Abstract
Seed oligosaccharides (OSs) and especially raffinose series OSs (RSOs) are hypothesized to play an important role in the acquisition of desiccation tolerance and consequently in seed storability. In the present work we analyzed the seed-soluble OS (sucrose, raffinose, and stachyose) content of several Arabidopsis accessions and thus identified the genotype Cape Verde Islands having a very low RSO content. By performing quantitative trait loci (QTL) mapping in a recombinant inbred line population, we found one major QTL responsible for the practically monogenic segregation of seed stachyose content. This locus also affected the content of the two other OSs, sucrose, and raffinose. Two candidate genes encoding respectively for galactinol synthase and raffinose synthase were located within the genomic region around this major QTL. In addition, three smaller-effect QTL were identified, each one specifically affecting the content of an individual OS. Seed storability was analyzed in the same recombinant inbred line population by measuring viability (germination) under two different seed aging assays: after natural aging during 4 years of dry storage at room temperature and after artificial aging induced by a controlled deterioration test. Thus, four QTL responsible for the variation of this trait were mapped. Comparison of the QTL genetic positions showed that the genomic region containing the major OS locus did not significantly affect the seed storability. We concluded that in the studied material neither RSOs nor sucrose content had a specific effect on seed storability.
In many plants species, including Arabidopsis (Ooms et al., 1993), seed maturation is accompanied by the accumulation of soluble oligosaccharides (OSs) (Horbowicz and Obendorf, 1994). These OSs, mainly Suc and raffinose series oligosaccharides (RSOs) are found in cotyledons, seed coats, and hypocotyls (Obendorf, 1997 and references therein). RSOs are derivatives of Suc to which Gal units are added to the Glc moiety of Suc through α-(1,6) bonds. Raffinose contains one Gal unit, whereas stachyose has two such units. RSOs appear during the later stages of seed development and they disappear upon germination (Obendorf, 1997). In Arabidopsis, seeds of the accession Landsberg erecta (Ler) accumulate raffinose and especially stachyose at the later stages of seed development, whereas the Suc content remains constant (Ooms et al., 1993). These three OSs together represent approximately 2% of the dry weight of mature seeds.
Studies in several species such as soybean (Glycine max), maize (Zea mays), and brassica (Brassica campestris [rapa]) have suggested that OSs might be involved in the protection of seeds against damage during seed dehydration and aging, and therefore in seed survival and storability (for review, see Obendorf, 1997; Sinniah et al., 1998). OSs have been speculated to be involved in the protection of membranes, proteins, and nucleic acids against damage that occurs during and upon the withdrawal of water in the drying seeds (Hoekstra et al., 1991). This protective role of OSs has been explained mainly by their capacity to retain the integrity of membranes through their interaction with the phospholipid headgroups, thus replacing water during dehydration. In addition, removal of water molecules from phospholipids can lead to membrane phase transitions at physiological temperatures (Hoekstra et al., 1989). When water is available, these transitions coincide with membrane leakage and cell death. Hoekstra et al. (1991) have shown that OS can depress the temperature of membrane phase transitions and prevent leakage of cellular solutes. Furthermore, free radicals may accumulate and cause damage to cellular components and structures during seed aging because scavenging systems are not operating at low moisture contents (McDonald, 1999). It has been suggested that OSs may form a viscous glassy state (condition in which a liquid achieves such high viscosity that it resembles a solid) (Leopold et al., 1994), which prevents molecular interactions, resulting in damage to membranes and macromolecules (Crowe et al., 1987). This glassy state in seeds seems to serve as a physical stabilizer protecting against deteriorative reactions. In particular, RSOs have been shown to have an excellent ability to form stable glasses, and therefore they have been considered essential components for the storability of seeds (Koster and Leopold, 1988).
Seed storability, defined as the longevity of seeds after storage, represents a trait important for the conservation of seed resources. However, to test this character seeds need to be stored for long times, and for that reason so-called controlled deterioration tests (CDT) have been developed (Powell and Matthews, 1984; Hampton and TeKrony, 1995) as an alternative to analyze this property more efficiently. In such tests, seed moisture content and temperature are increased to artificially accelerate seed aging. The viability of the seeds after CDT has been shown to be a reliable measurement for determining seed storability in a number of species, including Arabidopsis, where it has been tested using mutant seeds with storability defects (K. Tesnier, personal communication).
Despite substantial studies of both traits, seed OS composition and seed storability, their genetic analysis has been hampered due to their quantitative nature. It is only in the last decade with the advent of molecular marker technologies and the development of QTL-mapping procedures that genetic analyses and the identification of genomic regions controlling quantitative traits have become feasible (Tanksley, 1993; Jansen, 1996). The use of homozygous permanent mapping populations such as recombinant inbred lines (RILs) is very efficient in this respect, because it is possible to study an indefinite number of traits on the same experimental population, enabling the detection of loci with putative pleiotropic effects (i.e. one locus affecting different traits) (Prioul et al., 1997). The localization of QTL in plant species such as Arabidopsis, where molecular analyses can be performed efficiently, may eventually lead to the molecular identification of the respective genes (for review, see Alonso-Blanco and Koornneef, 2000).
In the present work we have analyzed the seed-soluble OS content of several Arabidopsis accessions and identified the genotype Cape Verde Islands (Cvi) as having a very low RSO content. A QTL-mapping approach has been used on an RIL population derived from a cross between Cvi and the laboratory accession Ler (Alonso-Blanco et al., 1998b) differing in the OS composition to identify and locate the loci responsible of OS variation. In addition, we have analyzed the same RIL population to locate QTL for seed storability to investigate if there are loci with pleiotropic effects on these two traits in Arabidopsis as it has been suggested for other species.
RESULTS
Variation for Seed-Soluble OS Content in Accessions of Arabidopsis, RILs, and Reciprocal Crosses of Ler and Cvi
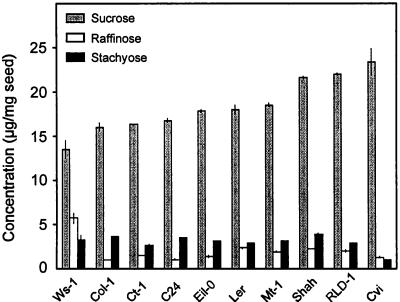
To determine genetic variation in seed-soluble OS content among Arabidopsis accessions, we have analyzed the seed-soluble OS composition of 10 accessions (Fig. 1). Suc appeared as the most abundant sugar in all of them, and raffinose and stachyose were the only detectable RSOs. Monosaccharides (Fru and Glc) were not detectable. The accession Wassilewksija (Ws-1) showed the highest RSO content and, opposite to all other analyzed accessions, had more raffinose than stachyose. In contrast, Cvi contained very low levels of RSOs. Nevertheless, the total OS content in Ws-1 and Cvi was not significantly different from that of other genotypes with the exception of Shahdara (Shah) and Rschew-1 (RLD-1) in which the total amount of Suc plus RSOs was significantly higher compared with the other accessions.
Figure 1.
Seed-soluble OS content of 10 Arabidopsis accessions, Wassilewksija-1 (Ws-1), Columbia-1 (Col-1), Catania-1 (Ct-1), C24, Eilenburg-0 (Eil-0), Ler, Martuba-1 (Mt-1), Shahdara (Shah), Rschew-1 (RLD-1), and Cape Verde Islands (Cvi). Columns correspond to means of two measurements of bulked seeds from six plants; vertical bars indicate the range of variation.
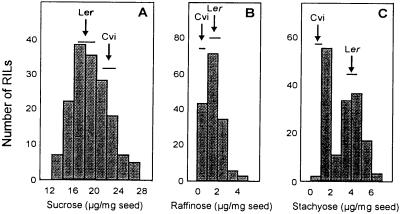
To identify and locate QTL responsible for the genetic variation observed for seed OS contents we have analyzed an RIL mapping population derived from the cross between Cvi, the lowest RSO content accession, and the laboratory accession Ler. Although, the analysis of other crosses such as Ws-1 × Cvi might show larger variation, the availability of a permanent mapping population between Cvi and Ler offers unique advantages (Alonso-Blanco and Koornneef, 2000). As shown in Figure 2, the RIL population showed transgression in both directions for Suc content. However, for raffinose and stachyose, transgression was only detected toward higher contents due to the low amount of RSOs in the Cvi parent. The levels of the three OSs were correlated among the RILs; raffinose content was positively correlated with stachyose content (r = 0.77), whereas Suc content correlated negatively with the raffinose and stachyose contents (r = −0.34 and r = −0.39, respectively).
Figure 2.
Frequency distributions of Suc, raffinose, and stachyose content in seeds of the Ler/Cvi RIL population. Arrows correspond to the parental line means and horizontal bars represent their ranges of variation.
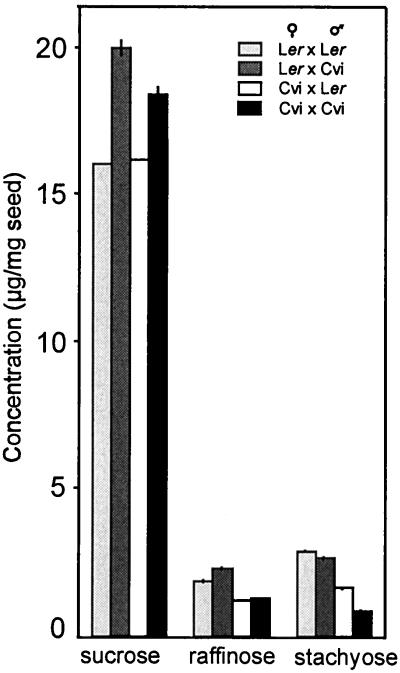
To investigate if the OS content was maternally affected we have analyzed hybrid seeds obtained from reciprocal crosses between the parental lines Ler and Cvi. As shown in Figure 3, the content of the three soluble OS was significantly different in the reciprocal hybrid seeds. The two sorts of hybrid seeds differed in their raffinose content, the contents being similar to those observed in seeds of the female parent, indicating maternal effects. The stachyose contents of the reciprocal hybrid seeds were different but intermediate between both parental values, suggesting both maternal and zygotic effects and partial dominance of the higher stachyose level. In contrast, the Suc content of the hybrid seeds was comparable with that in the seeds of the male parent, suggesting paternal effects.
Figure 3.
Soluble OS content in hybrid seeds obtained from reciprocal crosses between Ler and Cvi (means of two measurements; vertical bars indicate the range of variation).
Mapping QTL Controlling Seed-Soluble OS Content
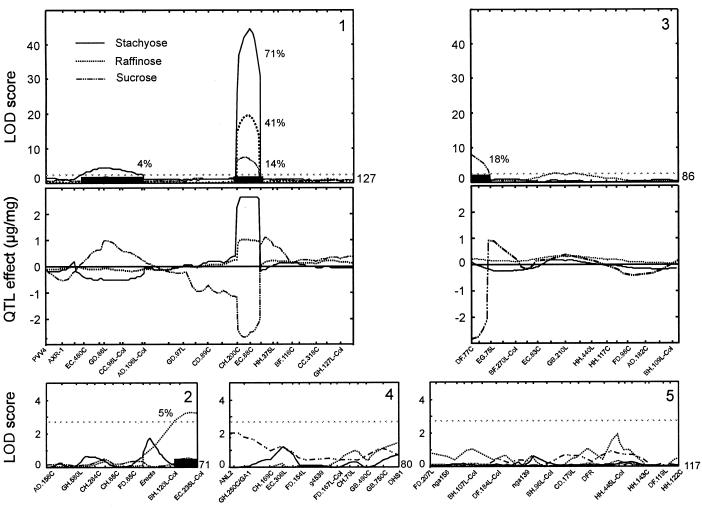
QTL mapping was performed for the quantity of the three major OSs (Suc, raffinose, and stachyose) (Fig. 4). In total, four genomic regions were detected, one affecting the content of the three OSs and an additional QTL specific for each OS. The additive effects of these QTL accounted for 72%, 45%, and 32% of the total variance for stachyose, raffinose, and Suc, respectively. The region on the lower arm of chromosome 1 near the amplified fragment length polymorphism marker EC.88C explained 71%, 41%, and 14% of the variation for stachyose, raffinose, and Suc, respectively. This QTL appears as the major locus responsible for the observed variation in RSO content, its Ler allele increasing the seed content of both RSOs and decreasing the content of Suc. The Ler allele for the additional QTL affecting raffinose (chromosome 2) and stachyose (chromosome 1) decreases the content of these OSs. The additional QTL affecting Suc (chromosome 3) is the largest QTL affecting this OS and it explained 18% of the variance of the lines. The Ler allele at both Suc QTL decreases the content, which indicates that other QTL controlling this trait remained undetected to explain transgression for higher Suc (Fig. 2).
Figure 4.
QTL likelihood maps for seed-soluble OS contents of the five linkage groups of Arabidopsis. The abscissas correspond to the genetic maps in cM; 1 through 5 indicate the linkage group number. The horizontal dotted lines correspond to the LOD score threshold of 2.8 used to declare the presence of a QTL. Two-LOD support intervals for the significant QTL are shown as black bars along the abscissas. QTL effects are shown for linkage groups 1 and 3 where the major QTL are located. These are given as twice the additive allele effects, i.e. as the mean differences between the two RIL genotypic groups carrying the Ler and Cvi alleles. A positive QTL effect represents that the Ler allele increases the content. The percentage of phenotypic variance explained by each QTL is reported close to the corresponding LOD score peak.
Analysis of QTL interactions detected significant epistasis between the two loci on chromosome 1 affecting stachyose content (P = 0.003). RILs carrying a Cvi allele at marker GD.86L and a Ler allele at marker EC.88C had a higher stachyose content than the high RSO content parent Ler, whereas RILs bearing the opposite allelic combination showed a similar content as Cvi. Therefore, the Cvi allele at the QTL near the marker GD.86L increases the stachyose content only when the QTL at EC.88C carries a Ler allele in agreement with the observed transgression in only one direction (Fig. 3).
Localization of RSOs Biosynthesis Genes
Since the QTL on chromosome 1 is the major locus controlling the OS variation in seeds, we searched for candidate genes in this genomic region. The genomic nucleotide sequences available in the databases as part of the International Arabidopsis Genome Project (http://www.arabidopsis.org) were analyzed to look for putative genes encoding known OS biosynthesis and degradation enzymes. The four putative enzymes were: galactinol synthase (GC; EC 2.1.4.123), raffinose synthase (RS; EC 2.4.1.82), stachyose synthase (SS; EC 2.4.1.67), and α-galactosidase (EC 3.2.1.22) (Krebbers et al., 1997). GS catalyzes the first committed step in the biosynthesis of RSO (Krebbers et al., 1997). RS and SS control subsequent steps in the biosynthesis of raffinose saccharides by adding a Gal unit to Suc or raffinose, respectively. The enzyme α-galactosidase degrades the RSOs. Two genes encoding, respectively, GS and RS were located on two different bacterial artificial chromosome (BAC) from a BAC contig in the lower arm of chromosome 1. Furthermore, homologous sequences of these genes were also found in other regions of the Arabidopsis genome, which suggests that both genes belong to gene families. We designed cleaved amplified polymorphic sequence (CAPS) markers specific for both genes and mapped them in the Ler/Cvi RIL population. The two genes appeared closely linked to the QTL around marker EC.88C on chromosome 1 and therefore, they are possible candidate genes for this major QTL.
Mapping QTL for Seed Storability
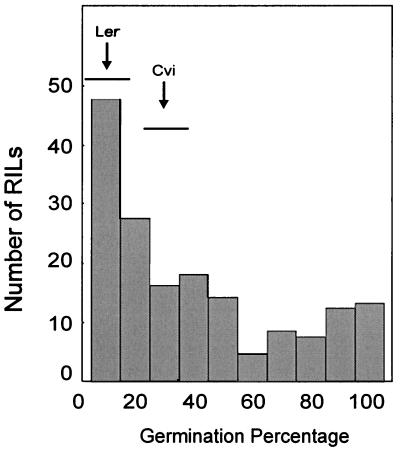
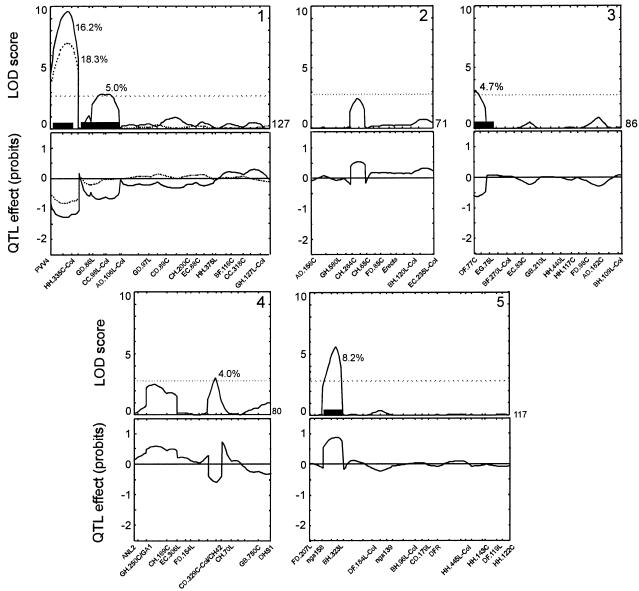
To determine whether there is a functional relationship between seed-soluble OS content and storability of seeds we have analyzed the same Ler/Cvi RILs to map QTL for this trait. Seed storability was measured as viability under two different seed aging assays, after natural aging following 4 years of storage and after artificial aging promoted by a CDT (see “Materials and Methods”). The two parental accessions Ler and Cvi were both relatively sensitive to the CDT, although they differed significantly (Fig. 5), Ler being more sensitive to the test conditions (lower storability) than Cvi. The RIL population showed transgression toward higher germination percentages, indicating the presence in the two parental lines of alleles increasing and reducing the tolerance to the given controlled stress (Fig. 5). Four putative QTL (Fig. 6) were identified affecting viability after controlled deterioration, their additive effects accounting for 56.5% of the phenotypic variance. These are QTL located on chromosome 1 (two closely linked QTL), chromosome 3, and chromosome 5. In addition to germination, the fraction of aberrant seedlings among germinating seeds is another parameter commonly used to measure seed storability (Coolbear, 1995) and was also analyzed. This trait showed a high correlation with the germination values after the CDT (r = 0.90) and QTL at similar positions were found to be responsible for its variation (data not shown).
Figure 5.
Frequency distribution of the seed viability (germination percentage) of the Ler/Cvi RIL after the CDT. Arrows correspond to the parental line means, and horizontal bars represent their ranges of variation.
Figure 6.
QTL likelihood maps for seed storability measured as viability after the CDT (solid line) and 4 years of natural aging (dashed line). The abscissas correspond to the genetic maps in cM (the linkage group number being indicated in the right upper corner of each panel). Horizontal dotted lines correspond to the LOD score threshold of 2.8 used to declare the presence of a QTL. Two-LOD support intervals for the significant QTLs are shown as black bars along abscissas. The additive QTL effects are expressed as probit units of the germination percentage after the CDT. These are estimated as the mean differences between the two RIL genotypic groups carrying the Ler and Cvi alleles (a positive value implies Ler increases the corresponding phenotypic value). The percentage of variance explained by each QTL is reported close to the corresponding LOD score peak.
The germination percentages after natural aging ranged from 61% to 100% (average 97). The QTL mapping for seed viability after 4 years of storage resulted in one QTL located on chromosome 1, which accounted for 18.3% of the phenotypic variance; a QTL at similar position was also the largest effect QTL detected after the CDT. QTL mapping for natural aging did not reveal any additional loci affecting storability.
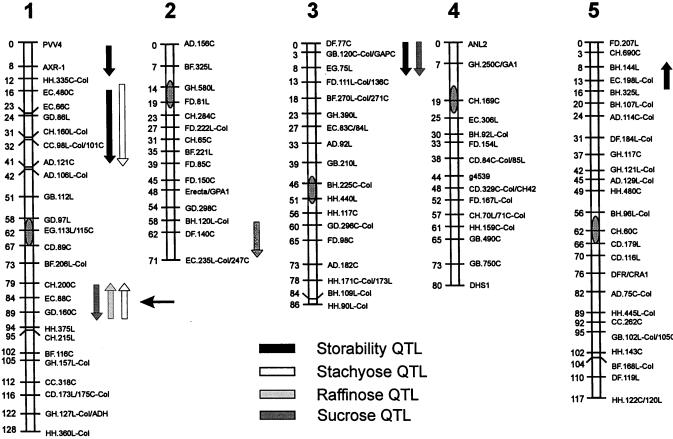
The comparison of map positions between the detected QTL for seed-soluble OS content and storability showed two genomic regions containing QTL for both traits (Fig. 7). The top of chromosome 1 affected stachyose content and storability and the locus on top of chromosome 3 affected Suc content and storability. No significant effect on seed storability was found in the vicinity of the OS QTL region around EC.88C on chromosome 1.
Figure 7.
Ler/Cvi linkage map showing the genetic location of QTL affecting seed-soluble OS contents and seed storability. Arrows indicate the direction of Ler allele phenotypic effect (up, increasing; down, decreasing). The length of the arrows depicts the 2-LOD support intervals. The horizontal black arrow points to the major OS QTL.
DISCUSSION
During the past decade the use of molecular marker technology and QTL mapping have contributed to a better understanding of the genetic basisof many agriculturally and biologically important quantitative traits, such as yield (Stuber et al., 1987) resistance/tolerance to biotic and abiotic stress (Koornneef and Peeters, 1999), and nutritional quality in numerous crops species (Paterson et al., 1991). In the present work we have used Arabidopsis for the analysis of seed content of the three main soluble OSs (Suc, raffinose, and stachyose) and showed that considerable variation exists among accessions (Fig. 1). By performing QTL mapping for seed OS content in a RIL population derived from a cross between the two accessions Ler and Cvi differing in seed OS composition, we identified one major QTL responsible for the practically monogenic segregation observed for seed stachyose content. It is very likely that this QTL affected pleiotropically the contents of the three detected OSs, showing opposite effects on RSOs and Suc contents. This locus might therefore be involved in the biosynthetic pathway of RSOs. Two candidate genes encoding for GS and RS were mapped within the 2-logarithm-of-odds (LOD) support interval of this QTL, the genetic distance between both genes being approximately 5 cM. Nevertheless this region has not been sequenced completely and other genes not found in this analysis and involved in RSO biosynthesis and degradation (i.e. SS and α-galactosidase) might locate within this interval. Further fine mapping using recombinants in this region, as well as complementation by plant transformation can be used in the future to determine whether any of these candidate genes corresponds to this QTL.
The maternal effects for stachyose and raffinose seed content detected by the analysis of OSs in hybrid seeds obtained in reciprocal crosses between the parental lines suggest either production of raffinose and stachyose by the testa or import of RSOs from maternal tissues. In agreement with this observation is the function that OSs and especially stachyose might have as transport sugars, as was described in cucurbits (Beebe and Turgeon, 1992). However, the apparent paternal effects observed on Suc content are rather unexpected and suggest an even more complex genetic control of the seed Suc content. Nevertheless, it should be noted that the parental and the reciprocal hybrids seeds differ considerably in size and that paternal effects on the seed size variation of these materials could not be excluded (Alonso-Blanco et al., 1999). The two seed Suc content QTL identified in this work colocated with seed size QTL reported previously (Alonso-Blanco et al., 1999). Since Suc seems to play an important role in the metabolic control of seed size (Weber et al., 1997), it is possible that some of these colocations are due to pleiotropic effects from the same gene on both traits.
The accessions Ler and Cvi were originally collected in nature, and therefore the observed variation in seed OS contents might be related to the adaptive properties of both genotypes. It has previously been shown (Liu et al., 1998) that cold increases the levels of GS mRNA, which suggests that this gene and RSOs may play a role in cold adaptation. Ler originates from northern Europe (Rédei, 1992), whereas Cvi comes from the Cape Verde Islands (Lobin, 1983) located at 16°N with a subtropical climate. Thus, the Ler and Cvi RSO contents and the allele effects at the major OS QTL were in agreement with this speculation, suggesting that seed RSOs might be involved in seed survival at low temperatures.
To determine whether OSs are important for seed storability, we have mapped QTL affecting this trait in the same RIL population analyzed for seed-soluble OS content. We have measured seed storability as viability (germination) after natural aging and after artificial aging induced by a CDT. The major QTL affecting storability was detected in both seed aging assays. However, the effect of the CDT on the viability of the seeds was much stronger than the effect of the natural aging, resulting in a more accurate mapping. Therefore, these results indicate that the CDT is a useful method to artificially accelerate seed aging. The comparison of map positions between the QTL identified controlling seed OS contents and the QTL affecting seed storability showed colocations in two regions (Fig. 7). The genomic region on top of chromosome 1 only marginally affected the stachyose content (its additive effects accounting for 4% of the variance) but considerably influenced seed storability (16.2% of the variance); the region on chromosome 3 strongly affected Suc content (18% of the variance) and had only a slight effect on seed storability (4.7% of the variance). In both cases a higher OS content co-segregates with better storability, and these colocations might be interpreted either as the presence of two closely linked genes or as a consequence of pleiotropy, higher seed content of either stachyose or Suc leading to increased viability after CDT. However, the major locus on chromosome 1 controlling RSOs and Suc in opposite directions did not show any significant effect on the germination ability after controlled deterioration. We conclude that in the studied material the variation observed for OS content does not evidently affect seed viability after CDT and that neither RSOs nor Suc had an apparent specific effect on seed storability. Nevertheless, this does not imply that OSs are not involved in seed protection and storability. Both RSOs and Suc might affect seed storability but due to the effect of the major OS QTL on both RSOs and Suc contents, their roles on storability might have been compensated and masked. Given the strong effect of the major OS locus on chromosome 1 on RSOs and the relatively smaller effect on Suc seed content, it is suggested that RSOs seem not to be substantially better than Suc in protecting the seeds against controlled deterioration.
In addition, several other loci appeared to affect seed storability independently from seed OS content, which shows genetic variation for other factors involved in this complex trait. Deterioration of seeds during storage has frequently been related to free radical-mediated oxidative damage of proteins, nucleic acids, and membranes (Coolbear, 1995). Factors controlling either the protection of cell structures or the recovery from damage might determine the difference between the two genotypes, Ler and Cvi. In particular, some of the QTL that we have identified for viability after CDT colocated with a cluster of genes related to stress tolerance on the top of chromosome 1 (Taji et al., 1999; Thorlby et al., 1999). Further fine mapping of these loci, combined with the analysis of other related traits, will allow the identification of the corresponding genes involved in the storability genetic variation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The following Arabidopsis accessions (the Nottingham Arabidopsis Stock Centre nos. are indicated in parentheses) were analyzed for OS content: C24 (N906), Col (N907), Ct-1 (N1094), Cvi (N8580), Eil-0 (N1133), Ler (NW20), Mt-1 (N1380), Shah (N929), RLD-1 (N913), and Ws-1 (N2223). F10 seeds of a set of 162 RILs, derived from crosses between the laboratory strain Ler originated from northern Europe (Rédei, 1992) and the accession Cvi, from the Cape Verde Islands (Lobin, 1983), were analyzed. These RILs have been previously characterized for amplified fragment polymorphism and CAPS markers (Alonso-Blanco et al., 1998b).
Plants were grown in an air-conditioned greenhouse supplemented with additional light (model TDL 58W/84, Philips, Eindhoven, The Netherlands) from middle September until the beginning of April, providing a day length of at least 14 h (temperature, 22°C–25°C). To reduce developmental and environmental effects on the various seed traits analyzed, we synchronized the onset of flowering and thereby of seed development. To do so the RILs were planted at three subsequent weeks according to their respective flowering times (Alonso-Blanco et al., 1998a), and the seeds of all genotypes were harvested on the same day. Twelve plants per RIL were grown in a two-block design to avoid environmental effects and their seeds harvested in a single bulk. For the accession analysis, six plants per genotype were grown, and their seeds were bulk harvested. Seed OS content analysis and the CDT were performed on seeds of the RILs and parental lines stored in the same open box for 2 years and 2 months at room temperature. To test the natural aging the seeds have been stored for 4 years under the same conditions. The seeds of the accessions and the crosses were stored under similar conditions during 4 months before analysis of their OS content.
Sugar Content Measurement
One hundred seeds from bulks of six to 12 plants were weighed and homogenized in 80% (v/v) methanol with the addition of 25 μg of melezitose as internal standard. The homogenate was heated for 15 min at 75°C and centrifuged 5 min at 10,000g. The supernatant was vacuum evaporated, and its residue was resuspended in 0.5 mL of pure water and injected into a Dionex HPLC system (Dionex, Sunnyvale, CA). Sugar content was determined with a high-pH-anion-exchange HPLC, using a gradient pump module (model GP40, Dionex) and an ED40-pulsed electrochemical detector (Dionex). Sugars were chromatographed on a CarboPac PA100 4- × 250-mm column (Dionex) preceded by a guard column (CarboPac PA100, 4 × 50 mm). Mono-, di-, and trisaccharides were separated by elution in increasing concentration of NaOH (50–200 mm) with a flow rate of 1 mL per minute. Peaks were identified by co-elution of standards. Sugar quantity was corrected by means of the internal standard and transformed to micrograms of sugar per milligram of seed.
Seed Storability Measurement
Seed storability was determined as viability (germination) after natural aging during 4 years and after the CDT. The CDT was performed as follows: bulked seeds from 12 plants, stored for 2 years and 2 months at room temperature, were equilibrated at 85% relative humidity (the obtained seed moisture content was approximately 10.5%). Thereafter the seeds were artificially aged during 4 d at 40°C because preliminary experiments showed that this deterioration is the most discriminative between Ler and Cvi (data not shown), and they were dried back at 32% relative humidity and 20°C during 3 d. The seeds were stored at 5°C, and thereafter germination was tested. Germination of 100 CDT-treated seeds (two replicates of 50 seeds) was tested on moist filter paper at 20°C and a 12-h-dark/12-h-light cycle by visually inspecting root tip emergence during 2 weeks. Non-germinating seeds were not viable as shown by the absence of staining in a tetrazolium viability test (Moore, 1985). In addition, the quality of the emerging seedlings was recorded by scoring the number of morphologically aberrant seedlings.
Natural aging was tested by performing a germination test on 4-year-old seeds (stored dry at room temperature without humidity control). Seeds were sown in triplicate (70–100 seeds per 6-cm Petri dish) on water-soaked filter paper (no. 595, Schleicher & Schull, Keene, NH) and exposed to cold (4°C) during 3 d. Thereafter seeds were transferred to a climate room (25°C, 16-h light/day; model TL57, Philips), and germination was scored after 7 d. The average germination percentages of the three replicates were calculated.
QTL Analysis
To map QTL using the RIL population, a set of 99 markers covering most of the Arabidopsis genetic map was selected from the previously published RIL Ler/Cvi map (Alonso-Blanco et al., 1998a). These markers spanned 482 cM with an average distance between consecutive markers of 5 cM and the largest genetic distance being 12 cM. Storability data (germination after 4 years of storage and viability after the CDT) were transformed to probit units to achieve normality.
The computer program MapQTL version 4.0 (Plant Research International, Wageningen University and Research Centre, Wageningen, The Netherlands) was used to identify and locate QTL linked to the molecular markers using both interval mapping and multiple-QTL model mapping (MQM) methods as described in its reference manual (http://www.cpro.wag-ur.nl/cbw/mapping/). The estimated additive effect and the percentage of variance explained by each QTL as well as the total variance explained by all of the QTL affecting a trait, were obtained with MapQTL in the final MQM model. For this, different cofactor markers were tested around the putative QTL positions (van Ooijen and Maliepaard, 1996), selecting as final cofactors the closest marker to each QTL, i.e. those maximizing the LOD score. A LOD score threshold of 2.8 was applied to declare the presence of a QTL, which corresponds to a general genome-wide significance P = 0.05 for normally distributed data, as was determined by extensive simulation experiments (van Ooijen, 1999). We verified this threshold for interval mapping by applying the permutation test to each data set (10,000 repetitions) and found a P = 0.05 LOD threshold of 2.6 for all traits. Two-LOD support intervals were established as ≈95% confidence intervals (van Ooijen, 1992).
For every trait, two-way QTL interactions were analyzed by ANOVA at a significance level of P < 0.005, using the general linear model module of the statistical package SPSS version 7.5 (SPSS, Inc., Chicago). For each analysis, the closest linked markers to the corresponding detected QTL were used as random factors in the ANOVA (the same markers used as cofactors in the MQM mapping with MapQTL).
Location of RSOs Biosynthetic Genes
CAPS markers (Konieczny and Ausubel, 1993) were developed to genetically map the genes encoding GS (EC 2.1.4.123) and RS (EC 2.4.1.82) in the Ler/Cvi RIL population. The map locations were determined with the software package JOINMAP (Plant Research International, http://www.cpro. wagur.nl/cbw/mapping/).
The primers for GS were based on the genomic nucleotide sequence of the BAC F8A5 (accession no. AC002292) located on chromosome one. The forward primer was 5′-TCG GTT ATT CTC CTT TGT TGT TTG-3′. The reverse primer was 5′-TTT CTA TGC CGT GAT GGA CTG TT-3′.
The primers for RS were based on the nucleotide sequence of BAC F20N2 (accession no. AC002328) located on chromosome one. The forward primer was 5′-GGG AGG AGT CAA ACC AGG TG-3′. The reverse primer was 5′-GGC ATC AAT GTC ACT GGT AAA G-3′.
PCR were carried out in 50-μL volumes containing 50 ng of genomic DNA, 100 μm each deoxynucleotide, 100 ng of both primers, and 0.2 units of Taq polymerase. Conditions for amplification were as follows: 30 s at 94°C, annealing for 2 min at 51°C or 61°C (GS and RS, respectively), and extension for 2 min at 72°C. The cycle was repeated 35 times. To detect the polymorphism, 10 μL of PCR product was cleaved with the restriction enzyme RsaI or BsaBI (for GS and RS, respectively) and analyzed in a 1.5% (w/v) agarose gel.
Footnotes
This work was supported by The Earth and Life Sciences Foundation subsidized by The Netherlands Organization for Scientific Research (to L.B.), by the Biotechnology Program of the European Union (grant no. BIO4CT965008 to C.A.-B.), and by the Agriculture and Fisheries Program of the European Community (grant no. FAIR–BM–98–4743 to K.T.).
LITERATURE CITED
- Alonso-Blanco C, Blankestijn-de-Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE-D, Coupland G, Koornneef M. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics. 1998a;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 2000;5:22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998b;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Beebe DU, Turgeon R. Localization of galactinol, raffinose, and stachyose synthesis in Cucurbita pepo leaves. Planta. 1992;188:354–361. doi: 10.1007/BF00192802. [DOI] [PubMed] [Google Scholar]
- Coolbear P. Mechanisms of seed deterioration. In: Basra AS, editor. Seed Quality: Basic Mechanisms and Agricultural Implications. New York: Food Product Press; 1995. pp. 223–277. [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Wistrom CA. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton JG, TeKrony DM. Handbook of Vigour Test Methods. Ed 3. Zurich: The International Seed Testing Association; 1995. [Google Scholar]
- Hoekstra FA, Crowe JH, Crowe LM. Membrane behavior in drought and its physiological significance. In: Taylorson RB, editor. Recent Advances in the Development and Germination of Seeds. New York: Plenum Press; 1989. pp. 71–88. [Google Scholar]
- Hoekstra FA, Crowe JH, Crowe LM. Effect of sucrose on phase behavior of membranes in intact pollen of Typha latifolia L., as measured with Fourier transform infrared spectroscopy. Plant Physiol. 1991;97:1073–1079. doi: 10.1104/pp.97.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbowicz M, Obendorf RL. Seed desiccation tolerance and storability: dependence on flatulence-producing oligosaccharides and cyclitols: review and survey. Seed Sci Res. 1994;4:385–405. [Google Scholar]
- Jansen RC. Complex plant traits: time for polygenic analysis. Trends Plant Sci. 1996;1:89–94. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Peeters AJM. Genetic approaches to abiotic stress responses. In: Shinozaki K, Yamaguchi-Shinozaki K, editors. Molecular Responses to Cold, Drought, Heat and Salt Stress in Higher Plants. R.G. Austin, TX: Landes Company; 1999. pp. 1–10. [Google Scholar]
- Koster KL, Leopold AC. Sugars and desiccation tolerance in seeds. Plant Physiol. 1988;88:829–832. doi: 10.1104/pp.88.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebbers E, Broglie R, Hitz B, Jones T, Hubbard N. Biotechnological approaches to altering seed composition. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 595–633. [Google Scholar]
- Leopold AC, Sun WQ, Bernal-Lugo I. The glassy state in seeds: analysis and function. Seed Sci Res. 1994;4:267–274. [Google Scholar]
- Liu J-JJ, Krenz DC, Galvez AF, de Lumen BO. Galactinol synthase (GS): increased enzyme activity and levels of mRNA due to cold and desiccation. Plant Sci. 1998;134:11–20. [Google Scholar]
- Lobin W. The occurrence of Arabidopsis thaliana in the Cape Verde Islands. Arabidopsis Inf Serv. 1983;20:119–123. [Google Scholar]
- McDonald MB. Seed deterioration: physiology, repair and assessment. Seed Sci Technol. 1999;27:177–237. [Google Scholar]
- Moore RP. Handbook on Tetrazolium Testing. Zurich: International Seed Testing Association; 1985. [Google Scholar]
- Obendorf RL. Oligosaccharides and galactosyl cyclitols in seed desiccation tolerance. Seed Sci Res. 1997;7:63–74. [Google Scholar]
- Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiol. 1993;102:1185–1191. doi: 10.1104/pp.102.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD, Lincoln SE, Lander ES, Tanksley SD. Mendelian factors underlying quantitative traits in tomato: comparison across species, generations and environments. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A, Matthews S. Application of the controlled deterioration vigour test to detect seed lots of Brussels sprouts with low potential for storage under commercial conditions. Seed Sci Technol. 1984;12:649–657. [Google Scholar]
- Prioul J-L, Quarrie S, Causse M, de Vienne D. Dissecting complex physiological functions through the use of molecular quantitative genetics. J Exp Bot. 1997;48:1151–1163. [Google Scholar]
- Rédei GP. A heuristic glance to the past of Arabidopsis genetics. In: Koncz C, Chua N, Schell J, editors. Methods in Arabidopsis Research. Singapore: World Scientific; 1992. pp. 1–15. [Google Scholar]
- Sinniah UR, Ellis RH, John P. Irrigation and seed quality development in rapid-cycling brassica: soluble carbohydrates and heat-stable proteins. Ann Bot. 1998;82:647–655. [Google Scholar]
- Stuber CW, Edward MD, Wendel JF. Molecular marker-facilitated investigations of quantitative trait loci in maize: II. Factors influencing yield and its component traits. Crop Sci. 1987;27:639–648. [Google Scholar]
- Taji T, Seki M, Yamaguchi-Shinozaki K, Kamada H, Giraudat J, Shinozaki K. Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:119–123. doi: 10.1093/oxfordjournals.pcp.a029469. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. Mapping polygenes. Annu Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- Thorlby G, Veale E, Butcher K, Warren G. Map positions of SFR genes in relation to other freezing-related genes of Arabidopsis thaliana. Plant J. 1999;17:445–452. doi: 10.1046/j.1365-313x.1999.00395.x. [DOI] [PubMed] [Google Scholar]
- van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- van Ooijen JW. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity. 1999;83:613–624. doi: 10.1038/sj.hdy.6886230. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. Sugar import and metabolism during seed development. Trends Plant Sci. 1997;2:169–174. [Google Scholar]