Abstract
Glaucoma is the leading cause of irreversible blindness globally.1 Despite its gravity, the disease is frequently undiagnosed in the community.2 Raised intraocular pressure (IOP) is the most important risk factor for primary open-angle glaucoma (POAG).3,4 Here we present a meta-analysis of 139,555 European participants that identified 112 genomic loci associated with IOP, 68 of which are novel. These loci suggest a strong role for angiopoietin-receptor tyrosine kinase signaling, lipid metabolism, mitochondrial function and developmental processes underlying risk for elevated IOP. In addition, 48 of these loci were associated with glaucoma in an independent cohort, 14 of which at a Bonferroni-corrected threshold. Regression-based glaucoma prediction models had an area under Receiving Operator Characteristic curve (AUROC) of 0.76 in USA NEIGHBORHOOD study participants and 0.74 in independent glaucoma cases from UK Biobank. Genetic prediction models for POAG offer an opportunity to target screening and timely therapy to individuals most at risk.
IOP is strongly associated with POAG, and population-based studies have suggested an increased risk of 16% for every mmHg increase in IOP.3 Lowering of IOP remains the only proven therapy to slow the progression of vision loss in POAG.5 IOP heritability is estimated at 55%6 and, to date, genome-wide association study (GWAS) meta-analyses have identified several loci associated with IOP7–9 and POAG10–12 which explain a minor proportion of disease heritability7 and provide only limited insight into its biological mechanisms. This relative lack of knowledge is partially due to insufficient statistical power of previous association works.
Here we present the largest genome-wide association study (GWAS) of IOP to date, in 139,555 participants of three cohorts: UK Biobank,13 EPIC-Norfolk 14 and the previously reported combined results from 14 European studies in the International Glaucoma Genetics Consortium (IGGC).8 Additionally, we examined associations of 120 significant IOP loci with glaucoma among independent UK Biobank participants (not included in the IOP discovery GWAS) and with clinically diagnosed POAG among participants of a large multicenter case-control study (NEIGHBORHOOD).10
First, a linear mixed model GWAS for IOP was carried out in UK Biobank participants (n=103,382). Results were replicated in and then meta-analyzed with results from EPIC-Norfolk (n=6,595) and the IGGC meta-analysis8 (n=29,578). Cohort summary details are presented in Supplementary Table 1. All participants were of European descent (Supplementary Fig. 1 and 2). The meta-analysis results had a genomic inflation factor of 1.28 (Supplementary Fig. 3), but an LD score regression intercept15 of 1.06 (SE=0.011) along with a (intercept-1)/(mean(χ2)-1)=0.12, consistent with IOP polygenicity rather than population structure.
The UK Biobank analysis alone identified 74 unique autosomal genomic regions meeting genome-wide significance (P<5x10-8), of which 45 were novel (not previously associated with IOP, glaucoma, or related endophenotypes). Results across the three studies were directionally consistent (Supplementary Table 2); 49 loci replicated in IGGC with a false discovery rate (FDR) of <0.05, and 27 loci replicated in either of the replication cohorts (IGGC or EPIC-Norfolk) at a Bonferroni-corrected threshold (P<6.8x10-4).
Combining the three separate study results into a meta-analysis of 139,555 participants revealed genome-wide significant associations for 112 unique autosomal genomic regions (Supplementary Fig. 4, Supplementary Table 2), of which 68 are novel (Table 1). A conditional analysis traced the origin of association signals to 133 SNP loci; when included together in a linear regression model, these SNPs collectively explain 17% of IOP variance in the EPIC-Norfolk cohort and 9% in UK Biobank. The difference in variance explained between the studies may be in part be due to less measurement error in EPIC-Norfolk where three measurements were taken per eye compared to just one measurement per eye in UK Biobank. Among the significant regions, there are previously reported IOP-associated loci,7,8 including TMCO1 (rs10918274, P=2.4x10-87), GAS7 (rs9913911, P=4.0x10-68), ABCA1 (rs2472493, P=6.2x10-59), and CAV1/CAV2 (P=2.5x10-56 for rs10281637). Additionally, 4 of the 10 previously reported POAG-associated loci not known to also be associated with IOP were among the significant regions: AFAP1 (rs28649910, P=8.9x10-41), FOXC1 (rs2745572, P=1.8x10-28), TXNRD2/GNB1L (rs17534001, P=5.2x10-12), and ATXN2/SH2B3 (rs10774624, P=3.4x10-10).10–12 These results strongly suggest these genes mediate POAG risk via raised IOP.
Table 1.
List of novel SNPs most significantly associated with IOP or POAG in our study. Results are presented for the IOP GWAS meta-analysis (UK Biobank, IGGC and EPIC-Norfolk; n=139,555) and for the association with POAG in the NEIGHBORHOOD study (3,853 cases and 33,480 controls). All IOP association P-values are genome-wide significant (P<5x10-8) and in bold if not previously reported as associated with IOP. POAG association P-values are in bold if novel and significant at a Bonferroni-corrected threshold of P<4.2x10-4. A full list of all genome-wide significant loci from the IOP GWAS is given in Supplementary Table 2 (including 68 novel loci) and their associations with POAG in NEIGHBORHOOD are shown in full in Supplementary Table 9.
| SNP ID | Chr | Position | Nearest gene | Effect allele | Effect allele frequency | IOP GWAS meta-analysis | NEIGHBORHOOD POAG association | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β | (95% CI) | P-value | OR | (95% CI) | P-value | ||||||
| rs4074961 | 1 | 38,092,723 | RSPO1 | C | 0.56 | -0.09 | (-0.11, -0.06) | 4.4x10-12 | 0.86 | (0.80, 0.92) | 8.9x10-6 |
| rs6781336 | 3 | 66,858,050 | KBTBD8, LRIG1 | A | 0.70 | 0.12 | (0.09, 0.15) | 2.7x10-18 | 1.06 | (0.98, 1.14) | 0.13 |
| rs9853115 | 3 | 186,131,600 | DGKG | T | 0.50 | 0.20 | (0.17, 0.22) | 8.9x10-52 | 1.17 | (1.09, 1.25) | 1.4x10-5 |
| rs368503 | 5 | 14,820,417 | ANKH | A | 0.72 | 0.11 | (0.08, 0.14) | 5.1x10-15 | 1.04 | (0.97, 1.12) | 0.30 |
| rs113985657 | 6 | 597,203 | EXOC2 | C | 0.85 | -0.15 | (-0.18, -0.11) | 1.2x10-15 | 0.83 | (0.75, 0.91) | 1.8x10-4 |
| rs17752199 | 6 | 51,406,848 | PKHD1 | A | 0.90 | 0.16 | (0.12, 0.20) | 2.2x10-14 | 1.34 | (1.20, 1.50) | 2.7x10-7 |
| rs9494457 | 6 | 136,474,794 | PDE7B | A | 0.62 | -0.12 | (-0.14, -0.09) | 3.7x10-19 | 0.91 | (0.84, 0.97) | 0.0063 |
| rs10230941 | 7 | 117,636,111 | CTTNBP2 | C | 0.64 | -0.09 | (-0.11, -0.06) | 4.6x10-11 | 0.88 | (0.82, 0.94) | 2.5x10-4 |
| rs62520913 | 8 | 124,614,322 | FBXO32 | T | 0.93 | 0.22 | (0.17, 0.27) | 3.6x10-17 | 1.13 | (0.98, 1.29) | 0.08 |
| rs12377624 | 9 | 129,373,110 | LMX1B | G | 0.63 | 0.15 | (0.13, 0.18) | 1.3x10-31 | 1.17 | (1.09, 1.25) | 2.4x10-5 |
| rs2433414 | 11 | 86,410,241 | ME3 | T | 0.80 | 0.13 | (0.10, 0.16) | 6.9x10-16 | 1.14 | (1.05, 1.24) | 0.0028 |
| rs7924522 | 11 | 128,380,742 | ETS1 | A | 0.34 | 0.11 | (0.08, 0.14) | 3.1x10-16 | 1.13 | (1.05, 1.21) | 7.4x10-4 |
| rs4775427 | 15 | 61,951,235 | VPS13C | T | 0.43 | 0.11 | (0.09, 0.14) | 4.1x10-18 | 1.11 | (1.03, 1.18) | 0.0032 |
| rs1874458 | 16 | 65,080,739 | CDH11 | A | 0.64 | -0.10 | (-0.13, -0.08) | 2.9x10-15 | 0.87 | (0.81, 0.93) | 8.0x10-5 |
| rs3743860 | 16 | 89,818,491 | FANCA | T | 0.58 | 0.10 | (0.08, 0.13) | 4.2x10-15 | 1.03 | (0.96, 1.10) | 0.39 |
Interestingly, four loci previously associated with primary angle-closure glaucoma, a form of glaucoma distinct from POAG, were also among the significant regions for IOP, namely HGF (rs327716, P= 6.1x10-13),16 PLEKHA7 (rs4141194, P=7.2x10-21), FERMT2 (rs8009633, P= 7.1x10-13) and GLIS3 (rs6476827, P= 1.2x10-10),17 suggesting that mechanisms underlying angle-closure also contribute to variation in IOP within the normal range. Three IOP-significant loci were in genes previously associated with optic disc cup area (a structural quantitative trait associated with glaucoma), but not with IOP or POAG, namely BCAS3 (rs3785855, P= 4.0x10-16), EFEMP1 (rs4672075, P= 1.9x10-11) and RARB (rs1286771, P= 4.7x10-9);8 this suggests that a proportion of optic disc structural variability in a population may be IOP-mediated.
Among the significant IOP loci, a strong association was observed for rs9853115 (P= 8.9x10-52), a SNP located in an ENCODE DNaseI hypersensitivity cluster region, 51kb upstream from the Diacylglycerol Kinase Gamma (DGKG) gene. Diacylglycerol is involved in adenosine receptor signaling, which is important in IOP regulation and a potential target for IOP-lowering therapy.18 More broadly, DGKG is involved in lipid metabolism, a function shared with other IOP-influencing genes.19 Very recently, DGKG has also been associated with IOP in a multi-ethnic study of individuals residing in the United States.9
Also, significantly associated were two loci harboring angiopoietin genes (ANGPT1, P=2.7x10-18 for rs4496939 and ANGPT2, P=1.7x10-13 for rs76020419); both are primary TEK (Receptor Tyrosine Kinase) ligands, mutations of which cause primary congenital glaucoma.20 In addition, significant association was also found for LRIG1 (rs6781336, P= 2.7x10-18), an endogenous feedback regulator of receptor tyrosine kinases, and FER Tyrosine Kinase (rs73220177, P=1.6x10-11). This suggests a critical role for angiopoietin-receptor tyrosine kinase (ANG-TEK) signaling in IOP regulation. ANG-TEK signaling is established as a key mediator of blood and lymphatic vessel development,21 and gene-set enrichment analysis of our meta-analysis results suggests a strong role for angiogenesis (Supplementary Table 3). TEK receptors are highly expressed in Schlemm’s canal endothelial cells,22 and disruption of ANG-TEK signaling in mice causes lack of Schlemm’s canal development.23 A locus near VEGFC was also strongly associated with IOP in our study (rs437376, P=5.8x10-9). VEGF-C stimulates VEGFR-3 tyrosine kinase signaling in lymphatic endothelial cells and a single injection of recombinant VEGF-C in the eyes of adult mice induced Schlemm’s canal growth with sustained reduction in IOP;24 this supports the hypothesis that Schlemm’s canal is a form of lymphatic vessel and that regulators of lymphangiogenesis are potential targets for glaucoma therapy.24
Some SNPs significantly associated with IOP in our healthy populations annotate near transcription factor-coding genes whose rare mutations cause congenital or childhood glaucoma (LMX1B,25 LTBP226). Several others are implicated in ocular development (MEIS127, SIX328, ADAMTS1829), axial length of the eye (RSPO130) and iris architecture (TRAF3IP131). Moreover, gene-enrichment analysis identified a key role for developmental processes (Supplementary Table 3). These results suggest that ocular developmental or anatomical variations insufficient to cause childhood glaucoma may manifest in later life with raised IOP and potentially POAG.
Supporting a mitochondrial contribution to POAG pathogenesis are four significant IOP loci at genes important for mitochondrial function. ME3 (rs2433414, P=6.9x10-16) has previously been implicated in POAG through a mitochondrial gene set analysis.32 VPS13C (rs4775427, P=4.1x10-18) is necessary for mitochondrial transmembrane potential, GCAT (rs6000889, P=2.2x10-12) regulates mitochondrial glycine production, and PTCD2 (rs10036789, P=7.7x10-10) is involved in mitochondrial RNA maturation.
Many of the IOP-associated SNPs we report have previously been associated with other ocular and systemic phenotypes (Supplementary Table 4). A subsequent systematic comparison of all significantly associated SNPs from the current IOP meta-analysis with all the previously published and currently public domain GWAS data33 revealed that IOP significantly shares genetic risk factors with other traits; the most significant correlations are with traits that have been previously linked epidemiologically to IOP or glaucoma such as heart rate,13 sleep duration,34 and cholesterol level34 (Supplementary Table 5).
Two of the IOP-associated SNPs are missense coding (rs12923138, ELMO3 and rs61755579, SOS2); the rest are outside gene-coding regions. Querying of eQTL effects on the GTEx database confirmed that many of these SNPs alter efficiency of transcription of genes in their immediate vicinity (Supplementary Table 6). Genes in the vicinity of the IOP-associated SNPs are highly expressed in human trabecular meshwork and ciliary body (Supplementary Table 7), tissues important in IOP homeostasis.35 Furthermore, S-PrediXcan analyses support a role for the IOP-associated SNPs in regulation of gene expression, especially for GAS7 (P=1.7x10-35) and AFAP1 (P=6.1x10-22) (Supplementary Table 8).
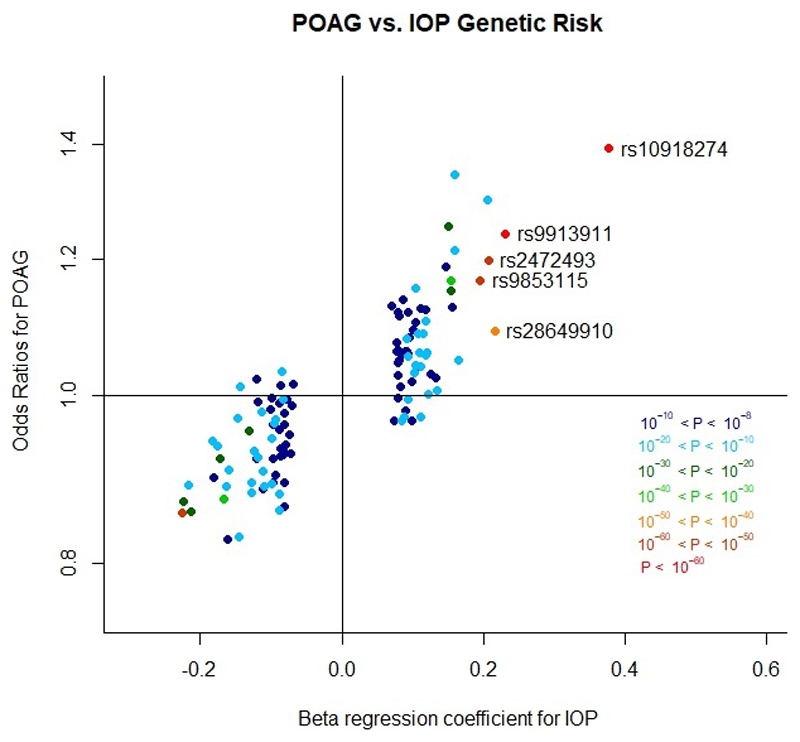
To evaluate the disease-relevance of the IOP-significant SNPs, we tested for association with clinically diagnosed POAG in participants of the NEIGHBORHOOD study10 (3,853 cases and 33,480 controls). In total, 48 SNPs were nominally associated with POAG (P<0.05), of which 14 SNPs were significant at a Bonferroni-corrected threshold of P<4.2x10-4. For all SNPs, we observed a remarkable correlation between the effect sizes for IOP and POAG (Fig. 1). Analysis of the high-tension (HTG) and normal-tension glaucoma (NTG) subgroups suggests that while the association is stronger in HTG, it is still evident in NTG despite IOP being within normal limits (Supplementary Table 9). Additionally, we identified similar associations between the IOP-significant SNPs and glaucoma (ascertained by self-report and hospital episode statistics data) among UK Biobank participants without IOP data available and therefore not part of the IOP GWAS (1,500 cases and 331,078 controls; Supplementary Table 10). There was no evidence of association between IOP-significant SNPs and age at glaucoma diagnosis in either cohort (Supplementary Tables 11 and 12).
Figure 1.
Scatter plot demonstrating the correlation of effect estimates for SNP associations with IOP in our GWAS meta-analysis with effect estimates for SNP associations with POAG in the NEIGHBORHOOD study. Each point represents one SNP from the 120 independent IOP-associated SNPs (derived from the conditional analysis of our IOP GWAS meta-analysis; 13 of 133 SNPs were not available in NEIGHBORHOOD). The color of each point represents the statistical significance of the SNP association with IOP (see key). Effect estimates are per risk allele.
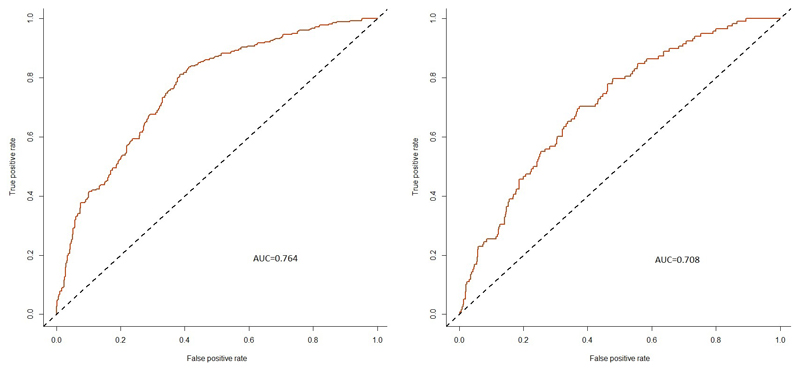
Using 120 significant variants from the conditional analysis (Supplementary Table 2) for which genotypes were available in NEIGHBORHOOD participants and three known POAG-associated polymorphisms showing no evidence of association with IOP in our meta-analysis (rs74315329 within MYOC, rs2157719 near SIX6, and rs8015152 within CDKN2B-As1), we built and evaluated the performance of a regression-based POAG prediction model that, in addition to the associated alleles’ predisposing or protective effects on glaucoma, also included age and sex. Despite being limited to a smaller number of significant SNPs, the prediction model performed well in a subset of the NEIGHBORHOOD study with individual-level genotype data available, in particular for HTG (AUROC=0.76) (Fig. 2). This model also performed well for predicting glaucoma in UK Biobank participants not previously included in the IOP GWAS, with an AUROC=0.74 (Supplementary Fig. 5).
Figure 2.
ROC curves for performance of the POAG-predictive model in HTG (left; n=1,298) and NTG (right; n=561) subsets versus controls (n = 2,606) from a subset of the NEIGHBORHOOD study with individual level genotype data available.
In summary, our analysis has identified 112 loci, 68 of which are novel, associated with IOP and the development of POAG. Several loci support an important role for ANG-TEK signaling in IOP regulation that may be a therapeutic target. Together with other genetic factors previously known to affect POAG risk, the loci explain and predict a substantial portion of POAG cases in two independent cohorts. Given there is currently no adequate population screening test for glaucoma,36 and half of glaucoma cases in the community are undiagnosed,2 genetic prediction models offer opportunity for improved case detection, earlier treatment, and preventing morbidity from the leading cause of irreparable blindness. The genetic loci identified in this study not only increase our understanding of the pathways involved in IOP and glaucoma, but also open up the possibility of using genetic markers to improve disease screening or even prediction of the natural history of disease in people at risk of glaucoma.
Online Methods
Study Methods
UK Biobank
The UK Biobank is a very large multisite cohort study established by the Medical Research Council, Department of Health, Wellcome Trust medical charity, Scottish Government and Northwest Regional Development Agency. Detailed study protocols are available online (see URLs section). A baseline questionnaire, measurements, and biological samples were undertaken in 22 assessment centres across the UK between 2006 and 2010. All UK residents aged 40 to 69 years who were registered with the National Health Service (NHS) and living up to 25 miles from a study centre were invited to participate. The study was conducted with the approval of the North-West Research Ethics Committee (ref 06/MRE08/65), in accordance with the principles of the Declaration of Helsinki, and all participants gave written informed consent.
Ophthalmic assessment was not part of the original baseline assessment and was introduced as an enhancement in 2009 for 6 assessment centres which are spread across the UK (Liverpool and Sheffield in North England, Birmingham in the Midlands, Swansea in Wales, and Croydon and Hounslow in Greater London). Participants completed a touch-screen self-administered questionnaire. The response options for ethnicity included White (English/Irish or other white background), Asian or British Asian (Indian/Pakistani/Bangladeshi or other Asian background), Black or Black British (Caribbean, African, or other black background), Chinese, mixed (White and Black Caribbean or African, White and Asian, or other mixed background), or other ethnic group (not defined). Self-reported glaucoma status was ascertained as participants who selected “glaucoma” from a list of eye disorders to the question, “Has a doctor told you that you have any of the following problems with your eyes?”
Participant IOP was measured once for each eye using the Ocular Response Analyzer (ORA; Reichert, Corp., Buffalo, NY). Participants who reported eye surgery within the previous 4 weeks or participants reporting an eye infection were precluded from having IOP measured. The ORA is a non-contact tonometer that measures the force required to flatten the cornea using a jet of air. Unlike conventional non-contact tonometry, the ORA measures two pressures; firstly, when the cornea flattens on inward motion, and secondly when the cornea is flattened on outward motion. The average of these two pressures has been calibrated to derive a Goldmann-correlated IOP (IOPg) and the difference between these two pressures has been shown to be related to the biomechanical properties of the cornea.37 A linear combination of these two pressures has been developed to derive a corneal-compensated IOP (IOPcc).38 We used IOPcc in analyses as it is thought to provide the most accurate assessment of true IOP and least affected by corneal properties.13
We excluded participants with a history of laser or surgery for glaucoma, eye injury, corneal graft surgery, or refractive laser surgery as these participants are likely to have IOP altered from physiological levels due to non-genetic causes. To handle extreme values of IOP, we excluded IOP measurements in the top and bottom 0.5 percentiles.
A significant proportion of participants with the highest IOPs in the cohort will have been diagnosed and treated with IOP-lowering medication in the community before entering the current study. Data for pre-treatment IOP were not available and excluding these participants would have truncated the study IOP distribution, thereby reducing statistical power for detecting associations with IOP. We therefore imputed pre-treatment IOP: in study participants reporting current IOP-lowering medication (n = 1,151), the measured IOP was divided by 0.7 based on the mean IOP reduction achieved by medication.39 This method has been used in previously published genome-wide association studies for IOP.7,40 Participant IOP was calculated as the mean of right and left eye values for each participant with data available for both eyes. If data were only available for one eye, we considered that value to be the participant’s IOP. Figures presenting the cleaning and derivation flow for IOP and glaucoma status are in the Supplementary Note.
Details for DNA extraction and genotyping of UK Biobank participants are given in the Supplementary Note.
The basic model tested was the average of IOP measured in the left and right eye as an outcome of a regression model whose predictor is the allele dosage at a given polymorphic locus, adjusted for age, sex and the first five principal components (see Supplementary Note for further details). Since there was, at the time of writing, evidence of cryptic relatedness among the UK Biobank participants, a linear mixed model that controls for population structure was used41 as implemented in the program BOLT-LMM (see URLs section).
International Glaucoma Genetics Consortium (IGGC)
The IGCC study was a meta-analysis of 37,930 participants from 19 studies of European (14 studies) and Asian (5 studies) descent.8 Similarly, to our study, mean IOP of right and left eyes was considered and pre-treatment IOP was imputed for participants using IOP-lowering medication. A variety of genotyping arrays were used across the different studies and genotypes were imputed using 1000 Genomes Phase 1 reference samples. SNPs with MAF<0.01 and imputation quality scores <0.3 were excluded. Linear regression analyses were adjusted for age, sex and the first five principal components for population-based studies, or family structure for family-based studies. For the purposes of the current study, we used publicly available summary results for the European subset of the IGGC study (n=29,578).
EPIC-Norfolk
The European Prospective Investigation into Cancer (EPIC) study is a pan-European prospective cohort study designed to investigate the aetiology of major chronic diseases.42 EPIC-Norfolk, one of the UK arms of EPIC, recruited and examined 25,639 participants between 1993 and 1997 for the baseline examination.43 Recruitment was via general practices in the city of Norwich and the surrounding small towns and rural areas, and methods have been described in detail previously.44 Since virtually all residents in the UK are registered with a general practitioner through the National Health Service, general practice lists serve as population registers. Ophthalmic assessment formed part of the third health examination and this has been termed the EPIC-Norfolk Eye Study.14
In total, 8,623 participants were seen for the Eye Study between 2004 and 2011, and IOP was measured using the ORA. Three measurements were taken per eye and the best signal value used. Mean IOPcc of right and left eyes was calculated and used in analyses. 99.7% of EPIC-Norfolk are of European descent and we excluded non-White participants. The EPIC-Norfolk Eye Study was carried out following the principles of the Declaration of Helsinki and the Research Governance Framework for Health and Social Care. The study was approved by the Norfolk Local Research Ethics Committee (05/Q0101/191) and East Norfolk & Waveney NHS Research Governance Committee (2005EC07L). All participants gave written, informed consent.
Details for genotyping and imputation of EPIC-Norfolk participants are given in the Supplementary Note.
Similarly to the UK Biobank GWAS, we examined the relationship between allele dosage and mean IOPcc using linear regression adjusted for age, sex and the first 5 principal components. Analyses were carried out using SNPTEST version 2.5.1.
NEIGHBORHOOD Study
All cases and controls met the clinical criteria used previously by the NEIGHBOR and GLAUGEN studies previously described.10,45,46 This study Subjects were enrolled using a protocol was approved by the Massachusetts Eye and Ear Infirmary institutional review board and all subjects signed consent forms approved by the local IRB prior to enrolling in the study.
Briefly, POAG cases were defined as individuals for whom reliable visual field (VF) tests showed characteristic VF defects consistent with glaucomatous optic neuropathy. Individuals were classified as affected if the VF defects were reproduced on a subsequent test or if a single qualifying VF was accompanied by a cup-disc ratio (CDR) of 0.7 or more in at least one eye. The majority of cases (over 90%) met this definition, including 96% of the NEIGHBOR cases;45 and all of the Massachusetts Eye and Ear Infirmary (MEEI), Nurses’ Health Study (NHS), Health Professionals Follow-up Study (HPFS), and Women’s Genomes Health Study (WGHS) cases. A small percentage (less than 10%) of the NEIGHBOR, Mayo, Marshfield and Iowa cases were defined by cup-to-disc ratio only because visual field data was not available, in some cases because of advanced disease (poor visual acuity) or other medical condition. The CDR definition was > 0.7 in both eyes or CDR asymmetry between the two eyes of 0.2 (Supplementary Table 2). In the OHTS study an alternative case definition based on progression of optic nerve degeneration was also used47 (see below). Patients with signs of secondary causes for elevated IOP such as exfoliation syndrome or pigment dispersion syndrome or critically narrow filtration structures were excluded. Elevation of IOP was not a criterion for inclusion of cases or controls; however, 1,868 cases did have a history of elevated IOP (≥22 mm Hg) measured in a clinical setting (typically between the hours of 8AM and 5PM) and were classified as high-tension glaucoma (HTG), while 725 cases did not have elevated IOP and were classified as normal-tension glaucoma (NTG). For 1,260, cases peak IOP data was not available. The controls were selected to be representative of the age range and gender of the cases. While the average age of cases and controls was not statistically different for any dataset included in the NEIGHBORHOOD, some datasets included cases and controls younger than age 55 which could reduce the power of the study. Controls had IOP < 21 mmHg, as measured in a clinical setting, CDR of less than 0.6 and did not have a family history of glaucoma.
Participants in the NEIGHBORHOOD used different genotyping chips and imputation methods as specified elsewhere.10
Imputed genotypes (1000 Genomes panel, March 2012, INFO score >0.9) for 3,853 cases and 33,480 controls from 8 independent datasets were used as the discovery cohort for the NEIGHBORHOOD genome-wide association study for POAG.10 Quality-control was performed for each data set as described in Bailey et al.10 Overall sample and genotype call rates were ≥ 95% for each site. Samples with Log R ratio (LRR) and B allele frequency (BAF) values suggestive of copy number variants were removed prior to analysis. Principal components (eigenvectors) were computed for all participants using EIGENSTRAT.48 For each dataset, logistic regression was performed in ProbABEL49 for all analyses (POAG overall, HTG, NTG), controlling for age, sex, and study-specific covariates including study-specific eigenvectors. Each analysis was evaluated separately for overall genomic inflation (implementing the R package GenABEL) (λ-value ≤ 1.05 for each dataset). Results were meta-analyzed in METAL50 implementing the inverse variance weighted method and applying genomic control correction.
For the prediction models and assessment of their performance, a balanced dataset of cases and controls (Ncases=Ncontrols=2,606) used were from only two subcohorts: NEIGHBOR and MEEI. The choice of the two largest subcohorts within NEIGBORHOOD assured that the prediction dataset was fully balanced and, as the genotyping and imputation pipelines followed for them were largely compatible, minimized the risk of stratification among the samples.
Statistical Analyses
Details of our statistical analyses are below and in the accompanying Life Sciences Reporting Summary.
Meta-analysis
Summary statistics from each strata (UK Biobank, the International Glaucoma Genetics Consortium meta-analysis8 and from the participants in the EPIC study that were not included in the IGGC meta-analysis) were combined using fixed-effects inverse variance weighted meta-analysis, using METAL.50 Random-effects meta-analyses results were also obtained using GWAMA,51 but results from this did not differ significantly from the fixed-effect model and the results shown are just from the latter. No genomic control adjustment was applied during the meta-analysis.
Conditional and explained heritability analyses
The program GCTA52 was used for the conditional analyses53 to identify independent effects within associated loci as well as the calculation of the phenotypic variance explained54 by all polymorphisms, genotyped or imputed, associated with the trait after the conditional analyses. The threshold of significance was set at 5x10-8 and the collinearity threshold was set at r2=0.9. The LD estimates were derived from the UKBB cohort.
Calculation of genomic inflation factor
To assess the potential inflation of association probabilities, genomic inflation factors55 were calculated and Q-Q plots were drawn using the package ‘gap’ in R (see URLs section).
Multiple testing correction
Two methods of correcting for multiple testing were used. The first was a classic Bonferroni, in which the threshold of significance (0.05) was divided by the number of experiments (n):
Given the large number of loci for which replication was needed, we additionally calculated the False Discovery Rates, using the Benjamini-Hochberg method.56
LD Score analyses
Inter-trait genetic correlation
Bivariate genetic correlations between IOP and other complex traits whose summary statistics are publicly available were assessed following previously described methodologies,57 using the program LD Score (see URLs section).
Regression intercept
To distinguish between the effect of polygenicity and those arising from sample stratification or uncontrolled population admixture, we followed previously suggested approaches15 to calculate the LD Score regression intercepts using the program LD Score (see URLs section).
Prediction analyses
To assess the potential value of the loci associated with IOP to predict POAG, regression-based models were deliberately trained and tested separately in two different groups. The first, is the set of UK Biobank participants for whom IOP measurements were not available (which made them ineligible to participate in the meta-analysis of the IOP regression analysis; see Supplementary Note). Since this information was questionnaire-derived, for these patients it was impossible to stratify the diagnosis of glaucoma into normal or high-tension glaucoma subgroups (NTG and HTG, respectively). This dataset was not balanced, since it included 1,500 cases of glaucoma and 331,078 individuals with no self-reported diagnosis of glaucoma. The second group was formed by the clinical cases and controls from two of the NEIGHBORHOOD subcohorts (NEIGHBOR AND MEEI). Patients and controls in this group were clinically characterized. They were a mixture of NTG and HTG cases (n=561 and n=1,298 respectively), a further 747 subjects of uncertain POAG type, and 2,606 controls).
We built the same model in all cases, which included age, sex, and the major genetic variants associated with IOP after the conditional analysis. We additionally included three known POAG-associated polymorphisms showing no evidence of association with IOP in our meta-analysis (rs74315329 within MYOC, rs2157719 near SIX6, and rs8015152 within CDKN2B-As1). To minimize bias, we did not use effect sizes observed for IOP to weigh the effects in other cohorts. Instead, in each group separately, logistic regressions were trained using a random subset of 80% of cases and controls. The ability of these trained models to correctly predict the presence of POAG (whether self-reported or doctor diagnosed, depending on the group), was assessed in the remaining 20% of the subjects. A Receiver Operating Characteristic (ROC) curve was drawn for each case and an Area Under the Curve (AUC) was calculated. R programming language and software environment for statistical computing (see URLs section) was used for both the logistic regression models (‘glm’) and to evaluate the performance of the model (‘ROCR’).
SNP and gene annotations
Polymorphisms associated at a GWAS level (P<5x10-8) were clustered within an “associated genomic region”, defined as a contiguous genomic region where GWAS-significant markers were within 1 million base pairs from each other. Significant polymorphisms were annotated with the gene inside whose transcript-coding region they are located, or alternatively, if located between two genes, with the gene nearest to it. The associated genomic regions were collectively annotated with the gene overlapping, or nearest the most significantly associated variant within that region. In addition, the polymorphic sites were functionally annotated using SNPnexus.58
GTEx
Due to unavailability of tissues extracted from human eyes, the influence of our significant SNPs on transcription of adjacent genes was assessed in all other tissues available to the GTEx Project59 and queried in the GTEx Portal (see URLs section).
Ocular gene expression
Gene expression in human trabecular meshwork and ciliary body tissue of genes at loci significant in the IOP GWAS were examined using results from a published RNA sequencing study.35 The expression level for each gene (adjusted for gene length and number of sequencing reads in a given sample) was presented in fragments per kilobase of transcript per million mapped reads (FPKM). Based on the overall gene expression distribution, genes with an FPKM≥1, an FPKM≥4.7 (33rd percentile) and an FPKM≥15.9 (67th percentile) were classified as lowly, moderately, or highly expressed, respectively.
S-PrediXcan
We used S-PrediXcan60 to estimate genetically regulated gene expression using whole-genome tissue-dependent prediction models trained with GTEx reference transcriptome data and then correlate this with IOP to identify genes involved in IOP regulation. S-PrediXcan is related to PrediXcan61 but uses GWAS summary statistics as input. Based on the GTEx analysis described above, we examined correlations using the following reference tissues: whole blood, adipose-omentum, brain-cortex, artery-aorta and artery-coronary. Results are presented in Supplementary Table 8 for all genes significant after Bonferroni correction for all genes tested in all tissues.
Gene-set enrichment
To identify pathways or other gene sets that were over or under-represented among our results, we used a Gene-Set Enrichment Analysis (GSEA) as implemented in the Meta-Analysis Gene Set Enrichment of Variant (MAGENTA) software.62 This program assigns scores to each gene based on the strength of association with IOP, adjusting for potential confounders such as gene length and linkage disequilibrium. Enrichment for any gene set was assessed within genes above the cut-off of the highest 75th centile of significant gene scores. For the current study, the most recent versions of Gene Ontology (GO), Panther, KGG, Biocarta and MSigDB databases were used. A permutational procedure and false-discovery rates were used to calculate significance of enrichment and control for multiple testing.
Data Availability Statement
UK Biobank data are available through the UK Biobank Access Management System (see URLs section).
The data sharing and preservation strategy in EPIC-Norfolk and full details about the study including contact information are on the study website (see URLs section). Investigators wishing to work with EPIC data should contact the EPIC management group through the website, letter, phone or fax, and proposals have to fulfil a number of criteria including that the work is within the bounds of consent given by participants and has been ethically reviewed and approved; there is no serious risk to the viability of continuing the cohort study e.g. through offence to the participants from use of the data supplied; the science of the proposal has been satisfactorily peer reviewed and the proposal does not duplicate work already being done.
URLs
UK Biobank protocols:
http://www.ukbiobank.ac.uk/resources/
http://biobank.ctsu.ox.ac.uk/crystal/docs.cgi
BOLT-LMM:
http://data.broadinstitute.org/alkesgroup/BOLT-LMM/downloads/
LD Score:
R programming language and software environment for statistical computing:
GTEx Portal:
https://www.gtexportal.org/home/
EPIC-Norfolk:
http://www.epic-norfolk.org.uk/
UK Biobank Access Management System:
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 17615. UK Biobank was established by the Wellcome Trust medical charity, Medical Research Council, Department of Health, Scottish Government, and Northwest Regional Development Agency. It also had funding from the Welsh Assembly Government, British Heart Foundation, and Diabetes UK.
EPIC-Norfolk infrastructure and core functions are supported by grants from the Medical Research Council (G1000143) and Cancer Research UK (C864/A14136). The clinic for the third health examination was funded by Research into Ageing (262). Genotyping was funded by the Medical Research Council (MC_PC_13048). We thank all staff from the MRC Epidemiology laboratory team for the preparation and quality control of DNA samples. AP Khawaja is supported by a Moorfields Eye Charity grant. PJ Foster has received additional support from the Richard Desmond Charitable Trust (via Fight for Sight) and the Department for Health through the award made by the National Institute for Health Research to Moorfields Eye Hospital and the UCL Institute of Ophthalmology for a specialist Biomedical Research Centre for Ophthalmology.
The NEIGHBORHOOD data collection and analysis is supported by NIH/NEI R01EY022305 (JL Wiggs) and NIH/NEI P30 EY014104 (JL Wiggs). Support for collection of cases, controls and analysis for individual datasets is as follows. Genotyping services for the NEIGHBOR study were provided by the Center for Inherited Disease Research (CIDR) and were supported by the National Eye Institute through grant HG005259-01 (JL Wiggs). Genotyping for the MEEI dataset and some NHS and HPFS cases (GLAUGEN) was completed at the Broad Institute and supported by GENEVA project grant HG004728 (LR Pasquale) and U01-HG004424 (Broad Institute). Genotype data cleaning and analysis for the GLAUGEN study was supported by U01HG004446 (C Laurie). Collecting and processing samples for the NEIGHBOR dataset was supported by the National Eye Institute through ARRA grants 3R01EY015872-05S1 (JL Wiggs) and 3R01EY019126-02S1 (MA Hauser). Funding for the collection of NEIGHBOR cases and controls was provided by NIH grants: EY015543 (RR Allingham),EY006827 (D Gaasterland); HL73042, HL073389, EY13315 (MA Hauser); CA87969, CA49449, UM1 CA186107,UM1 CA 167552, EY009149 (PR Lichter), HG004608 (C McCarty), EY008208 (FA Medeiros), EY015473 (LR Pasquale), EY012118 (M Pericak-Vance), EY015682 (A Realini), EY011671 (JE Richards), EY09580 (JE Richards), EY013178 (JS Schuman), RR015574, EY015872 (JL Wiggs), EY010886 (JL Wiggs), EY009847 (JL Wiggs), EY011008,EY144428 (K Zhang), EY144448 (K Zhang), EY18660 (K Zhang). The collection of Marshfield clinic cases and controls was supported by 1U02HG004608-01, 5U01HG006389-02 and NCATS/NIH grant UL1TR000427. In addition some NHS/HPFS cases and controls and analysis of GWAS data was supported by R01 CA131332. The WGHS is supported by HL043851 and HL080467 from the National Heart, Lung, and Blood Institute and CA047988 from the National Cancer Institute, the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen. POAG case identification in WGHS was supported by 3R01 EY15473-5S1 (LR Pasquale). JL Wiggs and LR Pasquale are supported by the Harvard Glaucoma Center for Excellence and an unrestricted grant from Research to Prevent Blindness. Dr. Pasquale is also supported by a Harvard Medical School Distinguished Scholar award.
M Simcoe is a recipient of a Fight for Sight PhD studentship. PGH the recipient of a FfS ECI fellowship. The statistical analyses were run in King’s College London Rosalind HPC LINUX Clusters and cloud server.
Footnotes
Author Contributions
A.P.K., J.N.C.B., M.S., R.P.I., Y.E.S. and P.G.H. conducted the analyses. A.P.K., J.N.C.B., P.J.F., J.L.W., C.J.H. and P.G.H. jointly wrote the manuscript. P.T.K. and P.J.F. designed the ophthalmic component of the UK Biobank study. N.J.W. and R.A.S. led genotyping of the EPIC-Norfolk study. R.W., C.Y.C., L.R.P. and J.L.H. critically appraised the analyses and critically reviewed the manuscript.
Competing Financial Interests
A.P.K. has received a lecturing honorarium from Grafton Optical.
P.J.F. reports personal fees from Allergan, Carl Zeiss, Google/DeepMind and Santen, a grant from Alcon, outside the submitted work
References
- 1.Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 2.Topouzis F, et al. Prevalence of open-angle glaucoma in Greece: the Thessaloniki Eye Study. Am J Ophthalmol. 2007;144:511–519. doi: 10.1016/j.ajo.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 3.de Voogd S, et al. Incidence of open-angle glaucoma in a general elderly population: the Rotterdam Study. Ophthalmology. 2005;112:1487–93. doi: 10.1016/j.ophtha.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Leske MC, et al. Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology. 2007;114:1965–72. doi: 10.1016/j.ophtha.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Garway-Heath DF, et al. Latanoprost for open-angle glaucoma (UKGTS): a randomised, multicentre, placebo-controlled trial. Lancet (London, England) 2015;385:1295–304. doi: 10.1016/S0140-6736(14)62111-5. [DOI] [PubMed] [Google Scholar]
- 6.Sanfilippo PG, Hewitt AW, Hammond CJ, Mackey DA. The heritability of ocular traits. Surv Ophthalmol. 2010;55:561–83. doi: 10.1016/j.survophthal.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Hysi PG, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46:1126–30. doi: 10.1038/ng.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Springelkamp H, et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017;26:438–453. doi: 10.1093/hmg/ddw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choquet H, et al. A large multi-ethnic genome-wide association study identifies novel genetic loci for intraocular pressure. Nat Commun. 2017;8:2108. doi: 10.1038/s41467-017-01913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey JNC, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48:189–94. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gharahkhani P, et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46:1120–1125. doi: 10.1038/ng.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014;46:1115–1119. doi: 10.1038/ng.3078. [DOI] [PubMed] [Google Scholar]
- 13.Chan MPY, et al. Associations with Intraocular Pressure in a Large Cohort: Results from the UK Biobank. Ophthalmology. 2016;123:771–82. doi: 10.1016/j.ophtha.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khawaja AP, et al. The EPIC-Norfolk Eye Study: rationale, methods and a cross-sectional analysis of visual impairment in a population-based cohort. BMJ Open. 2013;3:1–10. doi: 10.1136/bmjopen-2013-002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Z, et al. Hepatocyte Growth Factor Genetic Variations and Primary Angle-Closure Glaucoma in the Han Chinese Population. PLoS One. 2013;8:8–12. doi: 10.1371/journal.pone.0060950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khor CC, et al. Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nat Genet. 2016;48:556–562. doi: 10.1038/ng.3540. [DOI] [PubMed] [Google Scholar]
- 18.Agarwal R, Agarwal P. Newer targets for modulation of intraocular pressure: focus on adenosine receptor signaling pathways. Expert Opin Ther Targets. 2014;18:527–39. doi: 10.1517/14728222.2014.888416. [DOI] [PubMed] [Google Scholar]
- 19.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum Mol Genet. 2017;26:R21–R27. doi: 10.1093/hmg/ddx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souma T, et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest. 2016;126:2575–2587. doi: 10.1172/JCI85830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eklund L, Kangas J, Saharinen P. Angiopoietin–Tie signalling in the cardiovascular and lymphatic systems. Clin Sci. 2016;131:87–103. doi: 10.1042/CS20160129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kizhatil K, Ryan M, Marchant JK, Henrich S, John SWM. Schlemm’s canal is a unique vessel with a combination of blood vascular and lymphatic phenotypes that forms by a novel developmental process. PLoS Biol. 2014;12:e1001912. doi: 10.1371/journal.pbio.1001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson BR, et al. A lymphatic defect causes ocular hypertension and glaucoma in mice. J Clin Invest. 2014;124:4320–4324. doi: 10.1172/JCI77162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aspelund A, et al. The Schlemm’s canal is a VEGF-C/VEGFR-3-responsive lymphatic-like vessel. J Clin Invest. 2014;124:3975–86. doi: 10.1172/JCI75395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero P, Sanhueza F, Lopez P, Reyes L, Herrera L. c.194 A>C (Q65P) mutation in the LMX1B gene in patients with nail-patella syndrome associated with glaucoma. Mol Vis. 2011;17:1929–39. [PMC free article] [PubMed] [Google Scholar]
- 26.Ali M, et al. Null Mutations in LTBP2 Cause Primary Congenital Glaucoma. Am J Hum Genet. 2009;84:664–671. doi: 10.1016/j.ajhg.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcos S, et al. Meis1 coordinates a network of genes implicated in eye development and microphthalmia. Development. 2015;142:3009–20. doi: 10.1242/dev.122176. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh Y-W, Zhang X-M, Lin E, Oliver G, Yang X-J. The homeobox gene Six3 is a potential regulator of anterior segment formation in the chick eye. Dev Biol. 2002;248:265–80. doi: 10.1006/dbio.2002.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aldahmesh MA, et al. The syndrome of microcornea, myopic chorioretinal atrophy, and telecanthus (MMCAT) is caused by mutations in ADAMTS18. Hum Mutat. 2013;34:1195–1199. doi: 10.1002/humu.22374. [DOI] [PubMed] [Google Scholar]
- 30.Cheng CY, et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am J Hum Genet. 2013;93:264–277. doi: 10.1016/j.ajhg.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsson M, et al. GWAS findings for human iris patterns: Associations with variants in genes that influence normal neuronal pattern development. Am J Hum Genet. 2011;89:334–343. doi: 10.1016/j.ajhg.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khawaja AP, et al. Assessing the Association of Mitochondrial Genetic Variation With Primary Open-Angle Glaucoma Using Gene-Set Analyses. Invest Ophthalmol Vis Sic. 2016;57:5046–5052. doi: 10.1167/iovs.16-20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus DM, et al. Sleep disorders: a risk factor for normal-tension glaucoma? J Glaucoma. 2001;10:177–83. doi: 10.1097/00061198-200106000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Carnes MU, Allingham RR, Ashley-Koch A, Hauser MA. Transcriptome analysis of adult and fetal trabecular meshwork, cornea, and ciliary body tissues by RNA sequencing. Exp Eye Res. 2018;167:91–99. doi: 10.1016/j.exer.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Momont AC, Mills RP. Glaucoma Screening: Current Perspectives and Future Directions. Semin Ophthalmol. 2013;28:185–190. doi: 10.3109/08820538.2013.771200. [DOI] [PubMed] [Google Scholar]
- 37.Luce DA. Determining in vivo biomechanical properties of the cornea with an ocular response analyzer. J Cataract Refract Surg. 2005;31:156–62. doi: 10.1016/j.jcrs.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 38.Luce D. Methodology for Corneal Compensated IOP and Corneal Resistance Factor for An Ocular Response Analyzer. Invest Ophthalmol Vis Sci. 2006;47:2266. [Google Scholar]
- 39.van der Valk R, et al. Intraocular pressure-lowering effects of all commonly used glaucoma drugs: a meta-analysis of randomized clinical trials. Ophthalmology. 2005;112:1177–85. doi: 10.1016/j.ophtha.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 40.van Koolwijk LME, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riboli E, Kaaks R. The EPIC Project: rationale and study design. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol. 1997;26(Suppl 1):S6–14. doi: 10.1093/ije/26.suppl_1.s6. [DOI] [PubMed] [Google Scholar]
- 43.Day N, et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 44.Hayat SA, et al. Cohort Profile: A prospective cohort study of objective physical and cognitive capability and visual health in an ageing population of men and women in Norfolk (EPIC-Norfolk 3) Int J Epidemiol. 2013 doi: 10.1093/ije/dyt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiggs JL, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013;22:517–25. doi: 10.1097/IJG.0b013e31824d4fd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiggs JL, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feuer WJ, et al. The Ocular Hypertension Treatment Study: reproducibility of cup/disk ratio measurements over time at an optic disc reading center. Am J Ophthalmol. 2002;133:19–28. doi: 10.1016/s0002-9394(01)01338-1. [DOI] [PubMed] [Google Scholar]
- 48.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 49.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mägi R, Morris AP. GWAMA: Software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. S1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 57.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: A web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbeira A, et al. Integrating tissue specific mechanisms into GWAS summary results. bioRxiv. 2017 doi: 10.1101/045260. 45260. [DOI] [Google Scholar]
- 61.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayellet VS, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are available through the UK Biobank Access Management System (see URLs section).
The data sharing and preservation strategy in EPIC-Norfolk and full details about the study including contact information are on the study website (see URLs section). Investigators wishing to work with EPIC data should contact the EPIC management group through the website, letter, phone or fax, and proposals have to fulfil a number of criteria including that the work is within the bounds of consent given by participants and has been ethically reviewed and approved; there is no serious risk to the viability of continuing the cohort study e.g. through offence to the participants from use of the data supplied; the science of the proposal has been satisfactorily peer reviewed and the proposal does not duplicate work already being done.