Abstract
Background
Human retinal endothelial cells are employed increasingly for investigations of retinal vascular diseases. Analysis of gene expression response to disease-associated stimuli by reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR) is common. However, most reported work does not follow the minimum information for publication of qPCR experiments (MIQE) recommendation that multiple, stably expressed reference genes be used for normalization.
Methods
Two human retinal endothelial cell lines were treated with medium alone or containing stimuli that included: glucose at supraphysiological concentration, dimethyloxalyl-glycine, vascular endothelial growth factor, tumor necrosis factor-α, lipopolysaccharide and Toxoplasma gondii tachyzoites. Biological response of cells was confirmed by measuring significant increase in a stimulus-relevant transcript. Total RNA was reverse transcribed and analyzed by commercial PCR arrays designed to detect 28 reference genes. Stability of reference gene expression, for each and both cell lines, and for each and all conditions, was judged on gene-stability measure (M-value) less than 0.2 and coefficient of variation (CV-value) less than 0.1.
Results
Reference gene expression varied substantially across stimulations and between cell lines. Of 27 detectable reference genes, 11–21 (41–78%) maintained expression stability across stimuli and cell lines. Ranking indicated substantial diversity in the most stable reference genes under different conditions, and no reference gene was expressed stably under all conditions of stimulation and for both cell lines. Four reference genes were expressed stably under 5 conditions: HSP90AB1, IPO8, PSMC4 and RPLPO.
Conclusions
We observed variation in stability of reference gene expression with different stimuli and between human retinal endothelial cell lines. Our findings support adherence to MIQE recommendations regarding normalization in RT-qPCR studies of human retinal endothelial cells.
Keywords: Human, Retina, Endothelial Cell, Reference gene, PCR
Introduction
Diseases that involve the endothelium of the retinal vasculature are leading causes of impaired vision and blindness across the world: diabetic retinopathy; central retinal vein occlusion; retinopathy of prematurity; immune-mediated posterior uveitis; and ocular toxoplasmosis.1 In these conditions, endothelial cell dysfunction contributes to retinal vascular leakage, neovascularization and/or leukocytic or microbial infiltration. Consequently, human retinal endothelial cells are the subject of multiple in vitro studies of basic pathogenic mechanisms. Independent research groups – including our own – isolate primary retinal endothelial cells from human cadaveric eyes2,3 or purchase cells from commercial sources4,5 to undertake these studies, which commonly involve analysis by reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR).
Since its introduction over 20 years ago as a method for quantifying differences in gene expression between experimental conditions,6 RT-qPCR has become a standard and common tool in molecular research and diagnostics. Although RT-qPCR is a robust technique, methodological variations may profoundly impact output, with implications for interpretation and reproducibility of results across laboratories. The minimum information for publication of qPCR experiments (MIQE) guidelines were first proposed in 2009, to improve communication in reporting and also provide methodological standards for performing qPCR.7 One important consideration for compliance with the MIQE guidelines is the selection of endogenous reference genes – previously generically referred to as housekeeping genes – for normalization of mRNA concentrations. The guidelines require reference genes to be constitutively transcribed at the same level in all samples, regardless of source and/or environment, and further, strongly recommend multiple reference genes be employed for any normalization.7
To identify reference genes for MIQE-compliant RT-qPCR studies of human retinal endothelial cells, we exposed cell lines derived from retinae of two different human donors to stimuli commonly used to elicit disease-relevant responses: (1) glucose at supraphysiological concentration, which is the central abnormality in diabetes mellitus; (2) hypoxia simulator, dimethyloxalylglycine (DMOG); (3) key regulator of neovascularization, vascular endothelial growth factor (VEGFA); (4) master inflammatory cytokine, tumor necrosis factor (TNF)-α; (5) bacterial component, lipopolysaccharide (LPS); and (6) the cause of ocular toxoplasmosis, Toxoplasma gondii tachyzoites. We used PCR array profiling to interrogate expression of 28 reference genes in the cell lines. Our results show that human retinal endothelial cells transcribe a majority of reference genes stably under different conditions, but the exceptions to this finding vary with the stimulus and the cell population.
Materials and Methods
Overview of experimental design
Triplicate human retinal endothelial cell cultures were treated with medium only or one of six molecular or microbial stimuli that included: (1) glucose at supraphysiological concentration; (2) DMOG; (3) VEGFA; (4) TNF-α; (5) LPS; and (6) T. gondii tachyzoites. Total RNA was extracted from lysate prepared from each culture, and high quality of the RNA was verified. From every RNA preparation, cDNA was synthesized and separately analyzed by commercial PCR reference gene array. A biological response of cells to each stimulus was confirmed on the same RNA samples by RT-qPCR measurement of a stimulus-relevant transcript: (1) glucose at supraphysiological concentration, interleukin 1β (IL-1β); (2) DMOG, VEGFA; (3) VEGFA, Down syndrome critical region gene 1 (DSCR1); (4) TNF-α, intercellular adhesion molecule 1 (ICAM-1); (5) LPS, ICAM-1; and (6) T. gondii tachyzoites, suppressor of cytokine signaling 1 (SOCS1). Two human retinal endothelial cell lines, generated from eyes of two human cadaveric donors, were studied separately.
Endothelial cell and microbial culture
Our method for generating human retinal endothelial cell lines has been published.1 In summary, endothelial cells were isolated from paired human cadaver retinae (VisionGift, Portland, OR) by enzymatic digestion of tissue and selection with magnetic bead-conjugated anti-human CD31 antibody, and subsequently expanded by transduction with the mouse recombinant amphotropic retrovirus, LXSN16E6E7.8 These cell lines retain their endothelial phenotype, including expression of endothelial markers and formation of capillary-like tubes on Matrigel,1 and they have been used in multiple published studies of retinal vascular disease (e.g.9–11). Human retinal endothelial cells were cultured in MCDB-131 medium (Sigma-Aldrich, St. Louis, MO; catalogue number M8537), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (HyClone-GE Healthcare Life Sciences, Logan, UT) and endothelial growth factors (EGM-2 SingleQuots supplement, omitting FBS, hydrocortisone and gentamicin; Clonetics-Lonza, Walkersville, MD) at 37 °C and 5% CO2 in air.
GT-1 strain T. gondii were maintained in tachyzoite form by serial passage in confluent monolayers of human foreskin fibroblasts in Dulbecco’s modified Eagle’s medium (DMEM; Catalogue number, 12100; Life Technologies-Gibco, Grand Island, NY), supplemented with 44 mM sodium bicarbonate and 1% FBS, at 37 °C and at 5% CO2 in air. Plaque assays were performed using human fibroblast monolayers to verify parasite viability for each retinal endothelial cell infection: viability of 15% or greater was required, consistent with published measurements for a natural isolate.12
Reagents and proteins
D-Glucose was supplemented to a concentration of 30 mM. Dimethyloxalylglycine (Sigma-Aldrich; catalogue number D3695) was used at a working concentration of 1 mM. Recombinant human VEGFA (VEGF165) and TNF-α (both from R&D Systems, Minneapolis, MN; catalogue numbers 293-VE-010 and 210-TA-020) were used at working concentrations of 10 ng/mL and 25 ng/mL, respectively. Lipopolysaccharide from Escherichia coli 055:B5 (Sigma-Aldrich; catalogue number L6529) was used at a working concentration of 10 μg/mL.
Stimulation of human retinal endothelial cells
Human retinal endothelial cells were grown to confluence in 6-well plates, in modified MCDB-131 medium with 10% FBS and endothelial growth factors. Three monolayers on each plate were treated with one stimulus in fresh medium (i.e., glucose, DMOG, VEGFA, LPS, TNF-α and T. gondii: 2.5 × 106 freshly egressed tachyzoites per well) and the three remaining monolayers were treated with fresh medium alone. In the case of the T. gondii stimulations, FBS in the medium was reduced to 5%, to minimize effects of serum components on parasite viability. Cell monolayers were incubated for 4 hours at 37 °C and 5% CO2 in air ahead of RNA extraction, with the exception of cells treated with glucose at high concentration and respective controls; testing 11 potential targets of this stimulus indicated no biological response at 4 hours (data not shown), and therefore incubation was extended to 24 hours.
RNA isolation and reverse transcription
Retinal endothelial cell monolayers were covered with Buffer RLT (Qiagen, Hilden, Germany) containing 0.55 mM β-mercaptoethanol (Sigma-Aldrich), and cell lysates were frozen at −80 °C ahead of RNA extraction. Total RNA was extracted using the RNeasy mini kit (Qiagen), according to the manufacturer’s instructions and including the on-column DNase treatment. RNA concentration, purity and quality were determined by nanodrop spectrophotometry (NanoDrop 2000c, Thermo Scientific, Wilmington, DE) and chip-based capillary electrophoresis on the Agilent 2100 Bioanalyzer system (Agilent Technologies, Waldbronn, Germany). Only highly pure RNA, with A260/280 ratios greater than 1.8 and RNA integrity numbers over 9.0, was used for qPCR. Reverse transcription was performed using the iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA), with 500 ng RNA template per 20 μl RT reaction mix, yielding 25 ng/μl cDNA. For each RNA template, four 20 μl RT reactions were performed and cDNA was pooled, to minimize inefficiencies in individual RT reactions. This cDNA was diluted 1 in 10 with nuclease-free water, and 4 ul (containing 10 ng of cDNA) was used in subsequent qPCR.
Quantitative real-time polymerase chain reaction
To confirm induction of downstream targets of each stimulation, qPCR was performed on the CFX Connect Real-Time System (Bio-Rad, Hercules, CA) using 4 μL of cDNA, 4 μL of iQ SYBRGreen Supermix (Bio-Rad), 1.5 μL each of 20 uM forward and reverse primers (Supplementary Table 1), and 9 μL of nuclease-free water for each reaction.
Amplification consisted of: a pre-cycling hold at 95 °C for 5 minutes; 40 cycles of denaturation for 30 seconds at 95 °C; annealing for 30 seconds at 60 °C; extension for 30 seconds at 72 °C; and a post-extension hold at 72 °C for 1 second. A melting curve was produced by a 1-second hold at every 0.5 °C between 70 °C and 95 °C to confirm a single PCR amplicon. For each primer set, standard curves were generated with serially diluted amplicon to confirm an efficiency of at least 85%. Amplicon size was confirmed by electrophoresis on 2% agarose gel. Cycle threshold was measured with Cq determination mode set to regression. Relative expression was calculated in the Gene Expression Analysis module of CFX Manager v3.1 (Bio-Rad), which uses the 2−ΔΔCt method,13 normalizing to three reference genes that were stable for the specific condition and human donor by the criteria described below. Results corresponding to individual control and stimulated conditions were compared statistically by two-tailed Student’s t-test, and a significant difference between conditions was defined as one yielding a p-value less than 0.05.
Quantitative real-time polymerase chain reaction array
The qPCR arrays were performed with two PrimePCR Pathway Plates: Reference Genes H96 and Reference Genes Plus H96 (BioRad). Each of these plates contains validated and optimized primer pairs for 14 human reference gene transcripts in triplicate test and control samples (Supplementary Table 2), as well as primer pairs for detection of genomic DNA contamination, and for verifying cDNA quality and performance efficiency of qPCR. The manufacturer’s instructions were followed exactly, including the use of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and 4 μl (10 ng) of cDNA per reaction. The qPCR plates were run on CFX96 Connect Real-Time PCR Detection System, using a pre-programmed thermal cycling protocol consisting of: activation at 95 °C for 2 minutes; 40 cycles of denaturation at 95 °C for 5 seconds; annealing/extension at 60 °C for 30 seconds; and production of a melt curve from 65 °C to 95 °C, in 0.5 °C increments, for 5 seconds at each temperature.
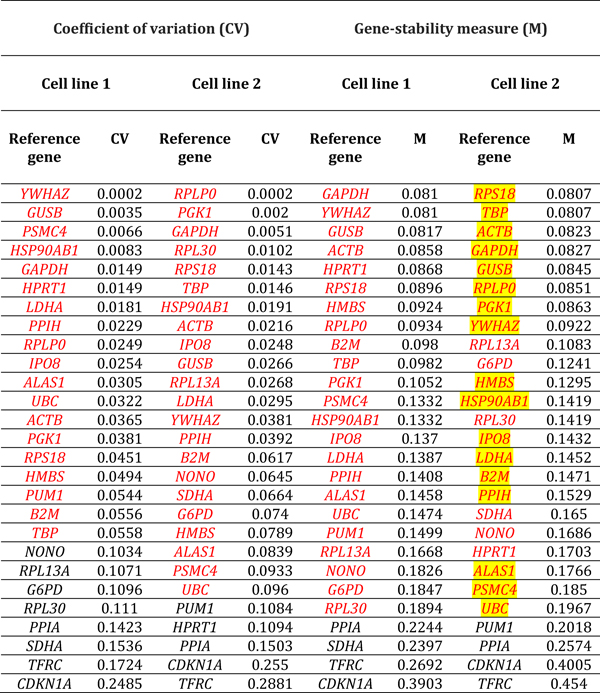
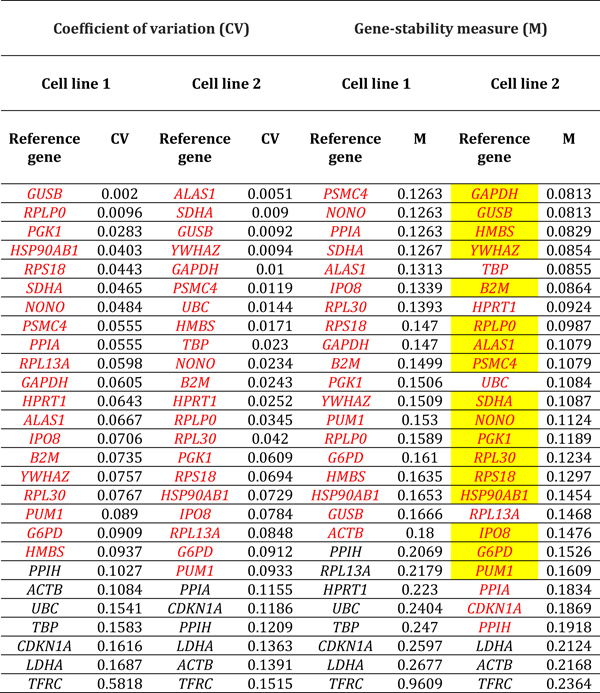
Reference gene expression was evaluated using the Gene Expression Analysis module of CFX Manager v3.1, as described above. The qBase program14 implemented within this module was used to calculate a gene-stability measure – or M-value – and a coefficient of variation – or CV-value – for each reference gene, by condition and donor. The M-value of a reference gene is the mean pairwise variation – defined as standard deviation of logarithm expression ratios – for that gene, in comparison to other reference genes tested for the same samples.15 Thus, a relatively low M-value equates with more stable expression. The CV-value represents a comparison of normalized relative quantities of reference gene across samples.14 Like the M-value, a relatively low CV-value indicates stability of expression. The CV- and M-values for each reference gene in the two retinal endothelial isolates and six conditions were ranked numerically. By applying stringent cut-offs of CV-value less than 0.1 and M-value less than 0.2, stably expressed reference genes were established for each donor and both donors, and for each condition and all conditions.
Results
Comparison of reference gene expression by human retinal endothelial cells treated with molecular or microbial stimuli
Two retinal endothelial cell lines established from paired eyes of two human cadaveric donors were treated with one of six molecular or microbial stimuli that are commonly used to study cell responses in disease: glucose at supraphysiological concentration; DMOG; VEGFA; TNF-α; LPS; and T. gondii tachyzoites; or medium alone as control for each treatment. All stimuli elicited significant increases in transcripts encoding known targets over baseline conditions, as determined by RT-qPCR, indicating biological responses by cells to all treatments: glucose, 2.3- and 1.6-fold increase in IL-1β transcript; DMOG, 7.8- and 18.8-fold increase in VEGFA transcript; VEGFA, 3.9- and 4.5-fold increase in DSCR1 transcript; TNF-α, 21.0- and 75.4-fold increase in ICAM-1 transcript; LPS, 12.5- and 12.7-fold increase in ICAM-1 transcript; and T. gondii tachyzoites, 6.4- and 8.6-fold increase in SOCS1 transcript (Figure 1).
Figure 1.
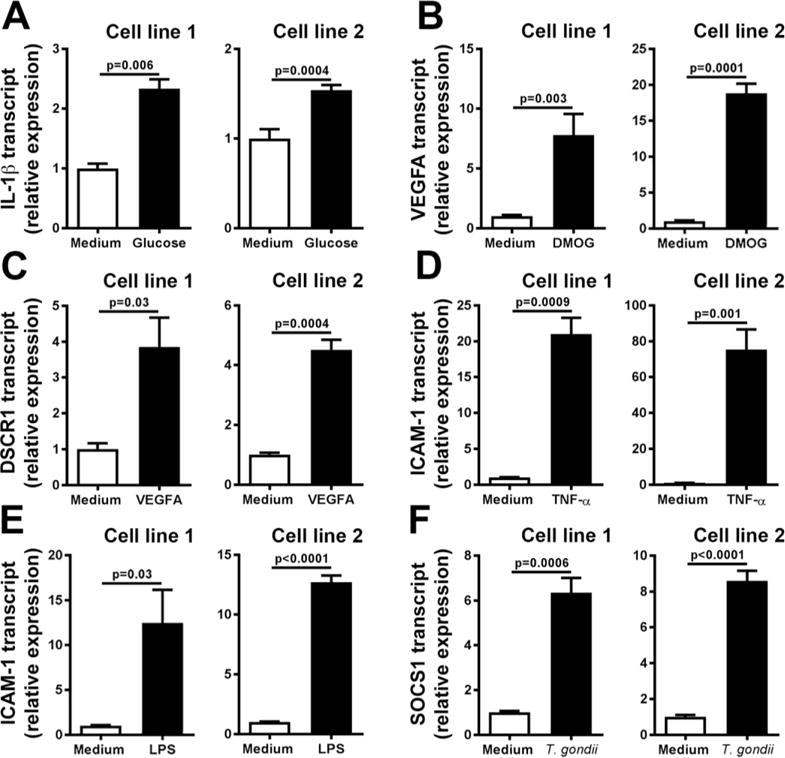
Expression of target molecules by human retinal endothelial cell lines following stimulation with: (A) glucose at supraphysiological concentration; (B) dimethyloxalylglycine (DMOG); (C) vascular endothelial growth factor (VEGFA); (D) tumor necrosis factor (TNF)-α; (E) lipopolysaccharide (LPS); and (F) Toxoplasma gondii tachyzoites, or medium alone. Three stable reference gene transcripts were applied for normalization of each result: ALAS1, SDHA and UBC (glucose); B2M, RPLP0 and YWHAZ (DMOG, VEGFA and T. gondii tachyzoites); ACTB, PPIA and RPLP0 (TNF-α); and ACTB, GAPDH and YWHAZ (LPS). Bars represent mean relative expression, with error bars showing standard error of the mean. n = 3 cultures/condition. Data were analyzed by two-tailed Student’s t-test. IL-1β = interleukin 1β; DSCR1 = Down syndrome critical region gene 1 (DSCR1); ICAM-1 = intercellular adhesion molecule 1; SOCS1 = suppressor of cytokine signaling 1.
Expression of 28 reference genes in stimulated versus control retinal endothelial cells was investigated by qPCR array, using two PrimePCR Pathway Plates, each spotted with primer pairs for 14 human reference gene transcripts in triplicate. The expression of 27 of the 28 reference genes was calculated as fold-difference for each molecular or microbial treatment relative to medium alone, following normalization to: PSMC4, IPO8 and SDHA (glucose); RPL13A, YWHAZ and G6PD (DMOG); HSP90AB1, UBC and HPRT1 (VEGFA); HSP90AB1, PSMC4 and ACTB (TNF-α); GAPDH, GUSB and YWHAZ (LPS); SDHA, ALAS1 and GUSB (T. gondii tachyzoites). The final reference gene, HBB, was removed from this analysis, based on PCR cycle threshold values that exceeded 35, indicating minimal to no expression in human retinal endothelial cells. Comparison of fold-changes revealed substantial differences in reference gene expression across stimulations. For the retinal endothelial cell line generated for donor 1, expression of reference genes varied: 0.5- to 1.1-fold for glucose at supraphysiological concentration, 0.8- to 1.7-fold for DMOG, 0.8- to 1.2-fold for VEGFA; 0.8- to 1.4-fold for TNF-α, 0.9 to 1.5-fold for LPS, and 0.8- to 2.6-fold for T. gondii tachyzoites (Figure 2). For the retinal endothelial cell line generated for donor 2, expression of reference genes varied: 0.8- to 1.3-fold for glucose at supraphysiological concentration, 0.04- to 1.7-fold for DMOG, 0.7- to 18.6-fold for VEGFA; 0.7- to 1.5-fold for TNF-α, 0.9- to 1.6-fold for LPS, and 0.8- and 1.3-fold for T. gondii tachyzoites (Figure 3). This comparison also revealed differences in reference gene expression between human retinal endothelial cell lines. Largest differences were observed for: LDHA under conditions of DMOG stimulation (1.70-fold versus 0.04-fold); RPLP0 under conditions of VEGFA stimulation (1.0-fold versus 18.6-fold); and GADPH under conditions of VEGFA stimulation (0.9-fold versus 4.3-fold), for retinal endothelial cell lines of donors 1 and 2, respectively.
Figure 2.
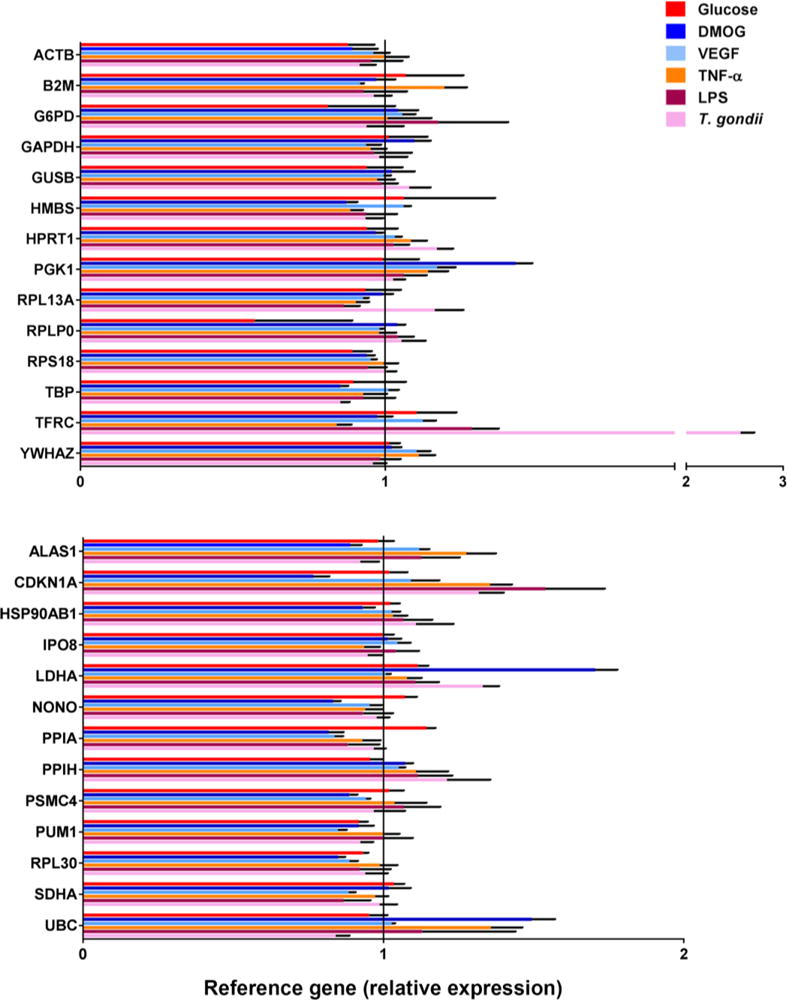
Expression of 27 reference genes by human retinal endothelial cell line 1 following treatments with: glucose at supraphysiological concentration; dimethyloxalyl-glycine (DMOG); vascular endothelial growth factor (VEGFA); tumor necrosis factor (TNF)-α; lipopolysaccharide (LPS); and Toxoplasma gondii tachyzoites. Three stable reference gene transcripts were applied for normalization of each result: PSMC4, IPO8 and SDHA (glucose); RPL13A, YWHAZ and G6PD (DMOG); HSP90AB1, UBC and HPRT1 (VEGFA); HSP90AB1, PSMC4 and ACTB (TNF-α); GAPDH, GUSB and YWHAZ (LPS); and SDHA, ALAS1 and GUSB (T. gondii tachyzoites). Colored bars represent mean expression by stimulated cells, calculated relative to expression by cells exposed to medium alone. Black error bars show standard error of the mean. n = 3 cultures/condition. Full names of reference genes with abbreviations appear in Supplementary Table 2.
Figure 3.
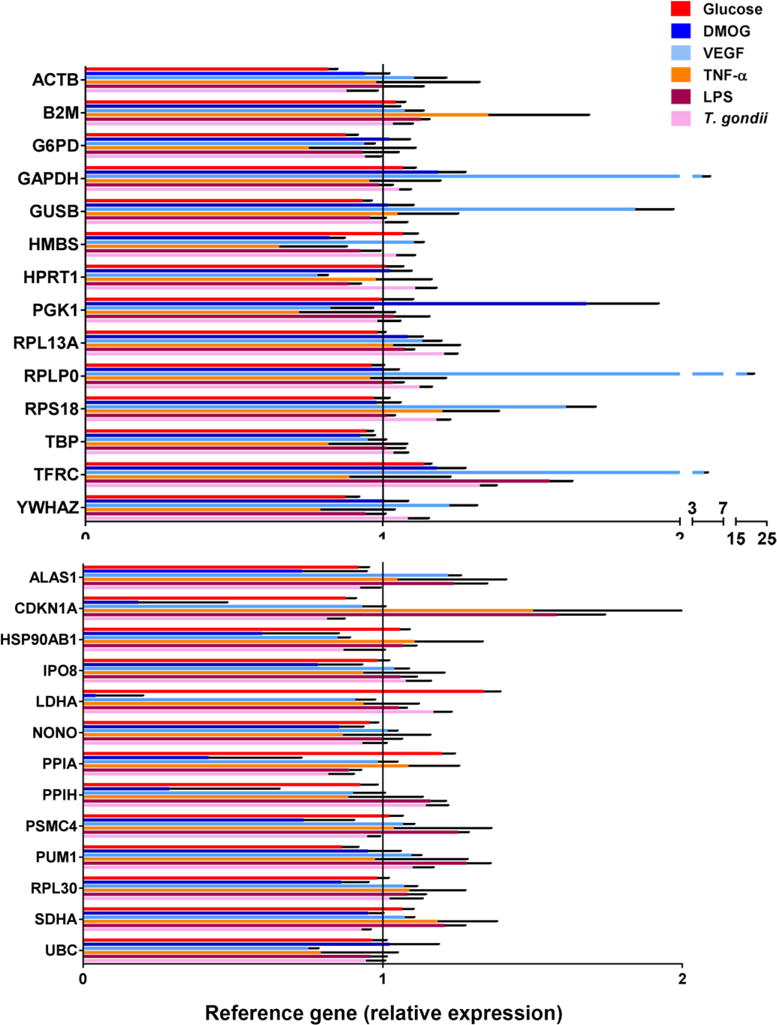
Expression of 27 reference genes by human retinal endothelial cell line 2 following treatments with: glucose at supraphysiological concentration; dimethyloxalyl-glycine (DMOG); vascular endothelial growth factor (VEGFA); tumor necrosis factor (TNF)-α; lipopolysaccharide (LPS); and Toxoplasma gondii tachyzoites. Three stable reference gene transcripts were applied for normalization of each result: PSMC4, IPO8 and SDHA (glucose); RPL13A, YWHAZ and G6PD (DMOG); HSP90AB1, UBC and HPRT1 (VEGFA); HSP90AB1, PSMC4 and ACTB (TNF-α); GAPDH, GUSB and YWHAZ (LPS); and SDHA, ALAS1 and GUSB (T. gondii tachyzoites). Colored bars represent mean expression by stimulated cells, calculated relative to expression by cells exposed to medium alone. Black error bars show standard error of the mean. n = 3 cultures/condition. Full names of reference genes with abbreviations appear in Supplementary Table 2.
Analysis of stability of reference gene expression in human retinal endothelial cells treated with molecular or microbial stimuli
Expression stability of reference genes in human retinal endothelial cells across stimulated and control conditions was assessed by M- and CV-values. Applying stringent criteria of CV-value less than 0.1 and M-value less than 0.2, we identified reference genes that demonstrated stable expression in retinal endothelial cell lines from the two human donors, under the six different conditions. By these criteria, overall 11–21 of 28 reference genes (41–78%) maintained expression stability following treatments, across stimuli and human retinal endothelial cell lines. For glucose at supraphysiological concentration, 22 and 14 references genes were stable by CV-value, and 25 and 13 references genes were stable by M-value in the two cell lines: 11 reference genes were stable by both criteria in each line (Table 1). For DMOG, 17 and 13 references genes were stable by CV-value, and 13 and 10 references genes were stable by M-value in the two cell lines: 11 reference genes were stable by both criteria in each line (Table 2). For VEGFA, 23 and 17 references genes were stable by CV-value, and 26 and 17 references genes were stable by M-value in the two cell lines: 17 and 15 reference genes were stable by both criteria (Table 3). For TNF-α, 21 and 16 references genes were stable by CV-value, and 22 and 15 references genes were stable by M-value in the two cell lines: 14 reference genes were stable by both criteria in each line (Table 4). For LPS, 18 and 21 references genes were stable by CV-value in the two cell lines, and 22 references genes were stable by M-value in each line: 21 and 16 reference genes were stable by both criteria (Table 5). For T. gondii tachyzoites, 20 and 19 references genes were stable by CV-value, and 18 and 24 references genes were stable by M-value, and in the two cell lines: 17 and 18 reference genes were stable by both criteria (Table 6).
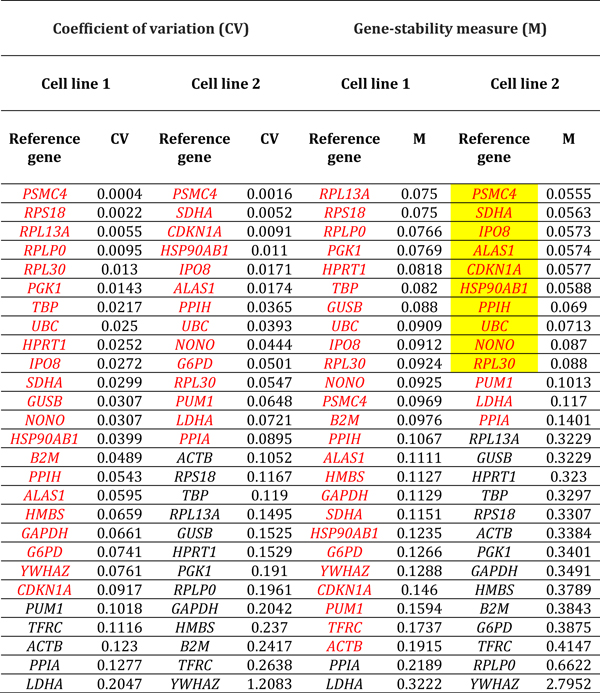
Table 1.
Reference genes expressed by human retinal endothelial cells following treatment with glucose at supraphysiological concentration. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
Table 2.
Reference genes expressed by human retinal endothelial cells following treatment with dimethyloxalylglycine. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
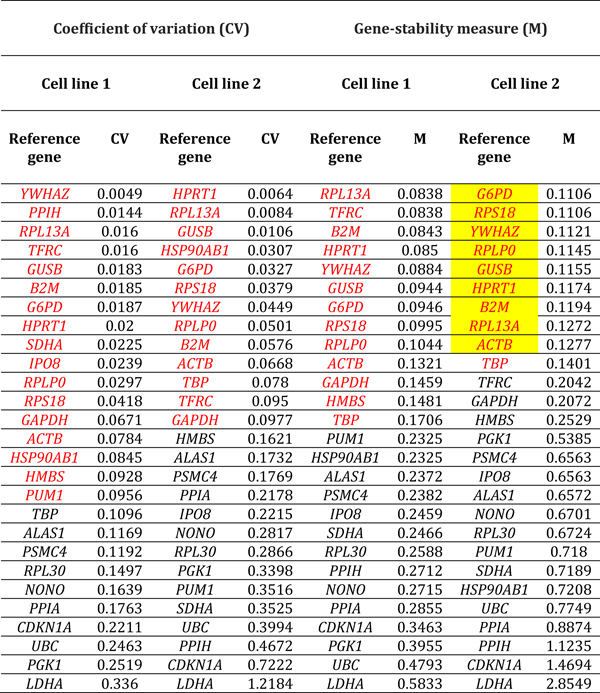
Table 3.
Reference genes expressed by human retinal endothelial cells following treatment with vascular endothelial growth factor. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
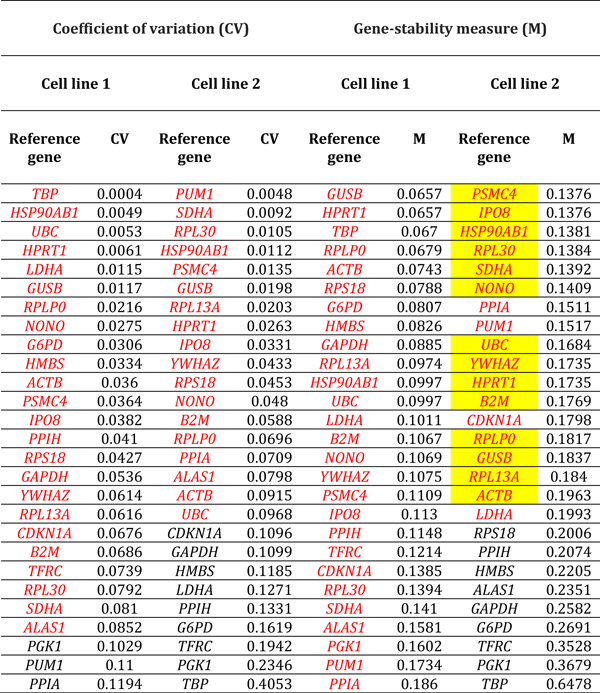
Table 4.
Reference genes expressed by human retinal endothelial cells following treatment with tumor necrosis factor-alpha. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
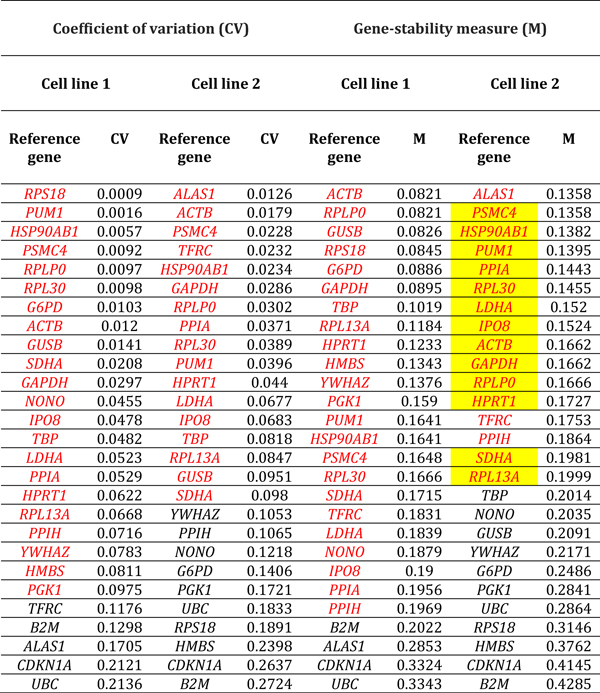
Table 5.
Reference genes expressed by human retinal endothelial cells following treatment with Escherichia coli 055:B5 lipopolysaccharide. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
Table 6.
Reference genes expressed by human retinal endothelial cells following treatment with GT-1 strain Toxoplasma gondii tachyzoites. Genes are ordered by coefficient of variation (CV) and gene-stability measure (M) (lowest to highest, indicating decreasing stability), calculated by comparison with control cells treated with medium alone (n = 3 samples/condition). Red indicates reference genes that were stably expressed according to CV-value below 0.1 or M-value below 0.2. Yellow highlight indicates reference genes that were stably expressed according to CV-value below 0.1 and M-value below 0.2, for both cell lines. Full names of reference genes with abbreviations appear in Supplementary Table 2.
Comparison of stable reference gene expressed in human retinal endothelial cells under different conditions of stimulation and by donor
Ranking of reference genes expressed in human retinal endothelial cells by lowest M- and CV-values for the two cell lines, indicated substantial diversity in the most stably expressed reference genes under different conditions of stimulation (Tables 1–6, summarized in Table 7). For glucose at supra-physiological concentration, the most stably expressed reference genes were PSMC4, IPO8 and SDHA. For DMOG, the most stably expressed reference genes were RPL13A, YWHAZ, GUSB and HPRT1. For VEGFA, the most stably expressed reference genes were HSP90AB1, UBC and HPRT1. For LPS, the most stably expressed reference genes were GAPDH, GUSB, YWHAZ and RPLP0. For TNF-α, the most stably expressed reference genes were HSP90AB1, PSMC4, and ACTB. For T. gondii tachyzoites, the most stably expressed reference genes were SDHA, ALAS1 and GUSB. Overall, no reference gene was expressed stably under all six conditions of stimulation for both donors. Fourteen reference genes were expressed stably under 3 or more conditions, including: 4 reference genes under 5 conditions (i.e., HSP90AB1, IPO8, PSMC4 and RPLPO); 6 reference genes under 4 conditions (i.e., ACTB, B2M, GUSB, RPL30, SDHA and YWHAZ); and 6 reference genes under 3 conditions (i.e., ALAS1, GAPDH, HPRT1, RPL13A, RPS18 and UBC) (Table 8).
Table 7.
Reference genes that demonstrated stable expression in human retinal endothelial cells following treatment with: glucose at supraphysiological concentration; dimethyloxalylglycine (DMOG); vascular endothelial growth factor (VEGFA); tumor necrosis factor (TNF)-α; lipopolysaccharide (LPS); and Toxoplasma gondii tachyzoites, as determined by gene-stability measure below 0.2 and coefficient of variation below 0.1, and ranked according to expression stability. Full names of reference genes with abbreviations appear in Supplementary Table 2.
| Rank | Reference gene | |||||
|---|---|---|---|---|---|---|
| Glucose | DMOG | VEGF | TNF-α | LPS | T. gondii | |
| 1 | PSMC4 | RPL13A | HSP90AB1 | HSP90AB1 | GAPDH | SDHA |
| 2 | IPO8 | YWHAZ, GUSB, HPRT1 | UBC | PSMC4 | GUSB | ALAS1 |
| 3 | SDHA | G6PD | HPRT1 | ACTB | YWHAZ, RPLP0 | GUSB |
| 4 | UBC | B2M | GUSB | RPLP0 | RPS18, ACTB | PSMC4, GAPDH |
| 5 | HSP90AB1 | RPS18 | PSMC4 | PUM1 | PGK1 | NONO, RPLP0 |
| 6 | ALAS1 | RPLP0 | RPLP0, NONO, IPO8 | GAPDH | HSP90AB1 | YWHAZ |
| 7 | RPL30 | ACTB | RPL30 | RPL30 | TBP | B2M |
| 8 | NONO | – | RPL13A | PPIA | IPO8 | PGK1 |
| 9 | PPIH | – | SDHA | HPRT1 | LDHA | HMBS |
| 10 | CDKN1A | – | YWHAZ | LDHA | HMBS | RPS18 |
| 11 | – | – | ACTB | IPO8 | PPIH | RPL30 |
| 12 | – | – | B2M | SDHA | PSMC4 | IPO8 |
| 13 | – | – | – | RPL13A | B2M | HSP90AB1 |
| 14 | – | – | – | – | ALAS1 | PUM1 |
| 15 | – | – | – | – | UBC | G6PD |
Table 8.
Reference genes demonstrating consistent expression stability in human retinal endothelial cells across treatments (ie. glucose at supraphysiological concentration; dimethyloxalyl-glycine (DMOG); vascular endothelial growth factor (VEGFA); tumor necrosis factor (TNF)-α; lipopolysaccharide (LPS); and Toxoplasma gondii tachyzoites), as determined by gene-stability measure below 0.2 and coefficient of variation below 0.1. Full names of reference genes with abbreviations appear in Supplementary Table 2.
| Reference gene | Condition | |||||
|---|---|---|---|---|---|---|
| Glucose | DMOG | VEGF | TNF-α | LPS | T. gondii | |
| Stable in 5 conditions | ||||||
| HSP90AB1 | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ |
| IPO8 | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ |
| PSMC4 | ✓ | ✕ | ✓ | ✓ | ✓ | ✓ |
| RPLPO | ✕ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Stable in 4 conditions | ||||||
| ACTB | ✕ | ✓ | ✓ | ✓ | ✓ | ✕ |
| B2M | ✕ | ✓ | ✓ | ✕ | ✓ | ✓ |
| GUSB | ✕ | ✓ | ✓ | ✕ | ✓ | ✓ |
| RPL30 | ✓ | ✕ | ✓ | ✓ | ✕ | ✓ |
| SDHA | ✓ | ✕ | ✓ | ✓ | ✕ | ✓ |
| YWHAZ | ✕ | ✓ | ✓ | ✕ | ✓ | ✓ |
| Stable in 3 conditions | ||||||
| ALAS1 | ✓ | ✕ | ✕ | ✕ | ✓ | ✓ |
| GAPDH | ✕ | ✕ | ✕ | ✓ | ✓ | ✓ |
| HPRT1 | ✕ | ✓ | ✓ | ✓ | ✕ | ✕ |
| RPL13A | ✕ | ✓ | ✓ | ✓ | ✕ | ✕ |
| RPS18 | ✕ | ✓ | ✕ | ✕ | ✓ | ✓ |
| UBC | ✓ | ✕ | ✓ | ✕ | ✓ | ✕ |
Discussion
Use of multiple stably expressed reference genes is a key implication of the MIQE guidelines. Two surveys conducted since the publication of these recommendations in 2009, reviewing diverse studies that used RT-qPCR in over 1700 articles, indicated normalization procedures were inadequate in the majority of published works.16 Indeed, the authors of these surveys highlighted “the problem of a huge body of literature that reports conclusions that may be meaningless and will cause research resources to be wasted”.
We identified articles published between 2009 and 2015 that described RT-qPCR studies involving human retinal endothelial cells specifically, using the National Library of Medicine of National Institutes of Health PubMed database (Figure 4, Table 9 and Supplementary Table 3). Our search yielded a total of 72 publications: the number of publications increased over time from 2 in 2009 to 23 in 2015. Consistent with current understanding of the pathogenesis of common retinal diseases, investigations most commonly involved treatment of human retinal endothelial cells with: glucose or advanced glycation end-products; hypoxia, hypoxia mimics or VEGF; and inflammatory stimuli. Overall, studies in just one of the 72 articles employed more than one reference gene, and stability of reference gene expression was reported in only one article also. The most common reference genes used in these works were ACTB (32 articles) and GAPDH (21 articles). This analysis of the relevant literature indicates that researchers performing RT-qPCR studies involving human retinal endothelial cells often do not follow the MIQE recommendations.
Figure 4.
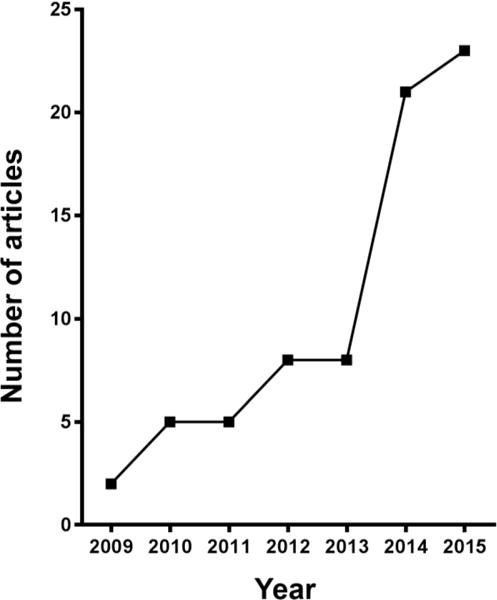
Number of articles published annually between 2009 and 2015, inclusive, which have reported studies of human retinal endothelial cells using RT-qPCR, as listed in the National Library of Medicine of National Institutes of Health PubMed database (www.ncbi.nlm.nih.gov/pubmed; search date = 28 February, 2017; search text = human retinal endothelial cell; languages filter = English).
Table 9.
Use of reference genes in RT-qPCR studies of human retinal endothelial cells in 72 articles published between 2009 and 2015 on the National Library of Medicine of National Institutes of Health PubMed database (www.ncbi.nlm.nih.gov/pubmed; search date = 28 February, 2017; search text = human retinal endothelial cell; languages filter = English). Some articles reported works that involved multiple treatments. Articles are listed in Supplementary Table 3.
| Treatment | Number of articles | Reference genes | |||||
|---|---|---|---|---|---|---|---|
| Identity | Number | Stable | |||||
| > 1 | 1 | NR | Yes | NR | |||
| Glucose/AGE | 25 | 18S rRNA, ACTB, GAPDH, HPRT1, U6 snRNA | 0 | 24 | 1 | 0 | 25 |
| Hypoxia & hypoxic mimics | 6 | ACTB, GAPDH | 0 | 5 | 1 | 0 | 6 |
| VEGF | 8 | ACTB, GAPDH | 0 | 8 | 0 | 0 | 8 |
| Cytokines | 12 | 18S rRNA, ACTB, GAPDH, TBP | 0 | 12 | 0 | 0 | 12 |
| Other proteins | 8 | 18S rRNA, ACTB, GAPDH, HPRT1 | 0 | 6 | 0 | 0 | 6 |
| Microbial mimics | 3 | GAPDH, HPRT1, PGK1, TBP | 1 | 2 | 0 | 1 | 2 |
| Enzymes | 2 | 18S rRNA, HPRT1 | 0 | 2 | 0 | 0 | 2 |
| Lipids | 2 | 18S rRNA, GAPDH, PPIA | 0 | 2 | 0 | 0 | 2 |
| Transfection of nucleic acid | 8 | 18S rRNA, ACTB, GAPDH | 0 | 8 | 0 | 0 | 8 |
| Drugs & chemicals | 9 | ACTB, GAPDH, PPIA | 0 | 9 | 0 | 0 | 9 |
| Shear stress | 2 | GAPDH | 0 | 2 | 0 | 0 | 2 |
| No treatment | 3 | ACTA, GAPDH, HPRT1 | 0 | 3 | 0 | 0 | 3 |
Abbreviations: AGE = advanced glycation end-products, VEGF = vascular endothelial growth factor, NR = not recorded. Full names of reference genes with abbreviations appear in Supplementary Table 2.
We used PCR array profiling to evaluate the potential application of 28 reference genes in RT-qPCR studies of human retinal endothelial cells. We examined reference gene expression in response to six stimuli, which are commonly used to elicit disease-relevant responses, in two retinal endothelial cell lines separately derived from eyes of two human donors, and we observed variation in expression of reference genes across stimuli and cell lines. Multiple statistical algorithms have been developed to evaluate the stability of reference genes in qPCR; a comparison of different methodologies indicates medium to high correlation.17 For this reason, and for real-world applicability, we chose to assess reference gene stability with two widely utilized measures – M-value and CV-value – and we set relatively stringent limits. Our investigation identified 10–15 of 28 reference genes (36–54%) expressed stably across different stimulated and control conditions. The group of stably expressed reference genes varied with condition, and no reference gene was stably expressed under every condition, although 4 of the 28 were stable for 5 of 6 conditions (i.e., HSP90AB1, IPO8, PSCM4 and RPLP0). Of direct relevance to published studies of human retinal endothelial cells, ACTB transcript was stable to treatments with DMOG, VEGFA, TNF-α and LPS, but not glucose and T. gondii tachyzoites, while GAPDH transcript was stable to treatments with TNF-α, LPS and T. gondii tachyzoites, but not glucose, DMOG and VEGFA.
Two independent groups have evaluated reference gene stability in human retinal endothelial cells under specific conditions. Wei et al18 stimulated human retinal endothelial cells with polyinosinic:polycytidylic acid, which activates innate immune responses via toll-like receptor 3, and evaluated expression of 10 reference genes by RT-qPCR. The authors identified HPRT1, TBP and PGK1 as the most stably expressed reference genes in that setting. Consistently, we observed stability of one or two of the same reference genes in human retinal cells treated with other immune mediators, TNF-α, LPS and T. gondii. Xie et al19 studied expression stability of 14 reference genes in human retinal endothelial cells treated with glucose at supraphysiological concentration and/or hypoxia by RT-qPCR. The investigators found an alternative group of reference genes to be most stably expressed: TBP, PUM1 and ALAS1. We also observed ALAS1 transcript level to be unaffected by treatment with highly concentrated glucose. However, none of these three molecules were represented in our list of stable reference genes in cells treated with the hypoxia mimic, DMOG. Reference gene expression has been studied in other endothelial cell populations, including human umbilical vein and brain endothelial cells, treated with metabolic and inflammatory stimuli.20–22 In each of these studies, different groups of reference genes were stably expressed, including ACTB, ALAS1, GAPDH, GUSB, RPLP0, RALBP1, RNU6-1 and TFRC. Interestingly, TFRC, which was represented on the PCR array we used, was not expressed stably under any treatment condition we studied.
Studies in different cell populations have identified a variety of experimental conditions, other than stimulations, that may impact reference gene expression stability. The time over which a stimulation is applied may alter the expression of reference gene transcripts substantially.23 Confluence of cells in culture may lead to changes in levels of reference genes,24 and the physical properties of the substrate on which cells are grown may influence reference gene expression.25 While it is clear that human subjects invariably display a range of responses to the same stimulus, and this is often reflected at the cellular level, in studies of gene expression by human cells, the possibility of inter-individual variation in expression of reference genes is often not considered. Previously we have demonstrated phenotypic differences in human retinal endothelial cell isolates from different donors by molecular profiling, as well as studies of function.2,26,27 In this current work, we observed differences in the expression stability of reference genes between retinal endothelial cell lines, generated from two human donors, for each of the six disease-associated stimuli that we tested.
Human retinal endothelial cells are being employed increasingly in studies of retinal vascular diseases, and many of these investigations are undertaken with an ultimate goal of developing new treatments. For such research, the importance of accurately normalized RT-qPCR data cannot be understated. Our work provides a guide to reference genes that may be suitable for normalization in RT-qPCR studies of human retinal endothelial cells subjected to different pathological stimuli. More importantly, however, our work highlights the variation in expression stability of reference genes that may be encountered with different stimuli and between human retinal endothelial cell isolates. Our findings provide strong evidence that researchers, who work in this field, should aim to follow MIQE recommendations for normalization of RT-qPCR data.
Supplementary Material
Highlights.
We investigated reference gene expression in human retinal endothelial cells
Gene expression varied substantially across stimulations and between cell lines
Normalization in RT-qPCR studies of gene expression should follow MIQE guidelines
Acknowledgments
This work was supported by grants from the National Eye Institute/National Institutes of Health (R01 EY019875), the National Health & Medical Research Council of Australia (APP1123684), the Australian Research Council (FT130101648) and the Ophthalmic Research Institute of Australia. The authors wish to thank Denise A. Galloway, PhD (Fred Hutchinson Cancer Institute Seattle, WA) for the gift of the LXSN16E6E7 virus.
Abbreviations List
- ACTB
Actin beta
- ALAS1
Aminoerulinate, delta-synthase 1
- B2M
Beta-2-microglobulin
- DMEM
Dulbecco’s modified Eagle’s medium
- DMOG
Dimethyloxalylglycine
- DSCR1
Down syndrome critical region gene 1
- FBS
Fetal bovine serum
- G6PD
Glucose-6-phophate dehydrogenase
- GAPDH
Glyceraldehyde-3-phophate dehydrogenase
- GUSB
Glucuronidase beta
- HBB
Hemoglobin, beta
- HMBS
Hydroxymethylbilane synthase
- HPRT1
Hypoxanthine phosphoribosyltransferase 1
- HSP90AB1
Heat shock protein 90kDA (cytosolic), Class B member 1
- ICAM-1
Intercellular adhesion molecule 1
- IL-1β
Interleukin 1β
- IPO8
Importin 8
- LDHA
Lactate dehydrogenase A
- LPS
Lipopolysaccharide
- MIQE
Minimum information for publication of qPCR experiments
- NONO
Non-POU domain containing, octamer-binding
- p21, Cip1
Cyclin-dependent kinase inhibitor 1A
- PGK1
Phosphoglycerate kinase 1
- PPIA
Peptidyl isomerase A (cyclophilin A)
- PPIH
Peptidyl isomerase H (cyclophilin H)
- PSMC4
Proteasome 26S subunit, ATPase 4
- PUM1
Pumilio RNA-binding family member 1
- RPL13A
Ribosomal protein L13a
- RPL30
Ribosomal protein L30
- RPLP0
Ribosomal protein, large, P0
- RPS18
Ribosomal protein S18
- RT-qPCR
Reverse transcription-quantitative real-time polymerase chain reaction
- SDHA
Succinate dehydrogenase complex flavoprotein subunit A
- SOCS1
Suppressor of cytokine signaling 1
- TBP
TATA box binding receptor
- TFRC
Transferrin receptor
- TNF-α
Tumor necrosis factor α
- UBC
Ubiquitin C
- VEGFA
Vascular endothelial growth factor
- YWHAZ
Tyrosine 3-mono-oxygenase/tryptophan 5-mono-oxygenase activation protein zeta
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bharadwaj AS, Appukuttan B, Wilmarth PA, et al. Role of the retinal vascular endothelial cell in ocular disease. Prog Retin Eye Res. 2013;32:102–180. doi: 10.1016/j.preteyeres.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JR, Choi D, Chipps TJ, et al. Unique gene expression profiles of donor-matched human retinal and choroidal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2007;48:2676–2684. doi: 10.1167/iovs.06-0598. [DOI] [PubMed] [Google Scholar]
- 3.Browning AC, Halligan EP, Stewart EA, et al. Comparative gene expression profiling of human umbilical vein endothelial cells and ocular vascular endothelial cells. Br J Ophthalmol. 2012;96:128–132. doi: 10.1136/bjophthalmol-2011-300572. [DOI] [PubMed] [Google Scholar]
- 4.Suarez S, McCollum GW, Bretz CA, et al. Modulation of VEGF-induced retinal vascular permeability by peroxisome proliferator-activated receptor-beta/delta. Invest Ophthalmol Vis Sci. 2014;55:8232–8240. doi: 10.1167/iovs.14-14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye EA, Steinle JJ. miR-146a attenuates inflammatory pathways mediated by TLR4/NF-kappaB and TNFalpha to protect primary human retinal microvascular endothelial cells grown in high glucose. Mediators Inflamm. 2016;2016:3958453. doi: 10.1155/2016/3958453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 7.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 8.Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashander LM, Appukuttan B, Ma Y, Gardner-Stephen D, Smith JR. Targeting endothelial adhesion molecule transcription for treatment of inflammatory disease: a proof-of-concept study. Mediators Inflamm. 2016;2016:7945848. doi: 10.1155/2016/7945848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharadwaj AS, Schewitz-Bowers LP, Wei L, Lee RW, Smith JR. Intercellular adhesion molecule 1 mediates migration of Th1 and Th17 cells across human retinal vascular endothelium. Invest Ophthalmol Vis Sci. 2013;54:6917–6925. doi: 10.1167/iovs.13-12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furtado JM, Bharadwaj AS, Chipps TJ, Pan Y, Ashander LM, Smith JR. Toxoplasma gondii tachyzoites cross retinal endothelium assisted by intercellular adhesion molecule-1 in vitro. Immunol Cell Biol. 2012;90:912–915. doi: 10.1038/icb.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan A, Behnke MS, Dunay IR, et al. Phenotypic and gene expression changes among clonal type I strains of Toxoplasma gondii. Eukaryot Cell. 2009;8:1828–1836. doi: 10.1128/EC.00150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Hellemans J, Mortier G, De Paepe A, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustin SA, Benes V, Garson J, et al. The need for transparency and good practices in the qPCR literature. Nat Methods. 2013;10:1063–1067. doi: 10.1038/nmeth.2697. [DOI] [PubMed] [Google Scholar]
- 17.Jacob F, Guertler R, Naim S, et al. Careful selection of reference genes is required for reliable performance of RT-qPCR in human normal and cancer cell lines. PLoS One. 2013;8:e59180. doi: 10.1371/journal.pone.0059180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei R, Stewart EA, Amoaku WM. Suitability of endogenous reference genes for gene expression studies with human intraocular endothelial cells. BMC Res Notes. 2013;6:46. doi: 10.1186/1756-0500-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Liu X, Li Y, Liu Y, Su G. Validation of RT-qPCR reference genes and determination of Robo4 expression levels in human retinal endothelial cells under hypoxia and/or hyperglycemia. Gene. 2016;585(1):135–142. doi: 10.1016/j.gene.2016.03.047. [DOI] [PubMed] [Google Scholar]
- 20.Bakhashab S, Lary S, Ahmed F, et al. Reference genes for expression studies in hypoxia and hyperglycemia models in human umbilical vein endothelial cells. G3. 2014;4:2159–2165. doi: 10.1534/g3.114.013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Diao H, Zhao L, et al. Identification of suitable reference genes for real-time quantitative PCR analysis of hydrogen peroxide-treated human umbilical vein endothelial cells. BMC Mol Biol. 2017;18:10. doi: 10.1186/s12867-017-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennani-Baiti B, Toegel S, Viernstein H, et al. Inflammation modulates RLIP76/RALBP1 electrophile-glutathione conjugate transporter and housekeeping genes in human blood-brain barrier endothelial cells. PLoS One. 2015;10:e0139101. doi: 10.1371/journal.pone.0139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 24.Greer S, Honeywell R, Geletu M, et al. Housekeeping genes; expression levels may change with density of cultured cells. J Immunol Methods. 2010;355:76–79. doi: 10.1016/j.jim.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Zhao L, Feng J, et al. Validation of reliable reference genes for real-time PCR in human umbilical vein endothelial cells on substrates with different stiffness. PLoS One. 2013;8:e67360. doi: 10.1371/journal.pone.0067360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamora DO, Riviere M, Choi D, et al. Proteomic profiling of human retinal and choroidal endothelial cells reveals molecular heterogeneity related to tissue of origin. Mol Vis. 2007;13:2058–2065. [PubMed] [Google Scholar]
- 27.Pan Y, Appukuttan B, Mohs K, Ashander LM, Smith JR. Ubiquitin carboxyl-terminal esterase L1 promotes proliferation of human choroidal and retinal endothelial cells. Asia Pac J Ophthalmol. 2015;4:51–55. doi: 10.1097/APO.0000000000000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.