouse model of Rett syndrome. J. Neurosci., 36, 11946–11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tian D., Stoppel L.J., Heynen A.J., Lindemann L., Jaeschke G., Mills A.A., Bear M.F. (2015) Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat. Neurosci., 18, 182–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hatton D.D., Sideris J., Skinner M., Mankowski J., Bailey D.B. Jr, Roberts J., Mirrett P. (2006) Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am. J. Med. Genet. A., 140a, 1804–1813. [DOI] [PubMed] [Google Scholar]
- 63. Roid G.H. (2003) Standford-Binet Intelligence Scales. Riverside Publishing, Itasca, IL. [Google Scholar]
- 64. Wechsler D. (2002) Wechsler Preschool and Primary Scale of Intelligence (WPPSI-III), 3rd edn.Pearson/PsychCorp, San Antonio, TX. [Google Scholar]
- 65. Wechsler D. (1997a) Wechsler Adult Intelligence Scale, 3rd edn.The Psychological Corporation, San Antonio, TX. [Google Scholar]
- 66. Sparrow S.S., Chicchetti D.V., Balla D.A. (2005) Vineland Adaptive Behavior Scales, 2nd edn.Pearson Assessments, Circle Pines. [Google Scholar]
- 67. Lord C., Risi S., Lambrecht L., Cook E.H. Jr, Leventhal B.L., DiLavore P.C., Pickles A., Rutter M. (2000) The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord., 30, 205–223. [PubMed] [Google Scholar]
- 68. Lord C., Rutter M., DiLavore P.C., Risi S., Gotham K., Bishop S. (2012) Autism Diagnostic Observation Schedule, 2nd edn). Western Psychological Services, Torrance. [Google Scholar]
- 69. Tassone F., Hagerman R.J., Ikle D.N., Dyer P.N., Lampe M., Willemsen R., Oostra B.A., Taylor A.K. (1999) FMRP expression as a potential prognostic indicator in fragile X syndrome. Am. J. Med. Genet., 84, 250–261. [PubMed] [Google Scholar]
- 70. Tassone F., Pan R., Amiri K., Taylor A.K., Hagerman P.J. (2008) A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J. Mol. Diagn., 10, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Filipovic-Sadic S., Sah S., Chen L., Krosting J., Sekinger E., Zhang W., Hagerman P.J., Stenzel T.T., Hadd A.G., Latham G.J. (2010) A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin. Chem., 56, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol., 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ferrari F., Mercaldo V., Piccoli G., Sala C., Cannata S., Achsel T., Bagni C. (2007) The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol. Cell. Neurosci., 34, 343–354. [DOI] [PubMed] [Google Scholar]
- 74. De Rubeis S., Pasciuto E., Li K.W., Fernandez E., Di Marino D., Buzzi A., Ostroff L.E., Klann E., Zwartkruis F.J., Komiyama N.H.. et al. (2013) CYFIP1 coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron, 79, 1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Todaro G.J., Green H. (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol., 17, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.