Abstract
The microenvironment is critical for stem cell maintenance and can be of cellular and non-cellular composition, including secreted growth factors and extracellular matrix (ECM)1–3. Although Notch and other signalling pathways have been reported to regulate quiescence4–9, the composition and source of niche molecules remain largely unknown. Here, we show that adult muscle satellite (stem) cells produce ECM collagens to maintain quiescence cell-autonomously. By ChIP-sequencing we identified NOTCH/RBPJ-bound regulatory elements adjacent to specific collagen genes, whose expression is deregulated in Notch mutant mice. Moreover, we show that satellite cell produced collagen V (COLV) is a critical component of the quiescent niche, as conditional deletion of Col5a1 leads to anomalous cell cycle entry and gradual diminution of the stem cell pool. Notably, the interaction of COLV with satellite cells is mediated by CALCR, for which COLV acts as a surrogate local ligand. Strikingly, systemic administration of a calcitonin derivative is sufficient to rescue the quiescence and self-renewal defects scored in COLV null satellite cells. This study unveils a Notch/COLV/CALCR signalling cascade that cell-autonomously maintains the satellite cell quiescent state and raises the possibility of a similar reciprocal mechanism acting in diverse stem cell populations.
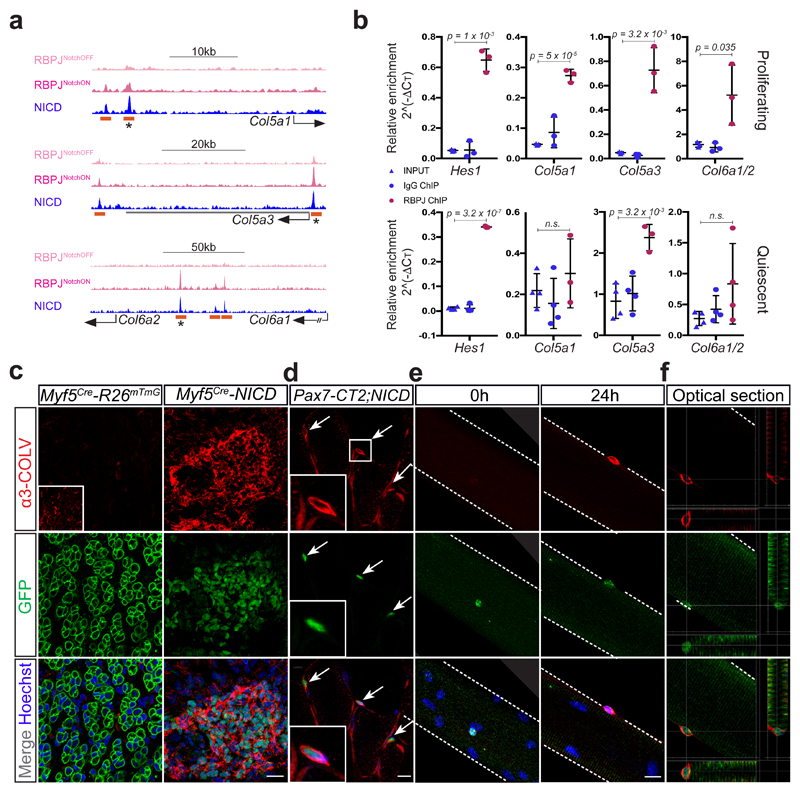
Notch activation antagonizes myogenesis by induction of transcriptional repressors (Hes/Hey family members) and sequestration of the co-activator Mastermind-like 1 from the muscle differentiation factor Mef2c10,11. However, Notch signalling has broader functions in muscle cells, including maintenance of quiescence4,5. To explore these functions, we exploited a ChIP-seq screening12 and observed that intracellular Notch (NICD) and its downstream effector RBPJ, occupied and regulated enhancers proximal to collagen genes Col5a1, Col5a3, Col6a1 and Col6a2, which are amongst the most abundant collagens produced by satellite cells (Fig. 1a, b; Extended data Fig. 1a-e). By analysing genetic models with altered Notch activity, we showed that the expression of these collagens tightly correlated with Notch activity in vivo (Extended data Fig. 2a-e). Moreover, transcriptional induction of Col5a1 and Col5a3 by NICD translated to elevated COLV protein levels, specifically the [(a1(V)a2(V)a3(V)] isoform (α3-COLV), in foetal forelimb (Fig. 1c) and adult hindlimb (Tibialis anterior, TA) myogenic cells (Fig. 1d; Extended data Fig. 2f for α3-COLV antibody specificity). Furthermore, we isolated collagen-depleted myofibres following treatment with collagenase to monitor de novo α3-COLV production. As Col5a1 and Col5a3 transcripts are downregulated upon exit from quiescence (Extended data Fig. 1a and Extended data Fig. 2g), no α3-COLV was detected in freshly isolated or activated satellite cells. Instead, genetic overexpression of NICD resulted in abundant, newly synthetized α3-COLV (Fig. 1e, f).
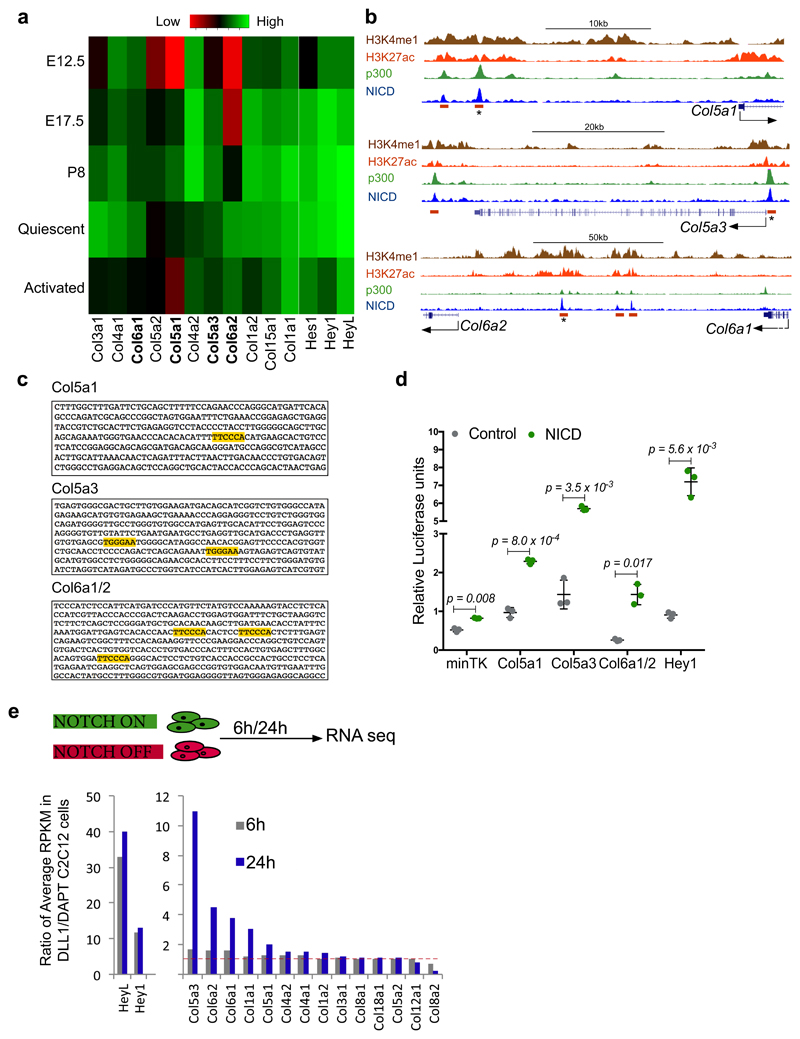
Figure 1. NICD/RBPJ regulates transcription of Col5 and Col6 genes by binding to distal regulatory elements.
(a) RBPJ/NICD ChIP-seq tracks from C2C12 cells indicating enhancers associated with the collagen-5a1, -5a3, -6a1 and -6a2 loci. Orange rectangle, RBPJ/NICD enhancers; asterisk, enhancers used for luciferase assays (Extended data Fig. 1c).
(b) Top: RBPJ ChIP in proliferating primary myogenic cells on Delta-like 1 (n=4 ChIPs). Bottom: RBPJ ChIP in quiescent satellite cells, fixed prior to isolation25 (n=3 ChIPs). Error bars, mean ± SD; two-sided unpaired t-test.
(c) Forelimb muscles of E17.5 Myf5Cre-NICD foetuses show upregulation of COLV. Inset shows low α3-COLV expression (higher exposure time). Note, membrane GFP-marked fibres in control and mononucleated NICD/PAX7+ cells in Myf5Cre-NICD26.
(d) Anti-GFP (satellite cells) and anti-α3-COLV immunostaining on transverse sections of quiescent adult TA muscles expressing NICD-ires-GFP (Pax7CT2-NICD). All GFP+ cells overexpressed COL5A3 (50 cells/mouse, n=3 mice).
(e) Freshly fixed single myofibres from Pax7CT2-NICD Extensor Digitorum Longus muscles, (t0h, left) or after 24h in culture (right) and stained for GFP and α3-COLV.
(f) Vertical and horizontal optical sections of myofibre presented in (e) from Pax7CT2-NICD mice (24h culture) showing COLV surrounding NICD-GFP+ satellite cells.
Scale bars: c, 50µm; d-f, 10 µm. Scale bar insets: c, 100 µm; d, 20 µm.
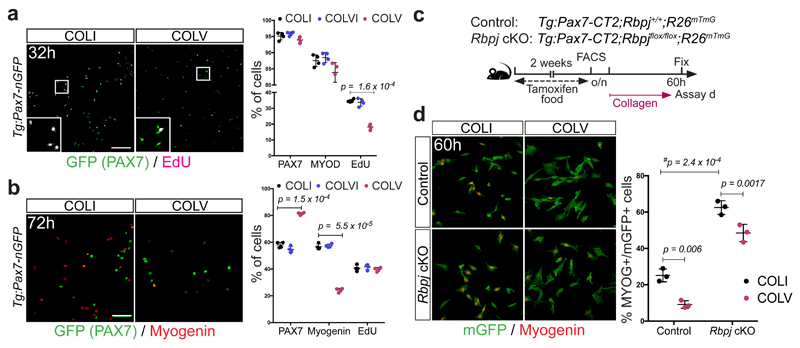
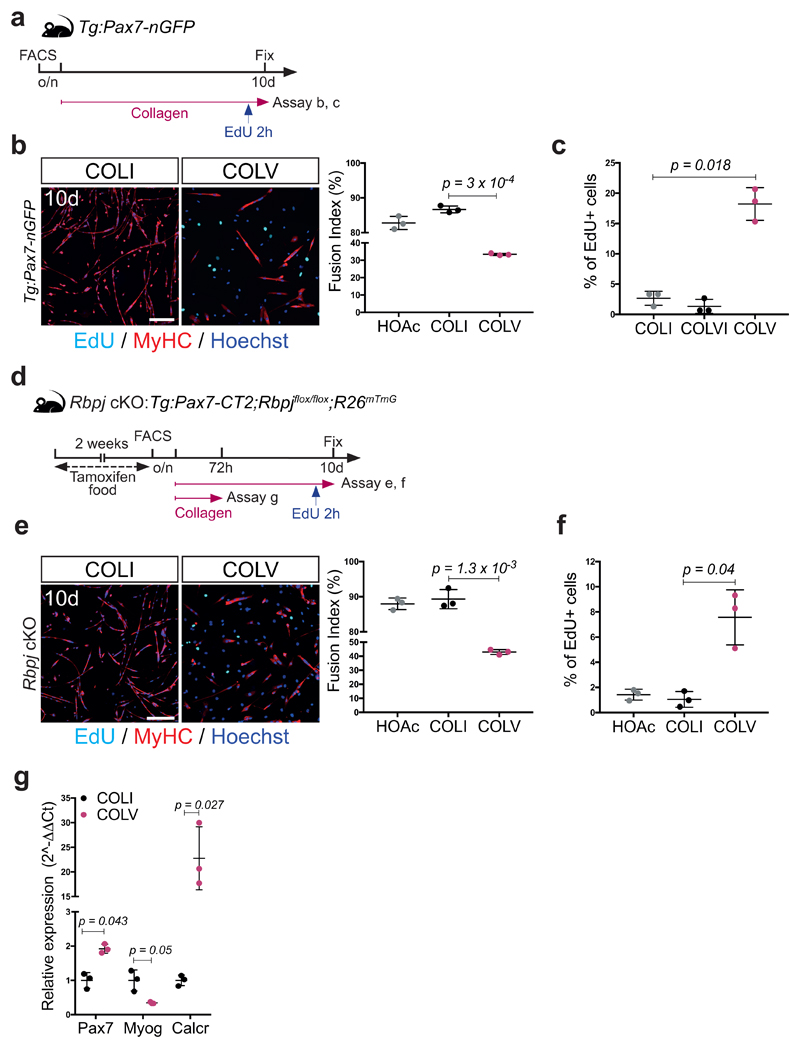
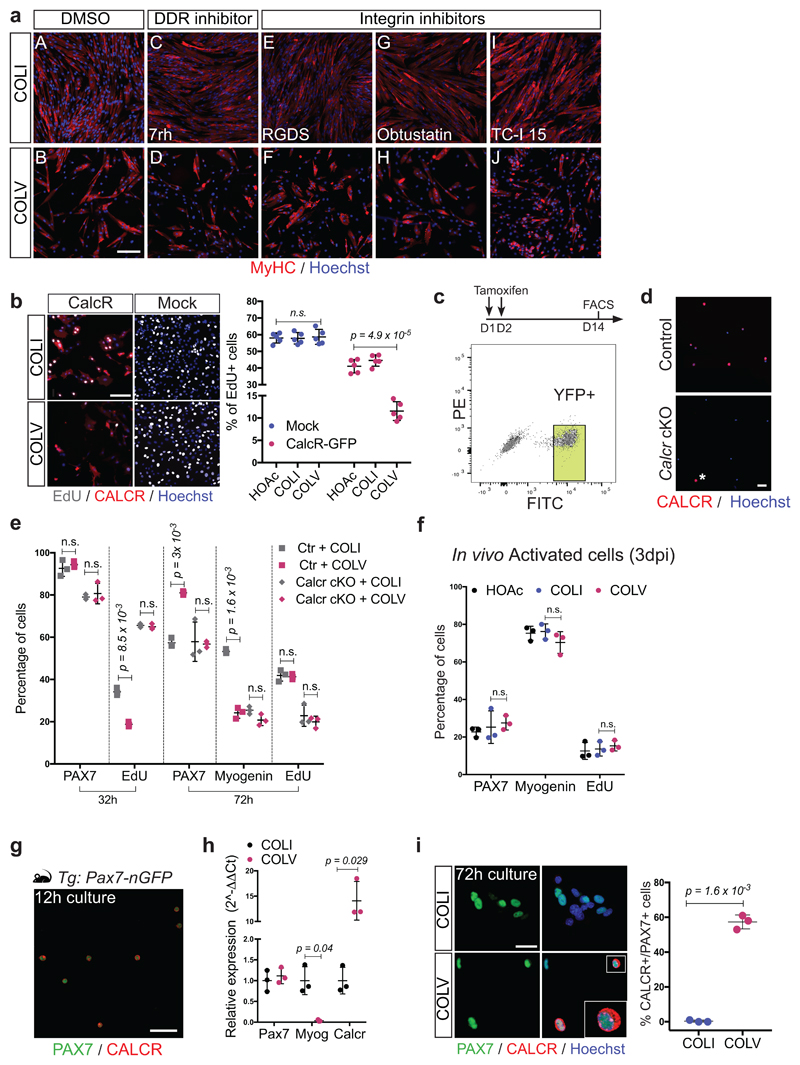
To assess the functional role of COLV, isolated satellite cells were incubated with COLI, COLV, or COLVI in the presence of EdU, and stained for PAX7, that marks muscle stem/progenitor cells, and the muscle commitment (MYOD) and differentiation (Myogenin). Strikingly, only the COLV-complemented medium delayed entry of quiescent cells into the cell cycle (32h, Fig. 2a) and consequently their proliferation and differentiation (72h, Fig. 2b; 10d Extended data Fig. 3a-c). As shown previously4,13, Rbpj-/- cells underwent precocious differentiation, and this was partially antagonized by COLV, consistent with the finding that Col5a1/3 genes are NICD/RBPJ targets (Fig. 2c, d and Extended data Fig. 3d-g). Taken together, these results show that COLV specifically sustains primary muscle cells in a more stem-like PAX7+ state, indicating that it could potentially play a role in the quiescent niche.
Figure 2. Collagen V delays proliferation and differentiation of satellite cells.
(a) EdU pulse (2h) of isolated satellite cells cultured for 32h: COLI (35%), COLVI (34%), COLV (18%); (n=4 mice, ≥250 cells, 2 wells/condition).
(b) Immunostaining of isolated satellite cells cultured for 72h. PAX7: 58%, 55% and 81%; Myogenin: 56%, 57% and 24% for COLI, COLVI and COLV, respectively (n=4 mice, ≥250 cells, 2 wells/condition).
(c) Experimental scheme for satellite cells plated overnight (o/n) before collagen treatment.
(d) Immunostainings of satellite cells incubated with collagens for 60h (n=3 mice, ≥200 cells, 2 wells/condition).
Error bars, mean ± SD; two-sided paired t-test; #p-value: two-sided unpaired t-test. Scale bar: 50μm.
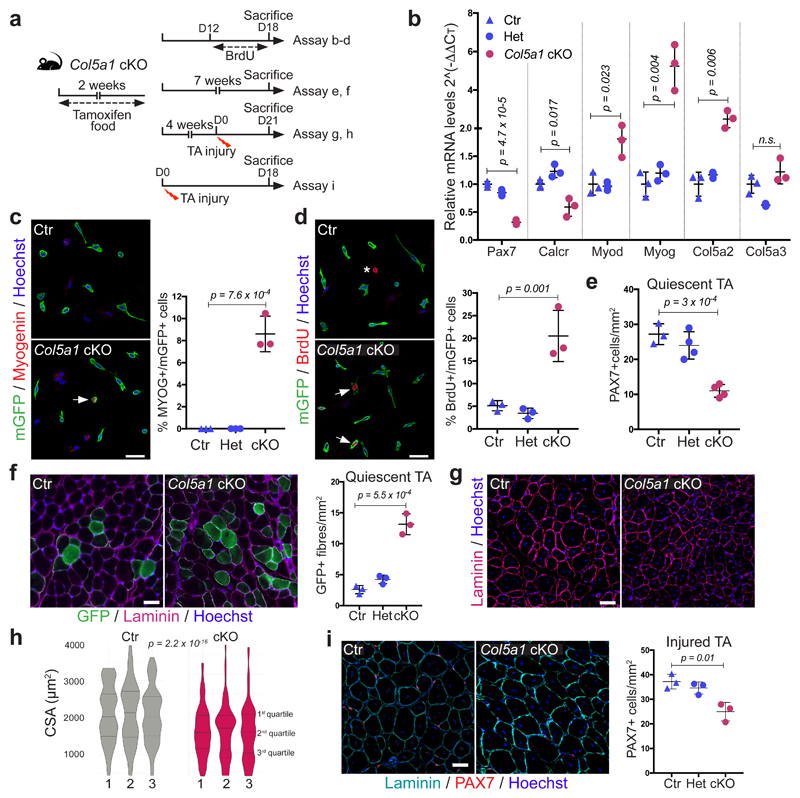
To determine if collagen V produced by satellite cells is a functional component of the niche, we generated compound Tg:Pax7-CreERT2;Col5a1flox/flox;R26mTmG (Col5a1 cKO) mice, in which COLV was depleted and simultaneously lineage-traced in GFP+ satellite cells4,14 (Fig. 3a and Extended data Fig. 4a). As the α1-chain of COLV is present in all COLV isoforms, which are trimeric, Col5a1 deletion produces complete COLV-deficient cells14. Remarkably, given the general stability of collagens, targeted deletion of Col5a1 resulted in upregulation of the differentiation markers Myod and Myog, and a concomitant reduction of the quiescence marker Calcr, as well as Pax7 only 18d after tamoxifen treatment (Fig. 3b). Mutant cells also showed ectopic expression of Myogenin (Fig. 3c), increased BrdU incorporation (Fig. 3d), and a significant decline in PAX7+ satellite cells (Fig. 3e). The Col5a1 cKO cells did not undergo apoptosis (data not shown), but fused to give rise to GFP-marked myofibres (Fig. 3f). Therefore, blocking de novo satellite cell-produced COLV resulted in their spontaneous exit from quiescence and differentiation, a phenotype reminiscent of Notch loss-of-function4,5.
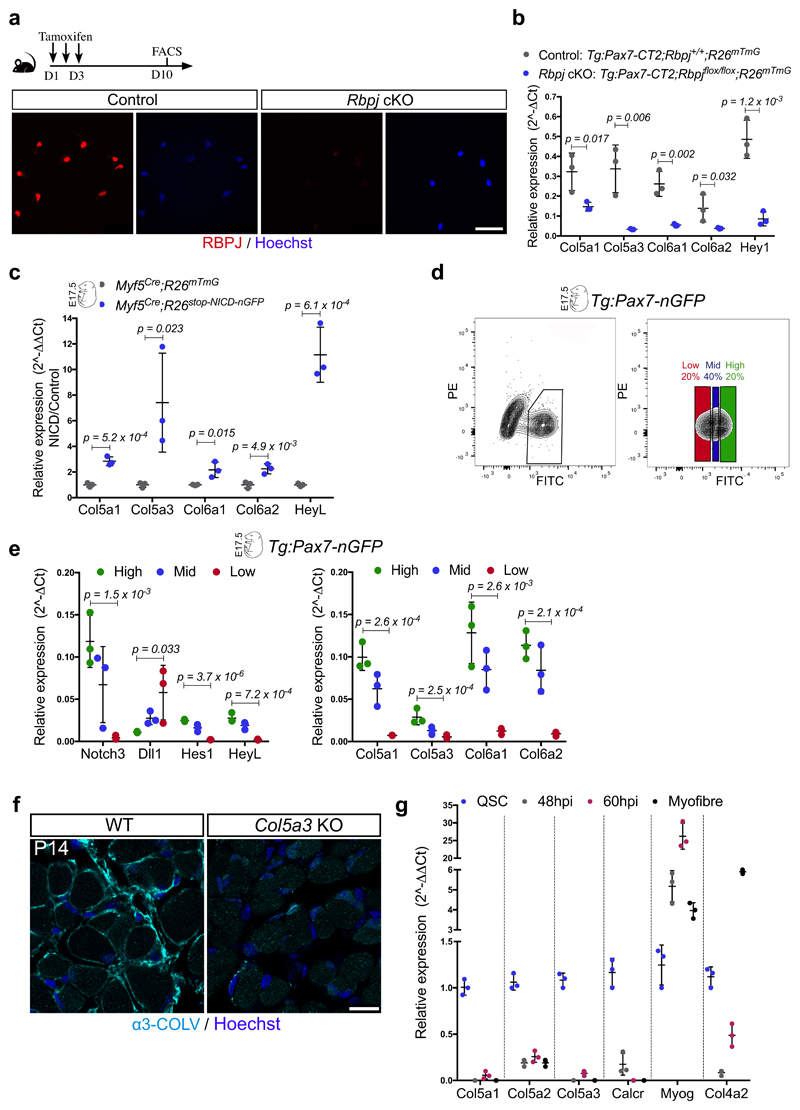
Figure 3. Satellite cell-produced COLV is required in vivo for self-renewal and maintenance of quiescence.
(a) Experimental schemes for Control (Ctr): Tg:Pax7-CT2;Col5a1+/+;R26mTmG, heterozygous (Het): Tg:Pax7-CT2;Col5a1flox/+;R26mTmG and conditional knock-out (cKO): Tg:Pax7-CT2;Col5a1flox/flox;R26mTmG mice.
(b) RT-qPCR of satellite cell (Pax7, Calcr) and differentiation (Myod, Myog) markers on Col5a1 mutant and control satellite cells isolated by FACS from resting muscle (n=3 mice/genotype).
(c) Representative images of membrane-GFP+ satellite cells from total muscle preparations from Control and Col5a1 null mice plated for 12h. Arrow, mGFP+/Myogenin+ cell (n=3 mice/genotype, ≥200 cells).
(d) GFP+ satellite cells from total muscle preparations plated for 12h. Asterisk, non-recombined BrdU+ cell; arrows, GFP+/BrdU+ cells (n=3 mice/genotype, ≥250 cells).
(e) Satellite cell quantification in Control, Het and Col5a1 cKO mice 7 weeks after tamoxifen treatment (n=3 (Ctr) and 4 (Het/cKO) TA muscle/genotype).
(f) Immunostaining of sections from Control and Col5a1 cKO TA muscles 7 weeks following tamoxifen treatment (n=3 mice/genotype).
(g) Immunostaining of sections from Control and Col5a1 cKO TA muscles 21d post-injury (n=3 mice/genotype).
(h) Muscle cross sectional area (CSA) distribution 21d post-injury showed as violin plots was significantly different in Control vs Col5a1 based on Kruskal-Wallis test (n=3 mice/genotype, 1000 fibres analysed/mouse).
(i) Immunostaining of sections 18 days post-cardiotoxin injury of Control and Col5a1 cKO TA muscles (n=3 mice/genotype).
Error bars, mean ± SD; two-sided unpaired t-test. Scale bar: 50μm, c and d; 100μm, f, g and i.
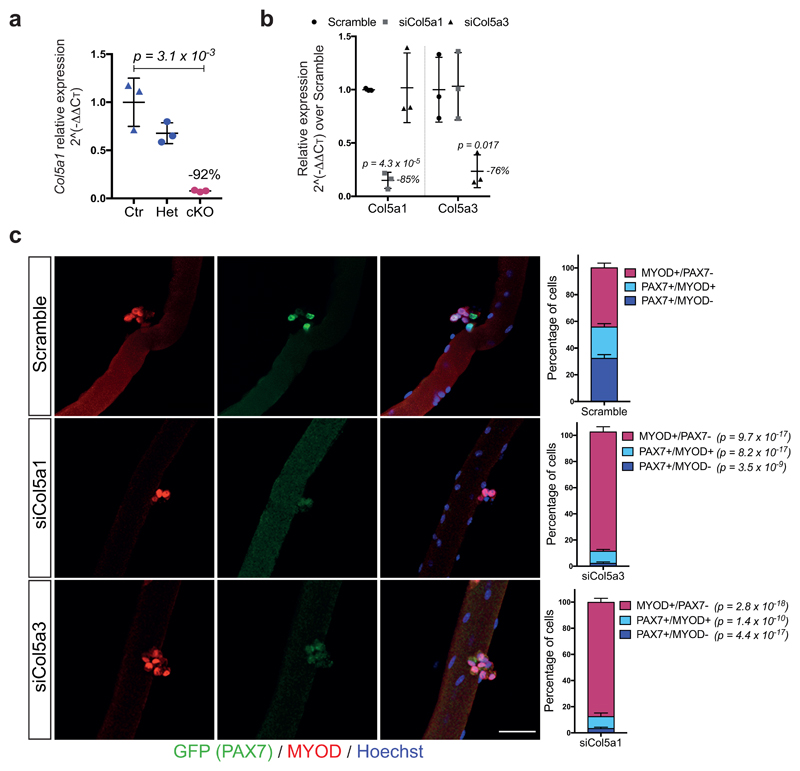
To investigate the role of Col5a1 in regeneration, we examined the morphology of TA muscles of Col5a1 cKO mice, 18d after cardiotoxin-mediated injury (Fig. 3a). Notably, mutant myogenic cells displayed smaller nascent myofibres compared to control (Fig. 3g, h). Unexpectedly, less self-renewing PAX7+ cells were observed in the Col5a1 cKO mice (Fig. 3i), in spite of abundant COLV in regenerating muscle (data not shown), likely produced by the resident fibroblasts, pointing to a cell-autonomous role for Col5a1. To investigate self-renewal in a more tractable system, we targeted COLV using short-interfering RNA (siRNA) on isolated myofibres in culture. Consistent with our in vivo observations, Col5a1 knock-down by siRNAs resulted in a drastic decrease in the number of the self-renewing PAX7+/MYOD– cells, compared to scramble control (Extended data Fig. 4b, c). Of note, siCol5a3 phenocopied siCol5a1, demonstrating that the active triple helix contains α3-COLV (Extended data Fig. 4c).
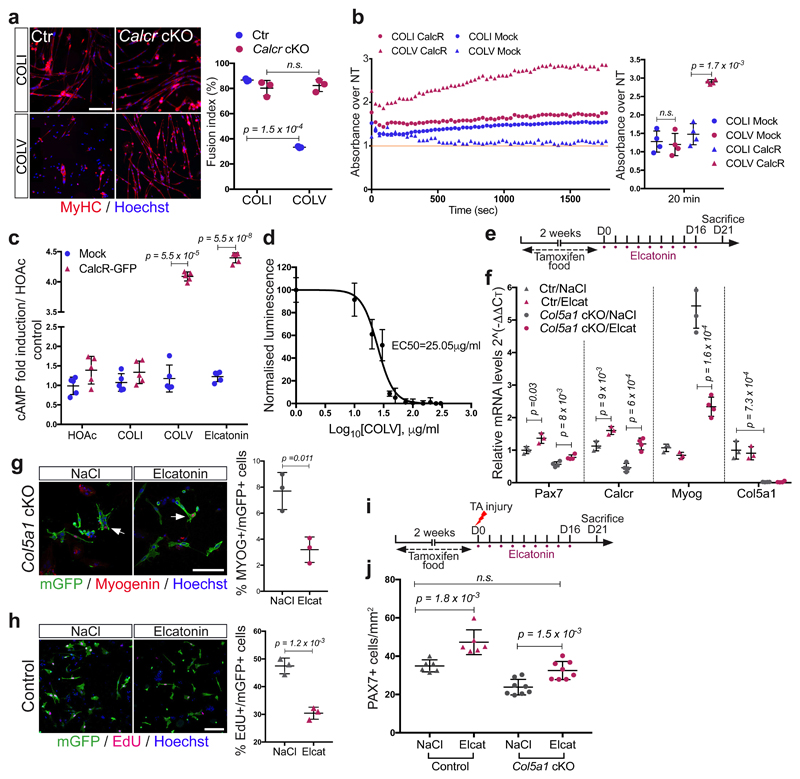
Substrate rigidity and geometry have been demonstrated to control stem cell properties, including differentiation and self-renewal15,16. However, we observed that COLV interacted with myogenic cells only when added in the medium, but not as a coating substrate (data not shown), leading us to speculate that it acted as a signalling molecule rather than a biomechanical modulator. To identify the cell surface receptor of collagen V on satellite cells, we used a myotube-formation assay (see Extended data Fig. 3b), coupled to inhibitors against known collagen receptors, including Integrins and the RTK receptor DDR17,18, but these did not obstruct the anti-myogenic activity of COLV (Extended data Fig. 5a). Since collagens have also been shown to bind G-protein coupled receptors (GPCR)19,20, we focused on Calcitonin receptor, a GPCR critical for maintenance of satellite cells21. Strikingly, only cells that expressed CALCR showed decreased proliferation in the presence of COLV (Extended data Fig. 5b), and Calcr-/- satellite cells isolated from cKO Pax7CreERT2;Calcrflox/flox mice failed to respond to COLV treatment (Fig. 4a and Extended data Fig. 5c-e), demonstrating that CALCR constitutes an essential mediator of the COLV signal (Extended data Fig. 4e). Accordingly, as CALCR is rapidly cleared following satellite cell activation21, COLV had no impact on cultured myogenic cells that had been activated in vivo (3 days post-injury; Extended data Fig. 5f). We note, however, that addition of COLV on freshly isolated satellite cells appeared to stabilise residual CALCR, and retain Calcr gene expression, thus allowing their prolonged interaction (Extended data Fig. 5g-i). In summary, we show that CALCR is a critical mediator of the effect of COLV on maintaining quiescence and stemness properties of satellite cells.
Figure 4. Interaction of COLV with satellite cells is mediated by the Calcitonin Receptor.
(a) Control (Pax7CT2/+; Calcr+/+; R26stop-YFP) and Calcr-deficient (Pax7CT2/+; Calcrflox/flox; R26stop-YFP) satellite cells incubated 10d with COLI or COLV and immunostained for differentiation (n=3 mice, ≥250 cells).
(b) Binding assay of COLV and CALCR by colorimetric on-cell ELISA based on the measurements of HRP absorbance. Runs test p-value <0.0001. Results presented as ratio of absorbance over non-treated cells (NT, orange line=1) at 20 min of HRP development.
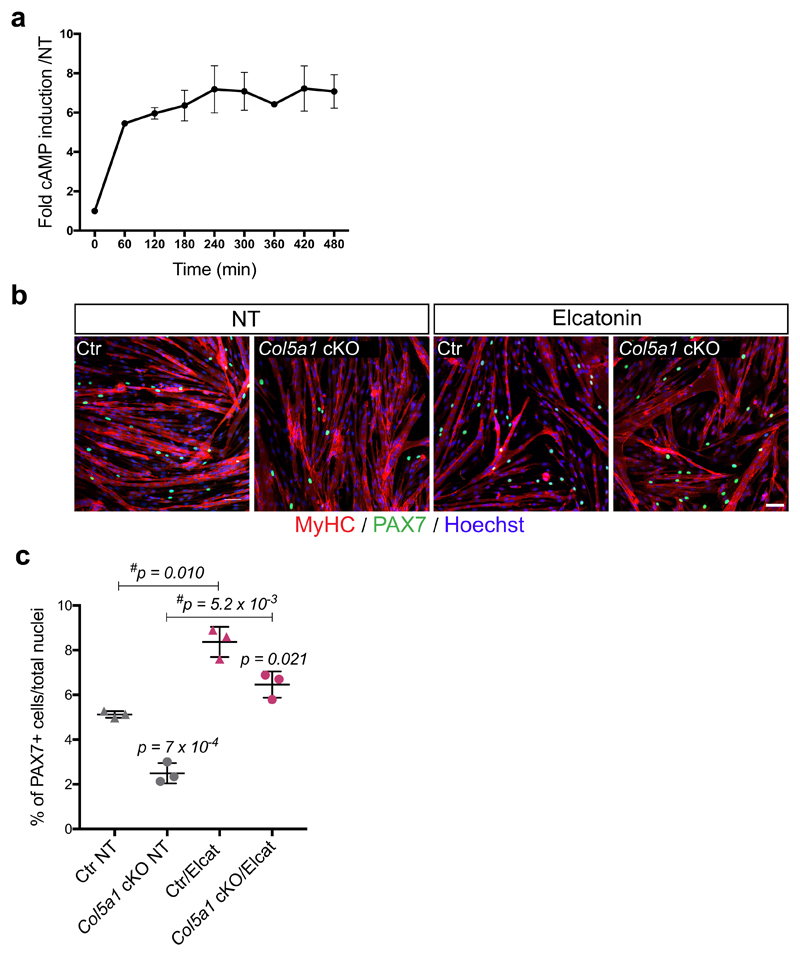
(c) cAMP measurements of CalcR-transduced C2C12 cells after 3h treatment with HOAc, COLI, COLV or Elcatonin. Graph represents fold cAMP induction over average of Mock cells treated with HOAc (n=4 independent assays).
(d) Dose-response: fold cAMP concentration in CalcR-transduced C2C12 cells treated for 3h with increasing concentrations of COLV. EC50=25.05μg/ml (n=4 independent assays).
(e) Experimental scheme of tamoxifen and Elcatonin administration to Col5a1 cKO and their corresponding control mice.
(f) RT-qPCR of satellite cells (Pax7, Calcr) and differentiation (Myog) markers on Elcatonin-treated Col5a1 cKO mutants and controls (n=3 mice/condition).
(g) Representative images of membrane-GFP+ satellite cells from total muscle preparations from Col5a1 null mice injected with saline or Elcatonin, plated for 12h. Arrows, mGFP+/MYOG+ cells (n=3 mice/condition, ≥200 cells).
(h) EdU (2h) and membrane-GFP staining of satellite cells from total muscle preparations from control mice treated with saline or Elcatonin, plated for 36h. Asterisk, mGFP-/EdU+ cell (n=3 mice/genotype, ≥400 cells).
(i) Experimental scheme of tamoxifen and Elcatonin administration to Control and Col5a1 cKO mice.
(j) PAX7+ cells on TA sections 21d post-injury in mice treated with saline or Elcatonin; (n=6 for Ctr and 8 mice for cKO mice/treatment).
Error bars, mean ± SD; (a-c) two-sided paired t-test; (f-j) two-sided unpaired t-test. Scale bar: 25μm.
To date, it has been assumed that CALCR in satellite cells is activated by circulating calcitonin peptide hormones, principally expressed by the parafollicular thyroid cells, pointing to systemic regulation of stem cell quiescence. Based on our findings, we reasoned that COLV serves as a local ligand for the CALCR receptor. Indeed, on-cell ELISA experiments showed that COLV, but not COLI, selectively bound to cells expressing CALCR (Fig. 4b). Significantly, this binding was functional with COLV, but not COLI, displaying rapid activation kinetics and upregulation of intracellular cAMP levels, a downstream reporter of CALCR activation22 (Fig. 4c, d and Extended data Fig. 6a). In vitro binding assays using the extracellular domain of CALCR did not result in robust interaction with COLV (data not shown). Therefore, we propose that the COLV/CALCR binding requires a specific configuration of the receptor, possibly involving the extracellular loops or co-factors. Taken together, these data demonstrate that COLV physically and functionally interacts with CALCR.
In this study, we showed that blocking COLV production from satellite cells resulted in rupture of quiescence and impaired self-renewal in vivo. The similarity of these phenotypes to Notch and Calcitonin receptor signalling abrogation, together with our ex vivo results, points to a cell-autonomous Notch/COLV/CALCR axis that sustains muscle stem cells in their niche. Consistent with this notion, administration of the CALCR ligand Elcatonin to control and Col5a1-null mice resulted in upregulation of the stem cell markers Pax7 and Calcr, indicating that the injected ligand was readily delivered to the quiescent satellite cells (Fig. 4e, f). Strikingly, Elcatonin mitigated the precocious Myogenin transcript and protein expression levels in Col5a1 mutant cells (Fig. 4f, g). Of interest, Elcatonin also prolonged the G0-to-S transition of control satellite cells exiting quiescence (Fig. 4h), suggesting that hyperactivation of CALCR could drive cells into a deeper, more dormant-like quiescent state, marked by higher Pax7 expression23. Therefore, CALCR activity appears to control quiescence quantitatively (loss of satellite cells in the absence of ligand COLV) and qualitatively (dormant-like satellite cells upon hyperactivation). Importantly, Elcatonin restored the number of PAX7+ satellite cells in regenerating Col5a1 cKO muscles to wild type levels (Fig. 4i, j), and in an ex vivo self-renewal reserve-cell model (Extended data Fig. 6b, c). Therefore, we show that endogenous calcitonin levels are not sufficient to maintain Col5a1 null cells, and that exogenous administration of a calcitonin derivative rescued the defects, likely via the activation of CALCR.
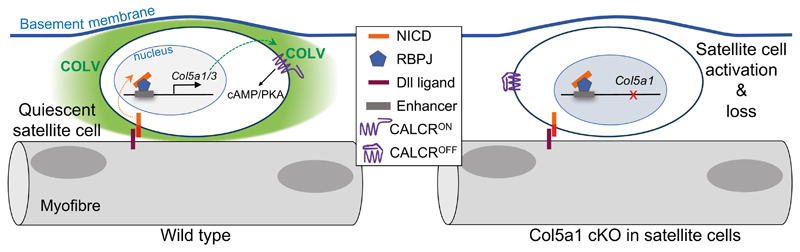
In this report, we describe a self-sustained signalling cascade, orchestrated by the Notch pathway and propagated by the ECM of the immediate skeletal muscle stem cell niche (Extended data Fig. 7). We propose that Notch acts as a sensor of the homeostatic environment, by reinforcing the niche with active collagen V that signals cell-autonomously and maintains stem cell quiescence. Upon disruption of the niche and physical separation of the ligands, Notch signalling is sharply downregulated and stem cells exit quiescence4,24. Based on our model, this halts further production of collagen V, thus favouring satellite cell activation (Extended data Fig. 7). It would be of interest to extend the novel Notch/COLV/CALCR signalling cascade described here to stem cells in other tissues and organisms where an extracellular matrix protein produced by the stem cell can act as a local ligand for cell-autonomous stability of the niche through a GPCR. The regulatory mechanism that we identify provides a framework to construct a more complete view of the stem cell niche, and to manipulate stem cell behaviour in a therapeutic context.
Methods
Mouse strains
Mouse lines used in this study have been described and kindly provided by the corresponding laboratories: Myf5Cre (27), Pax7CreERT2 (28) (used to recombine R26stop-NICD allele), R26stop-NICD-nGFP (29), R26mTmG (30) (membrane-Tomato floxed / membrane-GFP), Rbpjflox/flox (31), Pax7CT2/+; Calcrflox/flox; R26stop-YFP/stop-YFP (32) (triple mutant mice provided by Dr. Fukada) and Col5a1flox/flox (33). Tg:Pax7-CreERT2 (used to recombine Rbpj and Col5a1) and Tg:Pax7-nGFP lines were described previously34,35. All adult mice analysed were between 8 and 12 weeks old. Animals were handled according to national and European community guidelines, and protocols were approved by the ethics committee at Institut Pasteur.
Muscle injury, tamoxifen, BrdU and Elcatonin administration
For muscle injury, Tg:Pax7-CreERT2;Col5a1flox;R26mTmG mice were anesthetized with 0.5% Imalgene/2% Rompun and the Tibialis anterior (TA) muscle was injected with 50μl of Cardiotoxin (10µM; Latoxan). Tg:Pax7-CreERT2;Rbpjflox; R26mTmG mice were injected intraperitoneally with tamoxifen three times (250 to 300µl, 20mg/ml; Sigma T5648; diluted in sunflower seed oil/5% ethanol). Pax7CreERT2;Calcrflox;R26stop-YFP were injected intraperitoneally with tamoxifen twice (5mg/25g mouse) and sacrificed 2 weeks later. Pax7CreERT2;R26stop-NICD-ires-nGFP and Tg:Pax7-CreERT2;Col5a1flox; R26mTmG were fed tamoxifen containing diet for one and two weeks, respectively (Envigo, TD55125). Six days prior sacrifice Tg:Pax7-CreERT2;Col5a1flox; R26mTmG mice were given the thymidine analogue 5-Bromo-2’-deoxyuridine (BrdU, 0.5mg/ml, #B5002; Sigma) in the drinking water supplemented with sucrose (25mg/ml). Elcatonin (2.5ng/g mouse final concentration in 0.9% NaCl; Mybiosource, MBS143228) was injected subcutaneously 8 times every other day. Comparisons were done between age-matched littermates using 8-12 week-old mice.
Muscle enzymatic dissociation and stem cell isolation
Adult and foetal limb muscles were dissected, minced and incubated with a mix of Dispase II (Roche, 04942078001) 3U/ml, Collagenase A (Roche, 11088793001) 100ug/ml and DNase I (Roche, 11284932001) 10mg/ml in Hank’s Balanced Salt Solution (HBSS, Gibco) supplemented with 1% Penicillin-Streptomycin (PS; Gibco) at 37°C at 60rpm in a shaking water bath for 2h. The muscle suspension was successively filtered through 100µm and 70µm cell strainers (Milteny, 130-098-463 and 130-098-462) and then spun at 50g for 10min/4°C to remove large tissue fragments. The supernatant was collected and washed twice by centrifugation at 600g for 15min/4°C. Prior to FACS, the final pellet was resuspended in cold DMEM/1%PS supplemented with 2% FBS and the cell suspension was filtered through a 40µm strainer. Satellite cells were sorted with Aria III (BD Biosciences) using either the GFP (Tg:Pax7-nGFP or Tg:Pax-CreERT2;Rbpjflox;R26mTmG, or Tg:Pax7-CreERT2;Col5a1flox; R26mTmG) or the YFP (Pax7CT2; Calcrflox;R26stop-YFP) cell markers. Isolated, mononuclear cells were collected in DMEM/1%PS/2%FBS. Enzymatically dissociated muscle was also plated directly without FACS on Matrigel coated dishes (Corning, 354248; 30min at 37°C), and fixed 12h later with 4% paraformaldehyde (PFA)/PBS. Cells were immunostained following the protocol described above.
Chromatin immunoprecipitation
Cultured myoblasts
Satellite cells were isolated from adult Tg:Pax7-nGFP mice and plated on Delta-like1 coated dishes for 72h to maintain active Notch signalling, as described previously35,36. Cells were then processed for ChIP using a dual cross-linking protocol37, slightly modified. Briefly, cells were fixed on the dish with 2mM Di(N-succinimidyl) glutarate (Sigma, 80424) in PBS for 45min at RT. After two washes with PBS, cells were re-fixed with 1% formaldehyde/PBS for 10min at RT, before quenching the reaction with 1/20 volume of 2.5M Glycine for 5min at RT. The cells were then collected with a cell scraper in PBS supplemented with 1%BSA and protease inhibitors (Roche, 11697498001), and collected by spinning. Cell lysis and chromatin isolation were done using the Ideal ChIP-seq kit for histones (Diagenode, C01010051). Chromatin was sheared using a Bioruptor Pico (Diagenode B01060001) with 10 cycles of 30s on/off sonication. The samples were prepared in triplicates from different plates. 2x106 primary myogenic cells were used per ChIP and 2x104 per Input. The immunoprecipitations were performed following the manufacturer's guidelines using 6μl of anti-Rbpj antibody (Cell Signalling, #5313) or 1.5μl of rabbit control IgG antibody (Diagenode, C15410206) in a final volume of 300μl per ChIP. The purification of the immunoprecipitated DNA was performed using DiaPure columns (Diagenode, C03040001). RT-qPCR was performed using FastStart Universal SYBR Green Master mix (Roche, 04913914001) and analysis was performed using the 2-∆∆CT method38 normalised to the Neg16 region.
Quiescent satellite cells
Satellite cells were isolated from adult Tg:Pax7-nGFP mice using in situ fixation to preserve Notch signalling from dissociation-induced downregulation39. Cells were fixed as above in (2mM Di(N-succinimidyl) glutarate for 45min, followed by 10min with 1% formaldehyde at RT). Cell lysis and chromatin isolation were performed using Auto-TrueMicrochip kit (Diagenode, C01010140). Chromatin was sheared as above with 10 cycles of 30s on/off sonication using a Bioruptor Pico. 2x105 cells were used per ChIP and 2x103 per Input and IPs were performed using 2μl of anti-Rbpj antibody (Cell Signalling, 5313) or 0.5μl of rabbit control IgG antibody following the manufacturer guidelines. Immunoprecipitated chromatin preparations and input were purified using the Auto IPure kit v2 (Diagenode). RT-qPCR was performed using FastStart Universal SYBR Green Master mix (Roche, 04913914001) and analysis was performed using the 2-∆∆CT method38 normalised to the Neg16 region. Primers used for ChIP-qPCR are listed in Supplementary Table 1.
Cell culture and Collagen incubation
Satellite cells isolated by FACS were plated at 3x103 cells/cm2 on ibi-Treated μ-slides (Ibidi, 80826) pre-coated with 0.1% gelatin for 2h at 37°C. Cells were cultured in satellite cell growth medium (GM) containing Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco) supplemented with F12 (50:50; Gibco), 1% PS, 20% foetal bovine serum (FBS; Gibco) and 2% Ultroser (Pall; 15950-017) at 37°C, 3% O2, 5% CO2 for the indicated time. Twelve hours after plating, collagens (COLI rat tail, BD Biosciences, 354236; COLV human placenta, Sigma, C3657; COLVI human placenta, AbD Serotec 2150-0230) resuspended in HOAc acid at 1mg/ml, were added to the culture medium at a final concentration of 50μg/ml and cells were fixed with 4% PFA for 10min at RT. To assess proliferation, cells were pulsed with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU), 1x10-6M 2h prior to fixation (ThermoFisher Click-iT Plus EdU kit, C10640). Inhibitors used: Obtustatin (Integrin α1β1, Tocris, 4664, 100nM), TC-I 15 (Integrin α2β1 Tocris, 4527, 100μM), RGDS peptide (all integrins, Tocris, 3498, 100μM), 7rh40 (DDR1, kind gift from Dr. Ke Ding, 20nM).
Muscle fixation and histological analysis
Embryo forelimbs were fixed in 4% PFA/0.1% Triton for 2h, washed overnight with 1X PBS, immersed in 20% sucrose/PBS overnight, embedded in OCT, frozen in liquid nitrogen and sectioned transversely at 12-14μm. Isolated TA muscles were immediately frozen in liquid-nitrogen cooled isopentane and sectioned transversely at 8μm. For Pax7 staining on adult TA muscle, sections were post-fixed with 4%PFA, 15min at RT. After 3 washes with 1XPBS, antigen retrieval was performed by incubating sections in boiling 10mM citrate buffer pH6 for 10min. Sections were then blocked, permeabilised and incubated with primary and secondary antibodies as described in the “Immunostaining on cells, sections and myofibres” section.
Single Myofibre isolation and siRNA transfection
Single myofibres were isolated from Extensor Digitorum Longus (EDL) muscles following the previously described protocol41. Briefly, EDL muscles were dissected and incubated in 0.1% w/v collagenase (Sigma, C0130)/DMEM for 1h in a 37°C shaking water bath at 40rpm. Following enzymatic digestion, mechanical dissociation was performed to release individual myofibres that were then transferred to serum-coated petri dishes. Single myofibres were transfected with siCol5a1, siCol5a3 (Dharmacon SMARTpool Col5a1 (12831) L-044167-01 and Col5a3 (53867) L-048934-01-0005) or scramble siRNA (Dharmacon ON-TARGETplus Non-targeting siRNA #2 D-001810-02-05) at a final concentration of 200nM, using Lipofectamine 2000 (ThermoFisher, 11668) in Opti-MEM (Gibco). Four hours after transfection, 6 volumes of fresh satellite cell growth medium was added and fibres were cultured for 72h at 37°C, 3%O2. Myofibres were fixed for 15min in 4% PFA prior to immunostaining for proliferation, differentiation and self-renewal markers42.
Immunostaining on cells, sections and myofibres
Following fixation, cells and myofibres were washed three times with PBS, then permeabilised and blocked at the same time in buffer containing 0.25% Triton X-100 (Sigma), 10% goat serum (GS; Gibco) for 30min at RT. For BrdU immunostaining, cells were unmasked with DNaseI (1,000 U/ml, Roche, 04536282001) for 30 min at 37°C. Cells and fibres were then incubated with primary antibodies (Supplementary Table 2) for 4h at room temperature (RT). Samples were washed with 1X PBS three times and incubated with Alexa-conjugated secondary antibodies (Life Technologies, 1/1000) and Hoechst 33342 (Life Technologies, 1/5000) for 45min at RT. EdU staining was chemically revealed using the Click-iT Plus kit according to manufacturer’s recommendations (Life Technologies, C10640). For collagen staining, the myofibres and the muscle sections were incubated with 0.1% Triton X-100 for 30min at RT. Myofibres and sections were then washed 3 x 10min and incubated with 10% GS in PBS for 30min. After one wash, samples were incubated with primary antibodies and secondary antibodies as described in Supplementary Table 2. Confocal images were acquired with a Leica SPE microscope and Leica Application Suite or with Zeiss LSM 700 microscope and Zen Blue 2.0 software. 3D images were reconstructed from confocal Z-stacks using Imaris software. The Section view function was used to inspect the environment of the satellite cells by showing the cut in the x-, y-, and z-axes.
Reserve cell cultures
Enzymatically dissociated muscles were plated in gelatin-coated dishes (1/30 of total mouse muscle/cm2) in the satellite cell growth medium described above. When myotube formation was detected (day 7 to 10), recombination was induced by addition of 4-hydroxytamoxifen (4-OHT; Sigma, H6278) at final concentration of 1μM every other day. Seven days later, 4-OHT-containing medium was replaced every other day with fresh medium containing Elcatonin (0.1U/ml), for an additional 10 days. To assess proliferation, cells were pulsed with 1x10-6M EdU for 6h prior to fixation (10min, 4% PFA). Reserve cells were localised by immunofluorescence as PAX7+/EdU- 42. For each medium change, only half of the conditioned medium was removed and replaced by an equal volume of fresh medium.
Construction of luciferase reporters and luciferase assays
For the generation of luciferase reporters, candidate enhancers of Col5a1, Col5a3, Col6a1/2 (shared enhancer) and Hey1 were amplified by PCR from genomic DNA of C2C12 cells. The enhancers were then cloned into the firefly-luciferase pGL3-Basic vector (Promega, E1751) upstream of a minimal thymidine kinase promoter (minTK). The sequences of enhancers are listed in Supplementary Table 3. Transfected cells (Lipofectamine LTX, Life technologies, 15338030) were lysed and luciferase signal was scored using the Dual-Luciferase Reporter Assay System (Promega, E1910). For normalization, Renilla luciferase (pCMV-Renilla) was transfected at 1:20 ratio relative to firefly-luciferase constructs.
RNA isolation and Quantitative RT-PCR
Total RNA was extracted from satellite cells isolated by FACS using QIAGEN mini RNeasy kit and reverse transcribed using SuperScript III (Invitrogen, 18080093) according to manufacturer's instructions. RT-qPCR was performed using FastStart Universal SYBR Green Master mix (Roche, 04913914001) and analysis was performed employing the 2-∆∆CT method and using the average of the control values as a reference38. Specific forward and reverse primers used in this study are listed in Supplementary Table 1.
Stable cell line manipulations
Murine myoblast cell line C2C12 was cultured in DMEM/ 20% FBS/ 1% PS at 37°C, 5% CO2. Notch activation: Notch activation was achieved by plating cells on Dll1-coated dishes or by doxycycline inducible Notch constructs, as described previously12. Calcr retrovirus preparation and transduction: Calcitonin receptor C1a-type (pMXs-Calcr-C1a-IRES-GFP) and mock control (pMXs-IRES-GFP) retrovirus vectors were prepared as described previously32,43. Briefly, 48h after transfection of Platinum-E cells the supernatant was recovered and used to transduce C2C12. Two days later stably labelled GFP+ C2C12 cells were isolated by FACS. All stable cell lines used in this study are negative for mycoplasma contamination.
Quantification of cAMP
Transduced mock (IRES-GFP) and Calcr (CalcR-C1a-IRES-GFP) C2C12 cells were isolated by FACS based on GFP expression and seeded on 0.1% gelatin-coated, white culture 96-well plates (Falcon, 353296) at 3x103 cells/well. After overnight culture, the cells were incubated with the complete induction medium containing DMEM/1%PS/500μM IBMX (isobutyl-1-methylxanthine; Sigma, 17018)/100µM Ro 20-1724 ([4-(3-butoxy-4-methoxy-benzyl) imidazolidone]); Sigma, B8279)/MgCl2 40mM, collagen, solvant HOAc or Elcatonin (0.1U/ml) for 3h. The amount of intracellular cAMP was measured using cAMP-Glo Max Assay (Promega, V1681) following the manufacturer’s protocol. Luminescence was quantified with FLUOstar OPTIMA (BMG Labtech). EC50 value was determined with GraphPad Prism software using a sigmoid dose-response curve (variable slope).
Biotinylation of Collagens
Commercial collagen proteins (COLI rat tail, BD Biosciences, 354236; COLV human placenta, Sigma, C3657) were biotinylated using the Pierce EZ-Link Biotinylation Kit, with slight modifications. Briefly, 20µl of 1M Hepes was added to 0.5ml of 1mg/ml collagen dissolved in 0.5M HOAc. Then, 20µl of 100mM biotin reagent were added and incubated at room temperature for 1.5h. Biotinylated collagens were next dialyzed in 25mM HEPES, 2.5M CaCl2, 125mM NaCl, 0.005% Tween (Slide-A-Lyze MINI Dialysis Device, ThermoFisher 88401) overnight at 4°C.
On-cell Enzyme-Linked Immunosorbent Assay (ELISA)
Transduced mock and Calcr C2C12 were seeded on a clear bottom 96-well plate (TPP, 92096) at 3x103 cells/well density. After overnight culture, cells were treated with 50μg/ml of biotinylated collagens for 2h and fixed with 4%PFA/PBS for 15min. After 3x PBS washes, cells were blocked with a solution containing 10% GS, 2% BSA, PBS for 1h at RT, washed and incubated 1h/RT with goat anti-mouse biotin-HRP antibody (Jackson, 1/1000e, 115-035-003). After 3x PBS washes, the HRP signal was developed by addition of 3,3’,5,5’ tetramethylbenzidine (1-Step Ultra TMB-ELISA, Sigma, 34028). HRP substrate and absorbance at 650nm was measured once every 30sec for 30min with FLUOstar OPTIMA (BMG Labtech). The signal was normalized to the background signal (no secondary antibody) and to the number of cells assessed by Janus green staining (Abcam, ab111622).
Statistical analysis
No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. No animal has been excluded from analysis and no randomization method has been applied in this study. For comparison between two groups, two-tailed paired and unpaired Student’s t test were performed to calculate p values and to determine statistically significant differences (see Figure legends). Additional specific statistical tests are detailed in Figure legends. All experiments have been done twice with the same results. All statistical analyses were performed with Excel software or GraphPad Prism software; Kruskal-Wallis test was performed in R.
Extended Data
Extended data Figure 1. Identification of NICD/RBPJ-bound enhancers and response to activation of Notch signalling.
(a) Gene expression microarray data show that satellite cells express a specific subset of collagen types, which include the fibrillar COLI (Col1a1 and Col1a2), COLIII (Col3a1, possibly as [α1(III)]3 homodimer) and COLV (Col5a1, Col5a2 and Col5a3) and the non-fibrillar COLIV (Col4a1 and Col4a2), COLVI (Col6a1 and Col6a2) and COLXV (Col15a1, possibly as [α1(XV)]3 homodimer). Data are shown as a heatmap of normalized collagen transcripts expressed at different developmental time points (E12.5, E17.5, P08; Tg-Pax7-nGFP, GEO accession number GSE52192), quiescent and post-injury (t=60h post-BaCl2 injury).
(b) ChIP-seq tracks indicating RBPJ/NICD-occupied enhancers, associated to mouse Collagen-5a1, -5a3, -6a1 and -6a2 loci. H3K4me1, and H3K27ac, p300, and NICD are shown. Orange rectangles indicate RBPJ binding positions and asterisks the enhancers used for transcriptional activity assays for Extended data Fig. 1c.
(c) Core sequences of the selected RBPJ/NICD-bound enhancers (asterisked orange rectangle in Fig. 1a and Extended data Fig. 1b). The RBPJ consensus binding motif is highlighted in yellow.
(d) Transcriptional response of isolated enhancers to activation of Notch signalling in C2C12 cells. Firefly luciferase signal was measured in cells with doxycycline-inducible expressed hNotch1-GFP (NICD) and GFP-control cells treated with DAPT and were normalized to internal control (pCMV-Renilla). Data are expressed as Relative Luminescence Units (n=3 independent experiments). Error bars, mean ± SD; Two-sided paired t-test.
(e) RNA-seq based expression measurements of collagen genes in myogenic C2C12 cells, with active (DLL1-treated) or inhibited (DAPT-treated) Notch signalling for 6 or 24h (data available at GEO, no. GSE37184). Data are shown as DLL1/DAPT ratios of average RPKMs. Genes with low expression (RPKM <2) were eliminated. HeyL and Hey1 transcripts indicate Notch pathway activation. Red line designates no change (ratio=1). Abbreviation: RPKM= Reads Per Kilobase of exon model per Million mapped reads.
Extended data Figure 2. Notch signalling regulates Col5 and Col6 expression in vivo.
(a) Satellite cells isolated by FACS at day 10 post-tamoxifen injections from resting TA muscle from control (Tg:Pax7-CT2; Rbpj+/-; R26mTmG/+) and Rbpj null (Tg:Pax7-CT2; Rbpjflox/-; R26mTmG/+) mice immunostained for RBPJ.
(b) RT-qPCR of collagen genes in Rbpj conditional KO (cKO) and control satellite cells. Hey1 used as control for Notch signalling (n=3 mice/genotype).
(c) Induction of collagen genes in E17.5 control (Myf5Cre/+; R26mTmG/+) and Myf5Cre-NICD (Myf5Cre/+; R26stop-NICD-nGFP/+) cells isolated by FACS (RT-qPCR normalized to Gapdh, n=3 foetuses/genotype). HeyL reports Notch activity.
(d) FACS plots showing fractionation of GFP+ cells from E17.5 Tg:Pax7-nGFP foetuses into Pax7Hi (20% of population), Pax7Mid (40%), and Pax7Lo (20%). Intensity of GFP signal reflects activity of Pax7 promoter.
(e) Transcript levels of GFP+ cells isolated by FACS show a tight correlation between lineage progression, Notch signalling activity and collagen gene expression (n=3 foetuses/genotype).
(f) Specificity of α3-COLV antibody assessed by immunostaining of Tibialis anterior transverse section of WT and Col5a3 KO P14 postnatal pups (n=3 mice/genotype).
(g) Time course of gene expression performed by RT-qPCR on freshly isolated satellite cells (Quiescent), 48h or 60h after cardiotoxin injury of TA muscle (48hpi, 60hpi), and isolated single myofibres from EDL muscle of Tg:Pax7-nGFP mice. Col5a1/a3 were strongly downregulated in activated/differentiated cells. Quiescence (Pax7, Calcr) and differentiation (Myogenin) markers are indicated. Col4a2, a major component of the basement membrane, is expressed mainly by myofibres (n=3 mice /condition).
Error bars, mean ± SD; one-sided unpaired t-test. Scale bar: 50µm.
Extended data Figure 3. Collagen V delays proliferation and differentiation of satellite cells.
(a) Experimental scheme: isolated Tg:Pax7-nGFP satellite cells cultured overnight (o/n) before collagen treatment.
(b) Myosin heavy chain (MyHC)/EdU staining of satellite cells treated with COLI or COLV. Fusion index: 82%, 86%, and 33% for HOAc solvent, COLI, and COLV, respectively (n=3 mice, ≥250 cells, 2 wells/condition).
(c) Percentage of EdU+ primary myogenic cells after 10 days of culture with indicated collagens. EdU: 2.6%, 1.3% and 18.2% for COLI, COLVI and COLV respectively (n=3 mice, ≥250 cells, 2 wells/condition).
(d) Experimental scheme for Control and conditional knock-out mice. Satellite cells were plated overnight (o/n) before collagen treatment.
(e) GFP/Myogenin immunostaining of Control (Ctr) and Rbpj cKO satellite cells (n=3 mice/condition) incubated 60h in presence of COLI or COLV (n=3 mice, ≥200 cells, 2 wells/condition).
(f) Percentage of EdU+ cells (2h pulse) of Rbpj null primary myogenic cells, after 10 days of culture with HOAc or indicated collagens. EdU: 1.0 % and 7.6% for COLI and COLV respectively (n=3 mice, ≥150 cells, 2 wells/condition).
(g) RT-qPCR on GFP+ Rbpj null satellite cells isolated by FACS and cultured for 72h in the presence of COLI or COLV. Results are normalized to Tbp.
Error bars, mean ± SD; two-sided paired t-test; #p-value: two-sided unpaired t-test. Scale bar: 50μm.
Extended data Figure 4. COLV, and specifically the [α1(V)α2(V)α3(V)] isoform, is critical for satellite cell self-renewal.
(a) RT-qPCR of Col5a1 in Control (Ctr): Tg:Pax7-CT2; Col5a1+/+; R26mTmG, heterozygous (Het): Tg:Pax7-CT2; Col5a1flox/+; R26mTmG and conditional knock-out (cKO): Tg:Pax7-CT2; Col5a1flox/flox; R26mTmG mice 2 weeks after tamoxifen diet (n=3 mice/genotype).
(b) Transcript levels of the different Col5 mRNA chains in C2C12 after transfection of either control scramble, siCol5a1 or siCol5a3 showing the specificity of each siRNA for its given targeted mRNA. Data are normalized to Tbp gene expression (n=3 independent assays).
(c) siCol5a1 and siCol5a3 transfection of Tg:Pax7-nGFP isolated single myofibres cultured for 72h and immunostained for GFP and MYOD. Resident satellite cells enter the myogenic program and form clusters composed of proliferating (PAX7+/MYOD+/MYOG-), differentiated (PAX7-/ MYOG+) and self-renewed (PAX7+/ MYOD-) cells within 72h. Quantification of PAX7+/ MYOD-, PAX7+/ MYOD+ and PAX7-/ MYOD+ populations 72h after transfection. Scramble siRNA was used as negative control (n≥15 fibres counted from 3 mice).
Error bars, mean ± SD; (a) two-sided unpaired t-test; (b, c) two-sided paired t-test. Scale bar: 50µm.
Extended data Figure 5. Screening for COLV receptor candidates identifies CALCR.
(a) Screening for the COLV receptor: satellite cells from Tg:Pax7-nGFP mice were incubated for 10 days with COLV and candidate receptors were targeted with respective inhibitors: 7rh for DDR1 (C, D), the broad-spectrum integrin-binding competitor RGDS peptide (E, F), Obtustatin for integrin α1β1 (G, H), TC-I 15 for integrin α2β1 (I, J). DMSO solvent was used as a control for TC-I 15 and 7rh (A, B). Satellite cell differentiation was assayed by MyHC immunostaining.
(b) EdU (2h pulse) and CALCR staining of GFP+ C2C12 cells isolated by FACS and transduced with CalcR-GFP or Mock-GFP retrovirus and cultured for 24h with COLI (top) or COLV (bottom). Quantification of EdU positive cells of CalcR-C2C12 or Mock GFP cells treated for 24h with COLV or controls COLI and HOAc (n=5 independent experiments, ≥250 cells counted, 2 wells/condition). There was no significant difference between HOAc and COLI treated samples (data not shown).
(c) Experimental scheme of tamoxifen administration to Control (Ctr) (Calcr+/+) and cKO (Calcrflox/flox) mice. FACS plot of satellite cells from Pax7CreERT2/+; Calcrflox/flox; R26stop-YFP and Pax7CreERT2/+; Calcr+/+; R26stop-YFP sorted cells based on YFP expression.
(d) Control and Calcr cKO satellite cells isolated by FACS, fixed immediately after sorting and immunostained for CALCR to confirm the absence of CALCR protein from recombined cells. Asterisk shows a non-recombined, CALCR+ cell.
(e) Quantification of PAX7, Myogenin and EdU positive cells in Calcr-depleted satellite cells (Pax7CT2/+; Calcrflox/flox; R26stop-YFP) isolated by FACS and treated for 32h or 72h with COLI or COLV (n=3 mice, ≥250 cells counted, 2 wells/condition).
(f) Quantification of total PAX7 (GFP), Myogenin and EdU positive myogenic cells isolated by FACS from Tg:Pax7-nGFP mice 3 days after cardiotoxin injury of TA muscle, and incubated for 72h in presence of COLI or COLV in the culture medium (n=3 mice, ≥200 cells counted).
(g) CALCR protein in freshly isolated or 12h-cultured satellite cells from Tg:Pax7-nGFP mice, demonstrating that CALCR protein is still present when satellite cells are treated with different collagens (see Extended data Fig.2).
(h) Induction of Calcr transcript expression by RT-qPCR of Tg:Pax7-nGFP satellite cells isolated by FACS and cultured for 72h in the presence of COLI or COLV. Results are normalized to Tbp (n=3 mice).
(i) Immunostainings for CALCR protein of Tg:Pax7-nGFP satellite cells cultured for 72h in presence of COLI or COLV (n=3 mice, ≥50 cells, 2 wells/condition).
Error bars, mean ± SD; (b) two-sided unpaired t-test; (c-i) two-sided paired t-test. Scale bar: 50µm and 25µm in (g).
Extended data Figure 6. CALCR ligand Elcatonin can substitute the depletion of the surrogate ligand COLV.
(a) Intracellular levels of cAMP in CalcR-C2C12 cells treated with COLV for up to 480min (n=4 independent assays).
(b) Rescue of loss of COLV by Elcatonin in an ex vivo self-renewal reserve-cell model, where PAX7+ non-proliferative cells return to quiescence (see Methods). MyHC and PAX7 staining of Control (Ctr: Tg:Pax7-CT2; Col5a1+/+; R26mTmG) and Col5a1 null (Tg:Pax7-CT2; Col5a1flox/flox; R26mTmG) non-treated (NT) or treated with Elcatonin (Elcat). No GFP+/EdU+ cell (12h pulse) could be detected in any of the conditions indicating GFP+ cells are quiescent (data not shown).
(c) Quantification of percentage of reserve cells (PAX7+/total nuclei) (n=3 mice/genotype and condition, ≥350 cells counted).
Error bars indicate mean ± SD; two-sided paired t-test; #p value: two-sided unpaired t-test. Scale bar: 50µm.
Extended data Figure 7. Schematic model for Notch/COLV/CALCR axis in satellite cells.
A Notch/COLV/CALCR signalling cascade actively maintains muscle stem cell quiescence. satellite cells are in direct contact with the plasma membrane of the myofibre (black outline) and an overlying basement membrane (blue line). Activation of the Notch receptor is achieved by ligand (likely Dll-1 or Dll4) present on the muscle fibre. Induction of Col5a1 and Col5a3 (and also Col6a1/2) genes occurs via distal regulatory elements (blue box). Satellite cell-produced COLV is deposited under the basement membrane and acts as a surrogate ligand of the plasma membrane receptor CALCR, expressed also by the satellite cells, thereby propagating a cell-autonomous signalling system in the local niche. In the absence of COLV (deletion of Col5a1) the quiescent niche is disturbed, CALCR signalling is abrogated, and satellite cells spontaneously differentiate and fuse to myofibres, leading to exhaustion of the muscle stem cell pool.
Supplementary Material
Acknowledgments
We would like to thank H. Stunnenberg for the ChiP-seq and RNA-seq data, D. Castro for the RBPJ ChIP protocol, D. Greenspan for the anti-a3-COLV antibody and Col5a3 knock-out muscle samples, C. Moali for the SPR assay, F. Auradé and the Protein Core Facility, Institut Curie, for the production of CalcR proteins; K. Ding for the 7rh DDR1 inhibitor; F. Ruggiero for suggesting the on-cell Elisa experiment, and the CRT Cytometry platform of Institut Pasteur. F.R. was funded by the Association Française contre les Myopathies via TRANSLAMUSCLE (PROJECT 19507), Agence Nationale pour la Recherche grant Satnet (ANR-15-CE13-0011-01) and RHU CARMMA (ANR-15-RHUS-0003). S.T. was funded by Institut Pasteur, Centre National pour la Recherche Scientific and the Agence Nationale de la Recherche (Laboratoire d’Excellence Revive, Investissement d’Avenir; ANR-10-LABX- 73) and the European Research Council (Advanced Research Grant 332893). M.B.B. was funded by the Doctoral School grant and Fondation pour la Recherche Médicale.
Footnotes
Author Contributions
M.B.B., S.T. and P.M. proposed the concept, designed experiments and wrote the manuscript, F.R. oversaw revisions, and S.T. funded most of the study. P.M. and D.C. conducted initial experiments on enhancer analysis. D.C. and L.M. performed and analysed ChIP experiments. M.B.B. performed the remaining experiments and together with P.M. analysed the data. S.F and D.E.B. provided mouse models.
Author Information
Data availability
All data that support the findings of this study are available from the corresponding authors upon request.
Competing interests
The authors declare no competing financial interests.
References
- 1.Raymond K, Deugnier MA, Faraldo MM, Glukhova MA. Adhesion within the stem cell niches. Curr Opin Cell Biol. 2009;21:623–629. doi: 10.1016/j.ceb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 3.Watt FM, Huck WT. Role of the extracellular matrix in regulating stem cell fate. Nature reviews. Molecular cell biology. 2013;14:467–473. doi: 10.1038/nrm3620. [DOI] [PubMed] [Google Scholar]
- 4.Mourikis P, et al. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- 5.Bjornson CR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozo M, Li L, Fan CM. Targeting beta1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nature medicine. 2016;22:889–896. doi: 10.1038/nm.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung TH, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zismanov V, et al. Phosphorylation of eIF2alpha Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell. 2016;18:79–90. doi: 10.1016/j.stem.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H, et al. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buas MF, Kabak S, Kadesch T. The Notch effector Hey1 associates with myogenic target genes to repress myogenesis. J Biol Chem. 285:1249–1258. doi: 10.1074/jbc.M109.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castel D, et al. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasyutina E, et al. RBP-J (Rbpsuh) is essential to maintain muscle progenitor cells and to generate satellite cells. Proc Natl Acad Sci U S A. 2007;104:4443–4448. doi: 10.1073/pnas.0610647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun M, et al. Targeted deletion of collagen V in tendons and ligaments results in a classic Ehlers-Danlos syndrome joint phenotype. Am J Pathol. 2015;185:1436–1447. doi: 10.1016/j.ajpath.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yennek S, Burute M, Thery M, Tajbakhsh S. Cell adhesion geometry regulates non-random DNA segregation and asymmetric cell fates in mouse skeletal muscle stem cells. Cell reports. 2014;7:961–970. doi: 10.1016/j.celrep.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Leitinger B. Transmembrane collagen receptors. Annual review of cell and developmental biology. 2011;27:265–290. doi: 10.1146/annurev-cellbio-092910-154013. [DOI] [PubMed] [Google Scholar]
- 18.Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 19.Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Sci Signal. 2014;7:ra76. doi: 10.1126/scisignal.2005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo R, et al. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi M, et al. Calcitonin Receptor Signaling Inhibits Muscle Stem Cells from Escaping the Quiescent State and the Niche. Cell reports. 2015;13:302–314. doi: 10.1016/j.celrep.2015.08.083. [DOI] [PubMed] [Google Scholar]
- 22.Evans BN, Rosenblatt MI, Mnayer LO, Oliver KR, Dickerson IM. CGRP-RCP, a novel protein required for signal transduction at calcitonin gene-related peptide and adrenomedullin receptors. J Biol Chem. 2000;275:31438–31443. doi: 10.1074/jbc.M005604200. [DOI] [PubMed] [Google Scholar]
- 23.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 24.Mourikis P, Tajbakhsh S. Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev Biol. 2014 doi: 10.1186/1471-213X-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machado L, et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Reports. 2017 doi: 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 26.Mourikis P, Gopalakrishnan S, Sambasivan R, Tajbakhsh S. Cell-autonomous Notch activity maintains the temporal specification potential of skeletal muscle stem cells. Development. 2012;139:4536–4548. doi: 10.1242/dev.084756. [DOI] [PubMed] [Google Scholar]
- 27.Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 31.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi M, et al. Calcitonin Receptor Signaling Inhibits Muscle Stem Cells from Escaping the Quiescent State and the Niche. Cell reports. 2015;13:302–314. doi: 10.1016/j.celrep.2015.08.083. [DOI] [PubMed] [Google Scholar]
- 33.Sun M, et al. Collagen V is a dominant regulator of collagen fibrillogenesis: dysfunctional regulation of structure and function in a corneal-stroma-specific Col5a1-null mouse model. J Cell Sci. 2011;124:4096–4105. doi: 10.1242/jcs.091363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambasivan R, et al. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Mourikis P, et al. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012;30:243–252. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]
- 36.Hicks C, et al. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. Journal of neuroscience research. 2002;68:655–667. doi: 10.1002/jnr.10263. [DOI] [PubMed] [Google Scholar]
- 37.Vasconcelos FF, et al. MyT1 Counteracts the Neural Progenitor Program to Promote Vertebrate Neurogenesis. Cell Rep. 2016;17:469–483. doi: 10.1016/j.celrep.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Machado L, et al. In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Reports. 2017 doi: 10.1016/j.celrep.2017.10.080. [DOI] [PubMed] [Google Scholar]
- 40.Gao M, et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. Journal of medicinal chemistry. 2013;56:3281–3295. doi: 10.1021/jm301824k. [DOI] [PubMed] [Google Scholar]
- 41.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 42.Zammit PS, et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates 'reserve cells'. J Cell Sci. 1998;111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- 44.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.