Abstract
Fatty liver has been associated with unfavourable metabolic changes in circulation. To provide insights in fatty liver-related metabolic deviations, we compared metabolic association profile of fatty liver versus metabolic association profiles of genotypes increasing the risk of non-alcoholic fatty liver disease (NAFLD). The cross-sectional associations of ultrasound-ascertained fatty liver with 123 metabolic measures were determined in 1810 (Nfatty liver = 338) individuals aged 34–49 years from The Cardiovascular Risk in Young Finns Study. The association profiles of NAFLD-risk alleles in PNPLA3, TM6SF2, GCKR, and LYPLAL1 with the corresponding metabolic measures were obtained from a publicly available metabolomics GWAS including up to 24 925 Europeans. The risk alleles showed different metabolic effects: PNPLA3 rs738409-G, the strongest genetic NAFLD risk factor, did not associate with metabolic changes. Metabolic effects of GCKR rs1260326-T were comparable in many respects to the fatty liver associations. Metabolic effects of LYPLAL1 rs12137855-C were similar, but statistically less robust, to the effects of GCKR rs1260326-T. TM6SF2 rs58542926-T displayed opposite metabolic effects when compared with the fatty liver associations. The metabolic effects of the risk alleles highlight heterogeneity of the molecular pathways leading to fatty liver and suggest that the fatty liver-related changes in the circulating lipids and metabolites may vary depending on the underlying pathophysiological mechanism. Despite the robust cross-sectional associations on population level, the present results showing neutral or cardioprotective metabolic effects for some of the NAFLD risk alleles advocate that hepatic lipid accumulation by itself may not increase the level of circulating lipids or other metabolites.
Introduction
Non-alcoholic fatty liver disease (NAFLD) covers a range of liver disorders originating from excessive lipid, mainly triglyceride, accumulation in the liver (1). Fatty liver has been proposed to result in dyslipidemia due to increased secretion of very low-density lipoproteins (VLDL) and impaired clearance of intermediate and low-density lipoproteins (IDL, LDL) from circulation (2,3). Studies using detailed metabolic profiling have shown that fatty liver associates with multiple metabolic aberrations in circulation (4–6). In a study by Kaikkonen et al., fatty liver associated most prominently with triglycerides in the largest VLDL particle subfractions as well as with VLDL particle concentration, but robust associations were also seen with many non-lipid traits, such as amino acids leucine and isoleucine (4). Animal models and human studies have shown that liver fat content correlates with levels of blood lipids and glucose (7–9). However, fatty liver does not always seem to relate to dyslipidemia or changes in glycemic traits (10,11) and thus its effect on metabolic changes in circulation has remained unclear.
Genetic factors contribute to the pathogenesis of fatty liver. Four DNA sequence variants, Patatin-like phospholipase domain containing 3 (PNPLA3) rs738409-G, Glucokinase regulator (GCKR) rs780094-T, Neurocan (NCAN) rs2228603-T and Lysophospholipase-like 1 (LYPLAL1) rs12137855-C, were associated with computed tomography defined steatosis and biopsy-proven NAFLD involving lobular inflammation and fibrosis in a large-scale genome-wide association study (GWAS) (12). Other studies have shown that GCKR rs1260326-T and Transmembrane 6 superfamily member 2 (TM6SF2) rs58542926-T are the functional variants at the GCKR and NCAN loci, respectively (13–15). PNPLA3 rs738409-G is the strongest genetic risk factor for NAFLD (16) having an odds ratio of 3.24 for histologic NAFLD (12). GCKR rs1260326-T and TM6SF2 rs58542926-T are also recognized as important determinants of inter-individual variation in liver fat (13,17,18). Among these variants, the function of LYPLAL1 rs12137855-C is the least known.
In the present study, we perform metabolic profiling of ultrasound-ascertained fatty liver in young and middle-aged adults. To add insights into molecular mechanisms of fatty liver-related metabolic aberrations, we compare how the observational fatty liver associations match with metabolic association profiles of the aforementioned NAFLD risk alleles. We utilize publicly available summary statistics from a metabolomics GWAS to assess the detailed metabolic effects of the risk variants (19). Understanding the relation between fatty liver and changes in circulating lipids and metabolites is helpful in acquiring more opportunities for treatment and prevention of this complex condition and related cardiometabolic complications. Cardiovascular disease is the leading cause of death in patients with fatty liver (20) emphasizing the need of effective management of cardiovascular health in these individuals.
Results
Fatty liver and circulating metabolites
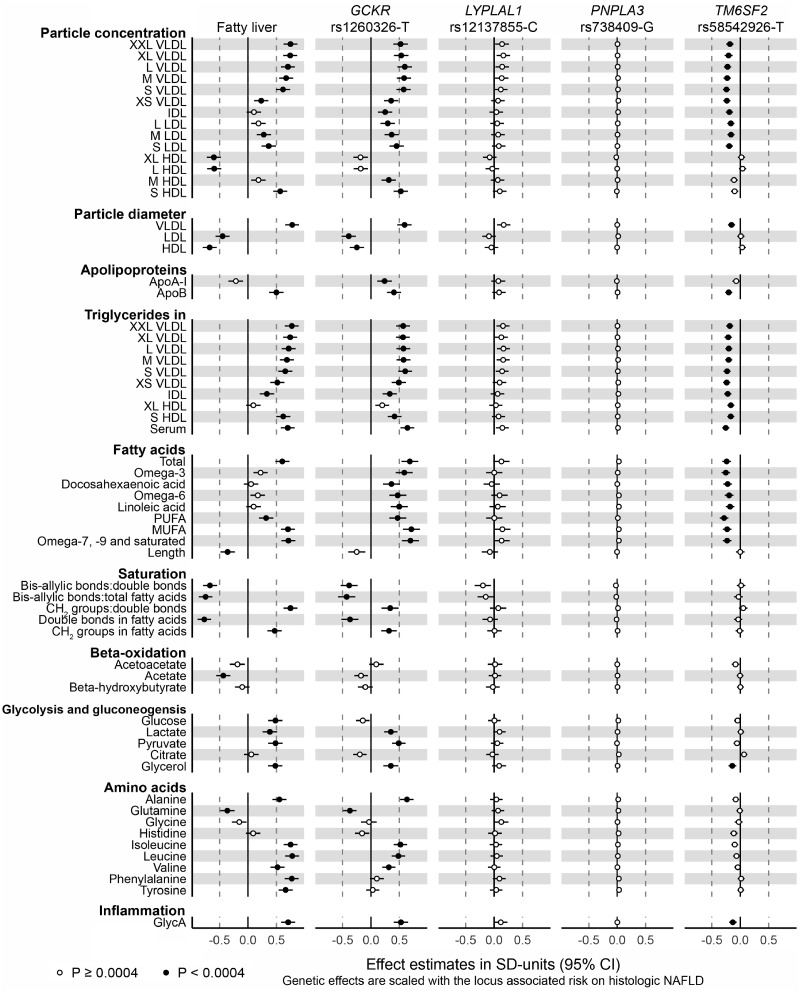
Characteristics of the YFS study population are shown in Table 1. Fatty liver showed association with 84 metabolic measures (P < 0.0004) when adjusted with age, sex and 10 genetic principal components (Fig. 1, panel 1; Supplementary Material, Figs S2 and S3; Table S2). Lipoprotein subclass concentrations showed a trend where the largest VLDL particles displayed the most pronounced associations that got weaker while the particle size decreased, and again stronger for medium and small LDL particles (Fig. 1, panel 1). Concentration of very large and large HDL particles showed negative association with fatty liver, while small HDL particles displayed a strong positive association. Fatty liver associated with increased VLDL diameter while the associations with LDL and HDL particle diameters were negative. Fatty liver showed positive association with apolipoprotein B concentration. Concentrations of serum total triglycerides and triglycerides in all lipoprotein subclasses except very large HDL were increased in association with fatty liver. Concentrations of circulating total fatty acids, PUFA, MUFA, and omega-7, -9, and other saturated fatty acids, and level of fatty acid saturation were increased in association with fatty liver, while its association with fatty acid length was negative. In addition, fatty liver displayed positive association with glucose, lactate, pyruvate and glycerol, as well as with amino acids alanine, isoleucine, leucine, valine, phenylalanine and tyrosine. Association between fatty liver and amino acid glutamine was negative.
Table 1.
Study population: YFS
| Fatty liver | No fatty liver | Total population | |
|---|---|---|---|
| N | 338 | 1472 | 1810 |
| Male (%) | 237 (70.1) | 630 (42.8) | 867 (47.9) |
| BMI, kg/m2 | 30.5 ± 5.3 | 25.5 ± 4.3 | 26.4 ± 4.9 |
| Age, years | 43.0 ± 4.7 | 41.6 ± 5.0 | 41.9 ± 5.0 |
| Smokers (%) | 55 (16.3) | 198 (13.5) | 253 (14.0) |
| Alcohol consumption per daya | 2.13 ± 1.8 | 1.87 ± 1.6 | 1.92 ± 1.6 |
One unit equals to 12 g of alcohol.
Values are mean ± SD, or absolute N count and corresponding percentage. YFS, Young Finns Study.
Figure 1.
Cross-sectional associations of fatty liver with lipoprotein particle subfraction concentrations, lipoprotein particle diameter, apolipoproteins, triglycerides, fatty acids, fatty acid saturation, beta-oxidation, glycolysis, amino acid-related metabolites and inflammation marker GlycA, and the corresponding associations with four NAFLD risk alleles. Cross-sectional associations were determined in 1, 810 adults aged 34–49 years of whom 338 were diagnosed with ultrasound-based fatty liver. The metabolic phenotypes were adjusted for age, sex, and 10 first genetic principal components prior to analysis. Genetic effects of the NAFLD risk alleles GCKR rs1260326-T, LYPLAL1 rs12137855-C, PNPLA3 rs738409-G and TM6SF2 rs58542926-T were acquired from a metabolomics GWAS including up to 24 925 Europeans (19). Genetic effect estimates were scaled with respect to the NAFLD risk associated with the corresponding locus (12). VLDL, very low-density lipoprotein; IDL, intermediate density lipoprotein; HDL, high density lipoprotein; PUFA, polyunsaturated fatty acids; MUFA, monounsaturated fatty acids; GlycA, glycoprotein acetylation.
The analysis further adjusted for alcohol consumption resulted in practically identical association profile with the primary analysis (Supplementary Material, Fig. S2). Adjustment for BMI attenuated the effect estimates, as expected, but the overall association pattern was similar to the primary analysis (Supplementary Material, Fig. S2).
NAFLD risk alleles in GCKR and LYPLAL1 tend to increase concentrations of circulating lipids
The studied NAFLD risk increasing alleles showed different association profiles on metabolic measures. The association profile of GCKR rs1260326-T was similar to the one of fatty liver especially in terms of circulating lipids and lipoproteins (Fig. 1, panel 2; Supplementary Material, Fig. S3; Table S3). However, some mismatches were observed. Glucose concentration showed a tenuous decrease in association with GCKR rs1260326-T while it was increased in association with fatty liver. Concentrations of amino acids phenylalanine and tyrosine showed no robust associations with GCKR rs1260326-T but were increased in association with fatty liver. Lipoprotein association profiles of the GCKR rs1260326-T and fatty liver differed in terms of very large and large HDL particle subfraction concentration and lipid composition: the particle concentration of the two HDL subfractions together with all lipids except triglycerides in these subfractions showed only negligible reduction in association with GCKR rs1260326-T but were highly decreased in association with fatty liver (Supplementary Material, Fig. S3). Concentration of apolipoprotein A-I was increased in the GCKR risk allele carriers, while it showed a small reduction in association with fatty liver. Some differences were also observed in concentrations of omega-3 fatty acids, docosahexaenoic acid, omega-6 fatty acids and linoleic acid. The results of GCKR rs780094-T, a variant originally identified as a NAFLD risk allele in the GCKR locus (12) being in linkage disequilibrium with the functional GCKR rs1260326-T (13) matched well with the results of GCKR rs1260326-T (Supplementary Material, Figs S1 and S3; Table S3).
The association profile of LYPLAL rs12137855-C was similar to the metabolic effects of GCKR rs1260326-T in terms of highly correlated point estimates of the two risk alleles (Supplementary Material, Fig. S4), but the effects of LYPLAL rs12137855-C were statistically less robust and the effect magnitudes were weaker than for the GCKR variant (Fig. 1, panel 3; Supplementary Material, Fig. S3; Table S3). The high correlation of the metabolic effects suggests that LYPLAL1 may function on a related biological pathway with GCKR.
PNPLA3 rs738409-G does not show association with circulating metabolic traits
PNPLA3 rs738409-G, the strongest genetic contributor to the hepatic fat content (16), displayed metabolic association profile close to null (Fig. 1, panel 4; Supplementary Material, Fig. S3; Table S3). When compared with the fatty liver effects, the NAFLD risk scaled metabolic effects of PNPLA3 rs738409-G were much closer to zero, and the confidence intervals for effect estimates of cross-sectional fatty liver and PNPLA3 rs738409-G were clearly separated.
TM6SF2 rs58542926-T associates with lower-risk lipoprotein lipid profile and lower glycoprotein acetylation
TM6SF2 rs58542926-T displayed strong association profile in particular on the lipoprotein lipid traits (Fig. 1, panel 5; Supplementary Material, Fig. S3; Table S3). However, the associations were negative indicating lower concentrations of the lipids and metabolites in relation to higher NAFLD risk. The TM6SF2 variant associated with lower concentrations of all the VLDL, IDL and LDL particle subclasses and all the lipid species in these subclasses. In addition, the variant associated inversely with serum total triglycerides and triglycerides in all the lipoprotein subclasses including the HDL subclasses. The variant associated with smaller VLDL particle diameter, while it did not show an effect on LDL nor HDL particle size. The TM6SF2 rs58542926-T associated with lower concentration of the apolipoprotein B but did not influence concentration of apolipoprotein A-I. It associated inversely also with all the fatty acid concentrations, while the qualitative measures such as fatty acid length or saturation measures were not influenced by this variant. In addition, the T allele associated with lower levels of glycerol and glycoprotein acetylation, a marker for inflammation (21). The results for NCAN rs2228603-T in the same NAFLD risk locus can be found in the supplement (Supplementary Material, Figs S1 and S3; Table S3).
Resemblance of the metabolic effects
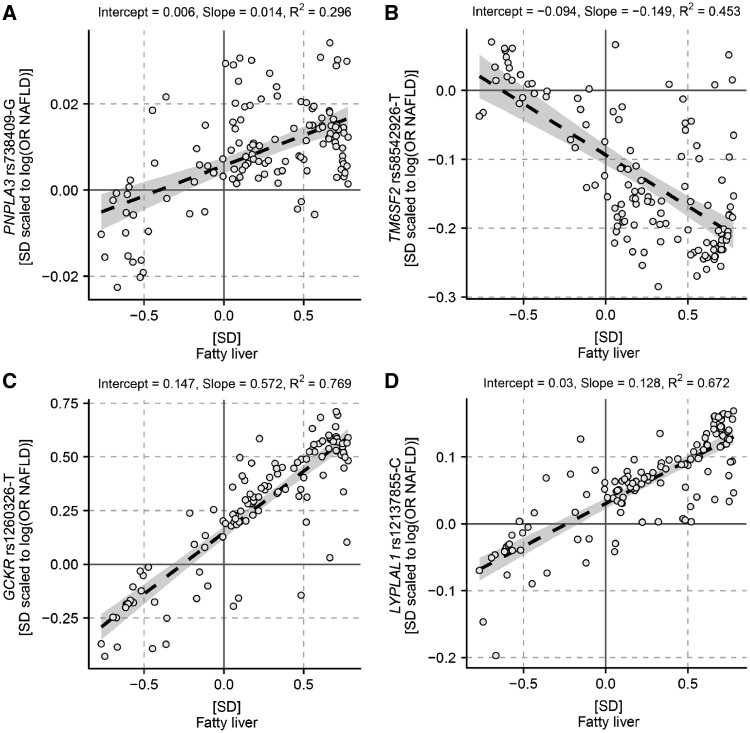
The correspondence between the metabolic effects of the risk alleles and observational fatty liver was the highest between the GCKR rs1260326-T and fatty liver (R2 = 0.77; Fig. 2C). The remaining coefficients of determination were R2 = 0.67 for LYPLAL1 rs12137855-C versus fatty liver (Fig. 2D), R2 = 0.45 for TM6SF2 rs58542926-T versus fatty liver (Fig. 2B) and R2 = 0.30 for PNPLA3 rs738409 G versus fatty liver (Fig. 2A).
Figure 2.
The overall match between the metabolic effects of the NAFLD risk alleles and fatty liver. The black dashed line shows the linear fit between metabolic changes associated with fatty liver and PNPLA3 rs738409-G (A), TM6SF2 rs58542926-T (B), GCKR rs1260326-T (C) and LYPLAL1 rs12137855-C (D). The grey area indicates the 95% confidence interval for the line. R2 is a measure of goodness of fit.
Pairwise comparisons of the risk allele association profiles indicated that the effects of GCKR rs1260326-T and LYPLAL1 rs12137855-C on circulating metabolites were highly similar (R2 = 0.71; Supplementary Material, Fig. S4). The overall pattern of metabolic effects of TM6SF2 rs58542926-T correlated inversely with effects of both GCKR rs1260326-T and LYPLAL1 rs12137855-C (R2 = 0.66, and R2 = 0.50, respectively; Supplementary Material, Fig. S4). The remaining correlations were weaker (0.19 ≤ R2 ≤ 0.24; Supplementary Material, Fig. S4).
To control for intercorrelation of the lipoprotein lipid traits, we further examined the correlations of metabolic effects using a subset of metabolic measures that represented independent clusters (k = 29) identified in the YFS metabolomics data (Supplementary Material, Fig. S5). The magnitudes of coefficients of determination were somewhat smaller than using the full data, as expected, but supported the findings of the primary analysis (Supplementary Material, Fig. S6).
Discussion
We determined fatty liver-related metabolic changes in 1810 young and middle-aged Finns using 123 circulating metabolic measures assessed by NMR-based platform. We further compared the cross-sectional observations with metabolic signatures of known NAFLD risk alleles obtained from a publicly available metabolomics GWAS including up to 24 925 individuals (19). The studied NAFLD risk alleles resulted in divergent metabolic association profiles. Despite PNPLA3 rs738409-G being the strongest genetic risk factor for NAFLD (16), it showed a null effect on circulating lipids and metabolites. Association profile of GCKR rs1260326-T was similar to the cross-sectional fatty liver associations, whereas TM6SF2 rs58542926-T provided evidence to the opposite direction. The present results provide metabolomic evidence supportive to the recent findings about worsened metabolic features seen in association with obesity-linked NAFLD, but not with NAFLD arising due to risk alleles in PNPLA3 and TM6SF2 (17,22,23). In turn, the more risk-prone metabolic association profile of the NAFLD risk allele in GCKR underlines the importance of proper regulation of glucose metabolism in both liver and cardiovascular health.
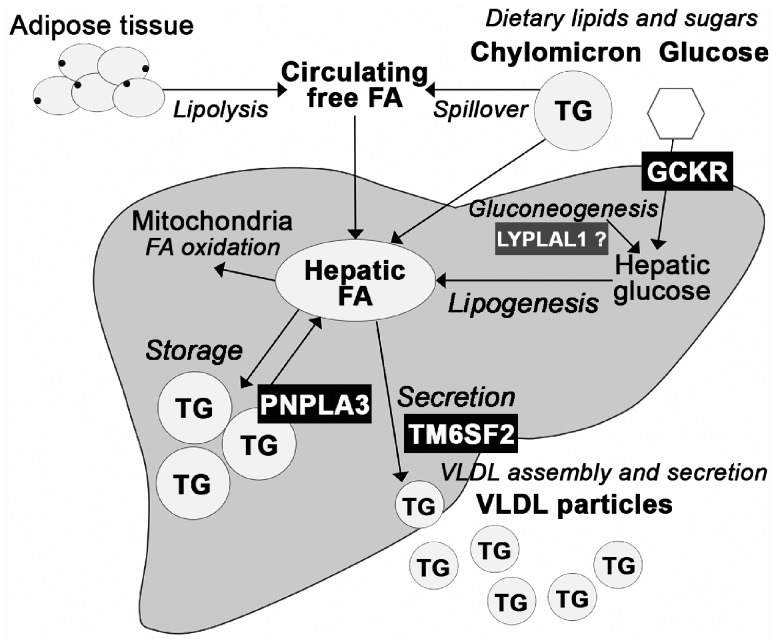
The metabolic association profiles reflect the biological functions of the risk alleles and suggest that the distinct molecular pathways giving rise to fatty liver may have divergent effects on circulating metabolome. The three pathways studied here are summarized in Figure 3. Pathway I, excessive hepatic glucose levels and amplified lipogenesis: GCKR rs1260326-T reduces GCKR ability to inhibit glucokinase resulting in enhanced hepatic glucose uptake, reduced fatty acid oxidation and increased lipogenesis (24). Hepatic fatty acids can be converted to triglycerides to be stored in hepatic lipid droplets or secreted in VLDL particles (25), where they can contribute respectively to development of steatosis or to levels of circulating lipids. Congruent with these observations, GCKR rs1260326-T increases risk of fatty liver (12,13) and raises concentrations of all the apolipoprotein B containing lipoprotein particles and triglycerides in these particles, while circulating glucose level is marginally reduced (Fig. 1, panel 2). GCKR rs1260326-T associates also with elevated levels of glycolysis-related metabolites, circulating fatty acids and increased fatty acid saturation (Fig. 1, panel 2) compatible with the enhanced glycolytic and lipogenic activities promoted by this variant (24,26,27). The amino acid association profile of GCKR rs1260326-T differs from the one of fatty liver in terms of concentrations of phenylalanine and tyrosine, both of which were increased in association with cross-sectional fatty liver but unaltered by the GCKR variant (Fig. 1, panel 1). Phenylalanine levels have been associated with cardiovascular disease risk (28) while tyrosine is linked with incidence of diabetes (29); the differences in the effects on the two biomarkers and on the two largest HDL subclasses and their lipids may be indicative to molecular pathways explaining why GCKR rs1260326-T rises concentrations of circulating lipids but does not seem to increase risk of cardiometabolic complications (30,31) as fatty liver does. LYPLAL1 rs12137855-C variant has a metabolically highly similar but statistically less robust effect than the GCKR rs1260326-T, as seen in highly correlated effect estimates of the two (Supplementary Material, Fig. S4). This finding advocates that LYPLAL1 may contribute to circulating and hepatic triglyceride levels by regulating hepatic glucose metabolism as does GCKR. This is supported by the discovery by Ahn et al. who showed that LYPLAL1 inhibition leads to increase in glucose production in human, rat and mouse hepatocytes (32).
Figure 3.
Relation of the studied NAFLD risk alleles to the main pathways in hepatic triglyceride partitioning. Liver converts carbohydrates to lipids in de novo lipogenesis. Newly synthesized fatty acids enter to the hepatic fatty acid pool which is also supplied by dietary fats and circulating free fatty acids derived mostly from adipose tissue lipolysis or lipoprotein lipase spillover (1,25). Fatty acids can be partitioned to oxidative pathway or esterified to triglycerides that can be stored in hepatic lipid droplets or used for VLDL production to be secreted from the liver (1,25). The hepatic fatty acid pool is located upstream from the lipid storage and secretion pathways, and thus abundance in hepatic fatty acids can contribute to both development of fatty liver and increased production of VLDL. In line with this, GCKR rs1260326-T that enhances the lipogenic pathway by providing more substrates for fatty acid biosynthesis (24) increases risk of fatty liver (12,13) and raises concentrations of all the apoB containing lipoproteins and lipids in these particles while circulating glucose level is marginally reduced (Fig. 1). LYPLAL1 may be functioning on the same hepatic glucose metabolism and lipogenesis related pathway, as the metabolic effects of GCKR rs1260326-T and LYPLAL1 rs12137855-C are highly similar (Supplementary Material, Fig. S4). On the contrary, TM6SF2 rs58542926-T impairs the secretory pathway leading to lipid accumulation into the liver (14) and reduction in levels of circulating lipids and lipoproteins (Fig. 1). PNPLA3 rs738409-G, in turn, enhances triglyceride accumulation to the storage pool by diminishing triglyceride hydrolysis to fatty acids (41,42), but does not directly contribute to VLDL secretion, and thus conveys no major consequences to circulation (Fig. 1).
Pathway II, reduced VLDL secretion: TM6SF2 contributes to VLDL secretion from the liver (33). In mice, knockdown of Tm6sf2 triplicates hepatic triglyceride content while blood lipids are decreased (14). Conversely, overexpression of human TM6SF2 in mice increases serum lipids levels (15). The NAFLD risk allele TM6SF2 rs58542926-T is a loss-of-function variant resulting in a misfolded protein undergoing accelerated degradation (14). The mechanism leading to impaired lipidation, subsequent degradation and thus reduced secretion of VLDL particles seem to be due to deficiency in polyunsaturated phosphatidylcholines in TM6SF2 rs58542926-T carriers (34). This allele associates with reduced concentrations of multiple lipid species in circulation while it has no influence on qualitative measures of circulating lipids, such as fatty acid saturation (Fig. 1, panel 5). The inverse associations between TM6SF2 rs58542926-T and all the circulating VLDL particle subclass concentrations, lipid species within the lipoprotein subclasses, as well as apolipoprotein B concentration are supportive for TM6SF2 rs58542926-T disturbing both lipidation and secretion of VLDL particles in humans. This differs from the observation of a study with Tm6sf2−/− mice, which provided evidence that mouse Tm6sf2 is required for lipidation of VLDL, but lack of it does not influence VLDL secretion (35). Our results are compatible with lipidomic effects of TM6SF2 rs58542926-T (36) and studies that associated this variant with a favourable plasma lipid profile and cardioprotective effect at the expense of increased risk for progressive NAFLD (17,37). Interestingly, TM6SF2 rs58542926-T associates with lower level of glycoprotein acetylation, a marker for inflammation (Fig. 1, panel 5), indicating that its cardioprotective effect is not limited only to lipoprotein and lipid metabolism. However, the same allele has been associated with increased risk of type 2 diabetes (38) underlining its complex effect on systemic metabolism.
Pathway III, impairment of triglyceride mobilization from hepatic lipid storage: PNPLA3 is located in the endoplasmic reticulum and lipid droplet membranes in human hepatocytes and shows both acyltransferase and hydrolase activities on glycerolipids (39–41). Despite the intensive research on the topic, the exact mechanism of how the common variant rs738409-G promotes development of liver disease is not clear. Several studies advocate that the variant inhibits triglyceride hydrolysis (39,41,42) possibly arising from accumulation of inactive PNPLA3 on lipid droplets disturbing mobilization of lipids (43,44). Some studies have shown that this risk allele does not influence common blood lipid traits, triglyceride profile, nor glucose homeostasis (10,11,22), while some studies have associated rs738409-G with lower lipid levels and protection from cardiovascular disease (45,46). The present results extend the findings of previous studies from common blood lipid traits to detailed fatty acid, amino acid and lipoprotein subfraction measures, and confirms the perception that rs738409-G has only minimal, if any, contribution to the metabolic traits in circulation (Fig. 1, panel 4). Our results are compatible with a mouse model where Pnpla3148M/M knock-in mice show no differences in levels of circulating lipids and glucose in comparison with wild-type mice regardless of the increase in liver triglycerides (43). These observations advocate that hepatic lipid accumulation can be neutral for circulatory changes and that the strong cross-sectional associations are largely due to lifestyle-related aspects such as dietary factors, excess energy intake or sedentary lifestyle. Dietary lipids and sugars and adipose tissue-derived fatty acids supply the fatty acid pool upstream from the lipid storage and secretion pathways (25), and consequently overnutrition could contribute to both fatty liver development as well as altered metabolic profile. The importance of nutritional factors is highlighted also when determining genetic risk for NAFLD, as adiposity has been shown to amplify the effect of the NAFLD risk alleles in PNPLA3, TM6SF2 and GCKR (47).
This study has some limitations. We examine a limited number of NAFLD risk alleles while there are multiple other genetic pathways contributing to pathogenesis of NAFLD (16,48,49). However, the risk alleles in PNPLA3, TM6SF2 and GCKR are important determinants of liver fat content and thus the current study setting covers some of the fundamental pathways involved in NAFLD pathogenesis (13,14). The role of the LYPLAL1 variant in NAFLD pathogenesis needs additional assessment; however, the current results provide a novel insight into LYPLAL1 function, which potentially helps in directing the focus of further studies. Regarding the cross-sectional associations, ultrasound lacks sensitivity to detect mild steatosis (50), leading to conservative association magnitudes. Despite the known deficiencies, ultrasound is a feasible method to be used for liver screening in a large population study. Also, the study populations compose of relatively young European individuals, and the present findings would need to be confirmed in other demographic groups. A recent study with ∼300 000 individuals detected a small effect for the PNPLA3 risk allele (46), and thus it is likely that with a larger sample size we would also see a small effect. However, when considering the great NAFLD risk increase associated with this variant (12), the relative effect on the circulating metabolic measures is minute. The strength of the metabolomics panel used is in detailed lipoprotein metabolism, while alternative techniques for metabolic profiling, such as a more detailed lipidomics platform (51), may reveal other biomarker changes associated with NAFLD induced by the studied risk alleles. The metabolic traits captured with the current panel include multiple known biomarkers of cardiometabolic diseases (52,53). Importantly, these biomarkers show strong associations with fatty liver, and the studied risk alleles show divergent effects on the corresponding measures underlining the heterogeneous nature of fatty liver.
The present study illustrates how open-access omics data can be utilized in evaluation of molecular mechanisms complex traits, such as NAFLD. The divergence in the direction of the risk allele association profiles advocates that different molecular pathways leading to NAFLD may have alternate effects on circulating lipids and other metabolites. We highlight the minimal metabolic effect of the strongest genetic determinant of NAFLD, PNPLA3 rs738409-G, and the cardioprotective effect of the TM6SF2 rs58542926-T on circulating lipids. Our findings suggest that, despite the strong population level associations, hepatic lipid accumulation by itself does not necessarily increase the risk of hyperlipidemia associated with cardiometabolic complications.
Materials and Methods
Study population
The Cardiovascular risk in Young Finns Study (YFS) is a population-based follow-up study started in 1980. In 2011, 2046 individuals aged 34–49 years participated to ultrasound imaging of the liver (Acuson Sequoia 512, Acuson, Mountain View, CA, USA). In total, fatty liver was diagnosed in 18.6% (N = 372) of the participants. The population and ultrasound imaging of the liver are described in more detail by Kaikkonen et al. (4) and Suomela et al. (54). After exclusion of pregnant women and individuals using lipid lowering medication or oral contraceptives, the total number of individuals included in the analysis was 1810 (Nfatty liver = 338). The study was approved by the local Ethics Committee and written informed consent was obtained from all the participants.
Metabolic effects of NAFLD risk alleles
Effect estimates of PNPLA3 rs738409-G, GCKR rs1260326-T, GCKR rs780094-T, TM6SF2 rs58542926-T, NCAN rs2228603-T and LYPLAL1 rs12137855-C on the metabolic measures were acquired from a GWAS performed using 14 European cohorts in up to 24 925 individuals (19). The mean age and BMI of the participants per cohort ranged from 23.9 to 61.3 years and from 23.1 to 28.2 kg/m2 with the whole sample means being 46.3 years and 26.0 kg/m2. The genotyping methods are described in the original publication (19).
Risk allele frequencies and odds ratios to NAFLD for the studied loci are described in Table 2. To facilitate comparison of the risk allele associated metabolic changes relative to NAFLD risk increase, the risk allele effects and related 95% confidence intervals were scaled with respect to the log(odds ratio) on histologic NAFLD associated with the corresponding locus in a large-scale GWAS (12). Scaling the metabolic associations with respect to a common factor allows evaluation of the effect similarities regardless of the magnitude of the absolute effect, which can be helpful when the study interest is on the metabolic pathways involved rather than in the absolute effects. The same principle has been applied for example in a study comparing metabolic effects of statin treatment versus genetic inhibition of HMGCR, where the large difference in the absolute effects of statins and HMGCR rs12916 genotype was overcome by scaling the metabolic effects to the magnitude on LDL-C lowering (55). P-values for the genetic effects were derived directly from the metabolomics GWAS (19).
Table 2.
Description of NAFLD risk increasing genotypes extracted from the open access data
|
NAFLD GWAS by Speliotes et al. (12) |
Metabolomics GWAS by Kettunen et al. (19) | ||||
|---|---|---|---|---|---|
| Gene | dbSNP reference | EA | Locus OR for histologic NAFLD (95% CI) | EAF | EAF |
| PNPLA3 | rs738409 | G | 3.24 (2.83–3.72) | 0.23 | 0.23 |
| NCAN | rs2228603 | T | 1.90 (1.55–2.34) | 0.07 | 0.07 |
| *TM6SF2 | rs58542926 | T | NA | 0.06 | |
| LYPLAL1 | rs12137855 | C | 1.21 (1.02–1.43) | 0.79 | 0.74 |
| GCKR | rs780094 | T | 1.18 (1.05–1.34) | 0.39 | 0.37 |
| *GCKR | rs1260326 | T | NA | 0.36 | |
DNA sequence variants studied in the present study were associated with computed tomography characterized steatosis and biopsy-proven NAFLD involving liver inflammation and fibrosis by Speliotes et al. (12). The functional variants explaining the NAFLD associations of NCAN rs2228603 and GCKR rs780094 are denoted separately (*). To achieve coherency, the NCAN locus is referred as TM6SF2 throughout the paper.
PNPLA3, Patatin-like phospholipase domain containing 3, NM_025225; NCAN, Neurocan, NM_004386; TM6SF2, Transmembrane 6 superfamily member 2, NM_001001524; LYPLAL1, Lysophospholipase-like 1, NM_001350628; GCKR, Glucokinase regulator, NM_001486; SNP, single-nucleotide polymorphism; EA, effect allele; OR, odds ratio; NAFLD, non-alcoholic fatty liver disease; EAF, effect allele frequency; NA, not available.
Metabolic profiling
Metabolic profiling of all samples included were assessed using a nuclear magnetic resonance (NMR) metabolomics platform described in (19) and reviewed in (52,53). To determine the cross-sectional metabolic associations of fatty liver, we utilized 123 metabolic measures representing a broad molecular signature of systemic metabolism (Supplementary Material, Table S1).
Statistical analyses
All analyses were done using R version 3.2.2. Due to the correlation of the metabolic measures, the number of independent tests performed is lower than the total number of 123 metabolic traits examined. We considered statistical significance at < 0.0004 (0.05/22/5), where 22 is the number of principal components explaining 95% of the variation in the NMR metabolomics data (19) and five is the number of analyses conducted.
Cross-sectional associations
Linear regression models were fitted to determine the cross-sectional associations of fatty liver with each of the metabolic measures in the YFS population. To enable the comparison of cross-sectional and genetic effect estimates, the data processing and analysis model were done correspondingly to Kettunen et al. (19): the metabolic phenotypes were adjusted for age, sex and 10 first genetic principal components preceding the analysis, and the resulting residuals were transformed to normal distribution by inverse rank-based normal transformation. The adjusted and transformed metabolic phenotypes were used as outcomes in the equations and fatty liver served as a categorical variable (fatty liver vs. no fatty liver). Secondary analyses were conducted with additional adjustment for alcohol consumption or BMI to evaluate the possible contribution of alcoholic fatty liver and obesity to the observed metabolic changes. The genetic principal components were calculated using PLINK software on linkage disequilibrium pruned markers after removing variants with more than 5% missing data and poorly genotyped samples with more than 5% missing data.
Comparisons of the metabolic association profiles
The overall resemblance between the risk alleles and fatty liver effects on metabolites was determined by a linear fit of each of the risk allele association profile versus cross-sectional fatty liver association profile (55). In addition, the linear fit of all the pairs of the risk allele association profiles were determined to identify alleles with similar metabolic effects informing about gene products functioning in related biological pathways. This method was also used to ensure that the metabolic profiles of the NAFLD risk alleles GCKR rs780094-T and NCAN rs2228603-T correspond to ones of GCKR rs1260326-T and TM6SF2 rs58542926-T that have been identified as the functional variants in GCKR and NCAN/TM6SF2 loci, respectively (13–15).
Due to the correlated nature of the lipoprotein lipid measures, we completed sensitivity analyses on the correlation tests with a subset of metabolic measures: we first assessed independent clusters of the metabolic measures in the YFS population using hierarchical clustering and gap statistics in factoextra R package, and then selected one metabolic measure per identified cluster for re-evaluation of the resemblance of the metabolic effects.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. P.W., A.J.K. and P.S. are employees and shareholders of Nightingale Health Ltd, a company offering NMR-based biomarker profiling. J.K. reports owning stock options of Nightingale Health Ltd.
Supplementary Material
Funding
This work was supported by University of Oulu Graduate School [E.S.]; Biocenter Oulu [S.S.]; European Commission [DynaHEALTH—H2020-633595, S.S.]; Academy of Finland [283045, 297338, 307247, J.K.] [312476, 312477, P.W.]; Novo Nordisk Foundation [P.W.] [NNF17OC0026062, J.K.]; Sigrid Juselius Foundation [M.A.K.]; and University of Bristol and UK Medical Research Council [MC_UU_12013/1, M.A.K.]. The Young Finns Study has been supported by Tampere and Turku University Hospitals [X51001, T.L.]; the Academy of Finland [286284, 134309 (Eye), 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), 41071 (Skidi)]; the Social Insurance Institution of Finland; Competitive State Research Financing of the Expert Responsibility area of Kuopio; Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation for Cardiovascular Research; Finnish Cultural Foundation; Tampere Tuberculosis Foundation; Emil Aaltonen Foundation; Yrjö Jahnsson Foundation; Signe and Ane Gyllenberg Foundation; and Diabetes Research Foundation of Finnish Diabetes Association. Funding to pay the Open Access publication charges for this article was provided by Novo Nordisk Foundation.
References
- 1. Cohen J.C., Horton J.D., Hobbs H.H., (2011) Human fatty liver disease: old questions and new insights. Science, 332, 1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adiels M., Taskinen M.R., Packard C., Caslake M.J., Soro-Paavonen A., Westerbacka J., Vehkavaara S., Hakkinen A., Olofsson S.O., Yki-Jarvinen H.. et al. (2006) Overproduction of large VLDL particles is driven by increased liver fat content in man. Diabetologia, 49, 755–765. [DOI] [PubMed] [Google Scholar]
- 3. Chatrath H., Vuppalanchi R., Chalasani N. (2012) Dyslipidemia in patients with nonalcoholic fatty liver disease. Semin. Liver Dis., 32, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaikkonen J.E., Wurtz P., Suomela E., Lehtovirta M., Kangas A.J., Jula A., Mikkila V., Viikari J.S., Juonala M., Ronnemaa T.. et al. (2017) Metabolic profiling of fatty liver in young and middle-aged adults: cross-sectional and prospective analyses of the Young Finns Study. Hepatology, 65, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mannisto V.T., Simonen M., Hyysalo J., Soininen P., Kangas A.J., Kaminska D., Matte A.K., Venesmaa S., Kakela P., Karja V.. et al. (2015) Ketone body production is differentially altered in steatosis and non-alcoholic steatohepatitis in obese humans. Liver Int., 35, 1853–1861. [DOI] [PubMed] [Google Scholar]
- 6. Mannisto V.T., Simonen M., Soininen P., Tiainen M., Kangas A.J., Kaminska D., Venesmaa S., Kakela P., Karja V., Gylling H.. et al. (2014) Lipoprotein subclass metabolism in nonalcoholic steatohepatitis. J. Lipid Res., 55, 2676–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dentin R., Benhamed F., Hainault I., Fauveau V., Foufelle F., Dyck J.R., Girard J., Postic C. (2006) Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes, 55, 2159–2170. [DOI] [PubMed] [Google Scholar]
- 8. Savage D.B., Choi C.S., Samuel V.T., Liu Z.X., Zhang D., Wang A., Zhang X.M., Cline G.W., Yu X.X., Geisler J.G.. et al. (2006) Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J. Clin. Invest., 116, 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seppala-Lindroos A., Vehkavaara S., Hakkinen A.M., Goto T., Westerbacka J., Sovijarvi A., Halavaara J., Yki-Jarvinen H. (2002) Fat accumulation in the liver is associated with defects in insulin suppression of glucose production and serum free fatty acids independent of obesity in normal men. J. Clin. Endocrinol. Metab., 87, 3023–3028. [DOI] [PubMed] [Google Scholar]
- 10. Speliotes E.K., Butler J.L., Palmer C.D., Voight B.F. GIANT Consortium, MIGen Consortium NASH C.R.N., Hirschhorn J.N. (2010) PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology, 52, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L.A., Boerwinkle E., Cohen J.C., Hobbs H.H. (2008) Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet., 40, 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J.. et al. (2011) Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet., 7, e1001324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Santoro N., Zhang C.K., Zhao H., Pakstis A.J., Kim G., Kursawe R., Dykas D.J., Bale A.E., Giannini C., Pierpont B.. et al. (2012) Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology, 55, 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjaerg-Hansen A., Vogt T.F., Hobbs H.H., Cohen J.C. (2014) Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet., 46, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holmen O.L., Zhang H., Fan Y., Hovelson D.H., Schmidt E.M., Zhou W., Guo Y., Zhang J., Langhammer A., Lochen M.L.. et al. (2014) Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat. Genet., 46, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anstee Q.M., Day C.P. (2013) The genetics of NAFLD. Nat. Rev. Gastroenterol. Hepatol., 10, 645–655. [DOI] [PubMed] [Google Scholar]
- 17. Goffredo M., Caprio S., Feldstein A.E., D'Adamo E., Shaw M.M., Pierpont B., Savoye M., Zhao H., Bale A.E., Santoro N. (2016) Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: a multiethnic study. Hepatology, 63, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dongiovanni P., Romeo S., Valenti L. (2015) Genetic factors in the pathogenesis of nonalcoholic fatty liver and steatohepatitis. Biomed. Res. Int., 2015, 460190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kettunen J., Demirkan A., Wurtz P., Draisma H.H., Haller T., Rawal R., Vaarhorst A., Kangas A.J., Lyytikainen L.P., Pirinen M.. et al. (2016) Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun., 7, 11122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Byrne C.D., Targher G. (2015) NAFLD: a multisystem disease. J. Hepatol, 62, 47.. [DOI] [PubMed] [Google Scholar]
- 21. Ritchie S.C., Wurtz P., Nath A.P., Abraham G., Havulinna A.S., Fearnley L.G., Sarin A.P., Kangas A.J., Soininen P., Aalto K.. et al. (2015) The biomarker GlycA is associated with chronic inflammation and predicts long-term risk of severe infection. Cell. Syst., 1, 293–301. [DOI] [PubMed] [Google Scholar]
- 22. Hyysalo J., Gopalacharyulu P., Bian H., Hyotylainen T., Leivonen M., Jaser N., Juuti A., Honka M.J., Nuutila P., Olkkonen V.M.. et al. (2014) Circulating triacylglycerol signatures in nonalcoholic fatty liver disease associated with the I148M variant in PNPLA3 and with obesity. Diabetes, 63, 312–322. [DOI] [PubMed] [Google Scholar]
- 23. Petaja E.M., Yki-Jarvinen H. (2016) Definitions of normal liver fat and the association of insulin sensitivity with acquired and genetic NAFLD-A systematic review. Int. J. Mol. Sci., 17, 10.3390/ijms17050633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beer N.L., Tribble N.D., McCulloch L.J., Roos C., Johnson P.R., Orho-Melander M., Gloyn A.L. (2009) The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet., 18, 4081–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hodson L., Frayn K.N. (2011) Hepatic fatty acid partitioning. Curr. Opin. Lipidol., 22, 216–224. [DOI] [PubMed] [Google Scholar]
- 26. Rees M.G., Wincovitch S., Schultz J., Waterstradt R., Beer N.L., Baltrusch S., Collins F.S., Gloyn A.L. (2012) Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia, 55, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santoro N., Caprio S., Pierpont B., Van Name M., Savoye M., Parks E.J. (2015) Hepatic de novo lipogenesis in obese youth is modulated by a common variant in the GCKR gene. J. Clin. Endocrinol. Metab., 100, 1125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wurtz P., Havulinna A.S., Soininen P., Tynkkynen T., Prieto-Merino D., Tillin T., Ghorbani A., Artati A., Wang Q., Tiainen M.. et al. (2015) Metabolite profiling and cardiovascular event risk: a prospective study of 3 population-based cohorts. Circulation, 131, 774–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tillin T., Hughes A., Wang Q., Würtz P., Ala-Korpela M., Sattar N., Forouhi N., Godsland I., Eastwood S., McKeigue P.. et al. (2015) Diabetes risk and amino acid profiles: cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall and Brent REvisited) study. Diabetologia, 58, 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaxillaire M., Cavalcanti-Proença C., Dechaume A., Tichet J., Marre M., Balkau B., Froguel P., (2008) The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general french population. Diabetes, 57, 2253–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bi M., Kao W.H.L., Boerwinkle E., Hoogeveen R.C., Rasmussen-Torvik L.J., Astor B.C., North K.E., Coresh J., Köttgen A. (2010) Association of rs780094 in GCKR with metabolic traits and incident diabetes and cardiovascular disease: the ARIC study. PLoS One, 5, e11690.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahn K., Boehm M., Brown M.F., Calloway J., Che Y., Chen J., Fennell K.F., Geoghegan K.F., Gilbert A.M., Gutierrez J.A.. et al. (2016) Discovery of a selective covalent inhibitor of lysophospholipase-like 1 (LYPLAL1) as a tool to evaluate the role of this serine hydrolase in metabolism. ACS Chem. Biol., 11, 2529–2540. [DOI] [PubMed] [Google Scholar]
- 33. Mahdessian H., Taxiarchis A., Popov S., Silveira A., Franco-Cereceda A., Hamsten A., Eriksson P., van't Hooft F. (2014) TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. U. S. A., 111, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luukkonen P.K., Zhou Y., Nidhina Haridas P.A., Dwivedi O.P., Hyotylainen T., Ali A., Juuti A., Leivonen M., Tukiainen T., Ahonen L.. et al. (2017) Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J. Hepatol., 67, 128–136. [DOI] [PubMed] [Google Scholar]
- 35. Smagris E., Gilyard S., BasuRay S., Cohen J.C., Hobbs H.H. (2016) Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J. Biol. Chem., 291, 10659–10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Y., Llaurado G., Oresic M., Hyotylainen T., Orho-Melander M., Yki-Jarvinen H. (2015) Circulating triacylglycerol signatures and insulin sensitivity in NAFLD associated with the E167K variant in TM6SF2. J. Hepatol., 62, 657–663. [DOI] [PubMed] [Google Scholar]
- 37. Pirola C.J., Sookoian S. (2015) The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: a meta-analysis. Hepatology, 62, 1742–1756. [DOI] [PubMed] [Google Scholar]
- 38. Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J.. et al. (2016) The genetic architecture of type 2 diabetes. Nature, 536, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Y., Cohen J.C., Hobbs H.H. (2011) Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem., 286, 37085–37093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jenkins C.M., Mancuso D.J., Yan W., Sims H.F., Gibson B., Gross R.W. (2004) Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem., 279, 48968–48975. [DOI] [PubMed] [Google Scholar]
- 41. He S., McPhaul C., Li J.Z., Garuti R., Kinch L., Grishin N.V., Cohen J.C., Hobbs H.H. (2010) A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem., 285, 6706–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pingitore P., Pirazzi C., Mancina R.M., Motta B.M., Indiveri C., Pujia A., Montalcini T., Hedfalk K., Romeo S. (2014) Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim. Biophys. Acta, 1841, 574–580. [DOI] [PubMed] [Google Scholar]
- 43. Smagris E., BasuRay S., Li J., Huang Y., Lai K.M., Gromada J., Cohen J.C., Hobbs H.H. (2015) Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology, 61, 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. BasuRay S., Smagris E., Cohen J.C., Hobbs H.H. (2017) The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology, 66, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Simons N., Isaacs A., Koek G.H., Kuc S., Schaper N.C., Brouwers M.C. (2017) PNPLA3, TM6SF2, and MBOAT7 genotypes and coronary artery disease. Gastroenterology, 152, 912–913. [DOI] [PubMed] [Google Scholar]
- 46. Liu D.J., Peloso G.M., Yu H., Butterworth A.S., Wang X., Mahajan A., Saleheen D., Emdin C., Alam D., Alves A.C.. et al. (2017) Exome-wide association study of plasma lipids in >300, 000 individuals. Nat. Genet., 49, 1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stender S., Kozlitina J., Nordestgaard B.G., Tybjaerg-Hansen A., Hobbs H.H., Cohen J.C. (2017) Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat. Genet., 49, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hooper A.J., Adams L.A., Burnett J.R. (2011) Genetic determinants of hepatic steatosis in man. J. Lipid Res., 52, 593–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R., Boren J., Montalcini T., Pujia A., Wiklund O.. et al. (2016) The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of european descent. Gastroenterology, 150, 1230.e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwenzer N.F., Springer F., Schraml C., Stefan N., Machann J., Schick F. (2009) Non-invasive assessment and quantification of liver steatosis by ultrasound, computed tomography and magnetic resonance. J. Hepatol., 51, 433–445. [DOI] [PubMed] [Google Scholar]
- 51. Kotronen A., Seppanen-Laakso T., Westerbacka J., Kiviluoto T., Arola J., Ruskeepaa A.L., Yki-Jarvinen H., Oresic M. (2010) Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity (Silver Spring), 18, 937–944. [DOI] [PubMed] [Google Scholar]
- 52. Würtz P., Kangas A.J., Soininen P., Lawlor D.A., Davey Smith G., Ala-Korpela M. (2017) Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol., 186, 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soininen P., Kangas A.J., Würtz P., Suna T., Ala-Korpela M. (2015) Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet., 8, 192–206. [DOI] [PubMed] [Google Scholar]
- 54. Suomela E., Oikonen M., Virtanen J., Parkkola R., Jokinen E., Laitinen T., Hutri-Kähönen N., Kähönen M., Lehtimäki T., Taittonen L.. et al. (2015) Prevalence and determinants of fatty liver in normal-weight and overweight young adults. The cardiovascular risk in Young Finns Study. Ann. Med., 47, 40–46. [DOI] [PubMed] [Google Scholar]
- 55. Wurtz P., Wang Q., Soininen P., Kangas A.J., Fatemifar G., Tynkkynen T., Tiainen M., Perola M., Tillin T., Hughes A.D.. et al. (2016) Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J. Am. Coll. Cardiol., 67, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.