Abstract
Germline poses unique challenges to gene expression control at the transcriptional level. While the embryonic germline maintains a global hold on new mRNA transcription, the female adult germline produces transcripts that are not translated into proteins until embryogenesis of subsequent generation. As a consequence, translational control plays a central role in governing various germ cell decisions including the formation of primordial germ cells, self-renewal/differentiation decisions in the adult germline, onset of gametogenesis and oocyte maturation. Mechanistically, several common themes such as asymmetric localization of mRNAs, conserved RNA-binding proteins that control translation by 3′ UTR binding, translational activation by the cytoplasmic elongation of the polyA tail and the assembly of mRNA-protein complexes called mRNPs have emerged from the studies on Caenorhabditis elegans, Xenopus and Drosophila. How mRNPs assemble, what influences their dynamics, and how a particular 3′ UTR-binding protein turns on the translation of certain mRNAs while turning off other mRNAs at the same time and space are key challenges for future work.
6.1. Introduction
Differential gene expression is at the core of development. Although most cells in our body possess the same genome, gene expression differs among the different cell types, which provides each cell type its unique identity. Gene expression can be controlled at any of the different stages starting from chromatin status to the posttranslational modification of proteins. Regulation at the transcriptional stage is energy efficient: when a gene product is not required, stopping transcription eliminates the need to degrade or store the gene product(s) in an inactive form. Consistent with this, most somatic development programs are controlled at the level of transcription. However, the transcriptional control as a sole mechanism of gene regulation is inadequate in certain important developmental and physiological situations. For example, the highly condensed metaphase chromatin is not readily accessible for transcription factors, yet specific proteins are required only at specific steps during the cell division cycle. Similarly, neurons need to rapidly produce new proteins at the synaptic ends of axons, which are often at considerable distance from the nucleus, where transcription occurs. Posttranscriptional mechanisms, such as translational control, play a key role in such circumstances. Several key decisions during germ cell development demand gene expression controls beyond the transcriptional step, and translational control plays a particularly crucial role at these decision points.
In the first part of this chapter, we discuss the germline-specific developmental contexts in which translational control of gene expression is preferred over the transcriptional control. The second part reviews our current understanding of the roles played by translational control during different stages of germ cell development, namely, specification, self-renewal/differentiation decision, meiotic progression, and oocyte maturation.
6.2. The Germline Challenges to Transcriptional Control of Gene Expression
6.2.1. Transcriptional Quiescence During Germline Specification
Transcriptional quiescence is a hallmark of the early embryonic germline in many organisms. In the early embryo, unlike the somatic blastomeres, which activate new mRNA transcription as soon as they are born, their germline counterparts do not initiate new mRNA transcription. These cells lack the actively transcribing form of RNA polymerase II, which indicates a genome-wide shutdown of mRNA transcription (Seydoux and Dunn 1997). Such a global suppression of transcription most likely helps the embryonic germline maintain totipotency by evading the influence of maternally inherited transcription factors, which would otherwise activate the various somatic differentiation programs. This is particularly well supported by the functioning of SKN-1 and PAL-1 transcription factors in the C. elegans embryo (see below). While the maternal SKN-1 protein is distributed equally to the somatic blastomere EMS and its germline sibling P2, it activates the transcription of genes that specify somatic fates in EMS, but not in P2 in which the PIE-1 protein maintains global transcriptional quiescence (Bowerman et al. 1993; Maduro et al. 2001; Seydoux et al. 1996). Similarly, PAL-1 activates muscle fate in the D blastomere but not in its germline sibling P4, where PIE-1 is present (Hunter and Kenyon 1996).
6.2.2. Transcriptional Quiescence During Meiosis and Gametogenesis
The highly condensed state of the meiotic chromatin is not readily accessible to transcription factors. In Drosophila male germ cells completely shut down their transcription as they enter meiosis (Olivieri and Olivieri 1965; Schafer et al. 1995). In other organisms spermatocytes are transcriptionally silent during meiotic entry (leptotene and zygotene) and again during spermatid elongation that involves chromatin compaction along with histone exchange (Sassone-Corsi 2002). Similarly, during oogenesis, de novo mRNA synthesis does not occur during early stages of meiosis. It is briefly activated during pachytene and diplotene before being globally silenced again through the final stages of maturation (Walker et al. 2007). This transcriptional quiescence of germ cells during gametogenesis and of the mature gametes necessitates that the transcripts be premade, stored in a translationally dormant state until their protein products are needed post fertilization in the embryo.
6.2.3. Making mRNAs for the Next Generation
Early embryonic development proceeds in the absence of new mRNA transcription. As a consequence, genes that control early embryogenesis are transcribed in the maternal germline, and the mRNAs are deposited in the oocyte. Premature translation of these maternal mRNAs—many of their protein products direct somatic differentiation—will be detrimental to the mother’s germline [e.g., see Ciosk et al. (2006)]. Therefore, translational control of these mRNAs is vital for gametogenesis.
6.2.4. Germ Cells Share a Common Cytoplasm
In many species, germ cells are connected by cytoplasmic bridges, which allow the sharing of cytoplasmic contents among germ cells during different developmental stages until they form mature gametes. This concept is stretched in certain species like C. elegans to an extent where all germ nuclei share a common cytoplasm—not an ideal condition for transcriptional control to be effective. Not surprisingly, translational control is the predominant mode of gene regulation in the C. elegans germline [(Merritt et al. 2008); see below].
6.3. Translational Control During Germ Cell Development
From the fate specification to gamete maturation, several key developmental events occur during the process of germ cell development. Germ cell fate determination is the first event in this long journey. In most species, germ cells are specified at a different location from the future gonad; as a consequence the primordial germ cells (PGCs) must migrate to, and get incorporated into, the somatic gonad. Since only a small number of PGCs are born in the early embryo, they enter a proliferative phase to establish a population of self-renewing germline stem cells (GSCs) in a micro-environment of the gonad called the GSC niche. Some of these GSCs then switch from the mitotic to the meiotic mode and finally differentiate into gametes. Translation regulation plays a central role in all these developmental stages.
6.3.1. Translational Control During Germ Cell Specification and PGC Development
There are two modes of germ cell specification in metazoans. In invertebrates and anuran amphibians, maternally inherited components specify germ cell fate. By contrast, no specific cell in the early embryo possesses germ cell fate in urodele amphibians and vertebrates; instead, inductive signals from neighboring cells induce it in certain cells (Extavour and Akam 2003; McLaren 1999). In organisms that follow the inheritance mode, maternally inherited germline components—principally mRNAs and proteins—reside in the same cytoplasm as the somatic factors at the zygotic stage. As a consequence, their asymmetric localization and translational activation of the localized transcripts are particularly crucial to germ cell fate specification in these organisms. The role of maternal mRNAs and their translational control during germ cell formation have been extensively studied in Drosophila and C. elegans.
6.3.1.1. Translational Activation of Localized mRNAs Determines Germ Cell Fate in Drosophila
Germ cell formation in Drosophila begins in the oocyte with the assembly of a special cytoplasm, called germplasm, at the posterior of the oocyte. Germplasm contains the polar granules consisting of proteins including Oskar, Tudor, Vasa, Valois, and Staufen and many mRNAs including oskar, which is required for PGC specification, and nanos, which is essential for PGC development (Forbes and Lehmann 1998; Kim-Ha et al. 1991; Markussen et al. 1995; Rongo et al. 1995). Although the mechanism of how the proteins get to the germplasm is not known, many of the mRNAs are transported by passive diffusion and localized by entrapment. One mechanism of entrapment involves partial base pairing between the mRNAs and piRNAs bound to the Piwi protein Aubergine (Forrest and Gavis 2003; Kugler and Lasko 2009; Vourekas et al. 2016). Oskar initiates germplasm formation; its expression alone is sufficient to induce germplasm formation at ectopic locations and thus serves as a central component of germ cell fate determination (Mahowald 2001). The oskar mRNA is transported in a translationally repressed state via microtubules to the oocyte posterior. This localization of oskar mRNA at the posterior end is essential for translational activation of Oskar (Jambor et al. 2014). Bruno, an RBP, inhibits oskar mRNA via both a cap-dependent and a cap-independent mechanism. For the cap-dependent function, Bruno binds to the 3′ UTR of oskar mRNA, resulting in the recruitment of Cup to the 5′ cap, which binds to eIF4E and prevents it from recruiting eIF4G, causing repression of oskar mRNA (Nakamura et al. 2004). Bruno also inhibits oskar translation in a cap-independent mechanism by packaging oskar mRNA into large particles that are inaccessible to the translational machinery (Chekulaeva et al. 2006; Nakamura et al. 2004). Further, Bicaudal-C, which generally regulates the polyA tail length of target transcripts, also contributes to the inhibition of oskar translation, possibly by inhibiting the association of cytoplasmic polyadenylation element binding protein (CPEB) Orb with oskar mRNA (Castagnetti and Ephrussi 2003; Chicoine et al. 2007; Saffman et al. 1998). This repression is alleviated at the posterior pole by a specific derepressing element in the 5′ UTR of oskar mRNA (Gunkel et al. 1998). Although the mechanism of derepression is not known, Vasa, Aubergine, Orb, and Staufen are required for the oskar mRNA’s efficient translation (Castagnetti and Ephrussi 2003; Chang et al. 1999; Harris and Macdonald 2001; Markussen et al. 1997; Micklem et al. 2000; Wilson et al. 1996).
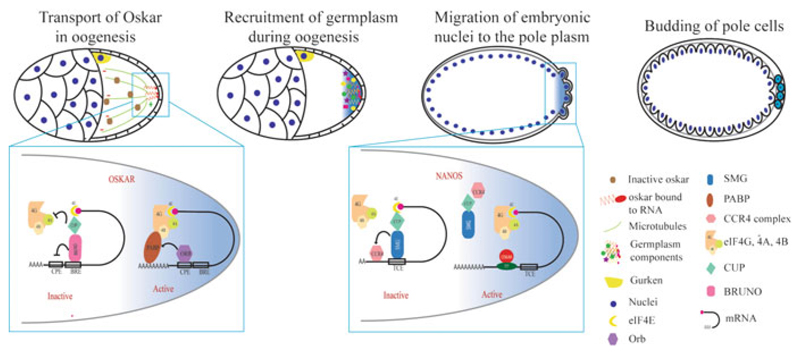
Oskar recruits other germplasm components such as Vasa and Tudor and about 200 maternal mRNAs to the germ granules (Frise et al. 2010; Trcek et al. 2015). Based on a few well-studied examples, such as nanos (nos), germ cell-less (gcl), polar granule component (pgc), and cyclin B, it is assumed that the translation of these mRNAs is specifically activated by the germ granule components and that their protein products direct PGC development. While the Gcl and Pgc proteins repress transcription in the Drosophila PGCs, Nos and its RNA-binding partner Pumilio prevent premature proliferation of PGCs by repressing the translation of cyclin B mRNA (Hanyu-Nakamura et al. 2008; Kadyrova et al. 2007; Leatherman et al. 2002). In addition, Nos and Pumilio are essential for PGC development and later GSC maintenance; they contribute to these processes possibly by suppressing other mRNAs (Forbes and Lehmann 1998; Harris et al. 2011). Like osk mRNA, nos mRNA is transported to the germplasm in a translationally silent state. The nos 3′ UTR contains several stem-loop structures which bind different RBPs at different stages of development. The RBPs Glorund and Smaug (Smg) suppress nos translation in the late-stage oocyte and embryo, respectively (Kalifa et al. 2006; Smibert et al. 1996). Smg recruits the deadenylase complex and targets nos mRNA for degradation in the bulk cytoplasm (Semotok et al. 2005). In addition, Smg recruits Cup and inhibits nos translation (Nelson et al. 2004). In the germplasm, Osk activates nos translation by dislodging Smg from its 3′ UTR (Dahanukar et al. 1999; Zaessinger et al. 2006). In summary, a complex interplay between various cis elements and trans-activating factors ensures localization-dependent translation of mRNAs that determine PGC formation and development in Drosophila in the transcriptionally quiescent early embryo (Fig. 6.1).
Fig. 6.1.
Translational control during germ cell specification in Drosophila. Germplasm is assembled during oogenesis by Oskar. oskar mRNA is transported in an inactive form and deposited at the posterior pole where its translation is activated. oskar translation is suppressed by Bruno with the help of Cup. At the posterior, oskar translation is activated by ORB; ORB recruits PABP and facilitates translational activation. Oskar further recruits other germplasm components during late stages of oogenesis. The early Drosophila embryo is a syncytium; at the start of cellularization, some of the posterior nuclei and the surrounding germplasm form the pole cells (PGCs) by budding. nanos mRNA is translationally suppressed in the anterior region by Smaug (Smg) which recruits Cup and prevents translation initiation. At the posterior end, nos mRNA is bound by Oskar, which prevents Smg from binding to nos-2 3′ UTR, which derepresses nos mRNA. Smaller cells diagrammed in the top left two cartoons represent the nurse cells
6.3.1.2. Asymmetric Segregation, Transcriptional Quiescence, and Sequential Translational Activation Lead to PGC Formation in C. elegans
In C. elegans, germ granules are present in the syncytial maternal germline even before oogenesis begins and are distributed initially throughout the zygotic cytoplasm (Strome and Wood 1982). Not surprisingly, an Oskar-like single factor that initiates germplasm assembly in the oocyte has not been identified in C. elegans. Although they are segregated to the germline during asymmetric cleavages (see below) and some of their protein and mRNA components are essential for PGC formation or development, the germ granules per se are not essential for the germ cell fate (Gallo et al. 2010). In sharp contrast to the syncytial beginning of the Drosophila embryo, the C. elegans embryo undergoes complete cytokinesis during embryonic cell divisions. The zygote undergoes an asymmetric cleavage generating a large anterior cell called AB and a small posterior cell called P1. While the Drosophila pole cells are committed to the germline as soon as they are formed, the P1 blastomere is not fully committed to the germline. Instead P1 undergoes three more rounds of asymmetric division generating one daughter cell at each of these divisions that acquires somatic fate (Fig. 6.2a), while the other daughter retains the germline (P) fate (Sulston et al. 1983). This process of asymmetry between the generation of the germline and somatic lineage necessitates (a) asymmetric segregation of maternally produced germline components at each of these divisions to the P lineage, (b) protection of germline components from degradation in the germline lineage, and (c) resistance of acquisition of somatic fate by the germline blastomeres P1, P2, P3, and P4 (Fig. 6.2b). These remarkable feats are accomplished by a combination of transcriptional quiescence and sequential translational activation of maternal mRNAs.
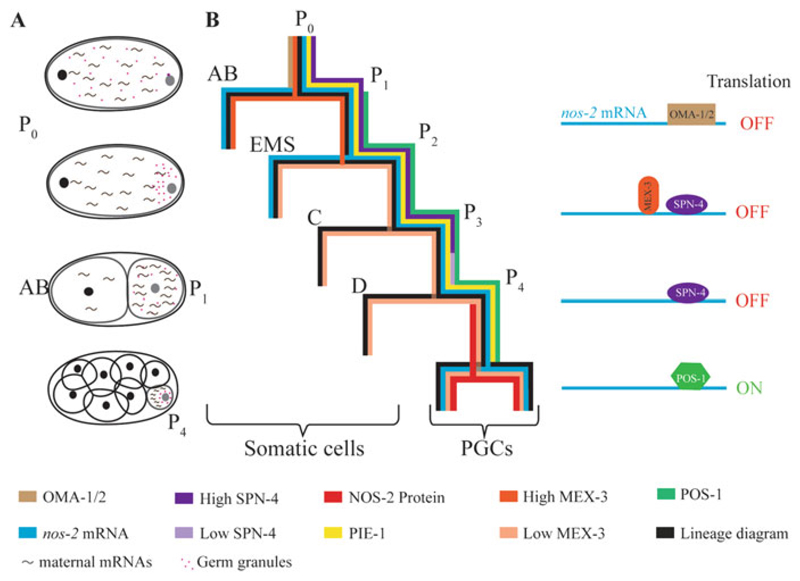
Fig. 6.2.
Translational control during germ cell specification in C. elegans. (a) Schematic representation of asymmetric cleavage and asymmetric distribution of maternal components. Pink dots and tilde-like structures represent the germ granules and the maternal mRNAs, respectively. (b) Distribution patterns of RBPs and nos-2 mRNA during early embryonic cleavages; colors representing the different components are indicated at the bottom. Translation of nos-2 mRNA is suppressed sequentially by OMA-1 and OMA-2 in oocytes (not shown here), by MEX-3 in the AB blastomere, and by SPN-4 in the P lineage until P3. The rapid decrease in the SPN-4 to POS-1 ratio in P4 enables POS-1 to compete out SPN-4 for binding to the nos-2 3′ UTR, which depresses nos-2 translation in P4
Transcriptional quiescence in the P lineage is maintained by the maternal protein PIE-1, which is segregated to, and maintained in, the P lineage (Mello et al. 1996; Seydoux et al. 1996). PIE-1 suppresses transcription by preventing phosphorylation of the carboxy-terminal domain (CTD) of RNA polymerase II, analogous to Drosophila Pgc (Batchelder et al. 1999; Seydoux and Dunn 1997; Zhang et al. 2003). Meanwhile, sequential translational activation of maternal mRNAs such as pos-1, apx-1, and pal-1 helps the somatic blastomeres arising from the P lineage to acquire their respective somatic fates; however, due to the presence of PIE-1, the corresponding germline siblings are protected from somatic differentiation (Hunter and Kenyon 1996; Mello et al. 1992; Tabara et al. 1999). The role of PIE-1 in the protection of germline identity is further underscored by the acquisition of somatic fate by P4 in the pos-1 mutant embryos. pos-1 mutant embryos exhibit a reduction in PIE-1 accumulation in the P4 blastomere simultaneous to PAL-1 inducing the muscle fate (Tabara et al. 1999).
Although the mechanisms remain unclear, the persistence of maternal mRNAs in the P lineage appears to be a consequence of transcriptional quiescence. The pie-1 mutant P blastomeres activate mRNA transcription but fail to maintain the maternal mRNAs. However, the degradation of maternal mRNAs can be suppressed by blocking RNA polymerase II activity (Tenenhaus et al. 2001). RNA-binding proteins produced from maternal mRNAs such as OMA-1, OMA-2, MEX-3, SPN-4, and POS-1 control the translation of other maternal mRNAs (Guven-Ozkan et al. 2010; Jadhav et al. 2008; Kaymak and Ryder 2013; Ogura et al. 2003; Pagano et al. 2009; Spike et al. 2014b; Tabara et al. 1999). A well-studied example is the translational control of nanos-2 (nos-2) mRNA, which encodes a C. elegans ortholog of the Drosophila Nanos (Subramaniam and Seydoux 1999). Translation of nos-2 mRNA is restricted to the P4 blastomere by the sequential actions of the repressors OMA-1/2, MEX-3, SPN-4, and the derepressor POS-1 (Fig. 6.2b) (Jadhav et al. 2008). Each of these proteins mediate their effect by binding to the respective recognition sequences in the nos-2 3′ UTR. In P4 the SPN-4/POS-1 ratio decreases enabling POS-1 to bind to nos-2 3′ UTR and derepressing nos-2 translation. The translationally competent nos-2 mRNA produces NOS-2 which promotes PGC development.
6.3.1.3. Role of Translational Control During PGC Development in Vertebrates
Akin to Drosophila oskar, the zebrafish buckyball (buc), a gene conserved across vertebrates, acts as the primary germplasm organizer in zebrafish oocytes. The buc mRNA is distributed throughout all the four blastomeres at the 4-cell stage, but the protein localizes to the germplasm at the cleavage furrows. However, it is not clear if translational regulation is responsible for this difference between the buc mRNA and protein distribution (Bontems et al. 2009). Similar to C. elegans nanos-2 mRNA, zebrafish nanos mRNA is translationally repressed via its 3′ UTR in early embryos and expressed only in PGCs (Koprunner et al. 2001). The zebrafish nanos mRNA is repressed in the somatic cells of early embryo by the mir-430 miRNA, which binds to the 3′ UTR and promotes deadenylation (Mishima et al. 2006). This repression is relieved in PGCs by a vertebrate-specific RBP called Dead end, which inhibits the miRNA binding to the nanos 3′ UTR (Kedde et al. 2007). Dead end is conserved in all vertebrates and has been shown to be a component of germplasm and required for germ cell development (Weidinger et al. 2003; Youngren et al. 2005).
Germ cell specification in mouse is an example for the induction mode. Although this mode of germ cell specification is primarily regulated at the transcriptional level, recently it has been shown that BlimP1, a transcriptional repressor crucial for PGC development, is negatively regulated by let-7 miRNA. The let-7-mediated suppression is eventually relieved by its negative regulator let-28 during PGC specification (West et al. 2009). Separate studies have shown that the BlimP1 3′ UTR contains target sites for let-7 binding and that the let-7 miRNA can suppress its translation (Nie et al. 2008).
6.3.2. Translational Control During Mitosis–Meiosis Decision
Genes that drive differentiation are often transcribed in the progenitor or stem cells. Once transcribed these mRNAs are stored in the stem cells until the onset of differentiation in a translationally silent state. The preeminence of translational control in mitosis–meiosis decision in C. elegans and Drosophila has been recognized for quite some time; recent studies support a similar role for translational regulation in the mouse testis as well (Zhou et al. 2015). Furthermore, translational control plays vital roles even in somatic adult stem cell maintenance (Crist et al. 2012). In both invertebrates and vertebrates, niche-dependent transcriptional regulation and germ cell-intrinsic translational regulatory networks control the mitosis–meiosis decision (Kimble 2011; Lehmann 2012).
6.3.2.1. A Complex RNA Regulatory Network Guides the Mitosis–Meiosis Decision in C. elegans
The C. elegans gonad exhibits distal-proximal polarity with GSCs at the distal end and mature gametes in the proximal region. The somatic cell called distal tip cell (DTC) signals GSCs via the LAG-2 Delta ligand localized on its cell membrane and the GLP-1 Notch receptor expressed in GSCs (Austin and Kimble 1987; Crittenden et al. 1994; Henderson et al. 1994; Yochem and Greenwald 1989). GLP-1, together with an “RNA regulatory loop” that is initiated by GLP-1 signaling, promotes self-renewal of the GSCs by inhibiting premature meiotic differentiation (Fig. 6.3). The RNA regulatory loop consists of the nearly identical PUF paralogs FBF-1 and FBF-2 (collectively called FBF), the KH domain RBP GLD-1, and the glp-1 mRNA. In the distal region, GLP-1 activates the transcription of FBF-2 (Lamont et al. 2004), which, along with FBF-1, represses the translation of gld-1 mRNA (Crittenden et al. 2002). As a result, GLD-1 expression is restricted to more proximal cells where it activates meiotic differentiation. In addition, GLD-1 represses glp-1 translation—thus completing the regulatory loop—and thereby reinforcing the fate switch (Marin and Evans 2003). Besides GLD-1, FBF inhibits differentiation by repressing the translation of meiotic entry promoters GLD-2 and GLD-3 and some of the meiotic machinery components (Eckmann et al. 2002; Merritt and Seydoux 2010; Millonigg et al. 2014). Further, FBF represses the translation of CKI-2, an inhibitor of CYE-1 cyclin E; CYE-1 and its kinase partner CDK-2, in turn, phosphorylate and inactivate GLD-1 (Jeong et al. 2011; Kalchhauser et al. 2011).
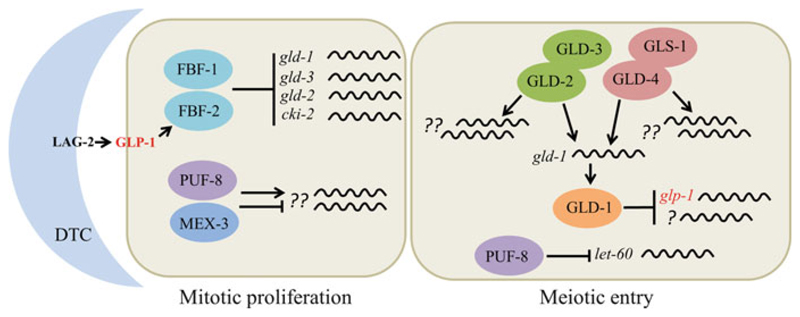
Fig. 6.3.
Translational control of the mitosis–meiosis decision in C. elegans. The LAG-2 ligand produced by the somatic cell called the distal tip cell (DTC) activates the GLP-1 receptor present on germ cells. This results in the transcriptional activation of the RBP FBF-2, which along with FBF-1 inhibits meiotic entry by suppressing the translation of gld-1, gld-3, gld-2, and cki-2 mRNAs. The RBPs PUF-8 and MEX-3 promote proliferation, possibly by regulating the translation of unknown mRNAs. Although a single proliferating cell is shown in this cartoon, the mitotic region extends to about 20-cell diameters from DTC. The entire proliferative zone comprises of a total of ~200 cells. The schematic on the right represents a cell from the transition zone. In the transition zone, GLP-1 activity and the levels of FBFs decrease, resulting in the expression of FBF targets such as GLD-1 and GLD-2. GLD-1 represses the translation of glp-1 and unknown mRNAs to promote meiotic entry. GLD-2 and GLD-4 (PAPs) promote GLD-1 expression. In addition, these two PAPs promote meiotic entry independently of GLD-1 by regulating the translation of unknown mRNAs. Furthermore, PUF-8 facilitates meiotic entry by repressing the translation of let-60, which encodes RAS, a well-known proliferation-promoting factor, in this zone
The size of the proliferation zone is controlled by both positive and negative regulation of glp-1 mRNA translation. While GLD-1 represses glp-1 translation (Marin and Evans 2003), the cytoplasmic polyA polymerase GLD-4 promotes it, in part, by extending the polyA tail length (Millonigg et al. 2014).
Preventing differentiation of the GSCs, however, is not sufficient for their self-renewal. PUF-8, another PUF family member, and the KH domain protein MEX-3 promote intrinsic GSC proliferation directly (Ariz et al. 2009). Germ cells lacking PUF-8 and MEX-3, although capable of meiotic entry in the absence of GLP-1 activity, do not enter meiosis; interestingly, they do not proliferate either. These observations suggest that the mRNAs regulated by PUF-8 and MEX-3 in the distal germline are likely to control mitotic proliferation independent of their effect on differentiation. The identities of these mRNAs are currently unknown. Interestingly, PUF-8 also represses the proliferative fate in the transition zone, where it functions redundantly with the GTPase-activating protein GAP-3 to inhibit the LET-60 RAS activity. While PUF-8 represses the translation of let-60 mRNA, GAP-3 inactivates LET-60 protein by promoting GTP hydrolysis. In the puf-8, gap-3 double-mutant adult germline, the active form of LET-60 RAS, increases, due to the loss of both mRNA- and protein-level controls, and activates the downstream MPK-1 ERK signaling. Consequently, cells continue to proliferate without entering differentiation (Vaid et al. 2013). However, the reasons underlying the yin-yang function of PUF-8 in promoting both proliferation and differentiation are currently an active area of investigation. Perhaps PUF-8 accomplishes this feat by interacting with different factor(s) in the different regions of the germline, which probably allows it to switch targets, analogous to its ortholog in Drosophila (see below).
At least three pathways dependent on translational control act redundantly to promote meiotic entry in C. elegans (Fig. 6.3). (1) The GLD-1 pathway, containing of the gene products GLD-1 and Nanos-3 (NOS-3) (Hansen et al. 2004; Kadyk and Kimble 1998). Here, NOS-3 promotes GLD-1 expression by an unknown mechanism. GLD-1 is a translational repressor and represses the translation of several mRNAs including cye-1, pal-1, mex-3, cep-1, tra-2, and rme-2 mRNAs; however, the targets that may be directly relevant for meiotic entry have not been identified (Biedermann et al. 2009; Jan et al. 1999; Lee and Schedl 2001; Mootz et al. 2004; Schumacher et al. 2005). Since GLD-1 is as a translational repressor, the most consistent hypothesis is that GLD-1 probably promotes meiotic entry by repressing mRNA(s) that inhibit meiotic entry. (2) The GLD-1 pathway is composed of the polyA polymerase (PAP) GLD-2 and its RNA-binding partner GLD-3 function (Eckmann et al. 2004; Kadyk and Kimble 1998; Wang et al. 2002). Unlike the canonical PAPs, GLD-2 lacks RNA-binding domain. Instead, it depends on other RBPs such as GLD-3 and RNP-8 to polyadenylate its target mRNAs. GLD-2 can promote meiotic entry independently of GLD-1; however, it has also been known to partner with GLD-3 and promote gld-1 mRNA translation, suggesting a complex interplay of these factors in maintaining the tight balance between mitosis and meiosis (Suh et al. 2006). In addition, another noncanonical PAP called GLD-4 and its RNA-binding partner GLS-1 also promote gld-1 mRNA translation and regulate GLD-1 protein levels. Thus, GLD-2 and GLD-4 act redundantly to promote the translation of gld-1 mRNA (Schmid et al. 2009). (3) The GLD-4–GLS-1 pathway promotes meiotic entry independent of GLD-1 and GLD-2, constituting the third meiotic entry pathway (Millonigg et al. 2014). Besides the gld-1 mRNA, GLD-2 promotes the stability and translation of many other mRNAs, including oma-2, egg-1, pup-2, and tra-2 mRNAs, in meiotic germ cells (Kim et al. 2010). However, the targets relevant to meiotic entry remain unknown for both GLD-2 and GLD-4. Since these cytoplasmic PAPs act as translational activators, one hypothesis could be that they promote the translation of mRNA(s) that promote meiotic entry, rather than repressing an interfering factor.
6.3.2.2. The GSC Niche Operates a Bistable Translational Switch to Control Self-Renewal and Differentiation Decisions in the Drosophila Ovary
Signaling from the GSC niche and an internal translational network is crucial to GSC maintenance in Drosophila ovary. Interestingly, different signaling cascades operate in the female and male germlines: BMP pathway in the ovary and JAK-STAT pathway in the testis [for a review see Lehmann (2012)]. Both promote the GSC fate by preventing precocious differentiation. In this chapter we will limit our discussion to the intrinsic factors acting downstream of the niche signaling in female GSCs as they are better understood. In the ovary, the BMP-type ligands Decapentaplegic (Dpp) and Glass bottom boat (Gbb) secreted by the niche repress the transcription of bag of marbles (bam), which is a differentiation-promoting factor, in GSCs (Chen and McKearin 2003; Song et al. 2004; Wharton et al. 1991; Xie and Spradling 1998). Separately, Nos and the PUF protein Pumilio (Pum) repress the translation of brain tumor (brat), mei-P26, and possibly a few other differentiation-promoting factors (Harris et al. 2011; Joly et al. 2013). The requirement of Nos and Pum is crucial for the maintenance of GSCs: in the absence of Nos or Pum, all GSCs differentiate, which depletes the ovary of GSCs (Forbes and Lehmann 1998; Gilboa and Lehmann 2004).
In the Drosophila ovary, GSCs divide such that one daughter cell (GSC) stays closer, and attached, to the niche, while the second daughter cell (cystoblast) forms away from the niche and receives less Dpp signal as a consequence, repression of bam is relieved in the cystoblast. Bam forms a complex with Sex-lethal, MeiP26, and BGCN and represses the translation of nos mRNA (Li et al. 2009, 2013). Reduction in Nos levels depresses brat translation, resulting in the accumulation of Brat in the cystoblast. Brat partners with Pum and represses the translation of Dpp signal transducers dMyc, Mad, Medea, and Schnurri, which reinforces the self-renewal-to-differentiation fate switch in the cystoblast (Harris et al. 2011; Newton et al. 2015). Thus, by changing its partner, from Nos in GSCs to Brat in cystoblasts, Pum is able to change its translational targets, from mei-P26 and brat mRNAs in the GSC to dMyc and mad mRNAs in the cystoblast, and enable these two cell types to maintain their respective fates (Fig. 6.4). However, identities of the other translational targets of Nos-Pum and Brat-Pum, which are likely to be directly involved in the execution of self-renewal or differentiation programs, are currently unknown. In addition, miRNAs and the general transcriptional machinery have also been suggested to play a role in GSC maintenance in the Drosophila ovary [for a review, see Slaidina and Lehmann (2014)]. The function of Brat-Pum pair appears to be restricted to enacting the self-renewal-to-differentiation fate switch; the continued presence of Pum is detrimental to the differentiation process, and its translation is repressed at later stages by Rbfox1 (Carreira-Rosario et al. 2016).
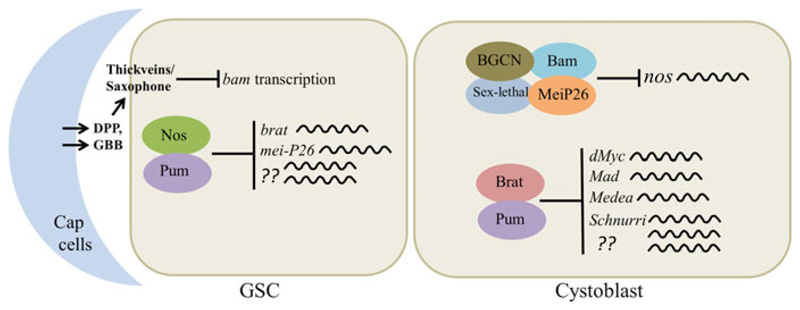
Fig. 6.4.
Translational control of the self-renewal/differentiation decision in the Drosophila ovary. In the Drosophila ovary, signaling from the niche (cap cells) suppresses bam transcription in the GSC. Apart from this, Nos-Pum pair represses the translation of brat, mei-P26, and other unknown mRNAs to prevent premature differentiation. The GSC divides such that one daughter cell is oriented away from the niche and does not receive sufficient niche signals to suppress bam. Bam forms a multi-protein complex with Mei-P26, Sex-lethal, and BGCN and suppresses nos translation. Absence of Nos derepresses the brat mRNA leading to Brat expression, which partners with Pum and inhibits the translation of dMyc, Mad, Medea, and Schnurri to promote differentiation
6.3.2.3. Assembly of Differentiation-Promoting mRNAs into Translationally Dormant mRNPs Maintains GSCs in the Mouse Testis
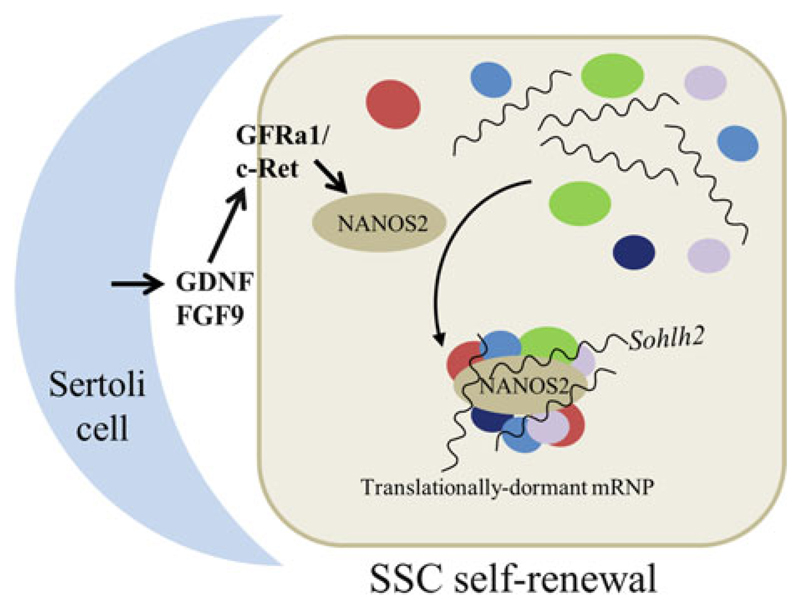
In mammalian females, germ cells enter meiosis soon after arriving at the gonad thus much of our current knowledge on mammalian GSC maintenance comes from studies on males. Spermatogonial stem cells (SSCs)—the GSCs in mouse testis— self-renew as well as differentiate amidst somatic cells called Sertoli cells. Fibroblast growth factor 9 (FGF9) and glial cell line-derived neurotrophic factor (GDNF) secreted by Sertoli cells are essential for the self-renewal of SSCs (Barrios et al. 2010; Bowles et al. 2010; Meng et al. 2000). Although these two signals induce the expression of a battery of transcription factors, one of the crucial downstream targets is Nanos2 (Fig. 6.5). Unlike in the Drosophila ovary, the mouse Nanos2 does not seem to associate with a PUF ortholog. Instead, Nanos2 interacts with another RBP, Dead end1 (DND1), to recruit differentiation-promoting mRNAs, such as Sohlh2, Dmrt1, Dazl, and Taf7l, into mRNP complexes, where it represses translation by interacting with the CCR4-NOT deadenylase complex and promoting the deadenylation of the target mRNAs (Suzuki et al. 2012, 2016; Zhou et al. 2015). Interestingly, not all mRNAs associated with the Nanos2 mRNPs are repressed at the posttranscriptional level. A few pre-mRNAs, such as Sohlh1 and Taf7l, increase in the absence of Nanos2, indicating a potential transcriptional control as well.
Fig. 6.5.
Nanos mRNP is crucial for spermatogonial stem cell maintenance in mice. In mouse, GDNF and FGF9 secreted by Sertoli cells activate Nanos2 expression in SSCs. Nanos2 sequesters differentiationpromoting mRNAs into mRNPs along with other RBPs
Furthermore, mere recruitment by Nanos2 alone is not sufficient; proper assembly of the mRNP, which requires the core mRNP component Rck, is also essential for translational suppression (Zhou et al. 2015). Apart from mRNAs, Nanos2 sequesters the mTOR protein as well to mRNPs and inhibits mTORC1 signaling pathway, which is known to negatively regulate SSC self-renewal (Zhou et al. 2015). In summary, maintenance of differentiation-promoting mRNAs in a translationally dormant state by assembling them into mRNPs appears to be the principal mechanism by which the SSCs are maintained in mice.
6.3.3. Translational Control of Meiotic Progression and Oocyte Maturation
Germ cells that enter differentiation execute two different developmental programs simultaneously: one, progression through the meiotic divisions and the other, gametogenesis. Exquisite control of translation, with great spatial and temporal precision, governs meiotic progression and oocyte maturation. In fact, much of our current knowledge of translational control mechanisms comes from studies on oocyte maturation. In most organisms, germ cells progress through meiotic prophase I and arrest at some point—the exact stage varies among species—prior to metaphase I (M-I). During this arrest period, oocytes grow enormously in size accumulating mRNAs and proteins required for the rest of oogenesis and early embryogenesis and the various factors required for translation. However, only a small fraction of the stored mRNAs is translated; others are stored in a translationally dormant state waiting for appropriate stimuli. Upon induction by hormonal (Xenopus and mammals) or sperm-derived (C. elegans) signals, the oocyte completes meiosis I and arrests at metaphase II (M-II) till fertilization. The progression through M-I to M-II, termed as oocyte maturation, is regulated by the translational activation of some of the stored mRNAs. Premature translation of mRNAs is detrimental to the development of fertilization-competent oocyte; hence strict translational control is crucial during both meiotic progression and oocyte maturation. In organisms ranging from C. elegans to mammals, regulation of the polyA tail by cytoplasmic polyadenylation is the principal mechanism of translation regulation during oogenesis. Nevertheless, other mechanisms are likely employed as well. The role of translational control during meiotic progression has been more thoroughly studied and better understood in the case of oogenesis in Xenopus and C. elegans; therefore, we will restrict ourselves to these two examples here.
6.3.3.1. Multiple RNA-Binding Proteins Act to Maintain Meiotic Commitment in C. elegans
Intriguingly, germ cells maintain mitotic potential even after initiating the meiotic program. In female germ cells, GLD-1 is the key factor that prevents mitotic reentry (Francis et al. 1995). Mutations in other genes, such as gld-2 and gld-4, also cause dedifferentiation (Kadyk and Kimble 1998; Schmid et al. 2009), but since these genes are known to promote gld-1 mRNA translation, the actual cause is possibly the reduction in GLD-1 expression (Fig. 6.6). GLD-1 inhibits the reentry, at least in part, by repressing the translation of cye-1 mRNA: while germ cells return to the mitotic mode after meiotic entry when GLD-1 is absent, they do not reenter mitosis if both GLD-1 and CYE-1 are absent (Biedermann et al. 2009). The translation of cye-1 mRNA is inhibited by at least one other factor, because the depletion of CYE-1 increases the number of meiotic cells in gld-1 gld-2 double-mutant germlines (Fox et al. 2011). Besides cye-1, GLD-1 represses a number of mRNAs whose protein products function at a later stage either during oocyte growth (RME-2) or embryonic patterning (MEX-3 and PAL-1). Premature translation of these mRNAs severely affects meiotic progression; for instance, when both GLD-1 and MEX-3 are absent, meiotic cells transdifferentiate into somatic lineages due to the ectopic translation of mRNAs encoding transcription factors such as PAL-1 (Ciosk et al. 2006). GLD-1 functions in spermatocytes as well, where it acts redundantly with PUF-8 to arrest dedifferentiation, but the downstream targets in this case are not known (Priti and Subramaniam 2015).
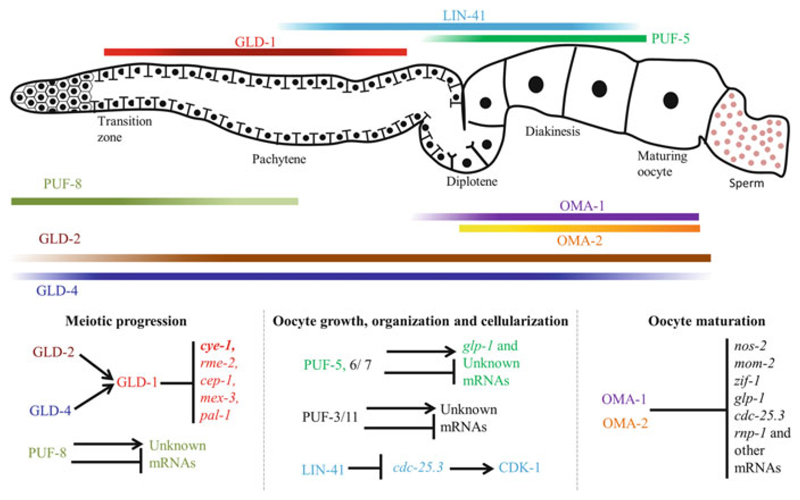
Fig. 6.6.
Translational regulation during meiotic progression, oocyte growth, and maturation in C. elegans. A cartoon of the adult hermaphrodite gonad is shown at the top. Orientation: distal to the left and proximal to the right. C. elegans gonad with different regions marked. Horizontal bars indicate expression patterns of the RBPs that regulate meiotic progression and/or oocyte maturation. Intensity variations of the color reflect the concentrations of the corresponding proteins. The RBPs and their corresponding target mRNAs in the different stages of meiotic development are shown at the bottom. See text for a more detailed description
6.3.3.2. mRNPs Control the Translation of Maternal mRNAs During Oocyte Maturation in C. elegans
Meiosis in developing C. elegans oocytes arrests at diakinesis of meiosis I. Oocytes grow rapidly in size by taking up cytoplasmic contents from the syncytial germline before closing off the cytoplasmic bridge. During this phase, while GLD-1 expression ceases, the expression of other RBPs such as LIN-41, MEX-3, OMA-1, OMA-2, PUF-5, and SPN-4 begins (Detwiler et al. 2001; Draper et al. 1996; Lublin and Evans 2007; Ogura et al. 2003; Spike et al. 2014a). These proteins together form mRNPs and potentially control the translation of a large number of stored mRNAs (Spike et al. 2014b). Specifically, LIN-41 controls oocyte growth by inhibiting premature cellularization and M-phase entry; it accomplishes this, at least in part, by repressing the translational of the CDK-1 activator CDC-25.3 (Spike et al. 2014a, b). In contrast, the nearly identical paralogs OMA-1 and OMA-2 promote cellularization and M-phase entry in the arrested diakinetic oocytes. In the oma-1 oma-2 double mutants, nuclear events such as the chromatin localization of the Aurora B kinase AIR-2 and nuclear envelope breakdown fail to occur, and the cytoplasmic streaming from the syncytial germline, which is required for oocyte growth, continues for longer than the wild type resulting in the formation of large oocytes (Detwiler et al. 2001; Govindan et al. 2009; Harris et al. 2006). The OMA proteins repress the translation of several mRNAs, including nos-2, glp-1, mom-2, zif-1, cdc-25.3, rnf-5, and rnp-1 mRNAs, in the oocyte (Guven-Ozkan et al. 2010; Jadhav et al. 2008; Kaymak and Ryder 2013; Oldenbroek et al. 2013; Spike et al. 2014b). How the OMA-mediated translational repression of these mRNAs contributes to regulating oocyte growth and maturation is yet to be explored. The PUF family proteins also contribute to oocyte development. PUF-5, PUF-6, and PUF-7 function redundantly to promote oocyte formation. Another set of PUF proteins, PUF-3 and PUF-11, limit oocyte growth. Although these PUF proteins control the translation of several mRNAs, the ones involved in oocyte formation and growth are unknown (Hubstenberger et al. 2012; Lublin and Evans 2007).
6.3.3.3. Precise Temporal Sequence of Translational Activation Directs Oogenesis in Xenopus
Since Xenopus produces large number of relatively big oocytes and oogenesis proceeds in the absence of transcription, the Xenopus oocyte has been an excellent model to investigate the mechanisms of translational control. During the arrest at diplotene, oocytes produce and accumulate large amounts of mRNAs and other translational accessories. Most of these mRNAs are stored in a translationally silent state until the oocyte resumes meiosis. Translational quiescence is maintained by the binding of an RBP called the cytoplasmic polyadenylation element binding protein (CPEB), which binds to a specific sequence element in the 3′ UTR termed the cytoplasmic polyadenylation element (CPE) (Lin et al. 2010). CPEB interacts with other proteins such as Maskin, which interacts with the initiation factor eIF4E and prevents translational initiation (Barnard et al. 2005). In addition, CPEB interacts with the PAP Gld2, the Xenopus ortholog of GLD-2 discussed above, and the polyA ribonuclease PARN. While Gld2 extends the polyA tail, PARN promptly shortens it through its function as a deadenylase (Barnard et al. 2004; Copeland and Wormington 2001; Kim and Richter 2006; Korner et al. 1998).
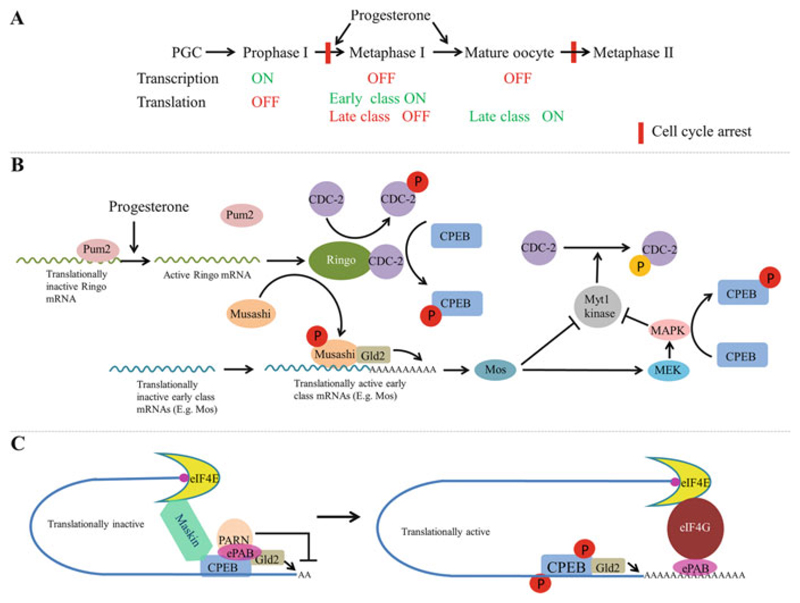
When the oocyte resumes meiosis, translation of the stored mRNAs is activated in a precise temporal sequence, which is crucial for the meiotic progression and oocyte maturation (Fig. 6.7). Based on the timing of activation, the mRNAs are classified as “early,” which are translated prior to the nuclear envelope breakdown [or the germinal vesicle breakdown (GBVD)] in Xenopus, and “late,” which are translated after GBVD. Unlike most mRNAs that are controlled by CPEB, translation of Ringo mRNA, a cyclin B-type protein, is under the control of the PUF protein Pum2 (Cao et al. 2010; Padmanabhan and Richter 2006). Upon stimulation by progesterone secreted by the follicle cells, Pum2 dissociates from the Ringo mRNA, resulting in its translation. Ringo then activates Cdc2/Cdk1, which phosphorylate several proteins including CPEB and Musashi, an RBP that binds to another 3′ UTR element called MPE (Fig. 6.7b) (Arumugam et al. 2012b; Kim and Richter 2007; Mendez et al. 2002). Activated by phosphorylation, Musashi recruits Gld2, which extends the polyA tail of the “early” mRNAs, such as the ones encoding Mos and cyclin B5, leading to their translational activation (Cragle and MacNicol 2014). Expression of Mos and cyclin B5 is thus crucial for the translational activation of the “late” mRNAs. Mos activates the mitogen-activated protein kinase (MAPK) pathway, which phosphorylates CPEB at sites distinct from the ones phosphorylated by Ringo-Cdc2 (Keady et al. 2007; Posada et al. 1993; Sagata et al. 1988). Thus, CPEB is phosphorylated by both the MAPK pathway and Ringo/Cdc2. Depending on the CPEB-containing repression complex assembled on a particular mRNA, any of several possible mechanisms respond to the CPEB phosphorylation and activate translation [for a review see Charlesworth et al. (2013)]. For instance, in immature stage VI oocytes, PARN is expelled from the CPEB complex in response to CPEB phosphorylation; in the absence of a competing deadenylase activity, the polyadenylation activity of Gld2 results in polyA tail elongation (Barnard et al. 2004; Kim and Richter 2006). In addition, phosphorylation of CPEB by Cdc2 leads to the dissociation of embryonic polyA binding protein (ePAB) from the CPEB-containing mRNP complex, which competes out Maskin for binding to eIF4E allowing the recruitment of eIF4G to the initiation complex, which enables translational initiation by promoting the binding of the 40S ribosomal subunit to the 5′ cap (Fig. 6.7c) (Barnard et al. 2005; Cao et al. 2006; Cao and Richter 2002; Kim and Richter 2007).
Fig. 6.7.
Sequential activation of polyadenylation during oocyte maturation in Xenopus. (a) Outline of the transcriptional and translational status at the key stages of oocyte maturation. (b) Summary of the signaling cascade activated by progesterone during oocyte maturation. Progesterone stimulation releases Ringo mRNA from Pum2-mediated repression. Ringo associates with Cdc-2, phosphorylates CPEB, Cdc-2, and Musashi. Phosphorylated Musashi recruits the PAP Gld2 and activates translation of early class mRNAs. One of them is Mos mRNA. Mos activates the MAPK pathway by phosphorylating MEK. MAPK in turn phosphorylates CPEB at a site distinct from the one phosphorylated by Ringo/cyclin B-Cdc-2. (c) Phosphorylation of CPEB activates translation of masked mRNAs. Translationally inactive (masked) mRNAs are bound by CPEB, ePAB, PARN, Gld-2, and Maskin. Maskin binds to CPEB and eIF4E, blocking association of eIF4E with eIF4G. Polyadenylation–deadenylation cycles by GLD2 and PARN keep the polyA tail short. Phosphorylation of CPEB expels PARN, leading to polyA tail extension. Another factor released from the complex is ePAB, which now binds to the polyA tail and associates with eIF4E by competing out Maskin and recruits eIF4G leading to translation activation
An important aspect of the activation of early mRNAs is the operation of at least two positive feedback loops. In one, the initial association of Ringo-Cdc2 sets up a positive feedback loop leading to rapid increase in the levels of active cyclin B-Cdc2 (MPF). In this loop, Ringo-Cdc2 activates the Cdc25 phosphatase and inactivates the Myt1 kinase (Karaiskou et al. 1999). Since Cdc25 activates Cdc2 by removing a phosphate added by the Myt1 kinase, this leads to an increase in the active form of Cdc2. The second positive feedback loop consists of two components: phosphorylated Musashi activates the translation of its own mRNA; in addition, the MAPK pathway, activated by Musashi via Mos, also phosphorylates Musashi (Arumugam et al. 2012a, b).
Phosphorylation and activation of Musashi are thus essential for oocyte maturation, while activation of either the cyclin B-Cdc2 or the MAPK pathway is sufficient for this process (Haccard and Jessus 2006). This leads to the interesting conundrum: how is it possible for the upstream activator (Musashi) of pathway 1 (Mos-MAPK) to be essential for a given process, while that pathway itself is redundant with a second pathway (Cdc2), which in turn activates the upstream activator (Musashi) of the first pathway.
6.4. Concluding Remarks
Translational control clearly plays a predominant role in germ cell development, all the way from specification to gametogenesis, in diverse organisms. Functions of some of the factors involved, notably the Nanos and PUF family proteins, are well conserved pointing to the ancient origin of the central role for translational regulation in the germline. Recent work has identified several key factors that regulate translation in the germline. Some new mechanisms have emerged as well. For instance, the formation of mRNPs as a way to store mRNAs in translationally inactive state has emerged as a common theme in several species. These findings have opened up several new questions for the immediate future. The mRNA targets directly responsible for some of the key processes are not known for many of the newly identified RBPs. Although we did not discuss the role of small RNA molecules extensively in this chapter, their role in the germline is just emerging. The mechanistic details of how mRNPs assemble and what influences their dynamics are other central questions that need further investigation.
Acknowledgments
Work in the KS laboratory is supported by grants from the Department of Science and Technology and the Department of Biotechnology, Government of India.
Contributor Information
Kumari Pushpa, Regional Centre for Biotechnology, Faridabad, Haryana, India.
Ganga Anil Kumar, Indian Institute of Technology-Kanpur, Kanpur, India; Indian Institute of Technology-Madras, Chennai, India.
Kuppuswamy Subramaniam, Indian Institute of Technology-Madras, Chennai, India.
References
- Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam K, Macnicol MC, Macnicol AM. Autoregulation of Musashi1 mRNA translation during Xenopus oocyte maturation. Mol Reprod Dev. 2012a;79:553–563. doi: 10.1002/mrd.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam K, MacNicol MC, Wang Y, Cragle CE, Tackett AJ, Hardy LL, MacNicol AM. Ringo/cyclin-dependent kinase and mitogen-activated protein kinase signaling pathways regulate the activity of the cell fate determinant Musashi to promote cell cycle re-entry in Xenopus oocytes. J Biol Chem. 2012b;287:10639–10649. doi: 10.1074/jbc.M111.300681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Barnard DC, Cao Q, Richter JD. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol Cell Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios F, Filipponi D, Pellegrini M, Paronetto MP, Di Siena S, Geremia R, Rossi P, De Felici M, Jannini EA, Dolci S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J Cell Sci. 2010;123:871–880. doi: 10.1242/jcs.057968. [DOI] [PubMed] [Google Scholar]
- Batchelder C, Dunn MA, Choy B, Suh Y, Cassie C, Shim EY, Shin TH, Mello C, Seydoux G, Blackwell TK. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 1999;13:202–212. doi: 10.1101/gad.13.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B, Wright J, Senften M, Kalchhauser I, Sarathy G, Lee MH, Ciosk R. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell. 2009;17:355–364. doi: 10.1016/j.devcel.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowles J, Feng CW, Spiller C, Davidson TL, Jackson A, Koopman P. FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev Cell. 2010;19:440–449. doi: 10.1016/j.devcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Kim JH, Richter JD. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat Struct Mol Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, Buszczak M. Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev Cell. 2016;36:562–571. doi: 10.1016/j.devcel.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development. 2003;130:835–843. doi: 10.1242/dev.00309. [DOI] [PubMed] [Google Scholar]
- Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA. 2013;4:437–461. doi: 10.1002/wrna.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin D. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragle C, MacNicol AM. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J Biol Chem. 2014;289:14239–14251. doi: 10.1074/jbc.M114.548271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Detwiler MR, Reuben M, Li X, Rogers E, Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 2001;1:187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Fox PM, Vought VE, Hanazawa M, Lee MH, Maine EM, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–2234. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frise E, Hammonds AS, Celniker SE. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol. 2010;6:345. doi: 10.1038/msb.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004;131:4895–4905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136:2211–2221. doi: 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel N, Yano T, Markussen FH, Olsen LC, Ephrussi A. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev. 1998;12:1652–1664. doi: 10.1101/gad.12.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Robertson SM, Nishi Y, Lin R. zif-1 translational repression defines a second, mutually exclusive OMA function in germline transcriptional repression. Development. 2010;137:3373–3382. doi: 10.1242/dev.055327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haccard O, Jessus C. Redundant pathways for Cdc2 activation in Xenopus oocyte: either cyclin B or Mos synthesis. EMBO Rep. 2006;7:321–325. doi: 10.1038/sj.embor.7400611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Harris JE, Govindan JA, Yamamoto I, Schwartz J, Kaverina I, Greenstein D. Major sperm protein signaling promotes oocyte microtubule reorganization prior to fertilization in Caenorhabditis elegans. Dev Biol. 2006;299:105–121. doi: 10.1016/j.ydbio.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell. 2011;20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Cameron C, Shtofman R, Gutman S, Evans TC. A network of PUF proteins and Ras signaling promote mRNA repression and oogenesis in C. elegans. Dev Biol. 2012;366:218–231. doi: 10.1016/j.ydbio.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- Jadhav S, Rana M, Subramaniam K. Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development. 2008;135:1803–1812. doi: 10.1242/dev.013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Mueller S, Bullock SL, Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA. 2014;20:429–439. doi: 10.1261/rna.041566.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Verheyden JM, Kimble J. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS Genet. 2011;7:e1001348. doi: 10.1371/journal.pgen.1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 2013;1:411–424. doi: 10.1016/j.stemcr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kalchhauser I, Farley BM, Pauli S, Ryder SP, Ciosk R. FBF represses the Cip/Kip cell-cycle inhibitor CKI-2 to promote self-renewal of germline stem cells in C. elegans. EMBO J. 2011;30:3823–3829. doi: 10.1038/emboj.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Karaiskou A, Jessus C, Brassac T, Ozon R. Phosphatase 2A and polo kinase, two antagonistic regulators of cdc25 activation and MPF auto-amplification. J Cell Sci. 1999;112(Pt 21):3747–3756. doi: 10.1242/jcs.112.21.3747. [DOI] [PubMed] [Google Scholar]
- Kaymak E, Ryder SP. RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J Biol Chem. 2013;288:30463–30472. doi: 10.1074/jbc.M113.496547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Kuo P, Martinez SE, Yuan L, Hake LE. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci. 2007;120:1093–1103. doi: 10.1242/jcs.03416. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Wilson TL, Kimble J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci USA. 2010;107:17445–17450. doi: 10.1073/pnas.1012611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3:a002683. doi: 10.1101/cshperspect.a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly (Austin) 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, Levin L, Boero J, Jongens TA. Germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr Biol. 2002;12:1681–1685. doi: 10.1016/s0960-9822(02)01182-x. [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 2001;15:2408–2420. doi: 10.1101/gad.915901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. Germline stem cells: origin and destiny. Cell Stem Cell. 2012;10:729–739. doi: 10.1016/j.stem.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci USA. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M. Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One. 2013;8:e58301. doi: 10.1371/journal.pone.0058301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Evans V, Shen S, Xing Y, Richter JD. The nuclear experience of CPEB: implications for RNA processing and translational control. RNA. 2010;16:338–348. doi: 10.1261/rna.1779810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Marin VA, Evans TC. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development. 2003;130:2623–2632. doi: 10.1242/dev.00486. [DOI] [PubMed] [Google Scholar]
- Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development. 1995;121:3723–3732. doi: 10.1242/dev.121.11.3723. [DOI] [PubMed] [Google Scholar]
- Markussen FH, Breitwieser W, Ephrussi A. Efficient translation and phosphorylation of Oskar require Oskar protein and the RNA helicase Vasa. Cold Spring Harb Symp Quant Biol. 1997;62:13–17. [PubMed] [Google Scholar]
- McLaren A. Signaling for germ cells. Genes Dev. 1999;13:373–376. doi: 10.1101/gad.13.4.373. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 2010;137:1787–1798. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Adams J, Grunert S, St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millonigg S, Minasaki R, Nousch M, Eckmann CR. GLD-4-mediated translational activation regulates the size of the proliferative germ cell pool in the adult C. elegans germ line. PLoS Genet. 2014;10:e1004647. doi: 10.1371/journal.pgen.1004647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton FG, Harris RE, Sutcliffe C, Ashe HL. Coordinate post-transcriptional repression of Dpp-dependent transcription factors attenuates signal range during development. Development. 2015;142:3362–3373. doi: 10.1242/dev.123273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, Chadburn A, Tuschl T, Knowles DM, Tam W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Kishimoto N, Mitani S, Gengyo-Ando K, Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. 2003;130:2495–2503. doi: 10.1242/dev.00469. [DOI] [PubMed] [Google Scholar]
- Oldenbroek M, Robertson SM, Guven-Ozkan T, Spike C, Greenstein D, Lin R. Regulation of maternal Wnt mRNA translation in C. elegans embryos. Development. 2013;140:4614–4623. doi: 10.1242/dev.096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri G, Olivieri A. Autoradiographic study of nucleic acid synthesis during spermato-genesis in Drosophila melanogaster. Mutat Res. 1965;2:366–380. doi: 10.1016/0027-5107(65)90072-2. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, Essien KI, Ryder SP. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc Natl Acad Sci USA. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada J, Yew N, Ahn NG, Vande Woude GF, Cooper JA. Mos stimulates MAP kinase in Xenopus oocytes and activates a MAP kinase kinase in vitro. Mol Cell Biol. 1993;13:2546–2553. doi: 10.1128/mcb.13.4.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priti A, Subramaniam K. PUF-8 functions redundantly with GLD-1 to promote the meiotic progression of spermatocytes in Caenorhabditis elegans. G3 (Bethesda) 2015 doi: 10.1534/g3.115.019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P. Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol. 1998;18:4855–4862. doi: 10.1128/mcb.18.8.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N, Oskarsson M, Copeland T, Brumbaugh J, Vande Woude GF. Function of c-mos proto-oncogene product in meiotic maturation in Xenopus oocytes. Nature. 1988;335:519–525. doi: 10.1038/335519a0. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermato-genesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Schafer M, Nayernia K, Engel W, Schafer U. Translational control in spermatogenesis. Dev Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Schmid M, Kuchler B, Eckmann CR. Two conserved regulatory cytoplasmic poly (A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 2009;23:824–836. doi: 10.1101/gad.494009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 1996;382:713–716. doi: 10.1038/382713a0. [DOI] [PubMed] [Google Scholar]
- Slaidina M, Lehmann R. Translational control in germline stem cell development. J Cell Biol. 2014;207:13–21. doi: 10.1083/jcb.201407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Song X, Wong MD, Kawase E, Xi R, Ding BC, McCarthy JJ, Xie T. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- Spike CA, Coetzee D, Eichten C, Wang X, Hansen D, Greenstein D. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics. 2014a;198:1535–1558. doi: 10.1534/genetics.114.168831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Coetzee D, Nishi Y, Guven-Ozkan T, Oldenbroek M, Yamamoto I, Lin R, Greenstein D. Translational control of the oogenic program by components of OMA ribonucleoprotein particles in Caenorhabditis elegans. Genetics. 2014b;198:1513–1533. doi: 10.1534/genetics.114.168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1982;79:1558–1562. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci USA. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS One. 2012;7:e33558. doi: 10.1371/journal.pone.0033558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Niimi Y, Shinmyozu K, Zhou Z, Kiso M, Saga Y. Dead end1 is an essential partner of NANOS2 for selective binding of target RNAs in male germ cell development. EMBO Rep. 2016;17:37–46. doi: 10.15252/embr.201540828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 2001;15:1031–1040. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid S, Ariz M, Chaturbedi A, Kumar GA, Subramaniam K. PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development. 2013;140:1645–1654. doi: 10.1242/dev.088013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature. 2016;531:390–394. doi: 10.1038/nature17150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Boag PR, Blackwell TK. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev Biol. 2007;304:382–393. doi: 10.1016/j.ydbio.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly (A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA, Thomsen GH, Gelbart WM. Drosophila 60A gene, another transforming growth factor beta family member, is closely related to human bone morphogenetic proteins. Proc Natl Acad Sci USA. 1991;88:9214–9218. doi: 10.1073/pnas.88.20.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Yochem J, Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- Zhang F, Barboric M, Blackwell TK, Peterlin BM. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 2003;17:748–758. doi: 10.1101/gad.1068203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Shirakawa T, Ohbo K, Sada A, Wu Q, Hasegawa K, Saba R, Saga Y. RNA binding protein Nanos2 organizes post-transcriptional Buffering system to retain primitive state of mouse spermatogonial stem cells. Dev Cell. 2015;34:96–107. doi: 10.1016/j.devcel.2015.05.014. [DOI] [PubMed] [Google Scholar]