Abstract
We describe a method to transform blood lymphocytes into human induced pluripotent stem cells by delivering 4 transcription factors with a non-integrative virus. Using human peripheral blood mononuclear cells (PBMCs) as the source cell type for hiPSC reprogramming is advantageous since blood samples are rapidly and safely obtained from nearly-all subjects. Reprogramming factors needed to make hiPSCs are introduced by infecting the PBMCs with non-integrating Sendai virus vectors. Reprogrammed cells can subsequently be quickly expanded for downstream use. In this unit, we present current protocols for the isolation of PBMCs from a small sample of human blood and subsequent viral reprogramming and expansion of PBMCs into hiPSCs.
Keywords: pluripotent stem cells, iPSCs, reprogramming, Sendai virus, cell fate, PBMC
INTRODUCTION
The development of human induced pluripotent stem cells (hiPSCs) unlocked new possibilities in stem cell biology and regenerative medicine (Takahashi et al., 2007). For the first time, human pluripotent stem cells could be mass-produced from somatic tissue without necessitating early stage human embryos. This contrasts with the requirements for derivation of human embryonic stem cells (hESCs), which have been available for two decades but are subject to significant restrictions and ethical controversies (Thomson et al., 1998). Importantly, hiPSCs are similar to hESCs in terms of gene expression and epigenetics (Chin, Pellegrini, Plath, & Lowry, 2010). Like hESCs, hiPSCs can be differentiated into most somatic lineages by modulating intracellular cell signaling pathways critical to human organogenesis (Burridge et al., 2014). Additionally, hiPSCs are genetically identical to the host tissue from which they were derived, allowing for the mass-production of patient-specific human tissues for the purposes of in vitro disease modeling, high throughput drug screening, and potentially for in vivo cellular replacement therapy (Sharma, Wu, & Wu, 2013). Thus, improved methods for effective hiPSC production remain in high demand due to the utility of hiPSC derivative cell types.
Various methods exist for producing hiPSCs, and multiple aspects of the cellular reprogramming process can be substituted with alternative approaches. First, the source somatic cell type to be reprogrammed can include skin fibroblasts, peripheral blood mononuclear cells, and epithelial cells (Churko, Burridge, & Wu, 2013; Park et al., 2008; Zhou et al., 2012). Most cells containing a nucleus can be reprogrammed to hiPSCs, although perhaps the most widely-used cells are peripheral blood mononuclear cells (PBMCs) such as lymphocytes or monocytes. This is partly because blood-draws are safe, accessible, and fast outpatient procedures that allow for the bulk isolation of PBMCs that could be used for reprogramming. This contrasts with skin biopsies needed to procure dermal fibroblasts, a procedure that can be more painful, less sterile, and leaves scars. Second, the reprogramming factors needed to initiate pluripotency can be modified. Traditionally, four “Yamanaka factor” genes (OCT4, SOX2, KLF4, and MYC (OSKM)) are overexpressed in somatic cells to activate the core signaling networks needed to revert somatic cells to a pluripotent state, although successful reprogramming has also been demonstrated with alternate transcription factors (Takahashi & Yamanaka, 2016). Perhaps the most variable aspect of the reprogramming process is the vector chosen to express the OSKM reprogramming factors. Indeed, a variety of vectors can be used during the procedure with older approaches using lentiviral OSKM overexpression vectors (Jared M. Churko et al., 2017; Takahashi et al., 2007). However, the lentivirus vector has the disadvantage of integrating virally-introduced genetic sequences into the hiPSC genome, making it suboptimal for downstream applications requiring Good Manufacturing Practice (GMP) conditions (Rao & Malik, 2012). Many current reprogramming methods use non-integrating vectors, such as Sendai virus, to express the core reprogramming and pluripotency factors (Schlaeger et al., 2015). Traditional plasmid-based expression vectors can also be used in the reprogramming process (Diecke et al., 2015). Finally, successful reprogramming has been reported by directly delivering the OSKM proteins or mRNAs into the recipient somatic cell (Kim et al., 2009; Rohani et al., 2016). Regardless of variability in the individual steps, the hiPSC reprogramming process takes approximately 1 month.
Ultimately, reprogrammed hiPSC lines can be rapidly expanded and differentiated for a variety of downstream purposes. Here, we provide current protocols for hiPSC generation and expansion from human peripheral blood mononuclear cells using non-integrating, Sendai virus-based reprogramming.
STRATEGIC PLANNING
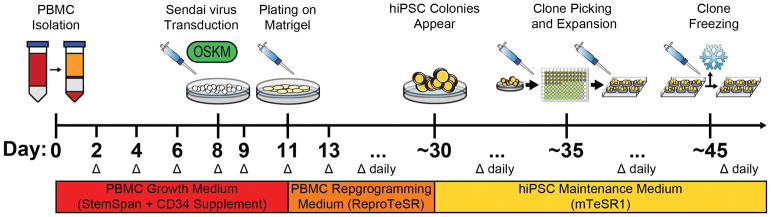
Generating hiPSCs by reprogramming PBMCs with non-integrating Sendai virus is a process that takes multiple weeks and requires careful planning. It is important to have all cell culture and reprogramming materials ready prior to starting the PBMC isolation and growth steps. It may be advantageous to create a schedule to keep track of the various steps in the reprogramming process. If many hiPSC lines are being produced, it may also be worthwhile to invest in an hiPSC clone picking microscope that can be placed in a designated cell culture hood to prevent contamination. PBMCs can be freshly isolated from whole human blood, or extracted from well-preserved blood samples. Although freshly isolated blood tends to be sterile, we still recommend designating an incubator for PBMC cell culture only and taking extra precautions by treating PBMCs as primary cells. Since whole human blood can be preserved, patient-specific blood samples can be shipped from distant locations if a patient with a genotype of interest is identified for hiPSC production. In terms of the amount of blood needed for PBMC isolation, we recommend 10 mL of whole blood, although others have reported success for PBMC isolation and hiPSC generation from as little as a single drop of human blood (Tan et al., 2014). During the PBMC isolation process, carefully extract the layer of cells containing PBMCs to prevent excess numbers of non-reprogrammable erythrocytes from contaminating the PBMCs to be reprogrammed. Additional of chemicals such as the fatty acid sodium butyrate (NaB) can further enhance reprogramming efficiency by facilitating epigenetic remodeling processes that occur during hiPSC formation (Mali et al., 2010). Feeder cells can be used to facilitate hiPSC reprogramming and growth, although most modern reprogramming methods do not require them (Nakagawa et al., 2014). Plan to freeze new hiPSC lines after production in the event of cell culture contamination or other anomalies. Figure 1 shows a timeline for hiPSC generation.
Figure 1.
Schematic depicting the timeline for hiPSC reprogramming from human peripheral blood using Sendai virus expressing the Yamanaka reprogramming factors. Expect this entire protocol to take more than one month from isolation to hiPSC line establishment. This timeline shows the days at which critical steps in the protocol are conducted and the media needed to maintain PBMCs and hiPSCs. A delta (Δ) indicates a media change.
BASIC PROTOCOL 1: ISOLATION AND EXPANSION OF HUMAN PBMCS FOR HIPSC GENERATION
This protocol will demonstrate how to isolate peripheral blood mononuclear cells (PBMCs) from human blood and subsequently expand PBMCs to prepare them for viral reprogramming to generate hiPSCs. Although a variety of somatic cell types can be used as the source material to make hiPSCs, PBMCs are easy to obtain via standard blood-draw and can be rapidly expanded when treated with the appropriate media containing PBMC-stimulating factors. A medium optimized for blood cell growth (StemSpan SFEM II + CD34+ Expansion Supplement) should facilitate proliferation of PBMCs. These mononuclear immune cells have nuclei that can be subjected to hiPSC reprogramming. This is in contrast with erythrocytes (red blood cells) which cannot be reprogrammed to hiPSCs due to the lack of a nucleus. The PBMC isolation and growth process will likely take up to 10 days, at which point an adequate number of PBMCs will have proliferated for use in downstream reprogramming. All cell culture should be conducted in a standard 37°C, 5% CO2 incubator using sterile technique. We recommend using a designated cell culture hood and centrifuge for PBMC preparation, isolation, and growth to prevent contamination. Extra care should be taken whenever handling human blood samples.
Materials
10 mL peripheral human blood
Phosphate-buffered saline (PBS; Life Technologies, cat. no. 20012-050)
500 mL cell culture media filters (Corning, cat. no. 431118)
StemSpan SFEM II blood medium (StemCell Technologies, cat. no. 09655)
StemSpan CD34+ Expansion Supplement (10X) (StemCell Technologies, cat. no. 02691)
0.5M EDTA (Thermo Fisher Scientific, cat. no. 15575020)
Ficoll Paque Plus (GE Healthcare, cat. no. 17-1440-03)
24-well tissue culture–treated plate
15- and 50-mL conical centrifuge tubes (e.g., BD Falcon)
Disposable plastic pipettes and tips
Standard cell culture laminar flow hood
Standard 37°C, 5% CO2 cell culture incubator
Cell culture centrifuge designated for centrifuging blood samples
Tissue culture microscope
Isolate human PBMCs from peripheral blood
Prepare PBMC growth media by adding 10 mL CD34+ Expansion Supplement to 500 mL StemSpan SFEM II. Filter and keep cool at 4°C until needed for use, at which point prewarm 25 mL of this PBMC growth media to 37°C.
Prepare 500 mL PBMC buffer solution by adding 2 mM EDTA to PBS. Filter and keep at to room temperature.
In a sterile cell culture hood, add 30 mL PBMC buffer solution to 10 mL human peripheral blood, obtained from a blood-draw, in a 50 mL conical tube. This is the buffered blood solution.
In a sterile cell culture hood, add 15 mL room temperature Ficoll Paque Plus stock to a new 50 mL conical tube.
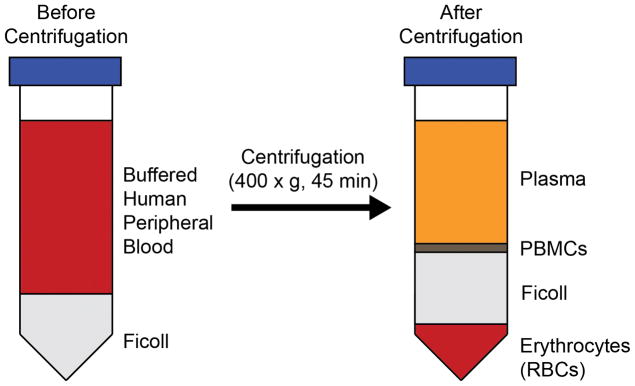
Very slowly add 35 mL buffered blood solution to the new 50 mL conical tube containing 15 mL Ficoll Paque Plus (Figure 2).
Centrifuge the 50 mL tube containing buffered blood and Ficoll Paque Plus at 400 × g for 45 minutes at room temperature.
After centrifuging, carefully place the 50 mL tube, containing the blood separated into its individual components, into a sterile cell culture hood (Figure 2).
Remove the top layer of the separated blood, which contains plasma, and discard it.
With a P1000 pipette, very carefully remove the interphase layer, which contains PBMCs, and transfer to a new 50 mL conical tube (Figure 2).
Add 45 mL PBMC buffer solution to the new conical tube containing PBMCs. Centrifuge at 250 × g for 15 minutes at room temperature.
After centrifuging, aspirate and remove supernatant, add 45 mL PBMC buffer solution, and again centrifuge PBMCs at 250 × g for 15 minutes at room temperature.
After centrifuging, aspirate and remove supernatant. Resuspend pelleted PBMCs in 1 mL prewarmed PBMC growth media.
Obtain a standard tissue culture-treated 24-well plate (no Matrigel precoating) and place it in the cell culture hood.
Add 1 mL PBMCs resuspended in PBMC growth media to one well of the 24-well plate. Return plate containing PBMCs to a designated 37°C, 5% CO2 cell culture incubator.
At 48 hours after plating, manually dissociate and resuspend all cells from the 24-well and add into a 15 mL conical tube. Centrifuge at 250 × g for 15 minutes at room temperature.
Aspirate supernatant and resuspend PBMCs in 1 mL fresh, prewarmed PBMC growth media.
Return 1 mL PBMCs resuspended in PBMC growth media to a new well in the tissue culture-treated 24-well plate. Return plate containing PBMCs to a designated 37°C, 5% CO2 cell culture incubator.
Continue conducting a PBMC growth media change (steps 15–17 above) every 2 days for approximately 6 more days. Cells should noticeably proliferate over the course of this PBMC culture period. When PBMCs have become approximately 95% confluent in the 24-well, they are ready for Sendai virus transduction.
Figure 2.
Schematic depicting separation of human peripheral blood before and after centrifugation in Ficoll. When subjected to a Ficoll gradient, whole blood will separate into layers containing plasma, PBMCs, Ficoll, and red blood cells. The PBMC layer will be thin and must be handled carefully.
BASIC PROTOCOL 2: TRANSDUCTION OF HUMAN PBMCS WITH SENDAI VIRUS HIPSC REPROGRAMMING VECTOR AND SUBSEQUENT HIPSC EXPANSION
This protocol will demonstrate how to properly transduce the PBMCs expanded in the previous protocol with a non-integrating Sendai virus vector expressing the transcription factors required for hiPSC reprogramming. Although a variety of viral, plasmid, protein, and mRNA-based reprogramming vectors exist, Sendai virus reprogramming is advantageous because it is highly effective for reprogramming PBMCs and because the transgenes expressed are not integrated into the hiPSC genome (Ban et al., 2011). The virally-transduced PBMCs will be grown in custom cell culture media optimized to promote the growth of PBMCs induced with hiPSC reprogramming factors. Over time, small hiPSC colonies will emerge from the reprogrammed cells, expressing the intracellular signaling networks required to maintain cellular pluripotency. Initially, expect these hiPSC colonies to grow slowly, and a moderate amount of cell death is likely following induction of hiPSC reprogramming. The hiPSC generation process could take up to one month or more, and we highly recommend diligent daily monitoring of transduced cells to quickly identify possible reprogrammed colonies. Ultimately, new hiPSC colonies can be picked and expanded to establish new hiPSC lines for downstream applications. All cell culture should be conducted in a standard 37°C, 5% CO2 incubator using sterile technique. Optionally, hiPSC reprogramming could also be conducted in a hypoxic incubator, as previous literature has demonstrated that hypoxia (5% oxygen instead of 20% at normoxia) may facilitate the reprogramming process (Yoshida, Takahashi, Okita, Ichisaka, & Yamanaka, 2009).
Materials
CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific, cat. no. A16517)
Matrigel (hESC-qualified; BD Sciences, cat. no. 354277)
Rho kinase inhibitor (ROCKi; Tocris cat. no. 1254)
500 mL cell culture media filters (Corning, cat. no. 431118)
Hemocytometer and trypan blue dye for cell counting
mTeSR1 medium (StemCell Technologies, cat. no. 05850)
ReproTeSR PBMC reprogramming medium (StemCell Technologies, cat. no. 05920)
DMEM/F12 medium (Thermo Fisher Scientific, cat. no. 11320033)
0.5M EDTA (Thermo Fisher Scientific, cat. no. 15575020)
6-, 24-, and 96-well tissue culture–treated plates
15- and 50-mL conical centrifuge tubes (e.g., BD Falcon)
Cell culture-grade water
Disposable plastic pipettes and tips
Standard cell culture laminar flow hood
Standard 37°C, 5% CO2 cell culture incubator
Cell culture centrifuge designated for centrifuging blood samples
Tissue culture microscope
Bambanker Cell Freezing Medium (Fisher Scientific, cat. no. NC9582225)
CoolCell LX (Sigma-Aldrich, cat. no. BCS-405)
Nunc™ Biobanking and Cell Culture Cryogenic Tubes (Fisher Scientific, cat. no. 375418)
Transduction of human PBMCs with Sendai virus hiPSC reprogramming vector
Prepare the Sendai virus cocktail in the CytoTune-iPS 2.0 Sendai Reprogramming Kit. Follow the manufacturer’s instructions to prepare a Sendai virus reprogramming cocktail volume so that 500,000 PBMCs will receive virus at a multiplicity of infection (MOI) of 5. The CytoTune-iPS 2.0 kit provides 3 separate viruses containing the Yamanaka reprogramming factors. Keep the cocktail on ice before use. A positive control GFP virus can also optionally be used to examine transduction efficiency.
Resuspend the PBMCs that were previously expanded in the PBMC growth media (in the designated 24-well from the previous protocol) and perform a cell count with a hemocytometer. Plate 500,000 PBMCs resuspended in 300 microliters total of PBMC growth medium into a new well of the 24-well plate.
Add the Sendai reprogramming viruses to the 300 microliters of PBMC growth medium containing PBMCs. Ideally, the total virus volume should not exceed 50 microliters. Return the plate to the 37°C, 5% CO2 incubator for 24 hours.
At 24 hours after transduction, remove all cells from the 24-well and transfer to a 15 mL conical tube.
Centrifuge the transduced PBMCs at 200 × g for 10 minutes. Remove the supernatant to remove the virus, and safely discard it. Resuspend the pelleted PBMCs in 500 microliters of fresh, prewarmed PBMC growth medium. Replate PBMCs into a new well in the 24-well plate. Return plate to 37°C, 5% CO2 incubator for 48 hours without media change.
Observe the cells daily after viral transduction. Significant cell death may occur, and morphological changes may begin.
Growth and expansion of Sendai virus-treated reprogrammed PBMCs
Prepare a 6-well plate coated with Matrigel extracellular matrix, on which reprogrammed PBMCs will grow after viral transduction. To coat plates with Matrigel, thaw Matrigel stock on ice and add 125 μL Matrigel to 50 mL of cold DMEM/F12 medium in a 1:400 dilution. Ensure that pipette tips are cooled to 4°C before pipetting Matrigel to prevent premature solidification of Matrigel. Two mL of Matrigel solution should be added per well of a 6-well dish. Matrigel should be allowed to coat the dish for at least 1 hour before use.
At 72 hours after viral transduction, collect the PBMCs being reprogrammed into a 15 mL conical tube.
Centrifuge the transduced PBMCs at 200 × g for 10 minutes. Remove the supernatant, and then resuspend the cells in 2 mL of fresh, prewarmed PBMC reprogramming medium (ReproTeSR).
Aspirate 1 well of the Matrigel pre-coated 6-well plate prepared previously. Replate all the PBMCs (suspended in ReproTeSR PBMC reprogramming medium) into the new well in the 6-well plate. Return the plate to the 37°C, 5% CO2 incubator for 48 hours.
After plating on Matrigel, continue observing the reprogrammed PBMCs for signs of morphology change. They should begin adhering to the Matrigel-coated well.
At 48 hours after plating reprogrammed PBMCs onto Matrigel, very gently aspirate and remove the old PBMC reprogramming medium. PBMCs may be loosely attached to the Matrigel at this point, so care must be taken not to dislodge them.
Gently add 2 mL fresh, pre-warmed PBMC reprogramming medium to the 6-well containing the reprogrammed PBMCs.
Continue gently replacing PBMC reprogramming medium daily for approximately 2–3 weeks, or until initial hiPSC colonies begin appearing. During this period, cells should begin proliferating and changing morphology, possibly beginning to adopt the tightly packed hiPSC colony morphology (Figure 3A).
When hiPSC colonies are observed, begin daily media change with 2 mL hiPSC maintenance medium (mTeSR1).
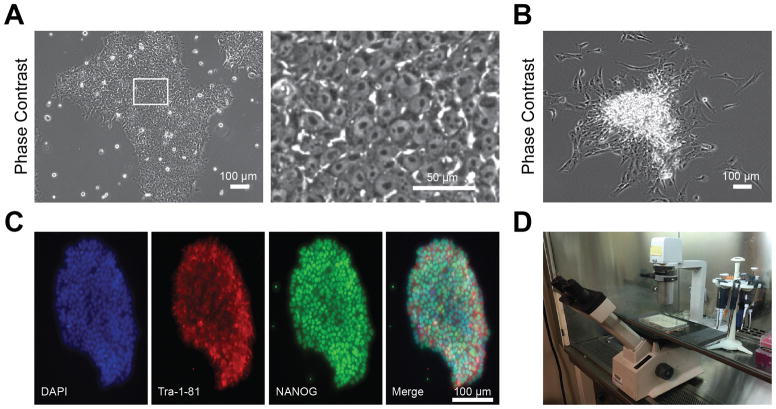
Figure 3.
Generation, characterization, and isolation of optimal hiPSCs from blood using Sendai virus reprogramming. A) Phase contrast images illustrating optimal hiPSC morphology following reprogramming. Note how hiPSCs are tightly packed into a colony. Inset depicts high magnification image, showing individual hiPSCs with characteristic large nuclei and small amounts of cytoplasm per cell. B) Phase contrast image illustrating suboptimal hiPSC morphology. Note how hiPSCs are spontaneously differentiating at the edges of the colony. C) Immunofluorescence staining for standard hiPSC markers including Tra-1-81 and NANOG. Optimal hiPSCs should express pluripotent stem cell markers such as these. D) Example microscope setup for hiPSC clone isolation. Microscope should be placed within a sterile cell culture hood.
Isolation, expansion, and storage of newly generated hiPSCs
Prepare new 96-, 24-, and 6-well plates coated with Matrigel extracellular matrix, as described above, into which new hiPSC colonies will be picked for isolation and subsequent expansion.
After hiPSC colonies appear in the 6-well plate containing reprogrammed cells and medium has been changed to hiPSC maintenance medium (mTeSR1), continue monitoring the colonies as they grow. Optimal hiPSC colonies will be tightly packed and contain cells with large nuclei and low amounts of cytoplasm, whereas suboptimal hiPSC colonies will often exhibit spontaneous differentiation (Figure 3A–B). Optimal hiPSC colonies will also tend to express characteristic marker proteins, such as NANOG and Tra-1-81, that are commonly associated with pluripotent stem cells (Figure 3C).
When an individual colony reaches approximately 200 cells in size or 200 microns in diameter, pick it carefully with a P10 pipette tip using a microscope in a sterile cell culture hood (Figure 3D). Place the colony into a Matrigel pre-coated 96-well containing 100 microliters of hiPSC maintenance medium with 10 μM rho kinase inhibitor diluted in water.
Continue picking reprogrammed hiPSC colonies from the 6-well dish into the 96-well dish until a desired number of colonies have been isolated for expansion. We recommend picking a minimum of 12 colonies.
At 24 hours after picking hiPSC clones into the 96-well plate, replace media with fresh hiPSC maintenance medium with 10 μM rho kinase inhibitor.
At 48 hours after picking hiPSC clones into the 96-well plate, replace media with fresh hiPSC maintenance medium without 10 μM rho kinase inhibitor. Keep replacing hiPSC maintenance medium for 3–4 days in the 96 well plate until hiPSC colonies have reached 1000 cells in size, at which point they can be seen with the naked eye.
-
When hiPSC clones in the 96-wells are visible to the naked eye, passage the colonies at a 1:1 dilution in hiPSC maintenance medium containing 10 μM rho kinase inhibitor into the same wells of the 96-well plate. This is to redistribute the hiPSCs evenly within the same well, allowing for homogenous growth of the clones without the center of a colony becoming overconfluent.
To conduct a standard hiPSC passage, first aspirate old hiPSC maintenance medium from a well. Then, add 0.5 mM EDTA diluted in PBS (100 microliters for a 96 well) for 5 minutes. After 5 minutes, aspirate EDTA gently from the well. Then, add 100 μM hiPSC maintenance medium containing 10μM rho kinase inhibitor to the well. Using a P200 pipettor, manually dislodge hiPSCs by repeatedly spraying and resuspending the cells off the well into this fresh medium.
At 24 hours after redistributing the hiPSC colonies in a 96-well plate, return media back to regular hiPSC maintenance medium without rho kinase inhibitor. Keep replacing hiPSC maintenance medium until hiPSCs in the 96-well plate achieve 95% confluency.
After hiPSCs in the 96-well have achieved 95% confluency, conduct a standard hiPSC passaging step, as described above, at a 1:1 dilution resuspended in hiPSC maintenance medium containing 10 μM rho kinase inhibitor. Move the entire cell suspension into a single well of a Matrigel pre-coated 24-well plate so that hiPSC clones have more surface area to grow.
At 24 hours post-passaging cells into a 24-well plate, return hiPSCs to regular hiPSC maintenance medium without rho kinase inhibitor. Replace hiPSC maintenance medium every 24 hours until hiPSCs reach approximately 90% confluency in a well of the 24-well plate.
After hiPSCs reach approximately 90% confluency in the 24 well, passage hiPSCs at 1:1 dilution in hiPSC maintenance medium containing 10 μM rho kinase inhibitor. Move the entire hiPSC suspension to a single well of a Matrigel pre-coated 6-well plate so that the hiPSC clones have more surface area to grow and proliferate.
At 24 hours after passaging hiPSCs to a 6-well place, return hiPSCs to regular hiPSC maintenance medium without rho kinase inhibitor. Replace regular hiPSC maintenance medium every day until hiPSCs reach approximately 90% confluency in the 6-well plate.
After hiPSCs reach approximately 90% confluency in the single well of the 6-well plate, passage hiPSCs at 1:10 dilution in hiPSC maintenance medium with 10 μM rho kinase inhibitor. Transfer 1:10 of the cell dilution into each well of a new, Matrigel pre-coated 6-well plate. These hiPSCs are now ready for long-term cell culture. Ideally, a 1:10 passage should be conducted every 4 days on 90% confluent hiPSCs using hiPSC maintenance medium with 10 μM rho kinase inhibitor. The new hiPSC lines may grow and differentiate best after passage 10.
-
Create a backup containing leftover hiPSCs from the previous 6-well passage step by freezing hiPSCs in Bambanker freezing solution.
Centrifuge the leftover hiPSCs for 5 minutes at 200 × g in a 15 mL conical tube.
After aspirating the supernatant, resuspend the hiPSC pellet in Bambanker at a target concentration of 1 million cells per mL Bambanker.
Add the Bambanker-hiPSC solution securely to a 1.8 mL cryo-tube, and put it into a CoolCell LX freezing apparatus.
Move the CoolCell LX containing the cryo-tubes to an −80°C freezer so that the cells can slowly and safely freeze at a rate of −1°C per minute.
After the temperature has dropped to −80°C, cryo-tubes should be removed from the CoolCell LX and kept at −80°C. If the cells are to be stored for a longer period, transfer cryo-tubes to a secure liquid nitrogen cell storage tank.
REAGENTS AND SOLUTIONS
Shelf life for all cell culture media is 1 month at 4°C.
Matrigel-diluted medium
Add Matrigel stock (hESC-qualified; BD Sciences, cat. no. 354277) to DMEM/F12 medium (Thermo Fisher Scientific, cat. no. 11320033) at a 1:400 dilution to coat plates for hiPSC culture. It is important to thaw Matrigel stock at 4°C and to dilute Matrigel in cold DMEM/F12 quickly to prevent premature Matrigel solidification. Once plates have been coated, store plates at 37°C until needed. When ready for use, aspirate Matrigel dilution from individual wells and add fresh cell culture media specific to hiPSCs, such as mTeSR1. We do not recommend repeatedly freeze-thawing Matrigel stock.
PBMC buffer solution
Phosphate-buffered saline (PBS; Life Technologies, cat. no. 20012-050) + 2 mM EDTA (Thermo Fisher Scientific, cat. no. 15575020)
PBMC growth medium
StemSpan SFEM II + CD34+ Expansion Supplement. Optimized for PBMC growth. (StemCell Technologies, cat. no. 09605 and 02691)
PBMC reprogramming medium
ReproTeSR PBMC reprogramming medium for hiPSC induction (StemCell Technologies, cat. no. 05920)
hiPSC maintenance medium (for daily hiPSC culture)
mTeSR1 feeder-free pluripotent stem cell maintenance medium (StemCell Technologies, cat. no. 05850)
hiPSC passaging medium (for passaging hiPSCs and maintaining small colonies after passaging)
mTeSR1 feeder-free pluripotent stem cell maintenance medium containing:
10 μM rho kinase inhibitor
Rho kinase inhibitor (ROCKi) stock solution
Add 3.12 mL cell culture-grade water to 10 mg rho kinase inhibitor (Tocris, cat. no. 1254) to achieve a 10 mM stock solution (1000x). Aliquot and store at −20°C.
COMMENTARY
Background Information
Human iPSCs have attracted significant interest for their potential to accelerate in vitro disease modeling, high-throughput drug screening, and potentially clinical cell therapy (Sharma et al., 2017; Sharma et al., 2014; Sharma et al., 2013). Almost any adult somatic cell type can be produced in vitro from hiPSC differentiation protocols in nearly unlimited quantities, since hiPSCs can be rapidly scaled up and quickly differentiated. This has proven to be quite useful for areas of research such as cardiovascular biology and neuroscience where some cell types of interest, such as cardiomyocytes and neurons, are either very difficult to procure from primary samples or are largely non-proliferative. An entire industry has developed around the efficient production, maintenance, and differentiation of hiPSCs, and training programs have been created to educate students and clinicians about the basic science and applications of this technology. Prior to the development of hiPSC technology, human pluripotent stem cells could only be derived by extracting cells from the early stage blastocysts of human embryos, a procedure fraught with ethical controversy (Thomson et al., 1998). In the United States, hESC research was struck with a federal funding moratorium during the 2000s, limiting the derivation of new hESC lines. With the seminal paper from Yamanaka and Takahashi in 2006 that described mouse iPSC production, new doors were unlocked that further enabled the progression of human regenerative medicine towards the aforementioned applications (Takahashi & Yamanaka, 2006). Their elegant work revealed that only four pluripotency-regulating factors are required to revert differentiated somatic cells back to a pluripotent state in mice, and human iPSC derivation was not far behind (Takahashi et al., 2007). Perhaps what made this study especially powerful was how easily reproducible it was. Indeed, labs around the world were able to derive hiPSCs in a matter of months after the initial iPSC study was published (Blelloch, Venere, Yen, & Ramalho-Santos, 2007). In addition to the bioethical advantages, hiPSCs could also be considered superior to hESCs because of their personalized nature. These hiPSCs exhibit the same genetic background as the adult individual from which they are derived, potentially allowing for new advances in personalized medicine and offering advantages in immunocompatibility for downstream cell therapies. Notably, the discovery of hiPSCs also accelerated research in direct cellular reprogramming, whereby terminally-differentiated somatic cell types such as fibroblasts, neurons, and cardiomyocytes can be directly interconverted without passing through a pluripotent state (Chuang et al., 2017; Ieda et al., 2010; Vierbuchen et al., 2010).
Vast technical improvements have been made over the past decade to enhance hiPSC reprogramming safety and efficacy, with one eye towards the future in hopes of generating GMP-grade hiPSCs and their derivatives for clinical therapies. The reprogramming vectors presented in the original hiPSC derivation papers were mostly lentivirus, which is effective but poses a risk for DNA integration into the newly created hiPSC lines. Of course, this is suboptimal for generating clinical grade hiPSCs, which ideally would be genetically identical to the individual from which they are derived. However, this shortcoming sparked further work into developing genetically optimal alternative vectors that could effectively express the Yamanaka pluripotency factors. A variety of plasmid, mRNA, and protein-based OSKM expression vectors have been developed over the past decade but tend to be less efficient than viral vectors in inducing reprogramming (Jared M. Churko et al., 2017; Diecke et al., 2015; Kim et al., 2009; Rohani et al., 2016). Perhaps the most widely-used of these second-generation OSKM expression vectors still utilizes a variety of virus, albeit a non-integrating one. The Sendai virus, a single-stranded RNA virus, was shown to be highly effective as a vector in transducing human somatic cells with the OSKM pluripotency factors, and unlike the DNA-integrating lentivirus in the retrovirus family, the expression of the pluripotency factors with Sendai virus is transient (Fusaki, Ban, Nishiyama, Saeki, & Hasegawa, 2009). OSKM-expressing Sendai virus vectors have also effectively been employed to generate iPSCs from non-human mammalian species, reflecting the conserved nature of the pluripotency-regulating gene networks (Fujie et al., 2014). This offers new possibilities in studying mammalian development, since embryonic stem cell generation from certain species has been notoriously difficult. Additionally, the ability to readily reprogram mononuclear cells in blood has only served to further expedite the reprogramming process, particularly for human patients hesitant to receive a potentially painful and scar-inducing skin biopsy. The use of blood also exhibits other advantages since blood can be readily procured within minutes in an outpatient setting, is largely sterile and devoid of contaminants that can interfere with cell culture, and contains a variety of blood cell types that can be quickly expanded when treated with mitotic stimulants. When taken in combination, these developments elicit a blood/Sendai virus hiPSC reprogramming and expansion protocol that we believe is safer and more effective in comparison to prior hiPSC production methods.
Critical Parameters and Troubleshooting
There are multiple critical parameters that will influence the success of the blood/Sendai virus hiPSC reprogramming protocol. We recommend freshly isolated blood for PBMC isolation to maximize PBMC survival after plating. The confluency of the PBMCs when subjected to the reprogramming Sendai virus could influence the efficacy of viral uptake. We recommend between 100,000–500,000 cells per reprogramming reaction, and cells should not be allowed to become overconfluent before initiating reprogramming. Overconfluency can be observed visually by seeing a media color change from red to yellow if cell culture media contains phenol red. When introducing the Sendai virus, it may be worthwhile to introduce a positive control GFP virus to examine transduction efficiency. We recommend using freshly aliquoted virus without multiple freeze/thaw cycles. If successful reprogramming is not achieved, consider using a hypoxic incubator (5% oxygen instead of 20% at normoxia) during the reprogramming process, as hypoxia has been reported to facilitate hiPSC conversion (Yoshida et al., 2009). Conducting a 50/50 instead of a complete medium switch when initially transitioning from different PBMC and hiPSC media types may further improve cell survival. The reprogramming process is particularly sensitive to standard operating procedures that should be routine for all cultured cells. For example, these cells usually will frequently die in expired culture media, or cold reprogramming media that would not kill other more robust human cell lines. Low levels of mycoplasma will also prevent successful reprogramming and frequent mycoplasma testing can improve hiPSC reprogramming efficiency. When picking hiPSC clones for cell line establishment after reprogramming, aim to pick colonies that are between 100–200 cells large. Larger colonies may spontaneously differentiate when replated, as portions of the replated large colony may fold on top of each other (Figure 3B). Very small colonies, when picked and replated, may not survive well. Addition of rho kinase inhibitor for multiple days after clone picking may dramatically improve hiPSC survival (Watanabe et al., 2007). It may be a good idea for beginners to practice picking and replating wild type colonies before moving to important reprogrammed cells. Newly-picked hiPSC colonies may proliferate slowly for a few passages, possibly up to passage 5. After thawing frozen hiPSC stocks, it is also normal to observe some cell death. Finally, regular karyotyping of hiPSC lines is recommended, as chromosomal abnormalities may arise in high-passage cells after long-term culture.
Interpreting Results
If followed appropriately, we believe that the protocols presented here will successfully generate hiPSCs from PBMCs using a non-integrating Sendai virus vector. Expect an hiPSC reprogramming efficiency ranging from 0.1–0.5% with this protocol. That is, if 100,000 PBMCs are subjected to reprogramming, expect approximately 100–500 colonies. However, this can highly vary based on the PBMC proliferation rate and survival. After successful reprogramming, ideal hiPSC colonies should resemble hESC morphology, with tightly packed cells devoid of fibroblast-like extensions (Figure 3A). The individual cells in an ideal hiPSC colony will have a large nucleus dominating the cell body and a round shape. Low quality hiPSC clones will not exhibit this ideal morphology and may spontaneously differentiate or proliferate rapidly, analogous to fibroblasts (Figure 3B). When immunostained, ideal hiPSC colonies will exhibit pluripotency markers such as NANOG, Tra-1-81, and SSEA4 (Figure 3C). After reprogramming, these hiPSCs can be grown on feeder cells. However, this is not required to maintain hiPSCs, and we recommend a feeder-free environment to reduce additional variables. The hiPSCs can be successfully frozen using a freezing medium such as Bambanker, stored for long-term in liquid nitrogen, and thawed with minimal cell death. Ultimately, successful hiPSC production will be verified by the ability to differentiate into cell types of interest from all three germ layers: mesoderm, endoderm, and ectoderm.
Time Considerations
Generation of hiPSCs from blood will require a significant amount of time and effort in cell culture with daily maintenance required. The entire process could take more than one month. During PBMC culture, hiPSC induction, hiPSC reprogramming, and hiPSC culture steps, the appropriate cell culture media will need to be changed regularly. Using the correct media is critical to the reprogramming process, especially during the first week after induction. Keep a close watch on reprogrammed cells during this critical period during the first week of reprogramming. Isolating individual hiPSC clones after the reprogramming process can be laborious, and using the appropriate cell culture microscope during clone picking can dramatically speed up the process. If many hiPSC lines are being created, it may be worthwhile to invest in a high-quality cell culture microscope for use in a sterile cell culture hood. Regular use of rho kinase inhibitor during the clone picking process can dramatically improve hiPSC survival after replating, especially if colonies are small or growing slowly. We also highly recommend expanding and freezing newly-picked clones at early passages (passage 5 is ideal) to prevent clone loss in case of cell culture contamination or other unforeseen scenarios.
SIGNIFICANCE STATEMENT.
Human induced pluripotent stem cells (hiPSCs) are custom, man-made pluripotent stem cells that can be differentiated into many cell types including those found in the heart, brain, liver, and other organs. hiPSCs have been used for research and clinical purposes. Unlike human embryonic stem cells (hESCs), hiPSCs can be created without harvesting cells from human embryos, a significant ethical and technical advantage. These hiPSCs can be rapidly produced from most human tissues, including blood, in a cellular reprogramming process that takes approximately one month. Although a variety of protocols exist for hiPSC production, here we describe the use of a non-integrating viral vector, which promotes highly efficient cellular reprogramming without leaving any copies of the transforming virus genome in the iPSCs.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH) Grants 1R01HL080494 and 1R01HL084553 (to C.E.S.), the Foundation Leducq (C.E.S.), the Howard Hughes Medical Institute (C.E.S.), and an NIH NHLBI Cardiovascular Development Consortium (CVDC) Postdoctoral Research Fellowship (A.S.).
Footnotes
CONFLICT OF INTEREST
None.
LITERATURE CITED
- Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, … Nishikawa S. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A. 2011;108(34):14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1(3):245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, … Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7(2):263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang W, Sharma A, Shukla P, Li G, Mall M, Rajarajan K, … Wu SM. Partial Reprogramming of Pluripotent Stem Cell-Derived Cardiomyocytes into Neurons. Sci Rep. 2017;7:44840. doi: 10.1038/srep44840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods in molecular biology. 2013;1036:81–88. doi: 10.1007/978-1-62703-511-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Lee J, Ameen M, Gu M, Venkatasubramanian M, Diecke S, … Wu JC. Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. Nature Biomedical Engineering. 2017;1(10):826–837. doi: 10.1038/s41551-017-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diecke S, Lu J, Lee J, Termglinchan V, Kooreman NG, Burridge PW, … Wu JC. Novel codon-optimized mini-intronic plasmid for efficient, inexpensive, and xeno-free induction of pluripotency. Sci Rep. 2015;5:8081. doi: 10.1038/srep08081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie Y, Fusaki N, Katayama T, Hamasaki M, Soejima Y, Soga M, … Era T. New type of Sendai virus vector provides transgene-free iPS cells derived from chimpanzee blood. PLoS One. 2014;9(12):e113052. doi: 10.1371/journal.pone.0113052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, … Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, … Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Taniguchi Y, Senda S, Takizawa N, Ichisaka T, Asano K, … Yamanaka S. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, … Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Rao MS, Malik N. Assessing iPSC reprogramming methods for their suitability in translational medicine. J Cell Biochem. 2012;113(10):3061–3068. doi: 10.1002/jcb.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani L, Fabian C, Holland H, Naaldijk Y, Dressel R, Loffler-Wirth H, … Stolzing A. Generation of human induced pluripotent stem cells using non-synthetic mRNA. Stem Cell Res. 2016;16(3):662–672. doi: 10.1016/j.scr.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, … Daley GQ. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33(1):58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, … Wu JC. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Science translational medicine. 2017;9(377) doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, … Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circulation research. 2014;115(6):556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Wu JC, Wu SM. Induced pluripotent stem cell-derived cardiomyocytes for cardiovascular disease modeling and drug screening. Stem Cell Res Ther. 2013;4(6):150. doi: 10.1186/scrt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17(3):183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tan HK, Toh CX, Ma D, Yang B, Liu TM, Lu J, … Loh YH. Human finger-prick induced pluripotent stem cells facilitate the development of stem cell banking. Stem Cells Transl Med. 2014;3(5):586–598. doi: 10.5966/sctm.2013-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, … Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25(6):681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5(3):237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, … Esteban MA. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012;7(12):2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]