Abstract
Neuroticism is a relatively stable personality trait characterised by negative emotionality (e.g., worry, guilt) 1; twin study heritability ranges 30 to 50% 2, and SNP-based heritability ranges 6 to 15% 3–6. Increased neuroticism is associated with poorer mental and physical health 7,8, translating to high economic burden 9. Genome-wide association (GWA) studies of neuroticism have identified up to 11 genetic loci 3,4. Here we report 116 significant independent loci from a GWA of neuroticism in 329,821 UK Biobank participants; 15 of these replicated at P<.00045 in an unrelated cohort (N = 122,867). Genetic signals were enriched in neuronal genesis and differentiation pathways, and substantial genetic correlations were found between neuroticism and depressive symptoms (rg = .82, SE=.03), major depressive disorder (MDD; rg = .69, SE=.07) and subjective wellbeing (rg = -.68, SE=.03) alongside other mental health traits. These discoveries significantly advance our understanding of neuroticism and its association with MDD.
Understanding why people differ in neuroticism will provide an important contribution to understanding people’s liability to poor mental health throughout the life course. The strong genetic correlation between neuroticism and mental health, especially anxiety and major depressive disorder 10,11, means that exploring the genetic contribution to differences in neuroticism is one way to understand more about these common and burdensome, but aetiologically intractable illnesses. In the largest GWA study of major depressive disorder (MDD; 130,664 cases vs 330,470 controls), 44 independent genetic loci were identified 12.
UK Biobank has health, medical and genetic information for over 500,000 individuals aged 39-73 years from the United Kingdom, assessed between 2006 and 2010 13,14. We performed a GWA analysis of trait neuroticism in 329,821 unrelated White British adults (152,710 male (46.3%)) with high-quality genotype data (Online Methods). Neuroticism was measured by the total score of the 12-item Eysenck Personality Questionnaire-Revised Short Form (EPQ-R-S) 15; missing item data (ranging 1.8% to 4.7%) were imputed with reference to age and sex, and individuals with greater than 4 missing items were excluded (Supplementary Note, Supplementary Table 1 and Supplementary Fig. 1). For analysis, the score was residualized for the effects of age, sex, assessment centre, genotype batch, array, and 40 genetic principal components. This score was tested against 18,485,882 bi-allelic single nucleotide polymorphism (SNP) variants, based on the Haplotype Reference Consortium panel 16, with a minor allele frequency ≥ 0.0005 and an information/imputation quality score of ≥ 0.1 under an additive model. The distribution of obtained versus expected results under the null hypothesis showed some genomic inflation, with a lambda of 1.15 (quantile-quantile plot shown in Supplementary Fig. 2). Univariate linkage disequilibrium score (LDSC) regression 17 estimates indicated that 96.8% of this inflation was due to the presence of a large polygenic signal with the intercept being close to 1 (1.02, SE = .01). SNP-based heritability of neuroticism was estimated at .108 (SE=.005) using LDSC.
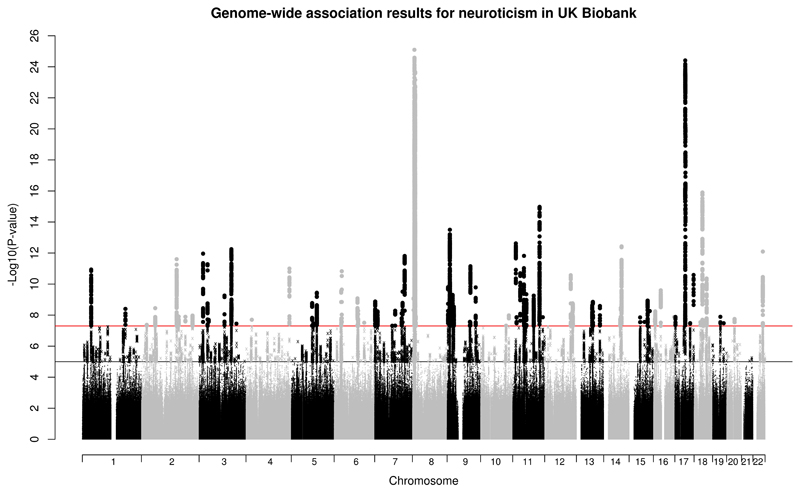
Genome-wide significance (P < 5 x 10-8) was demonstrated for 10,353 genetic variants with a further 17,668 variants at a suggestive level (P < 1 x 10-5) (Supplementary Table 2). The Manhattan plot is shown in Figure 1 and gene annotation for the significant SNPs in Supplementary Table 3. SNPs identified in previous neuroticism GWA studies were mostly significant in our sample (Supplementary Note and Supplementary Table 2) and substantial overlap with MDD SNPs (75%) and genes was found (Supplementary Note and Supplementary Table 4). The major histocompatibility complex (MHC) region has been previously linked to schizophrenia, a psychiatric comorbidity trait (including MDD) 18,19 and MDD 12. It contained 3 significant independent genetic loci associated with neuroticism, two were in genes (GABBR1, TNXB) connected with schizophrenia 20,21. The primary associated SNP, rs2021722, for schizophrenia 18, was present in our study and nominally significant (P = 9.42 x 10-5). Supplementary Figure 3 indicates the previous MHC associations in relation to our findings.
Figure 1.
GWA results for neuroticism in 329,821 UK Biobank individuals.
116 of the significant SNPs were independent (r2 > 0.1 and within 500kb of the significant index SNP); these lead SNPs are shown in Supplementary Table 5 with the number of associated SNPs, region size, and genes within the LD interval. 73 lead SNPs were located within genes, 5 were exonic (in MSRA, NOS1, PINX1, ZCCHC14, and C12orf49) and a further 2 were coding SNPs in RPP21 (a missense mutation) and AGBL1 (synonymous), 55 were intronic and 10 were noncoding RNA variants; 42 were intergenic. For the 116 independent SNPs, evidence of expression quantitative trait loci (eQTL) was explored using the GTEx database, 44 were eQTLs (Supplementary Table 5). A Regulome DB score was used to identify SNPs with a likely regulatory function. 33 of the 116 SNPs were included in the Regulome DB database and 8 of these had a score < 3, indicating that they are likely to be involved in gene regulation (Supplementary Table 5).
Replication of the significant association signals in UK Biobank was sought from the results of a GWA meta-analysis of neuroticism that we performed using 23andMe (N = 59,206) 22 and the Genetics of Personality Consortium (GPC-2; N = 63,661) 23. Of the 10,353 genome-significant SNPs in UK Biobank, 10,171 were available in the replication cohorts, and 8,774 of these increased in significance when the replication cohorts were meta-analysed with UK Biobank. This indicated a consistent direction of allelic effect (Supplementary Table 6).
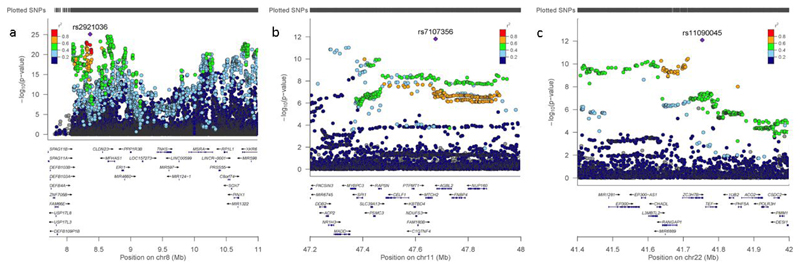
Of the 116 independent associated SNPs, 111 were present in the replication cohort, with 51 nominally significant (P < .05; Supplementary Table 5), and 15 at a Bonferroni-corrected level (P < .00045; Table 1). One of these, rs2953805, was previously associated with morning chronotype 24, a trait relating to lower neuroticism 25 and showing allelic effects in the expected direction. The low replication rate (13.5%) at a strict corrected level reflects the finding that effect sizes are extremely small (up to .02 of a SD increase in neuroticism score per allele) and will thus require similarly large replication samples to confirm their effects. Figure 2a-c shows the regional association plot for chromosomes 8, 11 and 22 in which multiple genes were present in the associated LD region. Of the five chromosome 8 loci only one lead SNP tagged a well-known inversion, previously linked to neuroticism (Supplementary Note and Supplementary Fig. 4), although associations in the broad region had been attributed to the inversion 4 and so might cautiously be considered as a single locus.
Table 1. Fifteen independent SNPs associated with neuroticism in UK Biobank most strongly replicated (with consistent allelic effect) in the meta-analysis of 23andMe and the GPC cohorts. Bolded genes were significant in the gene-based tests.
| Chr | SNP | MAF | Discovery P-value (N=329,821) | Replication P-value (N=122,867) | Nearest Gene | Distance to Gene | Genes within Range | Significant in Previous GWA Studies |
|---|---|---|---|---|---|---|---|---|
| 1 | rs169235 | .25 | 3.97×10-9 | 2.55E-05 | CACNA1E** | 0 | ||
| 5 | rs1422192^ | .17 | 1.68×10-9 | 6.54E-07 | LINC00461 | 0 | MEF2C** | |
| 8*ǂ | rs2921036 | .49 | 8.04×10-26 | 3.27E-07 | . | . | CLDN23, ERI1**, MFHAS1**, SGK223 | |
| 8*ǂ | rs2953805 | .47 | 3.02×10-22 | 1.26E-08 | U3 | 1292 | CLDN23, ERI1**, MFHAS1**, PPP1R3B | Morning vs Evening Chronotype 24 |
| 8*ǂ | rs6982308 | .49 | 6.46×10-21 | 2.26E-08 | MSRA** | 0 | ||
| 8*ǂ | rs7005884 | .45 | 1.92×10-23 | 1.34E-07 | XKR6 | 0 | C8orf74, PINX1**, PRSS55**, RP1L1, SOX7**, XKR6 | |
| 8*ǂ | rs10097870 | .47 | 2.18×10-24 | 6.51E-07 | LINC00208 | 5665 | BLK**, CTSB**, DEFB134**, DEFB135, DEFB136, FAM167A**, FDFT1, GATA4, LOC100133267, MTMR9, NEIL2, SLC35G5, XKR6 | |
| 9 | rs1521732 | .37 | 2.91×10-9 | 4.01E-06 | LINGO2 | 0 | ||
| 9 | rs72694263 | .08 | 2.12×10-8 | 0.000237 | . | . | ||
| 11* | rs7107356 | .50 | 1.52×10-12 | 1.34E-05 | AGBL2 | 4973 | ACP2, AGBL2, C1QTNF4, CELF1, DDB2**, FAM180B, FNBP4**, KBTBD4, MADD**, MTCH2**, MYBPC3, NDUFS3, NR1H3, NUP160**, PSMC3, PTPMT1, RAPSN, SLC39A13**, SPI1 | Neuroticism 4 |
| 11* | rs7111031 | .36 | 1.06×10-15 | 0.000215 | . | . | DRD2 | |
| 15* | rs7175083 | .48 | 1.16×10-9 | 0.000297 | LINGO1 | 0 | ||
| 17 | rs7502590 | .15 | 2.61×10-11 | 0.000146 | BAIAP2 | 0 | AATK, BAIAP2 | |
| 18* | rs11082011 | .34 | 1.25×10-16 | 2.05E-06 | CELF4 | 0 | ||
| 22* | rs11090045 | .30 | 8.04×10-13 | 5.40E-07 | ZC3H7B** | 0 | ACO2, C22orf46, CHADL, CSDC2**, DESI1, EP300, L3MBTL2**, MEI1, NHP2L1, PHF5A, PMM1, POLR3H, RANGAP1**, RBX1, TEF**, TOB2, XRCC6, ZC3H7B** |
Figure 2.
Regional association plot for suggestive/significant signals in UK Biobank on a) chromosome 8p (site of the inversion polymorphism), b) chromosome 11, and c) chromosome 22. The SNP association p-value is shown on the y-axis and the SNP position (with gene annotation) appears on the x-axis; for each SNP, the strength of LD with the lead SNP is colour coded based on its r2. Plots were produced in LocusZoom.
All 69 genes located within the 15 replicated loci were classified in terms of their molecular function, biological process and protein class using the Protein Analysis Through Evolutionary Relationships Classification System which includes 14,710 protein families categorised into 76,032 functionally distinct subfamilies 26. Supplementary Figure 4 shows that a large number of genes 1) coded for nucleic acid binding and transcription factors, 2) contributed to metabolic and cellular processes, and 3) had a role in binding and catalytic activity molecular functioning. Transcription factors, in particular, have been implicated in the aetiology of depression 27,28, and miRNAs—which have been linked with anxiety29 and depression 30—might target genes with roles in binding (e.g., POLR3H). The PsyGeNET (v2.0) database showed that of the 69 genes, four have been associated with psychiatric disorder (Supplementary Table 7): DRD2 (bipolar, depression, substance use/dependence, delirium), EP300 (alcoholic intoxication), TEF (depression) and MSRA (schizophrenia). Variants in CACNA1E have been associated with cross-psychiatric disorder overlap and migraine 18,31.
A GTEx database search for the 15 replicated SNPs showed that 9 were associated with significant regulation of 60 genes expressed in a variety of tissues (Supplementary Table 8). Of the 30 brain expression associations, half of these were in the cerebellum: 4 SNPs regulating 10 genes. Interestingly, MRI studies have shown associations between cerebellar volume and neuroticism, and cerebellar blood flow in response to negative emotional cues 32,33. In the BRAINEAC search, all SNPs were identified as eQTLs in at least one brain region at a nominal significance level (P < .05) and 10 were supported at a Bonferroni-corrected level of P < .0003 (Supplementary Table 9). Of potential interest, rs7107356, a novel SNP in an intergenic region of chromosome 11, regulates MTCH2 in the cerebellar cortex (P = 4.5x10-6). MTCH2 is involved in metabolic pathways and cell function 34 and variants of this gene have been associated with BMI 35.
Gene-based analysis of the GWA results was performed using MAGMA 36 ; 249 genes were significantly associated at a Bonferroni-corrected level (α = 0.05 / 18,080; P < 2.77 × 10-6 ; Supplementary Table 10). Three of these were genes (STH, HIST1H3J, HIST1H4L) containing a single SNP. Of the replicated independent GWA SNPs that were in genes, the following significant genes were corroborated in the gene-based results: CACNA1E, XKR6, MSRA, LINGO2, CELF4, ZC3H7B and BAIAP2. SNP rs6981523, previously identified in 23andMe for neuroticism 22, was an intergenic SNP near XKR6; this gene was the second most significant gene in our gene-based analysis (P = 6.55 × 10-32). L3MBTL2 and CHADL, wherein 23andMe’s other significant SNP, rs9611519, resided, showed respective gene-based p-values of 2.40 × 10-6 and 1.15 × 10-6.
Pathway analysis in MAGMA highlighted 5 significant gene ontology pathways (family-wise error P < 1.21 × 10-6): neuron spine (cellular), homophilic cell adhesion via plasma membrane adhesion molecules (biological), neuron differentiation (biological), cell cell adhesion via plasma membrane adhesion molecules (biological), and neurogenesis (biological). See Table 2 for further details. Of note is the neurogenesis pathway, a hypothesis of which exists for depression (and to a lesser extent, anxiety) based on stress reducing neurogenesis in the hippocampus and on the action of antidepressants on brain circuitry 37,38. Further, variants in PLXNA2, potentially involved in adult neurogenesis, have been associated with anxiety and neuroticism 39. Cell adhesion molecules have been implicated in neuropsychiatric disorder 40, and protocadherins specifically with neuroticism and risk of mood disorder 41, which supports the importance of cell adhesion pathways. A further gene-set analysis of genes expressing proteins that can bind to anti-depressant drug molecules was significant (P = .005) re-affirming the dependency of neuroticism and depression on shared biological pathways. This is consistent, for example, with findings for CRHR1 (highlighted in our SNP and gene-based analysis), a gene involved in normal hormonal responses to stress (the glucocorticoid pathway being a relevant and well-known target) and associated with anxiety, depression and neuroticism 3,42,43. That genes influencing neuroticism reveal pathways involved in currently prescribed and effective antidepressant action suggests that neuroticism could be a potentially useful clinical stratifying factor for effective antidepressant action. There may also be clinical utility in knowing a person’s level of neuroticism after the occurrence of a stressful life event and therefore pre-empting onset of depression via drug therapy in those high in neuroticism. Because our GWA of neuroticism reveals signals associated with the known biological action of existing antidepressants, it may be useful as a means of discovering (or re-purposing) new pharmacological interventions for MDD.
Table 2. Significant gene ontology pathways for neuroticism in UK Biobank.
| Pathway | Number of genes | Beta | SE | P-value | Corrected P | Definition |
|---|---|---|---|---|---|---|
| Neuron Spine | 147 | 0.560 | 0.107 | 7.77×10-8 | 0.0282 | A small membranous protrusion, often ending in a bulbous head and attached to the neuron by a narrow stalk or neck. |
| Homophilic Cell Adhesion Via Plasma Membrane Adhesion Molecules | 115 | 0.490 | 0.0938 | 8.81×10-8 | 0.0289 | The attachment of a plasma membrane adhesion molecule in one cell to an identical molecule in an adjacent cell. |
| Neuron Differentiation | 1341 | 0.145 | 0.0288 | 2.36×10-7 | 0.0357 | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| Cell Cell Adhesion Via Plasma Membrane Adhesion Molecules | 828 | 0.183 | 0.0364 | 2.72×10-7 | 0.0372 | The attachment of one cell to another cell via adhesion molecules that are at least partially embedded in the plasma membrane. |
| Neurogenesis | 195 | 0.419 | 0.0859 | 5.35×10-7 | 0.0439 | Generation of cells within the nervous system |
LD score regression 44 was used to estimate the genetic correlation between neuroticism and a variety of health traits (Supplementary Tables 11 and 12). The strongest correlation was observed for depressive symptoms (rg = .82, SE = .03). Major depressive disorder, subjective wellbeing, and tiredness showed moderate-to-strong correlations (.62-.69). The stronger correlation for depressive symptoms than depressive disorder might be indicative of improved sensitivity of continuous versus dichotomous traits but might also point to inventory item overlap (greater conceptual similarity) for depressive symptoms and/or noise in MDD diagnosis. Genetic correlations with neuroticism were moderate for self-rated health (.41), moderate-to-low for schizophrenia, ADHD, anorexia nervosa and educational attainment (~|.20|), and low for bipolar disorder and smoking status (|.11|). The genetic correlation of one between Eysenck neuroticism and other neuroticism scales (used by 23andMe and the GPC) confirms that GWA meta-analysis based on different measurement instruments is valid. Mendelian randomization was used to determine whether the genetic correlation between neuroticism and non-psychiatric variables (less likely to be influenced by pleiotropy), smoking status and educational attainment, represented a causal relationship from neuroticism. For smoking status, the beta of 0.23 was significant in the inverse variance weighted model (P = .00002) which is preferred in the presence of heterogeneity (P = .001); the MR Egger regression did not show significant directional pleiotropy (intercept = 0.02, P = .10) thus supporting a causal relationship. For educational attainment, the beta of -0.09 was significant (P = 8.35 × 10-6) in the inverse variance weighted model (heterogeneity P = 5.87 × 10-7), with no evidence of directional pleiotropy (intercept = 0, P = .23). Although theoretically less plausible, the reverse causal direction should be investigated in UK Biobank once a large number of significant SNPs influencing smoking status and educational attainment have been estimated in non-overlapping samples.
Polygenic profile analyses based on the SNP inclusion threshold with the optimal signal-to-noise ratio (P < .05) indicated that the neuroticism polygenic score explained 2.79% of the variance in neuroticism (β = .19, P = 2.65 × 10-47) and 0.8% of the variance in depression status (OR = 1.25, P = 1.53 × 10-8) in Generation Scotland (GS; N = 7,388) 45. Results for polygenic scores in GS based on other SNP significance inclusion thresholds (0.01, 0.05, 0.1, 0.5 and 1) from the UK Biobank GWA can be found in Supplementary Table 13.
The combination, in UK Biobank, of a large ethnically homogenous sample and a well-validated neuroticism scale has afforded the discovery of 15 stringently replicated genetic loci that influence neuroticism levels, four of them novel. Most lead variants were associated with gene regulation, with half of these expressed in the brain; single variant and gene associations overlapped substantially with MDD findings, and genes in antidepressant-targeted pathways were over-represented. There was also support for neuroticism having causal effects on socio-economic markers. These discoveries promise paths to understand the mechanisms whereby some people become depressed, and of broader human differences in happiness, and they are a resource for those seeking novel drug targets for major depression. After millennia in which scholars and researchers have sought the sources of individual differences in proneness to dysphoria 46, the present study adds significantly to explaining the (genetic) anatomy of melancholy.
Online Methods
Genome-wide association analysis in UK Biobank
An imputed dataset, including >92 million variants, referenced to the UK10K haplotype, 1000 Genomes Phase 3, and Haplotype Reference Consortium (HRC) panels was available in UK Biobank. The current analysis includes only those SNPs available in the HRC reference panel 47. Quality control filters were applied (see Supplementary Note) which resulted in 18,485,882 imputed SNPs for analysis in 329,821 individuals. The GWA of neuroticism was conducted using BGENIE 48, a program specifically developed to analyse UK Biobank data in a fast and efficient manner. Further information can be found at the following URL: https://jmarchini.org/bgenie/. A linear SNP association model was tested which accounted for genotype uncertainty. Neuroticism was pre-adjusted for age, sex, genotyping batch, genotyping array, assessment centre, and 40 principal components to speed up analysis.
The number of independent signals from the GWA analysis was determined using LD-clumping in PLINK v1.90b3i 49 (see URLs). The LD structure was based on SNPs with a p-value < 1 × 10-3 that were extracted from the imputed genotypes. Index SNPs were identified (P < 5 × 10-8) and clumps were formed for SNPs with P < 1 × 10-5 that were in LD (r2 > 0.1) and within 500kb of the index SNP. SNPs were assigned to no more than one clump.
Meta-analysis of GWA Results
Two meta-analyses were performed. Firstly, to check for replication of the significant (P < 5 × 10-8) GWA signals in UK Biobank, results from a meta-analysis of 23andme 50 (the full GWA summary statistics were made available from 23andMe) and the Genetics of Personality Consortium (GPC-2) 51 (the full GWA summary statistics were publicly available see URLs) were used. This meta-analysis was conducted using METAL 52 and due to the lack of phenotype harmonisation across the cohorts, a sample size weighted meta-analysis was preferred. A second meta-analysis of UK Biobank and the replication cohorts was performed using the same method, but only for the SNPs that were significant in UK Biobank.
Genome-wide Gene-based Analysis
Gene-based analysis of neuroticism was performed using MAGMA 53, which provides gene-based statistics derived using the results of the GWA analysis. Genetic variants were assigned to genes based on their position according to the NCBI 37.3 build, with no additional boundary placed around the genes. This resulted in a total of 18,080 genes being analysed. The European panel of the 1000 Genomes data (phase 1, release 3) was used as a reference panel to account for linkage disequilibrium. A genome-wide significance threshold for gene-based associations was calculated using the Bonferroni method (α=0.05/18,080; P < 2.77 × 10-6).
Functional annotation and gene expression
For the 116 independent genome-wide significant SNPs identified by LD clumping, evidence of expression quantitative trait loci (eQTL) and functional annotation were explored using publicly available online resources. The Genotype-Tissue Expression Portal (GTEx) (see URLs) was used to identify eQTLs associated with the SNPs. Functional annotation was investigated using the Regulome DB database 54 (see URLs). Further to GTEx searches, we investigated whether any of the 15 replicated SNPs were brain expression quantitative loci (eQTLs) by entering them into the brain eQTL database BRAINEAC (see URLs), which contains gene expression data across ten brain regions (cerebellar cortex, frontal cortex, hippocampus, medulla, occipital cortex, putamen, substantia nigra, temporal cortex, thalamus and intralobular white matter). The genes located in the region of replicated independent loci were investigated for protein function using the PANTHER database (Protein ANalysis THrough Evolutionary Relationships, see URLs) which stores data on the evolution and function of protein-coding genes from sequenced genomes of diverse species 55, our focus here on homo sapiens. Uncharacterized gene function is predicted via phylogenetic branching information and the resource enables biological pathway annotation.
Pathway Analysis
Biological pathway analysis was performed on the gene-based analysis results. This gene-set enrichment analysis was conducted utilising gene-annotation files from the Gene Ontology (GO) Consortium (see URLs) 56 taken from the Molecular Signatures Database (MSigDB) v5.2. The GO consortium includes gene-sets for three ontologies; molecular function, cellular components and biological function. This annotation file consisted of 5,917 gene-sets which were corrected for multiple testing correction using the MAGMA default setting correcting for 10,000 permutations.
To determine whether the genetic targets of antidepressants were enriched for neuroticism we performed a competitive gene-set analysis using MAGMA. Gene sets corresponding to the Anatomical Therapeutic Chemical Classification System code N06A Antidepressants (within the Psychoanaleptics class) were downloaded (see URLs). This resulted in a set of 110 unique genes corresponding to those that are the targets of the antidepressants. Enrichment for neuroticism was tested against a set of 5483 ‘druggable’ autosomal genes (see URLs), that is, they code for proteins which can bind to drug-like molecules. Of the 110 antidepressant genes 86 were found amongst the 5483 druggable genes.
Linkage Disequilibrium Score Regression
Univariate Linkage disequilibrium Score (LDSC) regression 57 was used to test for residual stratification in our GWAS summary statistics and to derive a heritability estimate. An LD regression was performed by regressing the GWA test statistics (χ2) on to each SNP’s LD score (the sum of squared correlations between the minor allele frequency count of a SNP with the minor allele frequency count of every other SNP). This regression allows for the estimation of heritability from the slope, and a means to detect residual confounders, the intercept. The percentage inflation in the test statistic due to polygenic signal can be derived by subtracting the LDSC ratio ((intercept - 1)/(mean χ2 - 1)), which represents inflation due to population stratification and other confounding, from 1 and multiplying by 100. Bivariate LDSC regression 58 was used to derive genetic correlations between neuroticism and 18 psychiatric and physical health phenotypes (see Supplementary Table 11). For Alzheimer’s disease, a 500-kb region surrounding APOE was excluded and the analysis re-run (Alzheimer’s disease (500kb)). The genetic correlation between neuroticism as measured by different inventories was also estimated. Further details, including source of GWA summary statistics can be found in the Supplementary Note. Sample overlap could not be controlled for in the LDSC analyses because the exact overlap between the UK Biobank data and the health traits was unknown. In such a case, constraining the intercept to a ‘wrong’ value could lead to biased estimates. Any sample overlap in the present analyses will only affect the intercept of the regression and could lead to inflated standard errors, but will not affect the genetic correlation 12.
Mendelian Randomization
Two sample Mendelian Randomization (MR) was performed using the TwoSampleMR 59 package implemented in R. GWA summary statistics from the GWA of smoking status in 74,053 Europeans 60 was used to create outcome data for the MR between neuroticism and smoking status. 77 independent SNPs associated with neuroticism were available in the smoking status GWA summary data to test for a causal effect of neuroticism on smoking status. There were no significant SNP signals for smoking status to test the reverse causation model. GWA summary statistics from the GWA of educational attainment in 126,559 Caucasians 61 was used to create outcome data for the MR between neuroticism and educational attainment. 75 independent SNPs associated with neuroticism were available in the educational attainment GWA summary data to test for a causal effect of neuroticism on educational attainment. There were too few significant SNPs available for educational attainment to test for a causal effect of educational attainment on neuroticism. Sensitivity analyses were performed to test for heterogeneity and a further test for horizontal pleiotropy was carried out.
Polygenic Prediction into Generation Scotland
Polygenic profile analyses were performed to predict neuroticism and depression status in Generation Scotland (GS) 62. Polygenic profiles were created in PRSice 63 using the UK Biobank neuroticism SNP-based association results, for 7,388 unrelated individuals in GS. SNPs with a MAF <0.01 were removed prior to creating the polygenic profiles. Clumping was used to obtain SNPs in linkage disequilibrium with an r2 < 0.25 within a 250kb window. Individuals were removed from GS if they had contributed to both UK Biobank and GS (n = 302). Polygenic profile scores were created based on the significance of the association in UK Biobank with the neuroticism phenotype, at p-value thresholds of 0.01, 0.05, 0.1, 0.5 and 1 (all SNPs). Linear regression models were used to examine the associations between the polygenic profile and neuroticism score in GS, adjusting for age at measurement, sex and the first 10 genetic principal components to adjust for population stratification. Logistic regression models were used to examine depression status, adjusting for the same covariates as in the neuroticism models. The false discovery rate (FDR) method was used to correct for multiple testing across the polygenic profiles for neuroticism at all five thresholds 64.
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource (Application Nos. 10279 and 4844). Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates [CZD/16/6] and the Scottish Funding Council [HR03006]. Ethical approval for the GS:SFHS study was obtained from the Tayside Committee on Medical Research Ethics (05/S1401/89 Tayside Committee on Medical Research Ethics A). We are grateful to all the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, healthcare assistants and nurses. This work was supported by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1); funding from the Biotechnology and Biological Sciences Research Council (BBSRC) and Medical Research Council (MRC) is gratefully acknowledged. This report represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. W.D.H. is supported by a grant from Age UK (Disconnected Mind Project). A.M.M. and I.J.D. are supported by funding from a Wellcome Trust Strategic Award (104036/Z/14/Z).
Footnotes
URLs
UK Biobank Resource: http://www.ukbiobank.ac.uk
BGENIE: https://jmarchini.org/bgenie/
BRAINEAC: http://www.braineac.org/
Druggable genome: http://dgidb.genome.wustl.edu/
Genotype-Tissue Expression Portal: http://www.gtexportal.org
Gene Ontology: http://geneontology.org
GPC-2 Summary Statistics: http://www.tweelingenregister.org/GPC/
Linkage Disequilibrium Score Regression: https://github.com/bulik/ldsc/wiki
METAL: http://csg.sph.umich.edu/abecasis/metal/index.html
PANTHER: http://pantherdb.org/
PLINK V2: https://www.cog-genomics.org/plink2
PsyGeNet: http://www.psygenet.org/web/PsyGeNET/menu/home
Regulome Database: http://www.regulomedb.org/
Data Availability
The GWA results generated by this analysis are publicly available at http://www.ccace.ed.ac.uk.
Author Disclosure
IJD was a participant in UK Biobank. The other authors declare no conflict of interest.
Author Contributions
M.L. drafted the manuscript with contributions from W.D.H. and I.J.D. G.D., D.C.L., R.E.M., M.J.A. and D.M.H. performed quality control of UK Biobank data and/or Generation Scotland. M.L, G.D, S.P.H., and M.S. analysed the data. T-K.C., C.F-R., W.D.H. and S.E.H. performed/assisted with downstream analysis. C.R.G, C.M.L., and A.M.M provided critical comments on the manuscript draft and analysis. M.L. and I.J.D. co-ordinated the work. All authors commented on and approved the manuscript.
References
- 1.Matthews G, Deary IJ, Whiteman MC. Personality Traits. Cambridge University Press; 2009. [Google Scholar]
- 2.Vukasovic T, Bratko D. Heritability of personality: A meta-analysis of behavior genetic studies. Psychological Bulletin. 2015;141 doi: 10.1037/bul0000017. [DOI] [PubMed] [Google Scholar]
- 3.Smith DJ, et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry. 2016;21:749–57. doi: 10.1038/mp.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okbay A, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Power RA, Pluess M. Heritability estimates of the Big Five personality traits based on common genetic variants. Transl Psychiatry. 2015;5:e604. doi: 10.1038/tp.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinkhuyzen AA, et al. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl Psychiatry. 2012;2:e102. doi: 10.1038/tp.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubzansky LD, Martin LT, Buka SL. Early manifestations of personality and adult health: a life course perspective. Health Psychol. 2009;28:364–72. doi: 10.1037/a0014428. [DOI] [PubMed] [Google Scholar]
- 8.Strickhouser JE, Zell E, Krizan Z. Does Personality Predict Health and Well-Being? A Metasynthesis. Health psychology: official journal of the Division of Health Psychology, American Psychological Association. 2017 doi: 10.1037/hea0000475. [DOI] [PubMed] [Google Scholar]
- 9.Cuijpers P, et al. Economic costs of neuroticism: a population-based study. Arch Gen Psychiatry. 2010;67:1086–1093. doi: 10.1001/archgenpsychiatry.2010.130. [DOI] [PubMed] [Google Scholar]
- 10.Few LR, et al. Genetic variation in personality traits explains genetic overlap between borderline personality features and substance use disorders. Addiction. 2014;109:2118–27. doi: 10.1111/add.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63:1113–20. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 12.Wray NR, Sullivan PF. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. bioRxiv. 2017 doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bycroft C, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv. 2017 [Google Scholar]
- 15.Eysenck SB, Eysenck HJ, Barrett P. A revised version of the psychoticism scale. Personality and individual differences. 1985;6:21–29. [Google Scholar]
- 16.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegyi H. GABBR1 has a HERV-W LTR in its regulatory region – a possible implication for schizophrenia. Biology Direct. 2013;8:5–5. doi: 10.1186/1745-6150-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei J, Hemmings GP. TNXB locus may be a candidate gene predisposing to schizophrenia. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;125B:43–49. doi: 10.1002/ajmg.b.20093. [DOI] [PubMed] [Google Scholar]
- 22.Lo M-T, et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet. 2017;49:152–156. doi: 10.1038/ng.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genetics of Personality, C et al. Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry. 2015;72:642–50. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duggan KA, Friedman HS, McDevitt EA, Mednick SC. Personality and Healthy Sleep: The Importance of Conscientiousness and Neuroticism. PLOS ONE. 2014;9:e90628. doi: 10.1371/journal.pone.0090628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulser F. The role of CREB and other transcription factors in the pharmacotherapy and etiology of depression. Annals of Medicine. 2002;34:348–356. doi: 10.1080/078538902320772106. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, et al. Forkhead box O transcription factors as possible mediators in the development of major depression. Neuropharmacology. 2015;99:527–537. doi: 10.1016/j.neuropharm.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Malan-Müller S, Hemmings SMJ, Seedat S. Big Effects of Small RNAs: A Review of MicroRNAs in Anxiety. Molecular Neurobiology. 2013;47:726–739. doi: 10.1007/s12035-012-8374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues in Clinical Neuroscience. 2014;16:43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambrosini A, et al. Possible Involvement of the CACNA1E Gene in Migraine: A Search for Single Nucleotide Polymorphism in Different Clinical Phenotypes. Headache: The Journal of Head and Face Pain. 2017;57:1136–1144. doi: 10.1111/head.13107. [DOI] [PubMed] [Google Scholar]
- 32.Schraa-Tam CKL, et al. fMRI activities in the emotional cerebellum: a preference for negative stimuli and goal-directed behavior. The Cerebellum. 2012;11:233–245. doi: 10.1007/s12311-011-0301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutter DJLG, Koolschijn PCMP, Peper JS, Crone EA. The Cerebellum Link to Neuroticism: A Volumetric MRI Association Study in Healthy Volunteers. PLOS ONE. 2012;7:e37252. doi: 10.1371/journal.pone.0037252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grinberg M, et al. Mitochondrial carrier homolog 2 is a target of tBID in cells signaled to die by tumor necrosis factor alpha. Mol Cell Biol. 2005;25:4579–90. doi: 10.1128/MCB.25.11.4579-4590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Computational Biology. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucassen PJ, Oomen CA, Schouten M, Encinas JM, Fitzsimons CP. Chapter 8 - Adult Neurogenesis, Chronic Stress and Depression A2 - Canales. In: Juan J, editor. Adult Neurogenesis in the Hippocampus. Academic Press; San Diego: 2016. pp. 177–206. [Google Scholar]
- 38.Schoenfeld TJ, Cameron HA. Adult Neurogenesis and Mental Illness. Neuropsychopharmacology. 2015;40:113–128. doi: 10.1038/npp.2014.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray NR, James MR, Mah SP, et al. Anxiety and comorbid measures associated with plxna2. Arch Gen Psychiatry. 2007;64:318–326. doi: 10.1001/archpsyc.64.3.318. [DOI] [PubMed] [Google Scholar]
- 40.Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Research. 2012;1470:130–144. doi: 10.1016/j.brainres.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Chang H, et al. The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorders. Mol Psychiatry. 2017 doi: 10.1038/mp.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeYoung CG, Cicchetti D, Rogosch FA. Moderation of the association between childhood maltreatment and Neuroticism by the corticotropin-releasing hormone receptor 1 gene. Journal of child psychology and psychiatry, and allied disciplines. 2011;52:898–906. doi: 10.1111/j.1469-7610.2011.02404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binder EB, Nemeroff CB. The CRF system, stress, depression and anxiety – insights from human genetic studies. Mol Psychiatry. 2010;15:574–588. doi: 10.1038/mp.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan BK, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith BH, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. International Journal of Epidemiology. 2013;42:689–700. doi: 10.1093/ije/dys084. [DOI] [PubMed] [Google Scholar]
- 46.Burton R. In: The Anatomy of Melancholy. Faulkner TC, Kiessling NK, Blair RL, editors. Oxford University Press; 1989. [Google Scholar]
- 47.Haplotype Reference, C. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bycroft C, et al. Genome-wide genetic data on ~ 500,000 UK Biobank participants. bioRxiv. 2017 [Google Scholar]
- 49.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo M-T, et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet. 2017;49:152–156. doi: 10.1038/ng.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genetics of Personality, C et al. Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry. 2015;72:642–50. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Computational Biology. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gene Ontology, C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic acids research. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bulik-Sullivan BK, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemani G, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016 [Google Scholar]
- 60.Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rietveld CA, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith BH, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. International Journal of Epidemiology. 2013;42:689–700. doi: 10.1093/ije/dys084. [DOI] [PubMed] [Google Scholar]
- 63.Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2014;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological) 1995:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.