Abstract
Commercial assays measuring HPV E6 viral oncoproteins, E6/E7 mRNA or DNA were used to test neck lymph node fine needle aspirates (FNA) and oropharyngeal samples (saliva and oral swabs) from 59 Canadian patients with oropharyngeal squamous cell carcinomas (OPSCC). Overall agreements of p16 antigen staining of tumors to FNA tested for OncoE6™, Aptima HPV E6/E7 mRNA and cobas HPV DNA were 81.4% (k 0.53), 94.9% (k 0.83) and 91.1% (k 0.73) respectively. Using HPV presence in a subset of 25 tumors as the comparator, overall agreement was 64.0% (k 0.08) with OncoE6™, 88.0% (k 0.65) with Aptima HPV E6/E7 mRNA and 91.7% (k 0.70) with cobas HPV DNA. HPV testing of oropharyngeal samples yielded lower agreements with tumor markers; 23.7–24.0% (k 0.02), 55.9–68.0% (k 0.24–0.37) and 78.9–86.9% (k 0.49–0.58) in the 3 respective tests. HPV 16 was present in 93.7–100% of the samples tested and showed 100% genotype agreement between FNA and tumors. The high rates for HPV E6 oncoproteins and E6/E7 mRNA suggests most patients were experiencing transcriptionally active HPV-related OPSCC. Results from these commercial assays performed on FNA but not oropharyngeal samples showed moderate to very good agreements with p16 and HPV testing of tumors.
1. Introduction
Head and neck squamous cell carcinomas (HNSCC) include malignancies in the oral cavity, oropharynx, nasopharynx, hypopharynx and larynx, among other sites [1]. Oropharyngeal squamous cell carcinomas (OPSCC) which arise primarily in palatine tonsils and at the base of the tongue are often related to infections by high-risk HPV (HR HPV), predominantly HPV16 [2]. HPV-related OPSCC is considered to be driven by transcriptionally active virus, with the production of HPV E6 and E7 oncoproteins; which are a prerequisite for viral driven oncogenesis to occur. E6 and E7 have been shown to interact with numerous cellular pathways including cell cycle regulation and apoptosis [3]. Immunohistochemistry to detect overexpression of the p16INK4a protein (IHC p16) has been used extensively as a surrogate tumor tissue marker for HPV-caused OPSCC [4]. Both p16 antigen and HPV DNA may be present in tumors with both transcriptionally active and transient HPV infections [5].
The presence of HPV E6/E7 mRNA and/or HPV E6 and E7 oncoproteins in tumor tissue have been used as a measurement of transcriptionally active HPV [6], [7]. HPV E6/E7 mRNA has been measured in frozen [8], [9] or formalin fixed paraffin embedded (FFPE) tumor tissue [10], [11] using PCR techniques. Elevated expression of both E6 and E7 oncoproteins have been measured in cervical cancer using a colorimetric immunographic assay [12]. Other measurements such as viral load, p16 upregulation, pRb and/or p53 degradation and serum antibodies to the E6/E7 proteins have been used to determine whether an HPV infection is transcriptionally active [6], [9]. The measurement of the presence of E6/E7 oncoproteins may represent the most direct approach to differentiate between transcriptionally active and transient HPV infections.
Commercial assays are available for the detection of HPV DNA (Hybrid Capture 2, cobas 4800, Cervista HPV) and HPV E6/E7 mRNA (Aptima HPV) and the identification of certain high-risk HPV genotypes and have been used extensively to detect HR HPV in cervical cancer. Because HPV-related OPSCC often metastasizes to cervical lymph nodes of the neck which are assessed for malignancy through the use of fine needle aspiration (FNA), these HPV tests have been evaluated on FNA [13], [14], [15], [16], [17], [18], [19]. Commercial cytological tests are available to measure changes in cellular protein expression (p16) and the presence HPV in tumor tissues by ISH [20], [21], [22]. Another approach has involved the detection of HPV in oral fluid and swabs [23], [24], [25], [26], [27]. The accuracy of testing non-primary tumor samples such as FNA and oral collections requires validation against the presence of p16 and/ or HPV in tumor tissue.
Because patients with HPV-related OPSCC have a better clinical outcome than those with HPV-negative tumors [28], [29], [30], an early and accurate laboratory diagnosis for HPV related tumorigenesis using convenient and/or minimally invasive samples would be useful.
2. Materials and methods
2.1. Study design
Study candidates were enrolled from a regional cancer clinic in Hamilton, Ontario, Canada. A total of 59 patients between the ages of 40 and 80 (the mean age was 59.8 years and 50 were males) had histopathologically-confirmed HNSCC and signed consent for the collection of an FNA, an oropharyngeal swab from the base of the tongue and tonsillar area, and saliva. These samples were tested for the presence of HPV using 3 separate commercial assays measuring OncoE6 proteins, HR HPV E6/E7 mRNA or HR HPV DNA. Each assay also provided HPV genotyping for HPV 16, 18/45 or other high risk types. The values for FNA testing were compared to the results of testing tumor tissues for the presence of IHC p16 antigens from all 59 patients and the presence of HPV mRNA from a subset of 25 extracted paraffin embedded tumors.
2.2. Sample collection
One ml of saliva was collected into an OMNIgene Discover saliva collection kit (OM-505; DNA Genotek) for mRNA testing. A second saliva sample was collected into an empty 15 ml conical tube for OncoE6 testing. The oropharyngeal swab was collected from the base of the tongue and tonsillar area by a study nurse and put into an empty 15 ml falcon tube and 5 ml of PreservCyt ThinPrep (TP) preservative fluid (Hologic) was added in the laboratory. The swabs and saliva were tested separately and results were pooled. FNA of the cervical lymph node was collected by a Head and Neck surgeon for HPV testing using several passes with a syringe and 22-gauge needle. The syringe was emptied directly into a vial containing 4 ml of TP liquid-based media and an additional 1 ml of TP was added when received in the laboratory.
2.3. OncoE6TM testing
The OncoE6TM Oral Test (Arbor Vita Corp) was performed according to the manufacturer's protocol. Aliquots of each sample type (1 ml) were pelleted, rinsed, lysed, and conditioned before 200 ul were transferred into vials containing Detector Reagent. A lateral flow test strip was placed into each vial for 55 min to allow migration upwards through the strip. Test units were then moved to vials containing Wash Solution, followed by immersion in vials containing Developing Solution. Test units were visually inspected and the appearance of one or more lines besides the control line indicated elevated expression of E6 oncoproteins of HPV 16 and/or 18.
2.4. Aptima HPV testing
Within 24–48 h of receipt in the laboratory, 1 ml of each sample was transferred to an Aptima tube containing specimen transport media (STM), and was tested with the Aptima HPV assay on a Panther automated system according the manufacturer's instructions (Hologic Inc). The Aptima HPV assay measures E6/E7 mRNA of 14 HRHPV genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68 in a transcription mediated amplification (TMA) assay. Assay results were interpreted based on the signal-to-cutoff ratio (S/CO) and values ≥ 0.5 were considered positive. Specimens positive by AHPV testing were genotyped using a second Aptima HPV 16, 18/45 genotyping assay.
2.5. cobas HPV PCR testing
One ml of each sample was transferred into an empty 13 ml Sarstedt screw-capped round based tube for cobas HPV 4800 DNA testing (Roche Molecular Systems) which was performed at the Newfoundland and Labrador Public Health Laboratory in St. John's, Newfoundland. The cobas assay identified genotypes 16 and 18, while simultaneously detecting 12 other HR HPV genotypes 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68 collectively. The cobas 4800 software interpreted the outcomes for each sample and reported the results as negative, HPV 16, HPV 18 or other HR HPV. Samples with insufficient volume (ISV) were repeated.
2.6. IHC p16 testing
IHC p16 was performed on 59 tumor tissues using a commercial kit (CINtec Histology Kit, Roche MTM Laboratories, Mannheim, Germany). Tumors were considered positive for p16 if ≥ 70% of tumor cells demonstrated strong diffuse nuclear and cytoplasmic staining. All p16-stained slides were blinded and reread by a study pathologist co-investigator.
2.7. Processing of tumor tissue for HPV testing
Retrieved FFPE blocks of tumor tissue from a subset of 25 patients (16 tonsillar, 7 BOT and 2 soft palate) were processed. Four curls of 25 µm were cut and placed into a 15 ml conical tube. For each block a new disposable blade was used, and a thorough wipe down of the microtome area was performed with RNAseZap/DNAZap decontamination solutions. Non-tissue paraffin blocks were used as a sentinel for contamination. The curls were then incubated in 2.9 ml of Aptima STM for 30 min at 60 °C with intermittent shaking to partially dissolve the paraffin wax, followed by 5-min incubation on ice. The sample was removed from each tube and pipetted into an empty Aptima STM tube, and tested for E6/E7 mRNA or DNA according to the HPV kit package inserts. A panel of positive and negative samples was shipped to the National Microbiology Laboratory for confirmatory testing; an in-house microsphere-based multiplex assay was used [31]. Extracted DNA was amplified by nested PCR using PGMY and GP5+/GPG+ primers and the products were typed by hybridization to probes specific for 46 mucosal HPV-types coupled to sortable microspheres (Luminex).
2.8. Analysis of data
Positive, negative and overall agreements between assay results on FNA and oral samples compared to results of testing tumor tissue by p16 cytology and/or HPV mRNA and DNA were calculated with kappa values for agreement beyond chance.
3. Results
Tumor tissues were tested by p16 IHC from all 59 patients (consisting of 33 with tonsillar tumors, 20 with tumors at the base of the tongue and 4 soft palate, 1 vallecula, 1 pharyngeal wall tumors (Supplemental table 1)). The IHC p16 test was positive in 81.4% (48/59) of the primary tumors. The 11 tumors negative in the p16 test consisted of 6 tonsillar, 2 base of tongue and 3 soft palate. Results of testing tumor tissue for HR HPV nucleic acids from the subset of 25 FFPE tissue blocks which were able to be retrieved and processed showed that 80.0% (20/25) contained HPV E6/E7 mRNA and DNA. The 5 tumors negative for HPV nucleic acids consisted of 2 tonsillar, 1 base of tongue and 2 soft palate. All of the HPV positive tumors were also IHC p16 positive and a single p16 positive was HPV negative (case 35 in Supplemental table 1).
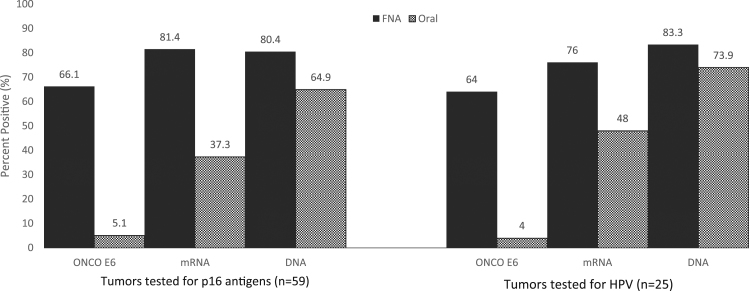
Testing lymph node FNA for HPV markers, positives were recorded by the OncoE6 test in 66.1% (39/59), by the Aptima E6/E7 mRNA assay in 81.4% (48/59) and by the cobas DNA test in 80.4% (45/56) (Fig. 1). Oral sample positivity rates were much lower; OncoE6 5.1% (3/59), Aptima E6/E7 mRNA 37.3% (22/59) and cobas DNA 64.9% (35/57) in the respective tests. For the subset of 25 patients, FNA positive results were recorded in 64% (16/25) by OncoE6, 76% (19/25) by Aptima and 83.3% (20/24) by cobas (Fig. 1). Oral positivity rates were 4% (1/25) for the OncoE6 test, 48% (12/25) for the Aptima E6/E7 mRNA test and 73.9% (17/23) for the cobas DNA test.
Fig. 1.
Detection HPV E6 oncoproteins, E6/E7, mRNA and DNA in fine needle aspirates (FNA) and oral samples from 59 patients with oropharyngeal squamous cell carcinomas tested for p16 antigens and a subset of 25 tested for p16 and HPV nucleic acids.
Genotyping from the OncoE6, cobas HPV DNA and Aptima HPV 16, 18/45 assays showed that most FNA positives were type 16 (Supplemental table 1). HPV genotyping between FNA and primary tumor tissue showed 100% agreement. The genotype 16 rates in samples from the oral cavity were lower than in FNA and there was a 23.8% disagreement of genotypes between oral and FNA samples in the Aptima HPV mRNA 16, 18/45 genotype test. Five patients with type 16 HPV in FNA samples had oral samples positive for HPV types other than 16, 18 or 45.
Comparing OncoE6, HPV E6/E7 mRNA and cobas HPV DNA testing showed that saliva had a higher rate of positives than orapharyngeal swabs in each test (OncoE6, 3 vs. 0; mRNA, 21 vs 10 and DNA, 32 vs 19) (Supplemental table 1).
Table 1 shows agreement and kappa calculations of the 3 testing methods on FNA and oropharyngeal samples, with tumors cytologically tested by p16 staining (n = 59) and by virological testing of extracted HR HPV nucleic acids (n = 25). Calculating positive, negative and overall agreements of p16 tested tumor tissue and FNA tested by the 3 tests, there were moderate to very good overall agreements (OncoE6 81.4% (48/59) k 0.53, Aptima E6/E7 mRNA 94.9% (56/59) k 0.83; and cobas DNA 91.1% (51/56) k 0.73). Agreements using oral samples were lower for each test and the kappa values were poor to moderate (OncoE6 23.7% (14/59) k 0.02, Aptima HPV mRNA 55.9% (33/59) k 0.24 and cobas HPV DNA 78.9% (45/57) k 0.49). When agreements were calculated using the 25 tumors that were HPV-tested as the comparator, overall agreements for FNA testing were as follows: OncoE6 64% (16/25) k 0.08; Aptima E6/E7 mRNA 88% (22/25) k 0.65 and cobas DNA 91.7% (22/24) k 0.70. Testing the oral samples showed agreements with the HPV tested tumors as follows: OncoE6 24% (6/25) k 0.02; Aptima E6/E7 mRNA 68% (17/25) k 0.37; cobas DNA 86.9% (20/23) k 0.58.
Table 1.
Agreements and kappa statistics of 3 assays on FNA and oral samples with tumors tested by IHC p16 and HR HPV nucleic acid tests.
| Sample | Test | Tumors tested for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| IHC p16 (n = 59) |
HR HPV mRNA and DNA (n = 25) |
||||||||
| % Pos. Agr. | % Neg. Agr. | % OA. Agr. | kappa | % Pos. Agr. | % Neg. Agr. | % OA. Agr. | kappa | ||
| FNA | OncoE6 | 79.2 (38/48) | 90.9 (10/11) | 81.4 (48/59) | 0.53 | 70 (14/20) | 40 (2/5) | 64 (16/25) | 0.08 |
| Aptima mRNA | 95.8 (46/48) | 90.9 (10/11) | 94.9 (56/59) | 0.83 | 90 (18/20) | 80 (4/5) | 88.0 (22/25) | 0.65 | |
| cobas DNA | 95.5 (42/44) | 81.8 (9/11) | 91.1 (51/56) | 0.73 | 100 (19/19) | 60 (3/5) | 91.7 (22/24) | 0.70 | |
| Oral | OncoE6 | 6.2 (3/48) | 100 (11/11) | 23.7 (14/59) | 0.02 | 5 (1/20) | 100 (5/5) | 24.0 (6/25) | 0.02 |
| Aptima mRNA | 45.8 (22/48) | 100 (11/11) | 55.9 (33/59) | 0.24 | 60 (12/20) | 100 (5/5) | 68 (17/25) | 0.37 | |
| cobas DNA | 76.1 (35/46) | 90.9 (10/11) | 78.9 (45/57) | 0.49 | 94.4 (17/18) | 60 (3/5) | 86.9 (20/23) | 0.58 | |
FNA- Fine needle aspirate
Oropharyngeal- Combined swab from base of tongue and saliva
IHC- Immunohistochemistry
HR HPV mRNA and DNA- (Aptima HPV E6/E7 mRNA test and cobas DNA test)
Pos. Agr.- Positive agreement
Neg. Agr.- Negative agreement
OA Agr.- Overall agreement K- Kappa statistic
4. Discussion
This study demonstrates that lymph node FNA samples from patients with metastatic OPSCC can be used to detect HPV16 E6 oncoproteins with the OncoE6 assay. This is the first report on the use of this test on lymph node FNA and oral samples for the diagnosis of OPSCC. A previous study demonstrated the presence of E6 oncoproteins in 7 freshly frozen oropharyngeal tumors containing HPV type 16 [9]. The original HPV E6 immunographic test was developed to measure E6 oncoproteins of genotypes 16, 18/45 in cervical samples for the diagnosis of cervical cancer [12] and was evaluated in large clinical trials in China [32], [33], showing sensitivity rates of approximately 70% for CIN3+ when compared to HPV DNA testing, and corrected for those lesions exhibiting HPV16, 18, 45. Agreement calculations in our study for the OncoE6 test with FNA samples varied from 81.4% when compared to tumors stained by IHC p16 to 64% when agreements were measured with tumor HPV nucleic acids (Table 1). These OncoE6 positivity rates are 10–25% lower than the Aptima mRNA or cobas DNA rates and are probably due to differences in test sensitivity, as the lower analytical sensitivity of the OncoE6 test is estimated at less than 5 × 103 HPV-E6 expressing cells [9] compared to approximately 48 copies of HPV16 mRNA in-vitro transcripts for the Aptima mRNA assays and 100 HPV16-infected cells for the cobas HPV DNA test, as indicated in the test package inserts. While it remains to be shown whether a higher analytical sensitivity of the OncoE6™ test would translate into a higher clinical sensitivity, it is also possible that larger quantitative studies might show that the very high analytical sensitivity of amplified HR HPV tests might have the potential to overcall HPV transcriptionally active tumors.
Testing metastatic lymph node FNA samples for HR HPV is a well-documented maneuver to determine whether the OPSCC may be HPV-related and several studies have evaluated commercial assays for this purpose [13], [14], [15], [16], [17], [18], [19]. In the current study using p16 positivity alone (n = 59), and HPV DNA or mRNA (n = 25) in tumor tissue for comparison, FNA testing agreements were poor to moderate for the OncoE6 test (64–81.4%; k 0.08–0.53), and good to very good for Aptima HPV mRNA (88.0–94.9%; k 0.65–0.83) and cobas HPV DNA (91.1–91.7%; k 0.70–0.73) respectively (Table 1).
Oral fluids are in close proximity to oropharyngeal tumors and collection of saliva and swabs are easy and non-invasive. The OncoE6 test performed on oropharyngeal samples was in agreement with p16 or HPV nucleic acid tested tumors at lower rates than HPV mRNA and HPV DNA (Table 1). These finding suggest that unlike FNA samples which contain metastasized HPV transcriptionally active cells and higher concentrations of E6 oncoproteins and E6/E7 mRNA, oropharyngeal samples contain lower concentrations of these analytes. These differences between tests may be due to analyte concentrations in the samples or our methods for processing the samples. We did not perform protein removal and nucleic acid extraction before processing the swabs and saliva samples into the 3 assays as recommended by D'Souza [34] and performed in a study by Agrawal [35], who showed similar HPV DNA oral rinse positivity rates to HPV DNA positive tumors. In the subset of patients from whom tumors were available for HPV testing in our study, HPV DNA was present in 73.9% (17/23) of the oral samples (Fig. 1). These rates are slightly lower than the 77.1% oral rate reported by Koslabova using a phosphate-buffered mouth wash to measure HPV DNA in patients with HPV DNA in FFPE extracted tumors [26]. The majority of the positives in our study were in the saliva sample compared to the swab (Supplemental table 1). This difference may be tumor site related or due to the use of the OMNIgene Discover collection kit for saliva, which is designed to preserve nucleic acids. Using the same commercial collection devices, Laprise et al. [36] observed higher HPV positivity rates in oral rinses compared to oral brushings in controls and cases of HNSCC.
It is estimated that about 90% of HPV-related cases of OPSCC are due to genotype 16 in FNA and in tumors [7], [8], [9], [11], [15]. The HPV genotype 16 positivity rates in our study for FNA and tumors ranged from 93.7% to 100% depending on the test used, and the genotype correlation between FNA and tumors showed 100% agreement. HPV 16 positivity rates were lower in oral samples than in FNA and this was more pronounced for the OncoE6 and the Aptima mRNA assay than the cobas DNA test. The Aptima mRNA assay showed more differences in genotypes between FNA and oral samples (Supplemental table 1). Five of 21 (23.8%) patients with HPV mRNA in FNA and oral samples had HRHPV genotype in their oral samples other than types 16, 18 or 45. This genotype difference between FNA and oral samples from the same patient was not observed by HPV DNA testing. These findings suggest that transient high risk genotypes may be found in the oral cavity not related to the HPV activity in the OPSCC, and FNA.
The HPV DNA detection rate in oral samples in our study was 73.9% (17/23) when compared to the DNA in tumors (Fig. 1). This finding agrees with previous studies [24], [25] showing lower HPV rates in saliva than in tumors. The oral DNA positivity rate for 14 patients with DNA positive tonsillar tumors was 92.9% (13/14) compared to 50% (2/4) for DNA positive tumors from the base of the tongue (Supplemental table 1). These findings can be compared to a previous study [27] showing saliva HPV DNA rates of 62% (18/29) for tonsillar and 44.4% (8/18) for tumors from the base of the tongue. The meaning of HPV DNA presence in oral samples is unclear. Most HPV infections are transient and HPV DNA is not considered a marker for oncogenically active HPV infection, although DNA viral load may be informative in this regard [9]. Previous studies have looked for indicators of oncogenically active HPV infections driving OPSCC. These have included somatic mutations of tumor DNA, and altered DNA sequences can be detected in saliva using multiplex PCR [37]. Tests for HPV16 mRNA, E6 oncoproteins, p53 or pRb downregulation and p16 upregulation in tumor tissue have been reported and combinations of these results have been used to determine whether the HPV infection is transcriptionally active [9]. A weakness of our study is that we did not incorporate more of these indicators of HPV transformation. In our study, none of the tumors were freshly frozen and available for testing for E6 oncoproteins. We used commercial tests for HPV E6 oncoproteins and HPV E6/E7 mRNA in FNA and oral samples from 59 patients with OPSCC. There was a strong correlation of IHC p16 positivity in tumors with the presence of HPV E6 oncoproteins and HPV mRNA and DNA in FNA. We were limited in the number of tumor tissues available for extraction and HPV testing but in the subset of tumors both HPV nucleic acid assays showed strong correlation with p16 positivity. In a single case (Tonsillar tumor 35) the tumor sample was p16 positive and negative for HPV mRNA and DNA (Supplemental table 1). Since the FNA was positive in the 3 HPV assays, we can assume that the tumor tissue was negative due to the inaccuracy of tissue extraction. Our observations suggest that the majority of these patients were experiencing transcriptionally active HPV infections as shown by the high HPV OncoE6 and E6/E7 mRNA positivity rates in FNA and tumors. The lower agreement values of E6 oncoproteins and E6/E7 mRNA in oral samples compared to tumor markers of p16 and HPV nucleic acids suggest that oral testing is not as informative with regard to HPV tumor transformation.
In summary the commercial tests used in this study performed well and enabled an accurate same-day diagnosis when testing neck lymph node FNA samples. Testing of oral samples was less informative towards the detection of an underlying tumor and may require further studies involving sample optimization.
Acknowledgements
This study was funded by Arbor Vita Corporation, DNA Genotek and Hologic Inc. J. Schweizer is an employee of Arbor Vita Corporation.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.pvr.2018.05.003.
Appendix A. Supplementary material
Supplementary material
References
- 1.Moutasim K.A., Robinson M., Thavaraj S. Human papillomavirus testing in diagnostic head and neck histopathology. Diagn. Histopathol. 2015;21(2):77–84. [Google Scholar]
- 2.Marur S., D'Souza G., Westra W.H., Forastiere A.A. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11(8):781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11(1):9. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 4.Lewis J.S. p16 Immunohistochemistry as a standalone test for risk stratification in oropharyngeal squamous cell carcinoma. Head. Neck Pathol. 2012;6(1):75–82. doi: 10.1007/s12105-012-0369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westra W.H. Detection of human papillomavirus (HPV) in clinical samples: evolving methods and strategies for the accurate determination of HPV status of head and neck carcinomas. Oral. Oncol. 2014;50(9):771–779. doi: 10.1016/j.oraloncology.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boscolo-Rizzo P., Pawlita M., Holzinger D. From HPV-positive towards HPV-driven oropharyngeal squamous cell carcinomas. Cancer Treat. Rev. 2016;42:24–29. doi: 10.1016/j.ctrv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Bishop J.A., Lewis J.S., Rocco J.W., Faquin W.C. HPV-related squamous cell carcinoma of the head and neck: an update on testing in routine pathology practice. Semin. Diagn. Pathol. 2015;32(5):344–351. doi: 10.1053/j.semdp.2015.02.013. (WB Saunders) [DOI] [PubMed] [Google Scholar]
- 8.Schache A.G., Liloglou T., Risk J.M., Filia A., Jones T.M., Sheard J. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin. Cancer Res. 2011;17(19):6262–6271. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baboci L., Holzinger D., Boscolo-Rizzo P., Tirelli G., Spinato R., Lupato V. Low prevalence of HPV-driven head and neck squamous cell carcinoma in North-East Italy. Papillomavirus Res. 2016;2:133–140. doi: 10.1016/j.pvr.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gondim D.D., Haynes W., Wang X., Chernock R.D., El-Mofty S.K., K S., Lewis J.S., Jr Histologic typing in oropharyngeal squamous cell carcinoma: a 4-year prospective practice study with p16 and high-risk HPV mRNA testing correlation. Am. J. Surg. Pathol. 2016;40(8):1117–1124. doi: 10.1097/PAS.0000000000000650. [DOI] [PubMed] [Google Scholar]
- 11.Smeets S.J., Hesselink A.T., Speel E.J.M., Haesevoets A., Snijders P.J., Pawlita M. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int. J. Cancer. 2007;121(11):2465–2472. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 12.Schweizer J., Lu P.S., Mahoney C.W., Berard-Bergery M., Ho M., Ramasamy V. Feasibility study of a human papillomavirus E6 oncoprotein test for diagnosis of cervical precancer and cancer. J. Clin. Microbiol. 2010;48(12):4646–4648. doi: 10.1128/JCM.01315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop J.A., Maleki Z., Valsamakis A., Ogawa T., Chang X., Pai S.I., Westra W.H. Application of the Hybrid capture 2 assay to squamous cell carcinomas of the head and neck. Cancer Cytopathol. 2012;120(1):18–25. doi: 10.1002/cncy.20175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith D.F., Maleki Z., Coughlan D., Gooi Z., Akpeng B., Ogawa T. Human papillomavirus status of head and neck cancer as determined in cytologic specimens using the hybrid-capture 2 assay. Oral. Oncol. 2014;50(6):600–604. doi: 10.1016/j.oraloncology.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo M., Khanna A., Dhillon J., Patel S.J., Feng J., Williams M.D. Cervista HPV assays for fine‐needle aspiration specimens are a valid option for human papillomavirus testing in patients with oropharyngeal carcinoma. Cancer Cytopathol. 2014;122(2):96–103. doi: 10.1002/cncy.21375. [DOI] [PubMed] [Google Scholar]
- 16.Kerr D.A., Pitman M.B., Sweeney B., Arpin R.N., Wilbur D.C., Faquin W.C. Performance of the Roche cobas 4800 high‐risk human papillomavirus test in cytologic preparations of squamous cell carcinoma of the head and neck. Cancer Cytopathol. 2014;122(3):167–174. doi: 10.1002/cncy.21372. [DOI] [PubMed] [Google Scholar]
- 17.Lastra R.R., Pramick M.R., Nakashima M.O., Weinstein G.S., Montone K.T., LiVolsi V.A., Baloch Z.W. Adequacy of fine-needle aspiration specimens for human papillomavirus infection molecular testing in head and neck squamous cell carcinoma. Cytojournal. 2013;10 doi: 10.4103/1742-6413.120789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baldassarri R., Aronberg R., Levi A.W., Yarbrough W.G., Kowalski D., Chhieng D. Detection and genotype of high-risk human papillomavirus in fine-needle aspirates of patients with metastatic squamous cell carcinoma is helpful in determining tumor origin. Am. J. Clin. Pathol. 2015;143(5):694–700. doi: 10.1309/AJCPCZA4PSZCFHQ4. [DOI] [PubMed] [Google Scholar]
- 19.Chernesky M., Gupta M., Jang D., Doerwald-Munoz L., Jackson B., Archibald S. Performance of Aptima E6/E7 mRNA HPV assay on fine-needle aspirates from cervical lymph nodes of patients with metastatic oropharyngeal squamous cell carcinoma. Otorhinolaryngol. Head Neck Surg. 2017;2(5):1–7. [Google Scholar]
- 20.Begum S., Gillison M.L., Nicol T.L., Westra W.H. Detection of human papillomavirus-16 in fine-needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2007;13(4):1186–1191. doi: 10.1158/1078-0432.CCR-06-1690. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M.Q., El‐Mofty S.K., Davila R.M. Detection of human papillomavirus‐related squamous cell carcinoma cytologically and by in situ hybridization in fine‐needle aspiration biopsies of cervical metastasis. Cancer Cytopathol. 2008;114(2):118–123. doi: 10.1002/cncr.23348. [DOI] [PubMed] [Google Scholar]
- 22.Schlecht N.F., Brandwein-Gensler M., Nuovo G.J., Li M., Dunne A., Kawachi N. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Mod. Pathol. 2011;24(10):1295–1305. doi: 10.1038/modpathol.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chai R.C., Lambie D., Verma M., Punyadeera C. Current trends in the etiology and diagnosis of HPV‐related head and neck cancers. Cancer Med. 2015;4(4):596–607. doi: 10.1002/cam4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao M., Rosenbaum E., Carvalho A.L., Koch W., Jiang W., Sidransky D., Califano J. Feasibility of quantitative PCR‐based saliva rinse screening of HPV for head and neck cancer. Int. J. Cancer. 2005;117(4):605–610. doi: 10.1002/ijc.21216. [DOI] [PubMed] [Google Scholar]
- 25.Tachezy R., Klozar J., Rubenstein L., Smith E., Saláková M., Šmahelová J. Demographic and risk factors in patients with head and neck tumors. J. Med. Virol. 2009;81(5):878–887. doi: 10.1002/jmv.21470. [DOI] [PubMed] [Google Scholar]
- 26.Koslabova E., Hamsikova E., Salakova M., Klozar J., Foltynova E., Salkova E., E Markers of HPV infection and survival in patients with head and neck tumors. Int. J. Cancer. 2013;133(8):1832–1839. doi: 10.1002/ijc.28194. [DOI] [PubMed] [Google Scholar]
- 27.Nordfors C., Vlastos A., Du J., Ährlund-Richter A., Tertipis N., Grün N. Human papillomavirus prevalence is high in oral samples of patients with tonsillar and base of tongue cancer. Oral. Oncol. 2014;50(5):491–497. doi: 10.1016/j.oraloncology.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Dayyani F., Etzel C.J., Liu M., Ho C.H., Lippman S.M., Tsao A.S. Meta-analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC) Head. Neck Oncol. 2010;2(1):15. doi: 10.1186/1758-3284-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K.K. Ang, E.M. Sturgis, Human papillomavirus as a marker of the natural history and response to therapy of head and neck squamous cell carcinoma. in: Proceedings of the Seminars in Radiation oncology (Vol. 22, No. 2, pp. 128–142). WB Saunders. [DOI] [PubMed]
- 30.O’rorke M.A., Ellison M.V., Murray L.J., Moran M., James J., Anderson L.A. Human papillomavirus related head and neck cancer survival: a systematic review and meta-analysis. Oral. Oncol. 2012;48(12):1191–1201. doi: 10.1016/j.oraloncology.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Zubach V., Smart G., Ratnam S., Severini A. Novel microsphere-based method for detection and typing of 46 mucosal human papillomavirus types. J. Clin. Microbiol. 2012;50(2):460–464. doi: 10.1128/JCM.06090-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao F.H., Jeronimo J.A., Qiao Y.L., Schweizer J., Chen W., Valdez M. An evaluation of novel, lower-cost molecular screening tests for human papillomavirus in rural China. Cancer Prev. Res. 2013;6(9) doi: 10.1158/1940-6207.CAPR-13-0091. (canprevres-0091) [DOI] [PubMed] [Google Scholar]
- 33.Qiao Y.L., Jeronimo J., Zhao F.H., Schweizer J., Chen W., Valdez M. Lower cost strategies for triage of human papillomavirus DNA‐positive women. Int. J. Cancer. 2014;134(12):2891–2901. doi: 10.1002/ijc.28616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Souza G., Sugar E., Ruby W., Gravitt P., Gillison M. Analysis of the effect of DNA purification on detection of human papillomavirus in oral rinse samples by PCR. J. Clin. Microbiol. 2005;43(11):5526–5535. doi: 10.1128/JCM.43.11.5526-5535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal Y., Koch W.M., Xiao W., Westra W.H., Trivett A.L., Symer D.E., Gillison M.L. Oral human papillomavirus infection before and after treatment for human papillomavirus 16–positive and human papillomavirus 16–negative head and neck squamous cell carcinoma. Clin. Cancer Res. 2008;14(21):7143–7150. doi: 10.1158/1078-0432.CCR-08-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laprise C., Madathil S.A., Schlecht N.F., Castonguay G., Soulières D., Nguyen-Tan P.F. Human papillomavirus genotypes and risk of head and neck cancers: results from the HeNCe Life case-control study. Oral. Oncol. 2017;69:56–61. doi: 10.1016/j.oraloncology.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Springer S., Mulvey C.L., Silliman N., Schaefer J., J, Sausen M. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015;7(293) doi: 10.1126/scitranslmed.aaa8507. (293ra104-293ra104) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material