Abstract
The in vivo gene networks involved in coordinating human fetal ovarian development remain obscure. In this study, quantitative mass spectrometry was conducted on ovarian tissue collected at key stages during the first two trimesters of human gestational development, confirming the expression profiling data using immunofluorescence, as well as in vitro modeling with human oogonial stem cells (OSCs) and human embryonic stem cells (ESCs). A total of 3,837 proteins were identified in samples spanning developmental days 47–137. Bioinformatics clustering and Ingenuity Pathway Analysis identified DNA mismatch repair and base excision repair as major pathways upregulated during this time. In addition, MAEL and TEX11, two key meiosis-related proteins, were identified as highly expressed during the developmental window associated with fetal oogenesis. These findings were confirmed and extended using in vitro differentiation of OSCs into in vitro derived oocytes and of ESCs into primordial germ cell–like cells and oocyte–like cells, as models. In conclusion, the global protein expression profiling data generated by this study have provided novel insights into human fetal ovarian development in vivo and will serve as a valuable new resource for future studies of the signaling pathways used to orchestrate human oogenesis and folliculogenesis.
Keywords: : development, germ cells, oocyte, embryonic stem cells
Introduction
Histological studies of fetal human ovarian tissue have produced a relatively well-defined time line of key steps in female gonadal development, beginning with primordial germ cell (PGC) colonization through to oocyte formation (oogenesis) and follicle formation (folliculogenesis). Briefly, within the first 4 weeks of human fetal development, the gonadal ridge is populated by migrating PGCs, and the area is subsequently filled with streams of somatic support cells from the mesonephros [1,2]. As the PGCs proliferate, they begin differentiation into oogonia, which are organized into germ cell nests. Between 9 and 11 weeks of development, the oogonia enter meiosis to generate oocytes. Folliculogenesis begins around 14–15 weeks of development, as the oocytes that are produced recruit a single layer of squamous pregranulosa cells to form primordial follicles; these structures then remain quiescent until receipt of undefined signals for growth activation to the primary stage of development [3–7].
Mechanistic studies of the gene networks and pathways that coordinate human fetal oogenesis and folliculogenesis have historically proven more difficult to pursue, primarily due to a paucity of available fetal tissue collected at defined stages. Many initial insights were provided through extrapolation of results from studies of fetal gonad development in animal models, primarily the laboratory mouse. However, key differences in how ovarian development is governed between human and mouse models have emerged over the years, limiting the relevance of mouse studies to human oogenesis and folliculogenesis. As one example, human PGCs express POU class 5 homeobox 1 (POU5F1; also known as OCT4) during the first trimester, but do not express DEAD-box helicase 4 (DDX4; also known as VASA) until the second trimester, when POU5F1 is downregulated. This pattern of expression is in stark contrast with that observed in the mouse, in which PGCs express both DDX4 and POU5F1 as they colonize the gonadal ridge [2]. Moreover, in human oocytes POU5F1 is expressed only until the point of follicle formation, contrasting the continued expression of Pou5f1 in mouse oocytes after meiotic prophase-I arrest [8,9]. Likewise, the classic germ cell marker, deleted in azoospermia-like (DAZL), also exhibits an expression profile in fetal human germ cells that differs significantly in timing and localization compared to fetal mouse germ cells [2]. Similarly, a developmentally timed switch in DAZL to protein boule-like (BOLL, also known as BOULE) function occurs during fetal oogenesis in humans but not in mice [10]. In addition to these species-specific differences in expression patterns of selected genes, striking differences exist in fundamentally important signaling pathways that govern germ cell development in humans versus mice, including the mechanisms that control retinoic acid availability for embryonic germ cell meiotic entry [11].
More recent efforts to map human female gonadal development have, however, been facilitated by the application of omics-based approaches to study human tissue samples, which then drive novel hypotheses and enable detailed comparisons across tissues within a species or the same tissue in different species. Most recently, a comprehensive microarray-based transcriptome analysis of developing human ovarian and testicular tissues reveals temporal gene expression patterns during sexual differentiation. Gene expression analysis targeted between 40 and 73 days of human development, an early developmental period covering the sex specification phase, revealed differential elevated expression of key genes in male and female gonads thought to have pivotal involvement in the sex specification of the bipotential gonad [12].
A study examining tissue from weeks 9 to 20 of human gestational development showed that expression of stimulated by retinoic acid gene 8 (STRA8), a germ cell-specific meiosis commitment gene [13], increases >50-fold in fetal ovaries after 11 weeks of development, relative to the mean baseline expression in testis samples across the same time course. Many other messenger RNA (mRNA) transcripts related to meiosis initiation and progression, including meiotic recombination protein SPO11 (SPO11), synaptonemal complex protein 3 (SYCP3), cohesin subunit SA-3 (STAG3), testis expressed 11 (TEX11), and testis expressed 14 (TEX14), followed this increased expression profile [13]. A similar approach has also been used to identify genes and pathways associated with the onset of primordial follicle formation in human fetal ovaries [14], the impact of maternal aging on oocyte quality [15–17], aberrations in oocytes of women with polycystic ovary syndrome [18], folliculogenesis [19], and in vitro oocyte maturation [20]. As insightful as these studies are, studies are limited by sample availability to small time frames of development, and the correlation between microarray and proteomic data can be limited.
In vitro modeling of female germline development and follicle formation using pluripotent stem cells (embryonic stem cells [ESCs] and induced pluripotent stem cells [iPSCs]) or unipotent stem cells (female germline or oogonial stem cells [OSCs]) has also provided new information on the factors that govern human ovarian development. Although studies of murine ESCs and iPSCs have successfully recapitulated the complete life cycle of female germ cells to the birth of live offspring [21], similar advances using human stem cell models have been much more challenging. Nonetheless, reports of germ cell formation from human ESCs and iPSCs have escalated from the generation of PGC-like cells in vitro [22,23] to the production of what appear to be immature oocytes enclosed in follicle-like aggregates from human iPSC cultures with enforced overexpression of DAZL and BOULE [24]. Paralleling this work, the discovery of OSCs in ovarian cortical tissue of reproductive-age women has provided yet another avenue for studying the processes of human female germ cell development and differentiation in vitro and in xenografts in vivo under defined experimental conditions [25–27].
To provide a detailed in vivo network analysis of human female gametogenesis and follicle formation, here we sought to generate an untargeted and quantitative protein expression profile of human oogenesis and folliculogenesis. Through liquid chromatography–mass spectrometry (LC-MS) based proteomic analysis of human fetal ovarian tissue collected between days 47 and 137 of development, as well as of adult human ovarian tissue, we established, for the first time, a protein expression reference set using bioinformatics analyses at several key stages of ovarian organogenesis, including PGC differentiation to oogonia during the first trimester through follicle assembly during mid gestation. In support of the validity of the proteomics data, we directly assessed selected gene products in human fetal ovaries, as well as in human OSC and human ESC models of in vitro germ cell formation. The resultant data sets should provide an invaluable community resource for future studies of the principal protein networks and signaling pathways involved in human oogenesis and folliculogenesis.
Materials and Methods
Human ovarian tissue samples
All procedures described herein have been reviewed and approved by the Institutional Review Board at Northeastern University, Saitama Medical University, and the University of Washington. Fetal ovarian tissues were collected after consented medical termination of pregnancy by the Laboratory of Developmental Biology at the University of Washington (supported by the NIH Resource-related Research Project Grant No. R24-HD000836) between 0.5 and 24 h postprocedure. Developmental age is based on date of conception, with estimation based on a combination of prenatal intakes, foot length, Streeter's stage, and crown-rump length. Whole ovary tissue samples were snap-frozen and stored at −80°C for processing for LC-MS or fixed in 4% neutral-buffered paraformaldehyde and stored at 4°C until processed for tissue sectioning and immunohistochemistry. Adult human ovarian cortical tissue was collected from reproductive-age women undergoing sex reassignment at Saitama Medical Center and then cryopreserved using vitrification until use [25].
Protein preparation, LC/MS/MS, and proteomics
For protein analysis, snap-frozen tissue samples were thawed in 1 mL of ice-cold RIPA Buffer per 100 mg of tissue. Lysates were then prepared by dounce homogenization and sonication on ice followed by centrifugation (10,000g) and protein concentration determined using the BCA Protein Assay Kit (Pierce). The lysates from fetal ovarian tissue and adult cortical tissue were normalized to 35 μg and run on a 10% Mini-PROTEAN TGX Gel (Bio-Rad) and prepared for LC-MS, performed at the University of Massachusetts Medical School Proteomics and Mass Spectrometry Facility. Raw data files were processed with Proteome Discoverer (version 1.4) (Thermo Fisher Scientific). Search parameters utilized were fully tryptic with two missed cleavages, parent mass tolerances of 10 ppm, and fragment mass tolerances of 0.05 Da. A fixed modification of carbamidomethyl cysteine and variable modifications of acetyl (protein N-term), pyroglutamic for N-term glutamine, and oxidation of methionine and serine/threonine phosphorylation were considered. Search results were loaded into the Scaffold Viewer (Proteome Software, Portland, OR) for assessment of protein identification probabilities and label-free quantitation. The peptide threshold was set as 95%, and the protein threshold was 99% with at least two identified peptides. Each sample set was then independently analyzed.
Quantitative comparative protein analysis between developmental time points
Common proteins were identified in tissue samples at each developmental time point (eg, “identified proteins”). Protein quantitative information was based on intensity-based absolute quantification (iBAQ) method provided by Scaffold [28]. In each sample, proteins with at least two valid iBAQ values out of the three technical replicates were determined as “quantified proteins,” and the average of the valid iBAQ values of the technical replicates for each protein was considered as the “protein abundance.” To compare the fetal and adult tissue samples, the log2-ratio of protein abundance of fetal tissue samples at days 47, 108, 122, and 137, respectively, to the adult tissue sample was calculated, and for each time point the median of the log2-ratio for fetal/adult tissue proteins was adjusted to zero (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/scd). The resultant log2-ratio represents the protein expression ratio of a developmental time point compared with adult. To determine the differentially expressed proteins between the developmental time point and adult, the proteins with log2 ratios (fetal tissue/adult tissue) >1 (fold change 2) or less than −1 (fold change 0.5) at all four time points were selected. A self-organizing map (SOM) clustering technique was then used as the secondary filtering strategy to categorize protein expression patterns across the four time points of the developmental samples [29]. The expression ratios of the proteins fitting the first criterion were submitted to the SOM clustering algorithm in GenePattern platform [30]. The number of clusters was set to four (Supplementary Fig. S1B). Initial random centroids were selected through random vectors. Neighborhood was selected as bubble. Other parameters used default values. Those were seed range as 42, iterations as 50,000, α initial and final as 0.1 and 0.005, respectively, and sigma initial and final were 5.0 and 0.5, respectively.
Comparison between developmental days 47 and 137
Analysis of developmental days 47 and 137 tissues was performed (1) to determine differentially expressed proteins present at both time points and (2) to determine proteins uniquely expressed between the two time points. Differentially regulated proteins at both developmental days 47 and 137 were determined by having at least two valid iBAQ values out of the three technical replicates in both time points, and the average of the valid iBAQ values of the technical replicates for each protein was considered the “protein abundance.” The log2-ratio of protein abundance at days 47–137 was calculated, and the median of these log2-ratios was adjusted to zero. A cutoff of 2.2 or 0.45 was used to define if the protein was upregulated or downregulated, respectively.
Proteins unique to ovarian tissue from developmental day 47 and not detected at day 137 were determined by three criteria as follows: (1) proteins must have three valid iBAQ values out of the three technical replicates at day 47; (2) based on the box and whiskers diagrams plotting with the coefficient of variation (CV%) calculated from the iBAQ values of the three technical replicates of the proteins in day 47, the proteins having greater CV% than third quartile plus the interquartile range times 1.5 were filtered. Notably, the lower bottom range (the values at the first quartile minus the interquartile range times 1.5) was negative and therefore not considered; and (3) the proteins had to have no valid iBAQ values in the three technical replicates in day 137. The proteins that were unique to day 137 and not observed in day 47 were determined using the same criteria.
Interaction network and pathway analysis
Ingenuity Pathway Analysis (IPA) was used to map significantly regulated proteins into canonical pathways. To compare each time point, upregulated and downregulated protein lists were submitted separately into IPA core analysis using default values. In the IPA “canonical pathway” analysis, the top 10 pathways ranked by P value were reported. To compare the fetal tissue samples at developmental days 47 and 137, the common proteins differentially regulated between days 47 and 137 were combined with the proteins uniquely expressed in either of the time point, and the overall protein list was uploaded for IPA core analysis using default values. The canonical pathways with a P value <0.05 and a z-value >2 or less than −2 were selected as significantly activated or deactivated pathways, respectively. InteractiVenn [31] was utilized to determine protein expression profiles between sample sets.
Tissue immunofluorescence
Paraformaldehyde-fixed ovarian tissue was dehydrated, embedded in paraffin, and 6 μm sections were placed onto slides. Following antigen retrieval and 1 h in blocking buffer (phosphate buffered saline [PBS], 2.0% bovine serum albumin [BSA], 0.1% Triton X-100, and 1.0% normal goat serum), tissue sections were incubated at 4°C overnight with 20 μg mL−1 rabbit anti-MAEL polyclonal antibody (Abcam; AB106713) or 2 μg mL−1 rabbit anti-TEX11 polyclonal antibody (Novus; NBP184359) prepared in blocking buffer. Slides were then washed and incubated in goat anti-rabbit DyLight 650 (Invitrogen; 84546) for 45 min at room temperature in the dark. Secondary only control sections were included in which primary antibody was omitted. Hoechst stain was used for visualization of nuclei. Slides were washed and cover-slipped with ProLong Gold Antifade Reagent. Images were acquired on a Zeiss Axioplan inverted fluorescent microscope.
Stem cell cultures
Human OSCs from two individuals were cultured as described previously [23,24]. Cells were plated, and cells in suspension were collected after 5 days, at which point suspension cells were pooled for analysis in comparison to the adherent cells. RNA was isolated by ReliaPrep RNA Mini Kit (Promega), and reverse transcription was carried out using the RevertAid First Strand cDNA Synthesis Kit (Thermo). Quantitative real-time PCR was performed with Fast SYBR Green (Thermo) on a StepOnePlus Thermocycler (Applied Biosystems) using the following primers: B2M Forward-GGCATTCCTGAAGCTGACAG, Reverse-TGGATGACGTGAGTAAACCTG; ZP3 Forward-AAGCCCACTGCTCTACTTCATGGT, Reverse-AGCAGGACCCAGATGAACTCAACA; MAEL Forward-AGAAATGGCTCGAGAATGGAG, Reverse-ACAAAGCTGACATATCTGGAGG; and TEX11 Forward-AGCCTTAAGTTTGGAGTCAAGAG, Reverse-TCAGAAGGACAAAACTGGGAC.
Human ESCs (WA09, WiCell) were cultured on human ESC-qualified Matrigel with Essential 8 Medium (Thermo Fisher Scientific). Cells were passaged routinely using ethylenediaminetetraacetic acid (EDTA) with daily media changes. For differentiation, cells were trypsinized and plated on Matrigel with 1X RevitaCell Supplement (Life Technologies) for 24 h to aid in single cell recovery. When cells reached 50% confluency, differentiation was initiated by transferring cells to differentiation media: Dulbecco's modified Eagle's medium/F12, 20% knockout serum replacement, 1X Non-Essential Amino Acids, and 0.1 mM β-mercaptoethanol. Cells were treated for the first 48 h with retinoic acid (Sigma, 10 μM) and subsequently received media changes every 48 h.
For imaging, cells were fixed in 4% paraformaldehyde at room temperature for 15 min and washed in PBS. Adherent human OSCs and human ESCs were imaged on glass coverslips, while suspension human OSCs were imaged in suspension in wells with glass coverslip bottoms. Cells were blocked in blocking buffer (PBS, 2.0% BSA, 0.1% Triton X-100, and 1.0% normal donkey or goat serum) for 1 h and were incubated at 4°C overnight with 20 μg mL−1 rabbit anti-MAEL polyclonal antibody (Abcam) or 2 μg mL−1 rabbit anti-TEX11 polyclonal antibody (Novus). Cells were incubated in goat anti-rabbit Alexa Fluor 650 (Abcam, 1:500) followed by costaining for DDX4 using goat anti-DDX4 (R&D Systems) and subsequently with donkey anti-goat Alexa Fluor 568 (Abcam, 1:500). Secondary only control sections were included in which primary antibody was omitted. Cells were imaged on a Zeiss Axioplan inverted fluorescent microscope.
Data availability
The proteomics data generated herein have been deposited into the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository [32], with PXD006898 assigned as the data set identifier. Expanded data tables for all mass spectrometry quantitative values and comparison analyses have been deposited at Mendeley Data under DOI:10.17632/x7zbyj6xh7.1.
Results
Global ovarian protein expression profiling
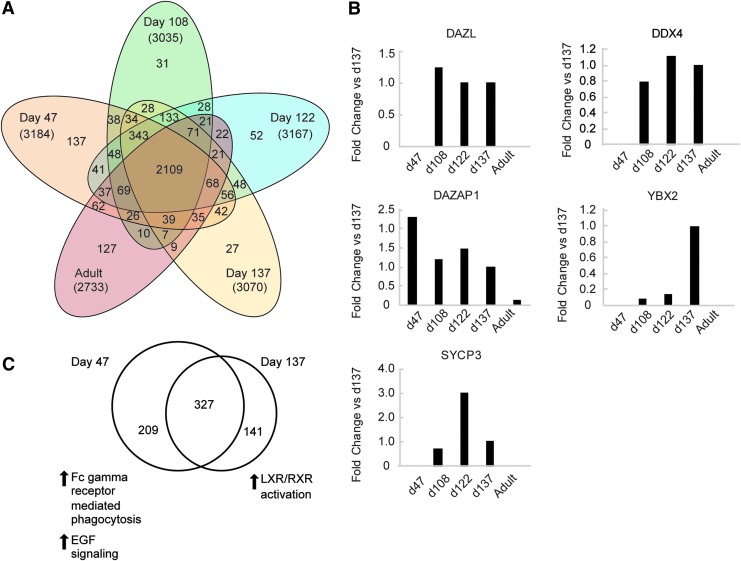
Ovarian tissue from days 47, 108, 122, and 137 of development was tested using LC-MS based quantitative proteomic analysis, with adult tissue (reproductive age) included as a reference (see Fig. 1 for developmental distribution of samples analyzed). Of note, the reproductive-age female ovarian tissue was obtained following gender reassignment surgery, which included the use of masculinizing hormones before surgery. As a result, the majority of the follicles are in the primordial and primary stages, with few secondary follicles. Between 2,700 and 3,200 proteins were identified for each time point in development (Fig. 2A). Among them, several known germ cell and oocyte markers were identified and then quantified using iBAQ value analysis. These included DAZL, DDX4, deleted in azoospermia-associated protein 1 (DAZAP1), Y-box protein 2 (YBX2; also referred to as MSY2 or CONTRIN), and SYCP3, which expectedly demonstrated variable expression levels throughout development, shown as fold change with respect to day 137 to normalize expression changes across the span of developmental time points studied (Fig. 2B).
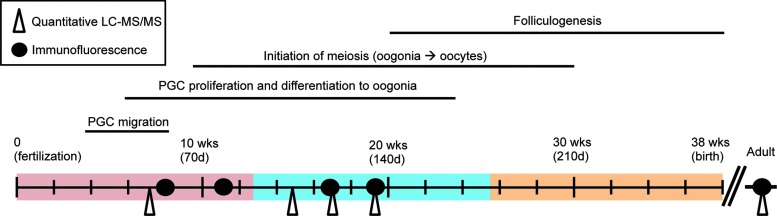
FIG. 1.
A timeline of human ovarian organogenesis from fertilization to birth indicating time points utilized to assess protein expression in whole ovarian tissue. The filled circles along the timeline represent samples used for immunofluorescence marker analysis, and the open triangles indicate samples used for proteomics analysis. Colored portions of the timeline indicate the trimesters of human fetal development: first-red, second-blue, and third-orange. LC-MS, liquid chromatography–mass spectrometry; PGC, primordial germ cell.
FIG. 2.
Quantitative mass spectrometry analysis of human ovarian tissue lysates. (A) A Venn diagram shows overlap of proteins detected by label-free quantitative mass spectrometry (3,837 total detected) for fetal tissue from developmental days 47, 108, 122, or 137 and adult reproductive aged tissue. (B) Quantitative mass spectrometry detection of germ cell proteins DAZL, DDX4, DAZAP1, YBX2, and SYCP3 at four stages of fetal development (days 47, 108, 122, and 137) and in a representative adult tissue sample presented as fold change compared to tissue from 137 days of development. (C) A Venn diagram illustrates the proteins detected in day 47 and day 137 fetal tissue with restrictive bioinformatics processing to detect only highly differentially expressed proteins; significantly upregulated pathways are specified. EGF, epidermal growth factor; LXR/RXR, liver X receptor/retinoic acid receptor pathway.
Protein expression profiles in fetal versus adult ovarian tissue
For quantified proteins, log2-ratios were calculated to compare protein expression levels across developmental time points and standardized to total protein input (see Materials and Methods for details). Proteins with log2-ratios >1 (twofold or more increase) or less than −1 (50% or more decrease) were selected for bioinformatic analysis. SOM clustering identified four clusters (Supplementary Fig. S1B), with two clusters (clusters 1 and 4) selected for further analysis based on the greatest number of differentially expressed proteins. Cluster 1 contained 71 proteins with lower expression in developmental versus adult ovarian tissue. Using IPA, we identified 20 canonical pathways represented within 71 proteins (P < 0.05), the top result being the acute phase response signaling pathway (P < 0.001) based on expression levels of plasminogen (PLG), hemopexin (HPX), fibronectin 1 (FN1), complement component 3 (C3), serpin family F member 1 (SERPINF1), ceruloplasmin (CP), and serpin family A member 3 (SERPINA3) (Table 1). Cluster 2 contained 72 proteins with lower expression in developmental versus adult ovarian tissue. IPA identified 13 canonical pathways (P < 0.05) (Table 2). The top pathway identified was DNA mismatch repair (P < 0.001) based on expression levels of proliferating-cell nuclear antigen (PCNA), MutS homolog 6 (MSH6), and flap structure-specific endonuclease 1 (FEN1). Expression levels of PCNA and FEN1 were also represented in the second canonical pathway identified that differed significantly between fetal and adult ovarian tissue, which was base excision repair (P < 0.001).
Table 1.
Canonical Pathways Observed in Cluster 1 (Top 10 of 20), Characterizing Proteins With Lower Expression at Four Observed Developmental Days (47, 108, 122, and 137) in Comparison to Adult Tissue
| Canonical pathways | P | Proteins |
|---|---|---|
| Acute phase response signaling | 4.37E-07 | PLG, HPX, FN1, C3, SERPINF1, CP, SERPINA3 |
| Caveolar-mediated endocytosis signaling | 4.90E-05 | HLA-A, ITGAV, CAV1, PTRF |
| Neuroprotective role of THOP1 in Alzheimer's disease | 2.75E-04 | PLG, HLA-A, SERPINA3 |
| Hepatic fibrosis/hepatic stellate cell activation | 1.74E-03 | FN1, COL6A1, COL6A3, COL12A1 |
| Coagulation system | 4.17E-03 | PLG, SERPINC1 |
| Complement system | 4.90E-03 | C3, CFH |
| LXR/RXR activation | 5.25E-03 | HPX, C3, SERPINF1 |
| FXR/RXR activation | 6.46E-03 | HPX, C3, SERPINF1 |
| Serotonin receptor signaling | 9.77E-03 | MAOB, SPR |
| Glioma invasiveness signaling | 1.12E-02 | PLG, ITGAV |
Table 2.
Canonical Pathways Observed in Cluster 2, Characterizing Proteins with Higher Expression at Four Observed Developmental Days (47, 108, 122, and 137) in Comparison to Adult Tissue
| Canonical pathways | P | Proteins |
|---|---|---|
| Mismatch repair in eukaryotes | 2.09E-05 | PCNA, MSH6, FEN1 |
| BER pathway | 6.92E-04 | PCNA, FEN1 |
| LPS/IL-1 mediated inhibition of RXR function | 4.68E-03 | ALDH1A2, CPT2, ALDH18A1, FABP5 |
| Glycine biosynthesis I | 1.82E-02 | SHMT2 |
| Clathrin-mediated endocytosis signaling | 1.95E-02 | CSNK2A2, CLTA, USP9X |
| AMPK signaling | 2.14E-02 | CPT2, SMARCC1, PPM1G |
| RAR activation | 2.19E-02 | CSNK2A2, ALDH1A2, SMARCC1 |
| Role of BRCA1 in DNA damage response | 2.34E-02 | MSH6, SMARCC1 |
| Thioredoxin pathway | 2.40E-02 | NXN |
| Cleavage and polyadenylation of pre-mRNA | 3.55E-02 | CSTF3 |
| Acyl-CoA hydrolysis | 4.20E-02 | ACOT7 |
| Proline biosynthesis I | 4.20E-02 | ALDH18A1 |
| dTMP De Novo biosynthesis | 4.20E-02 | SHMT2 |
Protein expression profiles in first- versus second-trimester fetal ovaries
In addition to identification of differentially expressed proteins throughout all fetal ovarian stages compared to adult tissue, our analysis also revealed significant differences in fetal ovaries associated with the timing of oogenesis and primordial follicle formation. A comparative analysis was performed between two important stages of ovarian development; day 47 (first trimester), when over 90% of the germ cells present are oogonia, and day 137 (second trimester), at which time roughly three quarters of the oogonia have differentiated into oocytes and folliculogenesis has begun [33,34]. A total of 327 proteins were expressed in fetal ovarian tissue at both stages examined, with 52 canonical pathways identified (P < 0.05; Fig. 2C). Of these, two pathways were notably elevated in tissue collected at day 47 compared to day 137, with activation z-scores >2.0: the Fc γ receptor-mediated phagocytosis pathway (P < 0.001) and the epidermal growth factor signaling pathway (P < 0.005) (Table 3). In contrast, the liver X receptor/retinoic acid receptor pathway (LXR/RXR) signaling pathway was elevated in ovarian tissue collected at day 137 compared to day 47 (P < 0.001; Table 3).
Table 3.
Activated Canonical Pathways (P < 0.05 and −2.2< z-Score >2.2) Observed in Human Ovarian Tissue from Developmental Days 47 and 137
| Canonical pathways | P | z-Scorea | Proteins |
|---|---|---|---|
| Fc γ receptor-mediated phagocytosis in macrophages and monocytes | 6.46E-04 | 3.16 | SRC, FYN, PRKCA, PLD3, YES1, CBL, HMOX1, DOCK1, ARPC1A, LYN |
| EGF signaling | 3.09E-02 | 2.24 | MAPK14, ITPR2, EGFR, SRC, PRKCA |
| LXR/RXR activation | 1.00E-04 | −3.46 | S100A8, TF, C3, FGA, FDFT1, AGT, CLU, SERPINF2, APOA1, SERPINA1, AHSG, TTR, ALB |
Positive z-scores denote samples with higher expression at 47 days of development, and negative values denote samples with higher expression at 137 days of development.
Meiosis-related protein profiles are enriched in second-trimester fetal ovaries
Analysis of days 47 versus 137 ovarian protein profiles by IPA identified >500 biological function categories (P < 0.05) from the 209 proteins unique to day 47 of development. Of these, the top results, not surprisingly, consisted of proteins that constitute cellular growth and proliferation and cellular movement categories. Conversely, proteins identified as being unique to fetal ovarian tissue collected on day 137 of development were associated with meiotic processes and synapsis. These included Ankyrin-repeat sterile alpha motif and basic leucine zipper domain-containing protein 1 (ASZ1), mitotic spindle assembly checkpoint protein MAD2A (MAD2L1), protein phosphatase 1 regulatory inhibitor subunit 2 (PPP1R2), Tudor, and KH domain-containing protein (TDRKH; Table 4). We identified several key meiosis-associated proteins that were significantly (P < 0.001) increased at day 137 of development (compared with day 47): SYCP3, STAG3, meiotic recombination protein REC8 homologue (REC8), maelstrom (MAEL), and TEX11.
Table 4.
Biological Function Categories Identified by Ingenuity Pathway Analysis to be Unique to Ovarian Tissue from Developmental Day 137 in Comparison to Developmental Day 47 (Top 10 of 500, P < 0.05)
| Categories | Diseases or functions annotation | P | Proteins |
|---|---|---|---|
| Cell cycle and reproductive system development and function | Meiosis of germ cells | 1.84E-10 | ASZ1, DDX4, MAD2 L1, MAEL, PPP1R2, REC8, SMC1B, STIM1, SYCP3, TDRKH, TEX11 |
| Cell cycle, DNA replication, recombination, and repair | Synapsis | 1.12E-07 | MAEL, REC8, SMC1B, STAG3, SYCE2, SYCP3 |
| Cell cycle and reproductive system development and function | Meiosis of male germ cells | 4.31E-07 | ASZ1, DDX4, MAEL, REC8, SMC1B, TDRKH, TEX11 |
| Cell cycle and reproductive system development and function | Meiosis of female germ cells | 1.65E-06 | MAD2 L1, PPP1R2, REC8, SMC1B, STIM1, SYCP3 |
| Cellular assembly and organization, DNA replication, recombination, and repair | Formation of synaptonemal complexes | 3.88E-06 | REC8, STAG3, SYCP3, TEX11 |
Targeted analysis of MAEL and TEX11 in human fetal ovaries
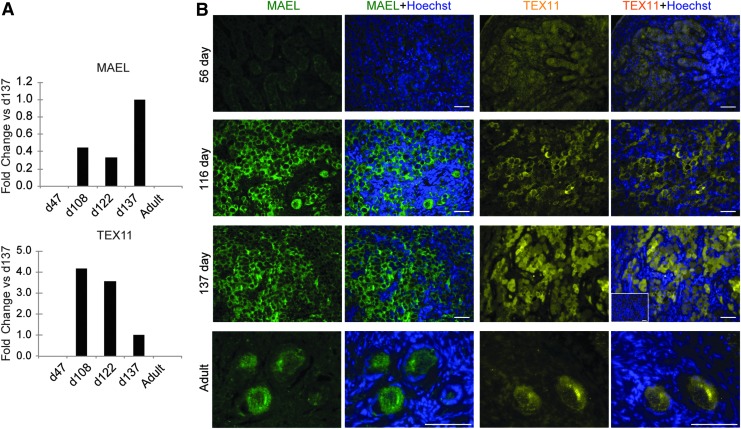
Of the proteins identified through bioinformatics, MAEL and TEX11 were selected for protein validation and further study. MAEL was selected based on its reported role in early oogenesis, while TEX11 has been implicated in meiotic progression [35,36], both of which are key functional biological processes in mouse ovarian development. We were specifically interested in validating MAEL and TEX11 in normal fetal human development due to the predicted differences of these proteins in temporal expression, which could prove valuable in assessing stages of germ cell differentiation, in vitro. Through our proteomic profiling data, we found that MAEL was detectable in human fetal ovaries at days 108 and 122 of development, and the levels of the protein were increased at day 137 (Fig. 3A). We also observed that TEX11 was detectable at both days 108 and 122 of development; however, TEX11 levels decreased at day 137 (Fig. 3A). Immunofluorescence analysis was then used to probe expression and localization of these proteins in human ovaries across development. At day 56, MAEL was not detectable in fetal ovaries, but was apparent in fetal ovaries at days 116 and 137 of development (Fig. 3B). Expression of MAEL was localized to the cytoplasm of germ cell nests, as well as the cytoplasm of oocytes in newly formed primordial follicles (Fig. 3B). By comparison, TEX11 was faintly detectable in ovarian tissue from day 56 of development, localized primarily to germ cell nests. By developmental day 116, TEX11 was strongly localized to the cytoplasm of germ cells and then exhibited a dramatic nuclear relocalization in germ cells by day 137 of development (Fig. 3B). Both MAEL and TEX11 remained detectable in the cytoplasm of oocytes contained within immature (primordial and primary) follicles in adult ovaries (Fig. 3B).
FIG. 3.
Protein detection of MAEL and TEX11 in fetal and adult ovarian tissue. (A) Quantitative mass spectrometry detection of MAEL and TEX11 at four stages of fetal development (developmental days 47, 108, 122, and 137) and in a representative adult tissue sample. Values presented are iBAQ values normalized as fold change with respect to the 137-day sample. (B) Immunofluorescence analysis of fixed and sectioned ovarian tissue reveals localization of MAEL (green) and TEX11 (yellow) to germ cell nests. MAEL and TEX11 were only detected in the oocytes of primordial and primary follicles in adult tissue, with representative follicles shown. Cells were counterstained with Hoechst (blue) to visualize DNA. Inset shows representative secondary only control image (merge). Scale bars, 100 μm. iBAQ, intensity-based absolute quantification.
Expression of MAEL and TEX11 in stem cell models of human female gametogenesis
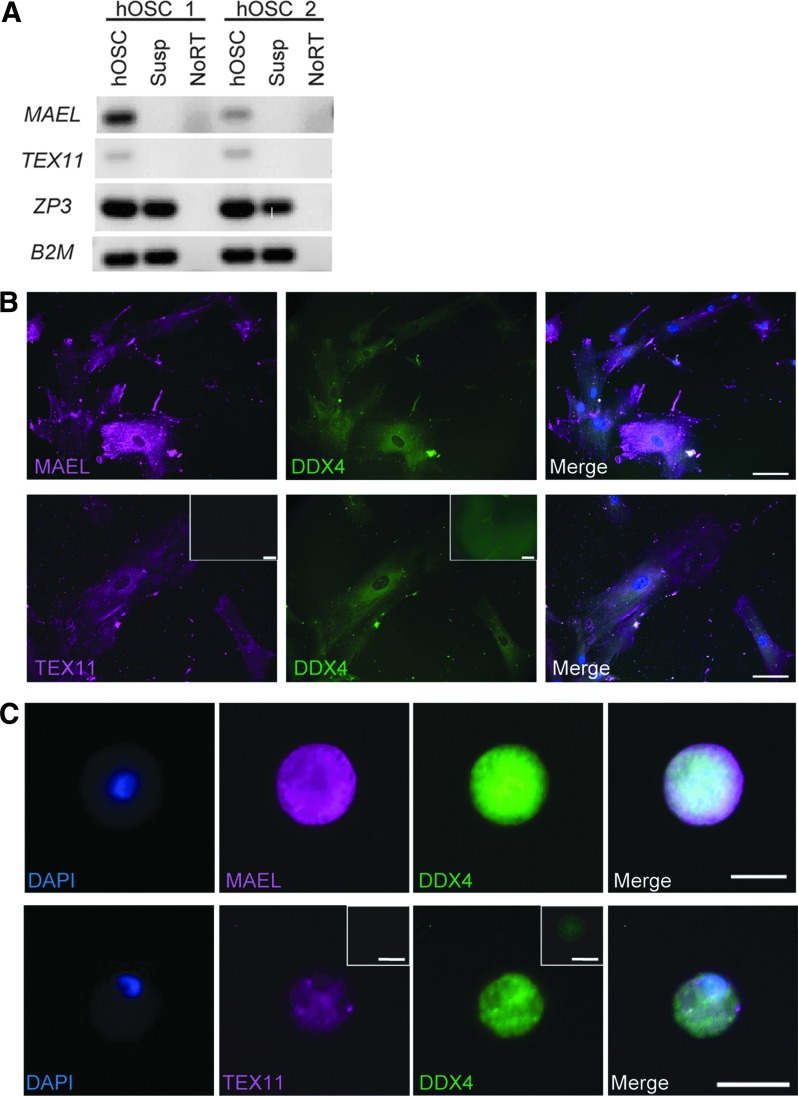
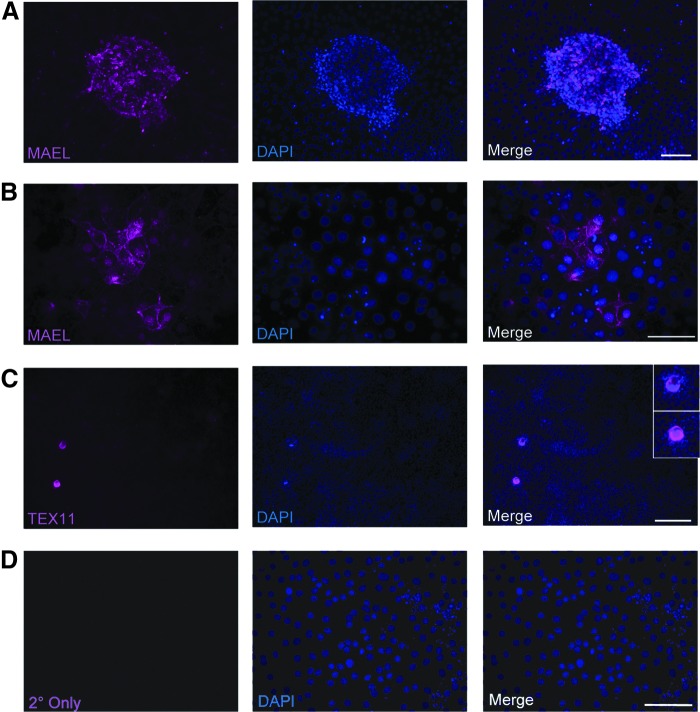
To extend our findings on MAEL and TEX11 expression in human fetal ovaries in vivo, in a final series of experiments, we evaluated the patterns of MAEL and TEX11 expression in two different in vitro model systems of human oogenesis from stem cells: human OSCs and human ESCs. In feeder cell-free cultures of human OSCs, both MAEL and TEX11 mRNA transcripts were detected in the adherent cell population which represents mitotically active (premeiotic) germ cells (Fig. 4A). As human OSCs exit the mitotic cycle and begin meiotic differentiation, the resultant in vitro-derived (IVD) oocytes are released into the culture supernatants [25,26,37]. Parallel analysis of this nonadherent cell population identified expression of SYCP3, consistent with prior studies [25], but an absence of MAEL and TEX11 mRNA transcripts (Fig. 4A). Interestingly, both adherent OSCs and IVD oocytes maintained expression of detectable levels of MAEL and TEX11 proteins, which were localized to the cytoplasm in both pools of cells (Fig. 4B, C). These latter observations aligned closely with the endogenous patterns of MAEL and TEX11 expression in immature oocytes present in adult ovarian tissue (Fig. 3B).
FIG. 4.
Analysis of MAEL and TEX11 expression in human oogonial stem cells and IVD oocytes. (A) Gene expression analysis by RT-PCR of MAEL, TEX11, and ZP3 in two hOSC lines derived from two patients. Cells released into suspension contained IVD oocytes and were analyzed separately from cultured cells. DNA was run on a 1.5% agarose gel and stained with ethidium bromide. (B) Immunofluorescence staining of cultured hOSCs for MAEL, TEX11, and DDX4 revealed cytoplasmic staining for all three markers in adherent hOSCs. Insets show representative secondary only control images. Scale bar, 50 μm. (C) Spontaneously generated IVD oocytes also stained positively for MAEL, TEX11, and DDX4. Insets show representative secondary only control images. Scale bar, 20 μm. hOSC, human oogonial stem cell; IVD, in vitro derived; RT-PCR, real-time PCR.
To complement and extend these studies further, we next used a differentiation strategy to track the expression of both MAEL and TEX11 in cultures of human ESCs that were differentiated toward the germ lineage. There are several published reports of PGC formation from human ESCs in spontaneously differentiating cultures [38–40]. Human ESCs were maintained in two-dimensional cultures on Matrigel and were cultured with or without retinoic acid for 48 h followed by an additional 8 days in culture in nonsupplemented ESC differentiation media. Immunofluorescence analysis of MAEL and TEX11 localization in differentiated ESC cultures at day 10 after treatment with retinoic acid revealed a variety of cellular morphologies of stained cells (Fig. 5A–D). MAEL-positive cells were distinct from neighboring differentiated cells by bright cytoplasmic staining (Fig. 5A). In other areas, large cells with round nuclei displayed nuclear and/or membrane localized MAEL (Fig. 5B). Differentiated ESCs at day 10 also revealed TEX11-positive cells that formed small spherical clusters amidst TEX11-negative cells (Fig. 5C).
FIG. 5.
hESC differentiations reveal hESC-derived cells positive for MAEL and TEX11. (A) MAEL-positive cells were identified by immunofluorescence in hESCs treated with retinoic acid (10 μM) for 48 h followed by 8 days of differentiation. Scale bar, 100 μm. (B) MAEL-positive cells were also identified with nuclear and/or membrane localized staining. Scale bar, 50 μm. (C) Differentiated hESCs at day 10 also resulted in small 3D aggregates that are TEX11 positive. Scale bar, 100 μm. (D) Differentiated hESCs at day 10 stained with secondary antibody only. Scale bar, 50 μm. 3D, three dimensional; hESC, human embryonic stem cell.
Discussion
Through the application of quantitative proteomics to the study of human fetal ovarian tissue collected during the first two trimesters of pregnancy, we have identified several specific protein-family networks and canonical signaling pathways that are selectively enhanced or reduced in expression during oogenesis and folliculogenesis. From a broad perspective, initial comparisons of our untargeted protein profiling results with those of prior studies of human fetal ovaries based on assessment of selected individual proteins or of genes represented in commercially available microarrays revealed some key similarities, which was important for confirmation of the approach used [13]. Examples of these include DAZL, an RNA-binding protein required for germ cell development [11], and DDX4, a germ cell-specific RNA helicase [2]. Our findings further demonstrated that in addition to DAZL, DDX4, YBX2, a marker for diplotene stage oocytes [41,42], and the meiosis-specific SYCP3 [43,44] proteins all follow a similar pattern of enhanced expression in human fetal ovaries during the periods associated with oogenesis and folliculogenesis, as expected. In addition, DAZAP1 was found to be more highly expressed in fetal ovaries during early developmental stages, which aligns developmentally with its role in mRNA translation initiation in spermatogenesis [45].
Digging deeper into our protein expression profiling data, we uncovered several additional findings through IPA that, based on extrapolation to other species and model systems, are logical in the context of the sequence of events occurring in female germ cells during fetal development. For example, our identification of DNA mismatch repair and DNA base excision repair as two of the most prominent canonical pathways whose components are expressed at high levels in first- and second-trimester fetal human ovaries reflects the critical roles that these pathways likely play in ensuring proper meiotic activation and progression in human female germ cells. In support of this, past studies have shown that meiotic failure in mouse models of female germ cell development is associated with aberrant function of DNA repair pathway genes [46–48] and that DNA repair genes are expressed at relatively high levels in human metaphase-II oocytes and blastocysts [49]. Likewise, our finding that the retinoid receptor signaling pathway was one of the only significantly scored canonical pathways in human fetal ovaries with higher expression during later developmental time points associated with active oogenesis compared with day 47 of development is concordant with prior evidence that retinoic acid controls germ cell meiotic entry in human fetal gonads [50].
In addition to retinoid receptor signaling, many other protein networks linked by IPA to meiotic commitment, recombination, and progression were found to be more highly expressed in fetal human ovarian tissue at days 137 versus 47 of development. Some of these proteins have been implicated previously in human oocyte development, including REC8 [51,52], STAG3 [52,53], and SYCP3 [54], whereas the involvement of several others in human female gametogenesis, such as ASZ1, MAD2A, PPP1R2, and TDRKH, is much more poorly defined [55].
Of the many meiosis-related proteins identified in our expression profile analyses, we elected to focus on MAEL and TEX11 for several reasons. In Drosophila, the mael gene product (maelstrom) is known to be crucial for coordinating microtubule organization during oogenesis [35]. In mice, disruption of the Mael gene leads to spermatogenic failure in males [56], as well as meiotic defects in fetal oocytes of females [57]. Although these studies in flies and mice underscore a critical role for this protein in ensuring error-free meiosis, most work on MAEL in humans has been confined to studies of cancer cell lines [58], with identification of MAEL in human germ cells limited to transcriptome screens of human PGCs [22,59]. Other gene knockout studies in mice have revealed that Tex11 is important for the successful repair of double-strand breaks that occur during meiosis [36]. In humans, TEX11 mRNA has been detected in fetal ovaries [13] and has been identified as a potential target of DAZL [60].
In addition to the quantitative proteomic mapping of changes in these two proteins in fetal ovarian tissue during early through mid development, we showed by immunohistochemical analysis that, while MAEL and TEX11 have somewhat different expression profiles during development, both are specifically localized to germ cell nests and to oocytes within follicles. Moreover, similar outcomes were observed using two different in vitro models of human oogenesis. Although correlative, we believe that these data, when considered with published studies in flies and mice [35,36,56,57], implicate MAEL and TEX11 as putative key players in the coordinated regulation of meiosis in human female germ cells. Future studies, including functional testing utilizing in vitro genetic manipulation in ESCs and OSCs, will be important in the investigation of the specific roles of these proteins. Interestingly, our expression analysis of TEX11 in human fetal ovarian tissue further revealed a distinct cytoplasmic to nuclear localization pattern of this protein during the developmental window associated with oogenesis followed by follicle formation. The timing of the nuclear translocation of TEX11 was consistent with the onset of the pachytene and diplotene phases during 16–20 weeks of development, concomitant with the initial formation of follicles. Together, these data suggest that TEX11 likely participates in meiosis-related DNA double-strand break repair in human female germ cells, akin to its function in mice [36]. In addition, in human spermatogenesis, TEX11 has been associated with the pachytene stage [61].
Before closing, two other points deserve mention. First, our comparison of fetal and adult ovarian tissue samples revealed that the acute phase response was the most prominent canonical pathway identified by IPA whose components were significantly higher in adult versus fetal ovaries. Acute phase response proteins are those involved in the body's response to inflammation, and this pathway has been proposed from mouse studies as playing a major role in repair of the ovarian surface epithelium after ovulatory rupture of each reproductive cycle during adult life (reviewed in [62]). This observation should therefore provide a strong impetus to further explore the roles of immune response signaling proteins in human follicular maturation and ovulation. Second, the proteomic datasets generated by this study are far more expansive than the selected outcomes discussed herein. While we intentionally focused our initial analyses on protein networks and signaling pathways that were not just prominent but are logical to consider in the context of oogenesis and folliculogenesis, such as DNA repair and meiosis, it is important to emphasize that the untargeted proteomic analyses performed herein have not been conducted previously with fetal human ovarian tissue. We both confirmed canonical signaling pathways involved in the process of ovarian organogenesis, as well as identified candidate proteins and pathways previously unlinked to ovarian development; from our analysis, it is clear that there are many processes important to the establishment of a healthy ovary that are under-investigated. Therefore, we hope that the data generated will serve as a valuable resource for the research community to generate new hypotheses and perhaps uncover unexpected roles for the vast number of proteins and pathways detected in fetal human ovaries during early to mid-gestational development.
Supplementary Material
Acknowledgments
The authors thank the Laboratory of Developmental Biology at the University of Washington (Seattle, WA; supported by NIH Resource-related Research Project grant no. R24-HD000836) for providing gestational ovarian tissue samples used for our studies and the University of Massachusetts Medical School Proteomics and Mass Spectrometry Facility for LC-MS/MS assistance. Funding was provided by NIH R37-AG012279 to J.L.T. and Northeastern University and OvaScience, Inc., to D.C.W.
Author Disclosure Statement
A.M.B., Y.G., Y.T., O.I., H.S., and B.K. declare no competing financial interests. J.L.T. discloses interest in intellectual property described in U.S. Patent 7,195,775, U.S. Patent 7,850,984, U.S. Patent 7,955,846, U.S. Patent 8,642,329, U.S. Patent 8,647,869, U.S. Patent 8,652,840, U.S. Patent 9,150,830, and U.S. Patent 9,267,111, is a scientific cofounder of OvaScience, Inc. (Waltham, MA), and is a member of the Scientific Advisory Board of OvaScience, Inc. D.C.W. discloses interest in intellectual property described in U.S. Patent 8,642,329, U.S. Patent 8,647,869, and U.S. Patent 9,150,830 and is a recipient of a corporate sponsored research award from OvaScience, Inc.
References
- 1.Baker TG. and Franchi LL. (1967). The fine structure of oogonia and oocytes in human ovaries. J Cell Sci 2:213–224 [DOI] [PubMed] [Google Scholar]
- 2.Anderson RA, Fulton N, Cowan G, Coutts S. and Saunders PT. (2007). Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol 7:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gondos B. (1987). Comparative studies of normal and neoplastic ovarian germ cells: 2. Ultrastructure and pathogenesis of dysgerminoma. Int J Gynecol Pathol 6:124–131 [DOI] [PubMed] [Google Scholar]
- 4.Bendsen E, Byskov AG, Andersen CY. and Westergaard LG. (2006). Number of germ cells and somatic cells in human fetal ovaries during the first weeks after sex differentiation. Hum Reprod 21:30–35 [DOI] [PubMed] [Google Scholar]
- 5.Motta PM, Makabe S. and Nottola SA. (1997). The ultrastructure of human reproduction. I. The natural history of the female germ cell: origin, migration and differentiation inside the developing ovary. Hum Reprod Update 3:281–295 [DOI] [PubMed] [Google Scholar]
- 6.Forabosco A. and Sforza C. (2007). Establishment of ovarian reserve: a quantitative morphometric study of the developing human ovary. Fertil Steril 88:675–683 [DOI] [PubMed] [Google Scholar]
- 7.Albamonte MS, Willis MA, Albamonte MI, Jensen F, Espinosa MB. and Vitullo AD. (2008). The developing human ovary: immunohistochemical analysis of germ-cell-specific VASA protein, BCL-2/BAX expression balance and apoptosis. Hum Reprod 23:1895–1901 [DOI] [PubMed] [Google Scholar]
- 8.Pesce M, Wang X, Wolgemuth DJ. and Schøler H. (1998). Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev 71:89–98 [DOI] [PubMed] [Google Scholar]
- 9.Stoop H, Honecker F, Cools M, de Krijger R, Bokemeyer C. and Looijenga LH. (2005). Differentiation and development of human female germ cells during prenatal gonadogenesis: an immunohistochemical study. Hum Reprod 20:1466–1476 [DOI] [PubMed] [Google Scholar]
- 10.He J, Stewart K, Kinnell HL, Anderson RA. and Childs AJ. (2013). A developmental stage-specific switch from DAZL to BOLL occurs during fetal oogenesis in humans, but not mice. PLoS One 8:e73996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Bouffant R, Guerquin MJ, Duquenne C, Frydman N, Coffigny H, Rouiller-Fabre V, Frydman R, Habert R. and Livera G. (2010). Meiosis initiation in the human ovary requires intrinsic retinoic acid synthesis. Hum Reprod 25:2579–2590 [DOI] [PubMed] [Google Scholar]
- 12.Mamsen LS, Ernst EH, Borup R, Larsen A, Olesen RH, Ernst E, Anderson RA, Kirstensen SG. and Andersen CY. (2017). Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Sci Rep 7:15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houmard B, Small C, Yang L, Naluai-Cecchini T, Cheng E, Hassold T. and Griswold M. (2009). Global gene expression in the human fetal testis and ovary. Biol Reprod 81:438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler PA, Flannigan S, Mathers A, Gillanders K, Lea RG, Wood MJ, Maheshwari A, Bhattacharya S, Collie-Duguid ES, et al. (2009). Gene expression analysis of human fetal ovarian primordial follicle formation. J Clin Endocrinol Metab 94:1427–1435 [DOI] [PubMed] [Google Scholar]
- 15.Steuerwald N, Cohen J, Herrera RJ, Sandalinas M. and Brenner CA. (2001). Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol Hum Reprod 7:49–55 [DOI] [PubMed] [Google Scholar]
- 16.Steuerwald NM, Bermúdez MG, Wells D, Munné S. and Cohen J. (2007). Maternal age-related differential global expression profiles observed in human oocytes. Reprod Biomed Online 14:700–708 [DOI] [PubMed] [Google Scholar]
- 17.Grøndahl ML, Andersen CY, Bogstad J, Nielsen FC, Meinertz H. and Borup R. (2010). Gene expression profiles of single human mature oocytes in relation to age. Hum Reprod 25:957–968 [DOI] [PubMed] [Google Scholar]
- 18.Wood JR, Dumesic DA, Abbott DH. and Strauss JF., 3rd (2007). Molecular abnormalities in oocytes from women with polycystic ovary syndrome revealed by microarray analysis. J Clin Endocrinol Metab 92:705–713 [DOI] [PubMed] [Google Scholar]
- 19.Markholt S, Grøndahl ML, Ernst EH, Anderse CY, Ernst E. and Lykke-Hartmann K. (2012). Global gene analysis of oocytes from early stages in human folliculogenesis shows high expression of novel genes in reproduction. Mol Hum Reprod 18:96–110 [DOI] [PubMed] [Google Scholar]
- 20.Jones GM, Cram DS, Song B, Magli MC, Gianaroli L, Lacham-Kaplan O, Findlay JK, Jenkin G. and Trounson AO. (2008). Gene expression profiling of human oocytes following in vivo or in vitro maturation. Hum Reprod 23:1138–1144 [DOI] [PubMed] [Google Scholar]
- 21.Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, Shimamoto S, Imamura T, Nakashima K, Saitou M. and Hayashi K. (2016). Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature 539:299–303 [DOI] [PubMed] [Google Scholar]
- 22.Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH. and Surani MA. (2015). Sox17 is a critical specifier of human primordial germ cell fate. Cell 160:253–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, et al. (2015). Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17:178–194 [DOI] [PubMed] [Google Scholar]
- 24.Jung D, Xiong J, Ye M, Qin X, Li L, Cheng S, Luo M, Peng J, Dong J, et al. (2017). In vitro differentiation of human embryonic stem cells into ovarian follicle-like cells. Nat Commun 8:15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White YAR, Woods DC, Takai Y, Ishihara O, Seki H. and Tilly JL. (2012). Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med 18:413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods DC. and Tilly JL. (2013). Isolation, characterization and propogation of mitotically active germ cells from adult mouse and human ovaries. Nat Protoc 8:966–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods DC. and Tilly JL. (2015). Autologous germline mitochondrial energy transfer (AUGMENT) in human assisted reproduction. Semin Reprod Med 33:410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W. and Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473:337–342 [DOI] [PubMed] [Google Scholar]
- 29.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES. and Golub TR. (1999). Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A 96:2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P. and Mesirov JP. (2006). GenePattern 2.0. Nat Genet 38:500–501 [DOI] [PubMed] [Google Scholar]
- 31.Heberle H, Vaz Meirelles G, da Silva FR, Telles GP. and Minghim R. (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Apli E, Birim M, et al. (2013). The proteomics identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41:D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurilo LF. (1981). Oogenesis in antenatal development in man. Hum Genet 57:86–92 [DOI] [PubMed] [Google Scholar]
- 34.Konishi I, Fujii S, Okamura H, Parmley T. and Mori T. (1986). Development of interstitial cells and ovigerous cords in the human fetal ovary: an ultrastructural study. J Anat 148:121–135 [PMC free article] [PubMed] [Google Scholar]
- 35.Sato K, Nishida KM, Shibuya A, Siomi MC. and Siomi H. (2011). Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Genes Dev 25:2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adelman CA. and Petrini JH. (2008). ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 4:e1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding X, Liu G, Xu B, Wu C, Hui N, Ni X, Wang J, Du M, Teng X. and Wu J. (2016). Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci Rep 6:28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT. and Pera RA. (2004). Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet 13:727–739 [DOI] [PubMed] [Google Scholar]
- 39.Chen HF, Kuo HC, Chien CL, Shun CT, Yao YL, Ip PL, Chuang CY, Wang CC, Yang YS. and Ho HN. (2007). Derivation, characterization and differentiation of human embryonic stem cells: comparing serum-containing versus serum-free media and evidence of germ cell differentiation. Hum Reprod 22:567–577 [DOI] [PubMed] [Google Scholar]
- 40.Tilgner K, Atkinson SP, Golebiewska A, Stojkovic M, Lako M. and Armstrong L. (2008). Isolation of primordial germ cells from differentiating human embryonic stem cells. Stem Cells 26:3075–3085 [DOI] [PubMed] [Google Scholar]
- 41.Gu W, Tekur S, Reinbold R, Eppig JJ, Choi YC, Zhenge JZ, Murray MT. and Hecht NB. (1998). Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol Reprod 59:1266–1274 [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Medvedex S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM. and Hecht NB. (2005). Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A 102:5755–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page SL. and Hawley RS. (2004). The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20:525–558 [DOI] [PubMed] [Google Scholar]
- 44.Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K. and Hoog C. (2002). Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296:1115–1118 [DOI] [PubMed] [Google Scholar]
- 45.Smith RW, Anderson RC, Smith JW, Brook M, Richardson WA. and Gray NK. (2011). DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA 17:1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, Kolodner R, Pollard JW. and Kucherlapati R. (1999). Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet 21:123–127 [DOI] [PubMed] [Google Scholar]
- 47.de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P. and te Riele H. (1999). Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev 13:523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Jr, Kolodner RD, Kucherlapati R, Pollard JW. and Edelmann W. (2000). MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev 14:1085–1097 [PMC free article] [PubMed] [Google Scholar]
- 49.Jaroudi S, Kakourou G, Cawood S, Doshi A, Ranieri DM, Serhal P, Harper JC. and SenGupta SB. (2009). Expression profiling of DNA repair gnes in human oocytes and blastocysts using microarrays. Hum Reprod 24:2649–2655 [DOI] [PubMed] [Google Scholar]
- 50.Childs AJ, Cowan G, Kinnell HL, Anderson RA. and Saunders PT. (2011). Retinoic acid signaling and the control of meiotic entry in the human fetal gonad. PLoS One 6:e20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoop-Myer C. and Amon A. (1999). Meiosis: Rec8 is the reason for cohesion. Nat Cell Biol 1:E125–E127 [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Cruz R, Brieño MA, Roig I, Grossmann M, Velilla E, Pujol A, Cabero L, Pessarrodona A, Barbero JL. and Garcia Caldés M. (2010). Dynamics of cohesin proteins REC8, STAG3, SMC1 beta and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum Reprod 25:2316–2327 [DOI] [PubMed] [Google Scholar]
- 53.Hopkins J, Hwang G, Jacob J, Sapp N, Bedigian R, Oka K, Overbeek P, Murray S. and Jordan PW. (2014). Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet 10:e1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Syrjanen JL, Pellegrini L. and Davies OR. (2014). A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife 3:E02963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saxe JP, Chen M, Zhao H. and Lin H. (2013). Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J 32:1869–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soper SF, van Der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P. and Bortvin A. (2008). Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15:285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malki S, van der Heijden GW, O'Donnell KA, Martin SL. and Bortvin A. (2014). A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell 29:521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao L, Wang Y, Zhou Y, Sun Y, Sun W, Wang L, Zhou C, Zhou J. and Zhang J. (2010). Identification of a novel human cancer/testis gene MAEL that is regulated by DNA methylation. Mol Biol Rep 37:2355–2360 [DOI] [PubMed] [Google Scholar]
- 59.Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu W, Guo H, et al. (2017). Single-cell RNA-Seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20:1–16 [DOI] [PubMed] [Google Scholar]
- 60.Rosario R, Smith RWP, Adams IR. and Anderson RA. (2017). RNA immunoprecipitation identifies novel targets of DAZL in human foetal ovary. Mol Hum Reprod 23:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yatsenko AN, Georgiadis AP, Röpke A, Berman AJ, Jaffe T, Olszewska M, Westernströer B, Sanfilippo J, Kurpisz M, et al. (2015). X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372:2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richards JS, Russell DL, Ochsner S. and Espey LL. (2002). Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu Rev Physiol 64:69–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data generated herein have been deposited into the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository [32], with PXD006898 assigned as the data set identifier. Expanded data tables for all mass spectrometry quantitative values and comparison analyses have been deposited at Mendeley Data under DOI:10.17632/x7zbyj6xh7.1.