Abstract
Synthetic biology aims to apply engineering principles to biology by modulating the behavior of living organisms. An emerging application of this field is the engineering of bacteria as a cancer therapy by the programming of therapeutic, safety, and specificity features through genetic modification. Here, we review progress in this engineering including the targeting of bacteria to tumors, specific sensing and response to tumor microenvironments, remote induction methods, and controllable release of therapeutics. We discuss the most prominent bacteria strains used and their specific properties and the types of therapeutics tested thus far. Finally, we note current challenges, such as genetic stability, that researchers must address for successful clinical implementation of this novel therapy in humans.
Introduction
Synthetic biology is a rapidly growing discipline that aims to rationally design the behavior of living organisms. Much of the field's focus has been on implementing genetic circuits, in which inputs are transformed in a cell into digital or analog outputs, in a manner analogous to a computer program executing an algorithm [1,2]. The field began with construction of the repressilator and toggle switch circuits in bacteria [3,4]. Shortly after the field's inception, researchers envisioned applications to cancer therapies by programming bacteria to sense and respond to a particular cancer disease state [5]. Since then, significant progress has been made in the design of genetic circuits for behavior, ranging from counting and pattern formation to oscillations and complex logic operations [2,6–8]. As advances in engineering bacteria behaviors emerge, more complex forms of bacterial cancer therapies can be developed.
Although the application of synthetic biology to cancer therapy is quite new, bacteria have been explored as cancer treatments for over a century. In 1890, William Coley induced tumor regression via administration of a cocktail of Streptococcus and other strains collectively known as “Coley's Toxins” [9]. This approach was thought to stimulate or activate the immune system and is considered one of the early forms of immunotherapy. Later, bacterial strains such as the obligate anaerobe Clostridium novyi were shown to grow selectively in hypoxic regions of solid tumors [10–12]. Several facultative anaerobic bacterial strains were also demonstrated to localize and grow in tumors rather than healthy tissue, presumably due to decreased immune surveillance in the necrotic core of tumors [13,14]. Interest in bacterial cancer therapy waned in the 20th century because of toxic side-effects resulting from inability to modify and control bacteria, as well as the advent of radiotherapy and chemotherapy [10]. Although these latter treatments became the mainstays of cancer treatment, more recently their toxicity and lack of specificity has become limiting, and more targeted approaches have gained traction [15]. With recent advances in cancer research and the newly available tools of synthetic biology, researchers are beginning to envision engineering bacteria to create potent cancer therapy. Improving upon the natural ability of bacteria to preferentially colonize tumors and implementing genetic circuitry can create a precisely controlled and highly specific delivery vehicle. A comprehensive review of bacterial therapies for cancer is given elsewhere [16]. Here, we focus specifically on the application of synthetic biology in bacteria to engineered bacterial cancer therapies, highlighting major instances of engineering in the last decade (Figure 1).
Figure 1. Overview of engineered bacterial cancer therapeutics.
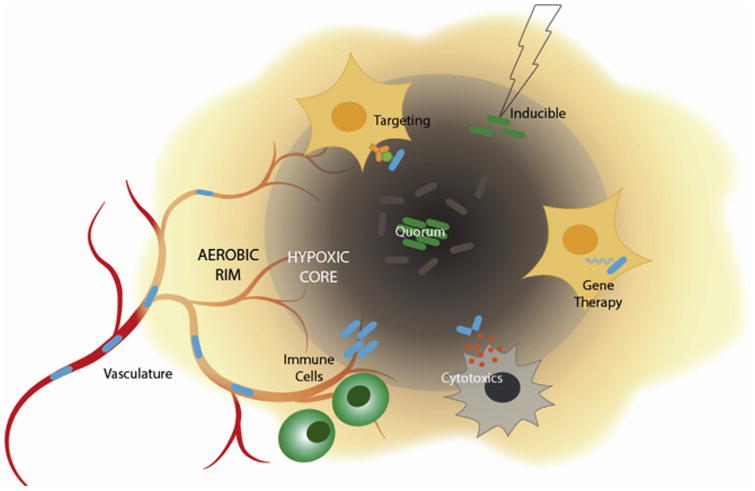
Synthetic biology is capitalizing on bacteria's natural ability to colonize immunoprivileged, hypoxic core regions of tumors through escaping from leaky vasculature. A variety of strategies such as targeting, inducing gene expression, quorum-sensing, expressing and releasing cytotoxics, and intracellular gene delivery have been engineered to control the behavior of these bacteria and produce anti-tumor effects.
Engineered bacterial cancer therapies
Traditional approaches to genetic engineering involve limited modification to natural bacteria functions. The synthetic biology approach utilizes bacteria as a modular platform for engineering, in which components like genes and promoters can be interchanged and combined to create nuanced and complex circuits. Here, we highlight several examples of genetic circuits designed to program bacteria for therapeutic benefits in cancer treatment.
Targeting and guiding
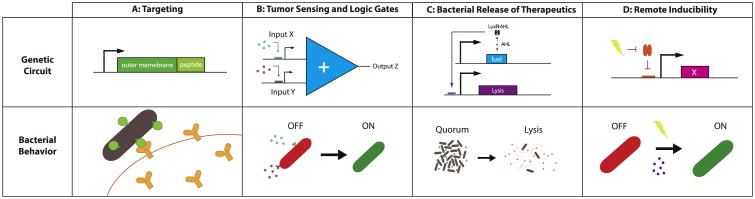
One way bacteria can naturally localize to tumors is by entering through the extensive tumor vasculature. Once inside, they can colonize the necrotic core, an immune-privileged environment protecting them from immune surveillance by macrophages and neutrophils [14,16]. This natural colonization process has the potential to be augmented by adding targeting or guiding mechanisms, which can also reduce the possibility of off-target colonization. One targeting method is engineering bacteria to express tumor homing proteins or peptides on the outer membrane (Figure 2A). Targeting motifs used so far include affibodies (proteins designed to bind targets such as upregulated receptors in cancer cells (e.g. HER2)), synthetic adhesion molecules which mimic immunoglobulin fragments and recognize antigen receptors, and known tumor-targeting peptides such as RGD [17–19]. In a recent example, Piñeero-Lambea et al. administered bacteria expressing synthetic adhesion molecules termed adhesins to tumor-bearing mice and observed more efficient tumor colonization than by wild type bacteria. In addition, they saw reduced off target colonization of the spleen or liver due to the lack of adhesion binding on those tissues compared to tumor cells. Their work created a modular platform, in which different synthetic adhesins can be used for targeted localization to a variety of tumors. Another approach to increase tumor localization is to use external cues to guide bacteria to the tumor site. Felfoul et al. engineered a strain of magnetotactic bacteria to carry drug-loaded nanoliposomes. After injection of grafted tumors in mice, bacteria were guided by the application of a magnetic field in the tumor [20]. Whether through engineering targeted or remotely guidable bacteria, enhancing tumor localization has the potential to improve colonization efficiency and off-target effects and may have a significant impact on achieving successful clinical use of these therapies.
Figure 2. Engineered circuits for bacterial cancer therapy.
(A) Targeting to tumor cells can be achieved by expression of tumor-specific peptides on the bacterial outer membrane. (B) Tumor microenvironments can be sensed specifically by AND logic gates, which expresses an output such as a therapeutic only if all required inputs or markers of the tumor microenvironment are present. (C) In many cases lysis of bacteria must be induced to release therapeutics into the tumor; one relevant circuit is the synchronized lysis circuit which uses quorum sensing. (D) Therapeutics or other actions can be driven by inducing promoters through chemical inducers or radiation.
Tumor sensing and logic circuits
The tumor microenvironment presents several unique chemical and physical signatures that bacteria can be programmed to sense, in order to limit their action to specific settings (Figure 2B). For example, groups have engineered bacteria to sense glucose gradients and hypoxia, known tumor environment cues. To sense glucose gradients, Panteli et al. utilized a previously established synthetic hybrid receptor, containing the periplasmic domain of the Trg chemotactic receptor, to induce GFP expression based on glucose concentration [21]. The programmed bacteria sensed glucose concentration gradients over tumor cell masses within a microfluidic chamber. Their approach can be used to characterize the glucose profiles and metabolic activity over different tumor types [22]. To sense hypoxia, researchers have utilized known oxygen sensitive promoters, such as the synthetic FF20 or the endogenous promoter pepT, which are activated by oxygen binding Fumarate and Nitrate Reduction (FNR) regulatory proteins [23]. These promoters were fused to either the production of a therapeutic molecule (for targeted delivery) or the expression of an essential gene such as asd (for containment of bacteria in the hypoxic tumor area) [24,25]. To find a collection of promoters that respond to tumor conditions, several groups have sequenced tumor-resident bacteria in mouse models [26,27]. Leshner et al. injected Salmonella typhimurium containing a promoter library driving GFP into mouse tumor models. By sorting and sequencing GFP expressing bacteria, they were able to identify tumor specific promoters [27]. In a follow-up paper, Deyneko et al. then adapted these promoters and incorporated hypoxia sensing elements to build a synthetic tumor specific hypoxic promoter [28].
Since bacteria can grow to a higher density in tumor environments than in healthy tissue, quorum (density) sensing can be used as a tumor sensitive switch (Figure 2C) [29–32]. In an application for cancer, Swofford et al. demonstrated activation of protein expression based on quorum sensing when bacteria population reached a critical density in mouse tumor models [31,33]. Quorum sensing has been multiplexed with additional input signals as well [29,30]. For example, Anderson et al. pioneered the use of AND gates for bacterial therapies in this context, sensing acylhomoserine lactone (AHL) and Mg+ [30]. In the broader context of synthetic biology, groups have built XOR, NAND, and more complex circuitry to tightly regulate microbial sensing and computing [1,2,34,35]. As knowledge of tumor conditions and biomarkers improve, and the ability to construct bacterial logic systems increases, bacterial cancer therapies will be able to utilize these frameworks for elevated safety and specificity.
Remote inducibility
Inducible systems use external cues for activation, allowing for an additional level of control over production of a therapeutic or genetic circuit of interest. Chemical inducers are commonly used in vitro to control bacterial promoters. In the context of bacterial cancer therapies, researchers have used chemical inducers such as l-arabinose, salicylic acid, Isopropyl β-d-1-thiogalactopyranoside (IPTG), AHL and tetracycline to remotely activate bacteria residing in tumors [36–41]. Chemical induction allows for actuation of dose and timing and requires little genetic modification. However, due to the unknown structure of tumor vasculature a priori and different tissue diffusivities of chemical inducers, local inducer concentration is often difficult to estimate and may result in inaccurate and non-uniform induction of bacteria inside of tumors. Additional challenges include being able to sustainably induce gene expression over time, although this challenge could be mitigated by toggle-switch circuits that require a single pulse of inducer [42].
In addition to chemical cues, inducible expression from electromagnetic or light waves has also been tested in the context of bacterial therapies (Figure 1). For instance, one approach is to use γ-irradiation to indirectly activate the inducible recA bacterial promoter [43–45]. The γ-irradiation causes DNA damage, which then promotes the degradation of the RecA repressor LexA. Removal of the repressor LexA allows transcription of downstream genes (Figure 2D) [43]. The advantage of using gamma waves is deep penetration of tumor tissue. However, the method also induces DNA damage, which can be toxic to nearby healthy cells and possibly introduce unwanted mutations in bacterial genes encoding therapeutics. Other types of wave modulation may include utilization of ultrasound waves or optogenetic approaches that control gene expression with visible light [46–48]. These modes of induction can provide more exact spatial and temporal control of microbes, although they may require specialized instruments or infrastructure for use.
Release of bacterial therapeutics
While bacteria can produce a wide variety of therapeutics, an ongoing challenge is the effective release of therapeutics from bacteria into the microenvironment. One method, lysis of bacteria, has been utilized by expressing specific phage lysis genes or changing culturing conditions [43,49]. Pijkeren et al. administered ampicillin to lyse bacteria and thus release plasmids for tumor cell uptake [50]. Instead of utilizing antibiotics directly for lysis, Chamacho et al. placed an adapted bacteriophage lambda lysis operon under a tetracycline inducible promoter to better control cell lysis. In the same bacteria, they also programmed a salicylic acid inducible protein production cascade to cause accumulation of therapeutics before lysis [51]. In addition to releasing therapeutics, lysis provides two other advantageous properties: (1) release of bacterial adjuvants that may stimulate immune responses, and (2) pruning of the population growth over time. Repeated lysis of bacteria can lead to oscillations in population growth. Din et al. engineered a circuit termed the synchronized lysis circuit (SLC) in which a lysis gene, as well as production of a therapeutic compound, was regulated by quorum sensing. The growth of SLC bacteria would trigger rhythmic bacterial death (Figure 2C) [29]. The SLC circuit led to reduction in tumor activity in vitro and in vivo, where it slowed tumor growth while controlling bacterial growth. Additionally, mice were healthier when treated with the lysis circuit bacteria than from bacteria with a constitutively produced therapeutic. Dynamic circuits such as these capable of driving periodic drug delivery may have unique implications, as the timing of drug administration has recently been shown to be important to therapeutic efficacy and chemoresistance development [52,53].
Bacterial secretion is another promising means of therapeutic delivery to tumor microenvironment [40,43,54]. Secretion can be achieved by use of a leader signal sequence, a short peptide fused to the N-terminus of the protein of interest [40,55,56]. These leader signal sequences are analogous to zip codes that traffic the translated protein to the bacterial periplasm followed by secretion outside of the cell [57]. The limitation of secretion methods is that the amount of protein delivered depends on the secretory pathway, which can be limited to certain organisms (e.g. Escherichia coli does not naturally secrete proteins) [58]. Interestingly, some groups have demonstrated successful delivery of cargo to tumor sites despite lysis or secretion, presumably due to basal lysis of bacteria in the tumor microenvironment [59–61]. In addition, mere release of therapeutics in extracellular space may not be therapeutically effective if the released protein has an intracellular anti-tumor effect. Utilizing cell-penetrating peptides (CPP), other cell invasion mediated strategies or type III secretion systems to traffic therapeutic cargo into cells are possible ways to overcome this challenge [5,62,63].
Strains and clinically relevant properties of bacteria in use
One important consideration for engineered bacterial therapies is the species and strain of bacteria used. Some of the different bacteria utilized so far include S. typhimurium, E. Coli, Bifidobacteria, lactic acid bacteria such as Streptococcus and Lactobacillus, Listeria, and Bacillus subtilis. Each of these strains has its own unique properties affecting its potential use for cancer therapy including its tumor colonization ability (Figure 1), ability to invade tissue, interaction with the immune systems, and ease of genetic manipulation.
Gram negative bacteria
Currently, the two most used bacteria for engineered bacterial cancer therapy are S. typhimurium and E. coli. Both are Gram-negative bacteria that have been shown to colonize tumors in mice at high ratios compared to normal tissues [13]. As Gram-negative bacteria, they naturally contain lipopolysaccharide (LPS, a 3-part phosphoglycolipid found in the cell wall), which can be responsible for immune system stimulation (Figure 1) [64]. However, these strains have several differences affecting their potential use for cancer therapy.
S. typhimurium (Salmonella enterica serovar Typhimurium) is the most widely studied engineered bacterial cancer therapy and has been used for a variety of applications, reaching as far as clinical trials. As a facultative anaerobe, it can grow in both the hypoxic core of tumors as well as the non-hypoxic regions. The most prominent strains of S. typhimurium, all genetically attenuated for safety, are VNP20009 and A1-R, and recently other strains have been investigated such as SL7207 and CRC2631 [4]. When administered, VNP20009 and other strains preferentially colonize tumors over healthy tissues at ratios greater than 1000:1 [15]. Interactions of the strain with the host immune system are also important; it has been shown that Salmonella is capable of recruiting immune cells such as T-cells, resulting in immune cell/T-cell-mediated tumor killing [65]. While pathogenic strains like S. typhimurium must be attenuated (modified) for safe clinical use, such as the mutation of an LPS gene in VNP20009, these modifications may reduce their clinical efficacy [64,66,67].
E. coli, a model organism extensively studied in synthetic biology, is the next most widely used bacteria in the field of cancer therapeutics. While it is also a Gram-negative facultative anaerobe, unlike S. typhimurium, it has non-pathogenic variants naturally found in the human gut (commensal strains, some of which are probiotics that have a positive effect on health when given) [68]. The ability to make use of these strains without further attenuation and their status as clinically approved probiotics makes them attractive candidates for use in therapies [69]. The most common probiotic strain in use is E. coli Nissle 1917, but alternative strains have been explored [41,69]. While E. coli has been used more than S. typhimurium in the overall field of synthetic biology, Prindle et al. demonstrated that significant genetic circuits built in E. coli could be transferred to the more clinically used S. typhimurium, noting that these two species have similar ease of engineering as they have fully sequenced genomes, knockout collections, and easily used tools for genetic manipulation [70].
Gram positive bacteria
Gram-positive bacteria have been explored as cancer therapies, but have been engineered to a lesser extent than S. typhimurium and E. coli. C. novyi was one of the early strains shown to have an anticancer effect, and recently attenuated C. novyi-NT has garnered renewed interest [50,54,71,72]. Other Gram-positive bacteria that have been used include lactic acid bacteria, Bifidobacteria, Listeria, and B. subtilis [50,73–75]. Bifidobacteria and lactobacillus have probiotic strains naturally found in the human gut and are already in use for other diseases, making them a popular choice for therapeutic delivery [76,77]. While Listeria has been investigated mostly as a vaccine, some groups have used it as a gene delivery vector for cancer therapy due to its intracellular life cycle [50,78,79]. Currently, engineering of Gram-positive bacteria is limited by the lack of synthetic biology tools available. Streptococcus, for example, is difficult to transform—a fundamental procedure in synthetic biology [80]. B. subtilis is naturally competent and its full genome has been sequenced, although genetic circuitry has not been as developed. New tools are being developed for genetic engineering in this species and others, though much work remains to be done [75,81–83].
Anti-cancer therapeutics delivered by bacteria
A major consideration in bacterial cancer therapy is the choice of therapeutic. While in some cases the bacterium itself is the therapeutic, bacteria are most often used to locally and specifically deliver a therapeutic molecule of interest, as controlled local delivery can mitigate unwanted off-target effects compared to delivering a therapeutic systemically [15,16,69]. Many reviews have detailed therapeutics used; they broadly fall under two types: proteins and nucleic acids [64,84,85]. Proteins include cytotoxic agents, immune-stimulatory molecules, growth pathway regulators, and prodrug enzymes [40,55,86]. One example of a cytotoxic agent is HlyE, a pore-forming toxin, which has been used by several groups [23,29]. Immunostimulatory molecules can include cytokines, antibodies and tumor-specific antigens. Recently, delivery in vivo of S. typhimurium expressing flagellin B, a structural component of the flagellum from Vibrio Vulnificus was shown to lead to recruitment of macrophages and tumor regression, possibly via activation of the toll like receptor pathways [40]. DNA encoding for cytokines and bacteria antigens can be delivered for gene transfer via a co-opted invasion system from Listeria, or shRNA can be delivered for RNAi-mediated gene knockdown [61,64]. Recently, some studies have also used bacteria as carriers for traditional chemotherapies, such as “bacteriobots” and “nanoswimmers” loaded with liposomes and nano-particles, respectively, of doxorubicin [87,88].
Conclusions
Engineered bacterial cancer therapy promises controllable, targeted delivery of therapeutics to tumors, mitigating some of the major issues of current therapies. There are several current challenges researchers must address to develop bacteria as a successful therapy.
An inherent issue in using engineered bacteria is the potential for mutation or plasmid loss, which could cause loss of therapeutic production or reversion of the safety modification. While antibiotics can be used to maintain selection for plasmids in vitro, use of such methods presents a challenge in vivo because they may lead to resistance development and microbiome dysbiosis. One approach to mitigate plasmid loss is genomic integration, which is known to be fairly stable. Clairmont et al. showed VNP20009 strain, with several chromosomal modifications for safe attenuation, was genetically stable over many generations in vitro and in vivo [91]. Integration is less convenient for rapid construction of variants, although many systems such as lambda red, phage integration, and CRISPR have been developed [89,90]. However, these systems typically have lower copy number and hence lower redundancy, leaving strains susceptible to loss of function mutations. Alternatively, stabilizing elements can be incorporated into engineered plasmids, such as plasmid segregation, toxin-antitoxin systems, and auxotrophy, including balanced-lethal systems [29,92,93]. In some applications, plasmid loss may in fact be desirable so that the toxin production function is not maintained.
Future applications in programming bacteria may include more complex feedback systems which could respond to cell death or self-regulate. As the field of cancer biology advances, more potential therapeutic targets will be discovered, allowing an ever-widening range of therapeutics to be engineered into bacteria. Finally, the efficacy of bacteria in clinical trials must be demonstrated for eventual use in patient treatment. While Bacillus Calmette-Guerin therapy is approved for use in bladder cancer, no engineered bacterial cancer therapy is clinically approved as of yet [94]. Currently, there are several clinical trials that utilize bacteria for cancer therapy. One example is Marina Biotech's CEQ508 bacteria, which delivers RNAi to treat a condition underlying colon cancer; additionally, Aduro is testing attenuated Listeria treatment and BioMed Valley Discoveries has a clinical trial for attenuated Clostridium [95–97]. Although clinical trials are in early stages, as more and more therapeutics reach the clinical trial phase, the successful use of engineered bacteria for cancer therapy may be just over the horizon.
Acknowledgments
This work was supported in part by the NIH Pathway to Independence Award (R00CA197649-02).
Footnotes
Author disclosure statement: No competing financial interests exist.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 2.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 3.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 5**.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. This work was the first to demonstrate the potential use of bacteria for cancer therapy, in which authors engineered bacteria to invade tumors by combining tumor-sensing circuits with an invasion molecule. [DOI] [PubMed] [Google Scholar]
- 6.Liu CL, Fu XF, Liu LL, Ren XJ, Chau CKL, Li SH, Xiang L, Zeng HL, Chen GH, Tang LH, et al. Sequential establishment of stripe patterns in an expanding cell population. Science. 2011;334:238–241. doi: 10.1126/science.1209042. [DOI] [PubMed] [Google Scholar]
- 7.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy EF. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop J. 2006;26:154–158. [PMC free article] [PubMed] [Google Scholar]
- 10.Malmgren RA, Flanigan CC. Localization of the vegetative form of Clostridium tetani in mouse tumors following intravenous spore administration. Cancer Res. 1955;15:473–478. [PubMed] [Google Scholar]
- 11.Moese JR, Moese G. Oncolysis by clostridia. I. Activity of Clostridium butyricum (M-55) and other nonpathogenic clostridia against the ehrlich carcinoma. Cancer Res. 1964;24:212–216. [PubMed] [Google Scholar]
- 12.Thiele EH, Arison RN, Boxer GE. Oncolysis by clostridia. Iii. Effects of clostridia and chemotherapeutic agents on rodent tumors. Cancer Res. 1964;24:222–233. [PubMed] [Google Scholar]
- 13.Yu YA, Zhang Q, Szalay AA. Establishment and characterization of conditions required for tumor colonization by intravenously delivered bacteria. Biotechnol Bioeng. 2008;100:567–578. doi: 10.1002/bit.21785. [DOI] [PubMed] [Google Scholar]
- 14.Yu YA, Shabahang S, Timiryasova TM, Zhang Q, Beltz R, Gentschev I, Goebel W, Szalay AA. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol. 2004;22:313–320. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- 15.Wang CZ, Kazmierczak RA, Eisenstark A. Strains, mechanism, and perspective: Salmonella-based cancer therapy. Int J Microbiol. 2016;2016:1–10. doi: 10.1155/2016/5678702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Forbes NS. Engineering the perfect (bacterial) cancer therapy. Nat Rev Cancer. 2010;10:785–794. doi: 10.1038/nrc2934. This review comprehensively discusses the field of bacteria used for cancer therapy, including the attributes of bacteria that suit them for this use and details on therapeutics produced by bacteria prior to the review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gujrati V, Kim S, Kim SH, Min J, Choy HE, Kim S, Jon S. Bio-engineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8 doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int. 2014;2014:852748. doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Piñero-Lambea C, Bodelón G, Fernández-Periáñez R, Cuesta AM, Álvarez-Vallina L, Fernández L. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol. 2015;4:463–473. doi: 10.1021/sb500252a. In this paper the authors created synthetic adhesin molecules expressed on the bacterial outer membrane which could recognize and bind to tumor antigens by way of an immunoglobulin domain, which significantly lowered off-target colonization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Felfoul O, Mohammadi M, Taherkhani S, de Lanauze D, Zhong Xu Y, Loghin D, Essa S, Jancik S, Houle D, Lafleur M, et al. Magneto-aerotactic bacteria deliver drug-containing nano-liposomes to tumour hypoxic regions. Nat Nanotechnol. 2016;11:941–947. doi: 10.1038/nnano.2016.137. In this work authors created a novel way of externally guiding bacteria to tumor sites by using magnetotactic bacteria and applying a magnetic field in the tumor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signaling by a hybrid protein – communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor envz. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panteli JT, Forbes NS. Engineered bacteria detect spatial profiles in glucose concentration within solid tumor cell masses. Biotechnol Bioeng. 2016;113:2474–2484. doi: 10.1002/bit.26006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, Kehoe SC, Lewis CE. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 2009;16:329–339. doi: 10.1038/gt.2008.188. [DOI] [PubMed] [Google Scholar]
- 24.Yu B, Yang M, Shi L, Yao Y, Jiang Q, Li X, Tang LH, Zheng BJ, Yuen KY, Smith DK, et al. Explicit hypoxia targeting with tumor suppression by creating an “obligate” anaerobic Salmonella typhimurium strain. Sci Rep. 2012;2:436. doi: 10.1038/srep00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mengesha A, Dubois L, Lambin P, Landuyt W, Chiu RK, Wouters BG, Theys J. Development of a flexible and potent hypoxia-inducible promoter for tumor-targeted gene expression in attenuated Salmonella. Cancer Biol Ther. 2006;5:1120–1128. doi: 10.4161/cbt.5.9.2951. [DOI] [PubMed] [Google Scholar]
- 26.Arrach N, Zhao M, Porwollik S, Hoffman RM, McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68:4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 27*.Leschner S, Deyneko IV, Lienenklaus S, Wolf K, Bloecker H, Bumann D, Loessner H, Weiss S. Identification of tumor-specific Salmonella typhimurium promoters and their regulatorylogic. Nucleic Acids Res. 2012;40:2984–2994. doi: 10.1093/nar/gkr1041. Using a promoter library, Leschner et al. were able to sort and sequence tumor colonizing bacteria in order to identify DNA motifs that correspond to activated promoters in the tumor microenvironment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Deyneko IV, Kasnitz N, Leschner S, Weiss S. Composing a tumor specific bacterial promoter. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155338. In this paper, Deyneko et al. constructed a tumor specific bacteria promoter by varying promoter length and DNA motif of transcription factor binding site. With their tumor specific promoter, they were able to demonstrate in vivo higher expression of reporter gene in tumor models versus healthy tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Din OM, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. Utilizing quorum sensing, the authors of this work were able to construct a synchronized lysis circuit that programs bacteria to lyse in a uniform oscillating behavior for in vivo therapeutic delivery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Swofford CA, Dessel N, Forbes NS. Quorum-sensing Salmo- nella selectively trigger protein expression within tumors. Proc Natl Acad Sci. 2015;112:3457–3462. doi: 10.1073/pnas.1414558112. In this paper, Swofford and coworkers integrated a quorum sensing reporter system in Salmonella and demonstrated density dependent colonization of tumor cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danino T, Prindle A, Hasty J, Bhatia S. Measuring growth and gene expression dynamics of tumor-targeted S. typhimurium bacteria. J Vis Exp. 2013:e50540. doi: 10.3791/50540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai Y, Toley BJ, Swofford CA, Forbes NS. Construction of an inducible cell-communication system that amplifies Salmonella gene expression in tumor tissue. Biotechnol Bioeng. 2013;110:1769–1781. doi: 10.1002/bit.24816. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 35.Stanton BC, Nielsen AA, Tamsir A, Clancy K, Peterson T, Voigt CA. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Royo JL, Becker PD, Camacho EM, Cebolla A, Link C, Santero E, Guzman CA. In vivo gene regulation in Salmonella spp. by a salicylate-dependent control circuit. Nat Methods. 2007;4:937–942. doi: 10.1038/nmeth1107. [DOI] [PubMed] [Google Scholar]
- 37.Loessner H, Endmann A, Leschner S, Westphal K, Rohde M, Miloud T, Hammerling G, Neuhaus K, Weiss S. Remote control of tumour-targeted Salmonella enterica serovar Typhimurium by the use of L-arabinose as inducer of bacterial gene expression in vivo. Cell Microbiol. 2007;9:1529–1537. doi: 10.1111/j.1462-5822.2007.00890.x. [DOI] [PubMed] [Google Scholar]
- 38.Stritzker J, Weibel S, Hlll PJ, Oelschlaeger TA, Goebel W, Szalay AA. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int J Med Microbiol. 2007;297:151–162. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Zheng JH, Nguyen VH, Jiang SN, Park SH, Tan W, Hong SH, Shin MG, Chung IJ, Hong Y, Bom HS, et al. Two-stepenhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. SciTransl Med. 2017;9 doi: 10.1126/scitranslmed.aak9537. In this paper, Zheng et al. engineered attenuated Salmonella typhimurium to express and secrete heterologous flagella in order to recruit immune cells via TLR4 and TLR5 pathway. They were able to demonstrate host anti-tumor response in vivo. [DOI] [PubMed] [Google Scholar]
- 41.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, Bhatia SN. Programmable probiotics for detection of cancerin urine. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, Silver PA. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci U S A. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganai S, Arenas RB, Forbes NS. Tumour-targeted delivery of TRAIL using Salmonella typhimurium enhances breast cancer survival in mice. Br J Cancer. 2009;101:1683–1691. doi: 10.1038/sj.bjc.6605403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nuyts S, Van Mellaert L, Theys J, Landuyt W, Lambin P, Anne J. The use of radiation-induced bacterial promoters in anaerobic conditions: a means to control gene expression in Clostridium-mediated therapy for cancer. Radiat Res. 2001;155:716–723. doi: 10.1667/0033-7587(2001)155[0716:tuorib]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Nuyts S, Van Mellaert L, Theys J, Landuyt W, Bosmans E, Anne J, Lambin P. Radio-responsive recA promoter significantly increases TNF alpha production in recombinant clostridia after 2 Gy irradiation. Gene Ther. 2001;8:1197–1201. doi: 10.1038/sj.gt.3301499. [DOI] [PubMed] [Google Scholar]
- 46.Kaberniuk AA, Shemetov AA, Verkhushawhereas VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods. 2016;13:591. doi: 10.1038/nmeth.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A, Shapiro MG. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat Chem Biol. 2017;13:75–80. doi: 10.1038/nchembio.2233. [DOI] [PubMed] [Google Scholar]
- 49.Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85–109. doi: 10.1128/MMBR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Pijkeren JP, Morrissey D, Monk IR, Cronin M, Rajendran S, O'Sullivan GC, Gahan CG, Tangney M. A novel Listeria monocytogenes-based DNA delivery system for cancer gene therapy. Hum Gene Ther. 2010;21:405–416. doi: 10.1089/hum.2009.022. [DOI] [PubMed] [Google Scholar]
- 51.Camacho EM, Mesa-Pereira B, Medina C, Flores A, Santero E. Engineering Salmonella as intracellular factory for effective killing of tumour cells. Sci Rep. 2016;6 doi: 10.1038/srep30591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levi F. Chronotherapeutics: the relevance of timing in cancer therapy. Cancer Causes Control. 2006;17:611–621. doi: 10.1007/s10552-005-9004-7. [DOI] [PubMed] [Google Scholar]
- 53.Levi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol. 2010;50:377–421. doi: 10.1146/annurev.pharmtox.48.113006.094626. [DOI] [PubMed] [Google Scholar]
- 54.Barbe S, Van Mellaert L, Theys J, Geukens N, Lammertyn E, Lambin P, Anne J. Secretory production of biologically active rat interleukin-2 by Clostridium acetobutylicum DSM792 as a tool for anti-tumor treatment. FEMS Microbiol Lett. 2005;246:67–73. doi: 10.1016/j.femsle.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 55.Loeffler M, Le'Negrate G, Krajewska M, Reed JC. IL-18-producing Salmonella inhibit tumor growth. Cancer Gene Ther. 2008;15:787–794. doi: 10.1038/cgt.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loeffler M, Le'Negrate G, Krajewska M, Reed JC. Attenuated Salmonella engineered to produce human cytokine LIGHT inhibit tumor growth. Proc Natl Acad Sci U S A. 2007;104:12879–12883. doi: 10.1073/pnas.0701959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei SP, Lin HC, Wang SS, Higaki P, Wilcox G. Characterization of the Erwinia carotovora peh gene and its product poly-galacturonase. Gene. 1992;117:119–124. doi: 10.1016/0378-1119(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 58.Lee CC, Wong DWS, Robertson GH. An E. coli expression system for the extracellular secretion of barley alpha-amylase. J Protein Chem. 2001;20:233–237. doi: 10.1023/a:1010904109747. [DOI] [PubMed] [Google Scholar]
- 59.Loeffler M, Le'Negrate G, Krajewska M, Reed JC. Salmonella typhimurium engineered to produce CCL21 inhibit tumor growth. Cancer Immunol Immunother. 2009;58:769–775. doi: 10.1007/s00262-008-0555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li YH, Guo KY, Chen H, Xie YM, Song CY, Tang X, Ren CM. Oral cytokine gene therapy against murine tumor using attenuated Salmonella typhimurium. Int J Cancer. 2001;94:438–443. doi: 10.1002/ijc.1489. [DOI] [PubMed] [Google Scholar]
- 61.Li X, Li Y, Wang B, Ji K, Liang Z, Guo B, Hu J, Yin D, Du Y, DJKopecko, Kim K, Lim D, Jeong K, Hong Y, Nguyen VH, Kim TH, Ryu S, Lim JA, Kim J, et al. Anti-tumoral effect of the mitochondrial target domain of noxa delivered by an engineered Salmonella typhimurium. PLoS One. 2013;9 doi: 10.1371/journal.pone.0080050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong JH, Kim K, Lim D, Jeong K, Hong Y, Nguyen VH, Kim TH, Ryu S, Lim JA, Kim J, et al. Anti-tumoral effect of the mitochondrial target domain of noxa delivered by an engineered Salmonella typhimurium. PLoS One. 2014:9. doi: 10.1371/journal.pone.0080050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reeves AZ, Spears WE, Du J, Tan KY, Wagers AJ, Lesser CF. Engineering Escherichia coli into a protein delivery system for mammalian cells. Acs Synth Biol. 2015;4:644–654. doi: 10.1021/acssynbio.5b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nallar SC, Xu DQ, Kalvakolanu DV. Bacteria and genetically modified bacteria as cancer therapeutics: current advances and challenges. Cytokine. 2017;89:160–172. doi: 10.1016/j.cyto.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Vendrell A, Mongini C, Gravisaco MJ, Canellada A, Tesone AI, Goin JC, Waldner CI. An oral Salmonella-based vaccine inhibits liver metastases by promoting tumor-specific T-cell-mediated immunity in celiac and portal lymph nodes: a pre-clinical study. Front Immunol. 2016;7 doi: 10.3389/fimmu.2016.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami T, DeLong J, Eilber FC, Zhao M, Zhang Y, Zhang N, Singh A, Russell T, Deng S, Reynoso J, et al. Tumor-targeting Salmonella typhimuriumA1-R in combination with doxorubicin eradicate soft tissue sarcoma in a patient-derived orthotopic xenograft (PDOX) model. Oncotarget. 2016;7:12783–12790. doi: 10.18632/oncotarget.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang MM, Swofford CA, Forbes NS. Lipid A controls the robustness of intratumoral accumulation of attenuated Salmonella in mice. Int J Cancer. 2014;135:647–657. doi: 10.1002/ijc.28700. [DOI] [PubMed] [Google Scholar]
- 68.Katouli M. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran J Microbiol. 2010;2:59–72. [PMC free article] [PubMed] [Google Scholar]
- 69.Kocijancic D, Felgner S, Frahm M, Komoll RM, Iljazovic A, Pawar V, Rohde M, Heise U, Zimmermann K, Gunzer F, et al. Therapy of solid tumors using probiotic Symbioflor-2 – restraints and potential. Oncotarget. 2016;7:22605–22622. doi: 10.18632/oncotarget.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prindle A, Selimkhanov J, Danino T, Samayoa P, Goldberg A, Bhatia SN, Hasty J. Genetic circuits in Salmonella typhimurium. ACS Synth Biol. 2012;1:458–464. doi: 10.1021/sb300060e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heap JT, Theys J, Ehsaan M, Kubiak AM, Dubois L, Paesmans K, Mellaert L, Knox R, Kuehne SA, Lambin P, et al. Spores of Clostridium engineered for clinical efficacy and safety cause regression and cure of tumors in vivo. Oncotarget. 2014;5:1761–1769. doi: 10.18632/oncotarget.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts NJ, Zhang L, Janku F, Collins A, Bai RY, Staedtke V, Rusk AW, Tung D, Miller M, Roix J, et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci Transl Med. 2014;6:249. doi: 10.1126/scitranslmed.3008982. ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bermúdez-Humarán LG, Aubry C, Motta JP, Deraison C, Steidler L, Vergnolle N, Chatel JM, Langella P. Engineering lactococci and lactobacilli for human health. Curr Opin Microbiol. 2013;16:278–283. doi: 10.1016/j.mib.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Yazawa K, Fujimori M, Nakamura T, Sasaki T, Amano J, Kano Y, Taniguchi Si. Bifidobacterium longum as a delivery system for gene therapy of chemically induced rat mammary tumors. Breast Cancer Res Treat. 2001;66:165–170. doi: 10.1023/a:1010644217648. [DOI] [PubMed] [Google Scholar]
- 75.Dong H, Zhang D. Current development in genetic engineering strategies of Bacillus species. Microb Cell Factories. 2014;13:63. doi: 10.1186/1475-2859-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 2014;11:4745–4767. doi: 10.3390/ijerph110504745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sciaranghella G, Lakhashe SK, Ayash-Rashkovsky M, Mirshahidi S, Siddappa NB, Novembre FJ, Velu V, Amara R, Zhou C, Li S, et al. A live attenuated Listeria monocytogenes vaccine vector expressing SIV Gag is safe and immunogenic in macaques and can be administered repeatedly. Vaccine. 2011;29:476–486. doi: 10.1016/j.vaccine.2010.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cory L, Chu C. ADXS-HPV: a therapeutic Listeria vaccination targeting cervical cancers expressing the HPV E7 antigen. Hum Vaccin Immunother. 2014;10:3190–3195. doi: 10.4161/hv.34378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salvadori G, Junges R, Morrison DA, Petersen FC. Overcoming the barrier of low efficiency during genetic transformation of Streptococcus mitis. Front Microbiol. 2016;7:1009. doi: 10.3389/fmicb.2016.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nyerges Á, Csörgő B, Nagy I, Bálint B, Bihari P, Lázár V, Apjok G, Umenhoffer K, Bogos B, Pósfai G, et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc Natl Acad Sci. 2016;113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu C, Huang J, Zhou R. Genomics of lactic acid bacteria: current status and potential applications. Crit Rev Microbiol. 2017:1–12. doi: 10.1080/1040841X.2016.1179623. [DOI] [PubMed] [Google Scholar]
- 83.Mugler A, Kittisopikul M, Hayden L, Liu J, Wiggins CH, Suel GM, Walczak AM. Noise expands the response range of the Bacillus subtilis competence circuit. PLoS Comput Biol. 2016;12:e1004793. doi: 10.1371/journal.pcbi.1004793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piñero-Lambea C, Ruano-Gallego D, Fernández L. Engineered bacteria as therapeutic agents. Curr Opin Biotechnol. 2015;35:94–102. doi: 10.1016/j.copbio.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 85.Dessel N, Swofford CA, Forbes NS. Potent and tumor specific: arming bacteria with therapeutic proteins. Ther Deliv. 2015;6:385–399. doi: 10.4155/tde.14.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lehouritis P, Stanton M, McCarthy FO, Jeavons M, Tangney M. Activation of multiple chemotherapeutic prodrugs by the natural enzymolome of tumour-localised probiotic bacteria. J Control Release. 2016;222:9–17. doi: 10.1016/j.jconrel.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 87.Zoaby N, Shainsky-Roitman J, Badarneh S, Abumanhal H, Leshansky A, Yaron S, Schroeder A. Autonomous bacterial nanoswimmers target cancer. J Control Release. 2016 doi: 10.1016/j.jconrel.2016.10.006. http://dx.doi.org/1016/j.jconrel.2016.10.006. [DOI] [PMC free article] [PubMed]
- 88.Nguyen VD, Han JW, Choi YJ, Cho S, Zheng S, Ko SY, Park JO, Park S. Active tumor-therapeutic liposomal bacteriobot combining a drug (paclitaxel)-encapsulated liposome with targeting bacteria (Salmonella typhimurium) Sensors Actuators B Chem. 2016;224:217–224. [Google Scholar]
- 89.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mosberg JA, Lajoie MJ, Church GM. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–799. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clairmont C, Lee KC, Pike J, Ittensohn M, Low KB, Pawelek J, Bermudes D, Brecher SM, Margitich D, Turnier J, et al. Bio-distribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 92.Dong WR, Xiang LX, Shao JZ. Novel antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD de novo synthesis pathway. Appl Environ Microbiol. 2010;76:2295–2303. doi: 10.1128/AEM.02462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kroll J, Klinter S, Schneider C, Voss I, Steinbuchel A. Plasmid addiction systems: perspectives and applications in biotechnology. Microb Biotechnol. 2010;3:634–657. doi: 10.1111/j.1751-7915.2010.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Griffiths TRL, Green WJF, Grice PT, Goddard JC, Kockelbergh RC. PD19-07 one-year follow-up results after sequential intravesical Bacillus calmette-guérin and device-assisted chemo-hyperthermia (mitomycin C delivered by the combat BRS system) for high risk non-muscle invasive bladder cancer patients…A Bacillus calmette-guérin-sparing strategy. J Urology. 2017;197:e367. [Google Scholar]
- 95.Toso JF, Gill VJ, Hwu P, Marincola FM, Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC, Stock F, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with meta-static melanoma. J Clin Oncol. 2002;20:142–152. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le DT, Brockstedt DG, Nir-Paz R, Hampl J, Mathur S, Nemunaitis J, Sterman DH, Hassan R, Lutz E, Moyer B, et al. A live-attenuated Listeria vaccine (ANZ-100) and a live-attenuated Listeria vaccine expressing mesothelin (CRS-207) for advanced cancers: phase I studies of safety and immune induction. Clin Cancer Res. 2012;18:858–868. doi: 10.1158/1078-0432.CCR-11-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vaze MB, Wu T, Seth S, Templin M, Polisky B. Engineering of trans kingdom RNAi (tkRNAi) against gastrointestinal polyps. Cancer Res. 2012;72 [Google Scholar]