Abstract
Background
Clinical genetic testing for heritable cardiovascular disease has become a widely-used tool to aid in the management of patients and their families. A five-category variant classification system is commonly used for genetic test results, but some laboratories further sub-classify variants of uncertain significance (VUS). How and whether patients perceive differences among the variant categories or sub-classifications of VUS is unknown.
Methods and Results
We tested whether participants perceived differences in genetic variant sub-classifications on outcomes including: risk comprehension, risk perception, worry, perceived uncertainty, and intentions. Order-randomized hypothetical cardiovascular genetic results were given to 289 participants enrolled in a genome sequencing study. Three categories of variants were presented to participants: VUS, Possibly Pathogenic, and Likely Pathogenic. Responses to the first variant presented were analyzed in a between-groups analysis, and responses to all three variants were analyzed in a within-groups analysis. When presented with all three results, participants distinguished among the sub-classifications on all outcomes (p<0.001). When given only a Possibly Pathogenic result, their risk perceptions were similar to those of a VUS, but they were more worried and intended to behave as if they had received a Likely Pathogenic result. Individuals depended more on their affective responses such as worry when they received only one result (p<0.05).
Conclusions
Participants are better able to distinguish pathogenicity sub-classifications when presented with multiple categories. Individuals who receive a single uncertain result in a cardiovascular disease gene may benefit from interventions to decrease worry, calibrate risk perceptions, and motivate variant-appropriate behaviors.
Keywords: genetic variation, genetic heart disease, genetic testing, classification, uncertainty, variant of uncertain significance
Journal Subject Terms: Genetics, Cardiovascular Disease, Behavioral/Psychosocial Treatment, Health Services, Diagnostic Testing
INTRODUCTION
Large multi-gene panels and exome and genome sequencing are being used increasingly to aid in the identification of the underlying genetic etiology of a variety of inherited conditions including familial cardiovascular disease1. This approach can be effective, particularly when the phenotype is equivocal, genetic heterogeneity high, or there is a family history suggestive of multiple co-existing conditions. In practice, commercial testing companies are offering ever-expanding cardiovascular panels2 to affected patients. Patients are also referred to cardiology clinics following identification of variants as secondary findings from genomic sequencing completed for clinical or research purposes as well as from direct-to-consumer testing. One of the greatest challenges in each scenario is determining whether an identified sequence variant is a cause of a disease3.
The ability to sequence has outpaced our ability to interpret the output, and thus sequencing many genes at once often leads to uncertain results4. Updated guidelines from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) recommended that sequence variants be divided into five categories: Pathogenic, Likely Pathogenic, Benign, Likely Benign, or Variants of Uncertain Significance (VUS)5. Some laboratories use classification schemes with different descriptors or more than five categories6,7,8,9. For example, some laboratories divide VUS results into two categories, “Possibly Pathogenic,” which is at the highest end of the ACMG/AMP VUS range, and VUS, which lies between Possibly Pathogenic and the ACMG/AMP Likely Benign classification. The clinical utility of this sub-classification is unclear.
In general, receiving a VUS from a laboratory can be challenging for both providers and their patients10. In one study, just over half of genetic counselors believed their patients understood their test results when given a VUS11. Patients’ interpretations of VUS have been shown to vary widely, with some reporting that they had an undetectable gene mutation and others believing that their disease diagnosis had no genetic basis12. Research is needed on how patients react to the five ACMG/AMP variant classifications as well as other classification schemes. Some laboratories find it useful to sub-classify VUS results because the category has such a broad range of pathogenicity associated with it as originally defined. In using sub-classifications of genetic variants, it could be assumed that participants are able to distinguish among them. This is the hypothesis we test in this study. It is important to explore outcomes of receiving these types of genetic results in cardiology with the goal of improving communication, understanding, and management not only in cardiology but for other health conditions.
In this study, we presented participants enrolled in a larger sequencing study with three hypothetical scenarios of cardiovascular genetic test results. One scenario asked them to imagine receiving a Likely Pathogenic result, and another a VUS, two of the ACMG/AMP classifications. In addition, they were asked to imagine receiving a “Possibly Pathogenic” variant, a sub-classification of the VUS category. Because participants were exposed to all three scenarios, it was possible to assess the extent to which they discerned differences among the three. Importantly, however, participants received the three scenarios in a randomized order, and doing so allowed us to assess responses to just the first scenario in a between-groups design. The latter analysis allowed us to determine if participants appreciated differences among the three variant classifications when only presented with one result, a common circumstance in clinical practice.
MATERIALS AND METHODS
Overview
The analytic methods and study materials will be made available to other researchers, but the data collected will not be made available to other researchers for purposes of reproducing the results or replicating the procedure. The authors confirm that some restrictions apply to the data underlying the findings as the consent signed by participants does not allow the public release of their data. We measured the following outcomes of receiving genetic test results: perceived risk, risk comprehension, worry, and perceived uncertainty13. We also examined how variant classifications influenced behavioral intentions. Specifically, our primary behavioral intention outcome was intention to share the genetic information with family members. We also measured secondary intentions including intentions to change health behaviors. These secondary intentions are more relevant to patients receiving cardiovascular variants as a secondary finding or through direct to consumer tests than for affected index cases. Finally, we determined whether the effects of the specific variant classification received on these behavioral intentions were explained by (or mediated by) the other outcomes that we measured (e.g., perceived risk).
Participants
Potential participants were recruited from two cohorts enrolled in the NIH ClinSeq® study14. The ClinSeq® study addresses the integration of genome sequencing into clinical research and care by assessing the technical, medical, and socio-behavioral aspects of implementing this technology. The first cohort primarily consisted of Caucasian individuals, largely of high socioeconomic status, who have been characterized as early adopters of a new health technology15. Roughly 25% of them were enrolled based on a diagnosis of atherosclerotic heart disease. The second cohort, which was actively enrolling at the time of this study, targeted individuals who self-identified as African-American, African or Afro-Caribbean and meet the same study eligibility criteria. Individuals in the current experiment must not have previously received genetic results that would affect their personal health or carrier results that included a VUS. These exclusionary criteria reduced the likelihood that reactions to uncertain genetic test results presented in this experiment would be influenced by past experiences receiving results. Our participants were recruited by phone, by postal mail, and/or by secure email and provided an online link which contained the consent document, hypothetical results, and survey questions. In addition, the survey asked the participants to provide their ClinSeq® identification numbers so that responses could be linked with demographic information collected in a previous ClinSeq® baseline survey.
This research was reviewed and approved by the National Human Genome Research Institute Institutional Review Board at the National Institutes of Health. Participants provided written informed consent for the overall protocol at the time of enrollment and indicated their consent for this specific ancillary study by checking a box on the first page of the survey. Participants were mailed a letter after the completion of the survey to thank them and to remind them that the test results were hypothetical.
Procedure
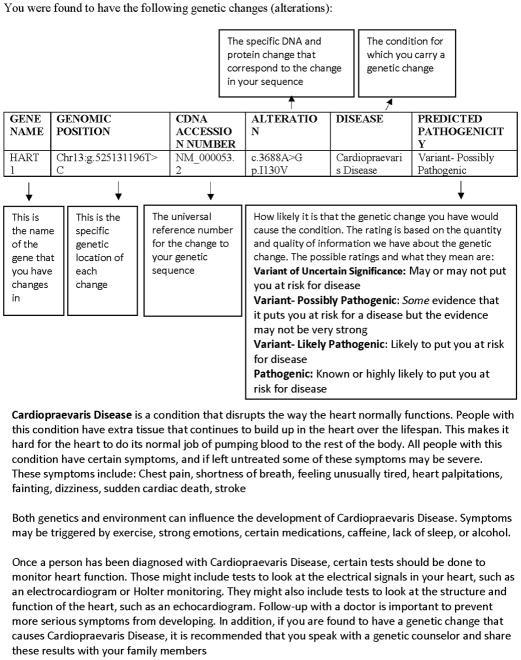
Participants received three hypothetical genetic test results via a web platform in a randomized order: “Variant of Uncertain Significance” (“may or may not put you at risk for disease”), “Variant- Possibly Pathogenic” (“some evidence that it puts you at risk for a disease but the evidence may not be very strong”), and “Variant- Likely Pathogenic” (“likely to put you at risk for a disease”) related to a hypothetical cardiovascular disease. To increase the reality of the hypothetical scenario, the result report (see Figure 1) included a brief description of the disease with which the gene variants are associated, an explanation of the variant classification system presenting a description of all possible types of variants, and screening recommendations for those with Pathogenic variants in this gene. As seen in Figure 1, participants were told that this disease disrupts the way the heart normally functions and could lead to symptoms such as chest pain, shortness of breath, feeling unusually tired, palpitations, fainting, dizziness, sudden cardiac arrest, or stroke. Participants were told that if they were diagnosed with this disease, certain tests such as ECG and Holter monitor testing should be done in addition to following up with specialty providers, speaking with a genetic counselor, and sharing the information with family members. After being presented with each hypothetical result, participants were asked to complete all dependent measures.
Figure 1. Hypothetical Results Report.
The hypothetical genetic results report presented to participants. Each participant was presented with three result reports in randomized order with predicted pathogenicity differing on each report: Variant of Uncertain Significance, Variant- Possibly Pathogenic, or Variant- Likely Pathogenic. All of the reports contained identical information regarding gene name, genomic position, CDNA accession number, alteration, disease, and disease description.
Measures
Outcome measures included risk comprehension, risk perception, worry, perceived uncertainty, and intentions to share the results with family members. The measurement scales can be seen in S1, which contains a copy of the full survey instrument used in this study.
Risk comprehension was measured by asking the participant to rate how harmful the genetic change was on a seven-point scale ranging from 1= “not harmful at all” to 7= “extremely harmful.” We measured risk perception with three items shown in past research to capture different aspects of perceived risk- absolute, experiential, and comparative risk16. Absolute risk perception refers to the cognitive conception of risk (e.g., how at risk participants think they are). Experiential risk represents intuitive or “gut feeling” response to risk (e.g., how at risk the participants feel, which may differ than their absolute risk perception). Comparative risk perception requires participants to compare their risk with others their same age and sex. Participants also reported how worried they were regarding three outcomes: 1) that your genetic finding puts you at an increased risk for developing heart disease, 2) that your existing health condition is caused by this genetic change, and 3) that your relatives could be affected by this genetic condition17.
Perceived uncertainty was measured using the Personal Uncertainty in Genomic Sequencing (PUGS) Scale18. This scale assesses practical uncertainties of genetic test results in three domains: clinical uncertainty (Cronbach’s α=0.86), affective uncertainty (Cronbach’s α=0.92), and credibility (Cronbach’s α=0.96).
The primary measure of intentions to share the results with family members was measured on a seven-point scale from 1= “extremely unlikely” to 7= “extremely likely.” In addition to the intentions to share results with family members, we also measured intentions to change lifestyle, undergo screening, seek genetic counseling, and seek specialty care19.
Statistical Analysis
Survey data were de-identified before analysis. Analyses were completed using STATA version 13. Variant classifications were treated as categorical variables and all of the dependent variables were treated as continuous. Frequency distributions of the main outcome variables were examined to ensure they were not heavily skewed and standard deviations were examined across conditions to ensure there were not substantial differences (homoscedasticity). To compare means, ANOVA and post-hoc Tukey tests were used. To assess if risk comprehension, risk perception, worry, and perceived uncertainty explained (“mediated”) the effects of variant received on behavioral intentions, we followed the conservative Baron and Kenny criteria for mediation followed by the Sobel-Goodman test to determine the significance of the differences in variance explained20. A within-groups analysis was performed using all participant responses, and a between-groups analysis focused on responses to the first variant received. For the purposes of our analyses, p-values <0.05 were considered significant. However, most analyses yielded significance much lower than that, and had we adopted p=0.01 as a more conservative criterion to account for multiple comparisons, only one of the many analyses would have become nonsignificant (at p=0.02).
Uncertain Responses
Our seven-point scales included a separate “uncertain” category. Such responses were removed from our main analyses. Of note, the number of uncertain responses in risk comprehension and absolute risk perception was not insubstantial, ranging from 8.8% for Possibly Pathogenic to 26% for VUS. To ensure that omission of uncertain responses did not have an appreciable effect on the findings, we also conducted both the within-groups and between-groups analyses in a way that integrated these responses. In particular, the uncertain responses were converted into the mid-point response (4’s on the seven-point Likert scales).
For all outcomes in the within-groups analyses, the results were unchanged when the uncertain responses were converted into the mid-point response. This was also true for the between-groups analyses with one exception: the difference in means for absolute risk perception when receiving a VUS vs. a Possibly Pathogenic variant became significant. We conclude that the findings are robust with respect to the inclusion or omission of uncertain responses.
RESULTS
Participant Characteristics
The response rate for the completion of the survey for the first type of variant received was 289 of 490 contacted (59%). As seen in Table 1, of the respondents who completed the survey, 63% were from the first ClinSeq® cohort, 54% were female, 83% were college graduates, and 98% identified as not Hispanic or Latino. 273 out of the 289 completed the entire survey for all three variant results and were included in the within-groups analyses.
Table 1.
Characteristics of Study Population
| Participants (N=289) | Total Participants Contacted (N=490) | p-value | ||
|---|---|---|---|---|
| Characteristic | Percentage (%) | Percentage (%) | ||
| Female | 54 | 55 | 0.78 | |
| First ClinSeq® Cohort | 63 | 56 | 0.07 | |
| Yearly Salary >$100,000/year | 63 | 60 | 0.38 | |
| Education | College Graduate or Above | 83 | 76 | 0.02 |
| Post-Graduate | 57 | 48 | 0.02 | |
Non-Participants are defined in this study as those who were contacted via email, phone, or postal mail and did not complete the survey. Of note, there is no complete way to differentiate between individuals who actually received their invitation to participate and chose not to participate and those who never received the invitation to participate. Of those who did not complete the survey, six from the first cohort and seven from the second cohort, comprising 5% of the total non-participant population, actively declined to participate.
Within-Groups Analysis
We begin by reporting within-group differences across the three classifications of genetic test results for the hypothetical cardiovascular disease. As seen in Table 2, risk comprehension and risk perceptions differed as predicted; participants appeared to understand that risk was lowest for the VUS result, higher for the Possibly Pathogenic result, and highest for the Likely Pathogenic result, with all pairwise comparisons statistically significant. This same pattern emerged for all outcomes including worry, perceived certainty, and intention to share the result with family. In short, when presented with all three types of genetic feedback, participants appreciated the differences among the three types and also expressed different intentions in response (in accordance with the same pattern).
Table 2.
Within Subjects Outcome Means for Each Variant Classification Received
| Outcome Variable (mean, standard deviation) | VUS | Possibly Pathogenic | Likely Pathogenic | F Value | p Value | Effect Size (Eta2) | |
|---|---|---|---|---|---|---|---|
| Risk Comprehension (4.52,1.58) | 3.55a | 4.39b | 5.52c | 122.97 | 0.0001 | 0.25 | |
| Risk Perception | Absolute (4.77, 1.23) | 3.93a | 4.70b | 5.62c | 169.00 | 0.0001 | 0.30 |
| Experiential (4.45, 1.43) | 3.64a | 4.36b | 5.35c | 130.66 | 0.0001 | 0.24 | |
| Comparative (5.22, 1.34) | 4.50a | 5.13b | 6.01c | 97.11 | 0.0001 | 0.21 | |
| Worry (3.49,1.58) | 2.67a | 3.39b | 4.43c | 115.52 | 0.0001 | 0.22 | |
| Perceived Uncertainty | Clinical Uncertainty (3.25,1.03) | 2.85a | 3.24b | 3.61c | 40.72 | 0.0001 | 0.09 |
| Affective Uncertainty (3.02,1.01) | 2.70a | 2.98b | 3.34c | 30.45 | 0.0001 | 0.07 | |
| Credibility (3.69,0.99) | 3.58a | 3.66b | 3.82c | 4.53 | 0.0111 | 0.01 | |
| Behavioral Intentions | Change Lifestyle (5.02,1.68) | 4.03a | 5.09b | 5.93c | 109.93 | 0.0001 | 0.21 |
| Screening (5.80,1.49) | 5.04a | 5.86b | 6.49c | 77.95 | 0.0001 | 0.16 | |
| Seek Specialty Care (5.42,1.74) | 4.53a | 5.48b | 6.27c | 79.39 | 0.0001 | 0.16 | |
| Share With Family (5.92,1.39) | 5.37a | 5.99b | 6.39c | 40.40 | 0.0001 | 0.09 | |
| Seek Genetic Counseling (5.06,1.79) | 4.30a | 5.11b | 5.75c | 50.01 | 0.0001 | 0.11 | |
Different subscripts indicate that the means were statistically significantly different based on Tukey post-hoc tests (p<0.05 for all comparisons). All analyses are significant at p=0.01 as well. Means and standard deviations for variables collapsed across conditions appear under each of the variable labels in the left columns.
Between-Groups Analysis
We next assessed reactions to just the first type of variant received in a between-groups ANOVA. As seen in Table 3, these analyses again yielded main effects of variant pathogenicity classification on experiential risk perception and worry with all three pairwise comparisons among the three classifications of genetic results obtaining statistical significance as in the within-groups analyses. We also observed an overall effect of variant classification on absolute risk perception, comparative risk perception, and risk comprehension; however, Tukey post-hoc tests showed that responses to VUS and Possibly Pathogenic results did not differ significantly from each other (although each was perceived as less risky than a Likely Pathogenic result). Participants also perceived more certainty in response to Likely Pathogenic results compared to VUS results, but there were no other differences between groups. Intentions to share the result with family members was significantly lower for those who had received a VUS as compared to Possibly Pathogenic and Likely Pathogenic variants which did not differ from each other. This pattern also emerged for the other behavioral intentions measured. In short, then, when participants had only seen one variant, they were less facile at discerning among the three types. Moreover, they were less likely to make distinctions between Possibly Pathogenic and Likely Pathogenic variants when expressing intentions regarding future actions (e.g., sharing results).
Table 3.
Between Subjects Outcome Means for Variant Received
| Outcome Variable (mean, standard deviation) | VUS | Possibly Pathogenic | Likely Pathogenic | F Value | p Value | Effect Size (Eta2) | |
|---|---|---|---|---|---|---|---|
| Risk Comprehension (4.83, 1.47) | 4.17a | 4.52a | 5.48b | 21.45 | 0.0001 | 0.14 | |
| Risk Perception | Absolute (4.88,1.24) | 4.21a | 4.61a | 5.53b | 33.49 | 0.0001 | 0.20 |
| Experiential (4.52,1.38) | 4.90a | 5.30a | 5.94b | 32.00 | 0.0001 | 0.18 | |
| Comparative (4.44, 1.33) | 3.77a | 4.38b | 5.20c | 14.56 | 0.0001 | 0.11 | |
| Worry (3.48,1.50) | 2.77a | 3.34b | 4.14c | 23.22 | 0.0001 | 0.14 | |
| Perceived Uncertainty | Clinical Uncertainty (3.40,0.98) | 3.18a | 3.37ab | 3.59b | 4.20 | 0.0160 | 0.03 |
| Affective Uncertainty (3.12,0.97) | 2.94a | 3.12ab | 3.26b | 2.52 | 0.0189 | 0.02 | |
| Credibility (3.69,0.96) | 3.55a | 3.75a | 3.74a | 1.26 | 0.2854 | 0.009 | |
| Behavioral Intentions | Change Lifestyle (5.29,1.67) | 4.46a | 5.28b | 5.93c | 20.21 | 0.0001 | 0.13 |
| Screening (6.28,1.19) | 5.83a | 6.40b | 6.51b | 8.82 | 0.0002 | 0.06 | |
| Seek Specialty Care (5.87,1.57) | 5.17a | 6.02b | 6.28b | 13.37 | 0.0001 | 0.09 | |
| Share With Family (6.28,1.12) | 5.91a | 6.47b | 6.42b | 6.71 | 0.0014 | 0.05 | |
| Seek Genetic Counseling (5.33,1.73) | 4.79a | 5.52b | 5.58b | 5.85 | 0.0033 | 0.04 | |
Different subscripts indicate that the means were statistically significantly different based on Tukey post-hoc tests (p<0.05 for all comparisons). All but one analysis is significant at p=0.01 as well, which suggests less concern about Type 1 error. Means and standard deviations for variables collapsed across conditions appear under each of the variable labels in the left columns.
Mediation
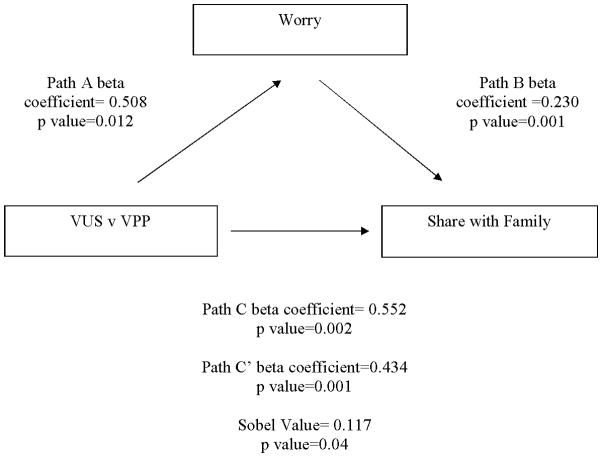
We used mediation analyses to understand if changes in worry, risk perception, risk comprehension, and perceived uncertainty, explained the relationship between receiving a particular variant classification and intention to share that result with family members. We conducted a separate mediation analysis for each pairwise comparison (VUS vs. Possibly Pathogenic, VUS vs. Likely Pathogenic, and Likely Pathogenic vs. Possibly Pathogenic). As seen in Table S2, most of the analyses suggested significant mediation. More specifically, differences in worry, risk comprehension, risk perception, and perceived uncertainty were found to explain the effect of variant classification on intentions to share the result with family members. This pattern was consistent across all of the secondary intentions we measured as well, and we illustrate this pattern in Figure 2 with the variables worry and intentions to share with family members for the comparison of VUS and Possibly Pathogenic results.
Figure 2. Example Mediation Figure.
Example of mediation demonstrated in this study using worry as a mediator of the relationship between receipt of a VUS vs. a Possibly Pathogenic variant and intentions to share with family members.
There were important differences, however, in the between-groups analyses of the first variant received. As seen in Table S3, worry and experiential risk perception explained differences in intentions between the Possibly Pathogenic and Likely Pathogenic variant classifications (p<0.05). This is in contrast to the within-groups analyses, where the mediation effects for worry and experiential risk perception when comparing Possibly Pathogenic and Likely Pathogenic variant classifications were not significantly different. In other words, emotional responses were more likely to explain effects of variant classification on intentions to share the result with family at the point in the experiment that participants had only viewed one variant (not all three). The same pattern emerged for all of the behavioral intentions measured in this study.
DISCUSSION
The main goal of this study was to assess whether individuals could discern differences among different classifications of genetic variants for a hypothetical cardiovascular disease. Two classifications were prescribed by ACMG/AMP guidelines (VUS and Likely Pathogenic), and an additional sub-classification is used by some laboratories (Possibly Pathogenic). We tested the hypothesis that individuals would be able to distinguish among sub-classifications of cardiac genetic variants, and found support for this hypothesis when participants received all three results, but the hypothesis was less supported when they received only one result. In the within-groups analyses, participants felt most at risk, felt most worried, perceived greater certainty, and expressed the highest intention to share the result with family members for a Likely Pathogenic result relative to a Possibly Pathogenic result, followed in turn by VUS. Most of the perceptions we measured explained the effects of the variant classification on intentions to share results with family members (as shown in mediation analyses).
The results have particular applicability for two populations: 1) the growing group of individuals referred to cardiology after receiving secondary findings related to cardiovascular disease following exome or genome sequencing and 2) patients (usually affected) who undergo genetic testing via large panels. Both research studies and clinical genomic sequencing are being used increasingly to aid in the identification of underlying genetic etiology of disease, and cardiovascular providers may receive increasing referrals for patients found to have secondary genomic findings in cardiovascular disease related genes. In addition, direct to consumer genetic testing is also becoming increasingly available and may lead to individuals receiving cardiovascular risk-related genetic results. In these contexts, individuals may not have any cardiovascular work-up. A background understanding of patient risk perception, affective responses, perceived uncertainty, and behavioral intentions is critical to effective communication of these results.
The other applicable population is more typical of those seen routinely in cardiovascular genetics settings. These are affected patients, typically probands, who undergo sequencing via large panels. For these patients, it is common to receive multiple types of genetic test results at one time, often including a VUS result. For example, the patient may receive a Likely Pathogenic variant consistent with his or her clinical phenotype, but also a VUS in another cardiovascular-related gene. Again, it is important to consider patient responses and how they distinguish among those results, in order to best facilitate discussions surrounding risk, negative affective responses, and variant-appropriate behaviors including sharing results with family members. Although some behavioral responses measured here are not relevant to this group (e.g., intention to seek cardiovascular care), our primary behavioral endpoint – intention to share results with family members- is important for this population.
It is also important to note, however, that there is movement toward a future in which many genetic testing results may be delivered outside of the traditional clinical settings. As the landscape of genetic testing evolves, the findings presented in this study may have additional salience. Furthermore, these results are applicable not just to cardiology, but also to genetic testing for other types of diseases.
In most clinical settings, individuals often only receive one type of result at a given time without the context of other results. A strength of our design was that participants received the three variants in an order that was randomized, allowing us to assess their reactions to the very first variant in a between-groups analysis. These analyses showed that participants were able to make distinctions among sub-classifications on some dimensions such as worry, but not on others such as perceived uncertainty. Although variant classification systems are designed to communicate the level of certainty regarding the meaning of the genetic change, they may be less effective in doing so when participants only receive one result than when receiving several results. In particular, when presented with only one genetic test result, individuals receiving a Possibly Pathogenic result comprehended and perceived risk of a Possibly Pathogenic result similarly to individuals who received a VUS result, but intended to disclosure their results to family members at rates similar to individuals who received a Likely Pathogenic result. Although we focused on intentions to share the results with family members in our study, other outcomes of genetic testing can carry significant risks, such as prophylactic surgeries or use of implantable devices, although at this time in the field of cardiology, these riskier types of outcomes are largely physician-directed and not based on genotype alone. For those receiving only one type of genetic test result, our results suggest that context may help in the effective communication of these results. Providers may consider methods of indicating levels of risk, certainty, etc. such as illustrative examples or visual diagrams to show how the patients’ result compares to other possible results.
In addition, our analyses showed that emotional variables served as significant mediators in the between-groups analyses comparing Possibly Pathogenic and Likely Pathogenic results, unlike the within-groups analyses. In other words, when participants had more of a negative emotional reaction to the first result they were given, they in turn showed higher intentions to share results with their family members. This suggests that their decision making regarding the first variant was more informed by emotion than when they were exposed to all three variants. This difference suggests that participants were being more deliberative when faced with all three variants, relying less on emotional responses such as worry. Accordingly, in addition to devoting more time to putting this variant in context, interventions aimed at better communicating genetic test results to patients might target different psychosocial processes depending on whether participants receive multiple variants simultaneously (in which case informational needs might be prioritized) or if they have previous experience receiving genetic test results to serve as context. For example, providers may consider devoting more session time to participant emotional responses when the participant only receives one genetic variant, but may focus on interventions related to absolute risk perception when participants receive multiple variants. In addition, this may offer further opportunities for interventional studies for return of results, specifically using context of other types of results during the results disclosure when genetic testing identifies one genetic variant.
Our results also support published findings that participants struggle with uncertain genetic test results11, as evidenced by the higher proportion of individuals who answered that they were “uncertain” and thus unable to rate their risk on a seven-point scale. Although some laboratories have attempted to mitigate some of this uncertainty by further stratifying VUS results, our results suggest that sub-classifications may be of less use to individuals than to clinicians and could lead to unintended consequences such as increasing unnecessary worry. Our findings are timely as there is debate about whether VUS results should be returned at all21. Although our findings do not fully address this issue, they suggest that sub-classification, which is potentially useful to clinicians, does not necessarily improve patient interpretation of VUS results. In the clinical setting, this task will likely fall to the clinicians interpreting the test results, who will need to integrate pathogenicity classifications with other patient risk factors to offer clear recommendations for health management.
Despite the novelty of the design and findings, this study has several limitations. Members of the ClinSeq® study cohort were relatively high in socioeconomic status, reducing generalizability given that they may not be representative of the larger population or of clinical populations. However, they tend to be similar to other populations that have engaged in genome sequencing15. Additionally, although some of our participants had cardiovascular disease, reactions of patients with cardiovascular disease diagnoses undergoing diagnostic genetic testing and receiving only one genetic test result may differ from these mostly healthy individuals who were presented a hypothetical result. In these cases, management and risk should be discussed in the context of the patient’s clinical picture and the genetic test result may have much more impact on family member medical management than for the individual undergoing genetic testing. It is unclear whether the effects of the different variant classifications would be sustained over time. Future studies should attempt to reproduce these outcomes in a more diverse clinical cohort using actual genetic test results over an extended time period. Finally, our experiment only investigates one example of a variant classification system that deviates from the ACMG/AMP recommendations. Different systems and descriptors used to sub-classify variants may yield different results than those in this study.
Conclusion
To our knowledge, this is the first experimental study to assess individual outcomes of receiving sub-classifications of uncertain genetic variants beyond the recommended five ACMG/AMP categories, and one of the few to assess outcomes of receiving uncertain genetic results in the context of cardiovascular disease. Given that a patient’s ability to distinguish among cardiac variant classification descriptors appears to depend on whether the result is received in conjunction with other results, cardiogenetics providers should consider the context in which the results are received and appropriate clinic time accordingly to enhance patient outcomes. For example, providers could practically devote more time to putting the variant in context (e.g., using illustrative examples of other variant types or using visual aids that describe the level uncertainty or other constructs relative to other variant classifications) as well as devoting more time to emotional responses of receiving genetic test results. In addition, these findings suggest that further expansion of the genetic variant classification system is likely to be of limited utility for patients who have experience with only one genetic result and may cause unintended consequences. These findings are also important for informing guidelines regarding variant classification systems and highlight potential limitations of using additional sub-classifications of variants to modify patients’ perceptions of risk and uncertainty.
Supplementary Material
Clinical Perspective.
Clinical genetic testing for heritable cardiovascular disease has become a widely-used tool to aid in the management of patients and their families. In the present study, we tested whether participants perceived differences among genetic variant classifications. To our knowledge, this is the first experimental study to assess individual outcomes of receiving sub-classifications of uncertain genetic variants beyond the five recommended categories, and one of the few to assess outcomes of receiving uncertain genetic results in the context of cardiovascular disease. Our results suggest that when participants were presented with multiple results, they appreciated differences among them as suggested by changes in risk perceptions, risk comprehension, worry, perceived uncertainty, and intentions to share the result with family members. In the clinical genetic testing context, however, patients may only receive one type of result at a time, without the context of other results. Given that a patient’s ability to distinguish among cardiac variant classification appears to depend on whether the result is received in conjunction with other results, cardiogenetics providers should consider the context in which the results are received and appropriate clinic time accordingly to enhance patient outcomes. Our study suggests that providers returning only one type of genetic variant to their patients might improve patient discernment of that variant by employing strategies such as using illustrative examples of other variant types to provide context as well as devoting more time to emotional responses of receiving genetic test results.
Acknowledgments
SOURCES OF FUNDING: This study was funded by the Intramural Research Program of the National Human Genome Research Institute, grant HG200387.03.
Footnotes
DISCLOSURES: Leslie Biesecker receives royalties from Genentech Corp., is an unpaid advisor to Illumina Corp. and receives honoraria from Wiley-Blackwell Inc. The other authors declare no conflict of interest. This study was funded by the National Human Genome Research Institute.
References
- 1.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 2.Cirino AL, Harris S, Lakdawala NK, Michels M, Olivotto I, Day SM, et al. Role of Genetic Testing in Inherited Cardiovascular Disease: A Review. JAMA Cardiol. 2017;2:1153–1160. doi: 10.1001/jamacardio.2017.2352. [DOI] [PubMed] [Google Scholar]
- 3.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han PK, Umstead KL, Bernhardt BA, Green RC, Joffe S, Koenig B, et al. A taxonomy of medical uncertainties in clinical genome sequencing. Genet Med. 2017:1–8. doi: 10.1038/gim.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S, Nazneen A, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The DNA Diagnostic Laboratory. [Accessed May 05, 2017];Understanding your Gene Sequencing Test Result. Available at http://www.hopkinsmedicine.org/dnadiagnostic/documents/patients_families/ddl_understanding_test_results.pdf.
- 7.LabCorp. [Accessed November 01, 2017];LabCorp: Variant Classification Specifications. https://www.integratedoncology.com/sites/integratedoncology/files/ClinVar%2Bclassification_Legal%2B%2BDST%2Bapproved%2B%2806-15-15%29.pdf.
- 8.Pepin MG, Murray ML, Bailey S, Leistritz-Kessler D, Schwarze U, Byers PH. The challenge of comprehensive and consistent sequence variant interpretation between clinical laboratories. Genet Med. 2016;18:20–24. doi: 10.1038/gim.2015.31. [DOI] [PubMed] [Google Scholar]
- 9.Vassy JL, Lautenbach DM, McLaughlin HM, Kong SW, Christensen KD, Krier J, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:1–12. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scherr CL, Lindor NM, Malo TL, Couch FJ, Vadaparampil ST. A preliminary investigation of genetic counselors’ information needs when receiving a variant of uncertain significance result: a mixed methods study. Genet Med. 2015;17:739–746. doi: 10.1038/gim.2014.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrucelli N, Lazebnik N, Huelsman KM, Lazebnik RS. Clinical Interpretation and Recommendations for Patients with a Variant of Uncertain Significance in BRCA1 or BRCA2: A Survey of Genetic Counseling Practice. Genetic Testing. 2002;6:107–113. doi: 10.1089/10906570260199357. [DOI] [PubMed] [Google Scholar]
- 12.Maheau C, Thorne S. Receiving Inconclusive Genetic Test Results: An Interpretive Description of the BRCA1/2 Experience. Research in Nursing and Health. 2008;31:553–562. doi: 10.1002/nur.20286. [DOI] [PubMed] [Google Scholar]
- 13.Heshka JT, Palleschi C, Howley H, Wilson B, Wells PS. A systematic review of perceived risks, psychological and behavioral impacts of genetic testing. Genet Med. 2008;10:19–32. doi: 10.1097/GIM.0b013e31815f524f. [DOI] [PubMed] [Google Scholar]
- 14.Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Research. 2009;19:1665. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis KL, Han PK, Hooker GW, Klein WMP, Biesecker LG, Biesecker BB. Characterizing Participants in the ClinSeq Genome Sequencing Cohort as Early Adopters of a New Health Technology. PLoS One. 2015;10:e0132690. doi: 10.1371/journal.pone.0132690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrer RA, Klein WMP. Risk perceptions and health behavior. Current Opinion in Psychology. 2015;5:85–89. doi: 10.1016/j.copsyc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer RA, Portnoy DB, Klein WMP. Worry and Risk Perceptions as Independent and Interacting Predictors of Health Protective Behaviors. Journal of Health Communication. 2013;18:397–409. doi: 10.1080/10810730.2012.727954. [DOI] [PubMed] [Google Scholar]
- 18.Biesecker BB, Woolford SW, Klein WMP, Brothers KB, Umstead KL, Lewis KL, et al. PUGS: A novel scale to assess personal uncertainties in genome sequencing. Clin Genet. 2017:1–8. doi: 10.1111/cge.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Facio FM, Eidem H, Fisher T, Brooks S, Linn A, Kaphingst KA, et al. Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet. 2013;21:261–265. doi: 10.1038/ejhg.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 21.King M, Levy-Lahad E, Lahad A. Population-Based Screening for BRCA1 and BRCA2 2014 Laskar Award. JAMA. 2014;312:1091–1092. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.