Abstract
We investigated the genetic, phenotypic, and interferon status of 46 patients from 37 families with neurological disease due to mutations in ADAR1. The clinicoradiological phenotype encompassed a spectrum of Aicardi–Goutières syndrome, isolated bilateral striatal necrosis, spastic paraparesis with normal neuroimaging, a progressive spastic dystonic motor disorder, and adult-onset psychological difficulties with intracranial calcification. Homozygous missense mutations were recorded in five families. We observed a p.Pro193Ala variant in the heterozygous state in 22 of 23 families with compound heterozygous mutations. We also ascertained 11 cases from nine families with a p.Gly1007Arg dominant-negative mutation, which occurred de novo in four patients, and was inherited in three families in association with marked phenotypic variability. In 50 of 52 samples from 34 patients, we identified a marked upregulation of type I interferon-stimulated gene transcripts in peripheral blood, with a median interferon score of 16.99 (interquartile range [IQR]: 10.64–25.71) compared with controls (median: 0.93, IQR: 0.57–1.30). Thus, mutations in ADAR1 are associated with a variety of clinically distinct neurological phenotypes presenting from early infancy to adulthood, inherited either as an autosomal recessive or dominant trait. Testing for an interferon signature in blood represents a useful biomarker in this context.
Keywords: Aicardi–Goutières syndrome, bilateral striatal necrosis, spastic paraparesis, dystonia, idiopathic basal ganglia calcification
Introduction
Adenosine deaminases acting on RNA (ADARs) catalyze the hydrolytic deamination of adenosine to inosine in double-stranded RNA, and thereby potentially alter the information content and structure of cellular RNAs.1 ADAR1 is encoded by a single-copy gene that maps to human chromosome 1q21 and is present in two main isoforms in mammalian cells. In mice, a loss of ADAR1 activity leads to a dramatic upregulation of interferon-stimulated gene (ISG) expression, which is dependent on the editing activity of ADAR1 and specific to the interferon-inducible full-length p150 isoform of the protein.2–4
In 2012, we reported mutations in ADAR1 to cause a phenotype consistent with the infantile encephalopathy Aicardi–Goutières syndrome (AGS), and demonstrated that, similar to the ADAR1 null mouse, the mutant genotype was associated with an upregulation of type I interferon signaling.5 Further to this, in 2014, we described both bilateral striatal necrosis (BSN), sometimes occurring after a trivial childhood infection, and otherwise nonsyndromic, slowly progressive spastic paraparesis associated with normal intellect occur due to ADAR1 dysfunction, again in association with the enhanced expression of type I interferon-induced gene transcripts.6–8 These data indicate that neurological disease can occur through inappropriate induction of the innate immune system by self-derived nucleic acids.
Here, we present an update of our experience of screening for ADAR1 mutations, describing the clinical, radiological, molecular, and interferon biomarker characteristics of a cohort of 46 patients from 37 families with neurological dysfunction due to mutations in ADAR1.
Materials and Methods
Patients and Methods
We ascertained clinical and molecular data through direct contact and/or via collaborating physicians. The study was approved by the Leeds (East) Research Ethics Committee (reference number 10/H1307/132), and the Comité de Protection des Personnes (ID-RCB/EUDRACT: 2014-A01017-40).
A diagnosis of AGS was suggested by characteristic clinical and neuroimaging features including cerebral atrophy, white matter disease, and intracranial calcification.9 BSN was diagnosed in the context of an acute or subacute onset of a dystonic/rigid motor disorder associated with magnetic resonance imaging features of bilateral striatal signal change with or without swelling. Spastic paraparesis/tetraparesis and spastic dystonia were diagnosed according to clinical signs, in the presence of either normal neuroimaging or mild nonspecific changes sometimes including calcification of the basal ganglia. Assessment of the motor and communication status of patients over the age of 1 year was made using the Gross Motor Function Classification System (GMFCS),10 the Manual Ability Classification System (MACS),11 and the Communication Function Classification System (CFCS).12
Mutational Analysis
Primers were designed to amplify the coding exons of ADAR1 (Supplementary Table S1, online-only). Purified polymerase chain reaction (PCR) amplification products were sequenced using BigDye terminator chemistry and an ABI 3130 DNA sequencer. Mutation description is based on the reference cDNA sequence NM_001111.4, with nucleotide numbering beginning from the first A in the initiating ATG codon. Variants were assessed using the in silico programs SIFT (http://sift.jcvi.org) and Polyphen2 (http://genetics.bwh.harvard.edu/pph2/), and population allele frequencies obtained from the ExAC (http://exac.broadinstitute.org) and gnomAD (http://gnomad.broadinstitute.org) databases.
Interferon Score
Whole blood was collected into PAXgene tubes, total RNA extracted using a PreAnalytix RNA isolation kit and RNA concentration assessed using a spectrophotometer (FLUOstar Omega, Labtech). Quantitative reverse transcription PCR (qPCR) analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems), and cDNA derived from 40 ng total RNA. Using TaqMan probes for IFI27 (Hs01086370_m1), IFI44L (Hs00199115_m1), IFIT1 (Hs00356631_g1), ISG15 (Hs00192713_m1), RSAD2 (Hs01057264_m1), and SIGLEC1 (Hs00988063_m1), the relative abundance of each target transcript was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18S (Hs999999001_s1), and assessed with the Applied Biosystems StepOne Software v2.1 and DataAssist Software v.3.01. For each of the six probes, individual data were expressed relative to a single calibrator. Relative quantification is equal to 2−ΔΔCt that is, the normalized fold change relative to the control data. The median fold change of the 6 genes compared with the median of 29 previously collected healthy controls is used to create an interferon score for each individual, with an abnormal interferon score being defined as greater than +2 standard deviations above the mean of the control group, that is, 2.466.
Results
Molecular Data
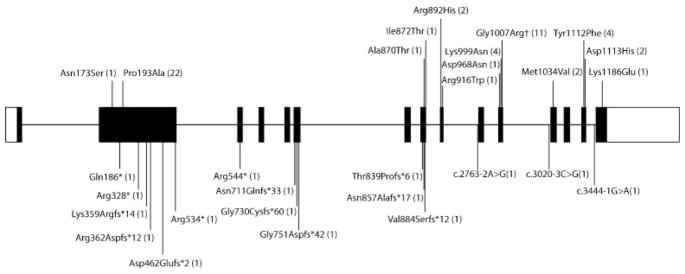
We collected data on 46 patients from 37 families of pan-ethnic origin with either biallelic mutations in ADAR1 (28 families) or the single known dominant-negative mutation p. Gly1007Arg (nine families) (Table 1; Fig. 1). In four families, the p.Gly1007A mutation was considered to have occurred de novo, while in three families, inheritance was confirmed or inferred (two paternal half-siblings born to an unaffected father unavailable for testing), with somatic mosaicism recorded in one case. In two families, inheritance could not be determined because DNA from both parents was not available. We observed three distinct homozygous mutations in five families (two families each sharing the same mutation), in four of which the parents were knowingly related. All of these mutations were missense. Of 23 families with compound heterozygous mutations, 22 carried the p. Pro193Ala mutation on one allele. In 13 of 22 families segregating this p.Pro193Ala substitution, the second molecular lesion was a null or splicing variant.
Table 1.
Family structure, ethnicity, and molecular data of ascertained ADAR1 mutation-positive cases
| AGS number |
Individuals tested |
Consanguinity | Ethnicity | cDNA | Protein | Allelic status |
Inheritance | SIFT | Polyphen2 | CADD Phred |
ExAc frequency |
gnomAD frequency |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGS081 | 3A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2675G>A | p.Arg892His | Het | Paternally inherited | Deleterious 0.01 | Probably damaging 1.000 | 35 | Novel | 1/252010 | ||||
| AGS093 | 1A, M, F | No | Italian | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2608G>A | p.Ala870Thr | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel | ||||
| AGS107 | 2A, M, F | Yes | Pakistani | c.3337G>C | p.Asp1113His | Hom | Both parents het | Deleterious 0.02 | Probably damaging 1.000 | 33 | Novel | Novel |
| AGS150 | 1A, M, F | No | Brazilian | c.3019G>A | p.Gly1007Arg | Het | De novo (paternity confirmed) | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS219 | 1A | Yes | Pakistani | c.3335A>T | p.Tyr1112Phe | Hom | Not tested | Tolerated 0.17 | Probably damaging 1.000 | 33 | Novel | Novel |
| AGS228 | 1A, M, F | No | Indian | c.2997G>T | p.Lys999Asn | Hom | Both parents het | Deleterious 0.03 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS251 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2615T>C | p.Ile872Thr | Het | Paternally inherited | Deleterious 0.01 | Probably damaging 1.000 | 26.9 | 1/121342 | 1/252270 | ||||
| AGS327 | 1A, M, F | No | Italian/ Hispanic | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.1076_1080del | p.Lys359Argfs*14 | Het | Paternally inherited | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS430 | 2A, M, F | No | Spanish | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2675G>A | p.Arg892His | Het | Paternally inherited | Deleterious 0.01 | Probably damaging 1.000 | 35 | Novel | 1/252010 | ||||
| AGS474 | 1A, M, F | No | White European | c.3019G>A | p.Gly1007Arg | Het | De novo (paternity confirmed) | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS530 | 2A, M | No | White European | c.3019G>A | p.Gly1007Arg | Het | Presumed inherited from asymptomatic Father | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS550 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2565_2568del | p.Asn857Alafs*17 | Het | Maternally inherited | Frameshift | Frameshift | Frameshift | Novel | 1/30224 | ||||
| AGS567 | 1A, M, F | No | Greek/ Lebanese | c.518A>G | p.Asn173Ser | Het | Paternally inherited | N/A | Probably damaging 0.999 | 24.3 | 34/121366 | 144/282658 1 hom |
| c.2515del | p.Thr839Profs*6 | Het | Maternally inherited | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS582 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2647_2648dup | p.Val884Serfs*12 | Het | Not known | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS663 | 2A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.1630C>T | p.Arg544* | Het | Maternally inherited | Stop | Stop | Stop | Novel | 2/252366 | ||||
| AGS679 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.3556A>G | p.Lys1186Glu | Het | Not known | Tolerated 0.11 | Probably damaging 0.999 | 31 | Novel | Novel | ||||
| AGS699 | 1A, M, F | No | White European | c.3019G>A | p.Gly1007Arg | Het | De novo (genotyping not undertaken) | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS703 | 1A | No | Asian/ White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.3100A>G | p.Met1034Val | Het | Not known | Deleterious 0.03 | Possibly damaging 0.760 | 25.8 | Novel | Novel | ||||
| AGS720 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2250del | p.Gly751Aspfs*42 | Het | De novo (genotyping not undertake n) | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS759 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2902G>A | p.Asp968Asn | Het | Maternally inherited | Tolerated 0.06 | Probably damaging 1.000 | 34 | Novel | Novel | ||||
| AGS765 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.556C>T | p.Gln186* | Het | Not known | Stop | Stop | Stop | Novel | Novel | ||||
| AGS788 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.1386_1390del | p.Asp462Glufs*2 | Het | De novo (paternity confirmed) | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS810 | 1A, MA, F | No | White European | c.3019G>A | p.Gly1007Arg | Het | Inherited from symptomatic mother | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS943 | 1A, M, F | No | North African | c.3019G>A | p.Gly1007Arg | Het | De novo (genotyping not undertaken) | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS1115 | 1A, M, F | Yes | Persian | c.2997G>T | p.Lys999Asn | Hom | Both parents het | Deleterious 0.03 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS1170 | 1A | No | Asian | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.3100A>G | p.Met1034Val | Het | Not known | Deleterious 0.03 | Possibly damaging 0.760 | 25.8 | Novel | Novel | ||||
| AGS1315 | 2A, M, F (mosaic) | No | White European | c.3019G>A | p.Gly1007Arg | Het | Father mosaic | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS1456 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.3020–3C>G | Splicing | Het | Not known | Splicing | Splicing | Splicing | Novel | Novel | ||||
| AGS1507 | 1A, M, F | No | Asian/ White European | c.577C>G | p.Pro193Ala | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2763–2A>G | Splicing | Het | Paternally inherited | Splicing | Splicing | Splicing | Novel | Novel | ||||
| AGS1537 | 1A | No | White European | c.3019G>A | p.Gly1007Arg | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
| AGS1542 | 2A, M, F | Yes | Asian | c.3335A>T | p.Tyr1112Phe | Hom | Both parents het | Tolerated 0.17 | Probably damaging 1.000 | 33 | Novel | Novel |
| AGS1824 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.1084_1085del | p.Arg362Aspfs*12 | Het | Maternally inherited | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS1980 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2130dupC | p.Asn711Glnfs*33 | Het | Not known | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS1989 | 1A, M, F | No | South American | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2187_2198delinsGT | p.Gly730Cysfs*60 | Het | Maternally inherited | Frameshift | Frameshift | Frameshift | Novel | Novel | ||||
| AGS2007 | 1A | No | White European | c.577C>G | p.Pro193Ala | Het | Not known | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.982C>T | p.Arg328* | Het | Not known | Stop | Stop | Stop | Novel | 1/252210 | ||||
| AGS2009 | 1A, M, F | No | White European | c.577C>G | p.Pro193Ala | Het | Paternally inherited | Deleterious 0 | Probably damaging 1.000 | 23.9 | 260/ 121402 | 602/282636 1 hom |
| c.2746C>T | p.Arg916Trp | Het | Maternally inherited | Deleterious 0 | Probably damaging 1.000 | 35 | Novel | Novel | ||||
| AGS2010 | 1A, M | No | Hispanic | c.3019G>A | p.Gly1007Arg | Het | M WT, F not tested | Deleterious 0 | Probably damaging 1.000 | 34 | Novel | Novel |
Abbreviations: A, affected; F, father; Het, heterozygous; Hom, homozygous; M, mother; MA, mother affected; WT, wild type.
Note: Nucleotide numbering based on transcript ADAR1 NM_001111.4. ExAc browser Beta version accessed on October 28, 2016 (http://exac.broadinstitute.org), gnomAD browser β version accessed on October 28, 2016 (http://gnomad.broadinstitute.org).
Fig. 1.
Schematic of ADAR1 gene showing mutations (according to protein nomenclature) ascertained in the present study. Missense and nonsense mutations are annotated above and below, respectively. Numbers in brackets indicate the number of families in which each mutation was observed. †Indicates mutation acting as a dominant negative.
Clinical Radiological Phenotype
Clinical radiological characteristics of all patients are summarized in Table 2, and characteristic radiological appearances are summarized in Fig. 2. Median age of disease onset was 14 months (range: birth–30 years). We observed 21 and 25 affected females and males, respectively. Although spasticity and dystonia were common features present in the majority of patients, clinically and radiologically distinct phenotypes could be defined, including classical AGS (15 patients), BSN (16 patients), apparently isolated spastic paraparesis (1 patient)/tetraparesis (2 patients), and a progressive spastic dystonic motor disorder (7 patients). In two of these latter cases, the initial presentation was of isolated lower limb spasticity, with a dystonic component and involvement of the upper limbs only becoming evident several years later. Four patients demonstrated radiological features of both AGS and BSN. The mother of a child with an AGS presentation was diagnosed at the age of 30 years with subtle psychological features and marked intracranial calcification. We identified three patients with significant neurological disease (a spastic/dystonic phenotype) in the absence of changes on brain imaging at presentation.
Table 2.
Clinical and radiological data relating to ascertained ADAR1 mutation-positive cases
| AGS num- ber |
Individual | Sex | Develop- mental status prior to onset |
Possible trigger |
Age at
in- itial ascertain- ment |
Features at presenta- tion |
Current age/ age at death (cause were known) |
Progres- sive course |
Status at last con- tact |
Neuroima- ging |
Interferon scores (age, deci- malized years) |
GMFCS | MACS | CFCS | Summary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AGS081 | P1 | F | Delayed | No | 5 mo | DD, dystonia, irritability | Died aged 17 y | Yes | SDT with severe ID | Characteristic of AGS | 24.267 (14.53); 53.356 (15.01); 45.676 (15.78) | V | V | V | AGS |
| P2 | F | Delayed | No | 5 mo | DD, dystonia, irritability, microcephaly | Died aged 23 mo | Yes | SDT with severe ID | Characteristic of AGS | NT | V | V | V | AGS | |
| P3 | M | Diagnosed at birth | No | Neonatal | Raised CSF IFN at birth with transient thrombocytopenia and petechiae | 9 y | Not obvious | SDT with severe ID | Characteristic of AGS | 37.822 (4.82); 21.590 (5.28) | V | V | V | AGS | |
| AGS093 | P1 | M | Delayed | No | 1 mo | DD, irritability, sleep and feeding disturbance | 20 y | Not obvious | SDT with severe ID | Characteristic of AGS and BSN | 25.608 (15.26); 46.665 (16.59) | V | V | V | AGS/BSN |
| AGS107 | P1 | F | Delayed | No | < 7 mo | DD, dystonia, irritability, microcephaly | Died aged 19 y | Not obvious | SDT with severe ID; AIHA | Characteristic of AGS | 64.22 (15.26) | V | V | V | AGS |
| P2 | F | Delayed | No | Neonatal | DD, dystonia, irritability, microcephaly | 14 y | Not obvious | SDT with severe ID; AIHA | Characteristic of AGS | NT | V | V | V | AGS | |
| AGS150 | P1 | F | Mild delay | No | 18 mo | Loss of head control, sitting and speech | 15 y | No | SDT with some ID | Some white matter disease and calcification of GP | 14.69 (10.88) | V | V | V | AGS |
| AGS219 | P1 | M | Delayed | No | < 6 mo | DD, poor head control | Died aged 6 y | Not obvious | SDT with severe ID; AIHA | Characteristic of AGS | NT | V | V | V | AGS |
| AGS228 | P1 | F | Delayed | No | Prenatal | IUGR, thrombocytopenia, HSM | Died aged 10 mo | Not obvious | SDT with severe ID | Characteristic of AGS | NT | < 1 year | < 1 year | < 1 year | AGS |
| AGS251 | P1 | F | Normal | Varicella infection | 9 mo | Loss of skills over a few weeks | 12 y | Not obvious | SDT with some ID; CB | BSN | 28.367 (8.1); 12.301 (9.27) | V | V | IV | BSN |
| AGS327 | P1 | M | Delayed | Possible viral infection (otitis media) | 8 mo | DD, encephalopathy, irritability | 8 y | Not obvious | SDT with severe ID; DSH | AGS with features of BSN | 23.382 (4.07); 9.402 (8.52) | V | V | V | AGS/BSN |
| AGS430 | P1 | F | Delayed | No | < 2 mo | DD, dystonia, irritability, microcephaly | Died aged 6 y | Uncertain | SDT with severe ID | Characteristic of AGS | 8.296 (4.75); 21.538 (5.53) | V | V | V | AGS |
| P2 | F | Delayed | No | < 2 mo | DD, dystonia, irritability, microcephaly | 9 y | Not obvious | SDT with severe ID | Characteristic of AGS | 12.444 (4.75); 14.306 (5.53) | V | V | V | AGS | |
| AGS474 | P1 | M | Normal | Vaccination | 4 mo | Nystagmus, gross and fine motor delay | 8 y | Yes, with worsening respiratory function and overall neurological deterioration | SDT with severe ID | Characteristic of AGS | 20.961 (5.42); 32.319 (5.88); 49.463 (6.02) | V | V | V | AGS |
| AGS530 | P1 | F | Normal | No | 5 y | Subacute loss of skills becoming rigid over a few months | 17 y | Yes | SDT with some understanding | BSN | 12.502 (13.41) | V | V | V | BSN |
| P2 | F | Normal | No | 1 y | Subacute loss of skills becoming rigid over a few months | 29 y | Yes | SDT with some understanding | BSN | 23.385 (26.21) | V | IV | IV | BSN | |
| AGS550 | P1 | M | Normal | D & V | 16 mo | Sudden-onset motor regression | Died aged 9 y (pneumonia) | Yes | SDT with some understanding | BSN | 6.429 (8.39) | V | V | V | BSN |
| AGS567 | P1 | M | Mild delay | Bronchiolitis | 9 mo | Sudden onset motor regression | 6 y | Yes | SDT with moderate ID; DSH | BSN | 36.387 (2.81) | V | V | V | BSN |
| AGS582 | P1 | M | Normal | No | 14 mo | Loss of skills | Died aged 10 y | Yes | SDT with moderate ID | BSN | NT | V | V | V | BSN |
| AGS663 | P1 | M | Normal | URTI | 11 mo | Sudden onset motor regression | 12 y | No | SDT moderate ID | BSN | NT | IV | III | III | BSN |
| P2 | M | Normal | URTI | 11 months | Sudden onset motor regression | Died age 18 years | Not obvious | SDT | BSN | 38.13 (17.53) | V | V | IV | BSN | |
| AGS679 | P1 | F | Normal | Unspecified viral infection | 18 mo | Sudden onset motor regression | 4 y | Yes, then some recovery | Dystonic gait and clumsy hand finger movements; intellectually normal | BSN | 3.802 (1.66) | II | III | III | BSN |
| AGS699 | P1 | M | Normal | No | 2 y | Falling | 8 y | Yes | Major LL spasticity; intellectually normal | Normal | 16.833 (4.91) | II | I | I | SP |
| AGS703 | P1 | M | Mild delay | No | 2 y | Loss of skills over a few weeks | 11 y | Yes | SDT with severe ID | Initially structurally normal MRI; BG calcification noted 2 years later | 20.427 (8.44); 29.817 (8.44) | V | IV | III | SDT |
| AGS720 | P1 | F | Normal | Unspecified viral infection | 18 mo | Rapid loss of skills | 9 y | No | SDT; intellectually normal; DSH | BSN | 12.057 (6.90) | V | V | III | BSN |
| AGS759 | P1 | F | Normal | URTI | 14 mo | Motor regression and speech arrest | 6 y | No | SDT; intellectually normal | Calcification of caudate and putamen | 11.048 (4.09); 18.633 (4.53) | III | II | III | SDT |
| AGS765 | P1 | F | Normal | URTI | 11 mo | Rapid loss of skills | 7 y | No | SDT with some ID | BG signal changes and atrophy with subcortical hypomyelination | NT | IV | V | IV | SDT |
| AGS788 | P1 | F | Normal | URTI/meningitis C vaccination | 15 mo | Acute regression, dystonia, extra-pyramidal movements, orofacial dyskinesia | 3 y | Not obvious | SDT with severe ID | BSN | 1.99 (1.29); 4.59 (2.46) | V | V | V | BSN |
| AGS810 | P1 (son to P2) | M | Mild delay | URTI | 12 mo | Rapid psychomotor regression, axial hypotonia, spastic dystonic tetraparesis | 9 y | Not obvious | SDT with severe ID | Characteristic of AGS | 40.571 (7.13); 14.851 (7.27) | V | V | V | AGS/BSN |
| P2 (mother to P1) | F | Normal | No | 30 y | Pain, fatigue, anxiety, sleep problems | 35 y | Possibly | Normal clinical examination; subtle psychological difficulties | Normal except for BG, WM, and Cb calcification | 25.743 (33.34); 12.836 (33.48) | I | I | I | ICC with psychiatric features | |
| AGS943 | P1 | M | Normal | Vaccination | 22 mo | SP | 13 y | Yes, developing asymmetric dystonia of upper limbs 7 y after initial presentation | SDT; intellectually normal | Some cortical atrophy with BG and WM calcification | 24.753 (11.75); 15.074 (12.11) | I | III | I | SP becoming SDT with preserved intellect |
| AGS1115 | P1 | M | Unknown | No | 4 mo | Hypotonia and dystonia | 2 y | Not obvious | SDT with severe ID | Characteristic of AGS | NT | V | V | V | AGS |
| AGS1170 | P1 | F | Normal | URTI | 9 mo | Acute regression with dystonia necessitating ICU admission | 2 y | Not obvious | SDT with severe ID | Initial bilateral high signal and swelling of BG progressing to extensive WM and cortical atrophy and severely atrophied putamina (no CT) | 17.627 (0.84); 1.158 (0.90); 3.578 (1.23) | V | V | V | AGS/BSN |
| AGS1315 | P1 (brother to P2, son of P3) | M | Normal | No | 2.5 y | ST with normal intellect | 6 y | Fluctuations | ST; intellectually normal; | Normal MRI (at 2 years) and CT (at 4 years) | 10.506 (5.53) | IV | II | I | ST |
| P2 (brother to P1, son of P3) | M | Delayed | No | DD obvious by 1 y | ST and speech delay | 4 y | Yes, age 2.5 y episode of definite regression | ST with severe ID | MRI normal, BG and PV calcification on CT | 17.147 (3.06) | IV | IV | IV | ST | |
| P3 (Father to P1 and P2; mosaic) | M | Always normal | NR | Always normal | Always normal | 31 y | No | Normal | No imaging | 2.692 (30.18) | I | I | I | Normal | |
| AGS1456 | P1 | M | Normal | Otitis media | 15 mo | Lethargy, dystonia, global regression | 17 y | Yes, with intermittent flares of encephalopathy and slowly progressive dystonia | SDT with severe ID; DSH | Mild hyper- intensity of the BG (no CT) | 9.063 (16.68) | V | V | V | SDT |
| AGS1507 | P1 | M | Moderate delay | No | 13 mo | Developmental arrest with onset of generalized dystonia | 9 y | Yes, episode of definite regression at age 4 years | SDT with some ID; DSH | BSN | 8.293 (8.56) | V | V | V | BSN |
| AGS1537 | P1 | F | Delayed | No | 15 mo | Motor delay with spastic tetraparesis | 11 y | No | SDT with some ID; AIHA | BG calcification (CT); normal MRI at age 10 years | 12.865 (10.33) | V | IV | III | SDT |
| AGS1542 | P1 | M | Normal | No | 21 mo | Rapidly progressive SP | 7 y | Yes, with progressive involvement of UL and spastic dystonia | SDT with some ID | Normal (no CT) | 12.24 (6.38); 18.051 (6.43) | IV | III | IV | SP becoming SDT |
| P2 | M | Likely delayed | No | 14 mo | Onset of dystonia and loss of skills | 19 mo | No | SDT with severe ID | No imaging | 7.031 (2.17) | V | V | V | Clinically AGS-like (but no imaging) | |
| AGS1824 | P1 | M | Normal | Unspecified viral infection | 11 mo | Acute regression with dystonia necessitating ICU admission | 5 y | No | SDT with severe ID; CB | BSN with BG calcification | 8.713 (5.55) | V | V | V | BSN |
| AGS1980 | P1 | M | Normal | Febrile illness | 14 mo | Left hemiparesis with loss of ambulation | 2 y | Yes, from uni- to bi- lateral; however, some skills (e.g., crawling, pulling to stand) subsequently reacquired | SDT with some ID | BSN (no CT) | NT | IV | IV | III | BSN |
| AGS1989 | P1 | M | Normal | Otitis media | 12 mo | Tremor and rapid loss of skills | 4 y | No | SDT with severe ID | BSN with BG calcification | NT | V | V | V | BSN |
| AGS2007 | P1 | M | Possible mild delay | Febrile respiratory illness | 15 mo | Developmental regression with loss of crawling and other skills | 3 y | No | SDT with severe ID | Characteristic of AGS | NT | V | V | V | AGS |
| AGS2009 | P1 | F | Normal | MMR and varicella vaccination | 13 mo | Developmental regression with loss of skills | 6 y | No | SDT with severe ID | BSN with BG calcification | NT | IV | IV | IV | BSN |
| AGS2010 | P1 | F | Normal | Vaccination | 6 mo | Lost all acquired skills over 6 mo period | 9 y | No | SDT with severe ID | Some white matter disease and calcification of GP | NT | V | V | V | AGS |
Abbreviations: AGS, Aicardi–Goutières syndrome; AIHA, autoimmune hemolytic anemia; BG, basal ganglia; BSN, bilateral striatal necrosis; CB, chilblains; CFCS, Communication Function Classification System; CSF, cerebrospinal fluid; CT, computed tomography; DD, developmental delay; DSH, dyschromatosis symmetrica hereditaria; D & V, diarrhea and vomiting; GMFCS, Gross Motor Function Classification System; GP, globus pallidus; HSM, hepatosplenomegaly; ICC, intracranial calcification; ICU, intensive care unit; ID, intellectual disability; IFN, interferon; IUGR, intrauterine growth retardation; LL, lower limb; MACS, Manual Ability Classification System; MRI, magnetic resonance imaging; NR, not relevant; NT, not tested; PV, periventricular; SD, spastic dystonia; SDT, spastic dystonic tetraparesis; SP, spastic paraparesis; ST, spastic tetraparesis; UL, upper limb; URTI, upper respiratory tract infection; WM, white matter.
Note: AGS1315_P3 (different shading) is not included in the patient data analysis because of mosaic status; disability scales were not calculated for AGS228 because of age < 1 year at last contact.
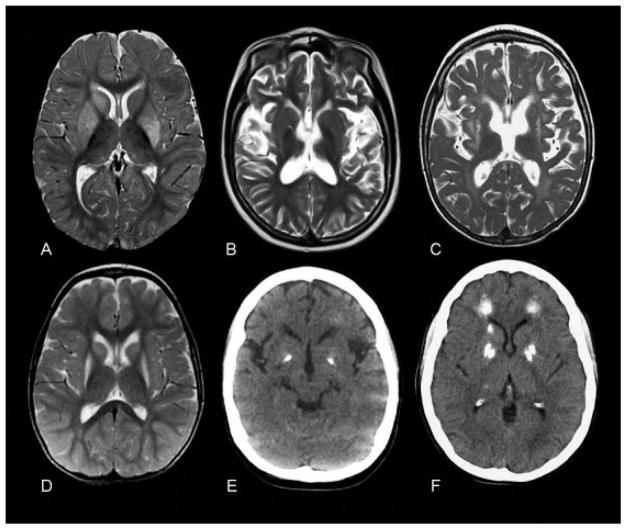
Fig. 2.
Characteristic neuroradiological features of ADAR1-related disease. Images (A) and (D) are axial T2 images of AGS251, presenting at 9 months of age with bilateral striatal necrosis following varicella zoster infection, showing characteristic high signal and swelling of head of caudate and putamen (A). (D) Follow-up at 35 months shows persisting signal change and shrinkage of caudate and putamen. Images (B) and (E) are from AGS150, a 10-year-old child presenting with an Aicardi–Goutières syndrome phenotype. (B) T2 axial MR shows cerebral atrophy with mildly increased signal in white matter. (E) CT shows dense bilateral globus pallidus calcification. Image (C) is of a patient presenting with an Aicardi–Goutières syndrome phenotype (AGS810_P1). (C) T2 axial MR at 5 years shows marked cerebral atrophy, white matter high signal, and signal change and shrinkage of the putamen. (F) CT scan of his mother (AGS810_P2) aged 34 years shows dense calcification of globus pallidus, head of caudate, and deep frontal white matter. Her MR (not shown) was normal. CT, computed tomography; MR, magnetic resonance.
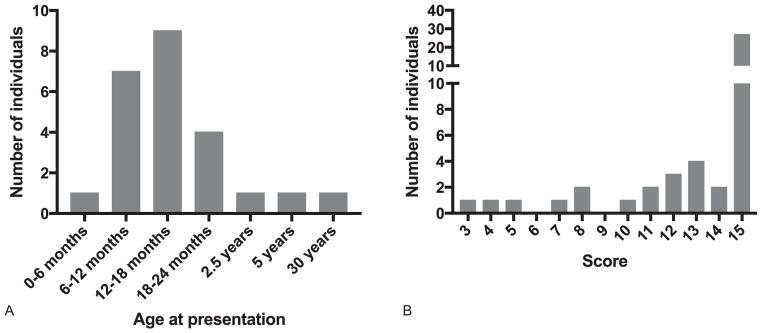
A total of 25 patients were considered to have demonstrated normal development prior to disease onset, in 18 of whom there was a history of either vaccination (4 patients) or a notable infectious episode (14 patients) in the period shortly preceding the development of clinical signs (Fig. 3A). Several patients experienced a rapid onset of dystonia/spasticity and loss of skills, with two patients being admitted to intensive care due to severe dystonic crisis. Others exhibited a more slowly progressive onset over weeks or months. Definite clinical progression beyond the initial presentation was recorded in 16 cases. Nine patients are deceased, between the ages of 10 months and 19 years, six of whom had early-onset disease consistent with AGS.
Fig. 3.
Age at presentation and associated disability. (A) Age at presentation in patients developing disease after a period of clearly normal development. (B) Assessment of gross motor function, manual ability, and communication status in living patients with mutations in ADAR1 over 1 year of age.
An assessment of gross motor function, manual ability, and communication status at last contact was made in 45 patients, of whom 27 were recorded to have none of any purposeful gross motor, hand and communication function (score of 5 on all three scales) (Fig. 3B). Five patients were able to walk with no or some support (GMFCS I–III). Eleven patients were capable of effective sender and receiver communication (CFCS I–III). Although formal testing was not undertaken, seven patients were considered to have normal intellectual function.
Five patients were reported to demonstrate hypo/hyper-pigmentation consistent with dyschromatosis symmetrica hereditaria (DSH) 1, and two patients were described with chilblain-like vasculitic lesions. Four patients were documented with autoimmune hemolytic anemia. Glaucoma was not recorded in any patient.
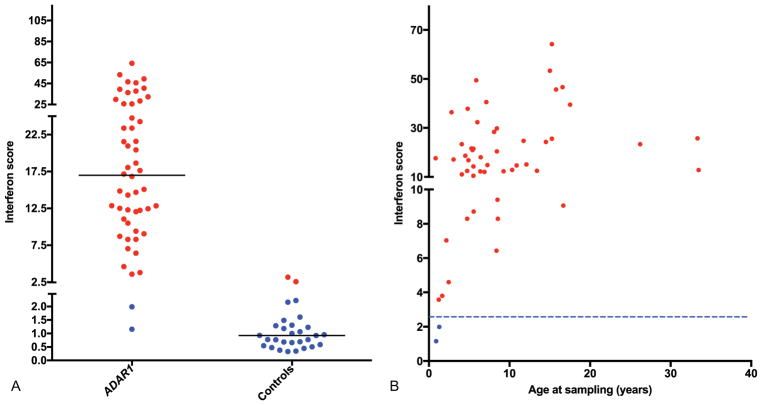
Interferon Status
We derived 52 interferon scores from 34 patients, 50 of which were abnormal, with a median interferon score across the group of 16.99 (interquartile range [IQR]: 10.64–25.71) compared with controls (median: 0.93, IQR: 0.57–1.30) (Fig. 4). Positive scores were observed up to 25 years after disease onset. We also tested 20 interferon scores from 16 parental carriers of a recessive mutation in ADAR1. Two samples from seven parents heterozygous for the recurrent p.Pro193Ala mutation demonstrated a positive interferon score, versus six samples from nine parents carrying a different mutation (Supplementary Fig. S1, online-only).
Fig. 4.
Interferon score data in ADAR1-mutated patients and controls. Summary of interferon score data (A) in ADAR1-mutated patients and controls and (B) in ADAR1-mutated patients by age. Circles indicate results above +2 SD of the mean of 29 controls (= 2.466, considered “positive”). Solid horizontal lines indicate median value of ADAR1-mutated and control groups. Dotted line indicates positive/negative boundary (2.466) of interferon score.
Discussion
In 2012, ADAR1 mutations were described in the context of the early-onset encephalopathy AGS, associated with the presence of intracranial calcification, white matter disease, and severe developmental delay.5 Subsequently, in 2014, mutations in ADAR1 were also shown to underlie cases of apparently nonsyndromic BSN, and of isolated spastic para-paresis with normal neuroimaging.6,7 Here, we confirm these associations, thus emphasizing the need to consider ADAR1-related disease in several distinct clinical scenarios triggering different investigative algorithms. Furthermore, we now describe a patient with a dominant-negative mutation in ADAR1 demonstrating an adult-onset phenotype evocative of “idiopathic” basal ganglia calcification characterized by intracranial calcification and subtle psychological disturbance. Our clinical and radiological findings highlight the propensity of ADAR1-related disease to incur basal ganglia dysfunction, and the value of basal ganglia calcification, frequently only appreciated on computed tomography, as a diagnostic indicator. In general, mutations in ADAR1 should be considered in the context of a motor disorder characterized by spasticity and dystonia. The onset of disease can occur after a period of normal development, sometimes associated with a rapid loss of skills, or a much slower progression over many years. Assessments using the GMFCS, MACS, and CFCS rating scales indicate that disease outcome in the cases that we have ascertained is frequently severe. It is of note that we observed cases with completely preserved intellect +/− normal neuroimaging in the face of significant motor disability.
Our own research focus is biased toward the ascertainment of pediatric disease. However, Tojo et al described a female patient with the dominant-negative p.Gly1007Arg mutation, presenting at the age of 17 years with gait disturbance and dystonic posturing of the legs, who experienced intellectual deterioration from 21 years of age, and became wheelchair bound a year later.13 Together with our observation of an adult female whose clinical phenotype only became evident at the age of 30 years, it is clear that later onset disease can occur due to ADAR1 deficiency. This latter case also illustrates the significant intrafamilial variability which can be seen in association with ADAR1 dysfunction, the mother presenting in adulthood with subtle psychological disturbance, while her son experienced a devastating early-onset encephalopathy.
ADAR1-related neurological disease can be inherited as either an autosomal recessive or autosomal dominant trait. We observed homozygosity for a missense mutation in five of 28 families segregating recessive disease. As previously suggested, the absence of patients with homozygous null mutations indicates that, as for the ADAR1 null mouse, complete loss of ADAR1 protein activity is likely embryonic lethal.5 Our molecular data reveal a remarkably high frequency of the p.Pro193Ala substitution, seen in 22 of 23 families with compound heterozygous molecular lesions in ADAR1. This mutation, which is recorded on 602 of 282,636 alleles in the gnomAD database, was not observed in the homozygous state in our cohort. That this variant was seen in combination with a null mutation in 13 families suggests that homozygosity for the p.Pro193Ala allele leads to a milder, later onset, or distinct phenotype not ascertained here, or may not be associated with disease. Perhaps of note, the gnomAD database includes one individual homozygous for this mutation. Finally, our molecular data highlight the dominant-negative p.Gly1007Arg mutation, which can occur de novo, or be inherited with variable expression and/or nonpenetrance at least into mid-adult life. The proximity of Gly1007 to the backbone of its RNA ligand, and the possibility for an arginine residue to make polyvalent interactions there suggests a mechanism whereby Arg1007 might bind more tightly to RNA and thus act as a competitive inhibitor of wild-type protein, while being itself catalytically inactive.14 In keeping with this model, we previously demonstrated that a plasmid expressing Gly1007Arg showed stronger inhibition of wild-type ADAR1 than equivalent amounts of a plasmid expressing catalytic inactive ADAR1.5
More than 130 different ADAR1 mutations have been documented in patients with DSH, an autosomal-dominant disorder characterized by the childhood onset of hypopigmented and hyperpigmented macules on the face and dorsal aspects of the extremities.15 DSH has only very rarely been reported outside of Japan and China, and even within identified families, a marked variability in expression is well recognized. In our series, five patients were noted to demonstrate pigmentary lesions consistent with DSH. The frequent observation of stop and frameshift variants in DSH indicates haploinsufficiency as the likely molecular pathology, consistent with the recent confirmation of our previous suggestion that two individuals with DSH would be at one in four risk of a pregnancy with ADAR1-related neurological disease.16
Loss-of-function mutations in ADAR1 have been classified within the so-called type I interferonopathy grouping, a novel set of inborn errors of immunity where it is proposed that an upregulation of type I interferon signaling is central to disease pathogenesis.17,18 The AGS phenotype can arise due to mutations in any one of seven genotypes within this grouping (AGS1–7: TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1), and apparently isolated spastic paraparesis has been reported in patients mutated in three of these genes (RNASEH2B, ADAR1, and IFIH1). In contrast, in an overview of 374 patients from 299 families with mutations in AGS1–7, BSN, the most frequently ascertained phenotype in the current series, was only recorded in the context of ADAR1-related disease, suggesting discrete factors relevant to gene/protein expression and disease mechanism consequent upon ADAR1 dysfunction.19 Also possibly reflective of this apparent specificity, in comparison to other genotypes, is the frequency of clinical progression, and the low risk of developing glaucoma and chilblain-like lesions (since we recorded no examples of the former and only two cases of the latter in our cohort).
The consistent finding of a positive interferon signature in peripheral blood in the series of patients reported here indicates the potential utility of this biomarker as a screening test for ADAR1-related disease, for the interpretation of ADAR1 genetic sequence variants of uncertain significance, and in the possible monitoring of treatment efficacy as anti-interferon therapies is developed.20,21 We emphasize that the interferon signature remains elevated many years after disease onset, providing evidence of ongoing pathology. ADAR1 is expressed throughout the brain including the basal ganglia (http://www.brain-map.org), and it has been shown that a loss of ADAR1 renders cells more susceptible to apoptosis following stress, including infection.22 We cannot rule out the possibility that the occurrence of fevers prior to frank neurological regression represents a prodrome in some cases. However, a history of vaccination or an apparently discrete infectious episode in several patients considered to be completely developmentally normal prior to disease onset, of whom 12 demonstrated BSN on neuroimaging, raises the possibility that the acute degeneration of striatal tissue seen in many patients with ADAR1 mutations might relate to a rapid induction of apoptosis triggered by viral infection/metabolic stress. Beyond this possibility, there is strong evidence that interferon is a neurotoxin,23–27 and we consider it likely that inappropriate and chronic exposure to type I interferons may be directly relevant to the ADAR1-related neurological phenotypes described here, perhaps induced by dsRNA species which are normally edited by ADAR1, thereby rendering them as immunology inert/marking them as self.1,3,4,28 These observations highlight the potential utility of treatments for ADAR1-related disease, which recent data suggest might be usefully targeted at antagonism of type I interferon signaling.29
Supplementary Material
Acknowledgments
Funding
Y.J.C. acknowledges funding from the European Research Council (GA 309449: Fellowship to Y.J.C.), ERA-NET Neuron (MR/M501803/1), and a state subsidy managed by the National Research Agency (France) under the “Investments for the Future” (ANR-10-IAHU-01). T.A.B. acknowledges funding from the NIHR. V.N. and K.M.R. acknowledge the clinical support of the C4RCD Research Group.
We are grateful to the affected families for their involvement in our research, and to all clinicians who contributed samples and clinical data not included here. We thank Marie-Louise Frémond for critical reading of the article. We would like to thank the Exome Aggregation Consortium (ExAC), the Genome Aggregation Database (gnomAD), and the groups that provided exome variant data for comparison.
Footnotes
Authors’ Contributions
J.H.L. and Y.J.C. collated and reviewed all clinical and radiological data. G.I.R. performed quantitative PCR analysis, with assistance from N.K., M.B., T.A.B., A.C.E.B., M.L.C., A. M.C., C.C.,R.C.D., F.R.D.,N.D., B.De A., V.De G., C.G.E.L. De G.,I. D., C De L., A.E., M.C.F., P.F., A.F., E.F., M.P.G., N.R.G., M.H., M.A. K., N.L., J.-P.S.-M.L., M.A.L., S.S.M., R.M., L.M.-S., G.M., M.M., V. N., S.O., J.D.O.-E., B.P.-D., F.P., K.M.R., M.R., F.R., P.R.-P., A.R., T.I. S., M.B.T., A.T., F.U., N.U., A.V., and A.W. provided clinical samples and critically reviewed clinical and immunological patient data. Y.J.C. conceived the study and wrote the initial draft with the assistance of G.I.R. All authors critically reviewed the article and agreed to its publication.
Financial Disclosure
None of the authors have any financial disclosure to report.
References
- 1.Liddicoat BJ, Chalk AM, Walkley CR. ADAR1, inosine and the immune sensing system: distinguishing self from non-self. Wiley Interdiscip Rev RNA. 2016;7(2):157–172. doi: 10.1002/wrna.1322. [DOI] [PubMed] [Google Scholar]
- 2.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10(1):109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liddicoat BJ, Piskol R, Chalk AM, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349(6252):1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43(5):933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice GI, Kasher PR, Forte GM, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet. 2012;44(11):1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livingston JH, Lin JP, Dale RC, et al. A type I interferon signature identifies bilateral striatal necrosis due to mutations in ADAR1. J Med Genet. 2014;51(2):76–82. doi: 10.1136/jmedgenet-2013-102038. [DOI] [PubMed] [Google Scholar]
- 7.Crow YJ, Zaki MS, Abdel-Hamid MS, et al. Mutations in ADAR1, IFIH1, and RNASEH2B presenting as spastic paraplegia. Neuropediatrics. 2014;45(6):386–393. doi: 10.1055/s-0034-1389161. [DOI] [PubMed] [Google Scholar]
- 8.La Piana R, Uggetti C, Olivieri I, et al. Bilateral striatal necrosis in two subjects with Aicardi-Goutières syndrome due to mutations in ADAR1 (AGS6) Am J Med Genet A. 2014;164A(3):815–819. doi: 10.1002/ajmg.a.36360. [DOI] [PubMed] [Google Scholar]
- 9.La Piana R, Uggetti C, Roncarolo F, et al. Neuroradiologic patterns and novel imaging findings in Aicardi-Goutières syndrome. Neurology. 2016;86(1):28–35. doi: 10.1212/WNL.0000000000002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palisano R, Rosenbaum P, Walter S, Russell D, Wood E, Galuppi B. Development and reliability of a system to classify gross motor function in children with cerebral palsy. Dev Med Child Neurol. 1997;39(4):214–223. doi: 10.1111/j.1469-8749.1997.tb07414.x. [DOI] [PubMed] [Google Scholar]
- 11.Eliasson AC, Krumlinde-Sundholm L, Rösblad B, et al. The Manual Ability Classification System (MACS) for children with cerebral palsy: scale development and evidence of validity and reliability. Dev Med Child Neurol. 2006;48(7):549–554. doi: 10.1017/S0012162206001162. [DOI] [PubMed] [Google Scholar]
- 12.Hidecker MJ, Paneth N, Rosenbaum PL, et al. Developing and validating the Communication Function Classification System for individuals with cerebral palsy. Dev Med Child Neurol. 2011;53(8):704–710. doi: 10.1111/j.1469-8749.2011.03996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tojo K, Sekijima Y, Suzuki T, et al. Dystonia, mental deterioration, and dyschromatosis symmetrica hereditaria in a family with ADAR1 mutation. Mov Disord. 2006;21(9):1510–1513. doi: 10.1002/mds.21011. [DOI] [PubMed] [Google Scholar]
- 14.Heale BS, Keegan LP, McGurk L, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28(20):3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi M, Suzuki T. Dyschromatosis symmetrica hereditaria. J Dermatol. 2013;40(5):336–343. doi: 10.1111/j.1346-8138.2012.01661.x. [DOI] [PubMed] [Google Scholar]
- 16.Kono M, Matsumoto F, Suzuki Y, et al. Dyschromatosis symmetrica hereditaria and Aicardi-Goutières syndrome 6 are phenotypic variants caused by ADAR1 mutations. J Invest Dermatol. 2016;136(4):875–878. doi: 10.1016/j.jid.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 17.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 18.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15(7):429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 19.Crow YJ, Chase DS, Lowenstein Schmidt J, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A(2):296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice GI, Forte GM, Szynkiewicz M, et al. Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 2013;12(12):1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice GI, Melki I, Frémond ML, et al. Assessment of type I interferon signaling in pediatric inflammatory disease. J Clin Immunol. 2017;37(2):123–132. doi: 10.1007/s10875-016-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284(43):29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavanagh D, McGlasson S, Jury A, et al. Type I interferon causes thrombotic microangiopathy by a dose-dependent toxic effect on the microvasculature. Blood. 2016;128(24):2824–2833. doi: 10.1182/blood-2016-05-715987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akwa Y, Hassett DE, Eloranta ML, et al. Transgenic expression of IFN-alpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol. 1998;161(9):5016–5026. [PubMed] [Google Scholar]
- 25.Campbell IL, Krucker T, Steffensen S, et al. Structural and functional neuropathology in transgenic mice with CNS expression of IFN-alpha. Brain Res. 1999;835(1):46–61. doi: 10.1016/s0006-8993(99)01328-1. [DOI] [PubMed] [Google Scholar]
- 26.Barlow CF, Priebe CJ, Mulliken JB, et al. Spastic diplegia as a complication of interferon Alfa-2a treatment of hemangiomas of infancy. J Pediatr. 1998;132(3 Pt 1):527–530. doi: 10.1016/s0022-3476(98)70034-4. [DOI] [PubMed] [Google Scholar]
- 27.Michaud AP, Bauman NM, Burke DK, Manaligod JM, Smith RJ. Spastic diplegia and other motor disturbances in infants receiving interferon-alpha. Laryngoscope. 2004;114(7):1231–1236. doi: 10.1097/00005537-200407000-00017. [DOI] [PubMed] [Google Scholar]
- 28.Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol. 2010;17(9):1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frémond ML, Rodero MP, Jeremiah N, et al. Efficacy of the Janus kinase 1/2 inhibitor ruxolitinib in the treatment of vasculopathy associated with TMEM173-activating mutations in 3 children. J Allergy Clin Immunol. 2016;138(6):1752–1755. doi: 10.1016/j.jaci.2016.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.