Abstract
Background
Genome-wide association studies (GWAS) identified single nucleotide polymorphisms (SNPs) involved in adult fat distribution. Whether these SNPs also affect abdominal and organ-specific fat accumulation in children is unknown.
Methods
In a population-based prospective cohort study among 1 995 children (median age: 9.8 years, 95% range 9.4;10.8), We tested the associations of six genetic risk scores based on previously identified SNPs for childhood BMI, adult BMI, liver fat, WHR, pericardial fat mass, visceral- and subcutaneous adipose tissue ratio (VAT/SAT ratio), and four individual SAT and VAT associated SNPs, for association with SAT (N=1 746), VAT (N=1 742), VAT/SAT ratio (N=1 738), liver fat fraction (N=1 950), and pericardial fat mass (N=1 803) measured by Magnetic Resonance Imaging.
Results
Per additional risk allele in the childhood BMI genetic risk score, SAT increased 0.020 standard deviation scores (SDS), (95% confidence interval (CI) 0.009;0.031, p-value:3.28*10-4) and VAT increased 0.021 SDS, 95% CI:0.009;0.032, p-value:4.68*10-4). The adult BMI risk score was positively associated with SAT (0.022 SDS increase, CI:0.015;0.029, p-value:1.33*10-9), VAT (0.017 SDS increase, CI:0.010;0.025, p-value:7.00*10-6), and negatively with VAT/SAT ratio (-0.012 SDS decrease, CI:-0.019;-0.006, p-value:2.88*10-4). The liver fat risk score was associated with liver fat fraction (0.121 SDS, CI:0.086;0.157, p-value:2.65*10-11). Rs7185735 (SAT), was associated with SAT (0.151 SDS, CI:0.087;0.214, p-value:3.00*10-6) and VAT/SAT ratio (-0.126 SDS, CI:-0.186;-0.065, p-value:4.70*10-5). After stratification by sex the associations of the adult BMI risk score with SAT and VAT and of the liver fat risk score with liver fat fraction remained in both sexes. Associations of the childhood BMI risk score with SAT, and the adult BMI risk score with VAT/SAT ratio were present among boys only, whereas the association of the pericardial fat risk score with pericardial fat was present among girls only.
Conclusion
Genetic variants associated with BMI, body fat distribution, liver and pericardial fat already affect body fat distribution in childhood.
Keywords: Body Mass Index, polymorphism, single nucleotide, pediatrics, Magnetic Resonance Imaging, Abdominal Fat
Introduction
Childhood overweight and obesity are related to short- and long-term complications, such as type 2 diabetes and cardiovascular disease (ref.1,2,3,4). Besides body mass index (BMI), body fat distribution is also considered to be important (ref.5). Especially abdominal fat, which can be stored as either subcutaneous (SAT) or visceral adipose tissue fat (VAT), is gaining interest (ref.6,7,8). In preadolescence on average lower levels of SAT and VAT are present than in adolescence and adulthood (ref.9). Previously, BMI was shown to be a relatively good measure for predicting SAT, but less so for VAT (ref.10,11). Also, fat accumulation in the liver and around the heart are suggested to play a role in metabolic disease (ref.12,13). A fatty liver is associated with dyslipidemia and dysglycemia, whereas pericardial fat is associated with coronary artery disease (ref.14,15). All four fat accumulation sites are heritable with heritability estimates of ranging from 30-60% (ref.11,16,17,18). Thus, fat distribution in particular areas, besides BMI, may affect the risk of metabolic disease and has a clear genetic component.
Recent large genome-wide associations studies (GWAS) have identified 97 single nucleotide polymorphisms (SNPs) associated with adult BMI and 15 SNPs associated with childhood BMI (ref.19,20,21). We have previously reported that genetic risk scores based on these SNPs were associated with infant growth and childhood adiposity measures determined using Dual-energy X-ray absorptiometry and ultrasound (ref.22). Although these 2-dimensional imaging techniques can estimate preperitoneal fat as a proxy of VAT, they are unable to accurately determine SAT, VAT, liver fat fraction, and pericardial fat (ref.23). Magnetic Resonance Imaging (MRI) can distinguish SAT, VAT, liver fat fraction, and pericardial fat more precisely and accurately by 3-dimensional measurements (ref.23,24). Other GWAS have identified SNPs for adult waist-hip ratio (WHR), SAT, VAT, VAT/SAT ratio, liver fat, and pericardial fat (ref.11,17,25,26,27). The genetic background of body fat distribution in children is largely unknown.
We hypothesized that genetic variants associated with childhood and adult BMI and more specific fat measures in adults are associated with fat accumulation in children. We tested in a population-based prospective cohort among 1 995 children whether genetic risk scores based on known variants are associated with SAT, VAT, liver fat fraction and pericardial fat assessed by MRI.
Methods
Study design and population
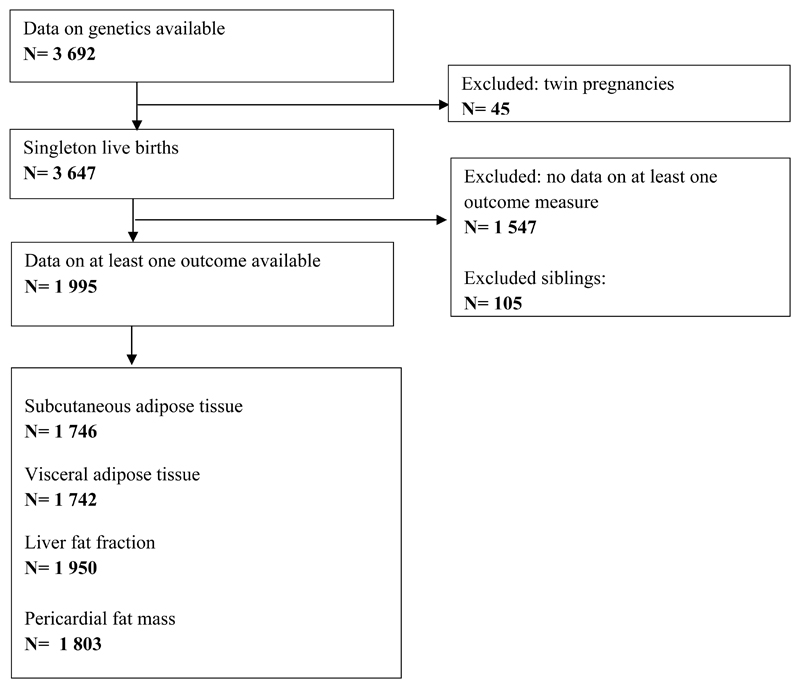
This study was embedded within the Generation R Study, a prospective population-based cohort in Rotterdam, the Netherlands (ref.24). All pregnant women residing in Rotterdam with a delivery date between April 2002 and January 2006 were invited to participate. The Medical Ethical Committee of Erasmus MC approved the study and an informed consent was obtained for all children. A total of 5 862 children participated in the follow-up at age ten years. GWA scans were available for 3 692 children (ref.28). Of these, MRI scans were available for 2 593 children (70%). All twins were excluded and only one non-twin sibling was selected per mother, based on data completeness or, if equal, randomly. The current study was limited to children with information on at least one of the outcomes (N= 1995). Figure 1 shows a participant flow chart.
Figure 1.
Flow chart of participants
Genetic risk scores and separate SNPs
DNA was isolated from cord blood or, for a small subgroup without cord blood samples, from blood samples taken at age 6 years (ref.28). For genome-wide association analysis the Illumina 610 and 660W Quad platforms were used (ref.24). Stringent quality checks were performed excluding individuals with low sample call rates or sex mismatches. Imputation of genotypes to the cosmopolitan panel of HapMap ii (release 22) was done using MACH software (ref.29,30). Prior to imputation, SNPs with a call rate <98%, significant deviations from Hardy-Weinberg equilibrium (P<1*10-6), or minor allele frequencies <0.1% were excluded. Information about the SNPs for the present study was extracted from our GWAS dataset. We constructed a total of four weighted and two unweighted risk scores. Each score summed the number of outcome increasing risk alleles from the GWA dosage data, and SNPs were weighted individually using effect sizes from the original GWAS (ref.22). For BMI, we constructed two weighted genetic risk scores, combining 15 childhood BMI SNPs in one and 97 adult BMI SNPs in the other (ref.20,21). For the 15 childhood SNPs, weights were recalculated from the previous GWAS meta-analysis excluding the Generation R Study data, as these were part of the discovery dataset (ref.21). For one of the 15 childhood BMI SNPs, rs1421085, no information was available in our GWA dataset. We used rs3751812 as a proxy (R2=0.93, D’=0.97). For four of the 97 adult BMI SNPs we used proxies (all R2>0.96, D’=1): rs13012571 was used as a proxy for rs13021737, rs1978487 for rs9925964, rs6445197 for rs2365389, and rs9636202 for rs17724992. The other two weighted risk scores were created for WHR and liver fat (ref.25,26). For 46 of the 49 WHR SNPs information was available in the GWA dataset. Rs4607103 was used as a proxy for rs2371767 (R2=0.90, D’=1). For the WHR SNPs rs8042543 and rs6556301 no good proxy was available leading to 47 WHR SNPs. As no effect estimates from previous GWAS were available for the pericardial fat mass and VAT/SAT ratio associated SNPs, unweighted risk scores were constructed based on three SNPs each for pericardial fat mass and VAT/SAT ratio (ref.27). A list of the SNPs included in the scores, a matrix listing the overlapping SNPs, and a matrix presenting the Pearson correlations between the risk scores are provided in the supplemental material (Supplemental material: Tables S1a, S1b, S1c). Previous GWAS identified one SNP for each of SAT (rs7185735), VAT and VAT adjusted for BMI (VATadjBMI) (rs2842895), SAT in women (rs2123685), and VAT and VATadjBMI in women (rs10060123) (ref.27). For these phenotypes, we could therefore not create risk scores and we tested these SNPs separately.
Measures of adiposity at 10 years
MRI has been described as an accurate and reproducible technique and is considered the gold standard for the measurement of intra-abdominal and organ fat deposition (ref.23,31,32,33). Adiposity measures were obtained from MRI scans as described previously (ref.24). All children were scanned using a 3.0 Tesla MRI (MR 750w, GE Healthcare, Milwaukee, WI, USA) using standard imaging and positioning protocols. Pericardial fat imaging in short axis orientation was performed using an ECG triggered black-blood prepared thin slice single shot fast spin echo acquisition (BB SSFSE) with multi-breath-hold approach. An axial 3-point Dixon acquisition for fat and water separation (IDEAL IQ) was used for liver fat and liver fat fraction imaging an axial abdominal scan from lower liver to pelvis and a coronal scan centered at the head of the femurs were performed with a 2-point DIXON acquisition (LavaFlex) (ref.34).
The obtained fat scans were analyzed by the Precision Image Analysis company (PIA, Kirkland, Washington, United States), using the sliceOmatic (TomoVision, Magog, Canada) software package. All extraneous structures and any image artifacts were removed manually (ref.23). Pericardial fat included both epicardial- and paracardial fat directly attached to the pericardium, ranging from the apex to the left ventricular outflow tract. Total subcutaneous and visceral fat volumes ranged from the dome of the liver to the superior part of the femoral head. Fat masses were obtained by multiplying the total volumes by the specific gravity of adipose tissue, 0.9 g/ml. Liver fat fraction was determined by taking four samples of at least 4 cm2 from the central portion of the hepatic volume. Subsequently, the mean signal intensities of these four samples were averaged to generate an overall mean liver fat fraction estimation. A more extensive description of the MRI measurement protocols can be found in the supplemental materials (Supplemental material; Additional file 2). BMI (kg/m2) was calculated from height and weight measured without shoes and heavy clothing (ref.35).
Statistical analysis
To examine whether the genetic risk scores were associated with the childhood adiposity measures we used linear regression analyses. To facilitate comparison of the effect estimates we created standard deviation (SD) scores for all outcomes. Sex- and age-adjusted SD scores (SDS) were constructed for BMI using the Dutch reference growth curves (Growth Analyser Research Calculation Tools, Version 4.0 http://www.growthanalyser.org). In order to make all outcomes except liver fat fraction independent of height, we estimated the optimal adjustment by log-log regression analyses (ref.36). All MRI adiposity measures except liver fat fraction and height were log-transformed, using natural logarithm (ln). Log-MRI adiposity measures were regressed on log-height. The regression slope then corresponds to the power to which height should be raised to calculate an index uncorrelated with height. Thus, we divided subcutaneous fat mass by height4, visceral fat mass by height3, and pericardial fat mass by height3. All height-adjusted outcomes approached a normal distribution after ln-transformation. All models were adjusted for sex and age except models with BMI as an outcome since BMI SDS were already adjusted for sex and age. All models included the first four genetic principal components to adjust for ancestry. A sensitivity analysis was performed adjusting all outcomes for BMI by conditional regression analysis to examine whether the associations were independent of BMI. Standardized residuals were obtained for each fat outcome from the regression of those outcomes on BMI. These standardized residuals were then used as outcome measures (ref.37). In addition, we repeated the analyses for SAT and VAT adjusting for VAT and SAT, respectively. Since body fat distribution may be different among boys and girls we planned a priori to stratify on sex (ref.38,39). Sex-specific associations were examined by adding the interaction term for the risk score with sex to the models. The variance explained by the risk scores was considered to be the increase in the unadjusted R2 between the model containing all covariates and the risk score or individual SNP, and the same model without the risk score/SNP. We applied Bonferroni correction to account for multiple testing, correcting for all ten exposures. We considered a p-value of smaller than 0.05/10=0.005 significant. All analyses were performed using the Statistical Package for the Social Sciences (SPSS) version 21.0 for Windows (IBM, Chicago, IL, USA). A power calculation was performed for all risk scores on the smallest and largest sample size used for analyses using a one sample, two sided test in R version 3.3.2, library ‘pwr’ (Supplemental material: Table S2).
Results
Participant characteristics
Characteristics of the participants are listed in Table 1. MRI scans were performed at a median age of 9.8 years (95% range 9.4;10.8). The median BMI of the children was 16.9 kg/m2 (95% range 14.0;24.0).
Table 1.
Characteristics of the study population. (N=1 995)
| Characteristics | |
|---|---|
| Birth | |
| Boys (%) | 979 (49.1) |
| Gestational age at birth (weeks) | 40.1 (36.3; 42.3) |
| Weight at birth (grams) a | 3 466 (510) |
| Childhood | |
| Age at visit (years) | 9.8 (9.4; 10.8) |
| Height (cm) a | 141.8 (6.6) |
| Weight (kg) a | 35.3 (7.0) |
| Body mass index (kg/m2) | 16.9 (14.0; 24.0) |
| Overweight (%) b | 273 (13.7) |
| Obese (%) b | 55 (2.8) |
| Subcutaneous adipose tissue (grams) | 1 291 (603; 5 246) |
| Visceral adipose tissue (grams) | 369 (161; 981) |
| Liver fat fraction (%) | 2.0 (1.3; 4.9) |
| Pericardial fat mass (grams) | 11 (5; 23) |
Values are medians (95% range) unless otherwise specified
Means (standard deviations)
The IOTF-classification was used to define overweight and obesity
Genetic risk scores and adiposity measures
The risk score for childhood BMI was associated with an increase in SAT, and VAT (Table 2). The adult BMI risk score was associated with an increase in SAT and VAT, and with a decrease in VAT/SAT ratio. The adult fatty liver risk score was solely associated with an increase in liver fat fraction, showing a relatively large increase of 0.121 SDS (95% CI 0.086;0.157) in liver fat fraction per additional average risk allele in the risk score. No associations were observed for the VAT/SAT ratio and pericardial fat risk scores (Table 2). Both the childhood and the adult BMI risk scores were associated with childhood BMI. Unweighted risk scores showed comparable results, except for the childhood BMI risk score for which the effect estimates were around twice as high as for the weighted risk score for all outcomes other than pericardial fat mass (Supplemental material, Supplemental table S3). Rs7185735 (SAT) was associated with an increase in SAT, and a decrease in VAT/SAT ratio and was also associated with childhood BMI (Table 2). The childhood and adult BMI risk scores, the WHR and VAT/SAT ratio risk scores and the VAT/SAT ratio and fatty liver risk scores were correlated (Supplemental material, Supplemental table S1c).
Table 2.
| Risk score | SAT (N=1 746) | VAT (N=1 742) | VAT/SAT ratio (N=1 738) | Liver fat fraction (N=1 950) | Pericardial fat mass (N=1 803) | BMI @ 10 (N=1 993) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (number of SNPs in risk score) | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P |
| Childhood BMI (N=15) | 0.020 | 3.28*10-4 | 0.021 | 4.68*10-4 | -0.005 | 0.335 | 0.009 | 0.098 | 0.004 | 0.541 | 0.030 | 1.60*10-7 |
| (0.009; 0.031) | (0.009; 0.032) | (-0.015; 0.005) | (-0.002; 0.020) | (-0.008; 0.015) | (0.019; 0.041) | |||||||
| Adult BMI (N=97) | 0.022 | 1.33*10-9 | 0.017 | 7.00*10-6 | -0.012 | 2.88*10-4 | 0.004 | 0.247 | 0.009 | 0.013 | 0.027 | 6.18*10-13 |
| (0.015; 0.029) | (0.010; 0.025) | (-0.019; -0.006) | (-0.003; 0.011) | (0.002; 0.017) | (0.019; 0.034) | |||||||
| Adult WHR (N=49) | -0.010 | 0.056 | -0.002 | 0.716 | 0.011 | 0.019 | -0.002 | 0.737 | -0.010 | 0.055 | -0.010 | 0.052 |
| (-0.019; 0.000) | (-0.012; 0.009) | (0.002;0.020) | (-0.011; 0.008) | (-0.020; 0.000) | (-0.020; 0.000) | |||||||
| VAT/SAT ratio (N=3)* | -0.006 | 0.768 | -0.008 | 0.694 | 0.001 | 0.966 | -0.012 | 0.554 | -0.013 | 0.513 | -0.008 | 0.702 |
| (-0.044; 0.032) | (-0.049; 0.033) | (-0.036; 0.037) | (-0.050; 0.026) | (-0.053; 0.027) | (-0.047; 0.031) | |||||||
| Adult fatty liver (N=5) | 0.003 | 0.853 | 0.003 | 0.863 | 0.001 | 0.966 | 0.122 | 2.54*10-11 | 0.010 | 0.590 | -0.008 | 0.702 |
| (-0.032; 0.039) | (-0.035; 0.042) | (-0.036; 0.037) | (0.086; 0.158) | (-0.028; 0.048) | (-0.047; 0.031) | |||||||
| Adult pericardial fat (N=3)* | -0.024 | 0.283 | 0.022 | 0.358 | 0.056 | 0.008 | -0.007 | 0.761 | 0.061 | 0.009 | -0.028 | 0.226 |
| (-0.068; 0.020) | (-0.025; 0.069) | (0.015; 0.098) | (-0.050; 0.037) | (0.015; 0.107) | (-0.072; 0.017) | |||||||
| rs7185735 (SAT) | 0.151 | 3.00*10-6 | 0.093 | 0.007 | -0.126 | 4.70*10-5 | 0.044 | 0.178 | 0.038 | 0.269 | 0.115 | 0.001 |
| (0.087; 0.214) | (0.025; 0.161) | (-0.186; -0.065) | (-0.020; 0.107) | (-0.029; 0.105) | (0.050; 0.181) | |||||||
| rs2123685 (SAT female) | -0.034 | 0.707 | -0.167 | 0.081 | -0.126 | 0.142 | 0.075 | 0.403 | -0.060 | 0.522 | 0.094 | 0.311 |
| (-0.210; 0.143) | (-0.355; 0.020) | (-0.293; 0.042) | (-0.101; 0.250) | (-0.243; 0.123) | (-0.088; 0.275) | |||||||
| rs2842895 (VAT and VATadjBMI) | 0.021 | 0.531 | -0.001 | 0.970 | -0.021 | 0.503 | -0.002 | 0.943 | 0.053 | 0.126 | 0.024 | 0.473 |
| (-0.044; 0.086) | (-0.071; 0.068) | (-0.083; 0.041) | (-0.067; 0.062) | (-0.015; 0.122) | (-0.042; 0.091) | |||||||
| rs10060123 (VAT and VATadjBMI female) | -0.049 | 0.185 | -0.068 | 0.081 | -0.011 | 0.758 | -0.028 | 0.444 | -0.068 | 0.079 | -0.055 | 0.141 |
| (-0.121; 0.023) | (-0.145; 0.009) | (-0.079; 0.058) | (-0.099; 0.044) | (-0.143; 0.008) | (-0.129; 0.018) |
Analyses were performed in children with complete data on genetic variants and covariates and at least one outcome under study
Values are linear regression coefficients for models adjusted for sex, age, and the first four principal components and represent the difference in standard deviation scores of the outcome measures for each additional average risk allele in the risk scores.
Bold values represent nominally significant outcomes
Unweighted risk scores since no information on the effect estimates was available. Bold values represent significant outcomes after Bonferroni correction for 10 analyses (P<0.005)
After adjusting the outcomes for BMI the associations of the BMI risk scores with the outcomes were no longer present (Supplemental material, Supplemental table S4). The adult fatty liver risk score remained associated with liver fat fraction (0.128 SDS increase in liver fat percentage per additional risk allele, CI:0.095;0.161) and a new association was observed for the adult pericardial fat risk score with pericardial fat (0.074 SDS increase, CI: 0.028;0.120). Rs7185735 (SAT) remained associated with SATadjBMI albeit with a lower effect estimate (Supplemental material, Supplemental table S4). Associations of the risk scores and individual SNPs with SAT adjusted for VAT and with VAT adjusted for SAT are shown in Supplemental table S5.
There was a nominally significant interaction with sex only for the associations of the liver fat risk score with SAT and VAT/SAT ratio, for rs7185735 (SAT) with VAT/SAT ratio, and for rs10060123 (VAT and VATadjBMI in girls) with liver fat (data not shown). After stratification by sex the associations that remained in both sexes were those of the adult BMI risk score with SAT and VAT, with a slightly higher effect estimate for SAT in boys compared to girls, and for the liver fat risk score with liver fat fraction, with a higher effect estimate in girls (Table 3). The childhood and adult BMI risk scores remained associated with BMI in both sexes, with a slightly higher effect estimate for the childhood BMI risk score in boys compared to girls. Associations in boys only were those of the childhood BMI risk score with SAT and of the adult BMI risk score with VAT/SAT ratio. Associations in girls only were those of the pericardial fat risk score with pericardial fat and of rs7185735 (SAT) with BMI (Table 3).
Table 3.
Associations of genetic risk scores with MRI adiposity measures in males (N=979) and females (N=1 016) separately a,b
| Risk score | SAT | VAT | VAT/SAT ratio | Liver fat fraction | Pericardial fat mass | BMI @ 10 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (number of SNPs in risk score) | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P | Beta (CI 95%) | P |
| Males | N=856 | N=851 | N=851 | N=958 | N=884 | N=977 | ||||||
| Childhood BMI (N=15) | 0.024 | 0.004 | 0.022 | 0.013 | -0.008 | 0.301 | 0.015 | 0.052 | -0.005 | 0.545 | 0.038 | 9.00*10-6 |
| (0.008; 0.040) | (0.005; 0.040) | (-0.023; 0.007) | (0.000; 0.031) | (-0.022; 0.012) | (0.021; 0.054) | |||||||
| Adult BMI (N=97) | 0.025 | 1.00*10-6 | 0.018 | 0.001 | -0.015 | 0.002 | 0.007 | 0.167 | 0.010 | 0.061 | 0.029 | 5.95*10-8 |
| (0.015; 0.035) | (0.007; 0.029) | (-0.025; -0.006) | (-0.003; 0.017) | (0.000; 0.021) | (0.018; 0.039) | |||||||
| Adult WHR (N=49) | -0.009 | 0.214 | -0.004 | 0.596 | 0.006 | 0.390 | -0.003 | 0.695 | -0.012 | 0.123 | -0.013 | 0.078 |
| (-0.024; 0.005) | (-0.020; 0.011) | (-0.008; 0.019) | (-0.016; 0.011) | (-0.027; 0.003) | (-0.028; 0.001) | |||||||
| VAT/SAT ratio (N=3)* | 0.028 | 0.329 | 0.019 | 0.549 | -0.017 | 0.525 | -0.014 | 0.598 | 0.006 | 0.838 | 0.023 | 0.442 |
| (-0.029; 0.085) | (-0.042; 0.080) | (-0.070; 0.036) | (-0.068; 0.039) | (-0.053; 0.065) | (-0.035; 0.081) | |||||||
| Adult fatty liver (N=5) | 0.033 | 0.202 | 0.024 | 0.398 | -0.034 | 0.166 | 0.094 | 1.65*10-6 | 0.001 | 0.976 | 0.003 | 0.904 |
| (-0.018; 0.084) | (-0.031; 0.078) | (-0.081; 0.014) | (0.045; 0.143) | (-0.053; 0.055) | (-0.050; 0.057) | |||||||
| Adult pericardial fat (N=3) * | 0.004 | 0.891 | 0.046 | 0.190 | 0.046 | 0.130 | -0.003 | 0.917 | 0.028 | 0.402 | -0.025 | 0.458 |
| (-0.059; 0.069) | (-0.023; 0.114) | (-0.013; 0.105) | (-0.064; 0.057) | (-0.038; 0.095) | (-0.090; 0.040) | |||||||
| rs7185735 (SAT) | 0.106 | 0.028 | 0.075 | 0.148 | -0.068 | 0.130 | 0.046 | 0.312 | 0.075 | 0.137 | 0.071 | 0.155 |
| (0.012; 0.200) | (-0.027; 0.176) | (-0.020; 0.156) | (-0.136; 0.043) | (-0.024; 0.174) | (-0.027; 0.168) | |||||||
| rs2123685 (SAT female) | -0.039 | 0.764 | -0.200 | 0.149 | -0.145 | 0.227 | -0.047 | 0.708 | -0.178 | 0.183 | 0.150 | 0.270 |
| (-0.293; 0.215) | (-0.471; 0.072) | (-0.380; 0.091) | (-0.190; 0.197) | (-0.439; 0.084) | (-0.117; 0.416) | |||||||
| rs2842895 (VAT and VATadjBMI) | 0.001 | 0.981 | -0.036 | 0.487 | -0.031 | 0.489 | -0.036 | 0.432 | 0.025 | 0.614 | -0.010 | 0.837 |
| (-0.093; 0.086) | (-0.138; 0.066) | (-0.119; 0.057) | (-0.054; 0.125) | (-0.073; 0.124) | (-0.107; 0.087) | |||||||
| rs10060123 (VAT and VATadjBMI female) | -0.070 | 0.191 | -0.071 | 0.220 | 0.023 | 0.640 | -0.111 | 0.028 | -0.055 | 0.322 | -0.070 | 0.206 |
| (-0.176; 0.035) | (-0.184; 0.042) | (-0.075; 0.122) | (-0.210; 0.012) | (-0.165; 0.054) | (-0.177; 0.038) | |||||||
| Females | N=890 | N=891 | N=887 | N=992 | N=919 | N=1 016 | ||||||
| Childhood BMI (N=15) | 0.016 | 0.031 | 0.019 | 0.014 | -0.002 | 0.731 | 0.003 | 0.715 | 0.011 | 0.147 | 0.023 | 0.002 |
| (0.001; 0.031) | (0.004; 0.035) | (-0.017; 0.012) | (-0.013; 0.018) | (-0.004; 0.027) | (0.008; 0.038) | |||||||
| Adult BMI (N=97) | 0.018 | 2.65*10-4 | 0.016 | 0.002 | -0.010 | 0.043 | 0.001 | 0.905 | 0.009 | 0.108 | 0.024 | 2.0*10-6 |
| (0.009; 0.028) | (0.006; 0.027) | (-0.020; 0.000) | (-0.010; 0.011) | (-0.002; 0.019) | (0.014; 0.034) | |||||||
| Adult WHR (N=49) | -0.010 | 0.149 | 0.0003 | 0.963 | 0.016 | 0.014 | -0.001 | 0.917 | -0.008 | 0.253 | -0.007 | 0.334 |
| (-0.023; 0.004) | (-0.014; 0.014) | (0.003; 0.029) | (-0.015; 0.013) | (-0.022; 0.006) | (-0.020; 0.007) | |||||||
| VAT/SAT ratio (N=3) * | -0.037 | 0.16 | -0.033 | 0.241 | 0.017 | 0.502 | -0.012 | 0.660 | -0.030 | 0.273 | -0.038 | 0.161 |
| (-0.088; 0.015) | (-0.087; 0.022) | (-0.033; 0.067) | (-0.066; 0.042) | (-0.085; 0.024) | (-0.090; 0.015) | |||||||
| Adult fatty liver (N=5) | -0.033 | 0.198 | -0.021 | 0.432 | 0.025 | 0.318 | 0.148 | 2.09*10-8 | 0.017 | 0.546 | -0.04 | 0.12 |
| (-0.084; 0.017) | (-0.075; 0.032) | (-0.024; 0.074) | (0.097; 0.200) | (-0.036; 0.070) | (-0.091; 0.011) | |||||||
| Adult pericardial fat (N=3) * | -0.045 | 0.147 | 0.004 | 0.905 | 0.063 | 0.038 | -0.007 | 0.834 | 0.100 | 0.002 | -0.024 | 0.434 |
| (-0.106; 0.016) | (-0.061; 0.069) | (0.004; 0.122) | (-0.070; 0.056) | (0.036; 0.164) | (-0.086; 0.037) | |||||||
| rs7185735 (SAT) | 0.197 | 6.00*10-6 | 0.110 | 0.018 | -0.186 | 1.20*10-5 | 0.041 | 0.368 | 0.006 | 0.900 | 0.160 | 3.71*10-4 |
| (0.112; 0.283) | (0.019; 0.201) | (-0.103; -0.269) | (-0.132; 0.049) | (-0.086; 0.098) | (0.072; 0.247) | |||||||
| rs2123685 (SAT female) | -0.034 | 0.788 | -0.138 | 0.298 | -0.103 | 0.397 | 0.193 | 0.132 | 0.085 | 0.621 | 0.034 | 0.787 |
| (-0.279; 0.212) | (-0.399; 0.122) | (-0.341; 0.136) | (-0.059; 0.445) | (-0.192; 0.322) | (-0.212; 0.280) | |||||||
| rs2842895 (VAT and VATadjBMI) | 0.035 | 0.449 | 0.031 | 0.525 | -0.005 | 0.917 | 0.022 | 0.642 | 0.082 | 0.092 | 0.050 | 0.283 |
| (-0.055; 0.124) | (-0.064; 0.126) | (-0.092; 0.082) | (-0.115; 0.071) | (-0.013; 0.177) | (-0.041; 0.141) | |||||||
| rs10060123 (VAT and VATadjBMI female) | -0.027 | 0.596 | -0.066 | 0.216 | -0.047 | 0.337 | 0.049 | 0.351 | -0.078 | 0.142 | -0.040 | 0.433 |
| (-0.125; 0.072) | (-0.170; 0.039) | (-0.142; 0.049) | (-0.054; 0.151) | (-0.182; 0.026) | (-0.140; 0.060) |
Analyses were performed in children with complete data on genetic variants and covariates and at least one outcome under study
Values are linear regression coefficients for models adjusted for sex, age, and the first four principal components and represent the difference in standard deviation scores of the outcome measures for each additional average risk allele in the risk scores.
Bold values represent nominally significant outcomes
Unweighted risk scores since no information on the effect estimates was available. Bold values represent significant outcomes after Bonferroni correction for 10 analyses (P<0.005)
The highest variance explained for the MRI fat measures was 2.2%, for the adult liver risk score with liver fat fraction in the full group. This was even higher for girls (3.1%) (Supplemental material: Tables S6, S7). A power calculation showed limited power to detect small effect estimates. The smallest effect that could be detected per additional risk allele with 80% power was 0.013 for the adult BMI risk score (N=1 993) (Supplemental material: Table S2).
Discussion
In this study a higher childhood BMI genetic risk score was associated with a higher SAT and VAT. The adult BMI genetic risk score was additionally associated with a lower VAT/SAT ratio. A higher adult liver fat risk score was associated with a higher liver fat fraction. Rs7185735, previously associated with SAT in adults, was associated with SAT and VAT/SAT ratio. The associations of the adult BMI risk score with SAT and VAT, and of the liver fat risk score with liver fat fraction were found in both boys and girls. The association of the adult BMI risk score with VAT/SAT ratio remained significant in boys only, whereas stratification by sex revealed an association for the pericardial risk score with pericardial fat mass in girls only. The associations of rs7185735 (SAT) with SAT and VAT/SAT ratio were significant in girls only.
Interpretation of main findings
Childhood overweight and obesity are risk factors for later cardiometabolic disease (ref.1,2,3,4). Previous studies have shown, that SNPs associated with BMI in adulthood already exert their effects during childhood, although sometimes smaller or even in the opposite direction (ref.20,21,22,40). In addition to BMI the distribution and storage of fat in specific locations may also contribute to the cardiometabolic risk (ref.5). Understanding the pathophysiology of body fat distribution from early life onwards may give insight into the mechanisms underlying cardiometabolic disease. The SNPs identified specifically for adult WHR, SAT, VAT, liver fat, and pericardial fat may also play a role in the distribution of body fat from early life onwards. To the best of our knowledge, our study is the first to investigate associations of adult body fat SNPs with childhood fat distribution assessed by MRI.
In this study we observed that the childhood and adult risk scores for BMI, a general adiposity measure, were associated with both SAT and VAT, but not with the more specific liver fat fraction or pericardial fat mass. This is not surprising, as a higher BMI is often accompanied by an increased SAT and/or VAT (ref.6). A stronger association was observed for the adult BMI risk score with BMI than with SAT and VAT, suggesting that not all SNPs identified to play a role in BMI necessarily play a role in SAT and/or VAT and that these represent different phenotypes. The risk of cardiometabolic disease is not the same for SAT and VAT. SAT is deemed less pathogenic than VAT (ref.41). The adult BMI risk score was additionally associated with VAT/SAT ratio. The direction of effect was opposite to that expected, suggesting that a genetic risk for increased BMI is associated with a lower VAT/SAT ratio. Possibly this inversed effect is caused by the stronger association of the adult BMI risk score with SAT than with VAT. This is also reflected in the analyses of SATadjVAT and VATadjSAT, where the adult BMI risk score showed an association with SATadjVAT, but not with VATadjSAT. It is known that SNPs in BMI-associated genetic regions, for example the FTO and MC4R regions, may have opposite or null effects on BMI in early childhood (ref.42). This is supported by the fact that the childhood BMI risk score was not associated with VAT/SAT ratio in our dataset, although we had limited power to detect a small effect size. Both BMI risk scores were associated with BMI, which is in line with previous results in younger children (ref.22). This indicates that the effect of the risk score on overall BMI is positive at this age, but that the relative effects on specific sites of fat accumulation in children may differ from those in adults.
The use of BMI as an overall measure of adiposity does not take into account body fat accumulation in specific locations (ref.5,7). The exact location and extent of fat accumulation in the body may provide a more precise determination of metabolic risk (ref.5). Waist-hip ratio is considered more representative than BMI for abdominal adiposity, which includes SAT and VAT (ref.38). We did not observe associations of the adult WHR risk score with the childhood adiposity measures, although the association with VAT/SAT ratio was nominally significant. Contrary to our expectations, we did not observe any associations for the VAT/SAT ratio risk score with any of the MRI measures. This may be because the effect of the SNPs included in this score is null in childhood, because our power to detect a small effect size was limited, or because only an unweighted risk score was available for this phenotype. However, the difference in effect estimates between weighted and unweighted risk scores was shown to be small in most analyses, except for the childhood BMI risk score. Therefore, we do not expect the use of the unweighted risk score to have a strong effect on the results. A higher adult fatty liver risk score was associated with an increased liver fat fraction indicating that at least some of the SNPs in the risk score affect liver fat fraction from childhood onwards. We could not draw this conclusion for the adult pericardial fat risk score with any of the MRI measures, which may again be due to the risk score being unweighted, the relatively young age of the children or limited power. After adjusting the outcomes for BMI the associations of the BMI risk scores with the abdominal fat measures were no longer present suggesting that the BMI associated SNPs affect overall fat accumulation and may not represent fat accumulation in these specific sites. By adjusting our outcomes for BMI we may also have lost some power which possibly hampered our ability to detect small effect sizes. We also found associations of the adult fatty liver risk score with liver fat fraction only and of the adult pericardial fat risk score with pericardial fat only, indicating that these risk scores seem to affect fat accumulation specifically in the liver and pericardium already in childhood and that these phenotypes are established via biological pathways distinct from those involved in more general adiposity measures. This is in line with the correlations between the genetic risk scores of the general adiposity measures. The weak but significant correlation of the VAT/SAT ratio and fatty liver risk scores is likely caused by one overlapping locus indicating some shared genetic background, which is biologically plausible but not reflected in the associations with the phenotypes (ref.43).
Multiple previous GWAS have identified SNPs for SAT and VAT (ref.11,27). For the current study we used the largest GWAS on SAT and VAT to date which revealed four separate SNPs for these phenotypes (ref.27). Rs7185735, associated with SAT in adults, was also associated with SAT and VAT/SAT ratio in children. After adjusting our outcomes for BMI the association with SAT remained but showed a lower effect estimate. The association with VAT/SAT ratio attenuated and remained borderline significant. Rs7185735 is located in the genetic region coding for FTO. This region was the first robustly identified region associated with BMI, which is also reflected with the observed association of rs7185735 with BMI (ref.44). More recently, it was suggested that SNPs located in FTO actually influence the expression of IRX3 and IRX5 that are involved in adipocyte lipid accumulation (ref.45,46).
Sex is associated with body fat distribution (ref.38,39). To examine this in more detail, we stratified our analyses on sex. Results showed a difference for the associations of the adult BMI risk score with VAT/SAT ratio, which remained in boys only, and for rs7185735 (SAT) which seemed to affect SAT, VAT/SAT ratio, and BMI in girls only. Previous work has shown sexual dimorphism for adiposity measures such as an increased VAT/SAT ratio, but this was not observed in children below the age of 16 (ref.47). We also showed that the association of the adult BMI risk score with VAT/SAT ratio in the full group was driven by the association in boys. The associations for the pericardial fat risk score with pericardial fat, and for rs7185735 (SAT) with VAT/SAT ratio were present in girls only. No associations were found for the individual SNPs identified by GWAS for VAT, or SAT and VAT in girls only. This may be due to limited power or because these specific SNPs have little or no effect in children. We also observed slightly higher effect estimates for some of our associations in either boys or girls, indicating that some adiposity associated SNPs may affect the accumulation of body fat differently in boys and girls.
Further research should be performed to examine whether the current associations can be replicated in other cohorts. Larger study populations may reveal additional associations. A larger study population will also provide more power to be able to investigate which individual SNPs in the risk scores are specifically affecting certain measures of body fat distribution.
Methodological considerations
Genetic information was available in 63% of our total sample size. Children without genetic information had a slightly lower VAT (p-value=0.001), and pericardial fat mass (p-value<0.001). We consider it unlikely that these differences have influenced our results. We did not find any differences regarding the other adiposity outcomes (p-value>0.05) (data not shown). Children without MRI measurements had a higher BMI and lower height than children with MRI measurements (p-value<0.05). Both children without genetic data and children without MRI measurements had a slightly lower gestational age at birth and a lower socio-economic status (p-value<0.05) than children with these data. These differences might reduce the generalizability of our findings. Not all SNPs were available in our GWAS dataset. We used a limited number of proxy SNPs for both BMI and the WHR risk scores in very high linkage disequilibrium (LD). Given the high number of SNPs available and the high LD for the proxies, both risk scores are considered a good representation of the original set of SNPs (ref.20,21,26). Although our population was relatively large, power may still have been limited, therefore our (lack of) findings with some of the adiposity outcomes should be interpreted with caution.
Conclusion
Our results suggest that genetic variants associated with childhood and adult BMI, adult body fat distribution, liver and pericardial fat already affect body fat distribution in childhood. We also found that adiposity associated genetic variants may regulate the distribution of fat in the body differently in boys and girls already before puberty.
Supplementary Material
Supplementary information is available at The International Journal of Obesity’s website.
Acknowledgements
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst and Artsenlaboratorium Rijnmond (STAR), Rotterdam. We gratefully acknowledge the contribution of participating mothers, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The generation and management of GWAS genotype data for the Generation R Study were done at the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, The Netherlands. We would like to thank Karol Estrada, Dr. Tobias A. Knoch, Anis Abuseiris, Luc V. de Zeeuw, and Rob de Graaf, for their help in creating GRIMP, BigGRID, MediGRID, and Services@MediGRID/D-Grid, (funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G) for access to their grid computing resources. We thank Mila Jhamai, Manoushka Ganesh, Pascal Arp, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters for their help in creating, managing and QC of the GWAS database. Also, we thank Karol Estrada for their support in creation and analysis of imputed data.
Funding
The general design of Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Netherlands Organisation for Scientific Research (NWO), the Ministry of Health, Welfare and Sport and the Ministry of Youth and Families. This research also received funding from the European Union’s Seventh Framework Programme (FP7/2007–2013), project EarlyNutrition under grant agreement no. 289346. VWJ received an additional grant from the Netherlands Organization for Health Research and Development (VIDI 016.136.361) and a Consolidator Grant from the European Research Council (ERC-2014-CoG-648916). JFF has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 633595 (DynaHEALTH). The study was supported by funding from the European Union’s Horizon 2020 research and innovation programme (733206, LIFECYCLE).
Abbreviations
- BMI
body mass index
- CI
Confidence interval
- GWAS
Genome-wide association studies
- Liver fat percentage adjBMI
Liver fat percentage adjusted for body mass index
- MRI
Magnetic resonance imaging
- Pericardial fat mass adjBMI
Pericardial fat mass adjusted for body mass index
- SAT
Subcutaneous adipose tissue
- SATadjBMI
Visceral adipose tissue adjusted for body mass index
- SATadjVAT
Subcutaneous adipose tissue adjusted for visceral adipose tissue
- SD
Standard deviation
- SDS
Standard deviation scores
- SNP
Single nucleotide polymorphism
- VAT
Visceral adipose tissue
- VATadjBMI
Visceral adipose tissue adjusted for body mass index
- VATadjSAT
Visceral adipose tissue adjusted for subcutaneous adipose tissue
- VAT/SAT ratio adjBMI
VAT/SAT ratio adjusted for body mass index
Footnotes
Conflict of Interest
None.
References
- 1.World Health Organisation. Obesity and Overweight. Fact sheet N°311. [accessed January 2017]; Available from: http://www.who.int/mediacentre/factsheets/fs311/en/
- 2.Reilly J, Methven E, McDowell Z, Hacking B, Alexander D, Stewart L, et al. Health consequences of obesity. Arch Dis Child. 2003;88:748–752. doi: 10.1136/adc.88.9.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker J, Olsen L, Sorensen T. Childhood body-mass index and the risk of coronary heart disease in adulthood. N Engl J Med. 2007;357:2329–2337. doi: 10.1056/NEJMoa072515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis C, McTigue K, Burke L, Poirier P, Eckel R, Howard B, et al. Mortality, health outcomes, and body mass index in the overweight range: a science advisory from the American Heart Association. Circulation. 2009;119:3263–3271. doi: 10.1161/CIRCULATIONAHA.109.192574. [DOI] [PubMed] [Google Scholar]
- 5.Tchernof A, Després J. Pathophysiology of human visceral obesity: an update. Physiol Rev. 2013;(1) doi: 10.1152/physrev.00033.2011. 2013. [DOI] [PubMed] [Google Scholar]
- 6.Bays H, González-Campoy J, Bray G, Kitabchi A, Bergman D, Schorr A, et al. Pathogenic potential of adipose tissue and metabolic consequences of adipocyte hypertrophy and increased visceral adiposity. Expert Rev Cardiovasc Ther. 2008;6:343–368. doi: 10.1586/14779072.6.3.343. [DOI] [PubMed] [Google Scholar]
- 7.Després J, Lemieux I, Bergeron J, Pibarot P, Mathieu P, Larose E, et al. Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler Thromb Vasc Biol. 2008;28:1039–1049. doi: 10.1161/ATVBAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- 8.Tang L, Zhang F, Tong N. The association of visceral adipose tissue and subcutaneous adipose tissue with metabolic risk factors in a large population of Chinese adults. Clin Endocrinol. 2016;85:46–53. doi: 10.1111/cen.13013. [DOI] [PubMed] [Google Scholar]
- 9.Hubers M, Geisler C, Plachta-Danielzik S, Muller MJ. Association between individual fat depots and cardio-metabolic traits in normal- and overweight children, adolescents and adults. Nutr Diabetes. 2017;7:e267. doi: 10.1038/nutd.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin K, Syme C, Abrahamowicz M, Leonard GT, Richer L, Perron M, et al. Routine clinical measures of adiposity as predictors of visceral fat in adolescence: a population-based magnetic resonance imaging study. PLoS One. 2013;8:e79896. doi: 10.1371/journal.pone.0079896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox C, Liu Y, White C, Feitosa M, Smith A, Heard-Costa N, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacks H, Fain J. Human epicardial adipose tissue: a review. Am Heart J. 2007;153:907–917. doi: 10.1016/j.ahj.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Fox C, Hickson D, Bidulescu A, Carr J, Taylor H. Fatty liver, abdominal visceral fat, and cardiometabolic risk factors: the Jackson Heart Study. Arterioscler Thromb Vasc Biol. 2011;31:2715–2722. doi: 10.1161/ATVBAHA.111.234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosito G, Massaro J, Hoffmann U, Ruberg F, Mahabadi A, Vasan R, et al. Pericardial fat, visceral abdominal fat, cardiovascular disease risk factors, and vascular calcification in a community-based sample: the Framingham Heart Study. Circulation. 2008;117:605–613. doi: 10.1161/CIRCULATIONAHA.107.743062. [DOI] [PubMed] [Google Scholar]
- 15.Speliotes E, Massaro J, Hoffmann U, Vasan R, Meigs J, Sahani D, et al. Fatty liver is associated with dyslipidemia and dysglycemia independent of visceral fat: the Framingham Heart Study. Hepatology. 2010;51:1979–1987. doi: 10.1002/hep.23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérusse L, Després J, Lemieux S, Rice T, Rao D, Bouchard C. Familial aggregation of abdominal visceral fat level: results from the Quebec family study. Metabolism. 1996;45:378–382. doi: 10.1016/s0026-0495(96)90294-2. [DOI] [PubMed] [Google Scholar]
- 17.Fox C, White C, Lohman K, Heard-Costa N, Cohen P, Zhang Y, et al. Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet. 2012;8:e1002705. doi: 10.1371/journal.pgen.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loomba R, Schork N, Chen C, Bettencourt R, Bhatt A, Ang B, et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149:1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speliotes E, Willer C, Berndt S. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke A, Kahali B, Berndt S, Justice A, Pers T, Day F, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felix J, Bradfield J, Monnereau C, van der Valk R, Stergiakouli E, Chesi A, et al. Genome-wide association analysis identifies four new susceptibility loci for childhood body mass index. Human Molecular Genetics. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monnereau C, Vogelezang S, Kruithof C, Jaddoe V, Felix J. Associations of genetic risk scores based on adult adiposity pathways with childhood growth and adiposity measures. BMC Genet. 2016;17:120. doi: 10.1186/s12863-016-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu H, Nayak K, Goran M. Assessment of abdominal adipose tissue and organ fat content by magnetic resonance imaging. Obes Rev. 2011;12:e504–e515. doi: 10.1111/j.1467-789X.2010.00824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kooijman M, Kruithof C, van Duijn C, Duijts L, Franco O, van IJzendoorn M, et al. The Generation R Study: design and cohort update 2017. Eur J Epidemiol. 2016;31:1243–1264. doi: 10.1007/s10654-016-0224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speliotes E, Yerges-Armstrong L, Wu J, Hernaez R, Kim L, Palmer C, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shungin D, Winkler T, Croteau-Chonka D, Ferreira T, Locke A, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu A, Deng X, Fisher V, Drong A, Zhang Y, Feitosa M, et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat Genet. 2016;49:125–130. doi: 10.1038/ng.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruithof C, Kooijman M, van Duijn C, Franco O, de Jongste J, Klaver C, et al. The Generation R Study: Biobank update 2015. Eur J Epidemiol. 2014;29:911–927. doi: 10.1007/s10654-014-9980-6. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Willer C, Ding J, Scheet P, Abecasis G. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuster A, Patlas M, Pinthus J, Mourtzakis M. The clinical importance of visceral adiposity: a critical review of methods for visceral adipose tissue analysis. Br J Radiol. 2012;85:1–10. doi: 10.1259/bjr/38447238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas E, Fitzpatrick J, Malik S, Taylor-Robinson S, Bell J. Whole body fat: content and distribution. Progress in nuclear magnetic resonance spectroscopy. 2013;73:56–80. doi: 10.1016/j.pnmrs.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Mitra S, Fernandez-Del-Valle M, Hill J. The role of MRI in understanding the underlying mechanisms in obesity associated diseases. Biochimica et biophysica acta. 2016 doi: 10.1016/j.bbadis.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Reeder S, Cruite I, Hamilton G, Sirlin C. Quantitative Assessment of Liver Fat with Magnetic Resonance Imaging and Spectroscopy. J Magn Reson Imaging. 2011;34 doi: 10.1002/jmri.22580. spcone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gishti O, Gaillard R, Manniesing R, Abrahamse-Berkeveld M, van der Beek E, Heppe D, et al. Fetal and infant growth patterns associated with total and abdominal fat distribution in school-age children. J Clin Endocrinol Metab. 2014;99:2557–2566. doi: 10.1210/jc.2013-4345. [DOI] [PubMed] [Google Scholar]
- 36.Wells JC, Cole TJ, steam As. Adjustment of fat-free mass and fat mass for height in children aged 8 y. Int J Obes Relat Metab Disord. 2002;26:947–952. doi: 10.1038/sj.ijo.0802027. [DOI] [PubMed] [Google Scholar]
- 37.Keijzer-Veen MG, Euser AM, van Montfoort N, Dekker FW, Vandenbroucke JP, Van Houwelingen HC. A regression model with unexplained residuals was preferred in the analysis of the fetal origins of adult diseases hypothesis. J Clin Epidemiol. 2005;58:1320–1324. doi: 10.1016/j.jclinepi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Schleinitz D, Böttcher Y, Blüher M, Kovacs P. The genetics of fat distribution. Diabetologia. 2014;57:1276–1286. doi: 10.1007/s00125-014-3214-z. [DOI] [PubMed] [Google Scholar]
- 39.Taylor R, Grant A, Williams S, Goulding A. Sex Differences in Regional Body Fat Distribution From Pre- to Postpuberty. Obesity (Silver Spring) 2010;18:1410–1416. doi: 10.1038/oby.2009.399. [DOI] [PubMed] [Google Scholar]
- 40.Vogelezang S, Monnereau C, Gaillard R, Renders C, Hofman A, Jaddoe V, et al. Adult adiposity susceptibility loci, early growth and general and abdominal fatness in childhood: the Generation R Study. Int J Obes. 2015;39:1001–1009. doi: 10.1038/ijo.2015.12. [DOI] [PubMed] [Google Scholar]
- 41.Blüher M. The distinction of metabolically 'healthy' from 'unhealthy' obese individuals. Curr Opin Lipidol. 2010;21:38–43. doi: 10.1097/MOL.0b013e3283346ccc. [DOI] [PubMed] [Google Scholar]
- 42.Hardy R, Wills A, Wong A, Elks C, Wareham N, Loos R, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19:545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frayling T, Ong K. Piecing together the FTO jigsaw. Genome Biol. 2011;12:104. doi: 10.1186/gb-2011-12-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claussnitzer M, Dankel S, Kim K, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brambilla P, Bedogni G, Moreno L, Goran M, Gutin B, Fox K, et al. Crossvalidation of anthropometry against magnetic resonance imaging for the assessment of visceral and subcutaneous adipose tissue in children. Int J Obes. 2006;30:23–30. doi: 10.1038/sj.ijo.0803163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.