Abstract
Cyp1b1 deletion and gestational vitamin A deficiency (GVAD) redirect adult liver gene expression. A matched sufficient pre- and post-natal diet, which has high carbohydrate and normal iron content (LF12), increased inflammatory gene expression markers in adult livers that were suppressed by GVAD and Cyp1b1 deletion. At birth on the LF12 diet, Cyp1b1 deletion and GVAD each suppress liver expression of the iron suppressor, hepcidin (Hepc), while increasing stellate cell activation markers and suppressing post-natal increases in lipogenesis. Hepc was less suppressed in Cyp1b1−/− pups with a standard breeder diet, but was restored by iron supplementation of the LF12 diet.
Conclusions
The LF12 diet delivered low post-natal iron and attenuated Hepc. Hepc decreases in Cyp1b1−/− and GVAD mice resulted in stellate activation and lipogenesis suppression. Endothelial BMP6, a Hepc stimulant, is a potential coordinator and Cyp1b1 target. These neonatal changes in Cyp1b1−/− mice link to diminished adult obesity and liver inflammation.
Keywords: vitamin A, vitamin A deficiency, cytochrome P450 1b1, hepcidin, lipogenesis
1. Introduction
Cytochrome P450 1b1 (Cyp1b1) is an atypical P450, demonstrating extra-hepatocyte expression and the capacity to metabolize endogenous substrates, such as estradiol, polyunsaturated fatty acids, and retinol (Chambers et al., 2007, Jennings et al., 2014, Larsen et al., 2015, Li et al., 2014). Cyp1b1 is expressed in many types of support cells, including mesenchymal progenitor cells, endothelia, pericytes, macrophage, and stellate cells (Choudhary et al., 2003, Piscaglia et al., 1999, Tang et al., 2009). Cyp1b1 acts in vascular cells to restrain local oxidative stress (Palenski et al., 2013) and metabolize estradiol for estrogen receptor-independent signaling mechanisms (Jennings et al., 2014, Malik et al., 2012, White et al., 2012). Cyp1b1 may exert developmental control over liver homeostasis, in part through changes in hypothalamic signaling (Bushkofsky et al., 2016, Larsen et al., 2015). Cyp1b1 deletion (Cyp1b1−/− mice) suppresses diet-induced obesity (DIO) by preventing adiposity from a post-weaning high fat diet and alters liver gene expression compared to wild type (WT) controls.
In adult Cyp1b1−/− male mice, we have characterized three clusters of diet-selective gene responses. These clusters demonstrate functional links to [1] growth hormone signaling through HNF4α, [2] leptin suppression of fatty acid synthesis genes, including stearoyl-coenzyme A desaturase 1 (Scd1), and [3] suppression of postprandial inflammation derived from a high carbohydrate diet (Bushkofsky et al., 2016, Larsen et al., 2015). Overexpression of Scd1 in Cyp1b1−/− mice restores adiposity and DIO on a post-weaning high fat diet (Li et al., 2014).
Cyp1b1−/− mice that retain DIO (R-Cyp1b1−/−) have been bred from outlier mice in the original colony. We show that these mice do not show the characteristic liver gene expression signature (Larsen et al., 2015). However, when R-Cyp1b1−/− mice were backcrossed with WT C57Bl/6J mice, the progeny reverted to the original DIO suppression phenotype. Here, we use R-Cyp1b1−/− mice to identify changes in neonatal liver gene expression responses that link neonatal development to the adult gene regulation that determines the DIO response.
Developmental expression of Cyp1b1 at embryonic day (E)9.5 is temporally localized to the hindbrain and the foregut, where liver development is initiating. Cyp1b1 expression corresponds to a key period of retinoid regulation of morphogenic patterning genes and overlaps with expression of the retinoid-responsive transcription factor, Hoxb1, in the hindbrain and the foregut (Chambers et al., 2007, Chambers et al., 2009, Huang et al., 1998, Stoilov et al., 2004). This connection between retinol and Cyp1b1 has led us to investigate neonatal gene expression patterns that may redirect adult liver function.
CYP1B1 metabolizes retinol to the bioactive retinoic acid (RA) (Chambers et al., 2007, Chambers et al., 2009). Vitamin A (VA)/retinol is obtained from the diet, absorbed in chylomicra, and stored as retinyl ester (RE) in stellate cells of the liver (Bonet et al., 2012, Harrison, 2012). Stellate cells comprise less than five percent of the total liver and are located between endothelia and hepatocytes in the space of Disse (Si-Tayeb et al., 2010). Retinol plays key roles in development, reproduction, and immunity that largely result from conversion to RA (Clagett-Dame and DeLuca, 2002, Napoli, 2012, See et al., 2008, Smith et al., 1987). Direct RA administration can decrease adiposity and improve glucose tolerance (Bonet et al., 2012).
Systemic retinol deficiency in mice requires initiation of a VA-deficient diet to the mother during mid-gestation, with continuation to maturity in the progeny (McCarthy and Cerecedo, 1952, Smith et al., 1987) (GVAD treatment). A VA-deficient diet can only be obtained by avoidance of unprocessed plant constituents that have appreciable retinoid sources (corn, grasses, etc) (Ross, 2010). The VA-deficient diet historically and currently used in these experiments contained cottonseed oil as a fat source (12 percent kcal from fat) and other defined carbohydrate, protein, and essential nutrient components (LF12 diet; Supplementary Table 1). When GVAD was combined with a post-weaning high fat diet (HFD), the typical obesity response is suppressed, much as seen with Cyp1b1 deletion. Here, we show that the growth hormone- and leptin-associated liver responses to Cyp1b1 deletion were largely unaffected by GVAD. We have recently shown that administration of the LF12 diet in gestation to the mother and post-weaning to the progeny produces major increases in a set of inflammatory markers (Maguire et al., 2017). This appears to be a postprandial response to high dietary carbohydrate that is suppressed by GVAD treatment. The gene expression responses overlapped extensively with changes seen with deficiency of the nuclear co-repressor, Nr0b2/Shp (Kim et al., 2014). We show that these changes also overlap with changes produced by Cyp1b1 deletion with the standard post-weaning low fat diet (Bushkofsky et al., 2016, Larsen et al., 2015).
We hypothesize that changes in liver development prior to weaning contribute to this shared adult obesity suppression response and to the overlapping gene expression changes. To study mice at birth and weaning under more defined conditions, we have moved from the standard breeder diet (BD) to the defined LF12 diet. Compared to the BD, the LF12 maternal diet has an increase in the balance of carbohydrate to fat and lacks the iron supplementation typically included to meet the demands of the progeny (Supplementary Table 1). We examined pups at birth and weaning with respect to genes that were changed similarly by retinol deficiency (GVAD) or Cyp1b1 deletion. Further insight into the relationship between effects of Cyp1b1 on neonatal and adult gene expression was provided by comparisons of normal Cyp1b1−/− with variant R-Cyp1b1−/− mice that lack the distinctive adult obesity suppression.
The overlapping gene expression responses in perinatal and neonatal livers exhibited an unusual signature: GVAD and Cyp1b1 deletion produced parallel changes, but the effect of Cyp1b1 deletion was reversed when combined with GVAD. This pattern was seen for suppression of multiple genes that convert acetyl CoA to lipogenic products, a response that is likely to contribute to adult obesity suppression. This response was preceded by stimulation of a cluster of stellate cell activation markers and by extensive suppression of Hamp/Hepc. Hamp/Hepc generates hepcidin, a 25 amino acid peptide that suppresses iron transfer into the circulation (Ganz, 2013) and Hamp2, a highly expressed gene duplication product that shares the Hepc locus. Hamp2 produces hepcidin 2, which has different activities that may include effects on metabolism (Lou et al., 2004). The unusual shared features of these responses points to a novel regulatory process that is impacted by retinol and Cyp1b1. This concept is reinforced by the fact that these responses are selectively absent in R-Cyp1b1−/− mice. The selective loss of many typical adult Cyp1b1 deletion responses in the R-Cyp1b1−/− variants is evaluated as a basis to link neonatal and adult regulation.
The unexpected Hepc findings have led us to adopt a new perspective on the relationship between Cyp1b1 and energy control through iron as a mediator. A dramatic loss of Hepc in Cyp1b1−/− and GVAD mice was first considered as a toxicity risk, based on the iron overload seen in adult Hepc-deficiency conditions when exposed to a normal adult diet (Ganz, 2013). The present work with perinatal and neonatal mice shows a distinct situation for iron/Hepc regulation during pregnancy, where regulation is primarily linked to iron deficiency caused by the developmental demands of the progeny. We establish, here, that Cyp1b1 deletion in mice fed the LF12 diet show exceptionally low Hepc expression, due to the need to enhance iron intake. These mice show remarkably high Hepc stimulation by elevated dietary iron. Differences between the BD and LF12 diets at constant iron content reveal a strong dependence of Hepc on other dietary components, notably carbohydrate and fat. This is complemented by the finding that carbohydrate metabolism and lipogenesis are also integrated through Cyp1b1 metabolism with iron homeostasis.
2. Materials and Methods
2.1 Animal care and husbandry
An in-house colony of wild type C57BL/6J (WT) (Jackson Labs, Bar Harbor, ME), Cyp1b1−/− (Buters et al., 1999), and an inbred sub-strain of Cyp1b1−/− mice that are resistant to DIO suppression, obtained by breeding the most obese mice, which led to a higher proportion of obese progeny, R-Cyp1b1−/− (Larsen et al., 2015) mice, were maintained in the AAALAC-accredited University of Wisconsin School of Medicine and Public Health facility. Mice were provided food and water ad libitum and maintained in a controlled 12-hour light/dark cycle. All protocols were approved by the School of Medicine and Public Health Animal Care and Use Committee (ACUC, Protocol number M005635). Nulliparous females aged 8–12 weeks were time mated, such that the presence of a vaginal plug was designated embryonic day (E)0.5. Prior to mating and until dietary administration, dams were maintained on a standard breeder diet (BD, Product Number 2019, Harlan Teklad, Madison, WI).
Offspring were examined at birth [post-natal day (PN)1], weaning (PN21), and 14 weeks of age.
2.2 Diet study - Adult (14wk) endpoint
Analysis of 14-week old adult male WT, Cyp1b1−/−, and R-Cyp1b1−/− mice offspring was as previously described (Bushkofsky et al., 2016, Larsen et al., 2015). The diets used for these mice comprised two maternal diets, a standard breeder diet (BD) with 22 percent kcal from fat derived from soybean oil, and the defined LF12 diet (Product Number TD.07291, Harlan Teklad) with 12 percent kcal from fat from cottonseed oil. The LF12 diet was also used in an equivalent VA-deficient formulation (VAD diet, Product Number TD.07655, Harlan Teklad). The VAD diet contains 220IU/kg of retinyl palmitate as a source of vitamin A, compared to the sufficient containing 24,000IU/kg.
The post-weaning diets were administered for 11 weeks, beginning at weaning (PN21), and comprised an approximately isocaloric low fat/high carbohydrate diet (LFD, Product Number D12450B, Research Diets, New Brunswick, NJ) or a high fat/low carbohydrate diet (HFD, Product Number D12492, Research Diets) subsequent to the BD (BD-LFD mice and BD-HFD mice, respectively), continuation of the LF12 diet (LF12-LF12), or the LF12 maternal diet followed by a post-weaning HFD (LF12-HFD) (Figure 1A).
Figure 1. GVAD and Cyp1b1 deletion each suppress diet-induced obesity, but R-Cyp1b1−/− mice do not.
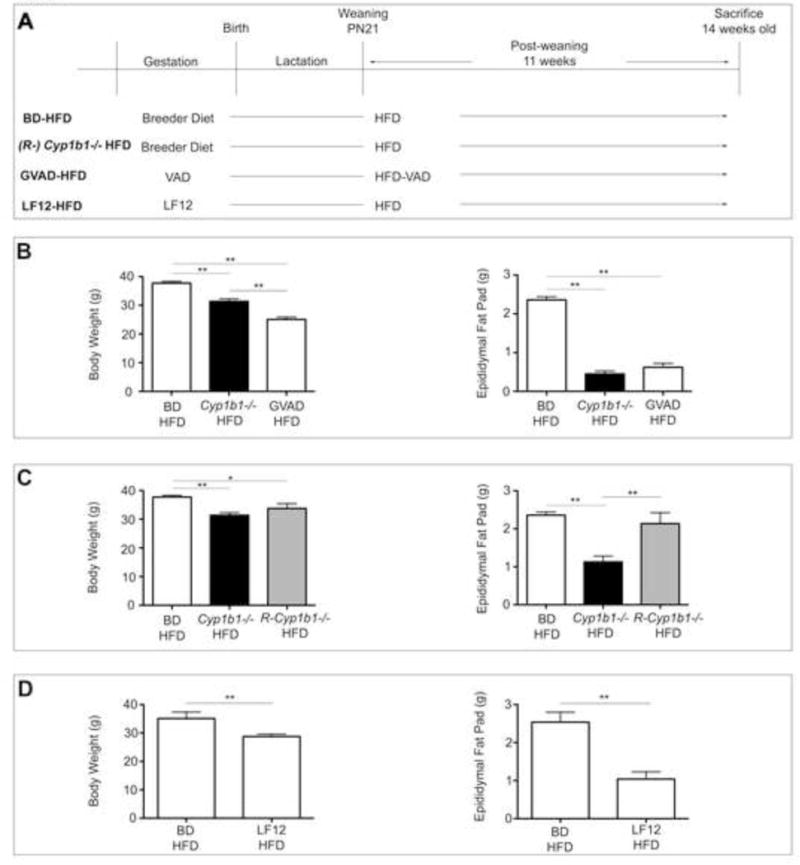
(A) Dietary administration for each group is shown. Body weight and epididymal fat pat pad weight in male offspring comparing obesity response on a control BD-HFD to (B) Cyp1b1−/− HFD and GVAD-HFD (C) R-Cyp1b1−/− HFD and (D) LF12 maternal diet (LF12-HFD). *p-value≤0.05, **p-value≤0.01
Retinol deficiency (GVAD) in adult offspring was initiated in the maternal diet at E10.5 by the VAD diet, followed by a VA deficient, high fat diet (HFD-VAD, Product Number TD.08210, Harlan Teklad) (GVAD-HFD mice) or continuation of the VAD diet (GVAD-LF12 mice) for 11 weeks post-weaning (Figure 1A). For each diet group, mice were euthanized individually by CO2, serum was collected by cardiac puncture, and tissues were individually weighed, flash frozen, and stored at −80°C until use.
2.3 Diet study – Birth and weaning endpoints
WT and Cyp1b1−/− offspring were examined at birth (PN1) and WT, Cyp1b1−/−, and R-Cyp1b1−/− offspring were examined at weaning (PN21). For each, pregnant dams were administered the LF12 or VAD diet (GVAD mice) at E4.5. Pups at birth were euthanized by extended CO2 exposure and decapitation on PN1. Liver was isolated and pooled by litter, regardless of sex, flash frozen and stored at −80°C until use. Weanling pups were euthanized individually by CO2 on PN21. Serum was collected by cardiac puncture and centrifugation, flash frozen and stored at −80°C until use. Tissues were individually weighed, flash frozen, and stored at −80°C until use.
2.4 Diet study – Iron supplementation
To examine the effects of dietary iron supplementation, WT and Cyp1b1−/− pregnant dams were administered the LF12 diet with added iron (LF12 + Iron, Product Number TD.160682, Harlan Teklad) at E4.5. Weanling male pups were euthanized individually by CO2 at PN21. Serum was collected by cardiac puncture and centrifugation, flash frozen and stored at −80°C until use. Tissues were individually weighed, flash frozen, and stored at −80°C until use.
2.5 Liver RNA isolation
Frozen liver was thawed in RNAlater Ice according to manufacturer’s instructions (Ambion, Foster City, CA). Total RNA was isolated from less than 20mg tissue by RNeasy Mini Kit accompanied by Qiashredder columns (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNA was spectrophotometrically measured for quantity and purity by A260/A280 on a Nanodrop, followed by visual inspection by gel electrophoresis.
2.6 Microarray analysis
Microarray analysis of livers at PN1 utilized liver samples from each of two pooled litters and weanling livers were performed as triplicate (WT and R-Cyp1b1−/−) or duplicate (Cyp1b1−/−) samples comprised of equal concentrations of pooled male biological replicates. Each used the Agilent Technologies 4x44k platform. Samples were prepared according to the manufacturer’s instructions for one-color labeling, including sample preparation, hybridization, and scanning. All samples were Cy3-labeled. Data was deposited in the NCBI Gene Expression Omnibus and can be accessed through GEO Series Accession Number GSE87844. Results were analyzed by the EDGE3 software using the Limma analysis, which assesses significance based on ANOVA statistics (Vollrath et al., 2009).
One adult liver microarray was analyzed from each of three male WT and Cyp1b1−/− LFD and HFD mice, as described previously (Bushkofsky et al., 2016, Larsen et al., 2015), using two-color labeling. GVAD-HFD (Cy3-labeled) liver gene expression was analyzed from each of three WT mice by two-color labeling relative to WT BD-HFD (Cy5) according to the manufacturer’s instructions. LF12-HFD, GVAD-LF12, and LF12-LF12 liver gene expression was assessed from each of two WT mice, as above for one-color labeling, and was deposited in the NCBI Gene Expression Omnibus and can be accessed through GEO Series Accession Number GSE87845. All samples used the Agilent Technologies 4x44k platform. Results were analyzed, as above, by the EDGE3 software using the Limma analysis for statistical significance. For reference, gene of interest names and abbreviations described herein are included in Supplementary Table 2.
2.7 Real time PCR
RNA (1.5μg) was reverse transcribed with Reverse Transcriptase (Promega, Madison, WI) and amplified by quantitative real time PCR (qPCR) in a 10μl reaction volume with Sybr Green (Promega). Expression was measured using a BioRad CFXConnect real time PCR detection system (Hercules, CA) and quantified using the standard curve method and normalized to GAPDH. Primer sequences are as follows:
-
Scd1
F 5′-CCGGAGACCCCTTAGATCGA-3′
R 5′-TAGCCTGTAAAAGATTTCAAA-3′
-
Fasn
F 5′-GCTGCGGAAACTTCAGGAAAT-3′
R 5′-AGAGACGTGTCACTCCTGGACTT-3′
-
Elovl6
F 5′-AGCAAAGCACCCGAACTAGGTGACACGATATTC-3′
R 5’- AGCGACCATGTCTTTGTAGGAGTACCAGGA-3’
-
Hamp
F 5′-AGAAAGCAGGGCAGACATTG-3′
R 5′-AGGTCAGGATGTGGCTCTAG-3′
2.8 Retinoid measurements
Serum retinol and liver retinol and retinyl esters were determined as previously stated (Riabroy and Tanumihardjo, 2014), with the following modifications. For weanling pups, serum was collected by cardiac puncture and pooled by litter, regardless of sex. 150μL serum was combined with an equal volume ethanol supplemented with 0.1% butylated hydroxytoluene and extracted three times with 500μL hexane. Liver (0.25 – 0.5g) was ground in sodium sulfate by mortar and pestle. Samples were extracted with dichloromethane to a final volume of 25mL and an aliquot (5mL) was dried under nitrogen. 50:50 (v:v) methanol:dichloroethane (0.1mL) was used for resuspension, 25μL was analyzed per sample. For each, retinyl burtyrate was added to measure extraction efficiency. Retinol and retinyl ester peaks were resolved on a Waters HPLC system with a C18 reverse-phase column (Milford, MA). Solvent A consisted of 92.5:7.5 (v:v) acetonitrile:water and solvent B consisted of 85:10:5 (v:v) acetonitrile:methanol:dichloroethane each with 0.05% triethylamine.
2.9 Serum Triglyceride Measurements
Serum triglyceride content of male and female, pooled serum samples (as for retinoid measurements) from weanling pups was determined using the Serum Triglyceride Determination Kit (Sigma-Aldrich, St. Louis, MO) according to manufacturer’s instructions.
2.10 Complete Blood Count (CBC)
Whole blood was collected by cardiac puncture at sacrifice of male pups at PN33 and diluted 1:4 in sterile PBS to a total volume of 300μl in EDTA-coated Microtainer™ tubes (Becton Dickinson, Franklin Lakes, NJ). Complete blood count (CBC) analysis was performed by the University of Wisconsin-Madison Veterinary Care Clinical Pathology Laboratory.
2.11 Statistical analysis
Comparisons were analyzed using Student’s t-test or one-way ANOVA with a Tukey post-test, where appropriate, to determine individual p-values (GraphPad Prism, La Jolla, CA). Data are represented as mean ± SEM.
3. Results
3.1 Cyp1b1 deletion and GVAD each suppress adiposity in mature mice, but the R-Cyp1b1−/− sub-strain was resistant
We have previously examined the effects of Cyp1b1 deletion on DIO, by measuring the effects of a post-weaning high fat/low carbohydrate diet (HFD) (Bushkofsky et al., 2016, Larsen et al., 2015) (Figure 1A). We have also examined the effects of GVAD on DIO with a post-weaning HFD (Figure 1A). Body weight and adiposity, used as a measure of DIO, were each lower in Cyp1b1−/− HFD and GVAD-HFD mice compared to the control BD-HFD (Figure 1B). R-Cyp1b1−/− mice on the same HFD protocol (Figure 1A) were resistant to DIO suppression (Figure 1C), but liver weight was not different from Cyp1b1−/− mice (data not shown). R-Cyp1b1−/− mice that retain DIO-responsiveness comprised approximately 10–20 percent of our original colony, but have been selectively crossed to provide mice in which this phenotype predominates (Larsen et al., 2015). The R-Cyp1b1−/− colony was restored to the original lean phenotype by a five generational backcross with WT mice (Larsen et al., 2015), according to rodent husbandry protocols (Lambert, 2007).
The GVAD protocol uses a maternal diet with a lower fat content (12 percent kcal from fat) than the standard breeder diet (22 percent kcal from fat). This basal diet (LF12) is similar to the low fat, post-weaning, control diet (LFD), sharing 20 percent refined protein content and approximately 70 percent kcal from carbohydrate (Supplementary Table 1). The combination of LF12 maternal diet with the HFD post-weaning (LF12-HFD) produced few liver gene expression differences when compared to the standard breeder diet combination (BD-HFD) (Maguire et al., 2017). Nevertheless, body weight and adipose were significantly decreased (Figure 1D).
3.2 Adult liver gene responses to diet exhibit both selective and shared responses to GVAD and Cyp1b1 deletion
Characterization of liver gene expression in WT and Cyp1b1−/− mice with a post-weaning, approximately isocaloric LFD or HFD has been described elsewhere (Bushkofsky et al., 2016, Larsen et al., 2015). Table 1 shows gene expression ratios that are the output from a set of Agilent microarrays following statistical processing by the EDGE3 software. Three clusters of genes are resolved based on differences in liver expression between Cyp1b1−/− and WT mice, each on the BD-HFD diet combination. These responses are compared to differences for the same genes using the GVAD-HFD combination with reference to the BD-HFD treatment in WT mice. Group A genes respond similarly to Cyp1b1 deletion on both the LFD and HFD and Group B genes respond selectively on the HFD. Neither set of genes responded in a similar way to GVAD treatment. A third group of genes, HF genes, shows similar expression differences between the HFD and LFD in WT and Cyp1b1−/− mice (Bushkofsky et al., 2016). The HFD/LFD differences were mostly prevented by the GVAD-HFD protocol, indicating a dependence on retinol. Additionally, these Cyp1b1-responsive genes were not responsive in R-Cyp1b1−/− mice.
Table 1.
Selective effects of Cyp1b1 deletion and GVAD on liver gene expression on the HFD in 14-week old mice
| Fold change upper versus | Cyp1b1−/− HFD | GVAD-HFD | R-Cyp1b1−/− LFD |
|---|---|---|---|
| lower | WT BD-HFD | WT BD-HFD | WT BD-LFD |
| Group A | |||
| Acot1 | −4.7** | n.s. | n.s. |
| Cd36 | −5.5** | n.s. | 2.3 |
| Cyp4a10 | −11.1** | −1.9** | n.s. |
| Lgals1 | −5.2** | −1.5* | 2.0 |
| Vnn1 | −6.1** | −1.5* | n.s. |
| Pparγ | −4.6** | n.s. | 1.6 |
| Egfr | 2.2* | n.s. | n.s. |
| Enho/Adropin | 2.0** | 2.4** | n.s. |
| Lifr | 3.4** | n.s. | n.s. |
| Selenbp1 | 2.5** | n.s. | n.s. |
| Cyp7b1 | 4.6* | n.s. | −2.6 |
| Nudt7 | 2.4** | n.s. | −1.9 |
| Group B | |||
| Scd1 | −6.0** | −1.5 | 1.5 |
| Me1 | −7.5** | −1.5* | −1.6 |
| Pdk4 | −3.7** | −2.0** | n.s. |
| Igfbp1 | −5.1* | n.s. | n.s. |
| Elovl5 | −2.2** | n.s. | n.s. |
| Osgin1 | 2.6** | n.s. | n.s. |
| HF genes | |||
| Cyp2c37 | n.s. | −2.9** | −2.7 |
| Gadd45 | n.s. | 5.1** | 3.7* |
| Lpl | 1.5 | 2.7** | 2.1 |
| Rbp1 | n.s. | 3.2** | 2.2 |
| Nr0b2/Shp | n.s. | −2.9** | n.s. |
| Nr1i3/Car | −1.5 | −1.8** | −1.5 |
| Sult1b1 | n.s. | −1.7* | n.s. |
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values where the net FC < 1.5 and p-value > 0.05
Diet: Carbohydrate/Fat/Protein; BD: 55/22/23; HFD: 20/60/20; LFD: 70/10/20
In Table 2, we use four expression ratios to compare the effects of Cyp1b1 deletion and GVAD on a low fat/high carbohydrate diet for a set of genes designated as 1B1-HF genes. Mice were fed ad libitum and sacrificed 6hr after nighttime food consumption. In Column 1, we measure the standard expression difference for WT mice on the HFD versus the LFD. The negative HFD/LFD ratio corresponds to stimulation on the LFD. The Column 2 expression ratio measures the impact of Cyp1b1 deletion on the LFD. Columns 3 and 4 provide data from a different set of experiments, in which we replaced the maternal breeder diet with the defined LF12 diet that is similar to the LFD (70 percent carbohydrate/12 and 10 percent fat, respectively). Comparison of Column 3 to Column 1 measures the impact of the change in maternal diet. The LF12 diet changes the fat content and source from the standard BD (58 percent carbohydrate/22 percent fat). The switch had little impact on the offspring response to the HFD, but appreciably increased the expression of 1B1-HF genes. Nearly all the ratios for genes in Column 3 are numerically larger than Column 1, corresponding to greater expression on the predominantly carbohydrate LF12 diet. The main difference arose from the switch from the maternal BD to LF12. The Column 4 ratio measures the impact of the GVAD treatment when applied to the same LF12-LF12 combination. This GVAD effect reversed 1B1-HF gene expression on the LF12-LF12 combination and produced an effect that is remarkably similar to the Cyp1b1 deletion response on the BD-LFD (Column 2 ratio).
Table 2.
GVAD and Cyp1b1 deletion produce similar responses on high carbohydrate diets
| Fold change upper versus | BD-HFD | Cyp1b1−/− LFD | LF12-HFD | GVAD-LF-12 |
|---|---|---|---|---|
| lower | BD-LFD | BD-LFD | LF12-LF12 | LF12-LF12 |
| 1B1-HF genes | ||||
| Anxa5 | −2.3* | −2.4* | −4.1** | −3.3** |
| Gas6 | −2.2** | −2.1** | −3.7** | −1.8* |
| Cd52 | −1.9** | −2.7** | −2.2 | −2.5** |
| H2-Ab1 | −1.5 | −2.1** | −3.5* | −1.9** |
| H2-Eb1 | −1.5 | −2.2** | −2.9* | −1.9** |
| Mmd2 | −4.6** | −3.9** | −5.5** | −9.5** |
| Klf6 | −2.4** | −2.9** | −4.8** | −2.2* |
| Cidec | −1.9 | −4.7 | −35.1** | −4.3 |
| Ly6d | −4.3** | −6.1** | −62.4** | −39.8** |
| Nupr1 | −5.5** | −5.1** | −4.2** | −3.3** |
| Prodh | 1.9* | 2.2* | 3.4** | 2.2** |
| Rhoc | −2.5** | −2.8** | −7.4** | −3.1** |
| Uap1l1 | −3.5** | −3.5* | −6.9** | −5.6** |
| Atf3 | −3.1* | −2.9* | −6.7** | −5.9** |
| Spp1 | −2.0 | −2.6* | −3.4** | −3.6** |
| Cd63 | −2.6* | −3.1* | −10.0* | −5.0** |
| Cxadr | −2.5* | −3.0** | −2.8** | −2.1** |
| Osbpl3 | −3.3** | −14.1** | −8.6** | −4.3** |
| Diet Related | ||||
| Gck | n.s. | n.s. | 1.9* | 1.5 |
| Ppp1r3g | −2.4 | −4.6** | −9.9** | −6.4* |
| Ppp1r3b | 1.8 | 2.2 | 2.4 | 3.5** |
| Ppp1r3c | 1.6 | 3.8** | n.s. | 4.5** |
| Hamp | n.s. | n.s. | −1.6 | −1.7 |
| Hamp2 | −1.6 | −2.3* | 1.6 | 6.5** |
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values where the net FC < 1.5 and p-value > 0.05
Diet: Carbohydrate/Fat/Protein; BD: 55/22/23; HFD: 20/60/20; LFD: 70/10/20; LF12: 69/12.2/18.8
We have previously shown that the changes in Cyp1b1−/− mice correlate with reported gene expression responses produced by deletion of the retinoid-responsive, metabolic co-repressor, Shp/Nr0b2 (Kim et al., 2014) (r2=0.78) (Bushkofsky et al., 2016). Similarly, GVAD effects correlated with the changes produced by Shp deletion (r2=0.74) (Supplementary Figure 1).
These 1B1-HF genes mostly correspond to genes that are markers of oxidative stress or inflammatory cells. They show high expression on the BD-LFD and on the LF12-LF12 protocols when compared to BD-HFD. We attribute this change to the high dietary carbohydrate and to postprandial oxidative stress caused by its metabolism (Oliveira et al., 2013). The 1B1-HF gene responses were not seen in R-Cyp1b1−/− mice (Supplementary Table 3).
While postprandial liver carbohydrate metabolism is regulated by Lrh/Nr5a2 (Bechmann et al., 2012), the major target gene, glucokinase (Gck) was not affected. However, three Ppp1r3 genes, which control the balance between glycogen synthase and phosphorylase activities (Luo et al., 2011), each responded extensively and similarly to GVAD and Cyp1b1 deletion (Table 2). We previously found in Cyp1b1−/− mice that increased Ppp1r3b and 3c and decreased Ppp1r3g each associate with a parallel increase in hepatocyte glycogen granules (Larsen et al., 2015). The LF12-LF12 protocol produced the largest increase in Ppp1r3g, which was also fully reversed by GVAD treatment.
3.3 GVAD and Cyp1b1 deletion decreased adipose and liver weight at weaning
We hypothesize that the overlap between Cyp1b1 deletion and GVAD in adult liver gene responses marks suppression of carbohydrate-induced oxidative stress and inflammation that derive from changes in the fetal/neonatal period. To test the effects of GVAD and Cyp1b1 deletion on late fetal and post-natal liver development, WT and Cyp1b1−/− pregnant dams were started on the GVAD protocol at E4.5, which preceded the period of major retinoid maternal transfer (Satre et al., 1992) (Figure 2A). Offspring were characterized at birth (PN1) and at weaning (PN21).
Figure 2. GVAD and Cyp1b1 deletion affect weanling pup physiological parameters.
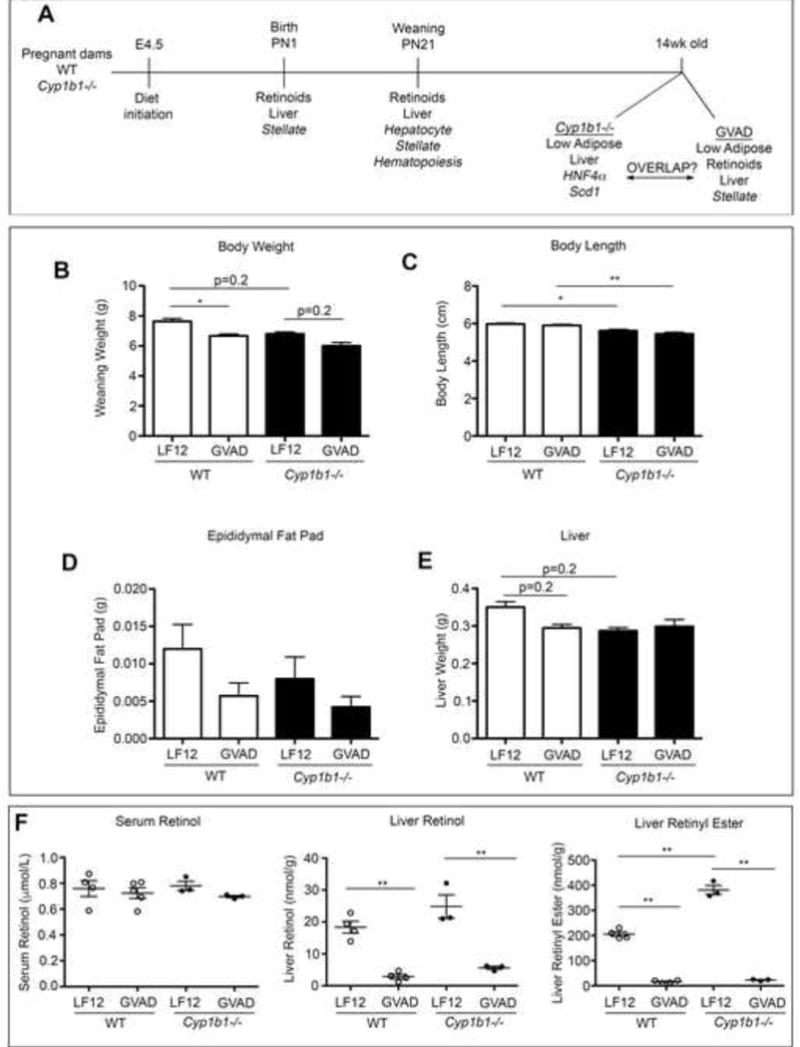
(A) Experimental design for C57Bl/6J (WT) and Cyp1b1−/− mice, each with GVAD examining offspring at birth (PN0), weaning (PN21), and 14weeks of age. At weaning, (B) Body weight, (C) nose to rump length, (D) epididymal fat pad, (E) liver weight, and (F) serum retinol, liver retinol, and liver retinyl ester content. *p-value≤0.05, **p-value≤0.01
There was no observable effect on the male to female ratio at weaning (data not shown), nor was there a difference in body weight between male and female offspring within a treatment group (data not shown). Unless otherwise noted, data does not distinguish males and females. Overt morphological signs of VA syndrome, including cleft palate, forelimb and ocular malformations (Clagett-Dame and DeLuca, 2002), were not observed.
At weaning, GVAD and Cyp1b1 deletion each decreased body weight by approximately 10 percent, which was further decreased by the combination (Cyp1b1−/− GVAD) (Figure 2B). The Cyp1b1 deletion effect on body length (Figure 2C) and decreased male epididymal fat pad suggest that body weight decreases were due to fat loss (Figure 2D). Liver weights were decreased by 15 percent with GVAD and Cyp1b1 deletion (Figure 2E), which represents a selective effect, as heart weight was unaffected (data not shown).
3.4 GVAD diet decreased liver RE content
Retinoid content prior to weaning has not previously been assessed after maternal dietary VA deficiency. Liver RE content relative to organ weight increased approximately five-fold between birth and weaning and a further four-fold between weaning and 14 weeks of age (Supplementary Figure 2A). Despite low accumulation at birth, liver retinol and RE content were decreased three-fold by GVAD (Supplementary Figure 2B). At weaning, serum retinol was unaffected by either GVAD or Cyp1b1 deletion. Hepatic RE content in Cyp1b1−/− mice was twice that of WT controls (p-value<0.01), while remaining susceptible to near complete retinoid depletion (Figure 2F).
3.5 Altered R-Cyp1b1−/− mice characteristics are established pre-weaning and link to retinol
R-Cyp1b1−/− mice showed appreciable differences at weaning from Cyp1b1−/− mice. Body weight of R-Cyp1b1−/− mice was larger than Cyp1b1−/− mice and increased with GVAD (Figure 3A). Some of the increased R-Cyp1b1−/− body weight derives from adipose with a two-fold increase on the LF12 diet and a four-fold increase with GVAD (Figure 3B). Parallel increases in body length, liver (Figure 3C and D), kidney, and heart weights (Supplementary Figure 3) indicate faster development.
Figure 3.
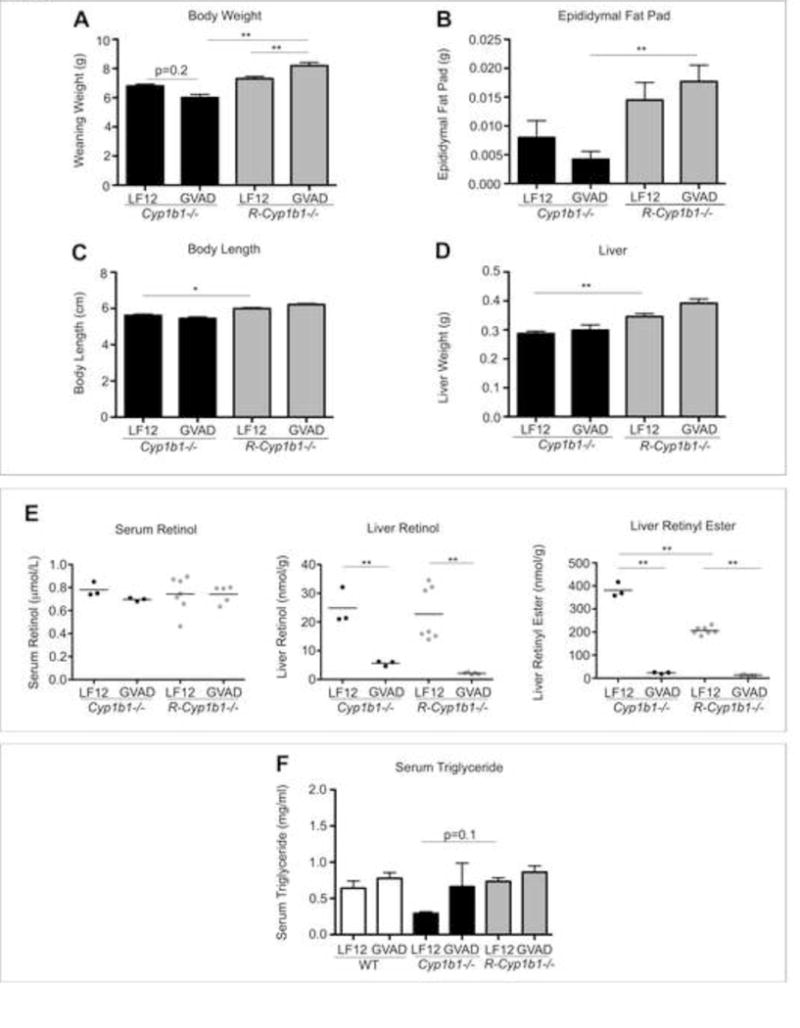
Cyp1b1−/− and R-Cyp1b1−/− pups respond differently to GVAD. (A) Body weight, (B) epididymal fat pad, (C) nose to rump length, and (D) liver weight in Cyp1b1−/− and R-Cyp1b1−/− weanling pups (PN21) administered a gestational vitamin a sufficient (LF12) or deficient (GVAD) diet. (E) Serum retinol, liver retinol, and liver retinyl ester content. (F) Serum triglyceride measurements of WT, Cyp1b1−/−, and R-Cyp1b1−/− LF12 and GVAD pups at weaning. *p-value≤0.05, **p-value≤0.01
Liver retinoid content in R-Cyp1b1−/− mice depleted similarly to the WT and Cyp1b1−/− with GVAD. However, the R-Cyp1b1−/− mouse liver did not replicate the increased RE storage evident in Cyp1b1−/− mice (Figures 2F and 3E).
Due to the changes in growth and adiposity, we measured serum triglyceride content. Serum triglyceride was not affected by GVAD. However, Cyp1b1 deletion caused a two-fold decrease (p-value>0.3), with an indication of reversal with the combination GVAD Cyp1b1−/− treatment. R-Cyp1b1−/− mice did not replicate the Cyp1b1−/− suppression of serum triglycerides, but rather trended to an increase, similarly to the effect on adipose (Figure 3F).
3.6 Ontogenic liver gene expression
The developmental growth period from birth through lactation is highly active and is a major period of adipocyte differentiation (Billon and Dani, 2012), retinoid storage (Satre et al., 1992), and liver maturation (Lee et al., 2012). Liver genes that demonstrated differential expression between birth and weaning (Weaning/Birth) in WT pups from LF12-fed dams were sorted by DAVID Functional Annotation Database (Huang da et al., 2009a, Huang da et al., 2009b). Erythropoiesis declined (Supplementary Table 4A) and xenobiotic/drug metabolism and energy homeostasis increased (Supplementary Table 4B) during this period.
3.7 Hamp suppression at birth by GVAD and Cyp1b1−/− did not affect erythropoiesis
Among the gene expression changes produced by GVAD and Cyp1b1 deletion, the appreciable suppression of Hamp mRNA stands out. Hamp mRNA generates precursor protein and iron suppressor hormone, hepcidin. For convenience, we will refer to Hamp mRNA, but hepcidin (Hepc) expression. Dietary VA impacts serum iron homeostasis through hepcidin (da Cunha et al., 2014). Hepc is expressed exclusively in hepatocytes and regulates iron homeostasis by inhibition of ferroportin, an iron transport channel, to prevent gut absorption and macrophage recycling of erythrocytes (Ganz, 2013). Fetal iron is delivered by transplacental transfer (Gambling et al., 2011), with less effect by Hepc (Feng et al., 2012). At birth, Hepc is expressed in the liver at approximately half of adult levels. Hepc exhibited 60 percent suppression by GVAD and 90 percent by Cyp1b1 deletion (Table 3). The combination Cyp1b1−/− GVAD removed the extra suppression linked to Cyp1b1 deletion (Figure 4A). This implicates two pathways for Hepc regulation, with opposing effects of retinol: direct retinol signaling and Cyp1b1-mediated signaling with retinol inhibition. In the 14-week old adult mouse, Hepc was no longer sensitive to Cyp1b1 deletion, while GVAD decreased Hepc by nearly two-fold (Table 2).
Table 3.
Hepcidin dysregulation affects iron homeostasis, lipid uptake, and stress responses
| PN0 - Birth | PN21 - Weaning | |||||
|---|---|---|---|---|---|---|
| Fold change upper versus | GVAD | Cyp1b1−/− LF12 | Weaning LF12 | GVAD | Cyp1b1−/− LF12 | Cyp1b1−/− GVAD |
| lower | LF12 | LF12 | Birth LF12 | LF12 | LF12 | Cyp1b1−/− LF12 |
| Iron Homeostasis | ||||||
| Hamp | −2.5* | −8.8** | −5.9** | −79.3** | −77.9** | 9.6** |
| Hamp2 | −1.5 | −1.9 | 187.1** | −50.2** | −28.0** | 3.7** |
| Hfe2 | n.s. | 4.1** | 1.7 | n.s. | 2.2 | n.s. |
| Ftl1 | −1.5 | 9.7** | 5.7* | −1.7 | 1.6 | n.s. |
| Ftl2 | −1.6 | 9.7** | 5.7 | −1.5 | 1.9 | n.s. |
| Gabarap | n.s. | 5.7** | 2.9 | n.s. | 2.0 | n.s. |
| Tfrc | n.s. | 1.5 | −1.9** | n.s. | 2.5** | −1.8** |
| Tfr2 | n.s. | −1.6* | −1.6* | n.s. | n.s. | n.s. |
| Trf | n.s. | 5.1** | 3.5* | n.s. | 2.2 | n.s. |
| Hmox1 | n.s. | 3.4** | −1.6 | n.s. | 1.5 | n.s. |
| Erythropoiesis | ||||||
| Hbb-bt | n.s. | n.s. | −3.8** | n.s. | 1.5 | n.s. |
| Gp9 | n.s. | −1.7* | −48.7** | n.s. | 3.2** | −2.1** |
| Epor | n.s. | −1.7 | −10.1** | n.s. | n.s. | n.s. |
| Metabolism | ||||||
| Rbp1 | n.s. | n.s. | −2.4** | n.s. | 1.5* | n.s. |
| Lpl | n.s. | n.s. | −1.6 | 1.5 | 1.9** | n.s. |
| Ppp1r3g | 2.1 | n.s. | 1.5 | −9.4** | −7.3** | n.s. |
| Scd1 | n.s. | n.s. | 86.9** | −4.3** | −12.2** | 1.9 |
| Stress Response | ||||||
| S100a9 | n.s. | n.s. | −135.7** | 1.5 | 4.0** | −4.1** |
| Krt23 | n.s. | n.s. | −4.0** | n.s. | 2.8** | −2.0** |
| Mt2 | 1.5 | n.s. | −6.7** | n.s. | 2.3** | −4.0** |
| Growth Regulation | ||||||
| Afp | n.s. | n.s. | −646.1** | n.s. | 16.6** | −4.1** |
| Igf2 | n.s. | −1.5 | −849.9** | n.s. | 33.3** | −14.5** |
| H19 | n.s. | 1.7* | −14.0** | n.s. | 7.5** | −4.0* |
| Igfbp2 | −1.5 | 1.5* | −3.5** | n.s. | 2.9* | n.s. |
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values where the net FC < 1.5 and p-value > 0.05
Figure 4. Developmental uncoupling of hepcidin genes, Hepc and Hamp2.
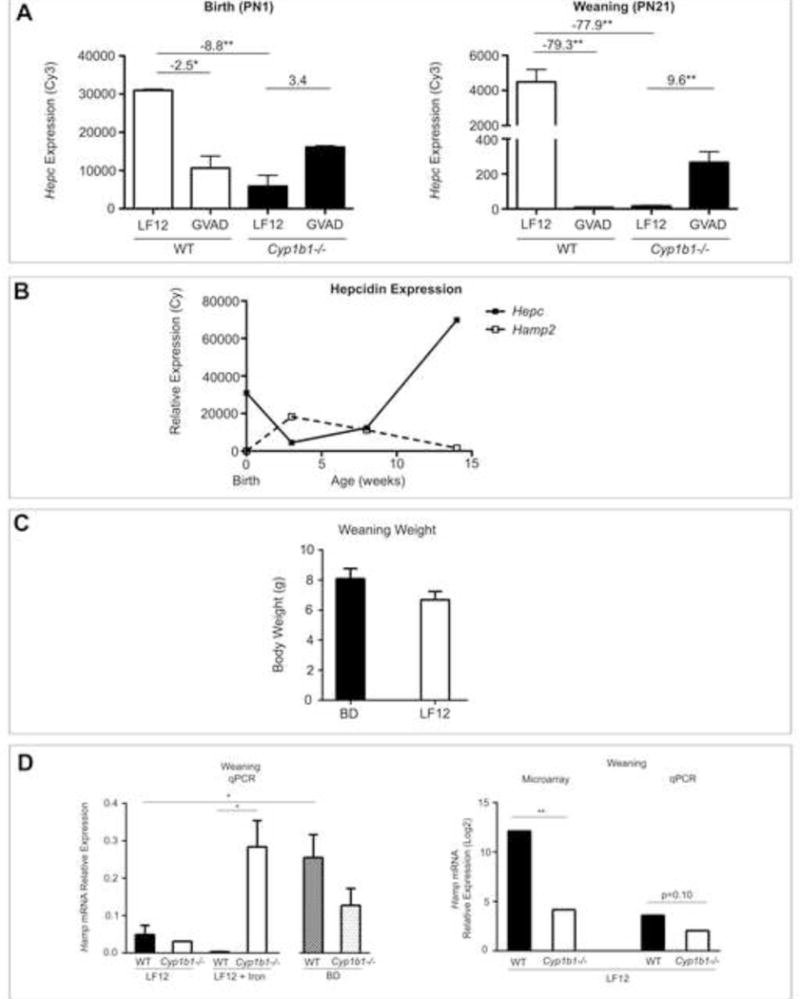
(A) Hepcidin (Hamp/Hepc) expression (Cy3) at birth (PN0) and at weaning (PN21). Fold change values are shown from EDGE3 processed data. (B) Raw relative expression (Cy3) microarray values of Hepc (solid line) and Hamp2 (dotted line) from birth through 14 weeks of age. (C) Body weight at weaning in WT and Cyp1b1−/− pups from dams fed the LF12 diet, LF12 + Iron diet or standard breeder diet (BD). (D) Hamp mRNA expression was measured by qPCR at weaning in WT and Cyp1b1−/− pups on the BD, LF12, and LF12 + Iron diet. Comparison between microarray and qPCR Hamp expression in WT and Cyp1b1−/− pups on the LF12 diet is highlighted. *p-value≤0.05, **p-value≤0.01
Large decreases in hepcidin elevate hepatic iron concentrations (Ganz, 2013) and the potential for uptake into hepatocytes. Iron storage is mediated by the interaction of transferrin (Trf) with transferrin receptors (Tfrc and Tfr2), which releases iron through endosomes (Anderson and Vulpe, 2009). The mRNA expression for these receptors was unaffected by GVAD at birth (Table 3). However, Trf expression at birth increased five-fold. There were increases in the intracellular storage protein, ferritin light chain (Ftl1 and Ftl2), which can also be exported to function as an iron transporter (Feng et al., 2012). Cyp1b1−/− pups at birth showed ten-fold increases in both Ftl1 and Ftl2 (Table 3). Heme oxygenase 1 (Hmox1), which liberates iron from heme, also increased substantially. Hemojuvelin (Hfe2) is a plasma membrane effector of Hamp mRNA transcription (Kautz et al., 2009) and was substantially increased in Cyp1b1−/− pups at birth (Table 3). Gabarap is another candidate regulator, a suppressor of Rac1, a potential attenuator of endosomal iron import (Green et al., 2002), and was highly stimulated in Cyp1b1−/− mice at birth. Gabarap, Hfe2, Trf, Hmox1 and the Ftl subunits showed increases in Cyp1b1−/− mice at weaning (Table 3).
In adult mice, iron typically controls ferritin activity by changing the translation rate, rather than at the level of mRNA (Milic et al., 2016). However, the Ftl mRNA expression at this early stage of development was over ten times lower than in mature livers. This response and the increase in Trf likely correspond to an advancement of development.
Despite the importance of Hepc in controlling the turnover of erythrocytes by macrophage (Ganz, 2013), the overall effects of GVAD and Cyp1b1 deletion at birth on major hematopoiesis markers, including hemoglobins (Hbb) and the erythropoietin receptor (Epor) were minimal (Table 3). Several genes more broadly associated with hematopoiesis showed small increases in Cyp1b1−/− mice at weaning, which were reversed by the combination Cyp1b1−/− GVAD, such as Tfrc and glycoprotein 9 (Gp9) (Table 3).
Small GVAD suppression effects on erythropoiesis were demonstrated shortly after weaning (PN33). The precursor reticulocytes and mature red blood cells each decreased 20–25 percent in GVAD mice, whereas Cyp1b1−/− mice reversed this trend. Leukocytes were decreased two-fold by GVAD in both WT and Cyp1b1−/− mice (Supplementary Figure 4).
3.8 Stellate cells are targeted by GVAD and Cyp1b1 deletion at birth
The few responses to GVAD at birth were mostly shared with Cyp1b1−/− mice and corresponded to markers of stellate cell activation (Table 4). GVAD and Cyp1b1 deletion increased the expression of smooth muscle actin (Acta2/Sma), Col1a1 and four other collagen genes, two matrix proteins [fibullin 2 (Fbln2), and lumican (Lum)], and Timp2. As with the Hepc expression pattern (Figure 4A), GVAD treatment of Cyp1b1−/− mice decreased the effect of Cyp1b1 deletion alone.
Table 4.
Stellate cell marker expression increases at birth
| PN0 - Birth | PN21 - Weaning | ||||
|---|---|---|---|---|---|
| Fold change upper versus | GVAD | Cyp1b1−/− LF12 | Cyp1b1−/− GVAD | GVAD | Cyp1b1−/− LF12 |
| lower | LF12 | LF12 | Cyp1b1−/− LF12 | LF12 | LF12 |
| Acta2/Sma | 1.5 | 1.7 | −1.6 | −2.2** | −2.7** |
| Col1a1 | 3.4** | 3.9* | −1.8 | n.s. | n.s. |
| Col1a2 | 2.6* | 4.6** | −1.5 | n.s. | 1.5. |
| Col3a1 | 2.8* | 2.5 | −1.8 | n.s. | 1.6* |
| Col8a1 | 1.6 | 3.7** | −1.5 | −1.6* | n.s. |
| Col5a2 | 1.5 | 1.6 | n.s. | n.s. | 1.5 |
| Timp1 | 2.0* | n.s. | n.s. | n.s. | 1.5 |
| Timp2 | 1.7* | 1.9* | n.s. | n.s. | n.s. |
| Cav1 | 2.5** | 2.0 | n.s. | n.s. | n.s. |
| Fbln2 | 2.5* | 2.1* | −1.5 | n.s. | n.s. |
| Lum | 2.4* | 2.0* | n.s. | n.s. | 1.8** |
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values, FC<1.5 and p-value>0.05
Early perinatal stellate cells may be different from those in adult livers. An approximately fourfold increase at birth in the expression of collagen (Col) and myofibroblastic transition genes (Mmp, Timp, Cav1, Fbln2, and Lum) is coupled with a three-fold decrease in the retinoid content by GVAD (Supplementary Figure 2). However, the Cyp1b1−/− livers were not depleted of retinoids when analyzed at weaning. This shift suggests an increase in a population of partially activated stellate cells that retain retinoid capacity (D’Ambrosio et al., 2011), perhaps corresponding to the active growth phase of the liver. Expression of these genes equilibrates at weaning, also paralleling the diminished increase in the iron-regulatory genes.
Activation of stellate cells is typically produced by oxidative stress. However, there are few signs of such stress in GVAD or Cyp1b1−/− mice at birth (Table 3).
3.9 Hamp2 increased after birth and was suppressed by GVAD and Cyp1b1 deletion
Hepc expression decreased six-fold during the lactation period and was also further suppressed by both GVAD and Cyp1b1 deletion (Figure 4A and B). At weaning, GVAD treatment of Cyp1b1−/− mice continued to diminish the effect of Cyp1b1 deletion. Despite the near complete removal of Hepc, which causes iron overload in adult mice, there was little sign of oxidative stress markers, such as NF-κB or NRF2-responsive genes (data not shown). The iron homeostasis adaptations seen at birth were not as prominent at weaning (Table 3). Dietary iron for the pre-weanling pup is transferred through lactation and most of the iron in milk is bound by lactoferrin, which delivers iron by a separate transfer process and bypasses ferroportin (Frazer et al., 2011).
Hamp2 represents a gene duplication at the same locus as Hepc, which shares control by Usf. Hamp2 shares 68 percent sequence similarity with Hepc. Each suppresses iron uptake, but Hamp2 is less effective and likely provides alternative regulatory features (Ilyin et al., 2003, Lou et al., 2004). Hamp2 showed very different developmental regulation, with low expression at birth, but a large post-natal increase that elevates expression to four times higher than Hepc at weaning (Figure 4B). The differential regulation suggests an alternative function for Hamp2 (Lou et al., 2004, Lu et al., 2015, Tjalsma et al., 2011). Hamp2 expression exhibited near complete suppression by GVAD and Cyp1b1 deletion and, again, GVAD reversed the extent of the effect of Cyp1b1 deletion (Table 5).
Table 5.
Neonatal metabolic gene stimulation is prevented by GVAD and Cyp1b1 deletion
| Fold change upper versus | Weaning LF12 | GVAD | Cyp1b1−/− LF12 | Cyp1b1−/− GVAD |
|---|---|---|---|---|
| lower | Birth LF12 | LF12 | LF12 | Cyp1b1−/− LF12 |
| Hamp2 | 187.1** | −50.2** | −28.0** | 3.7** |
| Fatty Acid Synthesis | ||||
| Srebp-1c | 4.9** | n.s. | n.s. | n.s. |
| Mlxipl/Chrebp | 4.4** | n.s. | −2.6** | n.s. |
| Srebp2 | 1.5 | n.s. | −1.5* | 2.0** |
| Acaca | 7.6** | −2.4** | −3.7** | 1.5** |
| Acacb | 7.9** | −3.0** | −12.3** | 1.7* |
| Me1 | 8.4** | −4.1** | −3.6** | 1.4* |
| Fasn | 28.3** | −3.3** | −12.2** | 2.7** |
| Scd1 | 86.9** | −4.3** | −12.2** | 1.9 |
| Elovl6 | 6.0** | −5.3** | −27.8** | 2.8** |
| Acly | 10.8** | −2.6** | −8.4** | 1.5* |
| Mup4 a | 73.8** | −3.9** | −7.3** | 2.2** |
| Thrsp | 136.6** | −1.7 | −10.0** | 4.6** |
| Cholesterol Metabolism | ||||
| Acss2 | 8.6** | −3.5** | −7.0** | 2.4** |
| Aacs | 10.4** | −2.7** | −17.5** | 4.5** |
| Idi1 | 4.7** | −1.6* | −10.6** | 10.1** |
| Sqle | 2.4* | −2.2** | −23.0** | 26.0** |
| Hmgcr | 4.0** | n.s. | −6.4** | 4.2** |
| Lss | 1.7* | n.s. | −3.4** | 5.0** |
| Hmgcs1 | 3.5** | n.s. | −4.7** | 3.8** |
| Cyp51 | 2.3* | −1.9* | −5.8** | 7.2** |
| Fdps | 4.1** | n.s. | −6.8** | 8.0** |
| Sc4mol | 3.5* | n.s. | −7.1** | 7.9** |
| Fdft1 | 3.1** | n.s. | −1.9 | 2.2** |
| Dhcr7 | 3.7** | −1.6 | −5.8** | 3.6** |
| Gadd45γ | 6.2** | −6.7** | −6.5** | 2.1 |
| Ppp1r3g | 1.5 | −9.4** | −7.3** | n.s. |
Additional Mup genes that follow the same expression pattern include: Mup1, Mup3, and Mup5
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values, FC<1.5 and p-value>0.05
3.10 Effects of iron and other components of maternal diet on Hepc expression
The high neonatal demand for iron in the developing tissues and the absence of stress gene expression markers suggest that the low expression of Hepc is due to the homeostatic response to limited availability of iron (Ramakrishnan et al., 2015). The six-fold decrease in Hepc in WT mice on LF12 between birth and weaning (Table 3 and Figure 4B) may represent an adaptation to maximize iron availability. By contrast, in adult animals fed a diet with normal iron content, comparable decreases in Hepc expression are accompanied by iron overload and severe oxidative stress (Ganz, 2013). Such decreases are produced by deletion of key stimulatory genes (Bmp6, Hfe, Hfe2).
To test this further, we consulted the suppliers on the iron content of the two maternal diets. The defined LF12 diet has three times less iron than the standard BD, which is supplemented to meet the extra demands of pregnancy (36ppm and 100ppm, respectively). Nevertheless, each diet produced pups in similar number and weight (Figure 4C). The LF12 diet produced much lower Hamp mRNA expression than the BD in WT male offspring (Figure 4D). In each case, the appreciable variation in Hamp expression within the BD and LF12 treatment groups correlated with post-natal body weight (BD r2=0.69 and LF12 r2=0.29) (data not shown). These correlations suggest that variations in lactational nutrition can diminish iron transfer with a consequent decrease in Hepc expression. Milk availability equally determines weight gain as a function of caloric intake. We find that weight differences at weaning are typically removed within 2-3 weeks of post-weaning ad libitum feeding.
Supplementation of the LF12 diet to match the iron content in the BD (LF12 + Iron, 100ppm) failed to increase mean Hamp mRNA expression in WT mice (Figure 4D). Differences in the Hamp expression between BD and LF12, therefore, derive from other components of the diet.
Equivalent analyses were carried out in male Cyp1b1−/− mice. Again, there was no impact of dietary iron on the weight or number of offspring (data not shown). The qPCR analyses confirmed the extensive decrease in Hamp mRNA expression for individual LF12 Cyp1b1−/− pups (Figure 4D). Remarkably, in contrast to WT mice, Hamp expression in Cyp1b1−/− pups was highly stimulated by the increase in dietary iron supplementation (LF12 + Iron) and substantially exceeded the expression for WT mice on this diet. This establishes that the lack of response to elevated iron in the WT mice on the LF12 diet is not due to poor bioavailability (Hubbard et al., 2013). The difference in Hamp mRNA expression between LF12 and BD evidently depends on Cyp1b1 and the impact of different diet components. The appreciable increase in the carbohydrate/fat ratio for the LF12 diet is a likely contributor, especially with respect to the impact of Cyp1b1 on fat homeostasis. These effects of diet and Cyp1b1 deletion in male pups were similar in litter-matched females (Larsen, data not shown).
3.11 Developmental increase in lipogenesis is delayed by GVAD and Cyp1b1 deletion
A role for hepcidin as a metabolic sensor has been suggested (Vuppalanchi et al., 2014). In the post-natal period, Hamp2 is expressed in parallel with a set of genes that convert acetyl-CoA to either oleate or cholesterol (Table 5). These pathways comprise seven genes for synthesis of oleic acid, including the key regulators of the divergence between mitochondrial oxidation and triglyceride synthesis (Acacb and Scd1) (Flowers and Ntambi, 2008) and 12 genes for synthesis of cholesterol, with HmgCoA reductase (Hmgcr) as a focal enzyme (Horton et al., 1998) (Figure 5A).
Figure 5. Pathways for synthesis and regulation of oleate and cholesterol from acetyl CoA and regulation.
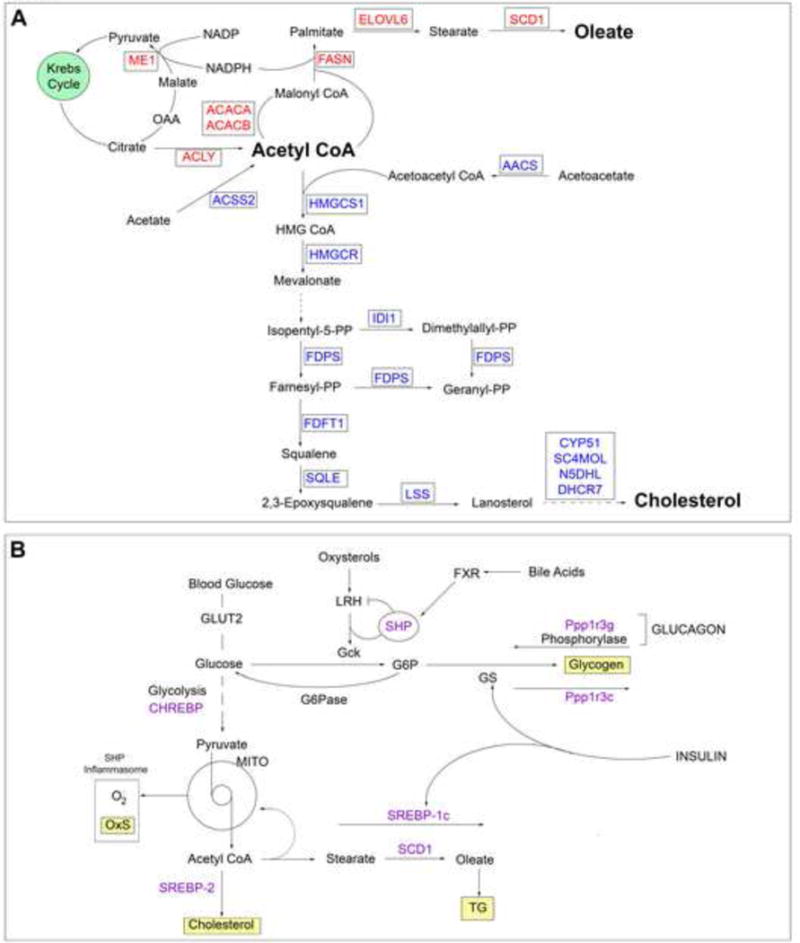
Pathways of synthesis (A) and regulation (B) highlight the role of the lipogenic genes shown in Table 5.
The regulation of these lipogenic genes by Cyp1b1 and retinol exhibits remarkable conservation of the response characteristics within each pathway, including a systematic difference in the pattern for gene response between the two branches of acetyl CoA metabolism. Like the Hepc and Hamp2 responses, the suppressions in Cyp1b1−/− mice were appreciably alleviated when combined with GVAD treatment. For the fatty acid synthesis genes, GVAD and Cyp1b1 deletion effects were similar and the reversal by the Cyp1b1−/− GVAD combination was modest. The cholesterol synthesis genes showed more effective suppression by Cyp1b1 deletion and near complete reversal by GVAD treatment of Cyp1b1−/− mice.
Sterol regulatory element binding proteins (SREBPs) control lipogenic gene expression (Eberle et al., 2004). Srebp-1c and Chrebp/Mlxipl control fatty acid synthesis (Caputo et al., 2014, Flowers and Ntambi, 2008), whereas Srebp2 is selective for cholesterol synthesis (Horton et al., 1998) (Figure 5B). Srebp-1c increased nearly five-fold between birth and weaning, but was unaffected by either GVAD or Cyp1b1 deletion. Chrebp and Srebp2 were each appreciably expressed and unaffected by GVAD, but moderately suppressed by Cyp1b1 deletion. Elsewhere, post-translational SREBP activity has been linked to iron homeostasis through AMPK-directed phosphorylation (Huang et al., 2013, Shah et al., 2016), possibly with different effectiveness for the selective pathway mediators.
Five other genes appear to be implicated in this lipogenic network based on their shared response. Four major urinary protein (Mup) genes, small molecule carrier proteins linked to fat homeostasis (Chen et al., 2015) and thyroid hormone responsive protein (Thrsp), a strongly expressed regulator of lipogenesis, each followed the oleate expression pattern (Table 5).
Based on parallel expression, the nuclear regulator growth arrest and DNA damage inducible γ (Gadd45γ) (Niehrs and Schafer, 2012), may mediate these lipogenesis changes (Table 5). Gadd45γ is highly expressed and exhibits a six-fold increase between birth and weaning. The same decrease by both GVAD and Cyp1b1 deletion show that each prevent the post-natal increase in Gadd45γ. The two-fold reversal of Cyp1b1 deletion suppression by GVAD fits the pattern of responses by oleic acid genes. Gadd45γ may mediate this global lipogenesis change by partnering with other transcription factors to direct DNA demethylation and transcription in development (Gierl et al., 2012, Johnen et al., 2013, Warr et al., 2012).
One further important gene that showed extensive shared suppression is the glycogen phosphorylase activator, Ppp1r3g, which was nearly completely suppressed by GVAD and Cyp1b1−/− mice (Table 5). This change suggests a redirection from lipid synthesis to glycogen storage, as we have previously described in adult Cyp1b1−/− mice (Larsen et al., 2015). This process is also sensitive to AMPK regulation (Zois and Harris, 2016).
We have used qPCR measurements on mRNA from livers of single pups to confirm the gene expression relationship between birth and weaning for a select set of lipogenic genes. Scd1, Fasn, and Elovl6 each displayed suppression by GVAD and Cyp1b1 deletion. The recovery and increased expression of coordinate Scd1 expression at 14 weeks of age is evident in GVAD mice (GVAD-LF12) (Figure 6). Most lipogenic genes suppressed at weaning recover their expression as adults (Supplementary Table 5).
Figure 6. Major fatty acid metabolism gene expression responses by GVAD and Cyp1b1 deletion.
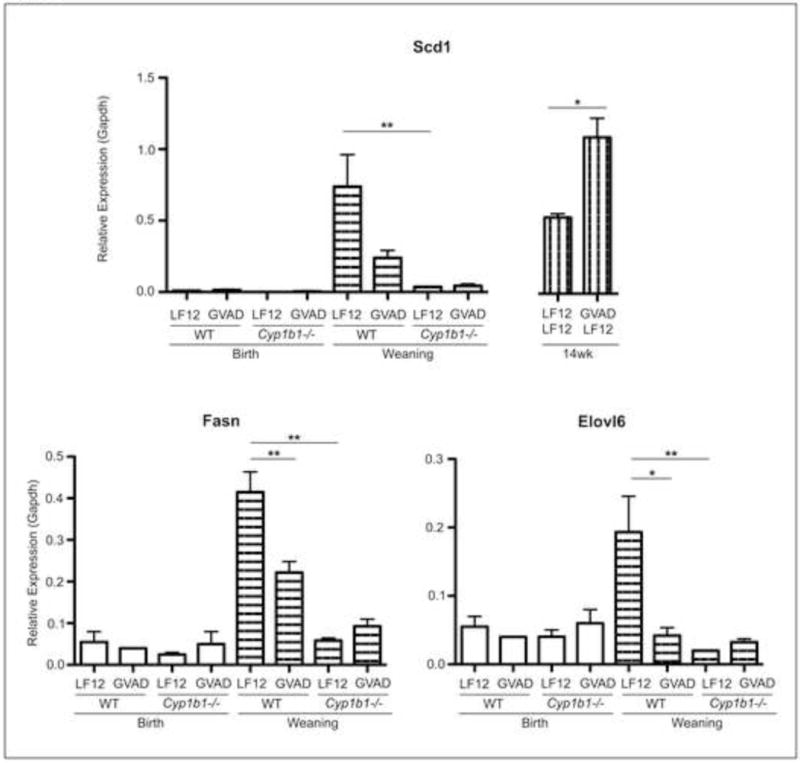
Lipogenic liver mRNA was measured in C57Bl/6J (WT) and Cyp1b1−/− vitamin A sufficient (LF12) or deficient (GVAD) offspring by qPCR, which replicated array results. *p-value≤0.05, **p-value≤0.01
3.12 Stimulation of Afp and Igf2 in Cyp1b1−/− mice is reversed by GVAD
Several other genes show a stimulatory response pattern at weaning, notably α-fetoprotein (Afp), insulin-like growth factor 2 (Igf2), and its opposite chromosome partner H19. Afp is typically regarded as a marker of hepatoblast development that initiates expression at approximately E8.5 and peaks at E17 then shows a post-natal decline through PN7 (Lee et al., 2012). Igf2 also shows very high perinatal expression with a similar acute post-natal decline. Expression of the major growth regulator, Igf2/H19, is highly dependent on epigenetic modulation of methylation status (Lee et al., 2014). Each matched the cholesterol pathway signature with increases in Cyp1b1 deletion and insensitivity to GVAD, except in combination (Table 3). In most respects, these large stimulations at weaning in Cyp1b1−/− mice represent a delay in the post-natal decreases. The suppression of lipogenic genes also corresponds to delays in the post-natal increase of these genes. An increase in the binding protein, Igfbp2, may balance the increase in Igf2. S100a9, Mt2, and Krt23, which also show similar stimulation patterns, are each markers of stress signaling (Table 3).
3.13 Liver gene expression is not maintained in R-Cyp1b1−/− mice at weaning
We have examined the GVAD and Cyp1b1 deletion responsive genes at weaning with respect to R-Cyp1b1−/− mouse liver gene expression. Most responses shared by GVAD and Cyp1b1 deletion were not seen in R-Cyp1b1−/− mice. However, some responses were completely or partially retained and some were selective to R-Cyp1b1−/− mice. The gene responses shared by Cyp1b1−/− and R-Cyp1b1−/− mice were genes that did not show a GVAD reversal under Cyp1b1 deletion conditions. The R-Cyp1b1−/− mice show normal liver retinoid depletion (Figure 3), but appear to lose gene responses.
The extensive suppression of the fatty acid and cholesterol synthesis genes was not retained in R-Cyp1b1−/− neonates. This resistance extended to Thrsp and the four Mup genes. Moderate suppression of modulatory gene expression (Ppp1r3g, Lpin1, Gadd45γ, Hepc, Hamp2) was observed (Table 6). None of these genes showed GVAD sensitivity when applied to R-Cyp1b1−/− pups (Table 6).
Table 6.
Lipogenic gene expression is not conserved between Cyp1b1−/− and R-Cyp1b1−/− at weaning
| Fold change upper versus | GVAD | Cyp1b1−/− LF12 | R-Cyp1b1−/− LF12 | R-Cyp1b1−/− GVAD |
|---|---|---|---|---|
| lower | LF12 | LF12 | LF12 | R-Cyp1b1−/− LF12 |
| GVAD and Cyp1b1−/− responsive | ||||
| Me1 | −4.1** | −3.6** | n.s. | −1.6 |
| Fasn | −3.3** | −12.2** | n.s. | −1.7 |
| Scd1 | −4.3** | −12.2** | 2.2** | n.s. |
| Elovl6 | −5.3** | −27.8** | n.s. | −2.2 |
| Idi1 | −1.6* | −10.6** | n.s. | −1.6 |
| Cyp51 | −1.9* | −5.8** | n.s. | n.s. |
| Gadd45 | −6.7** | −6.5** | −1.8** | −1.6 |
| Hamp2 | −50.2** | −28.0** | −5.3** | n.s. |
| Hamp | −79.3** | −77.9** | −5.0** | n.s. |
| Shared suppression | ||||
| Acta2 | −2.2** | −2.7** | −3.2** | n.s. |
| Ppp1r3g | −9.4** | −7.3** | −3.4** | n.s. |
| Rgs16 | −3.0** | −5.9** | −2.0* | n.s. |
| Lpin1 | −2.9** | −4.5** | −1.8** | n.s. |
| Egfr | −2.2** | −1.8** | −2.6** | 1.5 |
| Dmbt1 | −6.0** | −25.4** | −14.3** | n.s. |
| Dbp | −1.8 | −5.4** | −5.2* | 2.1 |
| Plk3 | −2.7* | −3.4** | −2.6** | n.s. |
| Ifi27l2b | −2.1* | −3.1** | −2.5** | n.s. |
| Chac1 | −2.7** | −3.0** | −2.3** | n.s. |
| Cyp2s1 | −2.1* | −2.1** | −2.6** | n.s. |
| Cyp1b1−/− selective | ||||
| Gp1ba | n.s. | 2.1** | 1.5* | n.s. |
| Gp9 | n.s. | 3.2** | n.s. | n.s. |
| Tfrc | n.s. | 2.3** | n.s. | n.s. |
| Afp | n.s. | 16.6** | n.s. | n.s. |
| Igf2 | n.s. | 31.9** | n.s. | n.s. |
| H19 | n.s. | 7.5** | 2.1 | −1.9* |
| R-Cyp1b1−/− selective | ||||
| Osgin1 | 1.7* | n.s. | 2.1** | −1.5 |
| Acot1 | n.s. | n.s. | 5.2** | n.s. |
| Cyp7a1 | n.s. | n.s. | −4.9** | 2.3* |
| Nnmt | n.s. | n.s. | −3.7** | n.s. |
p-value ≤ 0.05
p-value ≤ 0.01
n.s. Non-significant values, FC<1.5 and p-value>0.05
Three Cyp1b1 deletion stimulations, Tfrc, Afp, and Igf2, were not affected in R-Cyp1b1−/− mice. Again, the GVAD treatment of R-Cyp1b1−/− mice failed to produce the reversal effect seen with Cyp1b1−/− mice.
On the other hand, an appreciable number of changes at weaning were shared by Cyp1b1−/− and R-Cyp1b1−/− mice. These include Dbp, Dmbt1, Plk3, Chac1, and Cyp2s1. A notable feature of this cluster is that neither Cyp1b1−/− nor R-Cyp1b1−/− mice showed reversal of the response when administered the GVAD treatment. Lastly, several gene changes were specific to R-Cyp1b1−/− mice, including Acot1, Osgin1, Cyp7a1, and Nnmt1 (Table 6).
3.14 Many other markers of hepatocyte development were not impacted by GVAD or Cyp1b1 deletion at weaning
We addressed whether other gene pathways that activate in the post-natal period were affected. Phase I [P450 cytochromes (Cyp)] and phase II [glutathione S-transferases (Gst), N-acetyltransferases (Nat), sulfotransferases (Sult), and UDP-glucuronosyltransferases (Ugt)] xenobiotic metabolism gene expression increases from E16.5 to weaning and is a marker of liver hepatocyte maturation (Lee et al., 2012). None of these genes were affected by GVAD or Cyp1b1 deletion (Supplementary Table 6). Phase I Cyp genes respond extensively in adult Cyp1b1−/− mice through suppression of activity of lipid-sensitive receptors (PPARα, CAR, PXR, LXR) (Bushkofsky et al., 2016, Larsen et al., 2015). These genes are largely unaffected in this post-weaning period, thus emphasizing that these shifts in Cyp1b1−/− and GVAD mice do not represent general delays in liver development.
4. Discussion
Maternal retinol deficiency (GVAD) and Cyp1b1 deletion each prevented the obesity response to a post-weaning HFD (Figure 1B). Cyp1b1−/− mice demonstrate diet-selective stimulations of signaling by growth hormone and leptin (Bushkofsky et al., 2016, Larsen et al., 2015) that were insensitive to GVAD (Table 1). Here, we identified a cluster of adult liver genes that exhibit elevated expression on the low fat/high carbohydrate diets and that were similarly suppressed by Cyp1b1 deletion and GVAD (Table 2). These genes are mostly inflammation markers that are also suppressed by deletion of Shp/Nr0b2 (Kim et al., 2014) (Supplementary Figure 1). We have shown that the response changes in Cyp1b1−/− mice compared to WT mice correlate remarkably with the changes reported in Shp−/− mice (Maguire et al., 2017). Shp redirects postprandial metabolism from glycogen synthesis to glycolysis and lipogenesis (Garruti et al., 2012) (Figure 5B). Cyp1b1−/−livers show this increase in glycogen (Larsen et al., 2015). The response appears to derive from a high carbohydrate diet (LF12 or LFD) rather than a high fat diet (HFD). These expression changes correlate with suppression of Ppp1r3g, an activator of glycogen phosphorylase and, thereby, glycolysis (Luo et al., 2011, Zois and Harris, 2016). Ppp1r3g was suppressed extensively by both GVAD and Cyp1b1 deletion. Increased glycolysis activates mitochondrial oxidative stress, a potential mediator for this activation of inflammation markers (Gornicka et al., 2011).
We have hypothesized that Cyp1b1 and retinol initiate these changes early in hepatocyte development. This hypothesis is supported by four major inter-connected perinatal and neonatal liver changes. GVAD and Cyp1b1 deletion each suppress neonatal Ppp1r3g and lipogenesis pathways derived from acetyl CoA. These changes are preceded at birth by an extensive decrease in Hamp mRNA expression, which produces hepcidin, a peptide suppressor of iron release into the circulation. There is also a large coordinated increase in multiple markers of stellate cell activation. The lipogenic and stellate responses to Cyp1b1 deletion are retinol dependent. Each response is also selectively lost in variant R-Cyp1b1−/− mice. These mice were selected from the normal Cyp1b1−/− mice based on a failure to show the characteristic obesity suppression. They also lack the adult liver gene expression features of normal Cyp1b1−/− mice (Table 1). These specific response patterns point to a central perinatal retinol-sensitive change in Cyp1b1−/− mouse development that is absent in the R-Cyp1b1−/− variant. This change affects Hepc, stellate cells, and the onset of lipogenesis. Clearly, altered iron homeostasis can have a profound impact. The suppression effects on adiposity and serum triglycerides appear at weaning (Figures 2D, 3B, 3F) and not only match the lipogenesis changes, but also the adult changes in obesity (Figure 1B and C).
The connections between the 18 genes in the lipogenic pathways (Table 5, Figure 5A) are likely made through the three key regulators, Srebp-1c and Chrebp for fatty acid gene synthesis (Caputo et al., 2014, Eberle et al., 2004, Flowers and Ntambi, 2008) and Srebp-2 for cholesterogenic genes (Horton et al., 1998, Shimano et al., 1997). Each is primarily regulated by proteolytic cleavage by the activator Scap and inhibitory phosphorylation by AMPK (Eberle et al., 2004). The two pathways differ in their suppression selectivity, suggesting impact on their respective central mediators (Figure 5). Cholesterol synthesis genes are weakly suppressed by GVAD compared to the oleic acid pathway, but show a stronger reversal of the Cyp1b1 deletion effects by GVAD. This response signature is shared by Gadd45γ, a major regulator of DNA demethylation and developmental processes (Gierl et al., 2012, Johnen et al., 2013, Kaufmann et al., 2011, Warr et al., 2012), including brown fat regulation (Gantner et al., 2014).
These post-natal lipogenesis changes are paralleled by suppression of Hamp2, a duplicate gene at the hepcidin locus with few known functions. The predominant expression of Hamp2 (8/25 amino acid substitutions) at weaning compared to Hepc suggests a role in the linkage between iron homeostasis and metabolism, particularly as Hamp2 exhibits specific tissue distribution to the pancreas (Ilyin et al., 2003, Lou et al., 2004). In the adult mouse, Hepc is again more highly expressed, but is then relatively insensitive to GVAD and Cyp1b1 deletion, whereas Hamp2 mRNA is highly responsive (Table 2).
R-Cyp1b1−/− mice retain more Hamp/Hepc at weaning than Cyp1b1−/− mice, but lose all neonatal lipogenesis suppression and gene responses to GVAD (Table 6), despite normal liver retinol depletion (Figure 3E). While R-Cyp1b1−/− mice share some of the predominant Cyp1b1 deletion responses, they also exhibit changes that are unique to this variant. Elevated body and liver weights at weaning in R-Cyp1b1−/− mice are consistent with more efficient metabolism and the increased lipogenesis (Figure 3A and D; Table 6). R-Cyp1b1−/− responses that are not retained from the normal Cyp1b1−/− mice are highlighted by large increases in Afp and Igf2. Each response may be functionally important since Afp sequesters estradiol (Brock et al., 2012), a potential substrate for Cyp1b1, while Igf2 replaces Igf1 during early development (Lee et al., 2014).
The stellate cell activation signature seen at birth (Table 4) in GVAD and Cyp1b1−/− mice also occurs in adult WT mice when they are maintained on the maternal and post-weaning LF12 diet (Supplementary Table 7). This stellate cluster is then accompanied by the cluster of inflammatory markers that link to Shp and Ppp1r3g (Table 2) (Maguire et al., 2017). The stellate activation by GVAD may derive from the retinoid depletion (Supplementary Figure 1) (Wallace et al., 2015), but occurs without retinoid depletion in Cyp1b1−/− pups (Figure 2F). These stellate markers may, therefore, represent a semi-activated basal state that has been identified with appreciable retinyl ester content (D’Ambrosio et al., 2011). Thus, the stellate cells seem to be playing a distinctive developmental role in the expression of collagen and other markers in WT perinatal livers, which were four times higher than in the adult livers, while retinoid storage is ten times lower.
The perinatal suppression of Hamp/Hepc and aberrant iron homeostasis seemed at first to be a likely cause of these retinoid-sensitive changes in Cyp1b1−/− mice. However, there were few markers of oxidative stress typical of iron overload (Ganz, 2013). Other dietary factors are clearly involved, notably the different proportion of carbohydrate and fat that distinguish the LF12 diet from the BD. The maternal LF12 diet produced much lower Hepc expression than BD, even when dietary iron was elevated to equal levels. By contrast, Cyp1b1−/− pups showed even less Hepc expression than their WT counterparts, however, elevation of dietary iron reversed this effect. Thus, the effect of Cyp1b1 deletion on Hepc is dependent on the composition of the LF12 diet (i.e., high carbohydrate/fat ratio), and this effect is much greater when iron is in short supply. Iron homeostasis during pregnancy needs to sustain the high iron demand from multiple progeny (Feng et al., 2012, Ramakrishnan et al., 2015), involving Hepc control of ferroportin in the placenta (Gambling et al., 2011) or transfer from the maternal milk, which can bypass enterocyte ferroportin through lactoferrin (Frazer et al., 2011).
Endothelial Bmp6 and stimulation of Hepc in hepatocytes provides an alternative core target for Cyp1b1 (Canali et al., 2017). The key role of Cyp1b1 in the containment of oxidative stress in endothelia (Palenski et al., 2013, Tang et al., 2009), and the extensive effects of deletion on morphology and gene expression suggests that Bmp6 or endothelial-hepatocyte interactions are critically important. Bmp6 plays a key role not only in regulating Hepc, but also in coordinating iron regulation, stellate cells, and lipogenesis (Arndt et al., 2015). Bmp6 stimulates Bmpr1a/Bmpr2 receptors to signal through SMAD1/5/8 phosphorylation (Canali et al., 2017) (Figure 7) in partnership with the stimulation of transferrin receptors (Trfc, Tfr2) by iron-transferrin complexes (Kautz et al., 2009). These complexes also depend on the activator, hemojuvelin (Hfe2). Our data is consistent with an adaptation of hepatocytes to diminished Bmp6 signaling through large increases in Hfe2, ferritin (Ftl) and transferrin (Trf) in the Cyp1b1−/− pups at birth (Table 3). The low levels of Bmp6 mRNA were not affected by GVAD or Cyp1b1 deletion (data not shown), but the local sinusoidal endothelial-hepatocyte interactions may be more important. The susceptibility of Hfe2 to dietary differences provided by BD and LF12 offer a better test for involvement of the Bmp6 pathway.
Figure 7. Proposed mechanism for vitamin A and CYP1B1 modulation of lipogenic genes through hepcidin expression.
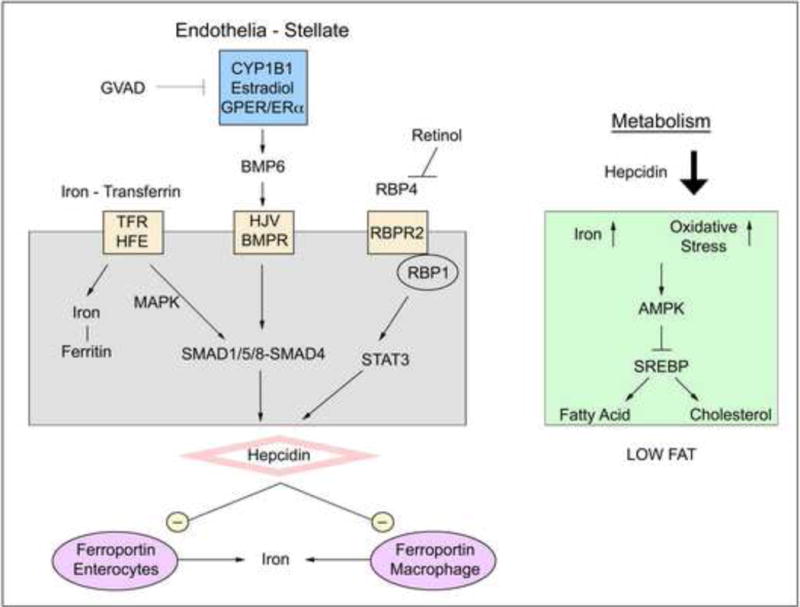
Hepcidin signaling is mediated by iron content, retinol, and CYP1B1 metabolism of estradiol, likely through GPR30/GPER/BMP6. Hepcidin acts to inhibit ferroportin, which releases iron into circulation from gut enterocytes or macrophage. Near depletion of hepcidin expression caused by GVAD, Cyp1b1 deletion, or the combination leads to increased iron concentrations and oxidative stress, which represses SREBP-mediated gene expression, including fatty acid and cholesterol synthesis genes.
A third contribution is delivered through STAT3 cytokine signaling when activated by Il6 (Armitage et al., 2011) and the serum binding protein, Rbp4 (Alapatt et al., 2013, Noy, 2016). This RBP4/STAT3 pathway may explain the reversal of Hepc formation and other activities by GVAD in Cyp1b1−/− mice (Figure 7).
These considerations leave the central question of how Cyp1b1 protects vascular cells from oxidative stress. Estradiol activation of GPER, an alternative estrogen receptor that acts in cell membranes of endothelial cells, has been implicated in BMP6 synthesis (Ikeda et al., 2012). ERα has similar activities at other sites. Vascular Cyp1b1 metabolism of estradiol to 16α-hydroxyestrone (Austin et al., 2012, White et al., 2012) or 2-methoxy estradiol generates these activities (Sharma et al., 2017). We have recently found that male and female pups from the same litter show remarkably similar WT and Cyp1b1 deletion effects on Hepc expression (Larsen, unpublished). However, estradiol generated by local aromatase activity may be more important here than systemic ovarian estradiol. Identification of estradiol metabolites in perinatal and post-natal WT and Cyp1b1−/− mice is therefore a priority.
Supplementary Material
Summary.
Hypothesis
Cyp1b1 deletion and retinol deficiency produce adult changes in hepatic metabolism through perinatal interventions that affect later development.
Cyp1b1 deletion and retinol deficiency share four broad expression changes:
Adult suppression of postprandial expression linked to glycogen metabolism and inflammation.
Neonatal suppression of 18 lipogenic genes controlled by Srebp forms.
Perinatal stimulation of 10 genes marking stellate activation.
Perinatal and Neonatal suppression of Hamp/Hepc, the source of the iron suppressor hepcidin.
Highlights.
Cyp1b1 and retinol maintain hepcidin expression in the neonatal liver.
Hepcidin suppression in Cyp1b1−/− mice is reversed by retinol depletion.
Cyp1b1−/− and GVAD activate stellate cells (PN0) and suppress lipogenesis (PN21).
These responses in Cyp1b1−/− mice are reversed by retinol depletion.
Each deficiency prevents glucose-induced oxidative stress by lowering Ppp1r3g.
Acknowledgments
We would like to thank Dr. Chad Vezina and Dr. Susan Smith for their comments and insight on the manuscript. We would also like to thank Dr. Jessica Flowers for her nutritional insight and dietary design.
Funding Sources
This work was supported by the National Institutes of Health [R01 DK090249, T32 HD041921] and by funding from the University of Wisconsin School of Medicine and Public Health Department of Cell and Regenerative Biology.
Abbreviations
- BD
standard breeder diet
- Cyp1b1
cytochrome P450 1b1
- DIO
diet-induced obesity
- E
embryonic day
- GVAD
gestational initiation of vitamin A deficient diet
- HFD
high fat diet
- LF12
novel low fat diet with 12 percent kcal from fat
- LF12 + Iron
LF12 diet supplemented with ferrous sulfate
- LFD
low fat diet
- PN
post-natal day
- R-Cyp1b1−/−
a substrain of Cyp1b1−/− mice that are resistant to DIO suppression
- RA
retinoic acid
- RE
retinyl ester
- VA
Vitamin A
- VAD diet
vitamin A deficient LF12 diet
- WT
wild type, C56Bl/6J
References
- Alapatt P, Guo F, Komanetsky SM, Wang S, Cai J, Sargsyan A, Rodriguez Diaz E, Bacon BT, Aryal P, Graham TE. Liver retinol transporter and receptor for serum retinol-binding protein (RBP4) J Biol Chem. 2013;288:1250–65. doi: 10.1074/jbc.M112.369132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GJ, Vulpe CD. Mammalian iron transport. Cell Mol Life Sci. 2009;66:3241–61. doi: 10.1007/s00018-009-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage AE, Eddowes LA, Gileadi U, Cole S, Spottiswoode N, Selvakumar TA, Ho LP, Townsend AR, Drakesmith H. Hepcidin regulation by innate immune and infectious stimuli. Blood. 2011;118:4129–39. doi: 10.1182/blood-2011-04-351957. [DOI] [PubMed] [Google Scholar]
- Arndt S, Wacker E, Dorn C, Koch A, Saugspier M, Thasler WE, Hartmann A, Bosserhoff AK, Hellerbrand C. Enhanced expression of BMP6 inhibits hepatic fibrosis in non-alcoholic fatty liver disease. Gut. 2015;64:973–81. doi: 10.1136/gutjnl-2014-306968. [DOI] [PubMed] [Google Scholar]
- Austin ED, Hamid R, Hemnes AR, Loyd JE, Blackwell T, Yu C, Phillips JA, III, Gaddipati R, Gladson S, Gu E, West J, Lane KB. BMPR2 expression is suppressed by signaling through the estrogen receptor. Biol Sex Differ. 2012;3:6. doi: 10.1186/2042-6410-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol. 2012;56:952–64. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Billon N, Dani C. Developmental origins of the adipocyte lineage: new insights from genetics and genomics studies. Stem Cell Rev. 2012;8:55–66. doi: 10.1007/s12015-011-9242-x. [DOI] [PubMed] [Google Scholar]
- Bonet ML, Ribot J, Palou A. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim Biophys Acta. 20121821:177–89. doi: 10.1016/j.bbalip.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Brock O, Keller M, Douhard Q, Bakker J. Female mice deficient in alpha-fetoprotein show female-typical neural responses to conspecific-derived pheromones. PLoS One. 2012;7:e39204. doi: 10.1371/journal.pone.0039204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushkofsky JR, Maguire M, Larsen MC, Fong YH, Jefcoate CR. Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4alpha and PPARalpha. Arch Biochem Biophys. 2016;597:30–47. doi: 10.1016/j.abb.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7, 12-dimethylbenz[a]anthracene-induced lymphomas. Proc Natl Acad Sci U S A. 1999;96:1977–82. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canali S, Zumbrennen-Bullough KB, Core AB, Wang CY, Nairz M, Bouley R, Swirski FK, Babitt JL. Endothelial cells produce bone morphogenetic protein 6 required for iron homeostasis in mice. Blood. 2017;129:405–414. doi: 10.1182/blood-2016-06-721571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo M, De Rosa MC, Rescigno T, Zirpoli H, Vassallo A, De Tommasi N, Torino G, Tecce MF. Binding of polyunsaturated fatty acids to LXRalpha and modulation of SREBP-1 interaction with a specific SCD1 promoter element. Cell Biochem Funct. 2014;32:637–46. doi: 10.1002/cbf.3067. [DOI] [PubMed] [Google Scholar]
- Chambers D, Wilson L, Maden M, Lumsden A. RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development. 2007;134:1369–1383. doi: 10.1242/dev.02815. [DOI] [PubMed] [Google Scholar]
- Chambers D, Wilson LJ, Alfonsi F, Hunter E, Saxena U, Blanc E, Lumsden A. Rhombomere-specific analysis reveals the repertoire of genetic cues expressed across the developing hindbrain. Neural Dev. 2009;4:6. doi: 10.1186/1749-8104-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Lee TY, Kwok CF, Hsu YP, Shih KC, Lin YJ, Ho LT. Major urinary protein 1 interacts with cannabinoid receptor type 1 in fatty acid-induced hepatic insulin resistance in a mouse hepatocyte model. Biochem Biophys Res Commun. 2015;460:1063–8. doi: 10.1016/j.bbrc.2015.03.155. [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Schenkman JB, Sarfarazi M, Stoilov I. Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Archives of Biochemistry and Biophysics. 2003;414:91–100. doi: 10.1016/s0003-9861(03)00174-7. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr. 2002;22:347–81. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio DN, Walewski JL, Clugston RD, Berk PD, Rippe RA, Blaner WS. Distinct populations of hepatic stellate cells in the mouse liver have different capacities for retinoid and lipid storage. PLoS One. 2011;6:e24993. doi: 10.1371/journal.pone.0024993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha MS, Siqueira EM, Trindade LS, Arruda SF. Vitamin A deficiency modulates iron metabolism via ineffective erythropoiesis. J Nutr Biochem. 2014;25:1035–44. doi: 10.1016/j.jnutbio.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–48. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Feng Q, Migas MC, Waheed A, Britton RS, Fleming RE. Ferritin upregulates hepatic expression of bone morphogenetic protein 6 and hepcidin in mice. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1397–404. doi: 10.1152/ajpgi.00020.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol. 2008;19:248–56. doi: 10.1097/MOL.0b013e3282f9b54d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer DM, Darshan D, Anderson GJ. Intestinal iron absorption during suckling in mammals. Biometals. 2011;24:567–74. doi: 10.1007/s10534-011-9429-2. [DOI] [PubMed] [Google Scholar]
- Gambling L, Lang C, McArdle HJ. Fetal regulation of iron transport during pregnancy. Am J Clin Nutr. 2011;94:1903S–1907S. doi: 10.3945/ajcn.110.000885. [DOI] [PubMed] [Google Scholar]
- Gantner ML, Hazen BC, Conkright J, Kralli A. GADD45gamma regulates the thermogenic capacity of brown adipose tissue. Proc Natl Acad Sci U S A. 2014;111:11870–5. doi: 10.1073/pnas.1406638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93:1721–41. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- Garruti G, Wang HH, Bonfrate L, de Bari O, Wang DQ, Portincasa P. A pleiotropic role for the orphan nuclear receptor small heterodimer partner in lipid homeostasis and metabolic pathways. J Lipids. 2012;2012:304292. doi: 10.1155/2012/304292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl MS, Gruhn WH, von Seggern A, Maltry N, Niehrs C. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev Cell. 2012;23:1032–42. doi: 10.1016/j.devcel.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Gornicka A, Morris-Stiff G, Thapaliya S, Papouchado BG, Berk M, Feldstein AE. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxid Redox Signal. 2011;15:437–45. doi: 10.1089/ars.2010.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green F, O’Hare T, Blackwell A, Enns CA. Association of human transferrin receptor with GABARAP. FEBS Letters. 2002;518:101–106. doi: 10.1016/s0014-5793(02)02655-8. [DOI] [PubMed] [Google Scholar]
- Harrison EH. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim Biophys Acta. 2012:70–7. doi: 10.1016/j.bbalip.2011.06.002. 1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–9. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Chen SW, Langston AW, Gudas LJ. A conserved retinoic acid responsive element in the murine Hoxb-1 gene is required for expression in the developing gut. Development. 1998;125:3235–46. doi: 10.1242/dev.125.16.3235. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Simcox J, Mitchell TC, Jones D, Cox J, Luo B, Cooksey RC, Boros LG, McClain DA. Iron regulates glucose homeostasis in liver and muscle via AMP-activated protein kinase in mice. FASEB J. 2013;27:2845–54. doi: 10.1096/fj.12-216929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard AC, Bandyopadhyay S, Wojczyk BS, Spitalnik SL, Hod EA, Prestia KA. Effect of dietary iron on fetal growth in pregnant mice. Comp Med. 2013;63:127–35. [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Tajima S, Izawa-Ishizawa Y, Kihira Y, Ishizawa K, Tomita S, Tsuchiya K, Tamaki T. Estrogen regulates hepcidin expression via GPR30-BMP6-dependent signaling in hepatocytes. PLoS One. 2012;7:e40465. doi: 10.1371/journal.pone.0040465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin G, Courselaud B, Troadec MB, Pigeon C, Alizadeh M, Leroyer P, Brissot P, Loréal O. Comparative analysis of mouse hepcidin 1 and 2 genes: evidence for different patterns of expression and co-inducibility during iron overload1. FEBS Letters. 2003;542:22–26. doi: 10.1016/s0014-5793(03)00329-6. [DOI] [PubMed] [Google Scholar]
- Jennings BL, George LW, Pingili AK, Khan NS, Estes AM, Fang XR, Gonzalez FJ, Malik KU. Estrogen metabolism by cytochrome P450 1B1 modulates the hypertensive effect of angiotensin II in female mice. Hypertension. 2014;64:134–40. doi: 10.1161/HYPERTENSIONAHA.114.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnen H, Gonzalez-Silva L, Carramolino L, Flores JM, Torres M, Salvador JM. Gadd45g is essential for primary sex determination, male fertility and testis development. PLoS One. 2013;8:e58751. doi: 10.1371/journal.pone.0058751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann LT, Gierl MS, Niehrs C. Gadd45a, Gadd45b and Gadd45g expression during mouse embryonic development. Gene Expr Patterns. 2011;11:465–70. doi: 10.1016/j.gep.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Kautz L, Meynard D, Besson-Fournier C, Darnaud V, Al Saati T, Coppin H, Roth MP. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114:2515–20. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- Kim SC, Kim CK, Axe D, Cook A, Lee M, Li T, Smallwood N, Chiang JY, Hardwick JP, Moore DD, Lee YKV. All-trans-retinoic acid ameliorates hepatic steatosis in mice by a novel transcriptional cascade. Hepatology. 2014;59:1750–60. doi: 10.1002/hep.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R. Breeding Strategies for Maintaining Colonies of Laboratory Mice: A Jackson Laboratory Resource Manual. The Jackson Laboratory 2007 [Google Scholar]
- Larsen MC, Bushkofsky JR, Gorman T, Adhami V, Mukhtar H, Wang S, Reeder SB, Sheibani N, Jefcoate CR. Cytochrome P450 1B1: An unexpected modulator of liver fatty acid homeostasis. Arch Biochem Biophys. 2015;571:21–39. doi: 10.1016/j.abb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Barraza-Villarreal A, Biessy C, Duarte-Salles T, Sly PD, Ramakrishnan U, Rivera J, Herceg Z, Romieu I. Dietary supplementation with polyunsaturated fatty acid during pregnancy modulates DNA methylation at IGF2/H19 imprinted genes and growth of infants. Physiol Genomics. 2014;46:851–7. doi: 10.1152/physiolgenomics.00061.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Ward WO, Knapp G, Ren H, Vallanat B, Abbott B, Ho K, Karp SJ, Corton JC. Transcriptional ontogeny of the developing liver. BMC Genomics. 2012;13:33. doi: 10.1186/1471-2164-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Jiang C, Larsen MC, Bushkofsky J, Krausz KW, Wang T, Jefcoate CR, Gonzalez FJ. Lipidomics reveals a link between CYP1B1 and SCD1 in promoting obesity. J Proteome Res. 2014;13:2679–87. doi: 10.1021/pr500145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou DQ, Nicolas G, Lesbordes JC, Viatte L, Grimber G, Szajnert MF, Kahn A, Vaulont S. Functional differences between hepcidin 1 and 2 in transgenic mice. Blood. 2004;103:2816–21. doi: 10.1182/blood-2003-07-2524. [DOI] [PubMed] [Google Scholar]
- Lu S, Seravalli J, Harrison-Findik D. Inductively coupled mass spectrometry analysis of biometals in conditional Hamp1 and Hamp1 and Hamp2 transgenic mouse models. Transgenic Res. 2015;24:765–73. doi: 10.1007/s11248-015-9879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Zhang Y, Ruan X, Jiang X, Zhu L, Wang X, Ding Q, Liu W, Pan Y, Wang Z, Chen Y. Fasting-induced protein phosphatase 1 regulatory subunit contributes to postprandial blood glucose homeostasis via regulation of hepatic glycogenesis. Diabetes. 2011;60:1435–45. doi: 10.2337/db10-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M, Bushkofsky JR, Larsen MC, Foong YH, Tanumihardjo SA, Jefcoate CR. Diet-dependent retinoid effects on liver gene expression include stellate and inflammation markers and parallel effects of the nuclear repressor, Shp. The Journal of Nutritional Biochemistry. 2017 doi: 10.1016/j.jnutbio.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik KU, Jennings BL, Yaghini FA, Sahan-Firat S, Song CY, Estes AM, Fang XR. Contribution of cytochrome P450 1B1 to hypertension and associated pathophysiology: a novel target for antihypertensive agents. Prostaglandins Other Lipid Mediat. 2012;98:69–74. doi: 10.1016/j.prostaglandins.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy PT, Cerecedo LR. Vitamin A deficiency in the mouse. J Nutr. 1952;46:361–76. doi: 10.1093/jn/46.3.361. [DOI] [PubMed] [Google Scholar]
- Milic S, Mikolasevic I, Orlic L, Devcic E, Starcevic-Cizmarevic N, Stimac D, Kapovic M, Ristic S. The Role of Iron and Iron Overload in Chronic Liver Disease. Medical Science Monitor. 2016;22:2144–2151. doi: 10.12659/MSM.896494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–67. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–7. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Noy N. Vitamin A in regulation of insulin responsiveness: mini review. Proc Nutr Soc. 2016;75:212–5. doi: 10.1017/S0029665115004322. [DOI] [PubMed] [Google Scholar]
- Oliveira MC, Menezes-Garcia Z, Henriques MC, Soriani FM, Pinho V, Faria AM, Santiago AF, Cara DC, Souza DG, Teixeira MM, Ferreira AV. Acute and sustained inflammation and metabolic dysfunction induced by high refined carbohydrate-containing diet in mice. Obesity (Silver Spring) 2013;21:E396–406. doi: 10.1002/oby.20230. [DOI] [PubMed] [Google Scholar]
- Palenski TL, Sorenson CM, Jefcoate CR, Sheibani N. Lack of Cyp1b1 promotes the proliferative and migratory phenotype of perivascular supporting cells. Lab Invest. 2013;93:646–62. doi: 10.1038/labinvest.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscaglia F, Knittel T, Kobold D, Barnikol-Watanabe S, Di Rocco P, Ramadori G. Cellular localization of hepatic cytochrome 1B1 expression and its regulation by aromatic hydrocarbons and inflammatory cytokines. Biochemical Pharmacology. 1999;58:157–165. doi: 10.1016/s0006-2952(99)00066-0. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan SK, Anderson ER, Martin A, Centofanti B, Shah YM. Maternal intestinal HIF-2alpha is necessary for sensing iron demands of lactation in mice. Proc Natl Acad Sci U S A. 2015;112:E3738–47. doi: 10.1073/pnas.1504891112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabroy N, Tanumihardjo SA. Oral doses of alpha-retinyl ester track chylomicron uptake and distribution of vitamin A in a male piglet model for newborn infants. J Nutr. 2014;144:1188–95. doi: 10.3945/jn.114.191668. [DOI] [PubMed] [Google Scholar]
- Ross AC. Diet in vitamin A research. Methods Mol Biol. 2010;652:295–313. doi: 10.1007/978-1-60327-325-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satre MA, Ugen KE, Kochhar DM. Developmental changes in endogenous retinoids during pregnancy and embryogenesis in the mouse. Biol Reprod. 1992;46:802–10. doi: 10.1095/biolreprod46.5.802. [DOI] [PubMed] [Google Scholar]
- See AW, Kaiser ME, White JC, Clagett-Dame M. A nutritional model of late embryonic vitamin A deficiency produces defects in organogenesis at a high penetrance and reveals new roles for the vitamin in skeletal development. Dev Biol. 2008;316:171–90. doi: 10.1016/j.ydbio.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Shah SA, Yoon GH, Chung SS, Abid MN, Kim TH, Lee HY, Kim MO. Novel osmotin inhibits SREBP2 via the AdipoR1/AMPK/SIRT1 pathway to improve Alzheimer’s disease neuropathological deficits. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Mauvais-Jarvis F, Prossnitz ER. Roles of G protein-coupled estrogen receptor GPER in metabolic regulation. J Steroid Biochem Mol Biol. 2017 doi: 10.1016/j.jsbmb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–54. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–89. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Smith SM, Levy NS, Hayes CE. Impaired immunity in vitamin A-deficient mice. J Nutr. 1987;117:857–65. doi: 10.1093/jn/117.5.857. [DOI] [PubMed] [Google Scholar]
- Stoilov I, Rezaie T, Jansson I, Schenkman JB, Sarfarazi M. Expression of cytochrome P4501b1 (Cyp1b1) during early murine development. Mol Vis. 2004;10:629–36. [PubMed] [Google Scholar]
- Tang Y, Scheef EA, Wang S, Sorenson CM, Marcus CB, Jefcoate CR, Sheibani N. CYP1B1 expression promotes the proangiogenic phenotype of endothelium through decreased intracellular oxidative stress and thrombospondin-2 expression. Blood. 2009;113:744–54. doi: 10.1182/blood-2008-03-145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Laarakkers CM, van Swelm RP, Theurl M, Theurl I, Kemna EH, van der Burgt YE, Venselaar H, Dutilh BE, Russel FG, Weiss G, Masereeuw R, Fleming RE, Swinkels DW. Mass spectrometry analysis of hepcidin peptides in experimental mouse models. PLoS One. 2011;6:e16762. doi: 10.1371/journal.pone.0016762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath AL, Smith AA, Craven M, Bradfield CA. EDGE(3): a web-based solution for management and analysis of Agilent two color microarray experiments. BMC Bioinformatics. 2009;10:280. doi: 10.1186/1471-2105-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi R, Troutt JS, Konrad RJ, Ghabril M, Saxena R, Bell LN, Kowdley KV, Chalasani N. Serum hepcidin levels are associated with obesity but not liver disease. Obesity (Silver Spring) 2014;22:836–41. doi: 10.1002/oby.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MC, Friedman SL, Mann DA. Emerging and disease-specific mechanisms of hepatic stellate cell activation. Semin Liver Dis. 2015;35:107–18. doi: 10.1055/s-0035-1550060. [DOI] [PubMed] [Google Scholar]
- Warr N, Carre GA, Siggers P, Faleato JV, Brixey R, Pope M, Bogani D, Childers M, Wells S, Scudamore CL, Tedesco M, del Barco Barrantes I, Nebreda AR, Trainor PA, Greenfield A. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38MAPK-mediated control of Sry expression. Dev Cell. 2012;23:1020–31. doi: 10.1016/j.devcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Johansen AK, Nilsen M, Ciuclan L, Wallace E, Paton L, Campbell A, Morecroft I, Loughlin L, McClure JD, Thomas M, Mair KM, MacLean MR. Activity of the estrogen-metabolizing enzyme cytochrome P4501B1 influences the development of pulmonary arterial hypertension. Circulation. 2012;126:1087–98. doi: 10.1161/CIRCULATIONAHA.111.062927. [DOI] [PubMed] [Google Scholar]
- Zois CE, Harris AL. Glycogen metabolism has a key role in the cancer microenvironment and provides new targets for cancer therapy. J Mol Med (Berl) 2016;94:137–54. doi: 10.1007/s00109-015-1377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


