Abstract
Common bean (Phaseolus vulgaris L.) is an important source of dietary protein and minerals worldwide. Genes conditioning variability for mineral contents are not clearly understood. Our ultimate goal is to identify genes conditioning genetic variation for Zn and Fe content. To establish mapping populations for this objective, we tested mineral content of 29 common bean genotypes. Chemical analyses revealed significant genetic variability for seed Zn and Fe contents among the genotypes. Genetic diversity was evaluated with 49 primer pairs, of which 23 were simple sequence repeats (SSR), 16 were developed from tentative consensus (TC) sequences, and 10 were generated from common bean NBS-LRR gene sequences. The discriminatory ability of molecular markers for identifying allelic variation among genotypes was estimated by polymorphism information content (PIC) and the genetic diversity was measured from genetic similarities between genotypes. Primers developed from NBS-LRR gene sequences were highly polymorphic in both PIC values and number of alleles (0.82 and 5.3), followed by SSRs (0.56 and 3.0), and markers developed from TC (0.39 and 2.0). genetic similarity values between genotypes ranged from 14.0 (JaloEEP558 and DOR364) to 91.4 (MIB152 and MIB465). Cluster analysis clearly discriminated the genotypes into Mesoamerican and Andean gene pools. Common bean genotypes were selected to include in crossing to enhance seed Zn and Fe content based on genetic diversity and seed mineral contents of the genotypes.
Keywords: Common bean, genetic diversity, mineral nutrients, breeding
Common bean (Phaseolus vulgaris L.) is consumed worldwide and is a staple food in many countries (Broughton et al. 2003). Beans are a rich source of protein, energy (approx. 380 kcal 100 g−1 of seeds), vitamins (thiamine, riboflavin, niacin, vitamin B6, folic acid), dietary fiber (especially soluble fraction), and minerals (calcium, iron, zinc, phosphorus, potassium, magnesium) (Reyes-Moreno and Paredes-Lopez 1993). The role of dry beans as functional foods in chronic disease risk reduction has been given increasing attention. Several recent literature reviews have highlighted the positive effects of dry bean on improving serum lipid profiles in patients with coronary heart disease or Type 2 diabetes (Anderson and Major 2002; Anderson et al. 2004; Winham et al. 2007).
Common bean (2n=2x=22) represents 50% of the grain legumes consumed worldwide. Common bean landraces exhibit wide genetic variability for seed color, shape, shininess, size, and have demonstrated adaptability to several environmental conditions (Rodiño et al. 2003). The genome size for common bean is estimated to be about 450 to 650 million base pairs (Mb)/haploid (Bennett and Leitch 1995) and is considered to be one of the smallest genomes among major crop species. Based on phaseolin seed storage protein variation (Gepts and Bliss 1986; Gepts 1990), marker diversity (Becerra-Velasquez and Gepts 1994), and morphology (Gepts and Debouck 1991), two major gene pools of wild common bean were identified. The Middle American gene pool extends from Mexico through Central America and into Colombia and Venezuela, meanwhile the Andean gene pool is found in Ecuador, Peru, Bolivia, and Argentina. The two domesticated gene pools appear to converge in Colombia (Gepts and Bliss 1986). A third, ancestral gene pool, based on a novel phaseolin type originated in southern Ecuador and northern Peru. The two major domestication events, Middle America (possibly west-central Mexico) and southern Andes (Andean) diverged into three races within each of the two major domesticated gene pools (Singh et al. 1991). The Middle American gene pool, consisting of races Durango, Jalisco, and Mesoamerica is represented by the medium and small seeded pinto, pink, black, white, and some snap beans. The Andean gene pool, consisting of races Nueva Granada, Peru, and Chile, is represented by the large-seeded kidney, cranberry, and many snap beans among others.
Knowledge of genetic diversity in a crop species is fundamental to its improvement. Evaluation of genetic diversity among adapted, elite germplasm can provide predictive estimates of genetic variation among segregating progeny for pure-line cultivar development (Manjarrez-Sandoval et al. 1997), and may estimate the degree of heterosis in progenies of some parental combinations (Cox and Murphy 1990; Barbosa-Neto et al. 1996).
The advancement of molecular biology has created new opportunities for estimating the genetic diversity at a molecular level; however, genetic studies are often restricted, both by the limited number of polymorphic markers and by the low level of variability within self-pollinated species. Assessing genetic diversity within a narrow genetic pool of novel breeding germplasm could lead to more efficient crop improvement by marker directed accumulation of desired alleles. Marker-assisted selection is likely to speed up the breeding process and decrease the amount of plant material that needs to be screened in such experiments. A variety of molecular, chemical, and morphological descriptors are used to characterize the genetic diversity among and within crop species. The development of gene- and/or expressed sequence tags-derived molecular markers provides opportunities for the assessment of molecular diversity at selected loci. Such gene-targeted markers can contribute to the study of genetic resources and to ecological studies (Van Tienderen et al. 2002). For all of these approaches, the availability of highly polymorphic and user friendly DNA markers is vital. Genetic variation in common bean has been reported based on restriction fragment length polymorphism (RFLP) (Becerra-Velasquez and Gepts 1994), random amplified polymorphic DNA (RAPD) (Duarte et al. 1999; Galván et al. 2001), simple sequence repeats (SSR) (Lioi et al. 2005; Diaz and Blair 2006; Blair et al. 2006, 2007; Benchimol 2007), and amplified fragment length polymorphism (AFLP) (Tohme et al. 1996; Beebe et al. 2001).
The first essential question regarding whether any species can be improved for a particular trait is to determine the degree of variability which exists for this trait within the species. The existing genetic diversity of a species in gene banks enables plant breeders to choose the most suitable strategy for improving the species (Lefort-Buson et al. 1988). Thus, the availability of diverse common bean accessions represents a valuable resource for the improvement of this species, since co-adapted genes of different accessions can convey similar response if selected for a specific trait (Harlan 1975). The knowledge of genetic diversity patterns can increase the efficiency for conservation, utilization and genetic improvement of common beans (Beebe et al. 2000b; Singh 2001).
Zinc (Zn) and iron (Fe) are essential micronutrients for human growth, development, and maintenance of the immune system. Zinc is needed for growth and for maintenance of immune function, which enhances both the prevention of and recovery from infectious diseases (Black 2003). Iron is needed for psychomotor development, maintenance of physical activity and work capacity, and resistance to infection (Stoltzfus 2001).
Common bean is an important source of protein and minerals, especially Zn and Fe. Genetic differences have been reported for seed Zn and Fe concentrations among genotypes and landraces (Singh et al. 1991; Graham et al. 1999; Moraghan and Grafton 2001). Beebe et al. (2000a) evaluated a core collection of over 1000 accessions of common beans in the field and found a range in Fe concentrations from 34 to 89 mg g−1, with an average of 55 mg g−1. Zinc concentrations in these same accessions ranged from 21 to 54 mg g−1, with an average of 35 mg g−1.
The genetics of Zn and Fe content appears to be complex. Analysis of three RIL populations at the International Center for Tropical Agriculture (CIAT) revealed that both Zn and Fe contents had continuous distribution, suggesting that these mineral contents were quantitatively inherited (Beebe et al. 2000a, Blair et al. 2005). Beebe et al. (2000a) and Blair et al. (2005) also observed that the parental accessions were very close to the extremes of the populations, with little evidence of transgressive segregation, suggesting that most of the favorable alleles came from the high-mineral content parent. A recent inheritance study (Cichy et al. 2005) demonstrated that a single gene controls seed Zn in navy bean and that high seed Zn is dominant over low seed Zn. Cichy et al. (2005) also observed transgressive segregation for seed Zn content in the F2 generation and speculated that this was the effect of additional minor genes. Forster et al. (2002) observed a similar result in a cross between two navy bean cultivars, Voyager and Albion. Singh and Westermann (2002) reported that a single dominant gene controls the resistance to soil Zn deficiency in common bean. Guzmán-Maldonado et al. (2003) reported two QTLs for seed Fe content and one for Zn content responsible for 25% and 15% phenotypic variation, respectively, from a cultivated×wild P. vulgaris cross. Almost all of these studies were based on seed mineral contents from plants grown in field conditions subject to various influences of soil characteristics and environments.
We conducted our study under greenhouse conditions with an array of common bean genotypes consisting of various genetic and geographical backgrounds. Our objectives were (1) to assess genetic diversity among 29 common bean genotypes using molecular markers developed from various genomic sources, and (2) to select parental genotypes suitable for mapping loci affecting mineral content in common bean.
MATERIALS AND METHODS
Plant Material
The 29 common bean cultivars/lines in this study consisted of 14 genotypes from CIAT, 13 from the United States of America, and one each from Brazil and India. Among the cultivars, 13 represent parents of one or more mapping populations developed for various interests, and vary in terms of growth habit and geographic origin. All common bean cultivars/lines, and country of origin, are listed in Table 1.
Table 1.
Common bean cultivars, gene pool group, origin, seed color, zinc and iron contents in the seeds
| No. | Cultivar/line | Gene pool | Origin | Market class | Seed color | Seed Zn (ppm) | Seed Fe (ppm) |
|---|---|---|---|---|---|---|---|
| 1 | Jalo EEP558 | Andean | Brazil | – | Cream | 59.5a–c | 8.9k |
| 2 | NUA 45 | Andean | CIAT breeding line | – | Purple mottled | 40.8f–h | 74.6cd |
| 3 | NUA 35 | Andean | CIAT breeding line | – | Purple mottled | 47.4c–g | 22.9i–k |
| 4 | NUA 56-1770 | Andean | CIAT breeding line | – | Purple mottled | 34.5gh | 110.6ab |
| 5 | NUA 59 | Andean | CIAT breeding line | – | Purple mottled | 30.9h | 34.2f–k |
| 6 | G 122 | Andean | Indian landrace | Cranberry | Red mottled | 41.1f–h | 42.0e–j |
| 7 | NY6020-4 | Andean | USA breeding line | Snap | White | 47.0c–g | 20.6i–k |
| 8 | Benton | Andean | USA | Snap | White | 53.6a–f | 74.5cd |
| 9 | Dorado | Mesoamerican | CIAT | Small red | Dark red | 42.4e–h | 84.3a–d |
| 10 | BAT 93 | Mesoamerican | CIAT breeding line | – | Cream | 54.7a–f | 42.1e–j |
| 11 | A 55 | Mesoamerican | CIAT breeding line | Black | Black | 52.6a–f | 22.2i–k |
| 12 | XAN 176 | Mesoamerican | CIAT breeding line | Black | Black | 44.0c–h | 17.2i–k |
| 13 | MIB 151 | Mesoamerican | CIAT breeding line | – | Light Cream | 58.9a–d | 64.4c–f |
| 14 | MIB 152 | Mesoamerican | CIAT breeding line | – | Black | 58.3a–e | 66.7c–e |
| 15 | MIB 154 | Mesoamerican | CIAT breeding line | – | Brown mottled | 48.6b–g | 92.8a–c |
| 16 | MIB 217 | Mesoamerican | CIAT breeding line | – | Black | 42.8e–h | 34.2f–k |
| 17 | MIB 465 | Mesoamerican | CIAT breeding line | – | Black | 64.4ab | 84.7a–d |
| 18 | MIB 466 | Mesoamerican | CIAT breeding line | – | Pink with black specks | 64.6a | 57.6d–h |
| 19 | ND88-106-04 | Mesoamerican | USA | Navy | White | 49.8a–g | 27.4h–k |
| 20 | Aztec | Mesoamerican | USA | Pinto | Cream mottled | 53.9a–f | 24.3i–k |
| 21 | Voyager | Mesoamerican | USA | Navy | White | 43.4d–h | 80.9b–d |
| 22 | Albion | Mesoamerican | USA | Navy | White | 53.9a–f | 28.8g–k |
| 23 | Mayflower | Mesoamerican | USA | Navy | White | 53.6a–f | 12.2jk |
| 24 | T-39 | Mesoamerican | USA | Black | Black | 45.0c–g | 29.0g–k |
| 25 | Jaguar | Mesoamerican | USA | Black | Black | 51.8a–f | 55.2d–h |
| 26 | Othello | Mesoamerican | USA | Pinto | Cream mottled | 45.1c–h | 58.9d–g |
| 27 | Ryder | Mesoamerican | USA | Small red | Red | 54.9a–f | 33.3g–k |
| 28 | Vista | Mesoamerican | USA | Navy | White | 49.2a–g | 112.9a |
| 29 | BelNeb-RR-1 | Mesoamerican | USA breeding line | Great Northern | White | 49.6a–g | 43.3e–i |
| Mean±SE | 49.5±1.49z | 50.4±5.46y | |||||
| LSD0.05 | 15.94 | 30.8 |
Significance at the 0.05 probability level on the basis of F tests with n−1 degrees of freedom.
Significance at the 0.01 probability level on the basis of F tests with n−1 degrees of freedom.
Means having common letter do not differ significantly.
Greenhouse Experiment
Common bean genotypes were grown in the greenhouse in 18–19 cm pots filled with Sunshine mix 1 (Sun Gro Horticulture Canada Ltd., formulated with Canadian sphagnum peat moss, coarse grade perlite, gypsum, and dolomitic lime) as substrate. The seeds were planted on 2007 Mar. 27 following a randomized complete block design with three replications. Two seeds for each entry were placed in each pot for germination, but one plant was allowed to grow until harvest of the seeds. Pots were watered periodically with tap water to the approximate field capacity to facilitate normal plant growth. No additional fertilizer or pesticide was applied during the period of experimentation.
Chemical Analysis
After harvesting, seeds from each pod of individual plant were mixed thoroughly, and 10 seeds were randomly taken for chemical analysis. Seeds were washed with deionized water containing Joy® detergent (Proctor and Gamble, Cincinnati, OH), and later rinsed with deionized water only. Samples were oven-dried at 70°C for 48 h, weighed, and ground in an agate mortar with an agate pestle (Brinkmann Instruments Co., Westbury, NY). A 300-mg aliquot of the ground material was processed for concentrated nitric acid digestion, followed by 30% hydrogen peroxide, before Fe and Zn concentrations were measured in parts per million (ppm) using atomic absorption spectroscopy (Moraghan and Grafton 2001).
DNA Isolation and PCR Primers
Total genomic DNA was extracted and purified from leaf tissue of 2- to 3-wk-old greenhouse-grown plants using the CTAB method as described by Doyle and Doyle (1987).
A total of 49 primer pairs from three different sources were used for genetic diversity analysis. The primer names, primer sequences and genomic locations in the Phaseolus core linkage map BAT 93/Jalo EEP558 (BJ) (Freyre et al. 1998) are presented in Table 2.
Table 2.
Primer name, primer sequence, polymorphic information content (PIC), amplified alleles, and linkage group of 49 primers
| Primer name | 5′ to 3′ Left primer sequence | 5′ to 3′ Right primer sequence | PIC | No. of alleles | Linkage group |
|---|---|---|---|---|---|
| SSR primers | |||||
| BM139 | TTAGCAATACCGCCA TGAGAG |
ACTGTAGCTCAA ACAGGGCAC |
0.68 | 5 | B02 |
| BM154 | TCTTGCGAC CGAGCTTCTCC |
CTGAATCTGAGGAA CGATGACCAG |
0.77 | 5 | B09 |
| BM157 | ACTTAACAAGGAATAG CCACACA |
GTTAATTGTTTCCAATA TCAACCTG |
0.43 | 2 | B10 |
| BM160 | CGTGCTTGGCGA ATAGCTTTG |
CGCGGTTCTGATCG TGACTTC |
0.57 | 3 | B07 |
| BM164 | CCACCACAAGGAGAA GCAAC |
ACCATTCAGGCCG ATACTCC |
0.44 | 2 | B02 |
| BM170 | AGCCAGGTGCAAGACC TTAG |
AGATAGGGAGCTGGT GGTAGC |
0.50 | 2 | B06 |
| BM175 | CAACAGTTAAAGGTCG TCAAATT |
CCACTCTTAGCA TCAACTGGA |
0.66 | 3 | B05 |
| BM181 | ATGCTGCGAGTTAA TGATCG |
TGAGGAGCAAACAGA TGAGG |
0.78 | 5 | B03 |
| BM211 | ATACCCACATGC ACAAGTTTGG |
CCACCATGTGCTCATGA AGAT |
0.50 | 2 | B08 |
| BMd2 | AGCGACAGCAAG AGAACCTC |
CAACAAACGGTGAT TGACCA |
0.76 | 7 | B02 |
| BMd9 | TATGACACCACTGGC CATACA |
CACTGCGACATGAG AGAAAGA |
0.49 | 2 | B04 |
| BMd16 | ATGACACCACTGGCC ATACA |
GCACTGCGACATGAG AGAAA |
0.74 | 4 | B04 |
| BMd17 | GTTAGATCCCGCCCA ATAGTC |
AGATA GGAAGGGCGTGGTTT |
0.66 | 3 | B02 |
| BMd25 | GCAGATCGCCTACTC ACAA |
CGTTGACGAGAAGCA TCAAG |
0.47 | 2 | B08 |
| BMd33 | TACGCTGTGAT GCATGGTTT |
CCTGAAAGTGC AGAGTGGTG |
0.72 | 5 | B11 |
| BMd36 | CATAACATCGAAGCC TCACAGT |
ACGTGCGTACGAATAC TCAGTC |
0.49 | 2 | B03 |
| BMd42 | TCATAGAAGATTTGT GGAAGCA |
TGAGACACGTA CGAGGCTGTAT |
0.48 | 2 | B10 |
| BMd53 | TGCTGACCAAGGAAAT TCAG |
GGAGGAGGCTTAAGC ACAAA |
0.50 | 2 | B05 |
| GATS91 | GAGTGCGGAAGCGA GTAGAG |
TCCGTGTTCCTCTGTCT GTG |
0.49 | 2 | B02 |
| K03289 | ATGACACCACTGGCC ATACA |
GCACTGCGACATGAG AGAAA |
0.36 | 2 | B04 |
| X74919 | GTTGCCACCGGTG ATAATCT |
GTGAGGCAAGAAGCC TTCAA |
0.43 | 2 | B05 |
| X80051 | AGTTAAATTATA CGAGGTTAGCCTAAATC |
CATTCCCTTCACACA TTCACCG |
0.49 | 2 | B09 |
| X96999 | CAAATCGCAACACCTC ACAA |
GTCGGAGCCA TCATCTGTTT |
0.49 | 2 | B03 |
| Primers developed from tentative consensus (TC) sequences | |||||
| 755-size | TCCATGTCCCCTACAGA TTAAGGA |
TCCAAAGCCTACAAA TGGAACCTA |
0.40 | 2 | B4 |
| 762-allele-indel | GGATTTGTACAGT TGAAGTGACTTCTG |
CAAGCCCTTGAATA CTGAGTGTGA |
0.29 | 2 | B3 |
| 822-SNP | GAAGGGAAGTAGC TCAAAGAGAAAGGCG |
AGAAGCCATGAAG ATGAAGAACTAGACC AATAGAAG |
0.43 | 2 | B1 |
| 909-SSR | GAGTGACATCAAGGA ACAGATTCTAAATTACC |
CACCCACCCCCGAA TTAAGTTGC |
0.47 | 2 | B1 |
| 1148-size | GACTGGAAACATGTTG AGAGAATGG |
AATGTGCAACTACAAC AGAGTGCAA |
0.43 | 2 | B2 |
| 1181-SNP | TTGTGGAGTTGG CGAGGGCG |
AATTTTGGTTTTGG TTATTGTTGTTATTTT CCCAG |
0.47 | 2 | B8 |
| 1286-SNP | ATGATGCCTTAT GGAATGCCCCGA |
TCCCCGTAGTA CTAATGTGCAGATCTCA AGAAT |
0.40 | 2 | B9 |
| 1564-CAPS | TGTTTCATTTGATC CCAAGCCTAT |
TTCAACCATACAGAGA ACACAGAAGG |
0.42 | 2 | B4 |
| 1639-SNP | AATCTTTGTTGGGTGT TGTAAAGTTTGGATA AAGA |
CAAGAAGTGTG AGACGTTCTTCCGCTG |
0.45 | 2 | B6 |
| 2285-SNP | GATATACAGGGAAGC ATTAGCACTCAATCC TGAT |
ACATGATGAGTTAC CCAAGCCCAAAAGAA |
0.19 | 2 | B11 |
| 2298-SSR | ATATCTACCGATC ATAAGCCTTATTAAG |
AAATGGAGAAGGG TTACCTTC |
0.37 | 2 | B7 |
| 2422-allele-indel | GGACCATTGGTACAT ATTACAGTATATA |
GACTACACAAGAAT GGACTATTAAGG |
0.45 | 2 | B5 |
| 2551-size-indel | GAATCTCGTTTTCCTG CTCAG |
GATCTTGTAAGACT TTACTTTTACATTAGGAG |
0.49 | 2 | B6 |
| 2647-allele-indel | TATTACCTTTTCATC CTATTTTACACCATG |
ATATGAAGCAGAA CCAATGACTTGTG |
0.46 | 2 | B1 |
| 2654-SSR | CTATGCTCTCAA GTACCGCAAATTGG |
GGTTCCTCCATTTCTTT GTCCAACTC |
0.33 | 2 | B3 |
| 2685-size-indel | GTGCTTGGAATT CGAATTGGAGGTG |
CGAAACCATGCTATCA ACACACATCAAC |
0.19 | 2 | B4 |
| Primers developed from nucleotide-binding site–leucine-rich repeat (NBS-LRR) sequence | |||||
| 0436 | GGCACCATCTC AGAAAAACAA |
TGATGAATGAA CGAATGGGA |
0.60 | 3 | – |
| 0536 | TTTAGGAAATTCAGA TTGAGATATTG |
TGATGAATGAA CGAATGGGA |
0.67 | 3 | – |
| 2353 | GCATCCATTCCGAA CAGAGT |
GCTGCATGTTT AGATTTTTCCA |
0.98 | 9 | – |
| 2555 | CAGCTCTTCCAAAGTTGGTG | GAATTTGTTGA GCAGTGCGA |
0.95 | 6 | – |
| 2356 | GCATCCATTCCGAA CAGAGT |
AAAGCATAAGC ACATTGTTCCA |
0.89 | 6 | – |
| 2657 | TGGATACAC CCATTCTTTCTTG |
GCAGATTGATTTTTA TTCAACGG |
0.98 | 7 | – |
| 2758 | GGAGAACCTCCT GCAACTGA |
TCTTTAAG AGAATTCGCGGC |
0.99 | 6 | – |
| 2859 | GTGAAGGTGGTTTG GCATTT |
TGCAGGAGGTTCTC CAGATT |
0.85 | 7 | – |
| 2960 | ATCTGGAGAACCTCCT GCAA |
ATTCGCGGCCTG TTAAATC |
0.84 | 4 | – |
| 3061 | GCATCCATTCCGAAG AGAGT |
AAATGCCGAAGCCT TGAATA |
0.47 | 2 | – |
Primers Collected from Reference Map
Twenty-three SSR primer pairs used in this study were developed from coding and non-coding sequences of common bean (Blair et al. 2003). McConnell et al. (2006) analyzed P. vulgaris TC (tentative consensus) sequences and identified 322 primer pairs with their polymorphism. In our study, we used 16 of these primer pairs.
Primers Developed from NBS-LRR Gene Sequences
Common bean NBS-LRR type disease resistance gene sequences were downloaded from NCBI GenBank and were aligned with multiple sequence alignment software CLUSTALX (1.81). Phylogenetic trees were generated to evaluate the relationship among different sequences using Neighbor-Joining options of the software. Initially, four NBS-LRR complete coding sequences were chosen to design a series of primers based on sequence alignment. The large (~5 to 6 kb) gene sequences were divided into small ordered overlapping fragments, and a total of 37 primer pairs were designed to amplify genomic DNA of ~500 bp using the web-based PCR-primer designing program ‘Primer 3’. Ten of these 37 primer pairs were selected and included in this study.
PCR Amplification
Each 20 μL amplification reaction consisted of 1 ×PCR buffer, 1.88 mM MgCl2, 200 μM of each deoxyribonucleotide triphosphate, 1 μM of primer, 1 unit of Taq polymerase, and 150 ng of template DNA. Amplification was performed in a 96-well BioRad thermal cycler.
The PCR program for SSR primers and primers developed from NBS-LRR gene sequences consisted of one cycle of 95°C for 3 min; 40 cycles of 95°C for 1 min, from 55 to 57°C for 1 min, and 72°C for 2 min; and one cycle of 72°C for 10 min. For primers developed from TC sequences, the PCR protocol was one cycle of 95°C for 3 min, followed by 40 cycles of 95°C for 20 s, from 52 to 58°C for 20 s, and 72°C for 2 min, one cycle of final extension for 72°C for 7 min. In all cases, the amplified products were separated on 2% agarose gel with 60 V and run for 200 min.
Data Collection and Statistical Analysis
Seed Zn and Fe content data were subject to analysis of variance (ANOVA) for a treatment effect. ANOVA were performed using general linear model of the statistical package, Minitab® Release 13.2 (Minitab Inc.). If a significant difference among the genotypes was detected with ANOVA at P ≤ 0.05, means comparison was performed (α=0.05) using Fisher’s least-significant-difference (LSD) test.
After gel electrophoresis of the PCR products the presence or absence of each single fragment was coded by 1 or 0, respectively, and scored for a binary data matrix. The resulting matrix was used to estimate genetic similarity (GS) among all pairs of lines by Dice coefficient of similarity (Nei and Li 1979) as follows:
Where Nij is the number of alleles (scored bands) shared by lines i and j, and Ni and Nj are the total number of scored bands in lines i and j, respectively.
Polymorphism information content provides an estimate of the discriminatory power of a locus, or loci, by taking into account not only the number of alleles that are expressed, but also the relative frequencies of those alleles. Polymorphism information content values were calculated by the algorithm as described by Anderson et al. (1993):
Where Pi is the frequency of the ith allele in the population.
Polymorphism information content values range from 0 (monomorphic) to 1 (very highly discriminative, with many alleles each in equal and low frequency).
Genetic diversity analyses were conducted using numerical taxonomy and the multivariate analysis system, NTSYSpc v. 2.2 (Rohlf 2000). Genetic similarity values were computed between all possible pairs with the SIMQUAL option and ordered in a similarity matrix. The similarity matrix was run by sequential, agglomerative, hierarchical, nested (SAHN) clustering (Sneath and Sokal 1973) with the unweighted pair group with arithmetic averaging (UPGMA) method as an option (Sokal and Michener 1958). The dendrogram and cluster groupings were constructed by the UPGMA clustering algorithm from the SAHN option of NTSYSpc v.2.2. Principal coordinate analysis was performed using EIGEN and graphs were plotted using MXPLOT module, again in the same software package.
An estimate of the confidence limits for the grouping produced by each dendrogram was obtained by performing 2000 bootstrap re-samplings in WinBoot (Yap and Nelson 1996).
RESULTS
The Zn and Fe contents in the seeds of the 29 common bean genotypes are presented in Table 1. There were significant differences for both Zn (P < 0.05) and Fe (P < 0.01) content of the seeds among genotypes. In general, the Zn and Fe content of the Middle American genotypes were 16.1 and 11.3%, respectively, higher than the Andean genotypes.
The variability for seed Fe content among the genotypes was high, ranging from 8.9 to 112.9 ppm. The highest seed Fe content was observed in the USA navy bean cultivar Vista and one of the CIAT lines, NUA 56-1770, bred for high iron content in the redmottled seed class of the Andean gene pool. The Brazilian breeding line, Jalo EEP558, a parent of the common bean core mapping population, contained the lowest seed Fe content from the set studied.
The genetic location of SSR primers and primers developed from TC sequences were known and used in this study to give a uniform coverage for the common bean genome. These markers detected a total of 153 polymorphic alleles (Table 2). The number of alleles per locus ranged from two to nine, with an average of 3.1. The PIC values ranged from 0.19 for the size-indel 2685 and SNP 2285 to 0.99 for pv 2758 (Table 2). In general, the magnitude of PIC value is higher in primers developed from NBS-LRR gene sequences followed by SSR primers and primers developed from common bean TC sequences.
A GS matrix based on all possible pairs of genotypes ranged from 14.0 to 91.4% (Table 3). The lowest pairwise GS was observed between the Andean common bean genotype Jalo EEP558 and the Mesoamerican common bean genotype Dorado. The highest pairwise GS was observed between two related Mesoamerican genotypes MIB 465 and MIB 152. Among the Andean genotypes, the GS values ranged between 32.7 and 89.1%, while they were between 31.7 and 91.4% among Mesoamerican genotypes. However, the mean GS within the Mesoamerican genotypes (60.5%) was higher than among the Andean genotypes (55.5%), indicating a higher overall diversity within the Andean group, similar to observations by Tofiño et al. (2007).
Table 3.
Similarity matrix (%) of 29 common bean genotypes based on 153 polymorphic bands
| 1. | Jalo EEP558 | 100 | ||||||||||||||||||||||||||||
| 2. | NUA 45 | 63.5 | 100 | |||||||||||||||||||||||||||
| 3. | NUA 35 | 61.1 | 73.3 | 100 | ||||||||||||||||||||||||||
| 4. | NUA56-1770 | 51.9 | 72.7 | 61.4 | 100 | |||||||||||||||||||||||||
| 5. | NUA 59 | 43.4 | 67.3 | 55.4 | 89.1 | 100 | ||||||||||||||||||||||||
| 6. | G 122 | 69.5 | 48.5 | 53.3 | 40.4 | 35.6 | 100 | |||||||||||||||||||||||
| 7. | NY6020-4 | 57.1 | 49.5 | 46.0 | 33.0 | 37.8 | 66.0 | 100 | ||||||||||||||||||||||
| 8. | Benton | 69.2 | 53.7 | 46.5 | 42.6 | 32.7 | 62.6 | 71.6 | 100 | |||||||||||||||||||||
| 9. | Dorado | 14.0 | 28.8 | 33.0 | 34.6 | 39.6 | 23.2 | 41.9 | 26.9 | 100 | ||||||||||||||||||||
| 10. | BAT 93 | 20.0 | 28.1 | 32.4 | 40.4 | 48.3 | 28.6 | 45.2 | 36.8 | 71.6 | 100 | |||||||||||||||||||
| 11. | A 55 | 43.0 | 47.8 | 38.5 | 46.0 | 44.2 | 37.3 | 44.6 | 48.6 | 59.8 | 63.2 | 100 | ||||||||||||||||||
| 12. | XAN 176 | 23.9 | 41.4 | 37.3 | 45.0 | 42.5 | 19.6 | 37.5 | 41.4 | 67.3 | 70.1 | 70.2 | 100 | |||||||||||||||||
| 13. | MIB 151 | 44.2 | 49.6 | 40.4 | 49.6 | 48.7 | 37.0 | 44.1 | 47.9 | 57.1 | 52.0 | 63.3 | 53.3 | 100 | ||||||||||||||||
| 14. | MIB 152 | 37.2 | 39.6 | 45.4 | 32.1 | 35.2 | 37.1 | 44.9 | 37.7 | 56.9 | 58.9 | 58.7 | 55.0 | 62.9 | 100 | |||||||||||||||
| 15. | MIB 154 | 51.4 | 55.7 | 50.5 | 57.4 | 53.9 | 44.2 | 40.4 | 44.2 | 40.7 | 47.1 | 50.8 | 36.2 | 67.2 | 55.9 | 100 | ||||||||||||||
| 16. | MIB 217 | 33.9 | 37.9 | 35.2 | 44.8 | 49.2 | 29.7 | 32.5 | 32.8 | 70.3 | 67.2 | 65.5 | 58.8 | 76.8 | 64.9 | 69.4 | 100 | |||||||||||||
| 17. | MIB 465 | 40.8 | 37.4 | 42.9 | 35.5 | 34.9 | 34.7 | 42.6 | 44.9 | 54.4 | 56.6 | 60.0 | 58.2 | 67.2 | 91.4 | 55.4 | 64.9 | 100 | ||||||||||||
| 18. | MIB 466 | 43.6 | 42.1 | 50.0 | 49.1 | 46.6 | 26.7 | 36.5 | 40.4 | 66.1 | 63.3 | 65.0 | 73.5 | 63.4 | 67.9 | 55.5 | 68.9 | 70.8 | 100 | |||||||||||
| 19. | ND88-106-04 | 31.2 | 46.8 | 42.7 | 54.1 | 56.6 | 25.5 | 41.1 | 41.4 | 62.3 | 59.8 | 61.4 | 62.1 | 68.3 | 45.9 | 65.5 | 72.3 | 50.9 | 58.1 | 100 | ||||||||||
| 20. | Aztec | 30.4 | 27.1 | 27.6 | 35.4 | 40.8 | 32.2 | 41.2 | 43.8 | 63.0 | 64.7 | 56.6 | 60.6 | 41.9 | 44.7 | 31.7 | 51.9 | 44.2 | 54.9 | 52.5 | 100 | |||||||||
| 21. | Voyager | 38.5 | 41.4 | 43.1 | 45.0 | 47.8 | 49.0 | 57.1 | 48.6 | 66.0 | 68.4 | 64.9 | 63.8 | 61.7 | 58.7 | 51.7 | 60.5 | 58.2 | 61.5 | 62.1 | 60.6 | 100 | ||||||||
| 22. | Albion | 29.1 | 37.4 | 36.7 | 41.1 | 42.2 | 34.7 | 50.0 | 44.9 | 66.0 | 63.7 | 61.8 | 67.3 | 62.1 | 59.0 | 41.1 | 57.4 | 67.9 | 61.9 | 56.4 | 53.6 | 69.1 | 100 | |||||||
| 23. | Mayflower | 43.2 | 38.3 | 41.5 | 34.8 | 37.6 | 32.1 | 41.4 | 47.0 | 57.7 | 61.2 | 76.3 | 67.8 | 61.3 | 61.9 | 50.0 | 66.7 | 64.9 | 67.8 | 57.6 | 52.4 | 57.6 | 57.9 | 100 | ||||||
| 24. | T-39 | 30.8 | 40.7 | 37.6 | 37.0 | 32.7 | 26.3 | 42.2 | 46.3 | 59.0 | 56.1 | 66.7 | 68.5 | 62.7 | 62.3 | 49.1 | 59.8 | 67.9 | 65.5 | 62.5 | 52.1 | 57.7 | 59.8 | 69.6 | 100 | |||||
| 25. | Jaguar | 42.3 | 40.7 | 42.4 | 40.7 | 40.0 | 32.3 | 45.9 | 48.1 | 53.8 | 59.6 | 77.5 | 66.7 | 61.5 | 56.6 | 49.6 | 58.6 | 57.9 | 70.2 | 50.4 | 58.3 | 57.7 | 56.1 | 88.7 | 72.2 | 100 | ||||
| 26. | Othello | 32.7 | 27.8 | 38.4 | 46.3 | 41.8 | 22.2 | 38.5 | 46.3 | 53.8 | 61.4 | 46.8 | 68.5 | 54.7 | 50.9 | 40.7 | 51.7 | 59.8 | 68.4 | 48.6 | 67.3 | 59.5 | 60.6 | 59.1 | 61.1 | 63.0 | 100 | |||
| 27. | Ryder | 27.5 | 53.8 | 52.6 | 59.6 | 58.5 | 27.4 | 40.0 | 32.7 | 60.0 | 65.5 | 56.1 | 62.4 | 58.4 | 60.8 | 53.2 | 69.6 | 54.4 | 69.1 | 59.5 | 46.8 | 62.4 | 59.0 | 55.9 | 55.8 | 62.3 | 62.3 | 100 | ||
| 28. | Vista | 34.9 | 33.6 | 40.4 | 46.0 | 45.2 | 30.8 | 42.1 | 38.9 | 58.7 | 67.2 | 74.1 | 69.0 | 62.3 | 66.7 | 49.2 | 61.2 | 64.3 | 72.3 | 51.7 | 51.5 | 62.1 | 62.5 | 78.3 | 69.0 | 85.0 | 65.5 | 71.6 | 100 | |
| 29. | BelNeb-RR-1 | 28.3 | 35.0 | 36.2 | 33.0 | 32.4 | 34.0 | 53.8 | 46.6 | 66.7 | 67.9 | 59.0 | 64.2 | 46.4 | 45.5 | 40.7 | 52.3 | 47.1 | 55.0 | 49.1 | 66.7 | 64.2 | 68.6 | 60.6 | 58.3 | 62.7 | 64.1 | 58.6 | 57.9 | 100 |
|
|
||||||||||||||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | ||
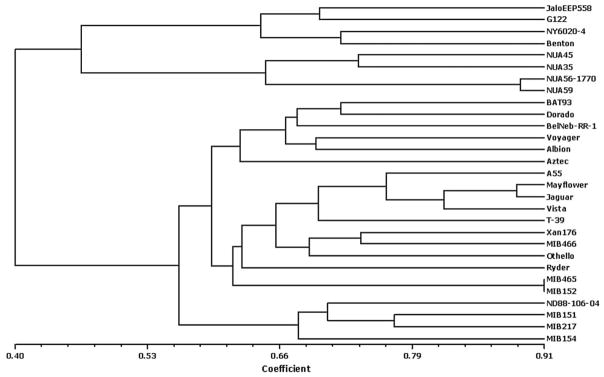
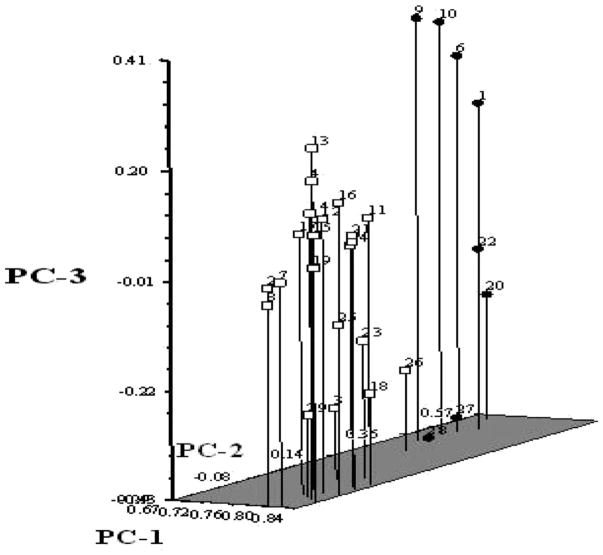
For a better understanding of the genetic relationship among common bean genotypes, the GS values of Table 3 were submitted to hierarchical clustering by UPGMA. The cluster analysis primarily separated the cultivars in close correspondence to two major clusters representing their primary gene pools (Fig. 1). Principal coordinates analysis revealed the global structure similar to the dendrogram analysis, but the distribution of these accessions was shown more clearly in three dimensions. Principal coordinate analysis showed that the first three eigen values explained 73.8% of the cumulative variation. These values were then plotted to identify the diversity of the genotypes (Fig. 2). Overall, the clustering pattern of the genotypes in the principal coordinates analysis corresponds with the dendrogram derived from UPGMA (Fig. 1). The Andean common bean cultivars were separated from the Middle American cultivars convincingly by PC1 (principal coordinate 1).
Fig. 1.
Dendrogram of common bean cultivars based on Dice coefficient of similarity matrix data using UPGMA.
Fig. 2.
Principal coordinate analysis of genetic diversity in 29 common bean cultivars. Open square symbols: Mesoamerican genotypes; closed circle symbols: Andean genotypes.
DISCUSSION
The highest seed Zn content was observed in our trial among genotypes MIB 465 and MIB 466, both belonging to the Middle American gene pool. The MIB lines were developed by the CIAT breeding program for improved nutritional quality. Beebe et al. (2000a) analyzed the seed Zn and Fe contents of over 1000 accessions in the CIAT core collection and suggested that the seed Fe content in the Andean gene pool tended to present higher values than those from the Mesoamerican pool under field grown conditions. The NUA lines were also developed by CIAT breeding program for high Fe content. These NUA lines previously observed with high Fe content at CIAT were variable in our trial. We conducted our study in greenhouse conditions, where there is no influence of soil differences as usually observed under field trial conditions. The variability for Zn and Fe contents in our study also could be due to the small sample size and inclusion of some genotypes selected for high Zn and Fe contents.
The UPGMA clustering analysis divided the genotypes into two major clusters. Two main branches of the dendrogram showed more than 50% GS, suggesting that genetic diversity is low between two distinct P. vulgaris gene pools. The genotypes Jalo EEP558, G122, NY 6020-4, Benton, and all four NUA breeding lines from CIAT originating from Andean gene pool, formed a distinct cluster separating from the other large group constituting the Middle American gene pool. Within the Andean group, the NUA lines, as expected, formed a subgroup. These CIAT lines were developed for high seed Fe content and share the same backcross pedigree, which includes one Mesoamerican bean parent, but seed class and plant selection for the Andean morphology. Likewise, within the large Mesoamerican group, the genotypes clearly formed three sub-groups. In the first sub-group, the genotypes BAT 93, Dorado, Aztec, BelNeb RR-1, Voyager and Albion, all are being used as parents of mapping populations developed for disease resistance and other trait attributes. Among these parents, the pinto bean cultivar Aztec and navy bean cultivar Voyager were bred for commercial cultivation in the United States of America. In the second sub-group, most of the black beans grouped together with a few pinto and navy bean cultivars. The six MIB accessions selected for nutritional quality traits in CIAT were placed into this second group and the other three formed a separate third group. MIB lines 465 and 152 grouped very closely, as they originated from the same pedigree. The other light-tan colored CIAT accession, MIB151 also has the same pedigree as of MIB465 and MIB152. However, a genetic shift appears to have occurred between these MIB lines, as these accessions clustered in two separate groups. This shift may be associated with selection for favorable alleles.
Several breeding strategies can be derived from the results of our genetic diversity analysis; however, our interest at present is to develop populations to map Zn and Fe content traits and to breed common bean cultivars with enhanced Zn and Fe contents. Substantial genetic variation existed among the genotypes under study that could be exploited for selection for mineral content. The clustering of the Andean accessions separate from the Mesoamerican accessions suggests that these common bean genotypes represent distinct germplasm and could be used to construct highly polymorphic populations. CIAT developed nutritionally enriched common bean lines clustered in both gene pools. Broad-based populations could be constructed by crossing selected cultivars from the two gene pools as well from combinations of several of the elite cultivars within each gene pool. We are currently using both approaches to develop common bean populations for further breeding and selection.
Selection of genotypes for breeding purpose based on micronutrient variability and genetic diversity has been suggested in many crop species including common bean (Beebe et al. 2000a), rice (Gregorio 2000), wheat (Monasterio and Graham 2000), and maize (Banziger and Long 2000). We have designed crossing programs based on seed Zn and Fe contents of the genotypes and their genetic divergence at the molecular level. The crossing program for seed Zn content was designed to cross genotypes with high Zn content, Jalo EEP558, MIB465, and MIB466, with the low Zn content genotypes, NUA56-1770 and NUA59 (Table 1). The high Zn content MIB lines belong to the Mesoamerican gene pool, while both the low Zn content NUA genotypes and the high Zn content genotype, Jalo EEP558, belong to the Andean gene pool. The GS values of genotypes NUA56-1770 with Jalo EEP558, MIB465, and MIB466 are 51.9, 35.5, and 49.1%, repectively, (Table 3) and those of NUA59 with Jalo EEP558, MIB465, and MIB466 are 43.4, 34.9, and 46.6%, respectively. These values indicate that the selected genotypes are very divergent at the molecular level and could result in better segregation and recombination of the desired alleles in successive generations during population development.
For the crossing plan to enhance seed Fe content, our choices for high Fe are genotypes Vista and NUA 56-1770 (Table 1), which belong to the two divergent gene pools (Fig. 1). The genotypes Jalo EEP558, NY6020-4, XAN176 and Mayflower have low seed Fe content (Table 1) and are included in the crossing program. Both the genotypes Jalo EEP558 and NY6020-4 are from the Andean gene pool, while the genotypes XAN176 and Mayflower represent the Mesoamerican gene pool. The selection of parents from both gene pools enables us to study inter- and intra-gene pool recombination events. The GS values of genotype Vista with the low Fe genotypes, Jalo EEP558, NY6020-4, XAN176 and Mayflower are 34.9, 42.1, 69.0, and 78.3%, respectively (Table 3). The high GS value between the genotypes Vista and Mayflower is obvious as both of them are navy bean cultivars adapted and selected for commercial cultivation in the United States of America. Although, these two genotypes belong to the same market class and are not genetically divergent, they differ significantly for seed Fe content. It can be expected that the introgression of a desired trait would be easier in this cross without sacrificing valuable trait(s) and/or adding unwanted trait(s) as a consequence of linkage drag. The GS of other high Fe genotype, NUA56-1770, with the low Fe genotypes, Jalo EEP558, NY6020-4, XAN176 and Mayflower, are 51.9, 33.0, 45.0 and 34.8%, respectively, (Table 3) indicating that the selected high Fe content Andean parent is genetically widely divergent from both Andean and Mesoamerican genotypes selected as low Fe content parents. The extent of genetic diversity present within the selected genotypes suggests that the introgression of genes for mineral nutrient might also benefit in the recombination of other economically important traits, especially when some of these selected genotypes (Jalo EEP558, NY 6020-4 and XAN 176) are being used as parents in mapping populations known to segregate for important biotic and abiotic resistance genes.
In order to generate advanced breeding lines with higher seed Zn and Fe contents, additional crossing programs may be considered. The Andean genotypes, Jalo EEP558 and Benton, and the Mesoamerican MIB genotypes, 151, 152, 465 and 466, have high Zn content in their seeds (Table 1). Similarly, the genotypes NUA45, NUA56-1770, Benton of the Andean gene pool and the genotypes MIB154, MIB465, Dorado, Voyager and Vista in the Mesoamerican gene pool have high Fe content in their seeds (Table 1). Crossing combinations with genotypes between and within gene pools having high genetic diversity and mineral content would be expected to accumulate positive alleles derived from unique sources and generate breeding lines with even higher seed mineral contents.
Acknowledgments
The authors thank Dr. Phillip E. McClean, Department of Plant Sciences, North Dakota State University, for providing SSR primers and primers developed from TC sequences. The project was supported by the USDA Cooperative State Research, Education and Extension Service; National Research Initiative-Plant Genome Program (Award#: 2006-03590).
Abbreviations
- CIAT
International Center for Tropical Agriculture
- GS
genetic similarity
- PIC
polymorphism information content
- SAHN
sequential, agglomerative, hierarchical, nested
- SSR
simple sequence repeats
- TC
tentative consensus
- UPGMA
unweighted pair group with arithmetic averaging
References
- Anderson JW, Randles KM, Kendall CWC, Jenkins DJA. Carbohydrate and fiber recommendations for individuals with diabetes: A quantitative assessment and meta-analysis of the evidence. J Am Coll Nutr. 2004;23:5–17. doi: 10.1080/07315724.2004.10719338. [DOI] [PubMed] [Google Scholar]
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Anderson JW, Major AW. Pulses and lipaemia, short- and long-term effect: potential in the prevention of cardiovascular disease. Br J Nutr. 2002;88:263–271. doi: 10.1079/BJN2002716. [DOI] [PubMed] [Google Scholar]
- Banziger M, Long J. The potential of increasing the iron and zinc density in maize through plant-breeding. Food Nutr Bull. 2000;21:397–400. [Google Scholar]
- Barbosa-Neto JF, Sorrells ME, Cisar G. Prediction of heterosis in wheat using coefficient of parentage and RFLP-based estimates of genetic relationship. Genome. 1996;39:1142–1149. doi: 10.1139/g96-144. [DOI] [PubMed] [Google Scholar]
- Becerra-Velasquez VL, Gepts P. RFLP diversity of common bean (Phaseolus vulgaris) in its centers of origin. Genome. 1994;37:256–263. doi: 10.1139/g94-036. [DOI] [PubMed] [Google Scholar]
- Beebe S, Gonzalez A, Rengifo J. Research on trace minerals in the common bean. Food Nutr Bull. 2000a;21:387–391. [Google Scholar]
- Beebe S, Rengifo J, Gaitan E, Duque MC, Tohme J. Diversity and origin of Andean landraces of common bean. Crop Sci. 2001;41:854–862. [Google Scholar]
- Beebe S, Sckroch PW, Tohme J, Duque MC, Pedraza F, Nienhuis J. Structure of genetic diversity among common bean landraces of Middle American origin based on correspondence analysis of RAPD. Crop Sci. 2000b;40:264–273. [Google Scholar]
- Benchimol LL, de Campos T, Carbonell SAM, Colombo CA, Chioratto AF, Formighieri EF, Gouvêa LRL, de Souza AP. Structure of genetic diversity among common bean (Phaseolus vulgaris L.) varieties of Mesoamerican and Andean origins using new developed microsatellite markers. Gen Res Crop Evol. 2007;54:1747–1762. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms. Ann Bot. 1995;76:113–176. [Google Scholar]
- Black RE. Zinc deficiency, infectious disease and mortality in the developing world. J Nutr. 2003;133:1485S–1489S. doi: 10.1093/jn/133.5.1485S. [DOI] [PubMed] [Google Scholar]
- Blair MW, Astudillo C, Beebe S. Analysis of nutritional quality traits in an Andean recombinant inbred line population. Bean Improv Coop. 2005;48:52–53. [Google Scholar]
- Blair MW, Pedraza F, Buendia HF, Gaitan-Solis E, Beebe SE, Gepts P, Tohme J. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.) Theor. Appl Genet. 2003;107:1362–1374. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Blair MW, Diaz JM, Hidalgo R, Diaz LM, Duque MC. Microsatellite characterization of Andean races of common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2007 doi: 10.1007/s00122-007-0644-8. (In press) [DOI] [PubMed] [Google Scholar]
- Blair MW, Giraldo MC, Buendia HF, Tovar E, Duque MC, Beebe S. Microsatellite marker diversity in common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2006;113:100–109. doi: 10.1007/s00122-006-0276-4. [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Hernández G, Blair M, Beebe S, Gepts P, Vanderleyden J. Beans (Phaseolus spp.) – model food legumes. Plant Soil. 2003;252:55–128. [Google Scholar]
- Cichy KA, Forster S, Grafton KF, Hosfield GL. Inheritance of seed zinc accumulation in navy bean. Crop Sci. 2005;45:864–870. [Google Scholar]
- Cox TS, Murphy JP. The effect of parental divergence on heterosis in winter wheat crosses. Theor Appl Genet. 1990;79:241–250. doi: 10.1007/BF00225958. [DOI] [PubMed] [Google Scholar]
- Diaz LM, Blair MW. Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theor Appl Genet. 2006;114:143–154. doi: 10.1007/s00122-006-0417-9. [DOI] [PubMed] [Google Scholar]
- Doyle J, Doyle J. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Duarte JM, dos Santos JB, Melo LC. Genetic divergence among common bean cultivars from different races based on RAPD markers. Genet Mol Biol. 1999;22:419–426. [Google Scholar]
- Forster SM, Moraghan JT, Grafton KF. Inheritance of seed-Zn accumulation in navy bean. Bean Improv Coop. 2002;45:30–31. [Google Scholar]
- Freyre R, Skroch P, Geffroy V, Adam-Blondon AF, Shirmohamadali A, Johnson W, Llaca V, Nodari R, Pereira P, Tsai SM, Tohme J, Dron M, Nienhuis J, Vallejos C, Gepts P. Towards an integrated linkage map of common bean. 4 Development of a core map and alignment of RFLP maps. Theor Appl Genet. 1998;97:847–856. [Google Scholar]
- Galván MZ, Aulicino MB, García Medina S, Balatti PA. Genetic diversity among Northwestern Argentinean cultivars of common bean (Phaseolus vulgaris L.) as revealed by RAPD markers. Gen Res Crop Evol. 2001;48:251–260. [Google Scholar]
- Gepts P. Genetic diversity of seed storage proteins in plants. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, editors. Plant population genetics, breeding, and genetic resources. Sinauer; Sunderland, MA: 1990. pp. 64–82. [Google Scholar]
- Gepts P, Bliss FA. Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia. Econ Bot. 1986;40:469–478. [Google Scholar]
- Gepts P, Debouck DG. Origin, domestication, and evolution of the common bean, Phaseolus vulgaris. In: Voysest O, Van Schoonhoven A, editors. Common beans: research for crop improvement. CAB; Oxford, UK: 1991. pp. 7–53. [Google Scholar]
- Graham RD, Senadhira D, Beebe S, Iglesias C, Monasterio I. Breeding for micronutrient density in edible portions of staple food crops: Conventional approaches. Field Crops Res. 1999;60:57–80. [Google Scholar]
- Gregorio GB, Senadhira D, Htut T, Graham RD. Breeding for trace mineral density in rice. Food Nutr Bull. 2000;21:382–386. [Google Scholar]
- Guzmán-Maldonado SH, Martínez O, Acosta JA, Guevara-Lara F, Parades-López O. Putative quantitative trait loci for physical and chemical components of common bean. Crop Sci. 2003;43:1029–1035. [Google Scholar]
- Harlan JR. Geographic patterns of variation in some cultivated plants. J Hered. 1975;66:184–191. [Google Scholar]
- Lefort-Buson M, Hebert Y, Damerval C. Tools for assessment of genetic and phenotypic diversity. Agronomie. 1988;8:173–178. [Google Scholar]
- Lioi L, Piergiovanni AR, Pignone D, Puglisi S, Santantonio M, Sonnante G. Genetic diversity of some surviving on-farm Italian common bean (Phaseolus vulgaris L.) landraces. Plant Breed. 2005;124:576–581. [Google Scholar]
- Manjarrez-Sandoval P, Carter TE, Jr, Webb DM, Burton JW. RFLP genetic similarity and coefficient of parentage as genetic variance predictors for soybean yield. Crop Sci. 1997;37:698–703. [Google Scholar]
- McConnell M, Mamidi S, Lee R, McClean PE. DNA sequence polymorphisms among common bean genes. Bean Improv Coop. 2006;49:143–144. [Google Scholar]
- Monasterio I, Graham RD. Breeding for trace minerals in wheat. Food Nutr Bull. 2000;21:392–396. [Google Scholar]
- Moraghan JT, Grafton KF. Genetic diversity and mineral composition of common bean seed. J Sci Food Agric. 2001;81:404–408. [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. PNAS. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Moreno C, Paredes-Lopez O. Hard-to-cook phenomenon in common bean-a review. Crit Rev Food Sci Nutr. 1993;33:227–286. doi: 10.1080/10408399309527621. [DOI] [PubMed] [Google Scholar]
- Rodiño AP, Santalla M, de Ron AM, Singh SP. A core collection of common bean from the Iberian peninsula. Euphytica. 2003;131:165–175. [Google Scholar]
- Rohlf FJ. NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Software; New York, N.Y: 2000. [Google Scholar]
- Singh SP. Broadening the genetic base of common bean cultivars: a review. Crop Sci. 2001;41:1659–1675. [Google Scholar]
- Singh SP, Westermann DT. A single dominant gene controlling resistance to soil zinc deficiency in common bean. Crop Sci. 2002;42:1071–1074. [Google Scholar]
- Singh SP, Gepts P, Debouck DG. Races of common bean (Phaseolus vulgaris, Fabaceae) Econ Bot. 1991;45:379–396. [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy: The principles and practice of numerical classification. W. H. Freeman and Company; San Francisco, CA: 1973. [Google Scholar]
- Sokal RR, Michener CD. A statistical method evaluating systematic relationship. Univ Kansas Sci Bull. 1958;38:1409–1438. [Google Scholar]
- Stoltzfus RJ. Iron-deficiency anemia: reexamining the nature and magnitude of the public health problem. Summary: implications for research and programs. J Nutr. 2001;131:S 697–700. doi: 10.1093/jn/131.2.697S. [DOI] [PubMed] [Google Scholar]
- Tofiño AP, Calderón JF, Palacio JD, Blair M. Variability study of 89 snap bean genotypes using the AFLP molecular technique. Bean Improv Coop. 2007;50:67–68. [Google Scholar]
- Tohme J, Gonzalez DO, Beebe S, Duque MC. AFLP analysis of gene pools of a wild bean core collection. Crop Sci. 1996;36:1375–1384. [Google Scholar]
- Van Tienderen PH, de Haan A, van der Linden CG, Vosman B. Biodiversity assessment using markers for ecologically important traits. Trends Ecol Evol. 2002;17:577–582. [Google Scholar]
- Winham DM, Hutchins AM, Johnston CS. Pinto bean consumption reduces biomarkers for heart disease risk. J Am Coll Nutr. 2007;26:243–249. doi: 10.1080/07315724.2007.10719607. [DOI] [PubMed] [Google Scholar]
- Yap I, Nelson RJ. WinBoot: A program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. International Rice Research Institute; Manila, Philippines: 1996. IRRI Discussion paper series no. 14. [Google Scholar]