Abstract
Transcriptome profiling is widely used to infer functional states of specific cell types, as well as their responses to stimuli, to define contributions to physiology and pathophysiology. Focusing on microglia, the brain macrophages, we report here a side-by-side comparison of classical cell sort-based transcriptome sequencing and the ‘RiboTag’ method that avoids cell retrieval from tissue context and yields translatome sequencing information. Conventional whole cell microglia transcriptomes were found to be significantly tainted by artifacts induced by tissue-dissociation, cargo contaminations and transcripts sequestered from ribosomes. Conversely, our data highlight the added value of RiboTag profiling to assess the accuracy of Cre transgenic mice. Collectively, this study indicates method-based biases, reveals observer effects and establishes RiboTag-based translatome profiling as a valuable complement to standard sort-based profiling strategies.
Cellular functions are defined by transcriptomes and proteomes. Global gene expression profiling can hence provide insights into specific contributions of distinct cell types to various physiological processes. Macrophages are myeloid immune cells that are strategically positioned to ingest and degrade dead cells, debris and foreign material, and orchestrate inflammation and immune defense. Moreover, emerging evidence supports additional critical tissue macrophage contributions to the establishment and maintenance of organ functions. Studies have highlighted the impact of the tissue environment on macrophage expression signatures and enhancer landscapes 1,2. Conversely, tissue macrophages lose distinct expression patterns once taken into culture 2, likely due to loss of original and exposure to novel environmental cues. Accurate expression profiling of cells in order to infer in vivo functions therefore requires methods that allow efficient and rapid retrieval of phenotypically specified cells or their RNA from intact organs.
Classically, the isolation of defined cell populations from their physiological tissue context involves the preparation of single cell suspensions followed by flow cytometry- or magnetic bead-based cell sorting. Depending on the cell type studied and its respective extent of tissue embedding, release of the cells can require mechanical processing and extensive enzymatic digests with prolonged incubations. Collectively, these manipulations bear the inherent risk of artifacts. Moreover, cell isolation protocols are often inefficient and likely prone to introduce bias towards subpopulations. Even optimized isolation protocols fail for instance to retrieve more than 10% of microglia cells from an intact mouse brain, estimated to comprise three million cells.
To circumvent the need for cell retrieval, alternative approaches were introduced that allow isolation of cell-specific translatomes by immuno-precipitation (IP) of epitope-tagged ribosomes from crude tissue extracts 3,4. In the 'RiboTag approach' developed by McKnight and colleagues 4, cell type-specific expression of Cre recombinase is used to activate expression of a hemagglutinin (HA) epitope-tagged ribosomal subunit (RPL22) by deletion of a 'floxed' wild-type exon. IP of the tagged ribosomes from whole tissue extracts with anti-HA antibody-coupled magnetic beads enables the pull-down of cell type-specific ribosome-attached mRNA, i.e. the translatome.
Here we report the application of the RiboTag approach to the study of microglia. Specifically, we compared previously reported Cx3cr1Cre and Cx3cr1CreER transgenic animals 5 for their potency and specificity to be used in microglia expression profiling using the RiboTag strategy. Side-by-side comparison of translatomes isolated by IP from crude tissue extracts and transcriptomes from sorted microglial cells highlights the advantages and disadvantages of the respective approaches. Whole cell transcriptomes were found to be contaminated by artifacts induced by tissue-dissociation, cargo contaminations and transcripts sequestered from ribosomes. Finally, we performed a translatome analysis on microglia from animals exposed to acute peripheral endotoxin challenge. Collectively, our results highlight the specifics of RiboTag profiling and establish this method as a valuable complement to standard sort-based profiling strategies.
Results
Definition of cell type specificity of CX3CR1Cre and CX3CR1CreER transgenic mice
The RiboTag strategy is a two-component approach relying on the combination of a floxed Rpl22HA allele 4 with a cell type-specific Cre recombinase transgene. Microglia display unique high expression of CX3CR1 6 and transgenic mice harboring a GFP reporter gene under the promoter of this chemokine receptor have been instrumental to study microglia morphology and dynamics, as GFP expression in adult mouse brains is restricted to microglia and non-parenchymal macrophages 6,7. More recently, we introduced two mouse strains that display Cre recombinase activity under the control of the Cx3cr1 promoter, either constitutively (Cx3cr1Cre mice), or following Tamoxifen (TAM) - mediated activation of an estrogen receptor-fused latent Cre recombinase (Cx3cr1CreER mice) 5,8 (Fig. 1a). To implement the RiboTag method for the study of microglia, we generated Cx3cr1Cre:Rpl22HA and Cx3cr1CreER:Rpl22HA mice. We then performed high-throughput RNA sequencing (RNAseq) on RNA isolated from whole brain tissue (input), RNA retrieved by a control IP (IP IgG) and by anti-HA IP (IP HA) of brain extracts from the two mouse strains (Fig. 1b). To assess the cell type specificity of the obtained translatomes, we compared them to published neuron and glia-specific gene expression signatures 9. Translatomes retrieved from Cx3cr1Cre:Rpl22HA and TAM-treated Cx3cr1CreER:Rpl22HA mice showed an enrichment for mRNAs encoding microglial proteins, such as Sall1, Csf1r, Trem2, Aif1 (Iba-1) and CD11b, that represent a small fraction in the total input, confirming rearrangement of the 'floxed' Rpl22HA allele in microglia (Fig. 1c). Conversely, key astrocyte and oligodendrocyte transcripts, such as Gfap, Aldh1l1, Aqp4 and Mbp, Mog, Olig1, Plp1, respectively, were de-enriched in both translatomes as expected (Fig. 1c). Translatomes retrieved by ribosome IP from brain homogenates of Cx3cr1Cre:Rpl22HA but not TAM-treated Cx3cr1CreER:Rpl22HA mice, also exhibited a prominent neuronal signature, including mRNAs encoding Calbindin 2, CX3CL1 and CELF (Fig. 1c, Supplementary Fig. 1). This suggested activation of RPL22-HA expression in neuronal cells of Cx3cr1Cre: Rpl22HA mice. Analysis of Cx3cr1Cre:Rosa26YFP animals that harbor a 'floxed' reporter allele revealed prominent neuronal labeling, comparable to that recently reported for LysM2Cre mice10 (Fig. 1d, Supplementary Fig. 2a). Moreover, immunohistochemical analysis of Cx3cr1Cre:Rpl22HA animals detected neuronal staining by anti-HA antibodies in spinal cord and brain sections, including Purkinje cells in the cerebellum, in line with the observed Calb2 expression (Fig. 1e, Supplementary Fig. 2b). In contrast, RPL22-HA expression in TAM-treated Cx3cr1CreER:Rpl22HA mice was restricted to microglia, as demonstrated by co-staining for IBA-1 (Fig.1e, Supplementary Fig. 2b). Since Cx3cr1gfp mice lack GFP labeling of adult neurons 6,11, rearrangements in Cx3cr1Cre mice are likely due to a transient and further-to-be-defined window of Cx3cr1 promoter activity during neuronal development. In support of this notion, one of the Cx3cr1Cre:Rpl22HA mice analyzed, also displayed astrocyte and oligodendrocyte transcripts, in line with the shared neuroectodermal origin of these glia cells and neurons (Fig. 1c). Of note, rearrangements in neurons of Cx3cr1Cre:Rpl22HA animals were observed irrespective of whether the floxed allele and the Cre transgene went together through the germline (data not shown).
Figure 1. RiboTag analysis reveals that Cx3cr1Cre mice but not Cx3cr1CreER animals display rearrangements in neurons.
(A) Scheme of Cx3cr1Cre and Cx3cr1CreER systems.
(B) Scheme describing the immuno-precipitation protocol, including brain homogenization, centrifugation to remove cell debris and incubation with magnetic beads and relevant antibodies.
(C) Heat maps of RNAseq data comparing IPs obtained from brains of Cx3cr1Cre:Rpl22HA and Cx3cr1CreER:Rpl22HA mice, represented by lists of genes of microglia (115), neurons (97), astrocytes (95) and oligodendrocytes (98) showing enrichment and de-enrichment of mRNAs of specific cell types in the different samples. Reference data sets 9. Each column represents an individual mouse, n=2 for Cx3cr1CreER no TAM, n=3 for Cx3cr1Cre and Cx3cr1CreER with TAM.
(D) Microscopic analysis of cortex brain sections from Cx3cr1Cre:R26-YFP mice (left panel) and Cx3cr1CreER:R26-YFP mice (TAM treated (right panel) or untreated controls (middle panel)), stained for IBA-1, YFP and DAPI, showing neuronal expression of YFP in Cx3cr1Cre brains and microglia-restricted YFP expression in Cx3cr1CreER brains. The animals analyzed are F1 offspring of the intercross of homozygote Cx3cr1CreER or Cx3cr1Cre animals and homozygote R26-YFP mice. Representative of 2 independent experiments.
(E) Immuno-fluorescent staining of tissue sections of Cx3cr1Cre:Rpl22HA (left) and TAM-treated Cx3cr1CreER:Rpl22HA (right) mice, stained for IBA1, HA and Hoechst, showing neuronal expression of HA in Cx3cr1Cre cortex, and microglia-restricted HA expression in Cx3cr1CreER spinal cord. Scale bars: 200µm (left), 50µm (right) Representative of 2 independent experiments.
(F) Flow cytometry analysis showing HA staining in microglia (CD11b+ CD45int, gated on Ly6C/G– DAPI–) of Rpl22HA TAM-treated mice (black line), Cx3cr1CreER:Rpl22HA mice, untreated (blue line) and TAM-treated (red line). Shadowed histogram represents isotype (IgG) control. Representative data of 3 repeats.
Cx3cr1CreER:Rpl22HA mice that were TAM-treated postnatally displayed brain macrophage-restricted activation of the RiboTag. Some enrichment for the microglia translatome was also observed without TAM treatment in these mice and rare YFP+ cells could be detected in non-treated Cx3cr1CreER:Rosa26YFP animals (Fig. 1c, d middle panel), corroborating reports of leakiness of the CreER system 12. However, as confirmed by flow cytometric analysis (Figure 1f), robust rearrangement and microglial expression of the HA epitope-tagged ribosome subunit in Cx3cr1CreER: Rpl22HA mice were dependent on TAM induction in our facility.
Collectively, these data illustrate the value of the RiboTag profiling approach to assess the accuracy of Cre transgenic mouse models, and to investigate specific cell types, including fate mapping and conditional mutagenesis.
Comparison of RiboTag profiling to cell sort-based transcriptomics
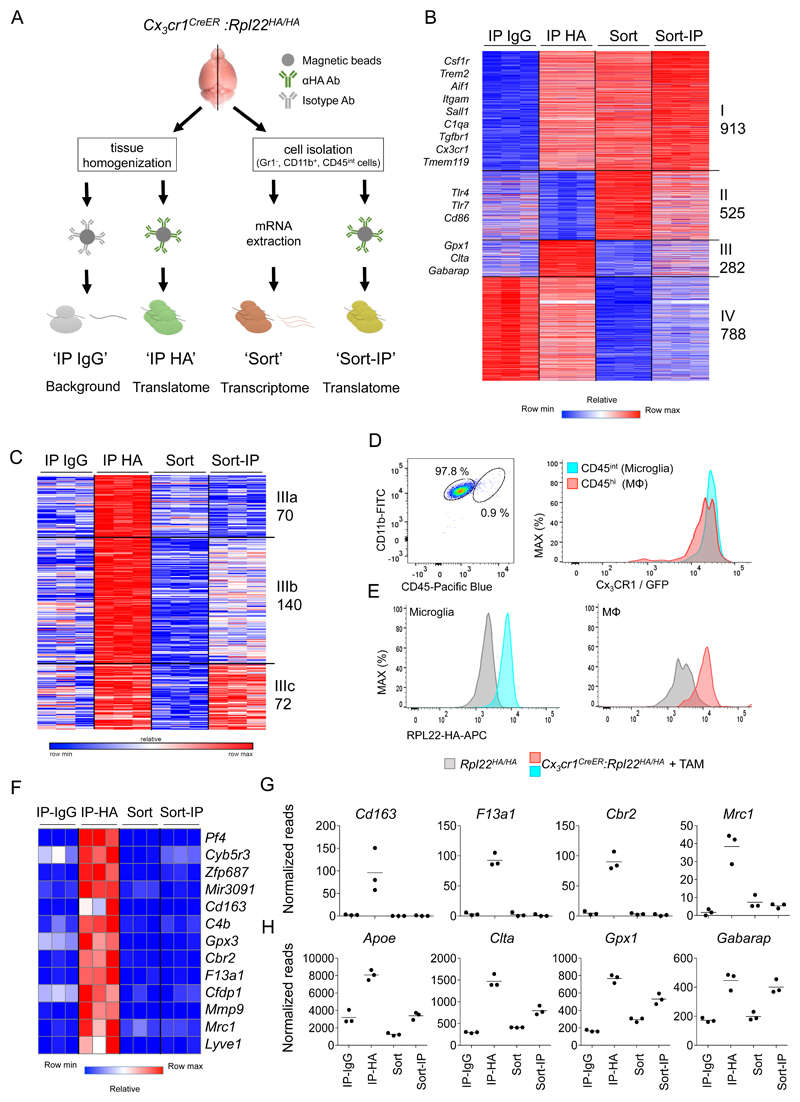
Having established the value of Cx3cr1CreER:Rpl22HA mice, we next compared translatomes and transcriptomes of sorted and unsorted microglia. Specifically, individual brains of TAM-treated Cx3cr1CreER:Rpl22HA mice were divided: one hemisphere underwent direct tissue homogenization followed by IP with anti-HA (IP-HA) or an isotype control antibody to define the method-related background (IP-IgG). The second hemisphere was subjected to the classical microglia isolation protocol involving tissue digestion followed by cell sorting of microglial cells (defined as DAPI– Ly6C/G (Gr1)– CD11b+ CD45int) (Fig. 2a, Supplementary Fig. 3). A fraction of the sorted microglia was taken for direct mRNA isolation to yield the whole transcriptome (Sort); another fraction was lysed and subjected to the anti-HA IP to retrieve the translatome of sorted cells (Sort-IP) (Fig. 2a). This experimental set-up allowed comparison of translatomes of sorted and unsorted microglia alongside whole transcriptomes of sorted microglia from the same brain, and thus to investigate the impact of the isolation protocol on gene expression.
Figure 2. Comparison of cell sort-based protocol and the RiboTag method to profile microglia.
(A) Scheme describing the experimental protocol comparing RiboTag and cell sort-based strategies.
(B) Heat map of RNAseq data of samples obtained in (A). Genes selected by maximum value>100 normalized reads (3,186 out of 17,406 genes), significantly changed (fold change>2, p-value<0.05) between: IP-HA vs IP-IgG, Sort vs IP HA, Sort-IP vs IP HA and Sort vs Sort-IP, Representing 2,508 genes. n=3, individual mice. Statistical test was part of the DESeq2 package, using p-adjusted.
(C) Heatmap representing K-means re-clustering of genes from cluster III from Figure 2B, showing genes high in IP-HA and low in Sort samples.
(D) FACS dot plot (left panel) showing separation of microglia (CD45int) from other brain macrophages (MΦ) (CD45hi) by flow cytometry. Histogram (right panel) of microglia and MΦ isolated from Cx3cr1GFP mice indicating high CX3CR1/GFP expression in both populations. Representative of 3 independent experiments.
(E) FACS histogram of HA staining in microglia (left panel) and other brain macrophages (MΦ) (right panel) in control Rpl22HA mice (grey) or TAM-treated Cx3cr1CreER:Rpl22HA mice (blue/red). Representative of 2 independent experiments.
(F) Heatmap of RNA-seq data of representative non-parenchymal brain macrophages genes, showing enrichment in IP-HA, but not in sorted samples.
(G) Graphs showing normalized reads of example genes from Figure 2F. Each dot represents an individual mouse, n=3, line represents mean.
(H) Graphs showing normalized reads of example genes from cluster III-b and III-c in Figure 2C, showing functional genes enriched in IP and Sort-IP. Each dot represents an individual mouse, n=3, line represents mean.
Unbiased K-means clustering of the significantly differentially expressed genes between at least two sample groups (IP-HA vs IP-IgG, IP-HA vs Sort, IP-HA vs Sort-IP and Sort vs Sort-IP, fold change>2, p<0.05), revealed 2508 differentially expressed genes, which could be divided into four clusters (Fig. 2b).
Cluster IV was discerned as RiboTag method-related background, since mRNA reads in the non-specific IP-IgG were higher than in the specific IP-HA and absent from sorted samples. IP specific genes were selected for being significantly higher in IP-HA than IP-IgG (fold change>2, p<0.05), and all genes below this threshold were removed from the analysis.
Cluster I comprised 913 mRNAs enriched in the specific IP-HA compared with IP-IgG and present in both samples of sorted microglia. This cluster includes established microglia signature genes, such as Aif1, Irf8, Sall1, Cx3cr1, Tgfbr, Tmem119 and Hexb (Fig. 2b; Supplementary Fig. 4), indicating that the retrieval methods we used are comparable. Cluster II was represented by 525 mRNAs that were highly abundant in sorted samples (both in translatome and transcriptome), but not present in the direct IP-HA. Cluster III comprised 282 mRNAs prominently enriched in the direct IP-HA, but less abundant in the sorted microglia. Cluster II and III highlight differences between the retrieval methods, as well as discrepancies between transcriptomes and translatomes and will be the focus of the remainder of this study.
Transcripts overrepresented in microglia translatomes
Cluster III (Fig. 2b) is defined by genes highly expressed in ‘IP-HA’ samples relative to ‘Sort’ samples and could be further subdivided according to transcript abundance in the ‘Sort-IP’ samples (Fig. 2c): Cluster III-a (72 mRNAs) were low in both samples of the sorted cells ('Sort' and 'Sort-IP'); Clusters III-b and III-c (210 mRNAs) were low in the ‘Sort’ samples but highly expressed in the ‘Sort-IP’ samples (Fig. 2c).
Cluster III-a can be largely explained by the presence of non-parenchymal macrophage mRNAs. Non-parenchymal brain macrophages, including perivascular, meningeal and choroid plexus macrophages, can be discriminated from CD11b+ CD45int microglia as CD11b+ CD45hi cells and therefore be excluded by fluorescence-activated cell sorting (Fig. 2d). CX3CR1 is expressed both in microglia and in non-parenchymal brain macrophages (Fig. 2d) 13. Accordingly, both CD45int microglia and CD45hi macrophages of TAM-treated Cx3cr1CreER:Rpl22HA mice express the HA-tagged Rpl22 isoform (Fig. 2e). Moreover, some of the non-parenchymal brain macrophages are long-lived similar to microglia and hence do not lose the rearranged alleles as monocytes do 13. Single cell transcriptomics have shown that non-parenchymal brain macrophages differ in gene expression from microglia 13,14. Accordingly, cluster III-a included Cd163, F13a1, Cbr2, Mrc1 and Lyve1 (Figure 2F, 2G).
The combined clusters III-b and III-c comprise mRNAs that are enriched in both IP-HA and Sort-IP translatomes over the whole transcriptomes, suggesting their functional importance for the cells. These include mRNAs encoding proteins related to metabolism (Gpx1, Sdhc), vesicular transport (Clta, Kdelr1, Ykt6), sphingolipid metabolism (Gm2a, Psap) and lipids (Apoe), as well as components of the GABA-receptor signaling cascade (Gabarap, Gnai2) (Figure 2H). Specific functions of these genes in microglia remain to be explored.
Collectively, these results highlight the value of a multifaceted approach, combination of the RiboTag and cell sorting strategies to improve cell type specificity. In addition, the RiboTag approach allows us to focus specifically on genes that are actively being translated and contributing to the cellular proteome at a particular time and location.
Transcripts overrepresented in transcriptome cluster I: Isolation artifacts
Cluster II (Fig. 2b) defines 525 mRNAs that were high in Sort, but low in IP-HA. This cluster can be further subdivided according to transcript abundance in the Sort-IP samples (Fig. 3a).
Figure 3. Microglia isolation results in cell activation.
(A) Heatmap representing K-means re-clustering of genes from cluster II from Figure 2B, showing mRNAs high in Sort and low in IP-HA samples.
(B) Graphs showing normalized reads of example genes from cluster II-a in Figure 3A, showing high level expression of immune-activation-related genes in sorted samples. Each dot represents an individual mouse, n=3, line represents mean.
(C) Ingenuity Pathway Analysis (IPA) of genes significantly higher (>2 fold change, p-value<0.05, according to DESeq2 statistical analysis) in Sort compared to IP-HA (n=3), showing activated pathways related to immune response represented by activation Z-score as calculated by IPA software.
Cluster II-a comprised 190 transcripts similarly expressed in Sort-IP and Sort samples and was found to include mRNAs that are related to immune activation, such as Cd86, Cd53, Tlr4, and Tlr7 (Fig. 3b). Ingenuity Pathway Analysis (IPA) of significantly upregulated genes in Sort vs. IP-HA (>2 fold change, p<0.05) showed upregulation of pathways such as 'Production of NOS and ROS', 'Phagocytosis' and 'TLR signaling' (Fig. 3c). Since these transcripts are high in both translatomes and transcriptomes of sorted cells, we assume that they reflect cell activation resulting from the isolation process, as recently reported from another system 15.
Commonly used macrophage isolation protocols, such as the one we applied for the microglia retrieval, include enzymatic tissue digestion at 37°C, i.e. a step that could cause cell activation and transcriptome alterations. Moreover, enzymes employed in these digests might contain endotoxin contaminations that could activate cells. To probe for the potential impact of these manipulations, we compared transcriptomes of sorted cells that were isolated from the same brain with or without collagenase and DNase digestion, and the RiboTag approach. Surprisingly though, both isolation procedures resulted in comparable transcriptional profiles, indicated by differential expression of 472 and 267 genes (cluster II and III, respectively), as compared to the relevant IP samples (Supplementary Fig. 5a). Global correlation of gene expression of the samples retrieved with or without incubation was high (r2 = 0.99), as compared to the correlation of sorted and IP samples (r2 = 0.1) (Supplementary Fig. 5b). Similarities were also apparent in a correlation matrix compiled from data of independent experiments (Supplementary Fig. 5c). Collectively, these data establish that the artifact is reproducible and suggest that it is introduced by extraction of the cells from their native environment, rather than subsequent manipulation.
Transcripts overrepresented in the transcriptome cluster II: Cargo contaminants and sequestered RNAs
Cluster II-b spanned 335 genes that were low in the Sort-IP compared with Sort samples (Fig. 3a), suggesting translatome-transcriptome differences.
Microglia are specialized phagocytes that like other macrophages take up dead cells and cell debris for clearance 16. Although we did not formally rule out other sources of contaminations, whole cell transcriptomes could hence include genetic material from recently ingested neighboring cells. Indeed, almost half of the mRNAs in cluster II-b (157 out of 335 genes) are likely to be derived from such external sources (Fig. 4a). Examples include Arhgap5, Son and Pisd-ps1, which are reportedly transcribed in astrocytes and neurons 9 (Fig. 4b).
Figure 4. Microglia transcriptomes, but not translatomes include cargo-derived mRNAs and mRNAs sequestered in nuclei and P bodies.
(A) Venn diagram of overlapping genes of cluster II-b (Figure 2C) (yellow) with genes of non-microglial cells, as selected according to 9. Non-microglia genes were selected by being with max value > 10, microglia reads < 30 and max value is > 3 fold change over microglia. Genes fitting these criteria represent cargo of ingested cells.
(B) Graphs showing normalized reads of example genes from list of shared genes from Figure 4A, showing high levels cargo contamination in Sort samples, but not in IP. Each dot represents an individual mouse, n=3 (upper panels); Expression of genes above in different brain cells, data obtained from 9 (lower panels). FKPM, fragments per kilobase of exon per million reads mapped.
(C) Graphs showing normalized reads of example genes of long non coding RNAs that reside within the nucleus and are presented only in Sort but not IP samples. Each dot represents an individual mouse, n=3, line represents mean.
(D) Violin plot representing splicing efficiency (left panel) and gene length (right panel) of genes in cluster II-b (orange) compared to genes not in II-b (green), showing that genes in cluster II-b are less efficiently spliced and have longer genes and longer 3’UTRs compared with the rest of genes in the dataset, suggesting nuclear retention. Splicing efficiency was computed by comparing intron-spanning and intron-crossing reads 28. Statistics was performed using Wilcoxon test, FDR correction was performed for right panel. * Splicing efficiency p=4.124*10-7, exon length p=6.636*10-63, Intron length p=1.575*10-27, UTR length p=1.865*10-35.
(E) Violin plot representing cellular localization of genes in other tissues (left panel Liver, right panel – MIN6 pancreatic beta cell line) with established nuclear and cytoplasmic fractions. Genes in cluster II-b (orange) are more nuclear compared to other genes (green). Statistics was performed using Wilcoxon test and FDR correction. * Liver p=7.731*10-49, MIN6 p=2.980*10-17. n (number of genes) = 316 (Liver 2b), 1970 (Liver not-2b), 306 (MIN6 2b), 1846 (MIN6 not-2b). Liver and MIN6 datasets were based on 2 independent experiments 22.
(F) Graphs showing normalized reads of immediate-early genes found in Sort but not in IP-HA (in cluster II-b, microglial genes), suggesting sequestration from translation in unsorted cells. Each dot represents an individual mouse, n=3, line represents mean.
(G) Diagram representing different states of mRNAs in the cell: nuclear retention, sequestration from translation in P-bodies or active translation in the ribosomes.
Long non-coding mRNAs (lncRNAs) were shown to be enriched in nuclei 17, where some of them act in transcriptional regulation. As expected, representatives of these lncRNAs, such as Malat1 and Neat1, were identified in the whole cell transcriptomes, while absent from translatomes, and appeared in Cluster II-b (Fig. 4c).
Gene expression is controlled at the level of transcription and translation. The latter comprises specific mechanisms that prevent mRNAs from their integration into ribosomes, including nuclear retention and sequestration into dedicated membrane-less cytosolic ribo-nucleoprotein complexes 18,19. The content of these organelles, such as processing bodies (P-bodies) and stress granules is only beginning to be defined 19, 20. However, sequestered mRNAs have been reported to be longer and to comprise extended 3’UTRs, as well as to display lower splicing efficiencies 19,21. When analyzed for these three parameters, mRNAs defined by Cluster II-b, showed significant presence of these hallmarks, as compared to all other clusters (Fig. 4d). Moreover, transcripts of cluster II-b also showed significant overlap with the list of nuclear retained mRNAs reported for other cellular systems 22 (Fig. 4e). Among the protein-coding mRNAs that seem sequestered from immediate translation, we found Fos, Jun, Egr1 and Zfp36l1 (Fig. 4f), which are immediate early genes that have been described to be induced within minutes after activation. Of note, these mRNAs appear also in the translatome of the sorted cells, suggesting that they move to the ribosomes during the isolation procedure. Collectively, discrepancies we observed between microglia translatomes and whole cell transcriptomes can be explained by the inclusion of cargo-derived transcripts and mRNAs sequestered to nuclei or P-bodies (Fig. 4g). These data highlight the value of the RiboTag approach to retrieve functionally relevant mRNAs.
RiboTag analysis of microglial response to peripheral LPS challenge
Arguably, method-related artifacts, like the ones associated with microglia isolation, could be neutralized, if controls and experimental samples are prepared using the same approach. However, this assumes that artifacts introduced by the isolation are not affected by biological treatments and challenges. To examine this issue, we performed the RiboTag protocol on TAM-treated Cx3cr1CreER: Rpl22HA mice following an intra-peritoneal lipopolysaccharide (LPS) injection (2.5 mg/kg). Brain hemispheres of individual LPS- and PBS-treated animals were subjected to either homogenization or microglia isolation and sorting, and processed (Fig. 2a) and used to generate a summary heat map of the RNAseq data (Supplementary Fig. 6). To define the effect of the isolation method on microglia of LPS-treated and PBS-treated mice, we performed separate analyses for differentially expressed genes between PBS and LPS treatment in each method and then assessed the respective overlap. The majority of genes detected as up- or down-regulated by the endotoxin challenge in the IP sample were shared with the sorted samples (Fig. 5a, 5c). Mutual genes correlated between methods, and showed a similar trend of up and downregulation (Fig.5b, 5d), indicating that bona fide LPS-induced changes are seen with both methods. Upregulated genes included Il4ra, Ch25h and Il1b (Fig. 5b), while microglia signature genes such as Tgfbr1, Sall1 and Cx3cr1, were down regulated (Fig. 5d).
Figure 5. Analysis of microglia isolated from mice challenged with LPS.
(A) Venn diagram of overlapping genes upregulated by LPS treatment in IP-HA (blue) and in Sort (yellow), showing 54 genes upregulated in IP-HA only, 247 genes shared between methods and 214 genes upregulated by sorting only.
(B) Correlation analysis of 247 shared genes (from Figure 5A) upregulated due to LPS in both methods, representing log2 fold change of significantly changed genes (log2 fold change>1, pValue<0.05, as calculated by DESeq2 statistical analysis) in LPS vs PBS in each of the methods (left panel); Graphs showing normalized reads of example genes detected as upregulated with LPS by both methods (right panels). Each dot represents an individual mouse, n=3 in PBS group, n=4 in LPS group, line represents mean. For IP-HA genes were selected by first being enriched over control IP-IgG, and then by LPS>PBS.
(C) Venn diagram of overlapping genes downregulated by LPS treatment in IP-HA (blue) and in Sort (yellow), showing 10 genes downregulated in IP-HA only, 104 genes shared between methods and 250 genes downregulated by sorting only.
(D) Correlation analysis of 104 shared genes (from Figure 5C) downregulated due to LPS in both methods, representing log2 fold change of significantly changed genes (log2 fold change<-1, p-value<0.05, as calculated by DESeq2 statistical analysis) in LPS vs PBS in each of the methods (Left panel); Graphs showing normalized reads of example genes detected as downregulated with LPS by both methods (Right panels). Each dot represents an individual mouse, n=3 in PBS group, n=4 in LPS group, lines represent mean. For IP-HA genes were selected by first being enriched over control IP-IgG, and then by LPS<PBS.
(E) Graphs showing normalized reads of example genes related to immune activation that were up- (upper panels) or down- (lower panels) regulated due to LPS treatment in sorted samples only but not detected in IP-HA, showing differential susceptibility of biologically treated samples to artifacts introduced by isolation method. Each dot represents an individual mouse, n=3 in PBS group, n=4 in LPS group, lines represent mean.
(F) Graphs showing normalized reads of example genes originated from ingested cargo that were up- (upper panels) or down- (lower panels) regulated due to LPS treatment in Sort only but not detected in IP-HA or Sort-IP, showing that the whole translatome includes LPS-dependent changes that do not originate in microglia thus introducing false information. Each dot represents an individual mouse, n=3 in PBS group, n=4 in LPS group, lines represent mean.
Notably, a considerable number of mRNAs changed upon LPS treatment only in the sorted samples, but not in the anti-HA IP from whole brain extract (46% of the upregulated and 71% of the downregulated genes) (Fig. 5a, 5c). Transcripts that were detected as changed upon LPS treatment in sorted samples (both Sort and Sort-IP) but not in IP-HA included genes related to immune activation like Il1a, Ccl2 and Vcam1 that were upregulated (Fig. 5e) and Tlr4, Siglech and Cd48 that were downregulated (Fig. 5e). Since they were found in the Sort-IP samples as well, these mRNAs are likely translated in sorted, but not in unsorted microglia. These data establish that the artifact introduced by the cell isolation is affected by the state of the animals from which the cells are retrieved. Moreover, mRNAs that are defined as 'cargo' contamination, due to their presence in the transcriptomes but absence from IPs of the sorted cells, changed following the LPS challenge (Fig. 5f). For example Pisd-ps1, Arhgap5 and Tia1 were downregulated and Ranbp2, Tet2 and Gas5 were upregulated upon LPS challenge (Fig. 5f). All of these genes were reported to be more highly expressed by other brain cells, rather than microglia (Supplementary Fig. 7) 9, and are absent from the translatomes of either sorted or unsorted microglia in our dataset.
Collectively, these results indicate that data retrieved from sorted microglia include false information that originates from sorting-related immune activation and cargo contamination of ingested cells. Importantly, our data establish that identical processing of control and test samples does not necessarily neutralize these artifacts, since they themselves are affected by the biological treatment. Taken together our study highlights the advantages and disadvantages of classical sort-based cell isolation protocols and the RiboTag approach (Supplementary Table 1).
Discussion
Here we compared experimental approaches to retrieve microglia expression signatures from brains of untreated and challenged animals. Specifically, we define strengths and weaknesses of the classical cell isolation and sort-based protocols and the RiboTag strategy 4, that relies on poly-ribosome immuno-precipitation from crude tissue extracts. Below we discuss the pros and cons of the respective techniques.
The RiboTag strategy was originally introduced by McKnight and colleagues and applied to expression profiling of neurons and Sertoli cells 4. Cell type-specificity of the approach depends on the accuracy of the Cre-drivers that is combined with the Rpl22HA allele. This aspect is highlighted in our study by the side-by-side comparison of Cx3cr1Cre and Cx3cr1CreER animals, which revealed the superiority of the inducible system to achieve brain macrophage specificity and exclude neurons. However, as reported earlier and confirmed in this study, even CX3CR1CreER mice not only target microglia, but also non-parenchymal macrophages 13. In situations where subpopulations can be phenotypically discriminated, as in the case of CD45int microglia and CD45hi perivascular macrophages, sort-based approaches, potentially combined with a RiboTag analysis like in this study, can hence be advantageous. This emphasizes the need for the development of novel Cre transgenic lines targeting microglia, including combinatorial approaches, such as the split-Cre strategy 23, to improve the cell type- or lineage-specificity. Importantly, current tests for accuracy of Cre transgenic lines are based on their combination with reporter alleles and the analysis of resulting double transgenic animals by flow cytometry.
As in the case of Cx3cr1Cre animals reported here, or LysMCre mice that were assumed to be myeloid cell-specific, but also target neurons 10, this approach is insufficient. Our results demonstrate that the RiboTag approach provides a useful complementary method to determine Cre line specificity, in particular for cell types such as neurons and endothelial cells that are notoriously difficult to isolate for flow cytometric analysis.
Macrophage expression signatures tend to be contaminated by material the phagocytes ingested from their surroundings. Since the RiboTag strategy only retrieves mRNAs that are associated with HA-epitope tagged host cell ribosomes, it excludes such exogenous material, and hence allows identification of bona fide macrophage mRNAs.
Although not directly addressed in this study, an additional benefit of the RiboTag approach is the fact, that it can be used to determine the impact of conditional mutations. Specifically, Cre-mediated rearrangements result in parallel mutagenesis and induction of the Rpl22HA allele. For instance this approach was used to define the impact of an Mecp2 deficiency on macrophages in brown adipose tissue (BAT), comparing translatomes retrieved from BAT of Cx3cr1CreER:Rpl22HA and Cx3cr1CreER:Rpl22HA:Mecp2fl/y mice 24.
Ribosome-associated mRNAs, as retrieved by the RiboTag approach are considered to reflect the translatome. Interestingly though, the microglial response to LPS using an analog experimental system, showed that mRNAs induced by the challenge can be prevented from translation by binding of a splicing factor to their 3'UTRs 25. Like other emerging layers of posttranscriptional expression control, this mechanism, which was revealed by a combined transcriptome and peptidome analysis on immunoprecipitated ribosomes, requires further study.
Above we listed advantages of the RiboTag approach, but an inherent weakness of this protocol is the fact that for confident assessment of gene expression in the targeted population, the method relies on an enrichment of the specific mRNA over the input, i.e. the whole tissue extract. The RiboTag approach hence precludes statements on the expression of genes that are equally expressed in the target cells and the surrounding tissue. For the assessment of these co-expressed genes, sort based strategies might be superior, although these also bear caveats, as outlined below.
Side-by-side comparison of the translatomes isolated by IP from crude tissue extracts and from sorted microglial cells with whole cell transcriptomes revealed a number of short-comings of the latter. First, we noted a prominent activation signature that is presumably introduced during the process of cell extraction from their tissue context. This artifact comprised proinflammatory genes, such as Cd86, Tlr4 and Tlr7, and will have to be considered in microglia profiling studies. Importantly, this robust and reproducible artifact could not be discerned when control and test samples were processed similarly. Rather we found that the isolation procedure had a differential impact on microglia retrieved from either challenged or unchallenged animals. Of note, artifacts like the once we report here that are introduced during the isolation procedure were shown to be significantly reduced, when the transcription inhibitor actinomycin D (ActD) was included during the cell preparation 26.
Taken together, our study shows that cell isolation coupled with sort-based methods and the RiboTag approach both have strengths and weaknesses, which should be considered when experiments are designed and conclusions are drawn. Cell isolation bears the risk of artifacts that might significantly confound transcriptome-based studies, including single cell analysis. Our study should hence caution experimentalists and make them aware of the 'observer effect' that is well established in physics 27, but often less appreciated in biology.
Methods
Animals
Mice were maintained on a 12 hr light/dark cycle, and food and water were provided ad libitum. All animals were on C57BL/ 6JOlaHsd background, maintained in specific pathogen-free (SPF) conditions and handled according to protocols approved by the Weizmann Institute Animal Care Committee (IACUC), as per international guidelines. The strains used included: Cx3cr1Cre mice (JAX stock # 025524 B6J.B6N(Cg)-Cx3cr1tm1.1(cre)Jung/J) 1; Cx3cr1CreER mice (JAX stock # 020940 B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J) 1; (JAX stock # 011029 B6N.129-Rpl22tm1.1Psam/J) 2; and Rosa26-YFP reporter mice 3. The presented Ribotag data were generated with animals homozygote for the Rpl22HA allele and heterozygote for the modified Cx3cr1 alleles. Mice heterozygote for the Rpl22HA allele yielded similar results (data not shown).
Microglia isolation protocols
Mice were anesthetized with Pental (1:2 in PBS) and were perfused with PBS. Brains were dissected, crudely chopped and incubated for 20 min at 37°C in a 1 ml HBSS solution containing 2% BSA, 1 mg/ml Collagenase D (Sigma) and 50µg/ml DNase1 (Sigma). In the middle of incubation homogenates were pipetted for further dissociation. Next, the homogenates were filtered through a 150μm mesh, washed with cold FACS buffer (2% FCS, 1mM EDTA in PBS-/-) and centrifuged at 2200 RPM, at 4°C, for 5 min. For the enrichment of microglia, the cell pellet was re-suspended with 3ml of 40% percoll solution and centrifuged at 2200 RPM, no acceleration and breaks, at RT for 15 min. Next, the cell pellet was resuspended, passed through 80µm mesh, washed with 5ml FACS buffer and centrifuged at 1400 RPM at 4°C for 5 min, followed by antibody (Ab) labeling and flow cytometry analysis. For the protocol excluding the digest, brains were chopped and then filtered through a 150μm mesh. The subsequent steps were as above, but without the enzymatic digest.
Tamoxifen (TAM) treatment
To induce gene recombination in CreER transgenic mice, tamoxifen (TAM) was dissolved in warm corn oil (Sigma) and administered orally via gavage for four times every other day. All animals were TAM-treated first at 4-6 weeks of age. Each oral application was of 5mg at a concentration of 0.1 mg / μl. Mice were examined 8 weeks following treatment. For the LPS treatment, mice were injected intraperitoneally (i.p.) with a single dose of LPS [2.5 mg/kg; E. coli 0111:B4; Sigma]; controls received or the same volume of vehicle solution (PBS).
Flow cytometry and cell sorting
Antibodies against CD11b (M1/70), Ly6C/G (Gr-1) (RB6-8C5), CD45 (30-F11) purchased from Biolegend or eBioscience were used. Samples were flow sorted using AriaIII (BD Biosciences, BD Diva Software) cell sorter. Analysis was performed on Fortessa (BD Biosciences, BD Diva Software) and analyzed with FlowJo software (Treestar).
Histology
Mice were anesthetized with Pental (1:2) and were perfused with PBS. Brains and spinal cords were excised and fixed for 4 hours in 2% paraformaldehyde. Brains were incubated for 72 hours in 30% sucrose and spinal cords in 18% EDTA prior to OCT (TissueTek) imbedding and freezing. Post fixed and stained 15-30 µm frozen sections were blocked in 2% horse serum for 2 hours and incubated overnight in 4°C with the primary antibody. Sections were washed 3 times in PBSX1 0.02% TritonX (Sigma) and conjugated with secondary antibody for 1 hour in 4°C. Before covering samples were washed 3 times and incubated for 5 minutes with Hoechst. Sections were analyzed by confocal laser scanning microscope Olympus BX51. Image acquisition was processed by Olympus image browser software. The following antibodies were used: Rat monoclonal anti-HA (1:100, Sigma), Rabbit polyclonal anti-Iba1 (1:250, Wako), mouse polyclonal anti-NeuN (1:100, Millipore) and Hoechst 33342 (1:10,000,Invitrogen).
Immunofluorescence
Mouse brains were fixed in 4% paraformaldehyde and subsequently paraffin-embedded by standard protocols. AT sections (7 µm thick) were deparaffinized in xylene and rehydrated through descending grades of ethanol to water. Antigen retrieval was performed in Tris/EDTA buffer at pH 9.5 twice for 5 minutes at 95 C. Sections were rinsed in PBS supplemented with 0.3% Triton-X (Sigma-Aldrich, St Lois, Missouri, USA) 3 times for 5 minutes. Unspecific binding sites were blocked using 1% bovine serum albumin in PBS supplemented with 0.3% Triton-X for 30 minutes at room temperature. The primary antibodies chicken anti-GFP (1:100; Abcam) and rabbit anti-Iba1 (1:200; WAKO; Richmond, VA) were incubated overnight at 4°C. To detect the primary antibodies, fluorochrome-conjugated secondary antibodies (1:200 each; all from Invitrogen; Karlsruhe, Germany) were applied for 1 h at RT. Autofluorescence of the tissue was quenched by using pre-warmed 0.3% sudan black for 2 minutes. Nuclear counterstain was performed by using 4,6-diamidin-2-phenylindol (DAPI; 1:10,000 in PBS) for 5 minutes, followed by 3 buffer rinses. Finally, sections were embedded with Dako immunofluorescence mounting medium (DAKO, Glostrup, Denmark). Images were taken using a FV1000 confocal laser scanning microscope (Olympus, Hamburg, Germany).
Ribosome Immunoprecipitation (IP)
Samples were extracted from mice, flash-frozen in liquid nitrogen and stored at −80 °C until use. Samples were homogenized on ice in ice-cold homogenization buffer (50 mM Tris, pH 7.4, 100 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT, 1:100 protease inhibitor (Sigma), 200 units/ml RNasin (Promega) and 0.1 mg/ml cycloheximide (Sigma) in RNase free DDW) 10% w/v with a Dounce homogenizer (Sigma) until the suspension was homogeneous. To remove cell debris, 1 ml of the homogenate was transferred to an Eppendorf tube and was centrifuged at 10,000g and 4°C for 10 min. Supernatants were transferred to a fresh Eppendorf tube on ice, then 10μl was removed for 'input' analysis and 5μl (=125µg) of anti-HA antibody (H9658, Sigma) or 5μl (=1µg) of mouse monoclonal IgG1 antibody (Sigma, Cat# M5284) was added to the supernatant, followed by 4 h of incubation with slow rotation in a cold room at 4 °C. (Subsequent further calibration of the amounts of the antibodies showed that 5µg antibody of anti-HA antibody and mouse monoclonal IgG1 antibody (Merck, Cat# PP100) yielded similar results, including transcripts enriched in the IgG IP samples seen in cluster IV of Fig. 2B (data not shown)). Meanwhile, Dynabeads Protein G (Thermo Fisher Scientific), 100 μl per sample, were equilibrated to homogenization buffer by washing three times. At the end of 4 h of incubation with antibody, beads were added to each sample, followed by incubation overnight in cold room at 4 °C. After not more than 12 h, samples were washed three times with high-salt buffer (50 mM Tris, 300 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT, 1:200 protease inhibitor, 100 units/ml RNasin and 0.1 mg/ml cycloheximide in RNase free DDW), 5 min per wash in a cold room on a rotator. At the end of the washes, beads were magnetized and excess buffer was removed, 150µl Lysis Buffer was added to the beads and RNA was extracted with Dynabeads mRNA Direct purification kit (Thermo Fisher). RNA was eluted in 6μl H2O and taken for RNA sequencing. For Sort-IP samples (RiboTag IP after sorting), ~50-100*103 cells were sorted into cold PBS, centrifuged 400g for 5 min at 4°C. Supernatant was removed and the pellet was re-suspended in 1ml of lysis buffer, the rest of IP was continued as above.
RNA sequencing
RNA-Seq of populations was performed as described previously 4. In brief, 5,000 microglia cells were sorted into 50μl of Lysis Buffer (Life Technologies) and stored at -80°C. mRNA was captured with Dynabeads oligo(dT) (Life Technologies) according to manufacturer’s guidelines. We used a derivation of MARS-seq 5. Library concentration was measured with a Qubit fluorometer (Life Technologies) and mean molecule size was determined with a 2200 TapeStation instrument. RNA-Seq libraries were sequenced using Illumina NextSeq-500.
Data Analysis
Raw reads were mapped to the genome (NCBI37/mm9) using hisat (version 0.1.6). Only reads with unique mapping were considered for further analysis. Gene expression levels were calculated using the HOMER software package (analyzeRepeats.pl rna mm9 -d <tagDir> -count exons -condenseGenes -strand + -raw) 6. Normalization and differential expression analysis was done using the DESeq2 R-package 30. Differential expressed genes were selected using a 2-fold change cutoff between at least two populations and adjusted pValue for multiple gene testing < 0.05. Gene expression matrix was clustered using k-means algorithm (matlab function kmeans) with correlation as the distance metric. Heatmaps were generated using Genee software.
Analysis of Gene Features
For comparison of features between genes in cluster IIb and other genes, we used gene models from GENCODE vM13. Splicing efficiency was computed as by Tilgner and colleagues 7 using polyA+ RNA-seq data from microglia 8. Cytoplasmic/nuclear expression levels in liver and MIN6 cells were obtained using RNA-seq data from Bahar Halpern and colleagues 9 quantified using RSEM with GENCODE vM13 gene models. Cytoplasmic/nuclear ratios were computed using DESeq2 30.
Statistical Analysis
In all experiments, data are presented as mean ± standard deviation, if not stated otherwise. Statistical tests were selected based on appropriate assumptions with respect to data distribution and variance characteristics. Statistical significance was defined as P<0.05. Sample sizes were chosen according to standard guidelines. Number of animals is indicated as 'n'. Of note, the sizes of the tested animal groups were also dictated by availability of the transgenic strains and litter sizes, allowing littermate controls. Pre-established exclusion criteria are based on IACUC guidelines. Animals of the same age, sex and genetic background were randomly assigned to treatment groups. The investigator was not blind to the mouse group allocation, although tested samples were assayed blindly.
Supplemental Information
Acknowledgements
We would like to thank all members of the Jung laboratory for helpful discussion. We further thank the staff of the Weizmann Animal facility, members of the FACS facility for expert advice and G. Friedlander for help with the bioinformatics. We would like to thank C. Glass for sharing sequencing data. The Jung laboratory was supported by the Israeli Science Foundation (887/11), an Infect-ERA grant, the European Research Council (Adv ERC 340345), and the Deutsche Forschungsgemeinschaft (CRC/TRR167 ‘NeuroMac’).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Accession numbers
The accession numbers for the RNA-seq datasets reported in this paper can be found at GEO: xxxx
Author contributions
Z.H. and S.J. conceived the project and designed the experiments; Z.H., A.V. and J.O. performed the experiments; L. C.-M. performed RNAseq and E.D. analyzed the data. I.U. and B.Z performed bioinformatics analysis. S.Y., A. S., D. V., S. B.-H., I.B. and M. G. advised on experiments; Z.H. and S.J. wrote the paper; S.J. supervised the project.
References
- 1.Lavin Y, et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gosselin D, et al. Environment Drives Selection and Function of Enhancers Controlling Tissue-Specific Macrophage Identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heiman M, et al. A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Molecular and Cellular Biology. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 8.Goldmann T, et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nature Publishing Group. 2013;16:1618–1626. doi: 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orthgiess J, et al. Neurons exhibit Lyz2 promoter activity in vivo: Implications for using LysM-Cre mice in myeloid cell research. Eur J Immunol. 2016;46:1529–1532. doi: 10.1002/eji.201546108. [DOI] [PubMed] [Google Scholar]
- 11.Kim K-W, et al. In vivo structure/function and expression analysis of the CX3C chemokine fractalkine. 2011;118:e156–67. doi: 10.1182/blood-2011-04-348946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonseca MI, et al. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. 2017:1–15. doi: 10.1186/s12974-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmann T, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015:1–8. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 15.van den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Meth. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 16.A-Gonzalez N, et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J Exp Med. 2017;214:1281–1296. doi: 10.1084/jem.20161375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braunschweig Ulrich, et al. Widespread Intron Retention in Mammals Functionally Tunes Transcriptomes. Genome Research. 2014;24(11):1774–86. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khong A, et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. MOLCEL. 2017:1–19. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubstenberger A, et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. MOLCEL. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Tebaldi T, et al. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012;13:220. doi: 10.1186/1471-2164-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halpern KB, et al. Nuclear Retention of mRNA in Mammalian Tissues. CellReports. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirrlinger J, et al. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS ONE. 2009;4:e4286. doi: 10.1371/journal.pone.0004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf Y, et al. Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat Immunol. 2017;137:495. doi: 10.1038/ni.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutej H, et al. Diverging mRNA and Protein Networks in Activated Microglia Reveal SRSF3 Suppresses Translation of Highly Upregulated Innate Immune Transcripts. CellReports. 2017;21:3220–3233. doi: 10.1016/j.celrep.2017.11.058. [DOI] [PubMed] [Google Scholar]
- 26.Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting Activated Cell Populations Using Single- Cell RNA-Seq. Neuron. 2017;96:313–328.e7. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Buks E, Schuster R, Heiblum M, Mahalu D, Umansky V. Dephasing in Electron Intererence by a ‘Which Path’ Detector. Nature. 1998;391:871. [Google Scholar]
- 1.Yona S, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavin Y, et al. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Research. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosselin D, et al. Environment Drives Selection and Function of Enhancers Controlling Tissue-Specific Macrophage Identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halpern KB, et al. Nuclear Retention of mRNA in Mammalian Tissues. CellReports. 2015;13:2653–2662. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.