Abstract
Niemann-Pick type C (NPC) disease is a rare lysosomal storage disease caused primarily by mutations in NPC1. NPC1 encodes the lysosomal cholesterol transport protein NPC1. The most common NPC1 mutation is a missense mutation (NPC1I1061T) that causes misfolding and rapid degradation of mutant protein in the endoplasmic reticulum. Cholesterol accumulates in enlarged lysosomes as a result of decreased levels of lysosomal NPC1I1061T protein in patient cells. There is currently no cure or FDA-approved treatment for patients. We sought to identify novel compounds that decrease lysosomal cholesterol storage in NPC1I1061T/I1061T patient fibroblasts using a high-content screen with the cholesterol dye, filipin and the lysosomal marker, LAMP1. A total of 3532 compounds were screened, including 2013 FDA-approved drugs, 327 kinase inhibitors and 760 serum metabolites. Twenty-three hits were identified that decreased both filipin and LAMP1 signals. The majority of hits (16/21) were histone deacetylase (HDAC) inhibitors, a previously described class of modifiers of NPC cholesterol storage. Of the remaining hits, the antimicrobial compound, alexidine dihydrochloride had the most potent lysosomal cholesterol-reducing activity. Subsequent analyses showed that alexidine specifically increased levels of NPC1 transcript and mature protein in both control and NPC patient cells. Although unsuitable for systemic therapy, alexidine represents a unique tool compound for further NPC studies and as a potent inducer of NPC1. Together, these findings confirm the utility of high-content image-based compound screens of NPC1 patient cells and support extending the approach into larger compound collections.
Introduction
Niemann-Pick type C disease (NPC, OMIM #257220 and #607625) is a rare [incidence ranging from ∼1: 89 000 based on projections (1), to 1: 120 000 live births based on clinical observations (2)] fatal neurodegenerative lysosomal storage disease that can manifest from infancy to adulthood depending on the genetic lesion (2,3). There are no FDA-approved treatments for NPC, though limited efficacy has been shown for miglustat, a glycosphingolipid synthesis inhibitor, which is approved for use outside the United States. The majority of NPC cases are caused by mutations in NPC1, the gene encoding the transmembrane lysosomal protein NPC1, while a small subset (<5%) are caused by mutations in the NPC2 gene that encodes a luminal lysosomal protein (2). Both genes encode proteins that are critical for cholesterol egress from lysosomes, though the exact role for each protein in cholesterol transport is not completely understood (4). While the hallmark of NPC is lysosomal cholesterol accumulation, disease severity depends on the type and position of the mutations in NPC1, as well as the genetic background of the individual (5–7). Most NPC patients have compound heterozygous NPC1 mutations, and the most common mutation, NPC1I1061T, typically leads to a juvenile disease onset and results in a misfolded protein that is rapidly degraded by the proteostasis machinery (5,8,9).
Previous studies identified HDAC inhibitors (HDACi) as negative modulators of lysosomal cholesterol storage (10), possibly through modulation of the proteostasis machinery (10). Early-stage clinical investigation of the HDACi, vorinostat (SAHA) is currently underway. Another drug proposed to modulate proteostasis, arimoclomol, is being tested in a phase III clinical trial for treatment of NPC1 patients (11). Finally, the cholesterol-reducing agent, 2-hydroxypropyl-ß-cyclodextrin (HPBCD), has shown efficacy in treatment of NPC1 patients a phase 1/2a trial (12), and is now in a pivotal phase 3 trial. Though promising, each therapy is associated with distinct off-target or adverse side effects, or limited blood–brain barrier permeability (13–15), the latter requiring direct delivery into the central nervous system (12,16). Further, although HDACi and arimoclomol are hypothesized to modulate the general chaperone machinery (10), neither of these drugs nor HPBCD directly addresses the protein misfolding defect present in the majority of NPC patients harboring missense mutations (17). Thus, there remains an unmet clinical need for more specific therapies that target the underlying molecular defect.
A number of large-scale compound screens designed to identify an effective NPC therapeutic have been reported. Screening strategies have varied. Early attempts utilized automated microscopy of NPC1 mutant CHO cells stained with the fluorescent cholesterol dye, filipin (18,19), while more recent screens have used fluorescent plate reader-based assays on patient-derived fibroblasts in conjunction with the acidophilic dye, lysotracker, to label acidic organelles such as late endosomes and lysosomes (20). Lysotracker staining is increased in both NPC patient fibroblasts and fibroblasts from other lysosomal storage disease patients relative to fibroblasts from healthy individuals, making it a useful screening reagent (20). Still others have utilized mutant cells from an Npc1 knockout mouse model (21). However, none of the previously described screens has considered lysosomal cholesterol content in NPC patient cells as a first-tier screen in a high-content fluorescent microscopy assay.
By examining both cholesterol content and lysosomes in patient cells using the dual markers filipin and anti-LAMP1, a lysosome/late endosome-specific marker, increased specificity is achieved. Moreover, a high-content phenotypic assay in patient-derived cells improves clinical relevance and reveals information about cell health and viability in response to test compounds, and has increased sensitivity and power compared to plate reader-based assays. In this report, we describe a high-content screen, based on filipin and LAMP1 readouts, of NPC patient fibroblasts. NPC patient fibroblasts homozygous for the most common NPC1 mutation, NPC1I1061T (5,8) were selected as a cell source given the prevalence of the mutation, the moderate degree of lysosomal cholesterol accumulation, and the potential to correct the protein misfolding phenotype with a proteostasis modulator (9). Using this assay, we screened 3532 small molecules, primarily FDA-approved drugs and well-characterized tool compounds that modulate known targets, affording opportunities for drug repurposing, and identification of specific molecular pathways and targets that impact the cellular phenotype. Among the hits identified were previously described modulators of NPC cholesterol accumulation as well as novel modulators.
Results
Development of a high-content image-based assay for lysosomal cholesterol accumulation in NPC patient cells
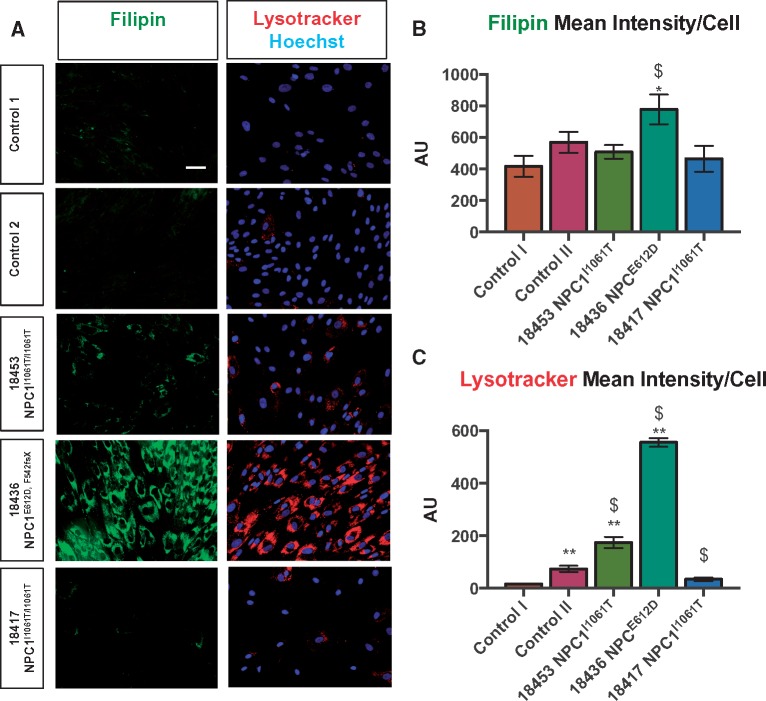
To develop a miniaturized high-content image-based assay in 384-well plates for monitoring lysosomal cholesterol accumulation, a panel of control and NPC patient cells representing a range of NPC1 genotypes was assembled (Table 1). These included fibroblasts from two patients homozygous for the most common NPC mutation, NPC1I1061T (5,8) and a severely affected patient with a compound heterozygous mutation (NPC1E612D/542fsX) (3). We first sought to validate the sensitivity and relevance of our assays by confirming the cellular phenotypes of the control and NPC patient cells. In addition, we aimed to determine the extent to which genotype and clinical phenotype correlate with cellular phenotype in our proposed system and the dynamic range of our assay. To that end, NPC patient lines, and two control fibroblast lines from unaffected individuals, were cultured in 384-well imaging plates, and characterized in parallel for cholesterol content, with the naturally fluorescent polyene antibiotic, filipin (19) and for lysosomes with lysotracker dye or by LAMP1 immunofluorescence as described in Supplementary Material, Figure S1.
Table 1.
Genotype of patient cells included in the study (NPC1 RefSeq cDNA NM_000271.4, NPC2 RefSeq cDNA NM_006432.3, genomic DNA NG_007117.1)
| Patient cells | NPC1 allele 1 | NPC1 allele 2 | |
|---|---|---|---|
| GM18453 (NPC1I1061T/I1061T) | c.3182 T>C (p.I1061T) | c.3182 T>C (p.I1061T) | |
| GM18417 (NPC1I1061T/I1061T) | c.3182 T>C (p.I1061T) | c.3182 T>C (p.I1061T) | |
| GM18436 (NPC1E612D/542fsX) | c.1836A>C (p.E612D) | c.1628delC (p.F542fsX) | |
| NPC1 null (NPC1−/−) | c.1628delC (p.F542fsX) | c.1628delC (p.F542fsX) | |
| Patient cells | NPC2 allele 1 | NPC2 allele 2 | |
| NPC2 null (NPC2−/−) | c.IVS2 + 5G→A (c.190 + 5G>A) (p.0) | c.IVS2 + 5G→A (c.190 + 5G>A) (p.0) | |
We observed a clear phenotypic spectrum for cholesterol content and lysosomal staining (Fig. 1). As observed in the literature (20), NPC1 patient cells had increased lysosomal signal, as inferred from LAMP1 staining, and cholesterol accumulation, based on filipin, compared with control cells, with GM18436 NPC1E612D/542fsX patient cells exhibiting higher levels than NPC1I1061T cells (Fig. 1). Notably, of the two fibroblast lines, GM18453 and GM18417, which harbor identical NPC1 mutations (NPC1I1061T/I1061T), GM18453 had a more pronounced phenotype (Fig. 1). Likewise, we observed variations in both filipin and lysotracker for control lines (Fig. 1), underscoring the importance of examining multiple patient and control lines, as modifier genes and epigenetic factors can impact phenotypes (5–7). Of the two NPC1I1061T patient lines, GM18453, which exhibited a stronger lysosomal cholesterol phenotype, was selected for further assay development and screening with the expectation that it should be amenable to rescue by proteostatic regulators as well as other molecular modulators.
Figure 1.
(A, B) Cholesterol accumulation and (A, C) lysosomes in control and NPC patient fibroblasts. Cholesterol content was assessed using the naturally fluorescent antibiotic, filipin [3]. Lysosomal morphology (size, distribution) was assessed using lysotracker-red. 20× magnification. Scale bar = 50 μm. Error bars: Mean ± SD, n = 3 wells/condition.*P < 0.05 versus Control I, **P < 0.01 versus Control I, $P < 0.05 versus Control II, one-way ANOVA followed by Tukey’s test.
HDACi are capable of ameliorating NPC patient cell lysosomal cholesterol storage, possibly through regulation of the proteostasis machinery (10). We tested the potent HDACi, trichostatin A (TSA) and panobinostat (Pb, LBH589) in our assay and found a robust reduction in lysosomal cholesterol content following 72 h treatment (Supplementary Material, Fig. S2A and B). HDACi treatment led to a reduction in cell number compared to DMSO-treated cells, likely due to both cytotoxicity and anti-proliferative effects of HDACi (22), but remaining cells appeared healthy according to nuclear parameters (Supplementary Material, Fig. S2C and D and data not shown). Taken together these data suggest that HDACi’s effects on lysosomal cholesterol are separable from its cytotoxicity. SSMD (strictly standardized mean difference) and Z′ values for screening were found to be acceptable for screening (23), with panobinostat exhibiting the most robust and reproducible effect on both filipin and LAMP1 immunofluorescence staining (Supplementary Material, Fig. S2A and B). Both SSMD and Z′ values provide information regarding effect size in screens; however, SSMD is more suitable for assessing quality control for strong or moderate effect differences compared to Z′, which is better suited to extremely strong effect differences (24). Therefore, SSMD was employed going forward for quality control assessment. Based on these results, GM18453 (NPC1I1061T/I1061T) patient fibroblasts and the control compound, panobinostat, were selected for a subsequent pilot screen.
To test the robustness of our assay on a smaller scale before screening a larger library, and to determine optimal timing of compound exposure, we performed a pilot screen of a kinase inhibitor library comprised of 327 well-annotated small molecule kinase inhibitors. The library was screened in duplicate at 5 μm, with 32 positive control wells and 32 negative control wells per plate (16 wells per side of plate). Parallel experiments were run at 48 and 72 h of exposure to compound. Although statistically significant decreases in both filipin and LAMP1 were observed at 48 h (Supplementary Material, Fig. S3), SSMD values were greater at 72 h (for filipin: 48 h, SSMD = −1.45, 72 h SSMD = −3.04). Of 327 compounds tested, 13 (4.0%) were cytotoxic based on cell counts. A compound was considered cytotoxic at a Z-score ≤ −5 for number of nuclei. A Z-score of −5 was chosen based on average Z-scores for the positive control, panobinostat, since HDACi are known to exhibit cytotoxic and anti-proliferative effects (22). Compounds were then filtered for hits that reduced filipin and LAMP1 without drastically affecting nuclear roundness (duplicate Z-score ≤ 3.5), intensity (−2.0 ≤ duplicate Z-score ≤ 2.0), or size (duplicate Z-score ≥ −2.0). A Z-score threshold of −2.0 was initially selected in order to identify compounds that fall outside of 95% of results in a normal distribution. This cut-off was then adjusted (e.g. nuclear roundness) to increase the number of hits for further characterization.
Of the remaining non-toxic compounds, 50/327 (15.3%) decreased filipin (duplicate Z-score ≤ −2) at 72 h, and 10/327 (3.1%) decreased LAMP1 (Z-score < −2.5 singleton as no compounds decreased LAMP1 in duplicate) but only four (1.2%) decreased both LAMP1 and filipin. These four represented three unique compounds: staurosporine (duplicate compounds were present in the library), the GSK3β inhibitor TWS-119, and MK-2 Inhibitor III (Table 2, Supplementary Material, Fig. S4 and data not shown). Thus, having demonstrated in our pilot screen that using filipin and LAMP1 as multiplexed readouts was an efficient approach that aided in hit detection and classification, we extended our screening efforts to larger compound collections described below.
Table 2.
Summary of libraries screened and plate statistics
| Library name | Compounds | SSMD FLP | SSMD LAMP1 | CV FLP | CV LAMP1 | FLP Hits | LAMP1 Hits | Dual hits | Cherry picked | Confirmed | Hits |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FDA repurposing | 2013 | −3.29 | −3.46 | 5.37 | 6.58 | 37 | 58 | 12 | 12 | 3 | SAHA (×2), alexidine |
| Kinase inhibitor | 327 | −3.04 | −1.61 | 5.43 | 7.23 | 50 | 10 | 4 | 2 | 1 | TWS-119 |
| Human metabolite | 760 | −2.07 | −3.45 | 14.92 | 4.77 | 31 | 35 | 3 | 3 | 0 | n/a |
| Metabolism | 127 | −4.05 | −3.55 | 3.35 | 6.65 | 1 | 6 | 1 | 1 | 1 | ChoKα inhibitor |
| Epigenetic | 260 | −4.28 | −5.25 | 4.58 | 3.80 | 50 | 51 | 27 | 27 | 16 | 14 HDACia, JFD00244, danusertib |
| Chaperone | 45 | −3.50 | −2.49 | 3.95 | 4.45 | 2 | 0 | 0 | 0 | 0 | |
| Total | 3532 | 171 | 160 | 47 | 45 | 21 | |||||
| Average | −3.15 | −3.39 | 7.11 | 5.89 |
SSMD, strictly standardized mean difference; CV, coefficient of variation.
HDACi: scriptaid, belinostat (PXD101), trichostatin A, HDACi III (M-344), oxamflatin, SAHA, BML-281, AR-42, CUDC-101, pracinostat (SB939), 4-iodo-SAHA, HC toxin, coumarin-SAHA, pyroxamide.
High-content screen for modifiers of lysosomal cholesterol in NPC patient cells
In an effort to identify a clinically tractable mediator of lysosomal cholesterol storage in NPC patient cells, a library of 2013 FDA-approved compounds was screened in duplicate using the dual LAMP1/filipin assay. In addition, we screened our assay against a collection of 260 compounds with epigenetic activity, 45 compounds with proteostatic activity, 760 human metabolites (25) and 127 compounds that target metabolic pathways (Table 2). Due to redundancy in the libraries, in total 3248 unique compounds were screened (Supplementary Material, Table S1). Average SSMD between DMSO and panobinostat-treated groups were −3.15 (filipin) and −3.39 (LAMP1) (26). Average coefficient of variation (CV) for DMSO and panobinostat-treated groups were 7.11% (filipin) and 5.89% (LAMP1) (Table 2).
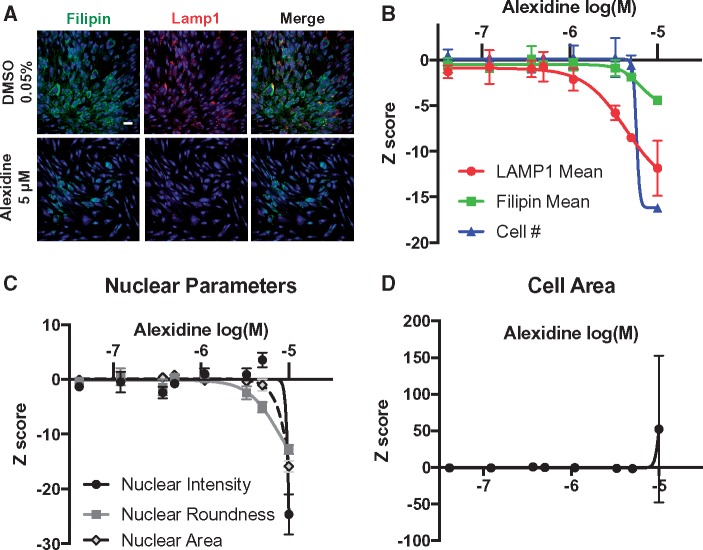
Seventy-five (2.1%) compounds were cytotoxic based on number of nuclei. Our criterion for hit selection was Z-score ≤ −2 in duplicate for both filipin and LAMP1 staining intensity. For initial hit selection, we considered compounds with evidence of cytotoxicity (either by nuclei number or morphology) given the high concentration at which compounds were screened and the small number of hits. From the primary screen, 171 compounds decreased filipin and 160 compounds decreased LAMP1 (Table 2, Supplementary Material, Table S1). Only 47 filipin hits (27%) were also LAMP1 hits (Table 2), an overall hit rate of 1.3%. The observed difference between filipin and LAMP1 hits may be related to differential flux rates for LAMP1 protein and cholesterol. Three of the 47 hits with decreases in both LAMP1 and filipin were cytotoxic based on number of nuclei. However, decreased lysosomal cholesterol was not generally correlated with cytotoxicity, as 68/75 (91%) of the cytotoxic compounds we identified did not exhibit decreases in both LAMP1 and filipin. Dose–response studies identified one compound, alexidine dihydrochloride (alexidine) as having dose-dependent activity, with half-maximal inhibitory concentrations (IC50) of 4.3 μm for 6.36 μm for LAMP1 and filipin readouts, respectively (Fig. 2A and B). Although alexidine was cytotoxic at the original dose screened (5 μm), its effect on lysosomal cholesterol storage was separable from cytotoxicity as indicated by nuclear measures (Fig. 2C) and cell area (Fig. 2D). Consistent with other reports (27), we did not identify any of the six statins included in our repurposing library as hits (Supplementary Material, Fig. S5).
Figure 2.
Alexidine treatment reduces lysosomal cholesterol accumulation. (A) Representative images of GM18453 NPC1I1061T/I1061T patient fibroblasts plated and treated 24 h later with DMSO or alexidine at indicated concentrations. Cells were fixed and stained with filipin, LAMP1 and Draq5 72 h after treatment. (B) Dose–response curves for alexidine-treated GM18453 patient fibroblasts. (C) Dose–response curves for nuclear parameters for alexidine-treated GM18453 patient fibroblasts. (D) Dose–response curve for cell area parameters for alexidine-treated GM18453 patient fibroblasts. Scale bar = 50 μm.
Alexidine is a novel enhancer of lysosomal cholesterol release through upregulation of NPC1- and NPC2-dependent and -independent pathways
Alexidine was then tested in secondary assays to determine whether the compound directly affected NPC1 expression. Western blot analysis revealed a ∼3-fold increase in NPC1 protein expression following 72 h treatment in both control and NPC1 mutant cells (NPC1I1061T/I1061T) (Fig. 3A and B). We did not observe an increase in NPC1 protein expression following 72 h HDACi (panobinostat) treatment (Fig. 3A and B), although HDACi treatment was effective as evidenced by robust induction of acetylated tubulin (Fig. 3C).
Figure 3.
Alexidine treatment induces NPC1 expression. (A) Western blot following 72 h drug treatment in fibroblasts from indicated genotypes, Anti-acetylated tubulin demonstrates appropriate HDACi activity. (B) Quantification of (A), n = 3 biological replicates, mean ± SD, ****P < 0.0001 one-way ANOVA followed by Tukey’s test. (C, D) EndoH digestion of NPC1 reveals an increase in both mature and immature NPC1 following alexidine treatment. NPC1I1061T/I1061T(C) or normal human fibroblasts (D) were plated and treated 24 h later with DMSO or alexidine. After 72 h, cells were harvested then subjected to Endo Hf or vehicle treatment. Western blot and normalized quantification for NPC1 in mutant (C) and normal (D) cell lysates. Open triangles mark Endo Hf resistant (Endo HR) species. Closed triangles mark Endo Hf sensitive (Endo HS) species. ****P < 0.0001, ***P < 0.001, **P < 0.01 versus glycospecies matched DMSO, two-way ANOVA followed by Dunnett’s test. (E) NPC1 expression by TaqMan qRT-PCR in indicated cell lines following 48 h drug treatment. n = 3 biological replicates, mean ± SD, ****P < 0.0001 versus DMSO, $versus Control alexidine two-way ANOVA followed by Dunnett’s test.
The NPC1I1061T missense mutation leads to rapid proteasomal degradation of immature NPC1 (9). Forced expression of NPC1-mutant protein allows some protein to escape the degradation pathway, resulting in increased delivery of mature, fully glycosylated mutant protein to the endosome/lysosome where NPC1 functions in lysosomal cholesterol trafficking (9). To assess the maturity of the increased NPC1 protein following alexidine treatment (Fig. 3A and B), we performed Endoglycosidase H (EndoH) digestion to remove immature N-linked glycan residues. Resistance to EndoH is indicative of protein trafficking beyond the endoplasmic reticulum in the secretory pathway. EndoH digestion revealed a dose-dependent increase in both EndoH-sensitive (immature) and EndoH-resistant (mature) NPC1 populations in alexidine-treated NPC1I1061T/I1061T fibroblasts, as well as an increase in EndoH-resistant NPC1 in alexidine-treated control fibroblasts (Fig. 3C and D).
qRT-PCR analysis of NPC1 transcript expression revealed that the alexidine-mediated change in NPC1 protein expression was accompanied by NPC1 gene expression changes (Fig. 3E). NPC1 transcript abundance was similar in vehicle-treated cells from control and mutant NPC1 cell lines, a finding that is in agreement with previously published studies (9).
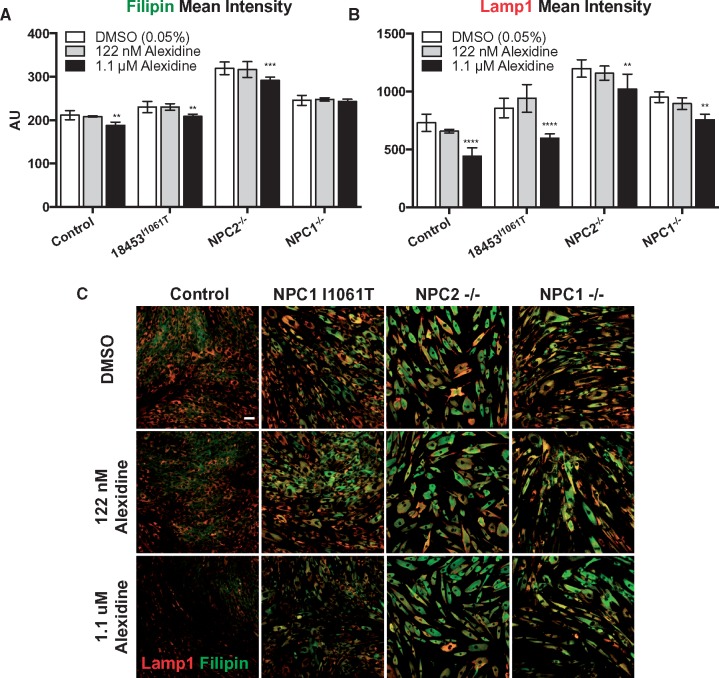
To determine whether alexidine acts in an NPC1- or NPC2-dependent manner, alexidine-mediated reduction of lysosomal cholesterol was next tested in control cells, and null mutant NPC1−/− and NPC2−/− cells (Table 1). Alexidine-reduced lysosomal cholesterol in control cells to a similar extent to NPC1I1061T/I1061T patient cells, and to a lesser extent in NPC1−/− and NPC2−/− cells (e.g. reduction in LAMP1 DMSO versus alexidine: control 40%, NPC1I1061T/I1061T 31%, NPC1−/− 15%, NPC2−/− 20%) (Fig. 4A–C). At the concentrations tested, alexidine was not cytotoxic, based on cell number, providing further evidence that alexidine’s effects on lysosomal cholesterol can be separated from its cytoxicity (Supplementary Material, Fig. S6). Thus, alexidine enhances both NPC1- and NPC2-dependent and -independent cholesterol release.
Figure 4.
NPC1/2 expression correlates with degree of alexidine-induced lysosomal cholesterol reduction. (A) Control, patient NPC1I1061T/I1061T, NPC1−/− and NPC2−/− fibroblasts were treated with DMSO (0.05%), 122 nm alexidine, or 1.1 μm alexidine for 72 h and then fixed, stained with filipin and anti-LAMP1 and imaged. Quantification of filipin (A) and LAMP1 (B) and representative images of treated cells (C). Scale bar = 50 μM. Bars: mean ± SD, n = 4 wells/condition, ****P < 0.0001, ***P < 0.001, **P < 0.01 versus genotype-matched DMSO, alexidine two-way ANOVA followed by Dunnett’s test.
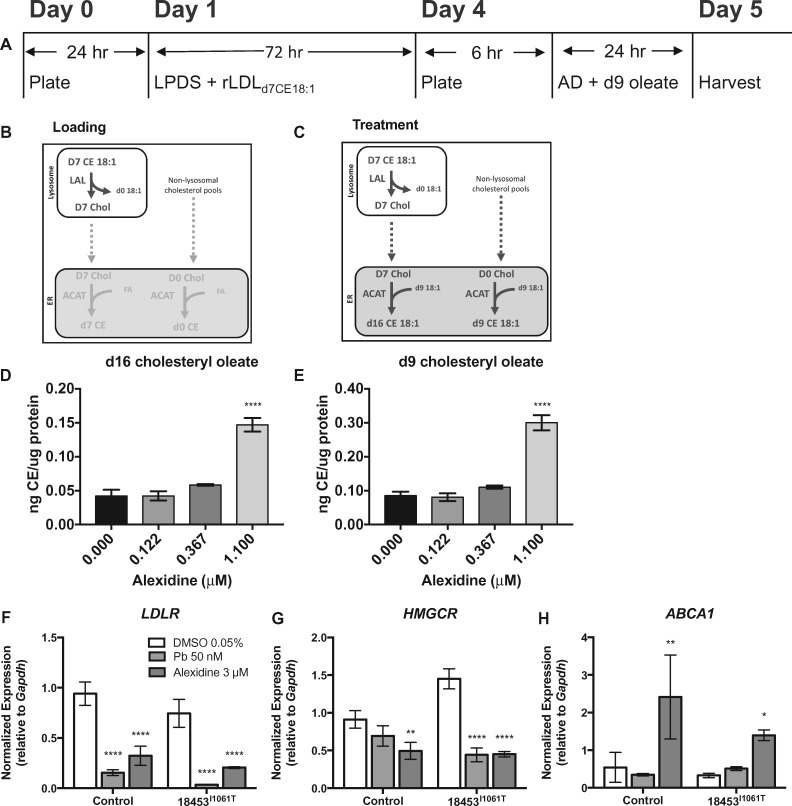
Finally, in an effort to understand the functional consequences of the observed alexidine-mediated NPC1 gene and protein expression changes, we performed LDL-stimulated cholesterol esterification assessments in alexidine-treated control and patient cells. To directly analyze the fate of lysosomal cholesterol, LDL was reconstituted with deuterated cholesteryl ester. We found an increase in esterification of LDL-derived cholesterol in NPC1I1061T/I1061T patient cells treated with alexidine (Fig. 5A–E). Increased esterification of non-deuterated cholesterol was also observed, suggesting trafficking of non-LDL-derived cholesterol to the endoplasmic reticulum was also enhanced. These data provide direct evidence for improved lysosomal cholesterol export following alexidine treatment. In support of this interpretation, we also found corresponding changes in gene expression related to improved cholesterol trafficking (Fig. 5F–H). We assayed by qRT-PCR expression of several genes known to be modulated by cholesterol movement from the lysosome. Alexidine reduced expression of sterol regulatory element-binding protein (SREBP2)-dependent targets like the cholesterol uptake transporter, LDLR and the cholesterol biosynthetic gene, HMGCR (28), similar to HDACi treatment in both control and patient cells (Fig. 5F and G). In addition, expression of the LXR-dependent cholesterol export gene, ABCA1 (29), was induced in alexidine-treated control and patient cells (Fig. 5H).
Figure 5.
Alexidine treatment stimulates esterification of lysosomal cholesterol. NPC1I1061T/I1061T fibroblasts were plated and treated 24 h later with lipoprotein-deficient media containing 50 g/mL LDL reconstituted with d7-cholesteryl oleate. After 72 h growth, cells were re-plated, then treated with alexidine or vehicle in media containing d9-oleate for 24 h. Cells were harvested and prepared for LC-MS. (A) Experimental work flow. (B, C) Cholesteryl esterification paradigms and cholesteryl-ester deuteration status in cells during the loading (B) and treatment (C) phase of the endoplasmic reticulum cholesterol trafficking assay. (D) Relative levels of d16-cholesteryl oleate and (E) d9-cholesteryl oleate after 24 h in DMSO, or alexidine-containing media. Values are normalized to total protein. Bars: mean ± SD, n = 3 wells/condition, ****P < 0.0001 versus DMSO, one-way ANOVA followed by Dunnett’s test. (F–H) Alexidine treatment modifies expression of cholesterol flux genes. Gene expression by TaqMan qRT-PCR in indicated cell lines following 48 h alexidine (3 μm) treatment. n = 3 biological replicates, mean ± SD, ****P < 0.0001, **P < 0.01, *P < 0.05 versus genotype-DMSO (two-way ANOVA followed by Dunnett’s test).
Discussion
We developed a phenotypic NPC patient cell-based assay for inhibitors of lysosomal cholesterol, employing tandem use of filipin and LAMP1 assays which aided in hit selection (Table 2) without substantially increasing assay time or labor input for this focused screen. We tested this assay in multiple control and patient lines and demonstrated in a high-throughput screen its utility for reproducibly identifying compounds that modulate levels of lysosomal cholesterol, including a novel lysosomal cholesterol-reducing molecule, alexidine.
Alexidine is an antibacterial bisbiguanide found in antimicrobial mouthwashes and contact lens solutions. Alexidine binds to bacterial lipopolysaccharides (LPS) and phospholipids and disrupts membranes by producing lipid phase separation and domain formation (30,31). In addition, alexidine has been demonstrated to affect metabolism in hematopoietic stem cells through activation of AMPK (32). Our data indicate that alexidine affects multiple cellular and molecular processes in control and NPC patient fibroblasts to reduce lysosomal cholesterol. Alexidine induces transcription of NPC1 (Fig. 3E) and in turn increases total NPC1 protein (Fig. 3A and B) as well as mature, EndoH-resistant NPC1 protein glycoforms (Fig. 3C and D). Increased mature NPC1 likely helps facilitate lysosomal cholesterol export (Fig. 5A–E), especially in NPC1I1061T patient cells where misfolded NPC1I1061T is rapidly degraded in the endoplasmic reticulum prior to processing in the Golgi and trafficking to the lysosome (9).
Not only does alexidine increase NPC1 expression, but it also affects expression of genes critical for cholesterol flux. Alexidine inhibits expression of SREBP2 targets that regulate cholesterol uptake and biosynthesis (28) while inducing expression of a key LXR target that regulates cholesterol export (29) (Fig. 5F–H). Alexidine may directly affect expression of cholesterol flux genes. More likely, these gene expression changes may result from the alexidine-mediated improvement in cholesterol trafficking in NPC patient cells, promoting increased cholesterol trafficking to mitochondria and conversion of cholesterol to oxysterol LXR ligands. The latter mechanism could explain the NPC1- and NPC2-independent reduction in lysosomal cholesterol observed in NPC1−/− and NPC2−/− cells. It is also possible that the NPC1/2-independent effect of alexidine on cholesterol storage may be mediated through activation of AMPK (32,33). Although alexidine demonstrated efficacy in NPC1−/− and NPC2−/− cells, the effect on lysosomal cholesterol reduction was blunted compared with control or NPC1I1061T cells (Fig. 4), suggesting that both NPC1 and NPC2 are required for full alexidine efficacy.
Forced overexpression of NPC1 was reported to contribute to phenotypic improvement in NPC1I1061T patient cells. It has been noted, for example, that transfection with an NPC1I1061T DNA construct is sufficient to rescue NPC1-mutant cells by forcing an adequate amount of mutant NPC1 to bypass the endoplasmic reticulum proteostasis control machinery (9). Since we observed a similar effect on lysosomal cholesterol accumulation in control cells and on NPC1 expression in patient cells, we hypothesize that the activity of alexidine was unlikely specific to the stabilization of misfolded-mutant NPC1. EndoH studies support this hypothesis as both EndoH-sensitive and EndoH-resistant pools of NPC1I1061T were increased in patient cells following alexidine treatment (Fig. 3C and D). This is further supported by an earlier study demonstrating that ∼50% of normal NPC1 protein is degraded within the ER (9).
The utility of a compound like alexidine for inducing NPC1 expression and maturation has been repeatedly explored using other approaches. HDACi have been shown to affect the proteostatic machinery in other lysosomal storage disease settings (34) and have demonstrated efficacy in decreasing lysosomal cholesterol storage for multiple different NPC1 patient mutations (17). This supports the hypothesis that HDACi modulate endoplasmic reticulum-associated degradation factors to allow more mutant NPC1 to reach the lysosome and improve cholesterol trafficking. Along similar lines arimoclomol is thought to act as a co-inducer, along with cellular stress, to upregulate the chaperone, HSP70 (11). Additional work by Ohgane et al. confirms the usefulness of a chaperone-based approach for improving the cholesterol storage defect of NPC patient cells (35). In this case, oxysterol derivatives were capable of binding and stabilizing mutant NPC1, thereby increasing expression and localization to the lysosome (35). Alternatively, the NPC1-binding protein TMEM97 was shown to be a negative regulator of NPC1 expression, making inhibition of TMEM97 an attractive, yet underexplored NPC therapeutic angle (36).
An alternate screening strategy, though technically challenging, might require assessment of the ratio of glycosylated, mature NPC1 relative to total cellular NPC1. NPC1 expression-inducing compounds like alexidine should increase total NPC1 expression (Fig. 3A) and moderately increase glycosylated NPC1, but a true NPC1 chaperone would likely robustly increase both, with the glycosylated fraction becoming the predominant species, as in control cells (Fig. 3C and D). This outcome could be screened against using NPC1 antibodies specific to immature and mature NPC1. To our knowledge, such reagents are not currently available. Another possible screening strategy raised by our findings is a transcription-based readout of endogenous NPC1. Although this approach would not enrich for mutant NPC1 chaperones, transcriptional activators for other protein misfolding diseases like Cystic Fibrosis have proved successful in preclinical studies (37).
In summary, we developed a sensitive high-throughput phenotypic screening assay for NPC patient fibroblasts sensitive to multiple genotypes and degrees of phenotypic severity. Based on a dual readout of lysosomal cholesterol, we screened ∼3500 compounds, including 2013 FDA-approved drugs and an additional ∼1500 bioactive compounds against NPC1I1061T patient cells. A number of HDACi, a class of compounds identified previously as modulators of cholesterol levels in NPC1 patient cells, were identified as hits. Of the non-HDACi hits, the strongest hit, alexidine, was confirmed as an inducer of both total and mature NPC1 protein expression that also modulates expression of genes critical for cholesterol flux, but is unlikely to act as a chaperone for misfolded NPC1I1061T. Although unsuitable for systemic therapy, alexidine has utility as a tool compound for further NPC studies and as a potent inducer of NPC1. Thus, our screen successfully identified alexidine as a novel enhancer of lysosomal cholesterol release through upregulation of NPC1- and NPC2-dependent and -independent pathways. These data demonstrate that this phenotypic assay has the capability to identify hits representing diverse molecular mechanisms that modulate cholesterol accumulation in NPC1-mutant cells, motivating extension to larger screens of more diverse compound collections.
Materials and Methods
Cell culture
NPC patient fibroblasts were obtained from the Coriell Institute for Medical Research (Camden, NJ) (Table 1). Control, normal human fibroblasts were obtained from Lonza (Allendale, NJ). NPC1-null fibroblasts (NPC11628delC) (38) and NPC2-null fibroblasts (NPC2IVS2 + 5G→A, National Institutes of Health, 94.85) (39) were obtained from archives in the Ory laboratory (Washington University, St. Louis, MO) (Table 1). Fibroblasts were cultured in DMEM high glucose with 10% fetal bovine serum (Hyclone GE Life Sciences, SH30396, Logan, UT), Penicillin-Streptomycin (Corning 30-002-CI, Manassas, VA), and 1× GlutaMAX (Gibco/Thermo Fisher, 35050-061, Waltham, MA). All experiments were performed on cells within five passages.
Compounds
Panobinostat was purchased from LC Labs (P-3703, Woburn, MA), and alexidine dihydrochloride from Cayman Chemical (13876, Ann Arbor, MI). The compound collections screened include Sanford Burnham Prebys’ internal FDA-approved repurposing collection (La Jolla, CA) which encompasses the Prestwick Chemical library (Prestwick Chemical, San Diego, CA), the Library of Pharmacologically Active Compounds (LOPAC) collection (Sigma-Aldrich) and 813 additional FDA-approved or ‘in clinical trial’ compounds assembled in-house. Additional libraries screened include the EMD-Millipore kinase inhibitor library composed of InhibitorSelect Libraries I, II, and III (San Diego, CA), an internally assembled epigenetic collection compiled from multiple libraries (Enzo Life Sciences, SelleckChem, Sigma-Aldrich and Cayman Chemical), a human metabolite library (25), metabolism collection (Focus Biomolecules, Plymouth Meeting, PA) and custom-assembled chaperone library. A complete list of compounds screened is provided in Supplementary Material, Table S1.
Filipin, LAMP1, Draq5 and lysotracker staining
Fibroblasts were plated (2500/well) for 72–96 h on a CellCarrier 384-well plate (Perkin Elmer, 6007550, Waltham, MA). Fibroblasts were fixed in 4% formaldehyde for 15 min at room temperature. Cells were then washed with PBS and permeabilized with 0.2% saponin in PBS and incubated overnight at 4°C with 1: 800 anti-LAMP1 (Abcam, 24170, San Francisco, CA) diluted in 5% normal donkey serum. Following PBS wash, cells were incubated with secondary antibody and 50 μg/ml filipin (Sigma, F9765, St. Louis, MO) diluted in 5% normal donkey serum for 1.5 h at room temperature. Fibroblasts were then washed with PBS and stained with 1: 1000 Draq5 (Thermo Fisher, 62251, Waltham, MA).
Lysotracker (Life Technologies, L-7528, Carlsbad, CA) staining was performed on live cells. Cells were incubated in 50 nm lysotracker in growth media for 30 min at 37°C and washed and imaged in phenol red-free live-cell imaging media (Thermo Fisher, A14291DJ) with 2 μg/ml Hoechst (Thermo Fisher, H3570).
Image acquisition and analysis
Following filipin/LAMP1 staining, cells were immediately imaged with a Perkin-Elmer Opera Phenix or QEHS (Waltham, MA) high-content automatic imaging system with a 20× air objective in confocal mode. Due to field size differences, five fields per well were imaged for each experiment analyzed with the QEHS and two fields per well were imaged for each experiment analyzed on the Opera Phenix such that >200 cells were analyzed from each well. Images were analyzed on a per cell basis and then values were averaged across all cells and all fields acquired for each well. Images were analyzed and quantified using Perkin-Elmer Harmony® image analysis software (Supplementary Material, Fig. S1). Using Perkin Elmer’s proprietary image analysis tools, cells were segmented first by identifying nuclei (‘Find Nuclei’ building block) (Supplementary Material, Fig. S1C), and then surrounding cytoplasmic area (‘Find Cytoplasm’ building block) (Supplementary Material, Fig. S1D). To determine cell health, cell number was determined along with three nuclear parameters: nuclear roundness, area, and staining intensity (‘Calculate Morpholopgy Properties’) (Supplementary Material, Fig. S1E). Mean cytoplasmic LAMP1 and filipin intensities (‘Calculate Intensity Properties’ building block) were then measured in the segmented cytoplasm (Supplementary Material, Fig. S1F and G) such that the intensity of each was normalized to total cell area on a per cell basis. No background correction was performed, however the cell segmentation algorithm was adjusted and validated for individual plates to account for staining differences between experiments. Cell number was counted for each well and compared with the average for control wells by Z-score to determine cytotoxicity.
Gene expression
Fibroblasts were plated on 60 mm dishes (250 000 cells/dish) and treated with compounds for 48 h. RNA was isolated from fibroblasts using the Qiagen RNeasy Kit (74104, Hilden, Germany). A total 8.5 ng of RNA per target probe were added to the Taqman RNA to CT 1 step kit (Applied Biosystems, 4392938, Foster City, CA) with the appropriate Taqman probe (Applied Biosystems, 4331182) (GAPDH: _g1, NPC1: Hs00264835_m1, ABCA1: Hs01059137_m1, LDLR: Hs01092524_m1, HMGCR: Hs00168352_m1). Gene expression analysis was performed using a ViiA™ 7 Real-Time PCR System (Thermo Fisher, Waltham, MA) using the 2-Δ(ΔCT) method.
Western blot
Fibroblasts (250 000/plate) were plated and treated with compounds for 72 h. Cells were lysed in 1× RIPA buffer (Cell Signaling Technologies, 9806S, Danvers, MA) with 1× protease inhibitors (Roche, 11836170001, Basel, Switzerland) and protein was quantified using Bio-Rad DC Protein Assay (5000112, Hercules, CA). A total of 20 μg of lysate were resolved on a 4–12% Bis/Tris NuPage protein gel (Thermo Fisher, NP0322PK2) and transfered to a nitrocellulose membrane (Thermo Fisher, LC2006). Membranes were probed for β-Actin 1: 10 000 (Sigma, A5441), NPC1 1: 1000 (in-house antibody, Ory Laboratory, St. Louis, MO) (40), and acetylated Tubulin 1: 5000 (Sigma, T7451) diluted in 5% non-fat milk.
EndoH assay
On day 0, NPC1I1061T/I1061T or normal human skin fibroblasts were plated in 2 ml culturing media at 1 × 105 or 0.5 × 105 cells/well in a 6-well dish, respectively. On day 1, 1 ml of media was removed and replaced with 1 ml complete media containing Alexidine or DMSO vehicle. On day 4, cells were harvested by adding 50 l RIPA buffer to each well. After 30 min, cell lysate was collected and centrifuged at 18 400g for 20 min at 4. The protein concentration of the clarified lysate was determined by BCA assay. A total of 10–20 g total protein/condition were combined with 1× glycoprotein denaturing buffer (New England Biolabs) and 1× GlycoBuffer (New England Biolabs) in water then incubated at 50 for 30 min. To each tube, 10 000 U/ml Endo Hf (New England Biolabs) or an equal volume of water was added. Samples were incubated overnight at 37. Proteins were separated on a 4–12% Bis/Tris gradient gel, transferred to nitrocellulose, then probed for -tubulin (Sigma, T6199) or NPC1 (40) diluted in 5% non-fat milk.
Esterification assay
On day 0, NPC1I1061T/I1061T fibroblasts were plated at 5.8 × 105 cells/10 cm dish. On day 1, media was changed to lipoprotein-depleted media with 50 μg/ml LDL reconstituted with deuterated-cholesteryl oleate (d7 CE 18: 1). On day 4, cells were replated at 1.2 × 105 cells/well in a 6-well dish in 2 ml complete media. Once adhered, 1 ml of media was removed and replaced with 1 ml Alexidine treatment media spiked with 10 μL/mL deuterium-labeled oleate (d9 18: 1, Avanti, 861809) complexed to BSA (prepared at 12% in 150 mm NaCl). After 24 h, cells were washed, collected by scraping, pelleted and stored at −20. For analysis, cell pellets were needle homogenized in PBS. Homogenate was combined with 2: 2: 1 CHCl3/H2O/5M NaCl and 50 ng d7 cholesteryl-linoleate as an internal standard. Samples were vortexed, then centrifuged 5 min 100g 4. The CHCl3 layer was collected, dried, then reconstituted in 9: 1 MeOH/CHCl3. Ion transitions for d7-cholesteryl linoleate (666.619/369.400), d9-cholesteryl oleate (677.619/369.400) and d16-cholesteryl oleate (684.619/369.400) were monitored in positive mode by liquid chromatography-mass spectrometry (LC-MS). Protein concentration was determined by BCA.
Data and statistical analysis
Data are presented as mean ± standard deviation (SD) unless otherwise stated. Data were evaluated for statistical significance using methods described in figure legends.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank members of the Conrad Prebys Center for Chemical Genomics at Sanford Burnham Prebys for their technical support and advice, including Haowen Zhou, Sylvia Kim, Drs Sean Sherman, Ian Pass, FuYue Zeng, Debbie Chen, Sumeet Salaniwal, and Susanne Heynen-Genel.
Conflict of Interest statement. The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was supported by the Hide and Seek Foundation and SOAR-NPC (SOAR-NPC-2015-01) to A.G.B and R.K. and by the National Institutes of Health (T32 HL134635) to M.F. and (NIDDK113171) to R.J.K.
References
- 1. Wassif C.A., Cross J.L., Iben J., Sanchez-Pulido L., Cougnoux A., Platt F.M., Ory D.S., Ponting C.P., Bailey-Wilson J.E., Biesecker L.G.. et al. (2016) High incidence of unrecognized visceral/neurological late-onset Niemann-Pick disease, type C1, predicted by analysis of massively parallel sequencing data sets. Genet. Med., 18, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vanier M.T. (2010) Niemann-Pick disease type C. Orphanet. J. Rare Dis., 5, 16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ory D.S. (2000) Niemann-Pick type C: a disorder of cellular cholesterol trafficking. Biochim. Biophys. Acta, 1529, 331–339. [DOI] [PubMed] [Google Scholar]
- 4. Gong X., Qian H., Zhou X., Wu J., Wan T., Cao P., Huang W., Zhao X., Wang X., Wang P.. et al. (2016) Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and ebola infection. Cell, 165, 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park W.D., O'Brien J.F., Lundquist P.A., Kraft D.L., Vockley C.W., Karnes P.S., Patterson M.C., Snow K. (2003) Identification of 58 novel mutations in Niemann-Pick disease type C: correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum. Mutat., 22, 313–325. [DOI] [PubMed] [Google Scholar]
- 6. Benussi A., Alberici A., Premi E., Bertasi V., Cotelli M.S., Turla M., Dardis A., Zampieri S., Marchina E., Paghera B.. et al. (2015) Phenotypic heterogeneity of Niemann-Pick disease type C in monozygotic twins. J. Neurol., 262, 642–647. [DOI] [PubMed] [Google Scholar]
- 7. Bauer P., Knoblich R., Bauer C., Finckh U., Hufen A., Kropp J., Braun S., Kustermann-Kuhn B., Schmidt D., Harzer K.. et al. (2002) NPC1: complete genomic sequence, mutation analysis, and characterization of haplotypes. Hum. Mutat., 19, 30–38. [DOI] [PubMed] [Google Scholar]
- 8. Millat G., Marcais C., Rafi M.A., Yamamoto T., Morris J.A., Pentchev P.G., Ohno K., Wenger D.A., Vanier M.T. (1999) Niemann-Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am. J. Hum. Genet., 65, 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gelsthorpe M.E., Baumann N., Millard E., Gale S.E., Langmade S.J., Schaffer J.E., Ory D.S. (2008) Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J. Biol. Chem., 283, 8229–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pipalia N.H., Cosner C.C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., Maxfield F.R. (2011) Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc. Natl. Acad. Sci. U. S. A., 108, 5620–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirkegaard T., Gray J., Priestman D.A., Wallom K.L., Atkins J., Olsen O.D., Klein A., Drndarski S., Petersen N.H., Ingemann L.. et al. (2016) Heat shock protein-based therapy as a potential candidate for treating the sphingolipidoses. Sci. Transl. Med., 8, 355ra118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ory D.S., Ottinger E.A., Farhat N.Y., King K.A., Jiang X., Weissfeld L., Berry-Kravis E., Davidson C.D., Bianconi S., Keener L.A.. et al. (2017) Intrathecal 2-hydroxypropyl-beta-cyclodextrin decreases neurological disease progression in Niemann-Pick disease, type C1: a non-randomised, open-label, phase 1-2 trial. Lancet, 390, 1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyseng-Williamson K.A. (2014) Miglustat: a review of its use in Niemann-Pick disease type C. Drugs, 74, 61–74. [DOI] [PubMed] [Google Scholar]
- 14. Crumling M.A., Liu L., Thomas P.V., Benson J., Kanicki A., Kabara L., Halsey K., Dolan D., Duncan R.K. (2012) Hearing loss and hair cell death in mice given the cholesterol-chelating agent hydroxypropyl-beta-cyclodextrin. PloS One, 7, e53280.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siegel D., Hussein M., Belani C., Robert F., Galanis E., Richon V.M., Garcia-Vargas J., Sanz-Rodriguez C., Rizvi S. (2009) Vorinostat in solid and hematologic malignancies. J. Hematol. Oncol., 2, 31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vite C.H., Bagel J.H., Swain G.P., Prociuk M., Sikora T.U., Stein V.M., O’Donnell P., Ruane T., Ward S., Crooks A.. et al. (2015) Intracisternal cyclodextrin prevents cerebellar dysfunction and Purkinje cell death in feline Niemann-Pick type C1 disease. Sci. Transl. Med., 7, 276ra26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pipalia N.H., Subramanian K., Mao S., Ralph H., Hutt D.M., Scott S.M., Balch W.E., Maxfield F.R. (2017) Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J. Lipid Res., 58, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pipalia N.H., Huang A., Ralph H., Rujoi M., Maxfield F.R. (2006) Automated microscopy screening for compounds that partially revert cholesterol accumulation in Niemann-Pick C cells. J. Lipid Res., 47, 284–301. [DOI] [PubMed] [Google Scholar]
- 19. Bornig H., Geyer G. (1974) Staining of cholesterol with the fluorescent antibiotic “filipin”. Acta Histochem., 50, 110–115. [PubMed] [Google Scholar]
- 20. Xu M., Liu K., Swaroop M., Sun W., Dehdashti S.J., McKew J.C., Zheng W. (2014) A phenotypic compound screening assay for lysosomal storage diseases. J. Biomol. Screen., 19, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liscum L., Arnio E., Anthony M., Howley A., Sturley S.L., Agler M. (2002) Identification of a pharmaceutical compound that partially corrects the Niemann-Pick C phenotype in cultured cells. J. Lipid Res., 43, 1708–1717. [DOI] [PubMed] [Google Scholar]
- 22. Sambucetti L.C., Fischer D.D., Zabludoff S., Kwon P.O., Chamberlin H., Trogani N., Xu H., Cohen D. (1999) Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol. Chem., 274, 34940–34947. [DOI] [PubMed] [Google Scholar]
- 23. Zhang J.H., Chung T.D., Oldenburg K.R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen., 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 24. Zhang X.D. (2008) Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J. Biomol. Screen., 13, 363–377. [DOI] [PubMed] [Google Scholar]
- 25. Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S.. et al. (2007) HMDB: the human metabolome database. Nucleic Acids Res., 35, D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang X.D., Ferrer M., Espeseth A.S., Marine S.D., Stec E.M., Crackower M.A., Holder D.J., Heyse J.F., Strulovici B. (2007) The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen., 12, 497–509. [DOI] [PubMed] [Google Scholar]
- 27. Xu M., Liu K., Swaroop M., Porter F.D., Sidhu R., Finkes S., Ory D.S., Marugan J.J., Xiao J., Southall N.. et al. (2012) delta-Tocopherol reduces lipid accumulation in Niemann-Pick type C1 and Wolman cholesterol storage disorders. J. Biol. Chem., 287, 39349–39360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horton J.D., Goldstein J.L., Brown M.S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest., 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Repa J.J., Turley S.D., Lobaccaro J.A., Medina J., Li L., Lustig K., Shan B., Heyman R.A., Dietschy J.M., Mangelsdorf D.J. (2000) Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science, 289, 1524–1529. [DOI] [PubMed] [Google Scholar]
- 30. Lobene R.R., Soparkar P.M. (1973) The effect of an alexidine mouthwash on human plaque and gingivitis. J. Am. Dent. Assoc., 87, 848–851. [DOI] [PubMed] [Google Scholar]
- 31. Chawner J.A., Gilbert P. (1989) Interaction of the bisbiguanides chlorhexidine and alexidine with phospholipid vesicles: evidence for separate modes of action. J. Appl. Bacteriol., 66, 253–258. [DOI] [PubMed] [Google Scholar]
- 32. Liu X., Zheng H., Yu W.M., Cooper T.M., Bunting K.D., Qu C.K. (2015) Maintenance of mouse hematopoietic stem cells ex vivo by reprogramming cellular metabolism. Blood, 125, 1562–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dai S., Dulcey A.E., Hu X., Wassif C.A., Porter F.D., Austin C.P., Ory D.S., Marugan J., Zheng W. (2017) Methyl-beta-cyclodextrin restores impaired autophagy flux in Niemann-Pick C1-deficient cells through activation of AMPK. Autophagy, 13, 1435–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang C., Rahimpour S., Lu J., Pacak K., Ikejiri B., Brady R.O., Zhuang Z. (2013) Histone deacetylase inhibitors increase glucocerebrosidase activity in Gaucher disease by modulation of molecular chaperones. Proc. Natl. Acad. Sci. U. S. A., 110, 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohgane K., Karaki F., Dodo K., Hashimoto Y. (2013) Discovery of oxysterol-derived pharmacological chaperones for NPC1: implication for the existence of second sterol-binding site. Chem. Biol., 20, 391–402. [DOI] [PubMed] [Google Scholar]
- 36. Ebrahimi-Fakhari D., Wahlster L., Bartz F., Werenbeck-Ueding J., Praggastis M., Zhang J., Joggerst-Thomalla B., Theiss S., Grimm D., Ory D.S.. et al. (2016) Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-Pick type C1 disease cells. Hum. Mol. Genet., 25, 3588–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hutt D.M., Herman D., Rodrigues A.P., Noel S., Pilewski J.M., Matteson J., Hoch B., Kellner W., Kelly J.W., Schmidt A.. et al. (2010) Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat. Chem. Biol., 6, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun X., Marks D.L., Park W.D., Wheatley C.L., Puri V., O’Brien J.F., Kraft D.L., Lundquist P.A., Patterson M.C., Pagano R.E.. et al. (2001) Niemann-Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am. J. Hum. Genet., 68, 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Millat G., Chikh K., Naureckiene S., Sleat D.E., Fensom A.H., Higaki K., Elleder M., Lobel P., Vanier M.T. (2001) Niemann-Pick disease type C: spectrum of HE1 mutations and genotype/phenotype correlations in the NPC2 group. Am. J. Hum. Genet., 69, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Millard E.E., Srivastava K., Traub L.M., Schaffer J.E., Ory D.S. (2000) Niemann-Pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J. Biol. Chem., 275, 38445–38451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.