Abstract
The interplay with bacteria is of crucial importance for the interaction of multicellular organisms with their environments. Studying the associations between the nematode model organisms Caenorhabditis elegans and Pristionchus pacificus with bacteria constitutes a powerful system to investigate these interactions at a mechanistic level. P. pacificus is found in association with scarab beetles in nature and recent studies revealed the succession and dynamics of this nematode and its microbiome during the decomposition of one particular host species, the rhinoceros beetle Oryctes borbonicus on La Réunion Island. However, these studies were performed using culture-free methods, with no attempt made to establish bacterial cultures from the beetle-nematode ecosystem and to investigate the effects of these microbes on life history traits in P. pacificus. Here, we establish and characterize a collection of 136 bacterial strains that have been isolated from scarab beetles and figs, another Pristionchus-associated environment, as a resource for studying their effect on various nematode traits. Classification based on 16S sequencing identified members of four bacterial phyla with the class of Gammaproteobacteria representing the majority with 81 strains. Assessing the survival of P. pacificus on individual bacteria allowed us to propose candidate groups of pathogens such as Bacillaceae, Actinobacteria, and Serratia. In combination with chemoattraction data, it was revealed that P. pacificus is able to recognize and avoid certain groups of pathogens, but not others. Our collection of bacterial strains forms a natural resource to study the effects of bacterial diet on development and other traits. Furthermore, these results will form the basis of future studies to elucidate the molecular mechanisms of recognition and pathogenicity.
Introduction
Bacteria form an integral part of the ecology of all living beings and the influence of the gut microbiota on human health has been increasingly recognized during the last decade [1]. Nematodes like C. elegans are an excellent model to study the interactions between bacteria and their hosts [2], because they are easy to grow using monoxenic bacterial cultures, eg. Escherichia coli OP50 as food source. In addition, worms as well as bacteria are genetically tractable, which can provide detailed mechanistic insights into the interaction between host and bacteria and their impact on development and behavior [3,4]. We study the nematode Pristionchus pacificus a close relative of the rhabditid C. elegans, but belonging to the Family Diplogastridae [5]. P. pacificus and C. elegans have been estimated to have diverged 280–430 million years ago [6]. P. pacificus is found in a necromenic association with scarab beetles [7], i.e. nematodes are maintained as growth-arrested dauer larvae on the beetle and upon the beetle’s death resume development and reproduce. They feed on the microorganisms growing on the beetle’s carcass and recent decomposition studies using the rhinoceros beetle Oryctes borbonicus from La Réunion Island as host have indicated that the decaying beetles and P. pacificus have largely overlapping microbiomes [7]. While P. pacificus and C. elegans share many biological features, such as the mode of reproduction, the presence of an alternative developmentally arrested dauer stage, and the same chromosome number, nematodes of the P. pacificus lineage have gained the ability to form tooth-like structures that allow them to predate on other nematodes [8–11]. Interestingly, these feeding structures represent an example of phenotypic plasticity because P. pacificus can form two alternative mouth forms with stenostomatous animals being strict bacterial feeders, whereas eurystomatous animals are omnivorous feeders that can also kill other nematodes [11]. Whether or not these predatory structures are formed during development is environmentally controlled. Thus, Pristionchus mouth-form plasticity represents a developmental decision similar to other examples of phenotypic plasticity in animals, such as the caste system in social insects [12] or color patterns in butterfly wings [13]. To explore the full range of environmental variables that potentially influence developmental decisions, we have recently started to modify culture conditions [14] and tested food sources other than E. coli OP50 bacteria [15]. Specifically, these studies have shown that growth of worms on yeasts or in liquid culture conditions has an effect on mouth-form plasticity [14,15]. The association of Pristionchus nematodes with scarab beetles is stable over millions of years of evolution and has resulted in more than 30 Pristionchus species that are found worldwide, often in species-specific interactions with scarab beetles [16]. In addition, a recent study discovered a second branch of the Pristionchus genus that is found in association with figs and fig wasps [17]. Strikingly, fig-associated Pristionchus species are even capable of producing up to five different mouth morphotypes, whereas beetle associated Pristionchus are usually dimorphic [17]. Evidence supported that distinct morphotypes were associated with the degree of maturity of figs and it was hypothesized that the presence of certain bacteria may trigger these developmental decisions. Unfortunately, we failed to cultivate fig-associated Pristionchus nematodes permanently under laboratory conditions, which prohibited the further elucidation of environmental cues controlling the development of individual morphs.
The interactions between Pristionchus nematodes and bacteria have been studied in the last decade largely by exploring beetle-derived bacteria from Germany and other European sampling sites [18]. These original studies had indicated differences in the response of P. pacificus and C. elegans to Bacillus ssp., which have initiated large-scale studies of Bacilli and their effect on both nematodes [19–22]. However, P. pacificus does not normally occur in Europe and the original bacterial isolates mentioned above were obtained from P. maupasi and P. entomophagus.
To further study the interactions between P. pacificus with its natural bacteria, we have carried out a culture-based approach to isolate and investigate nematode associated bacteria from three different locations in Asia, Africa and the Indian Ocean and used both hosts, beetles and figs. Specifically, we isolated bacteria from figs retrieved from Vietnam, South Africa, and La Réunion Island and from scarab beetles from La Réunion Island, which forms a hotspot of P. pacificus diversity [7,23]. In total, we classified 136 bacterial isolates based on their 16S ribosomal RNA sequences. We test the nematodes´ capability to survive on these bacteria and their chemoattractive potential relative to standard E. coli OP50 cultures. Our data show that most of the isolated bacteria support growth of P. pacificus worms and most bacteria seem to be more attractive than the standard E. coli OP50 strain. Furthermore, the finding of a weak correlation between survival and chemotaxis data raises the question to what extent nematodes can sense which food sources are suitable for them.
Materials and methods
Nematode and bacterial culture conditions
The wild type strain of P. pacificus (PS312) was grown at 20°C on nematode growth medium (NGM) seeded with E. coli OP50 before use in experiments. Bacterial strains were cultured on following growth media: LB agar (1% tryptone, 0.5% yeast extract, 1% NaCl and 1.5% agar), NGM [24], YPD agar (2% bacteriological peptone, 1% Yeast extract, 2% Glucose and 1.5% Agar), NA (Thermo scientific, Oxoid, CM0003), TSA (1.5% tryptone, 0.5% soytone –enzymatic digest of soybean meal, 0.5% sodium chloride, 1.5% agar), PDA (Difco™ Potato Dextrose Media, BD, 1.5% Agar).
Sample collection and isolation of wild bacteria
We collected different beetle species (Oryctes borbonicus, Adoretus sp., Hoplia sp. and Amneidus godefroyi) from La Réunion Island (Fig 1A) using sweeping nets, black light traps and pitfall traps baited with dung [25]. Only adult beetles were collected before being transferred to the laboratory alive. To avoid contamination by human associated bacteria, all sample collections were done wearing gloves. Under sterile conditions animals were sacrificed by cutting them in half transversely and all body parts were placed on LB agar plates. Bacteria were only isolated from beetles that also showed the presence of Pristionchus nematodes. Plates that were negative for Pristionchus but that were positive for other nematodes were discarded. Isolated bacteria were spotted on LB plates and colonies were singled out for two rounds to get pure bacterial strains. For genotyping, bacterial strains were sub-cultured and then prepared for sequencing using PCR amplification of 16S ribosomal RNA genes. Permits for beetle samplings on La Réunion Island were provided by Sylvain Leonard from the Office National des Forets and Benoit Lequette from the Parc National de La Réunion between 2012 and 2017. Note that the research permits did not allow the disclosure of the exact sampling localities because several beetle species are endangered (i.e. Oryctes borbonicus).
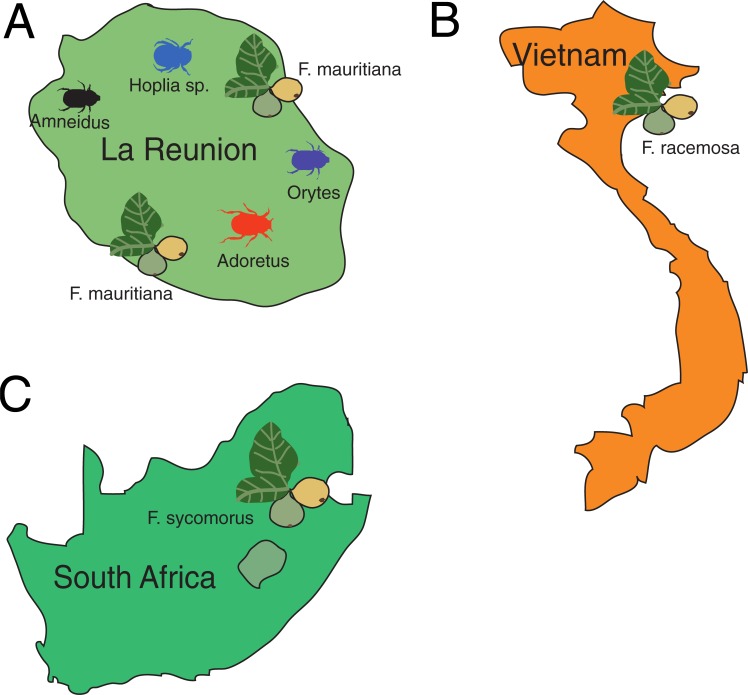
Fig 1. Regional maps of beetle and fig sampling sites.
(A) Map of La Réunion Island showing the approximate beetle and fig sampling sites. O. borbonicus, Adoretus sp., Hoplia sp. and A. godefroyi beetles were collected to isolate Pristionchus-associated bacteria. Similarly, F. mauritiana figs were sampled and processed for bacteria isolation. (B) and (C) F. sycomorus (Brummeria, Pretoria, South Africa) and F. racemosa (Hanoi, Vietnam) figs were dissected under sterile conditions from Pristionchus-positive specimens to isolate bacteria.
To isolate bacteria from figs, we collected several fig species including F. mauritiana (La Réunion), F. sycomorus (South Africa), and F. racemosa (Vietnam) (Fig 1B and 1C). Individual figs were dissected under sterile conditions and the presence of Pristionchus nematodes was confirmed. 500 μl of fig juice was extracted with a sterile pipette and suspended in sterile PBS, and aliquots were spread on LB, NGM, YPD, NA, TSA, and PDA agar plates and then grown for 1–2 d at 30°C. Single colonies were isolated from plates, grown in LB (shaking at 180 rpm, 30°C) or until significant growth was achieved, and frozen at −80°C in 25% glycerol stocks. Permits for fig samplings on La Réunion Island were provided by Benoit Lequette from the Parc National de La Réunion between 2014 and 2016.
Bacteria identification
Each bacterial colony was grown overnight in LB broth and DNA was extracted using Epicenter MasterPure DNA purification kit (Illumina, San Diego, USA). Polymerase chain reaction (PCR) amplification of bacterial 16S rRNA genes was carried out in 25 μl reactions using a universal primer set SSU 27f (5’-AGAGTTTGATCMTGGCTCAG-3’) and SSU 1492r (5’-TACGGYTACCTTGTTACGACTT-3’) [18]. Thermal cycling conditions were as follows: 3 min at 95°C followed by 30 cycles of 15 s at 95°C, 30 s at 55°C, 1.5 min at 72°C, and a final step of 8 min at 72°C. A typical reaction contained 2 μl 10x PCR buffer, 2 μl 2·mmol·l–1 dNTPs, 1μl 10 μmol·l–1 27f, 1μL10 μmol·l–1 1492r, one unit of Taq DNA polymerase, 12.8·μl H2O and 1μL of bacterial DNA. PCR amplicons were visualized by standard agarose gel electrophoresis [26]. All high quality 16S rRNA gene sequences of bacteria were classified by the SILVAngs webservice of the SILVA database [27].
Survival assays
Bacterial liquid cultures were established by inoculating 5ml LB with a single bacterial colony. Subsequently, cultures were grown overnight at 30°C. Bacterial suspensions (50μl) were spread with an L-shaped spreader on NGM medium petri dishes with diameter of 6cm and were incubated overnight. Twenty young adult P. pacificus worms that were well fed on E. coli OP50, were washed five times with PBS and picked to intermediate plates seeded with test bacteria to reduce contamination, a standard procedure in nematode survival assays. One hour later, worms were picked to the final assay plates seeded with test bacteria. Each plate was kept at 20°C. Survival of worms was monitored daily for 5 days. Nematodes were transferred every two days to fresh plates to prevent misidentification of original worms from offspring. Mortality was determined by prodding worms with a metal pick and nematodes that did not respond were considered dead. In total, we performed three biological replicates per bacterial strain.
Chemotaxis assays
Chemotaxis assays were modified from previous studies [28,29]. Briefly, 20 μl of overnight bacterial suspension was placed 0.5 cm away from the edge of a 6 cm Petri dish filled with NGM medium. The same amount of E. coli OP50 was placed on the opposing side acting as the counter attractant. Approximately 50–200 J4/adult stage P. pacificus individuals were placed at the edge of the plate, equidistant to each of the bacterial spots. All nematodes were previously fed on E. coli OP50. Plates were incubated at room temperature. After 3h the number of nematodes found in each bacterial spot was recorded. A chemotaxis index was used to score the response of the nematodes, which consisted of: number of nematodes in the region spotted with test bacteria minus the number of nematodes in control bacteria spots, the result was divided by the total number of nematodes counted [29]. This gave a chemotaxis score ranging from –1.0 (repulsion) to 1.0 (attraction). Three plates were used per replicate, and the procedure was repeated four to six times for each bacterium.
Statistical analysis
For each bacteria analysed, we averaged the survival and chemotaxis values from all replicates and employed a Wilcoxon rank-sum test to test for significant differences between taxonomic groups. P-values were corrected for multiple testing with the Bonferroni method. Correlation between survival and chemotaxis data was calculated as Spearman correlation. All analyses and plots were done using the statistical program R.
Results
A bacterial strain collection from Pristionchus-associated environments
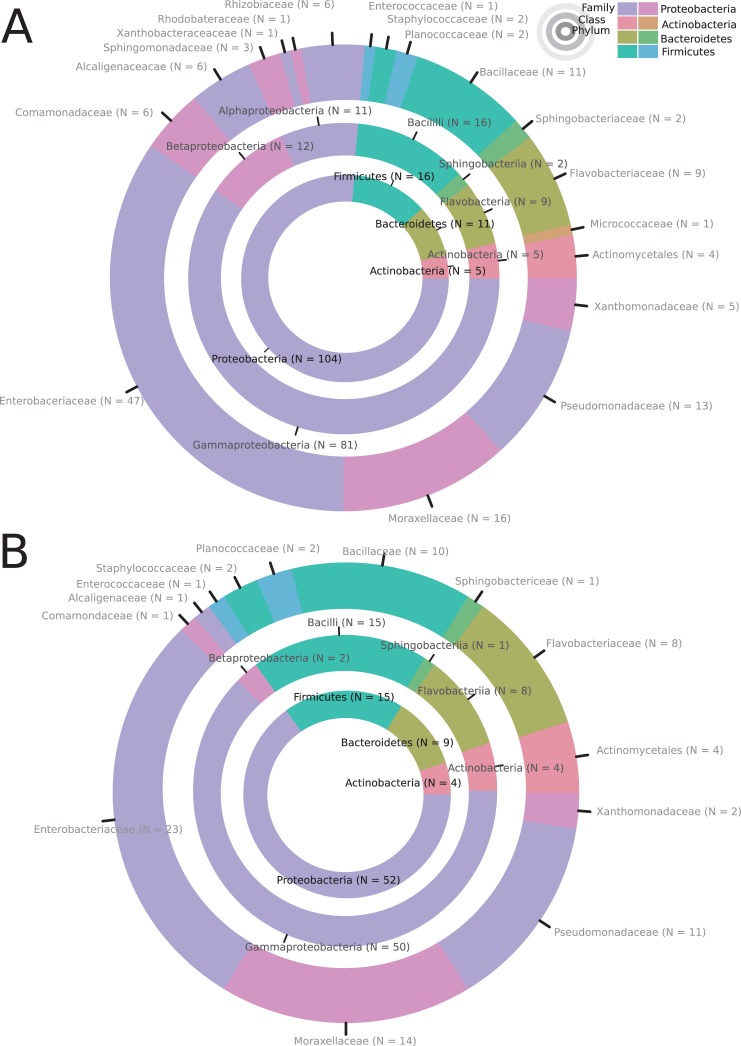
We used fig and beetle samples provided by our collaborators, to isolate and cultivate a total of 136 bacterial strains (S1 Table). Specifically, 80 different bacterial strains were isolated from beetle species (O. borbonicus, Adoretus sp., Hoplia sp. and A. godefroyi collected from La Réunion Island) (Fig 1A). In addition, we isolated 26 bacterial strains from F. mauritiana (La Réunion), 21 strains from F. racemosa (Vietnam) and nine strains from F. sycomorus (South Africa)(Fig 1A). Each strain was classified after sequencing a fragment of the 16S ribosomal RNA gene. Overall, we isolated strains belonging to four bacterial phyla, Proteobacteria, Firmicutes, Bacteriodetes, and Actinobacteria (Fig 2A). The same four phyla were also present when only considering bacterial isolates from beetles (Fig 2B). These findings are consistent with the recent high throughput sequencing of the microbiome of O. borbonicus and P. pacificus [7]. Out of the 104 isolated strains of Proteobacteria, 47 are representatives of the family Enterobaceriaceae, which has also been found to be by far the most abundant family of bacteria in decaying beetles [7]. Thus, our culture-based method is largely consistent with the culture-free results of O. borbonicus-associated microbes and provides a collection of 136 bacterial strains for laboratory studies.
Fig 2. Taxonomic distribution of bacterial isolates.
(A) Circles show the distribution of bacterial strains at the level of phyla (innermost circle), class (middle circle), and family (outermost circle) based on classification of the 16S ribosomal RNA gene [27]. Proteobacteria are by far the largest group (N = 104 strains). (B) Distribution of bacterial strains that were isolated from beetles on La Réunion Island.
Most bacterial isolates are not pathogenic to P. pacificus
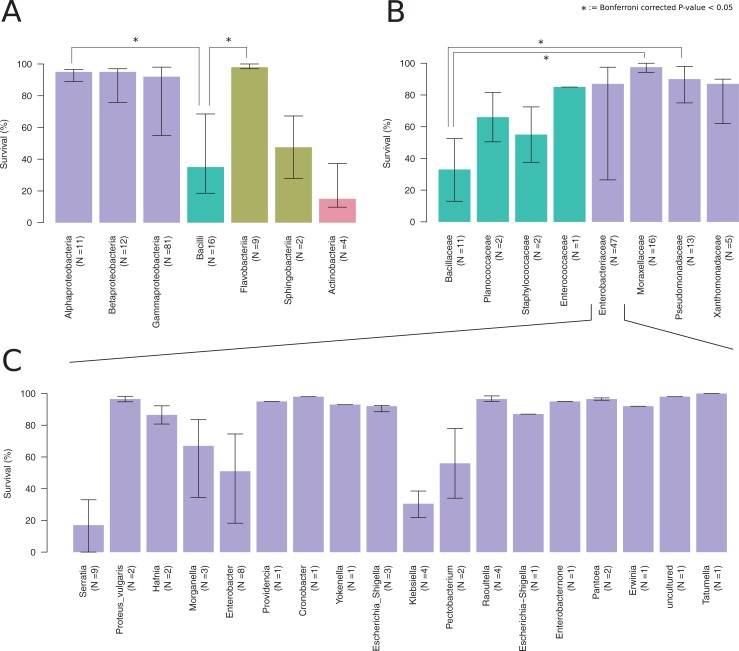
Next, we tested how well P. pacificus strains can survive on the isolated bacteria. To this end, we exposed 20 young adults to individual bacterial strains and counted the number of surviving worms after five days. Control experiments performed on E. coli OP50 showed a survival rate of between 95–100%. In our experimental setup, worms could survive on most of the tested bacteria (Fig 3). Fig 3 shows the result of the survival tests at the level of bacterial classes and selected families. Survival on Bacilli strains was significantly lower (Wilcoxon Rank-Sum test, Bonferroni corrected p-value < 0.05) than on Flavobacteria and Alphaproteobacteria (Fig 3A). In contrast, sample sizes of Actinobacteria and Sphingobacteria were too low to reveal statistically significant differences in nematode survival in comparison to other bacterial classes. However, investigating the survival patterns at higher taxonomic resolution, we found that the lower survival on Bacilli strains is largely due to members of the family Bacillaceae (Fig 3B) and the high variability in Gammaproteobacteria can be attributed to variability in the family Enterobacteriaceae. At the level of genera, the variability within Enterobacteriaceae appears to be caused by lower survival on individual strains belonging to Serratia, Morganella, Enterobacter, Klebsiella and Pectobacterium (results were not statistically significant after multiple testing correction, Fig 3C). While some Serratia strains have previously been described to be pathogenic to P. pacificus [30], it also seems that some Enterobacter and the human pathogen Klebsiella strains can also be pathogenic to P. pacificus.
Fig 3. Survival of P. pacificus nematodes on different bacteria.
(A) Bars show median and interquartile ranges of survival of P. pacificus worms in response to various bacterial classes. All pairwise tests for significantly different survival were done by a Wilcoxon rank-sum test. Results that remained significant after Bonferroni correction are highlighted. (B) Distribution of survival rate for deeply sampled and highly variable bacterial classes (Bacilli and Gammaproteobacteria) at higher phylogenetic resolution (family level). Within the class Bacilli, decreased survival is mostly due to strains of the family Bacillaceae. Within the class Gammaproteobacteria, the largest variability in nematode survival is observed in the family Enterobacteriaceae. (C) Distribution of survival within Enterobacteriaceae at genus level.
P. pacificus nematodes are attracted to most bacterial isolates
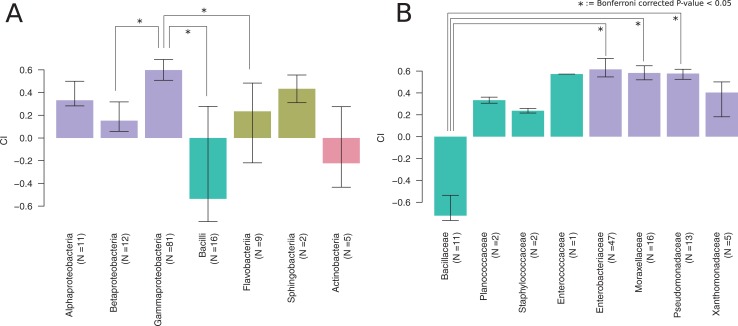
To test whether P. pacificus nematodes are attracted towards the isolated bacteria, we performed chemotaxis assays by giving worms the choice between two alternate food sources. Specifically, we used one spot of the target bacteria opposite one spot of an equal volume E. coli OP50 and counted the number of worms in each of the spots after three hours. Subsequently, we then calculated a chemotaxis index (CI). A CI of -1 indicates repulsion from the test bacterium, whereas a CI of 1 indicates attraction. Our results show that most bacterial isolates are preferred by P. pacificus as opposed to control spots. However, individual strains of the classes Actinobacteria, Flavobacteria, and Bacilli showed negative CIs (Fig 4A). Within Bacilli, the repulsive effect was mostly due to the family of Bacillaceae (Fig 4B), whereas other families of Bacilli frequently showed positive CIs indicating strong strain specificity.
Fig 4. Chemoattraction towards different bacteria.
(A) While most bacterial strains seem to be preferred over control spots, the class Bacilli shows a significantly repulsive effect (Wilcoxon test, Bonferroni corrected p-value<0.05) in comparison to three other bacterial classes. (B) Within the class Bacilli, the repulsive effect is largely due to strains of the family Bacillaceae.
Weak correlation between survival and chemotaxis
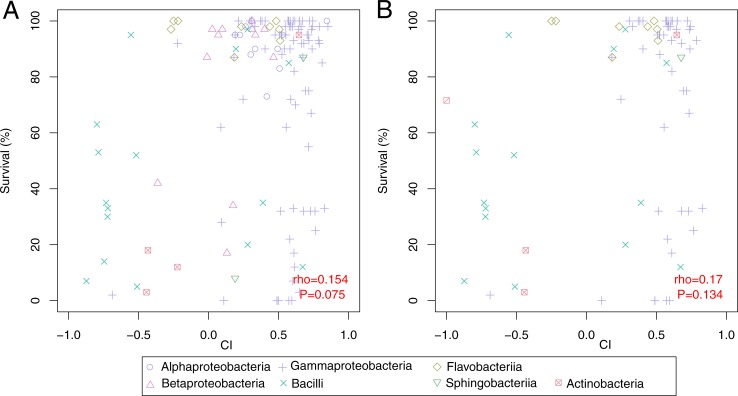
Combining the results of the survival and chemotaxis assays it appears as if P. pacificus can recognize and escape from certain pathogens. For example, certain strains of Bacillus and Actinobacteria show low survival of P. pacificus and strong repulsion in chemoattraction assays (Fig 5). In addition, most non-pathogenic strains seem to be preferred by worms over E. coli OP50 control spots. To test to what extent P. pacificus can distinguish suitable food sources from pathogens, we calculated the correlation coefficients between chemotaxis and survival data (Fig 5A), which revealed only a weak trend for the whole data set (Spearman’s rho = 0.154, P = 0.075). Even restricting the analysis to bacterial strains that were isolated from beetles and are therefore more likely to be seen by P. pacificus worms in the wild did not result in a higher correlation (Fig 5B). In particular, P. pacificus is attracted to multiple strains of the genus Serratia, which are known pathogens of this nematode species (Figs 3C and 4). Thus, P. pacificus can recognize and avoid certain but not all pathogens.
Fig 5. Correlation between survival and chemotaxis.
(A) Testing for the correlation between survival and chemotaxis data we found a weak trend (Spearman’s rho = 0.154, P = 0.075) for bacteria resulting in higher survival rates to also have higher chemotaxis indices, compared to strains that do not support growth or are pathogenic. (B) Similar correlation tests for those bacterial strains isolated from beetles where again, no strong signal was observed (Spearman’s rho = 0.17, P = 0.134).
Discussion
In this study we have isolated and characterized 136 bacterial strains from Pristionchus-associated environments, scarab beetles and figs. As ecologically relevant results of microbial-animal interactions are most likely to be obtained when microbes from the same environment are used in which the test organism lives, we only isolated bacteria from samples that showed the presence of Pristionchus nematodes. Despite the fact that our culture conditions most likely only allow isolation of a small percentage of the total bacterial community, the cultivable strains will form a powerful resource to study how Pristionchus nematodes interact with their environment and in particular, how bacterial diet can influence developmental decisions, such as the mouth form dimorphism [9]. Previous work on Cryptococcus yeast has demonstrated shifts in mouth form ratios of P. pacificus nematodes upon altered diet [15]. Thus, it is highly likely that some of the isolated bacteria induce similar effects.
We have screened P. pacificus survival on all isolated bacterial strains and found that multiple strains of diverse taxonomic groups are candidates for nematode pathogens. Among these, the genus Serratia has been previously described as potent killer of P. pacificus and C. elegans [30] and our survival assays showed that one of the Stenotrophomonas sp. isolates can be a potential pathogen to P. pacificus. Note that survival of nematodes on individual bacteria does not necessarily indicate the ability of the nematode to grow and reproduce on these strains. However, during the course of experiments, we kept nematodes on various bacterial strains for several generations indicating that P. pacificus can complete its life cycle on many of the isolated bacteria. It is important to note that in nature P. pacificus is exposed to a mixture of bacteria and therefore, survival assays performed with monoxenic cultures of test bacteria are partially artificial. Future studies will aim to study combinations of bacteria simultaneously, thereby mimicking more closely the situation seen in nature.
Complementary experiments of chemotaxis showed that bacterial classes like Bacilli and Actinobacteria that caused reduced survival are avoided by P. pacificus, which preferred feeding on E. coli OP50 when having the choice. However, explicitly testing for a correlation between survival and chemotaxis data did not allow the conclusion that P. pacificus can broadly recognize and avoid pathogenic bacteria. Overall, our chemotaxis experiments may suggest that most isolates are preferred over E. coli OP50 control spots. This finding came as a surprise given that the strain PS312 is in permanent culture since 1988 and has been exclusively fed on E. coli OP50 [5], but apparently has not developed a preference for it. Nevertheless this finding is consistent with observations from C. elegans showing that other bacteria such as Comamonas are much better food sources than E. coli OP50 [5,31]. However, the statement that P. pacificus prefers many bacteria over E. coli OP50 is to be regarded with care because the assay conditions utilized can not control for the exact bacterial concentration. In many of the assays, we observed that thicker colonies are not necessarily preferred by nematodes, suggesting that even if differences in bacterial concentrations exist, they seem to have a minor effect on the results of the chemotaxis assays. This might be due to the strong P. pacificus perception of oxygen [32]. Taken together, the survival and chemotaxis data showed substantial phylogenetic signal indicating that related bacteria give rise to a similar response in terms of nematode survival and chemoattraction. This may suggest that the overall biochemical composition of bacteria causes the observed effect on P. pacificus nematodes. Interestingly both potential groups of pathogens (Bacilli and Actinobacteria) that can be recognized and avoided by P. pacificus are Gram-positive and spore forming bacteria suggesting that one or multiple features associated or correlated with the Gram-positive life style and/or spore formation are responsible for the response in nematodes.
In summary, the collection of bacterial strains that has been described in this study constitutes a resource for future studies of interactions between nematodes and bacteria. Our findings raise a number of interesting questions for future investigations, e.g. which bacterial factors are recognized by worms and how do they sense them? Given the substantial variability in survival, how are these patterns reflected in terms of development and other life history traits? Which of the isolated bacteria is the best food source for P. pacificus? Given that these associations can be studied in very controlled conditions and nematodes and bacteria are genetically tractable, combined investigation of nematodes and bacteria forms a powerful experimental system to study the effect of microbiota on organisms at a mechanistic level.
Supporting information
(XLSX)
Acknowledgments
We thank Drs. Vladislav Susoy, Matthias Herrmann, Eduardo Moreno for providing beetle and fig material.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) Graduiertenkolleg "Molecular principles of bacterial survival strategies" (GRK 1708) to R.J.S.
References
- 1.Althani AA, Marei HE, Hamdi WS, Nasrallah GK, El Zowalaty ME, Al Khodor S, et al. Human microbiome and its association with health and diseases. J Cell Physiol. 2016. August; 231(8):1688–94. doi: 10.1002/jcp.25284 [DOI] [PubMed] [Google Scholar]
- 2.Yilmaz LS, Walhout AJM. Worms, bacteria, and micronutrients: an elegant model of our diet. Trends Genet. 2014. November 26;30(11):496–503. doi: 10.1016/j.tig.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, et al. Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell. 2014. February 13;156(4):759–70. doi: 10.1016/j.cell.2014.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacNeil L, Watson E, Arda HE, Zhu LJ, Walhout AJM. Diet-induced developmental acceleration independent of TOR and Insulin in C. elegans. Cell. 2013. March 28;153(1): doi: 10.1016/j.cell.2013.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer RJ, Carta LK, Kim S, Sternberg PW. Morphological, genetic and molecular description of Pristionchus pacificus sp. n.(Nematoda: Neodiplogasteridae) Fundam. Appl. Nematol., 1996, 19(6),511–521. [Google Scholar]
- 6.Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I,et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008. October;40(10):1193–8. doi: 10.1038/ng.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer JM, Baskaran P, Quast C, Susoy V, Rödelsperger C, Glöckner FO, et al. Succession and dynamics of Pristionchus nematodes and their microbiome during decomposition of Oryctes borbonicus on La Réunion Island. Environ Microbiol. 2017;19(4):1476–89. doi: 10.1111/1462-2920.13697 [DOI] [PubMed] [Google Scholar]
- 8.Bento G, Ogawa A, Sommer RJ. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature. 2010. July 22;466(7305):494–7. doi: 10.1038/nature09164 [DOI] [PubMed] [Google Scholar]
- 9.Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell. 2013. November 7;155(4):922–33. doi: 10.1016/j.cell.2013.09.054 [DOI] [PubMed] [Google Scholar]
- 10.Rödelsperger C, Meyer JM, Prabh N, Lanz C, Bemm F, Sommer RJ. Single-molecule sequencing reveals the chromosome-scale genomic architecture of the nematode model organism Pristionchus pacificus. Cell Rep. 2017. October 17;21(3):834–844. doi: 10.1016/j.celrep.2017.09.077 [DOI] [PubMed] [Google Scholar]
- 11.Wilecki M, Lightfoot JW, Susoy V, Sommer RJ. Predatory feeding behaviour in Pristionchus nematodes is dependent on phenotypic plasticity and induced by serotonin. J Exp Biol. 2015. May;218:1306–13. doi: 10.1242/jeb.118620 [DOI] [PubMed] [Google Scholar]
- 12.Weitekamp CA, Libbrecht R, Keller L. Genetics and evolution of social behavior in insects. Annu Rev Genet Annual Reviews; 2017. November 27;51(1):219–39. [DOI] [PubMed] [Google Scholar]
- 13.Beldade P, Mateus ARA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol. 2011;20(7):1347–63. doi: 10.1111/j.1365-294X.2011.05016.x [DOI] [PubMed] [Google Scholar]
- 14.Werner MS, Sieriebriennikov B, Loschko T, Namdeo S, Lenuzzi M, Dardiry M, et al. Environmental influence on Pristionchus pacificus mouth form through different culture methods. Sci Rep. 2017. August 3;7:7207 doi: 10.1038/s41598-017-07455-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanghvi G V, Baskaran P, Röseler W, Sieriebriennikov B, Rödelsperger C, Sommer RJ. Life history responses and gene expression profiles of the nematode Pristionchus pacificus cultured on Cryptococcus yeasts. PLoS One. 2016. October 14;11(10):e0164881 doi: 10.1371/journal.pone.0164881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragsdale E J, Kanzaki N, Herrmann M. Taxonomy and natural history: the genus Pristionchus. In: Pristionchus pacificus A nematode model for comparative and evolutionary biology. (Ed.: Sommer R J) Brill, Leiden. [Google Scholar]
- 17.Susoy V, Herrmann M, Kanzaki N, Kruger M, Nguyen CN, Rödelsperger C, et al. Large-scale diversification without genetic isolation in nematode symbionts of figs. Sci Adv. 2016. January 15;2(1):e1501031 doi: 10.1126/sciadv.1501031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rae R, Riebesell M, Dinkelacker I, Wang Q, Herrmann M, Weller AM, et al. Isolation of naturally associated bacteria of necromenic Pristionchus nematodes and fitness consequences. J Exp Biol. 2008;211(12):1927–36. [DOI] [PubMed] [Google Scholar]
- 19.Iatsenko I, Boichenko I, Sommer RJ. Bacillus thuringiensis DB27 produces two novel protoxins, Cry21Fa1 and Cry21Ha1, which act synergistically against nematodes. Appl Environ Microbiol. 2014. May;80(10):3266–75. doi: 10.1128/AEM.00464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iatsenko I, Sinha A, Rödelsperger C, Sommer RJ. New role for DCR-1/dicer in Caenorhabditis elegans innate immunity against the highly virulent bacterium Bacillus thuringiensis DB27. Infect Immun. 2013. October;81(10):3942–57. doi: 10.1128/IAI.00700-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lightfoot JW, Chauhan VM, Aylott JW, Rödelsperger C. Comparative transcriptomics of the nematode gut identifies global shifts in feeding mode and pathogen susceptibility. BMC Res Notes. 2016. March 5;9:142 doi: 10.1186/s13104-016-1886-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rae R, Witte H, Rödelsperger C, Sommer RJ. The importance of being regular: Caenorhabditis elegans and Pristionchus pacificus defecation mutants are hypersusceptible to bacterial pathogens.Int J Parasitol. 2012. July;42(8):747–53. doi: 10.1016/j.ijpara.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 23.McGaughran A, Rödelsperger C, Grimm DG, Meyer JM, Moreno E, Morgan K, et al. Genomic profiles of diversification and genotype-phenotype association in island nematode lineages. Mol Biol Evol. 2016;33(9):2257–72. doi: 10.1093/molbev/msw093 [DOI] [PubMed] [Google Scholar]
- 24.Stiernagle T. Maintenance of C. elegans.WormBook. 2006. February 11:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann M, Mayer WE, Hong RL, Kienle S, Minasaki R, Sommer RJ. The nematode Pristionchus pacificus (Nematoda: Diplogastridae) is associated with the oriental beetle Exomala orientalis (Coleoptera: Scarabaeidae) in Japan. Zoolog Sci. 2007;24(9):883–9. doi: 10.2108/zsj.24.883 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch E. F. and Maniatis T. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Press; 1989. [Google Scholar]
- 27.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013. January 27;41(Database issue):D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438(7065):179–84. doi: 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
- 29.Hong RL, Sommer RJ. Chemoattraction in Pristionchus nematodes and implications for insect recognition. Curr Biol. 2006;16(23):2359–65. doi: 10.1016/j.cub.2006.10.031 [DOI] [PubMed] [Google Scholar]
- 30.Sinha A, Rae R, Iatsenko I, Sommer RJ. System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus. PLoS One. 2012;7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol. 2006. January; 209(Pt 1)89–102. doi: 10.1242/jeb.01955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno E, McGaughran A, Rödelsperger C, Zimmer M, Sommer RJ. Oxygen-induced social behaviours in Pristionchus pacificus have a distinct evolutionary history and genetic regulation from Caenorhabditis elegans. Proc Biol Sci. 2016. February 24;283(1825):20152263 doi: 10.1098/rspb.2015.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.