Abstract
Intelligence — the ability to learn, reason and solve problems — is at the forefront of behavioural genetic research. Intelligence is highly heritable and predicts important educational, occupational and health outcomes better than any other trait. Recent genome-wide association studies have successfully identified inherited genome sequence differences that account for 20% of the 50% heritability of intelligence. These findings open new avenues for research into the causes and consequences of intelligence using genome-wide polygenic scores that aggregate the effects of thousands of genetic variants. In this Review, we highlight the latest innovations and insights from the genetics of intelligence and their applications and implications for science and society.
Introduction
Life is an intelligence test. During the school years, differences in intelligence are largely the reason why some children master the curriculum more readily than other children. Differences in school performance predominantly inform prospects for further education, which in turn lead to social and economic opportunities such as occupation and income. In the world of work, intelligence matters beyond educational attainment because it involves the ability to adapt to novel challenges and tasks that describe the different levels of complexity of occupations. Intelligence also spills over into many aspects of everyday life such as the selection of romantic partners and choices about health care1. This is why intelligence — often called general cognitive ability2 — predicts educational outcomes3, occupational outcomes4,5 and health outcomes6 better than any other trait. It is also the most stable psychological trait, with a Pearson correlation coefficient of 0.54 from age 11 years to age 90 years7. Box 1 describes what intelligence is and how it is assessed.
Box 1. What is intelligence?
Intelligence can be broadly defined as the ability to learn, reason and solve problems74. It is a latent trait that cannot be directly observed but is inferred from a battery of diverse cognitive test scores, as in widely used ‘intelligence tests’ that yield a so-called ‘IQ score’, which is an acronym for an outdated concept of an ‘intelligence quotient’. Psychometric tests of cognitive abilities differ widely in form and content. For example, some assess verbal ability and others non-verbal ability, some give strict time limits and some are untimed (see figure for examples). Notwithstanding these differences, cognitive test scores are positively inter-correlated75, suggesting that any differences in test scores that occur within an individual are smaller than test score differences that exist between individuals. In other words, a person who scores high on one type of cognitive test relative to other people will also do comparatively well on other cognitive tests. This phenomenon is known as the positive manifold, or simply g, the general factor of intelligence, which emerges from the test scores' covariance, discovered by Spearman in 190476, about the same time that Mendel’s laws of inheritance were rediscovered. The g-factor exemplifies the generalist nature of intelligence as a complex trait that penetrates many behavioural and psychological outcomes, including educational attainment, occupational status, health and longevity77,78.
Individual differences in intelligence are fairly stable across the lifespan, especially from teenage years onwards, with correlations of 0.6 and above38,79. However, intelligence is also subject to change, both within and between individuals. For example, scores from timed cognitive tests tend to peak in young adulthood and decline thereafter80. But more importantly, intelligence has been shown to be malleable, especially in children, through major systematic interventions, such as education81, dietary supplementation82 or adopting children away from impoverished home environments83. That said, identifying ways to effectively improve intelligence remains a key challenge for intelligence research, with many interventions failing to produce reliable and long-term positive effects82,84,85.
Box 1 fig.
During the past century, genetic research on intelligence was in the eye of the storm of the nature–nurture debate in the social sciences8,9. In the 1970s and 1980s, intelligence research and its advocates were vilified10–12. The controversy was helpful in that it raised the quality and quantity threshold for the acceptance of genetic research on intelligence. As a result, bigger and better family studies, twin studies, and adoption studies amassed a mountain of evidence that consistently showed substantial genetic influence on individual differences in intelligence13. Meta-analyses of this evidence indicate that inherited differences in DNA sequence account for about half of the variance on measures of intelligence14.
These studies and applications in neuroscience15 were already pushing intelligence research towards rehabilitation when it was thrust to the forefront of the DNA revolution 4 years ago by genome-wide association studies (GWAS) focused on a very different variable; years of education. In this Review, we discuss early attempts to find the inherited DNA differences that account for the substantial heritability of intelligence, and a twist of fate involving GWAS on years of education, before discussing the results of recent large GWAS of intelligence. The second half of this Review focuses on genome-wide polygenic scores (GPS) for intelligence that aggregate the effects of thousands of DNA variants associated with intelligence across the genome (see Box 2 for how GPS are constructed). We illustrate how GPS for intelligence will transform research on the causes and consequences of individual differences in intelligence, before ending with a discussion of societal and ethical implications. We do not discuss other important research related to intelligence such as evolutionary research16,17 and neuroscience research15 in order to focus on the role of GPS in the new genetics of intelligence.
Box 2. Creating GPS.
Thousands of SNPs are needed to account for the heritability of intelligence and other complex traits because the effect sizes of SNP associations are so small. Aggregating thousands of these miniscule effects in a GPS is the crux of the new genetics of intelligence. There are at least a dozen labels to denote GPS. Most involve the word ‘risk’, such as polygenic risk scores. We prefer the term genome-wide polygenic score (GPS) because ‘risk’ does not apply to quantitative traits, such as intelligence, that have positive as well as negative poles86. The ‘genome-wide’ addition to ‘polygenic score’ distinguishes GPS from polygenic scores that aggregate candidate genes or just the top hits from GWAS. Finally, another reason for using the acronym GPS is that we cannot resist the metaphor of the other ‘GPS’, global positioning system. We see IQ GPS as a system to triangulate on the genetics of intelligence from all domains of the life sciences.
An intelligence test score is a composite of several tests, often with each test weighted by its contribution to general intelligence. In the same way, a GPS is a composite of SNP associations, weighted by their correlation with the trait. The table shows how a GPS could be constructed for one individual for 10 SNPs. GWAS results are used to determine which of the two alleles for a SNP is positively associated with the trait, called the increasing allele. For each SNP, a genotypic score is created by adding the number of increasing alleles. A GPS sums the number of increasing alleles across SNPs — hence, why this is called an additive model. In this example, the individual’s GPS is 9. Because there are 10 SNPs, the possible range of the GPS is from 0 to 20.
A more predictive GPS can be constructed by weighting each genotypic score by the effect size of the SNP (beta for quantitative traits, odds ratio for qualitative traits) as gleaned from GWAS results (see table). For instance, for SNP 1, the correlation with the trait is five times greater than for SNP 10. Multiplying the genotypic score by the correlation gives a weighted genotypic score (see table, last column). Summing these weighted genotypic scores gives this individual a GPS of 0.023 for intelligence. Other ways to improve the predictive power of GPS include taking into account expected SNP effect sizes, the genetic architecture of the trait and specifically modelling linkage disequilibrium87. Programmes including LDpred88 and PRSice89 provide pipelines for the construction of GPS.
How many SNPs should be included in a GPS? The goal is to maximize predictive power in samples independent from the GWAS samples. Using only genome-wide significant hits does not predict nearly as well as using tens of thousands of SNPs. LDpred uses all SNPs, imputed as well as measured, although most SNPs are given near-zero weights.
Once a GPS for intelligence is created for each individual in a sample, it can be used like any other variable in analyses. For example, it can be used to investigate the extent to which this genetic index of intelligence mediates or moderates effects on variables of primary interest to the researcher.
| SNP | Increasing allele | Allele 1 | Allele 2 | Genotypic score | Correlation with trait | Weighted genotypic score |
|---|---|---|---|---|---|---|
| SNP 1 | T | A | T | 1 | 0.005 | 0.005 |
| SNP 2 | C | G | G | 0 | 0.004 | 0.000 |
| SNP 3 | A | A | A | 2 | 0.003 | 0.006 |
| SNP 4 | G | C | G | 1 | 0.003 | 0.003 |
| SNP 5 | G | C | C | 0 | 0.003 | 0.000 |
| SNP 6 | T | A | T | 1 | 0.002 | 0.002 |
| SNP 7 | C | C | G | 1 | 0.002 | 0.002 |
| SNP 8 | A | A | A | 2 | 0.002 | 0.004 |
| SNP 9 | A | T | T | 0 | 0.001 | 0.000 |
| SNP 10 | C | C | G | 1 | 0.001 | 0.001 |
| Polygenic score | 9 | 0.023 |
Finding the heritability of intelligence
Similar to many other complex traits, early results for intelligence were disappointing for more than 100 candidate gene studies18 and for seven GWAS19–25. From the 1990s until 2017 no replicable associations were found. GPS from these early GWAS, which we refer to as ‘IQ1’, predicted only 1% of the variance of intelligence in independent samples. It became clear that the problem was power: the largest effect sizes of associations between individual single nucleotide polymorphisms (SNPs) and intelligence were extremely small, accounting for less than 0.05% of the variance of intelligence. The average effect size of the tens of thousands of SNPs needed to explain the 50% heritability of intelligence is of course much lower. If the average effect size is 0.005%, 10,000 such SNP associations would be needed to explain the 50% heritability of intelligence. To achieve sufficient power for detecting such tiny effect sizes (that is, power of 80%, P = 0.05 one-tailed), sample sizes greater than 250,000 are required. IQ1 GWAS had sample sizes from 18,000 to 54,000, which seemed large at the time but were not sufficiently powered to detect such small effects.
Breakthrough for years of education
A breakthrough for intelligence research came from the unlikely variable of the number of years spent in full-time education, often referred to as educational attainment. Because ‘years of education’ is obtained as a demographic marker in nearly every GWAS, it was possible to accumulate sample sizes with the necessary power to detect very small effect sizes26. Its relevance to intelligence is that years of education is highly correlated phenotypically (0.50) and genetically (0.65) with intelligence27.
In 2013, a meta-analytic GWAS analysis of years of education yielded three genome-wide significant SNP associations in a sample of 125,000 individuals from 54 cohorts28. These associations could be replicated in independent samples29. The largest effect size associated with an individual SNP accounted for a meagre 0.02% of the variation, equivalent to about 2 months of education. Although individual SNPs of such miniscule effect size are fairly useless for prediction, a GPS based on all SNPs regardless of the strength of their association with years of education predicted 2% of the variance in years of education in independent samples28,29. We refer to this GPS as EA1.
Spurred on by this success, in 2016, a second meta-analytic GWAS analysis with a sample size of 294,000 identified 74 significant loci30. This analysis produced a GPS, EA2, that predicted 3% of the variance in years of education on average in independent samples30. Surprisingly, GPS for years of education predicted more variance in intelligence than they predicted for the GWAS target trait of years of education27. For example, EA2 GPS predicts 3% of the variance in years of education but it predicts 4% of the variance in intelligence30. A third GWAS currently in progress includes more than one million participants, making it the largest GWAS for any trait to date. Preliminary results from this GWAS have identified more than 1,000 significant associations and a GPS, EA3, that predicts more than 10% of the variance in years of education in independent samples (Philipp D. Koellinger, personal communication)31. Hence, EA3 GPS is expected to predict more than 10% of the variance in intelligence. The effect size of EA3 GPS for predicting intelligence is likely to rival that of family socioeconomic status, which is indexed by parents' years of education. Across studies, parents' education correlates 0.30 with children’s intelligence, implying that it accounts for 9% of the variance in children’s intelligence38. This association is, however, confounded by genetics, because children inherit the DNA differences that predict their intelligence from their parents. Furthermore, parental phenotypes, such as education, only estimate an average association for offspring, whereas GPS predict intelligence for each individual.
Large-scale GWAS of intelligence
In 2017, the largest GWAS meta-analysis of intelligence, which included ‘only’ 78,000 individuals, yielded 18 genome-wide significant regions32. A GPS (IQ2) derived from these GWAS results finally broke the 1% barrier of previous GWAS of intelligence by predicting 3% of the variance of intelligence in independent samples. However, IQ2 still has less predictive power than the 4% of the variance explained by the EA2 GPS.
A follow-up GWAS meta-analysis reached a sample size of 280,000 with the inclusion of cognitive data from the UK Biobank (www.ukbiobank.ac.uk). This GWAS analysis increased the number of identified genome-wide significant regions from 18 to 20633. A GPS derived from these GWAS analyses, IQ3, predicts about 4% of the variance of intelligence in independent samples33. Other meta-analytic GWAS using the UK Biobank data, which were released in June 2017 and are publicly available, yield similar results34.
These IQ and EA GPS results are summarized in Figure 1. It might seem disappointing that the increase of the intelligence GWAS sample sizes from 78,000 to 280,000 only boosted the predictive power of the IQ GPS from 3% to 4%. However, this result is parallel to GWAS results for years of education: after increasing sample sizes from 125,000 to 294,000, the variance in years of education predicted by the EA GPS only grew from 2% to 3%. Note that the predictive power of EA GPS jumped to more than 10% of the variance in preliminary analyses of the latest meta-analytic GWAS (EA3) with a sample size of over one million (Philipp D. Koellinger, personal communication). We can expect a similar jump in predictive power of the IQ GPS when the sample size for GWAS meta-analyses of intelligence exceeds one million. However, it is more difficult to obtain huge sample sizes for intelligence, which has to be tested, than for years of education, which can be assessed with a single self-reported item.
Figure 1. Variance explained by IQ GPS and by EA GPS in their target traits as a function of GWAS sample size.
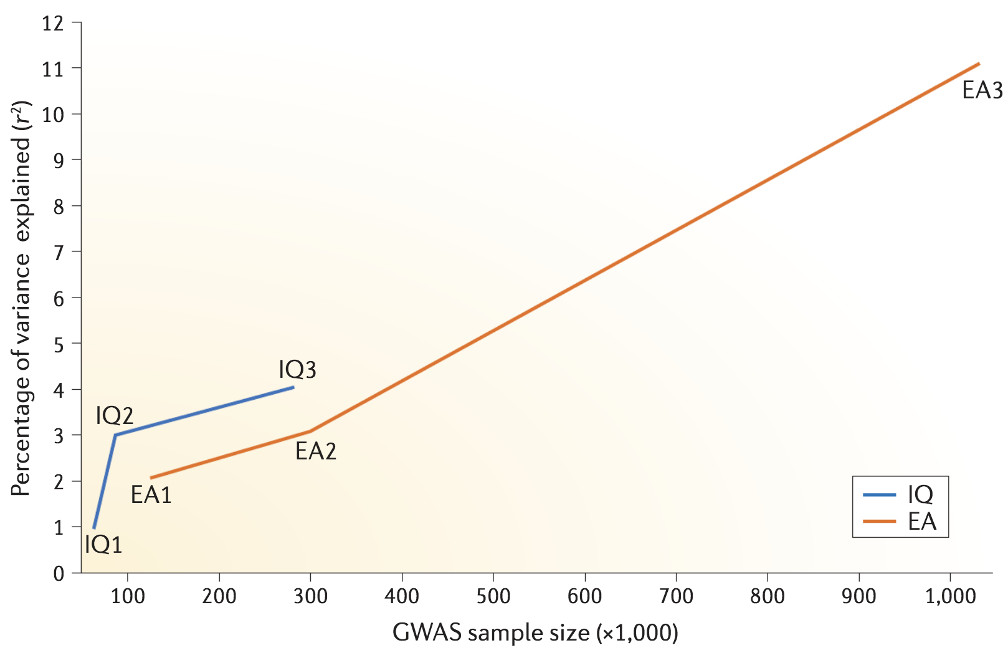
GPS prediction of intelligence and educational attainment has increased linearly with sample size. The predictive power of GPS derived from GWAS of intelligence has risen in the last 2 years from 1% to 4%. The latest EA3 GPS predicts more than 10% of the variance in intelligence (P. D. Koellinger, personal communication), more than twice as much as the latest IQ3 GPS. Extrapolating from the results of EA3 with a sample size of over one million, we predict that more than 10% of the variance in intelligence will be predicted from an IQ GPS derived from a GWAS of intelligence with a sample size of one million. IQ122: n = 54,000, r2 = 0.01. IQ232: n = 78,000, r2 = 0.03. IQ333: n = 280,000, r2 = 0.04. EA128: n = 125,000, r2 = 0.02. EA230: n = 294,000, r2 = 0.03. EA3: n = 1,100,000, r2 > 0.10.
Missing heritability
It is possible to use multiple GPS to boost the power to predict intelligence by aggregating GPS in a way analogous to aggregating SNPs to produce GPS (Box 3). Including EA2 GPS, IQ2 GPS and other GPS in this multivariate way can already predict up to 7% of the variance in intelligence35,36. Multivariate GPS analyses that incorporate multiple GPS in addition to EA2 GPS and IQ2 GPS will explain substantially more than 10% of the variance in intelligence, which is more than 20% of the 50% heritability of intelligence.
Box 3. Using multiple GPS to predict a trait.
Aggregating thousands of SNP associations in GPS has been key to predicting individual differences in complex traits such as intelligence. In an analogous manner, it is possible to aggregate many GPS to exploit their joint predictive power. For example, multiple GPS were used to predict intelligence in a sample of 6,710 unrelated 12-year-olds35. This approach is a multiple regression prediction model that accommodates multiple correlated predictors while preventing overfitting based on training in one sample and testing in another sample in a repeated cross-validation design. This approach predicted 4.8% of the variance in intelligence. Although EA2 GPS alone accounted for most of the variance, other GPS added significantly to the prediction of intelligence, especially GPS derived from GWAS of high-IQ individuals 57, childhood IQ19 and household income90. More than 7% of the variance in intelligence was predicted36 using another approach called Multi-Trait Analysis of GWAS (MTAG)91, which performs a meta-analysis from summary statistics for a few correlated GPS and produces new summary statistics that can be used to create a multivariate GPS.
The success of GWAS came from its atheoretical approach that analyses all SNPs in the genome rather than selecting candidate genes. In the same way, an atheoretical approach can be used in analyses of multiple GPS by incorporating as many GPS as possible rather than selecting a few candidate GPS. For example, the first study of this sort, mentioned above35, included a total of 81 GPS from well-powered GWAS of cognitive, medical and anthropometric traits available in LD Hub92 that together predicted 4.8% of the variance in intelligence. Although EA2, IQ and income GPS drove most of the predictive power of this multiple-GPS analysis, significant independent contributions to the prediction of intelligence were also found for major depressive disorder GPS and autism spectrum disorder GPS. These latter associations were in the direction expected based on the negative genetic correlation between intelligence and depression and the surprising positive genetic correlation between intelligence and autism (see Multivariate genetic research).
Nonetheless, 10% is a long way from the heritability estimate of 50% obtained from twin studies of intelligence14. This lacuna is known as the ‘missing heritability’, which is a key genetic issue for all complex traits in the life sciences39 (Box 4). The current limit for the variance that can be predicted by GPS is SNP heritability, which estimates the extent to which phenotypic variance for a trait can be explained by SNPs across the genome without identifying specific SNP associations. For intelligence, SNP heritability is about 25%33,40. It is safe to assume that GPS for intelligence using current SNP chips can approach the SNP heritability limit of 25% by amassing ever-larger GWAS samples and by using multi-trait GWAS that include traits related to intelligence such as years of education. However, breaking through this ceiling of 25% SNP heritability to the 50% heritability estimated from twin studies — assuming that twin studies yield accurate estimates of the total variance explained by inherited DNA differences — will require different technologies, such as whole-genome sequencing data that include rare variants, not just the common SNPs used on current SNP chips.
Box 4. Twin, SNP and GPS heritabilities.
Heritability is the proportion of observed (phenotypic) differences among individuals that can be attributed to genetic differences in a particular population. Broad heritability involves all additive and nonadditive sources of genetic variance, whereas narrow heritability is limited to additive genetic variance. Additive genetic variance refers to the independent effects of alleles or loci that ‘add up’. Nonadditive genetic variance involves effects of alleles or loci that interact.
Twin heritability compares the resemblance of identical and fraternal twins to estimate genetic and environmental components of variance. For intelligence, twin estimates of broad heritability are 50% on average14. Adoption studies of first-degree relatives yield similar estimates of narrow heritability of intelligence, suggesting that most genetic influence on intelligence is additive.
SNP heritability is estimated directly from SNP differences between individuals. It does not specify which SNPs are associated with a trait. Instead, it uses chance genomic similarities across hundreds of thousands of SNPs genotyped on a SNP chip for thousands of unrelated individuals to estimate the extent to which genomic covariance accounts for phenotypic covariance in these individuals. For intelligence, SNP heritability is about 25%22,33,40.
GPS heritability is the proportion of variance that can be predicted by the GPS. For intelligence, GPS heritability is currently about 10% (P. D. Koellinger, personal communication).
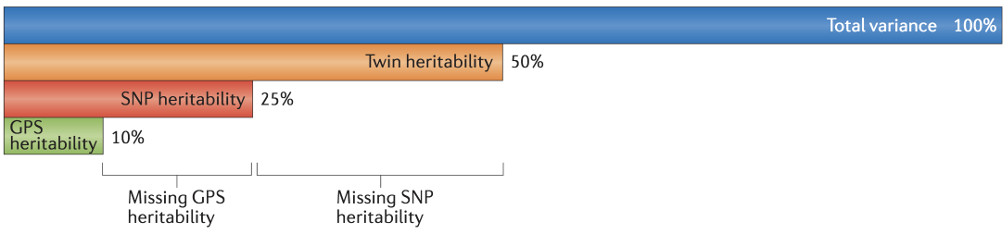
These three types of heritability denote two types of ‘missing heritability,’ as shown in the figure. SNP heritability is the ceiling for GWAS and for GPS heritability because all three rely on the additive effects of SNPs genotyped on SNP chips93. The missing heritability gap between GPS heritability (10%) and SNP heritability (25%) can be narrowed by increasing GWAS sample size. Narrowing the missing heritability gap between SNP heritability (25%) and twin heritability (50%) will require different technologies that consider, for example, rare variants, gene-gene interaction, and gene-environment interaction.
Box 4 fig.
GPS in intelligence research
The extremely polygenic nature of intelligence means that it will be a long slog from genome-wide significant ‘hits’ found across GWAS of intelligence. A bottom-up approach focused on specific genes will be difficult for two reasons. First, many hits are in intergenic regions, which means that there are no ‘genes’ to trace through the brain to behaviour. Second, the biggest hits have miniscule effects — less than 0.05% of the variance — which means that hundreds of thousands of SNP associations are needed to account for the 50% heritability estimated by twin studies. A systems biology approach to molecular studies of the brain is needed that is compatible with this extreme polygenicity41.
By contrast, the top-down approach of GPS that aggregate thousands of these tiny effects is already transforming research on intelligence42. Unlike quantitative genetic studies that require special samples such as twins, or GWAS that require huge samples in the hundreds of thousands, GPS can be used to add a genetic dimension to any research with modest sample size. For example, a GPS for intelligence that predicts 10% of the variance only needs a sample size of 60 to detect its effect with 80% power (P =.05, one-tailed).
GPS are unique predictors in the behavioural sciences. They are an exception to the rule that correlations do not imply causation in the sense that there can be no backward causation when GPS are correlated with traits. That is, nothing in our brains, behaviour or environment changes inherited differences in DNA sequence. A related advantage of GPS as predictors is that they are exceptionally stable throughout the life span because they index inherited differences in DNA sequence. Although mutations can accrue in the cells used to obtain DNA, like any cells in the body these mutations would not be expected to change systematically the thousands of inherited SNPs that contribute to a GPS.
In other words, a GPS derived from a GWAS of any trait at any age would be expected to correlate near 1.0 when the GPS is constructed from DNA obtained at birth and in adulthood for the same individual, although we are not aware of any empirical evidence relevant to this prediction. If GPS for individuals do not change during the life span, a GPS derived from GWAS of intelligence in adulthood will predict adult intelligence just as well from DNA obtained at conception or birth as from DNA obtained in adulthood. By contrast, intelligence tests at birth cannot predict intelligence at age 18 years. At 2 years of age, infant intelligence tests predict less than 5% of the variance of intelligence in late adolescence37,38.
GPS are unbiased in the sense that they are not subject to training, faking or anxiety. They are also inexpensive, costing less than US$100 per person. This expense would not be incurred specifically to predict intelligence; the same SNP chip genotype information used in GWAS can be used to create GPS for hundreds of disorders and traits, one of which is intelligence.
GPS for intelligence will open new avenues for research into the causes and consequences of intelligence. Three examples are developmental change and continuity, multivariate links between traits, and gene–environment interplay. A critical requirement for capitalizing on these opportunities is to make the ingredients for GPS publicly available — that is, GWAS summary-level statistics (Box 5).
Box 5. Make GWAS summary-level statistics publicly available.
It is essential for continued rapid scientific advances using GPS that summary-level statistics from GWAS are made publicly available for all SNPs following publication. The reason why public access to summary statistics is important is that the construction of GPS requires an effect size indicator and P value for each SNP in the GWAS. GWAS summary-level statistics are also necessary for other analyses, most notably LD score regression, which is used to estimate genetic correlations among traits54.
Until 2017, GWAS summary-level data were stored in different databases using different formats, which made it difficult to use the data to investigate traits across studies. This problem has been solved with LD Hub, a centralized database and web interface that provides an automated pipeline for entering and using GWAS summary-level data92.
However, only about 10% of published GWAS results are publicly available on LD Hub. Some GWAS consortia are exemplars for making GWAS summary-level data immediately upon publication, or even before publication, such as the Psychiatric Genomics Consortium94. In intelligence research, a paragon is the Social Science Genetic Association Consortium27, which is responsible for five of the six GWAS for which summary statistics are publicly available in the intelligence section of LD Hub, although three of the five GWAS were for years of education rather than for intelligence itself.
In contrast, some authors apply conditions for the use of the summary statistics from their published GWAS paper. Others refuse to share these statistics altogether. A worrying trend is that several commercial organizations do not allow summary GWAS statistics from their samples to be used in open-access summary-level statistics for all SNPs when their samples are included in meta-analytic GWAS. Concerns about privacy have been put forth as an explanation, but these fears should be allayed as it is not possible to re-construct individual-level data from summary-level GWAS statistics in large heterogenous samples95.
Such asymmetrical data-sharing policies between industry and academia will hold back research in the field. If a group does not want their summary-level GWAS statistics to be freely available for a published meta-analytic GWAS, their data should not be used in ‘publication’, true to its Latin origin publicare, which means ‘to make public’.
Developmental research
One of the most interesting developmental findings about intelligence is that its heritability as estimated in twin studies increases dramatically from infancy (20%) to childhood (40%) to adulthood (60%), while age-to-age genetic correlations are consistently high43,44. What could account for this increasing heritability despite unchanging age-to-age genetic correlations? Twin studies suggest that genetic effects are amplified through gene–environment correlation as time goes by45. That is, the same large set of DNA variants affects intelligence from childhood to adulthood, resulting in high age-to-age genetic correlations, but these DNA variants increasingly have an impact on intelligence as individuals select environments correlated with their genetic propensities, leading to greater heritability of intelligence.
Developmental hypotheses about high age-to-age genetic correlations and increasing heritability can be tested more rigorously and can be extended using GPS. Does the variance explained by GPS for intelligence increase from childhood to adolescence to adulthood? Are the correlations between GPS at these ages consistently high?
High age-to-age genetic correlations for intelligence imply that GWAS of adults should predict intelligence in childhood. The EA2 GPS30, currently the best genetic predictor of intelligence until EA3 GPS becomes available, was derived from a GWAS meta-analysis of years of education in adults who had completed their education. Nonetheless, the EA2 GPS predicts 2% of the variance in intelligence at age 7 years, 3% at age 12 years, and 4% at age 16 years in our longitudinal study46.
Multivariate genetic research
Multivariate genetic research focuses on the genetic covariance between traits rather than the variance of each trait. A specific multivariate question for intelligence research is why EA GPS predict twice as much variance in intelligence as do GPS for intelligence itself. This question raises interesting methodological and conceptual issues (Box 6).
Box 6. EA GPS and intelligence.
EA GPS predict intelligence because the genetic correlation between years of education and intelligence is greater than 0.50 in twin studies97 and LD score regression studies98. The genetic correlation of 0.50 also sets a limit on the extent to which EA GPS can predict intelligence.
But why do EA GPS predict intelligence to a greater extent than they predict EA itself? That is, EA2 GPS predicts 3% of the variance in years of education 30 but it predicts 4% of the variance in intelligence46. Moreover, EA GPS predict intelligence much better than IQ GPS predict intelligence themselves. The IQ3 GPS from the most recent GWAS of intelligence predicts 4% of the variance of intelligence33 but the EA3 GPS predicts more than 10% of the variance in intelligence (P. D. Koellinger, personal communication).
There are two likely reasons why EA GPS currently predict intelligence to a greater extent than EA GPS predict years of education itself. First, intelligence may be more heritable (60% in adults) than years of education (40%) in twin studies99. Second, years of education is a coarse measure, primarily indicating whether an individual completed university. Years of education is largely bimodal, with a spike at the end of secondary school and another peak for individuals who attended university. By contrast, intelligence is a more refined measure than years of education that captures the commonalities among diverse tests of cognitive abilities and is normally distributed. That is, educational achievement is not just a proxy for intelligence. It is also predicted by personality traits such as conscientiousness and well-being and having fewer mental health problems such as depression. Together, these non-cognitive traits account for as much of the heritability of educational achievement as intelligence96. The EA GWAS incorporates SNPs associated with any of these traits, not just with intelligence14.
Of note, the current GWAS sample sizes for EA are three times larger than for intelligence. The GPS effect sizes for intelligence are similar to those for EA GPS for comparable effect sizes (that is, IQ2 as compared to EA1, and IQ3 as compared to EA2; see Fig. 1). For this reason, we predict that an IQ GPS derived from a GWAS of intelligence with a sample size of one million, such as EA3, will predict at least as much variance in intelligence as does the current EA3 GPS. In other words, intelligence is not actually predicted to a greater extent by EA GPS than by intelligence GPS when the powers of the discovery GWAS are similar.
Multivariate genetic research is especially important for intelligence because genetic effects in the cognitive domain have been shown in twin studies to be general. That is, genetic effects correlate highly across most cognitive abilities such as verbal and spatial abilities as well as most educational skills such as reading and mathematics47. A recent multivariate finding is that the EA2 GPS predicts 5% of the variance in comprehension and efficiency of reading48. This is by far the most powerful GPS predictor of reading ability because there have as yet been no large GWAS of reading with replicable results49. EA GPS are also likely to predict other educational skills such as mathematics and other cognitive abilities such as spatial ability.
EA GPS do not only predict reading. They are correlated genetically with a wider range of variables than any other trait50. This pervasive genetic influence of EA GPS extends to a negative genetic correlation with schizophrenia and positive genetic correlations with height51, myopia52, and surprisingly with autism53. Linkage disequilibrium (LD) score regression analysis54, which uses summary GWAS statistics rather than GPS for individuals, finds a similar pattern of results for intelligence using the IQ2 GWAS: the negative genetic correlation with schizophrenia (–0.20) and the positive genetic correlations with height (0.10) and autism (0.21)32. The same LD score regression analysis32 found that intelligence significantly correlated genetically with many other traits, including Alzheimer disease (–0.36), smoking cessation (–0.32), intracranial volume (0.29), head circumference in infancy (0.28), depressive symptoms (–0.27), attention-deficit–hyperactivity disorder (–0.27), ever smoked (–0.23), longevity (0.22) and, of course, years of education (0.70).
Despite this evidence for ability-general genetic effects, genetic correlations across cognitive abilities and educational skills are not 1.0, which implies that there are ability-specific SNP associations. An important direction for research is to identify ability-specific GPS derived from large GWAS analyses focused on specific cognitive abilities independent of general intelligence. Preliminary analyses of this sort would be possible using existing GWAS of intelligence because most of these studies assessed multiple measures of specific cognitive abilities, which were combined to index intelligence. These data could be re-analysed in meta-analytic GWAS that focus on specific abilities included in multiple studies. However, what is needed are large GWAS focused on well measured specific cognitive abilities such as verbal, spatial and memory abilities and specific cognitive skills taught in schools such as reading, mathematics and language. The pay-off from these studies will be GPS that predict specific abilities independent of general intelligence. These ability-specific GPS could be used to create profiles of genetic strengths and weaknesses for individuals that could be the target for personalized prediction, prevention and intervention.
In addition to investigating links between different traits, multivariate genetic research can examine genetic links between dimensional and diagnostic measures of the ‘same’ domain. For example, EA2 GPS predicts reading disability just as much as reading ability, from slow readers to speed-readers48. Because GPS are always normally distributed, they will show that there are no etiologically distinct common disorders, only continuous dimensions55. This is also true for very low and for very high intelligence46. Even extremely high intelligence is only quantitatively, not qualitatively, different genetically from the normal distribution56,57. The exception is severe intellectual disability, which is genetically distinct from the rest of the distribution of intelligence58 and affected by rare, often de novo, mutations of relatively large effect59.
Research on gene–environment interplay
The high heritability of intelligence should not obscure the fact that heritability is significantly less than 100%. Research using genetically sensitive designs has led to one of the most important findings about environmental influence on intelligence. Intelligence has always been known to run in families but it was assumed that this family resemblance was due to nurture, called shared family environmental influence. That is, siblings were thought to be similar in intelligence because they grew up in the same family and attended the same schools. Twin and adoption studies consistently support this assumption but only up until adolescence. After adolescence, the effect of shared family environmental influence on intelligence is negligible, which means that family environments have little effect on individual differences in the long run45,60. Family resemblance for intelligence is due to nature rather than nurture, although it should be emphasized that we are referring to the normal range of environmental influence, not the extremes such as neglect or abuse. However, little is known about the specific environmental factors that make children growing up in the same family different14.
The importance of both genetics and environment for cognitive development recommends investigating the interplay between them. GPS for intelligence will greatly facilitate this research because they offer, for the first time, the possibility of directly assessing genetic propensities of individuals to investigate their interplay with aspects of the environment. Gene–environment (GE) interplay refers to two different concepts, GE interaction and GE correlation.
GE interaction denotes a conditional relationship in which the effects of genes on intelligence depend on the environment. For example, some twin research suggests that heritability of intelligence is lower in low socioeconomic status family environments and higher in high socioeconomic status family environments61. This hypothesis predicts that GPS for intelligence will correlate less with intelligence in environments of low socioeconomic status compared to those with high socioeconomic status. The first test of this hypothesis using the EA2 GPS found no evidence for such an interaction46. That is, EA2 GPS were just as much correlated with intelligence in low socioeconomic status as in high socioeconomic status family environments. GPS provide a particularly powerful approach to test for GE interaction as compared to twin studies62.
In contrast to GE interaction, GE correlation refers to the correlation between genetic propensities and experiences. GE correlation is the reason why most environmental measures used in the behavioural sciences show genetic influence in twin studies63. Associations between environmental measures and behavioural traits such as intelligence are also mediated in part by genetic differences. Research using GPS is beginning to confirm these twin study findings about the ‘nature of nurture’ by showing, for example, that EA GPS correlate with social mobility64 and capture covariation between environmental exposures and children’s behaviour problems and educational achievement65. GE correlation provides a general model for how genotypes become phenotypes — how children select, modify and create environments correlated with their genetic propensities. GPS will greatly advance research on GE correlation by providing an individual-specific index of the ‘G’ of GE interplay. GPS will also make it possible to assess environmental influences on intelligence while controlling for genetic influences.
Implications for society
The most exciting aspect of GPS is their potential for addressing novel, socially important questions, which we will illustrate with three recent examples from our own research. First, children in public and private schools differ in their EA2 GPS scores because private schools select pupils based on genetic differences in intelligence66. Second, intergenerational educational mobility reflects EA2 GPS differences67. Finally, the EA2 GPS predicts twice as much variance in educational attainment and occupational status in the post-Soviet era as compared to the Soviet era in Estonia, a finding compatible with the hypothesis that heritability is an index of equality of opportunity and meritocracy68.
Understanding ourselves
IQ GPS will be used to predict individuals’ genetic propensity to learn, reason and solve problems, not only in research, but also in society, as direct-to-consumer genomic services provide GPS information that goes beyond single-gene and ancestry information. We predict that IQ GPS will become routinely available from direct-to-consumer companies along with hundreds of other medical and psychological GPS that can be extracted from genome-wide genotyping on SNP chips. Using GPS to predict individuals’ genetic propensities requires clear warnings about the probabilistic nature of these predictions and the limitations of their effect sizes (Box 7).
Box 7. Using GPS to predict outcomes in individuals.
GPS must be used with caution when predicting outcomes in individuals. We illustrate the probabilistic nature of GPS predictions using data on EA2 GPS and school achievement from the Twins Early Development Study100. School achievement was assessed by scores from a UK-wide examination, the General Certificate of Secondary Education (GCSE), administered at the end of compulsory education at age 16 years. GCSE scores were age- and gender-regressed, and EA2 GPS were constructed as described elsewhere46. We used the EA2 GPS prediction of GCSE scores as an example because the effect size of this association is currently the strongest in the behavioural sciences, accounting for 9% of the variance46. It will soon be possible to explain a similar amount of variance in intelligence, and with that GPS will become available to predict intelligence for individuals.
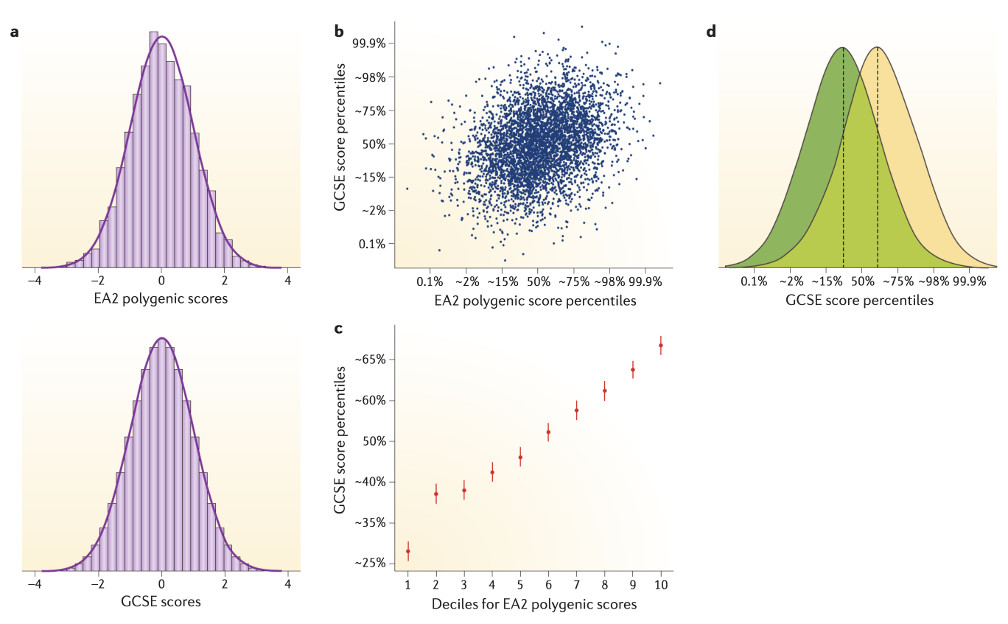
The starting point for prediction is the distribution of individual differences (see the figure, part a). The EA2 GPS is normally distributed, as GPS always are. The measure of school achievement is also normally distributed. GPS prediction of individual differences is based on its covariance with the target trait, school achievement in this example. The scatterplot between EA2 GPS and GCSE scores (see the figure, part b) indicates the difficulty of predicting individual outcomes when the correlation is modest, 0.30 in this example. Squaring this correlation indicates that EA2 GPS predicts 9% of the variance in GCSE scores. Although higher EA2 GPS can be seen to predict higher GCSE scores on average, there is great variability between individuals. For example, the individual with the second highest EA2 GPS has a GCSE score only slightly above the average. Conversely, an individual with the eighth lowest EA2 GPS has a GCSE score above the 75th percentile.
Despite this variability, powerful predictions can be made at the extremes. For example, when the sample was divided into ten equal-sized groups (deciles) on the basis of their EA2 GPS, a strong relationship between average EA2 GPS and average GCSE scores emerged that was most evident at the extremes (see the figure, part c). Specifically, the average school achievement of individuals in the lowest EA2 GPS decile is at the 28th percentile. For the highest EA2 GPS decile, the average school achievement is at the 68th percentile.
Nonetheless, individuals within the lowest and highest EA2 GPS deciles vary widely in school achievement (see the figure, part d). The overlap in the two distributions is 61%. These issues of variability in prediction are the same for any predictor that accounts for 9% of the variance in the target trait. As bigger and better GPS emerge, the predictive power will increase.
In summary, GPS are useful for individual prediction as long as the probabilistic nature of the prediction is kept in mind.
Box 7 fig.
Although simple curiosity will drive consumers’ interests, GPS for intelligence are more than idle fortune-telling. Because intelligence is one of the best predictors of educational and occupational outcomes, IQ GPS will be used for prediction from early in life before intelligence or educational achievement can be assessed. In the school years, IQ GPS could be used to assess discrepancies between GPS and educational achievement, that is, GPS-based over-achievement and under-achievement. The reliability, stability and lack of bias of GPS make them ideal for prediction, which is essential for the prevention of problems before they occur. A ‘precision education’ based on GPS could be used to customize education, analogous to ‘precision medicine’.
A novel, socially important direction for research using IQ GPS is to understand differences within families. First-degree relatives are on average only 50% similar genetically, which means they are on average 50% different genetically. A major impact of GPS will be to recognize and respect these large genetic differences within families.
For scores on an intelligence test standardized to have a mean of 100 and a standard deviation of 15, the average difference between pairs of individuals who are selected randomly from the population is 17 IQ points. The average difference between parents and offspring and between siblings is 13 IQ points69. IQ GPS might help parents understand why their children differ in school achievement. Because GPS are probabilistic, a low IQ GPS does not mean that a child is destined to go no further in education than secondary school. But it does mean that the child is more likely to find academic learning more difficult and less rewarding than a sibling with a high IQ GPS.
Ethical implications
Genomic research and studies of intelligence face four principal ethical concerns: the notion of biological determinism, the potential for discrimination and stigmatization, the question of ownership of information, and the emotional impact of knowledge about one's personal genomics and intelligence. These and other ethical issues are explored in detail by the programme of ethical, legal and social implications (ELSI), which is an integral part of the Human Genome Project70. Also, recent books discuss ethical as well as scientific issues about personal genomics specifically in relation to education71 and occupation72. Much of these ethical discussions focus on single-gene disorders, for example Huntington disease, which has 100% penetrance. By contrast, GPS are 'less dangerous’ because they are intrinsically probabilistic, not hard-wired and deterministic like single-gene disorders. It is important to recall here that although all complex traits are heritable, none is 100% heritable. A similar logic can be applied to IQ scores: although they have great predictive validity for key life outcomes1–6, IQ is not deterministic but probabilistic. In short, an individual is always more than the sum of their genes or their IQ scores.
Issues of discrimination and stigmatization have accompanied research into genetics and intelligence from the beginning, typically because findings from both fields of study were applied to justify policies that served socio-political ideologies. For example, IQ testing was infamously used to differentiate European immigrants to the United States of America who arrived at Ellis Island in the early 1900s, and to guide eugenic ideas about sterilization in Britain and the United States of America throughout the 20th century11. It is important to acknowledge the risk of discrimination that occurs on the back of scientific findings about individual differences. It is, however, equally important to realize that research does not lead directly to any policy recommendations. We must be careful not to blame the scientists or entire disciplines when their findings are used wrongly9.
Who 'owns' our genetic information? And who should decide who can access it? The question of ownership of personal data has become pivotal but also increasingly complex in our current age of information. At the same time, understanding and managing the emotional impact that stems from knowledge about our genomics and intelligence has emerged as a new societal responsibility. It is beyond the scope of this paper to elucidate these issues in the depth that they deserve but we expect that the discussions of ethical issues that surround personal genomics will consolidate the DNA revolution.
Conclusions
Genetic association studies have confirmed a century of quantitative genetic research showing that inherited DNA differences are responsible for substantial individual differences in intelligence test scores. A reachable objective shared with all complex traits in the life sciences is to close the gap between the 10% variance in intelligence scores explained by GPS and its SNP heritability of about 25%. A more daunting challenge is to break through the ceiling of 25% SNP heritability to reach the 50% heritability estimated by twin studies.
Until 2016, GPS could only predict 1% of the variance in intelligence. Progress has been rapid since then, reaching our current ability to predict 10% of the variance in intelligence from DNA alone. GPS will soon be available that can predict more than 10% of the variance in intelligence, that is, more than 20% of the 50% heritability of intelligence estimated from twin studies, and more than 40% of the 25% SNP heritability of intelligence. This is an important milestone for the new genetics of intelligence because effect sizes of this magnitude are large enough to be “perceptible to the naked eye of a reasonably sensitive observer”73. With these advances in the past few years, intelligence steps out of the shadows and takes the lead in genomic research.
In addition to investigating traditional issues about development, multivariate links between traits and gene-environment interplay, IQ GPS will open new avenues for research into the causes and consequences of intelligence. The new genetics of IQ GPS will bring the omnipotent variable of intelligence to all areas of the life sciences, without having to assess intelligence.
Key points.
Until 2017, genome-wide polygenic scores derived from GWAS of intelligence were only able to predict 1% of the variance in intelligence in independent samples.
Polygenic scores derived from GWAS of intelligence can now predict 4% of the variance in intelligence.
More than 10% of the variance in intelligence can be predicted by multi-polygenic scores derived from GWAS of both intelligence and years of education. This accounts for more than 20% of the 50% heritability of intelligence.
Polygenic scores are unique predictors in two ways: First, they predict psychological and behavioural outcomes just as well from birth as later in life. Second, polygenic scores are causal predictors in the sense that nothing in our brains, behaviour or environment can change the differences in DNA sequence that we inherited from our parents.
Polygenic scores for intelligence can bring the powerful construct of intelligence to any research in the life sciences without having to assess intelligence using tests.
Acknowledgements
We gratefully acknowledge the ongoing contribution of the participants in the Twins Early Development Study (TEDS) and their families. TEDS is supported by a programme grant to RP from the UK Medical Research Council (MR/M021475/1 and previously G0901245), with additional support from the US National Institutes of Health (AG046938). The research reported here has also received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ grant agreement 602768 and ERC grant agreement 295366. RP is also supported by a Medical Research Council Professorship award (G19/2). SvS is supported by a Jacobs Foundation Research Fellowship award (2017-2019).
Glossary
- Twin studies
Comparing the resemblance of identical and fraternal twins to estimate genetic and environmental components of variance.
- Variance
An index of how spread out scores are in a population, which is calculated as the average of the squared deviations from the mean.
- Genome-wide association studies
(GWAS). Studies that aim to identify loci throughout the genome associated with an observed trait or disorder.
- Heritability
The proportion of observed differences among individuals that can be attributed to inherited differences in genome sequence (Box 4).
- Candidate gene studies
Studies that focus on genes whose function suggests that they might be associated with a trait, in contrast to genome-wide association studies.
- Genome-wide polygenic scores
(GPS). A genetic index of a trait for each individual that is the sum across the genome of thousands of SNPs of the individual’s increasing alleles associated with the trait, usually weighted by the effect size of each SNP’s association with the trait in GWAS.
- Linkage disequilibrium (LD) score regression analysis
For each SNP in a GWAS, regresses chi-square statistic from GWAS summary statistics against linkage disequilibrium scores.
- Effect size
The proportion of variance of a trait in the population accounted for by a particular factor such as a GPS.
- Single-nucleotide polymorphisms
(SNPs). Single base-pair differences in inherited DNA sequence between individuals.
Biographies
Author biographies
Robert Plomin is a Professor of Behavioural Genetics at King’s College London. His research spans molecular and quantitative genetic approaches to complex traits, with a focus on cognitive abilities and disabilities. He established and directs the Twins Early Development Study, one of the world's premier studies of how genes and environments shape development.
Sophie von Stumm is an Associate Professor at the London School of Economics and Political Science. Her research centers on the causes and consequences of individual differences in psychological and behavioural development across the life course. She takes an inter-disciplinary approach to observing behaviour and environments, primarily through the application of new assessment technologies that enable collecting big, high-quality data.
Footnotes
Author contributions
The authors contributed equally to all aspects of the manuscript.
Competing interests statement
The authors declare no competing interests.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Subject categories
Biological sciences / Neuroscience / Cognitive neuroscience / Intelligence [URI /631/378/2649/1579]
Biological sciences / Genetics / Genetic association study / Genome-wide association studies [URI /631/208/205/2138]
Biological sciences / Genetics / Genome / Genetic variation [URI /631/208/726/649]
Biological sciences / Genetics / Behavioural genetics [URI /631/208/1515]
Scientific community and society / Social sciences / Education [URI /706/689/160]
ToC blurb
Recent genome-wide association studies have catapulted the search for genes underlying human intelligence into a new era. Genome-wide polygenic scores promise to transform research on individual differences in intelligence, but not without societal and ethical implications, as the authors discuss in this Review.
References
- 1.Gottfredson LS. Why g matters: The complexity of everyday life. Intelligence. 1997;24:79–132. [Google Scholar]
- 2.Deary IJ, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482:212–214. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- 3.Deary IJ, Strand S, Smith P, Fernandes C. Intelligence and educational achievement. Intelligence. 2007;35:13–21. [Google Scholar]
- 4.Schmidt FL, Hunter J. General mental ability in the world of work: occupational attainment and job performance. J Pers Soc Psychol. 2004;86:162–173. doi: 10.1037/0022-3514.86.1.162. [DOI] [PubMed] [Google Scholar]
- 5.Strenze T. Intelligence and socioeconomic success: A meta-analytic review of longitudinal research. Intelligence. 2007;35:401–426. [Google Scholar]
- 6.Calvin CM, et al. Childhood intelligence in relation to major causes of death in 68 year follow-up: prospective population study. Brit Med J. 2017;357:2708. doi: 10.1136/bmj.j2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deary IJ, Pattie A, Starr JM. The stability of intelligence from age 11 to age 90 years: the Lothian birth cohort of 1921. Psychol Sci. 2013;24:2361–2368. doi: 10.1177/0956797613486487. [DOI] [PubMed] [Google Scholar]
- 8.Editorial. Intelligence research should not be held back by its past. Nature. 2017;545:385–386. doi: 10.1038/nature.2017.22021. [A landmark in the acceptance of genetic influence in intelligence, concluding that ‘It’s well established and uncontroversial among geneticists that together, differences in genetics underwrite significant variation in intelligence between people.’] [DOI] [PubMed] [Google Scholar]
- 9.Pinker S. The blank slate: The modern denial of human nature. Penguin; 2003. [Google Scholar]
- 10.Block NJ, Dworkin GE. The IQ controversy: Critical readings. New York: Pantheon; 1976. [Google Scholar]
- 11.Gould SJ. The mismeasure of man. WW Norton; 1982. [Google Scholar]
- 12.Kamin LJ. The science and politics of IQ. Potomac, Maryland: Erlbaum; 1974. [Google Scholar]
- 13.Bouchard TJ, McGue M. Familial studies of intelligence: A review. Science. 1981;212:1055–1059. doi: 10.1126/science.7195071. [DOI] [PubMed] [Google Scholar]
- 14.Knopik VS, Neiderheiser J, DeFries JC, Plomin R. Behavioral Genetics. 7th edn. New York: Worth; 2017. [Google Scholar]
- 15.Haier RJ. The neuroscience of intelligence. Cambridge: Cambridge University Press; 2016. [Google Scholar]
- 16.Hare B. Survival of the friendliest: Homo sapiens evolved via selection for prosociality. Annu Rev Psychol. 2017;68:155–186. doi: 10.1146/annurev-psych-010416-044201. [DOI] [PubMed] [Google Scholar]
- 17.Sternberg RJ, Kaufman JC. The evolution of intelligence. Psychology Press; 2013. [Google Scholar]
- 18.Chabris CF, et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23:1314–1323. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benyamin B, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19:253–258. doi: 10.1038/mp.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher LM, Davis OS, Craig IW, Plomin R. Genome-wide quantitative trait locus association scan of general cognitive ability using pooled DNA and 500K single nucleotide polymorphism microarrays. Genes Brain Behav. 2008;7:435–446. doi: 10.1111/j.1601-183X.2007.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies G, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies G, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N= 53 949) Mol Psychiatry. 2015;20:183–192. doi: 10.1038/mp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies G, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N= 112 151) Mol Psychiatry. 2016;21:758–767. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plomin R, et al. A genome-wide scan of 1842 DNA markers for allelic associations with general cognitive ability: a five-stage design using DNA pooling and extreme selected groups. Behav Genet. 2001;31:497–509. doi: 10.1023/a:1013385125887. [DOI] [PubMed] [Google Scholar]
- 25.Trampush J, et al. GWAS meta-analysis reveals novel loci and genetic correlates for general cognitive function: a report from the COGENT consortium. Mol Psychiatry. 2017;22:336. doi: 10.1038/mp.2016.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cesarini D, Visscher PM. Genetics and educational attainment. Science of Learning. 2017;2:1–7. doi: 10.1038/s41539-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rietveld CA, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [Used EA1 SNPs to predict intelligence, although less than 1% of the variance was predicted.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietveld CA, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [The GWAS origin of EA1, which yielded a GPS that predicted 1% of the variance in years of education.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rietveld CA, et al. Replicability and robustness of genome-wide-association studies for behavioral traits. Psychol Sci. 2014;25:1975–1986. doi: 10.1177/0956797614545132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [The GWAS origin of EA2 GPS, which increased the prediction of educational attainment from 1 to 3% of the variance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okbay A, et al. GWAS of educational attainment – Phase 3: Main results. Paper presented at the 47th Behaviour Genetics Annual Meeting; Oslo, Norway. 2017. Jun 29, [The largest GWAS of educational attainment (N- 1,100,000) increased the power of its GPS (EA3) to predict more than 10% of the variance in the targete trait of educational attainment.] [Google Scholar]
- 32.Sniekers S, et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nature Genet. 2017;49:1107–1112. doi: 10.1038/ng.3869. [The GWAS origin of IQ2 GPS, which increased the prediction of intelligence from 1% to 3%.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savage JE, et al. GWAS meta-analysis (N= 279,930) identifies new genes and functional links to intelligence. BioRxiv. 2017 doi: 10.1101/184853. [The largest GWAS of intelligence to date, which yielded a GPS (IQ3) that predicted 4% of the variance in intelligence.] [DOI] [Google Scholar]
- 34.Davies G, et al. Ninety-nine independent genetic loci influencing general cognitive function include genes associated with brain health and structure (N= 280,360) BioRxiv. 2017 doi: 10.1101/176511. [DOI] [Google Scholar]
- 35.Krapohl E, et al. Mol Psychiatry. Advance online publication; 2017. Multi-polygenic score approach to trait prediction. [Using a multiple-GPs approach, 81 GPS derived from well-powered GWAS predicted 5% of the variance in intelligence.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill WD, Davies G, McIntosh AM, Gale CR, Deary IJ. A combined analysis of genetically correlated traits identifies 107 loci associated with intelligence. BioRxiv. 2017 doi: 10.1101/160291. [Using Multiple-Trait Analysis of GWAS for intelligence, educational attainment, and income predicted 7% of the variance in intelligence in an independent sample.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honzik MP, Macfarlane JW, Allen L. The stability of mental test performance between two and eighteen years. J Exp Educ. 1948;17:309–324. [Google Scholar]
- 38.von Stumm S, Plomin R. Socioeconomic status and the growth of intelligence from infancy through adolescence. Intelligence. 2015;48:30–36. doi: 10.1016/j.intell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plomin R, et al. Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychol Sci. 2013;24:562–568. doi: 10.1177/0956797612457952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plomin R. Blueprint: how DNA shapes personality. London: Allen Lane/Penguin; In press. [Describes genetic research on behaviour from twin studies to the DNA revolution and its implications for science and society.] [Google Scholar]
- 43.Haworth CM, et al. A twin study of the genetics of high cognitive ability selected from 11,000 twin pairs in six studies from four countries. Behav Genet. 2009;39:359–370. doi: 10.1007/s10519-009-9262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plomin R, Deary IJ. Genetics and intelligence differences: five special findings. Mol Psychiatry. 2015;20:98–108. doi: 10.1038/mp.2014.105. [Highlights five genetic findings that are special to intelligence differences, including one not mentioned in this review: assortative mating is much greater for intelligence than for other traits.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Briley DA, Tucker-Drob EM. Explaining the increasing heritability of cognitive ability across development: A meta-analysis of longitudinal twin and adoption studies. Psychol Sci. 2013;24:1704–1713. doi: 10.1177/0956797613478618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selzam S, et al. Predicting educational achievement from DNA. Mol Psychiatry. 2017;22:267–272. doi: 10.1038/mp.2016.107. [Showed that EA2 predicted 9% of the variance in tested educational achievement at age 16, which was the strongest GPS prediction of a behavioural trait at that time.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plomin R, Kovas Y. Generalist genes and learning disabilities. Psychol Bull. 2005;131:592–617. doi: 10.1037/0033-2909.131.4.592. [DOI] [PubMed] [Google Scholar]
- 48.Selzam S, et al. Genome-wide polygenic scores predict reading performance throughout the school years. Sci Stud Read. 2017;21:334–349. doi: 10.1080/10888438.2017.1299152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrion-Castillo A, et al. Evaluation of results from genome-wide studies of language and reading in a novel independent dataset. Genes Brain Behav. 2016;15:531–541. doi: 10.1111/gbb.12299. [DOI] [PubMed] [Google Scholar]
- 50.Krapohl E, et al. Phenome-wide analysis of genome-wide polygenic scores. Mol Psychiatry. 2015;21:1188–1193. doi: 10.1038/mp.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marioni RE, et al. Common genetic variants explain the majority of the correlation between height and intelligence: the generation Scotland study. Behav Genet. 2014;44:91–96. doi: 10.1007/s10519-014-9644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams KM, et al. Phenotypic and genotypic correlation between myopia and intelligence. Sci Rep. 2017;7 doi: 10.1038/srep45977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill WD, et al. Age-dependent pleiotropy between general cognitive function and major psychiatric disorders. Biol Psychiatry. 2016;80:266–273. doi: 10.1016/j.biopsych.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nature Rev Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 56.Spain SL, et al. A genome-wide analysis of putative functional and exonic variation associated with extremely high intelligence. Mol Psychiatry. 2016;21:1145–1151. doi: 10.1038/mp.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zabaneh D, et al. A genome-wide association study for extremely high intelligence. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.121. Advance online publication. [This GWAS of intelligence used a novel strategy to increase power -- a case-control design in which the cases were individuals with extremely high IQ from the top 0.0003 of the population (mean IQ of 170)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reichenberg A, et al. Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proc Natl Acad Sci. 2016;113:1098–1103. doi: 10.1073/pnas.1508093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nature Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 60.Plomin R, Daniels D. Why are children in the same family so different from one another? Behav Brain Sci. 1987;10:1–16. [Google Scholar]
- 61.Tucker-Drob EM, Bates TC. Large cross-national differences in gene× socioeconomic status interaction on intelligence. Psychol Sci. 2016;27:138–149. doi: 10.1177/0956797615612727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanscombe KB, et al. Socioeconomic status (SES) and children's intelligence (IQ): In a UK-representative sample SES moderates the environmental, not genetic, effect on IQ. PLoS ONE. 2012;7:e30320. doi: 10.1371/journal.pone.0030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plomin R, Bergeman CS. The nature of nurture: Genetic influence on “environmental” measures. Behav Brain Sci. 1991;14:373–386. [Google Scholar]
- 64.Belsky DW, et al. The genetics of success. Psychol Sci. 2016;27:957–972. doi: 10.1177/0956797616643070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krapohl E, et al. The nature of nurture: Widespread covariation of early environmental exposures and trait-associated polygenic variation. Proc Natl Acad Sci. doi: 10.1073/pnas.1707178114. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith-Woolley E, et al. Differences in exam performance between pupils attending different school types mirror the genetic differences between them. Science of Learning. doi: 10.1038/s41539-018-0019-8. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayorech Z, Krapohl E, Plomin R, von Stumm S. Genetic influence on intergenerational educational attainment. Psychol Sci. 2017:1302–1310. doi: 10.1177/0956797617707270. [Both twin analyses and EA2 GPS show genetic influence on intergenerational educational attainment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rimfeld K, et al. Genetic meritocracy during and after the Soviet era in Estonia. Nature Human Behavior. doi: 10.1038/s41562-018-0332-5. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plomin R, DeFries JC. Genetics and intelligence: Recent data. Intelligence. 1980;4:15–24. [Google Scholar]
- 70.McEwen JE, et al. The ethical, legal, and social implications program of the National Human Genome Research Institute: reflections on an ongoing experiment. Annu Rev Genomics Hum Genet. 2014;15:481–504. doi: 10.1146/annurev-genom-090413-025327. [DOI] [PubMed] [Google Scholar]
- 71.Bouregy S, Grigorenko EL, Latham SR, Tan M. Genetics, ethics and education. Cambridge University Press; 2017. [Google Scholar]
- 72.Conley D, Fletcher J. The Genome Factor: What the social genomics revolution reveals about ourselves, our history, and the future. Princeton University Press; 2017. [Google Scholar]
- 73.Cohen J. Statistical power analysis for the behavioral sciences, rev. Lawrence Erlbaum Associates; 1977. [Google Scholar]
- 74.Gottfredson LS. Mainstream science on intelligence. Wall St J. 1994 [Google Scholar]
- 75.Carroll JB. Human cognitive abilities: A survey of factor-analytic studies. Cambridge University Press; 1993. [Google Scholar]
- 76.Spearman C. " General Intelligence," Objectively Determined and Measured. Am J Psychol. 1904;15:201–292. [Google Scholar]
- 77.Jensen AR. The g factor: The science of mental ability. Praeger Publishers; 1998. [Google Scholar]
- 78.Deary IJ. Intelligence. Annu Rev Psychol. 2012;63:453–482. doi: 10.1146/annurev-psych-120710-100353. [authoritative overview of intelligence research] [DOI] [PubMed] [Google Scholar]
- 79.Gow AJ, et al. Stability and change in intelligence from age 11 to ages 70, 79, and 87: the Lothian Birth Cohorts of 1921 and 1936. Psychol Aging. 2011;26:232–240. doi: 10.1037/a0021072. [DOI] [PubMed] [Google Scholar]
- 80.Schaie KW. Developmental influences on adult intelligence: The Seattle longitudinal study. Oxford University Press; 2005. [Google Scholar]
- 81.Brinch CN, Galloway TA. Schooling in adolescence raises IQ scores. Proc Natl Acad Sci. 2012;109:425–430. doi: 10.1073/pnas.1106077109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Protzko J. Does the raising IQ-raising g distinction explain the fadeout effect? Intelligence. 2016;56:65–71. [Google Scholar]
- 83.Duyme M, Dumaret A-C, Tomkiewicz S. How can we boost IQs of “dull children”?: A late adoption study. Proc Natl Acad Sci. 1999;96:8790–8794. doi: 10.1073/pnas.96.15.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melby-Lervåg M, Hulme C. Is working memory training effective? A meta-analytic review. Dev Psychol. 2013;49:270–291. doi: 10.1037/a0028228. [DOI] [PubMed] [Google Scholar]
- 85.Puma M, et al. Head Start Impact Study. Final Report. Administration for children & families. 2010 [Google Scholar]
- 86.Plomin R, Simpson MA. The future of genomics for developmentalists. Dev Psychopathol. 2013;25:1263–1278. doi: 10.1017/S0954579413000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nature Rev Genet. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vilhjálmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Euseden J, et al. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. https://github.com/choishingwan/PRSice/wiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hill WD, et al. Molecular genetic contributions to social deprivation and household income in UK Biobank. Current Biology. 2016;26:3083–3089. doi: 10.1016/j.cub.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turley P, et al. MTAG: Multi-Trait Analysis of GWAS. bioRxiv. 2017 doi: 10.1101/118810. [DOI] [Google Scholar]
- 92.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang J, et al. Concepts, estimation and interpretation of SNP-based heritability. Nature Genet. 2017;49:1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
- 94.Sullivan PF, et al. Psychiatric genomics: an update and an agenda. Am J Psychol. Advance online publication. [Google Scholar]
- 95.Bacanu SA. Sharing extended summary data from contemporary genetic studies is unlikely to threaten subject privacy. PLoS One. 2017;12:e0179504. doi: 10.1371/journal.pone.0179504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krapohl E, et al. The high heritability of educational achievement reflects many genetically influenced traits, not just intelligence. Proc Natl Acad Sci. 2014;111:15273–15278. doi: 10.1073/pnas.1408777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Calvin CM, et al. Multivariate genetic analyses of cognition and academic achievement from two population samples of 174,000 and 166,000 school children. Behav Genet. 2012;42:699–710. doi: 10.1007/s10519-012-9549-7. [DOI] [PubMed] [Google Scholar]
- 98.Marioni RE, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence. 2014;44:26–32. doi: 10.1016/j.intell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Branigan AR, McCallum KJ, Freese J. Variation in the heritability of educational attainment: An international meta-analysis. Soc Forces. 2013;92:109–140. [Google Scholar]
- 100.Haworth CM, Davis OS, Plomin R. Twins Early Development Study (TEDS): a genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Res Hum Genet. 2013;16:117–125. doi: 10.1017/thg.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]


