Abstract
The tryptophan (Trp) transport system has a high affinity and selectivity toward Trp, and has been reported to exist in both human and mouse macrophages. Although this system is highly expressed in interferon-γ (IFN-γ)–treated cells and indoleamine 2,3-dioxygenase 1 (IDO1)–expressing cells, its identity remains incompletely understood. Tryptophanyl-tRNA synthetase (TrpRS) is also highly expressed in IFN-γ–treated cells and also has high affinity and selectivity for Trp. Here, we investigated the effects of human TrpRS expression on Trp uptake into IFN-γ–treated human THP-1 monocytes or HeLa cells. Inhibition of human TrpRS expression by TrpRS-specific siRNAs decreased and overexpression of TrpRS increased Trp uptake into the cells. Of note, the TrpRS-mediated uptake system had more than hundred-fold higher affinity for Trp than the known System L amino acid transporter, promoted uptake of low Trp concentrations, and had very high Trp selectivity. Moreover, site-directed mutagenesis experiments indicated that Trp- and ATP-binding sites, but not tRNA-binding sites, in TrpRS are essential for TrpRS-mediated Trp uptake into the human cells. We further demonstrate that the addition of purified TrpRS to cell culture medium increases Trp uptake into cells. Taken together, our results reveal that TrpRS plays an important role in high-affinity Trp uptake into human cells.
Keywords: aminoacyl tRNA synthetase; tryptophan; amino acid transport; interferon; indoleamine-pyrrole 2,3-dioxygenase (IDO1); tryptophanyl-tRNA synthetase; mutagenesis
Introduction
Tryptophan (Trp) depletion plays important roles in the regulation of immunosuppressive function (1–4). Trp catabolism mediated by the enzyme indoleamine 2,3-dioxygenase 1 (IDO1) is emerging as an important pathway in immune regulation (1–4). IDO1 is a heme-containing enzyme that is expressed in various organs, particularly the placenta and immune cells (1–3), and is induced by interferon-γ (IFN-γ), which orchestrates a multisignal response to fight pathogens and neoplasia (5, 6). It is the rate-limiting enzyme in the kynurenine pathway, catabolizing Trp to kynurenine (7–9). This protein has been implicated in maternal tolerance toward the allogeneic fetus, control of T cell proliferation, and regulation of antitumor immunity (1–4). IDO1-mediated immunosuppression at the maternal–fetal interface of the placenta might play an important role in the maternal tolerance of the allogeneic fetus (1). It helps to protect fetuses from rejection by the maternal immune system and can promote tolerance and immunosuppression (1). In addition, many human tumors constitutively express IDO1, and its expression by immunogenic mouse tumor cells prevents their rejection by preimmunized mice (4). In humans, this enzyme can be expressed by monocyte-derived macrophages (MDMs)2 and certain monocyte-derived dendritic cells (DCs) (2, 3). IDO1-mediated Trp catabolism in MDMs and DCs arrests T cell proliferation, thereby providing a molecular basis for its immunosuppressive function (2, 3). Indeed, loss of IDO1 expression or activity has been shown to enhance several autoimmune diseases (10).
Mammalian cells cannot synthesize Trp and because it does not diffuse into cells, this amino acid must be transported across the plasma membrane of MDMs and DCs by a specific transmembrane transport system to be catabolized by IDO1. In mammalian cells, transporter-mediated Trp uptake occurs mainly via the ubiquitously expressed neutral amino acid transporter System L that transports large hydrophobic amino acids with branched or aromatic side chains such as His, Ile, Leu, Met, Phe, Trp, Tyr, and Val (11–13). It is heterodimeric and composed of a surface antigen 4F2 heavy chain plus one of two catalytic L chains LAT1 or LAT2 (11–13). The Km of Trp for System L transporter is 20–60 μm (11, 12) and normal plasma concentrations are ∼50 μm (14). A novel amino acid transport activity with high affinity and selectivity for Trp was reported to be expressed in IFN-γ–treated or IDO1-expressing cells (15–17). Previous studies have shown that Trp depletion to concentrations less than 1 μm inhibited T cell proliferation (2, 18). Because MDMs readily deplete Trp present in the extracellular medium to nanomolar levels via IDO1 activity in vitro, the additional high-affinity and high-selective Trp transport system enables efficient uptake of Trp at low concentrations. It thereby becomes increasingly important for Trp uptake as the local extracellular concentration decreases (15). IDO1 induces expression of this novel Trp transport system in mouse and human tumor cells (16). IFN-γ up-regulates IDO1 expression and in turn induces the additional Trp-selective transport system with a higher affinity for Trp than System L transporter to promote IDO1 substrate availability (15, 17). The decreased availability of Trp in the extracellular medium suppresses T cell proliferation and causes immune tolerance. The high-affinity Trp-specific transport system is expressed at low levels in fresh monocytes but undergoes selective induction during MDM differentiation. This transport system allows MDMs to take up Trp efficiently under conditions of low substrate concentration (15).
Human tryptophanyl-tRNA synthetase (TrpRS) is the only aminoacyl-tRNA synthetase whose expression is induced by IFN-γ (19–25). Aminoacyl-tRNA synthetases catalyze the ligation of specific amino acids to their cognate tRNAs (26). Aminoacylation occurs in two steps: first is the formation of an aminoacyl-AMP from an amino acid and ATP, and second is the transfer of the aminoacyl group of aminoacyl-AMP to a particular tRNA to form aminoacyl-tRNA (27). Noncanonical functions distinct from aminoacylation have been reported, such as the cell-signaling functions of human TrpRS and tyrosyl-tRNA synthetase (TyrRS) in pathways connected to the immune system or angiogenesis (28–34). TrpRS is highly expressed during differentiation from monocytes to MDMs or DCs (35, 36). Vertebrate TrpRS has an NH2-terminal appended domain. In normal cells, human TrpRS exists in two forms: the major full-length (FL) TrpRS and a less abundant mini TrpRS, in which the extra NH2-terminal domain is deleted because of alternative splicing of the pre-mRNA such that Met-48 becomes the NH2-terminal residue (37, 38). We previously found that human mini, but not FL, TrpRS functions as an angiostatic factor (31). The expression of human FL and mini TrpRSs is increased following stimulation of human cells by IFN-γ (19–25). Moreover, we recently showed that the expression of TrpRS is also enhanced by exposure of mouse cells to IFN-γ (39). Human FL TrpRS (amino acids 1–471) is cleaved by elastase to produce T2 TrpRS (amino acids 94–471), which also acts as an angiostatic factor (31, 40). Whereas human FL and mini TrpRSs retain aminoacylation activity, T2 TrpRS is inactive (31, 40).
It has been reported that a novel Trp transport system that has a high affinity and selectivity toward Trp exists in human and mouse macrophages (15–17). This Trp transport system is expressed in cells treated with IFN-γ or IDO1-expressing cells (15–17). It should be noted that TrpRS, which is highly expressed in IFN-γ–treated cells, also has a high affinity and selectivity for Trp (41, 42). In the present study, the mechanism of Trp uptake into cells was investigated by focusing mainly on TrpRS. Trp uptake was measured by using cells, in which TrpRS mutant of Trp-, ATP-, or tRNA-binding site was overexpressed. In addition, the effect of adding purified TrpRS mutant into cell culture medium on Trp uptake was analyzed. We provide herein a comprehensive biochemical characterization of a novel transport system with nanomolar affinity and high selectivity for Trp.
Results
IFN-γ treatment of human THP-1 and HeLa cells increase Trp uptake into the cells
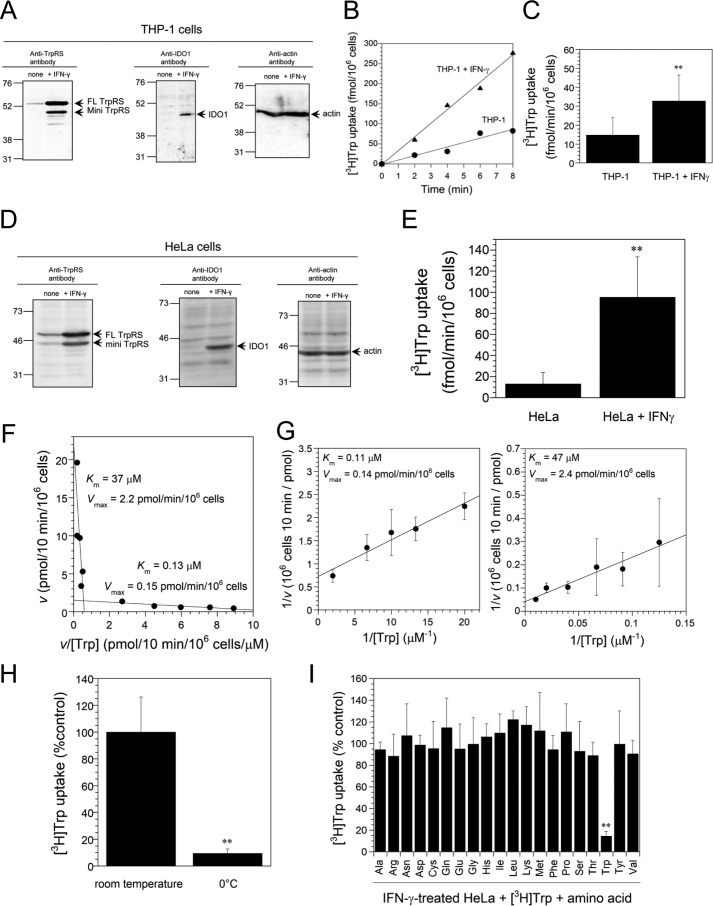
The expression levels of TrpRS, IDO1, and β-actin proteins in untreated and IFN-γ–treated THP-1 cells were analyzed by Western blotting using anti-TrpRS, anti-IDO1, or anti–β-actin antibodies. β-actin (predicted molecular size 42 kDa) was employed as a loading control (Fig. 1A). As shown in Fig. 1A, IFN-γ treatment of THP-1 cells enhanced the expression of human FL TrpRS (53 kDa), mini TrpRS (48 kDa), and IDO1 (45 kDa) proteins (Fig. 1A). Previous reports demonstrated that low concentration of [3H]Trp (150 nm) uptake was not mediated by System L transporter but by a novel Trp transport system (14). Indeed, as shown in Fig. 1, B and C, Trp (150 nm) uptake rates were enhanced by treatment of IFN-γ. Moreover, IFN-γ treatment of HeLa cells induced the expression of IDO1 and TrpRS proteins (Fig. 1D) and enhanced Trp uptake into HeLa cells (Fig. 1E).
Figure 1.
[3H]Trp uptake into IFN-γ–treated human THP-1 and HeLa cells. A, Western blot analyses of IFN-γ–treated THP-1 cells by using anti-TrpRS, anti-IDO1, or anti-actin antibodies. The arrow indicates the expected position for FL TrpRS, mini TrpRS, IDO1, or β-actin. The sizes in kilodaltons of molecular markers are indicated at the left. B, representative plots of [3H]Trp uptake into IFN-γ–treated THP-1 cells or nontreated THP-1 cells. Black circles, THP-1; black triangles, THP-1 + IFN-γ. C, initial rates of [3H]Trp uptake into IFN-γ–treated THP-1 cells or nontreated control. All data from Trp uptake experiments are expressed as mean ± S.D. of nine independent experiments. **, p < 0.01 versus nontreated THP-1 control. D, Western blot analyses of IFN-γ–treated HeLa cells. The sizes in kilodaltons of molecular markers are indicated at the left. E, [3H]Trp uptake into nontreated control or IFN-γ–treated HeLa cells. All data are expressed as mean ± S.D. of 12 independent experiments. **, p < 0.01 versus nontreated HeLa control. F, Eadie-Hofstee plots of Trp uptake into IFN-γ–treated HeLa cells over a broad concentration range (50 nm–100 μm). All data are expressed as mean from at least three independent experiments. G, Lineweaver-Burk plots of Trp uptake into IFN-γ–treated HeLa cells analyzed separately for low (50–500 nm) and high (8–100 μm) ranges of Trp concentrations. All data are expressed as mean ± S.D. of at least three independent experiments. H, energy dependence of Trp transport. [3H]Trp uptake into IFN-γ–treated HeLa cells at 0 °C was compared with that at room temperature. Results are expressed as a percentage of the value of the control at room temperature, and they represent mean ± S.D. of three independent experiments. **, p < 0.01 versus room temperature control. I, the high-affinity transport system of IFN-γ–treated HeLa cells is selective for Trp. Competition studies were performed using a low concentration of [3H]Trp (150 nm) in the absence or presence of 20-fold excess of unlabeled amino acids (3 μm). Results are expressed as a percentage of the value of the control without any unlabeled amino acid. All data are expressed as mean ± S.D. of at least three independent experiments. **, p < 0.01 versus the control without any unlabeled amino acid.
To confirm the observation that IFN-γ induces a novel Trp-selective transport system (15), the Michaelis-Menten constant Km and maximum velocity Vmax of Trp transport system were determined. Eadie-Hofstee plots of Trp uptake data over a broad concentration range (50 nm–100 μm) revealed that Trp uptake did not follow a simple pattern for a single transport system but occurs via two transport systems (Km values of 0.13 and 37 μm) (Fig. 1F). Lineweaver-Burk plots of the uptake data contained two best fit lines which further demonstrate that two independent transport systems were involved in Trp transport. Each line followed classical Michaelis-Menten kinetics but displayed markedly different substrate affinities when analyzed separately for low (50–500 nm) and high (8–100 μm) substrate concentrations ranges (Km values of 0.11 and 47 μm) (Fig. 1G). The parameters determined in this study were almost the same as previously published data that also display two-component Km values of 0.33 and 24 μm (15). The Km value of the low-affinity system was consistent with values reported for System L transporter (Km value of 20–60 μm) (11, 12). Therefore, the high-affinity system is distinct from System L.
Subsequently, the effect of reducing the temperature on Trp uptake was tested. As shown in Fig. 1H, lowering the temperature from room temperature to 0 °C decreased Trp uptake by ∼90%. This result indicates that Trp uptake was performed via an energy-dependent transmembrane transport mechanism rather than surface-receptor binding because membrane trafficking is inhibited at 0 °C. Furthermore, uptake of Trp was measured at a low concentration of [3H]Trp (150 nm, chosen to be near the Km of the high-affinity system) in the absence or presence of 20-fold excess unlabeled amino acids as competitors. Under these conditions, only unlabeled Trp significantly inhibited [3H]Trp uptake (85% inhibition) (Fig. 1I). Taken together, these findings demonstrate that IFN-γ induces a Trp-selective transport system.
Inhibition of human TrpRS expression by TrpRS-specific siRNA decreased and overexpression of human FL TrpRS increased Trp uptake into cells
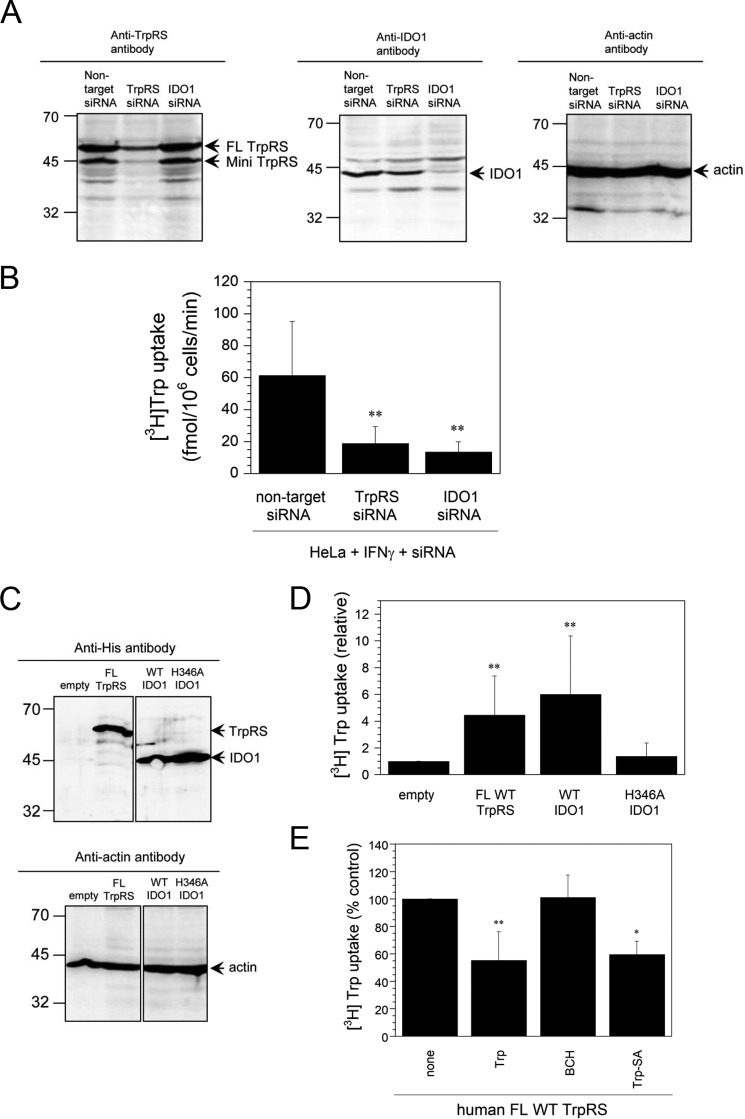
The effect of [3H]Trp uptake into HeLa cells treated with IFN-γ and siRNA was examined. Suppression of TrpRS or IDO1 expression by TrpRS- or IDO1-specific siRNA and overexpression of His6-tagged human FL TrpRS (56 kDa) or IDO1 (48 kDa) in HeLa cells were confirmed by Western blot analyses (Fig. 2, A and C). Suppression and overexpression of these two proteins, respectively, decreased and increased Trp uptake rates into HeLa cells (Fig. 2, B and D).
Figure 2.
[3H]Trp uptake into IFN-γ– and siRNA-treated or human FL TrpRS- or IDO1-overexpressing HeLa cells. A, Western blot analyses of HeLa cells treated with IFN-γ and nontarget, TrpRS-, or IDO1-specific siRNA. The sizes in kilodaltons of molecular markers are indicated at the left. B, initial rates of [3H]Trp uptake into HeLa cells treated with IFN-γ and siRNA. All data are expressed as mean ± S.D. of at least eight independent experiments. **, p < 0.01 versus nontarget siRNA control. C, Western blot analyses of His6-tagged human FL TrpRS- or IDO1-overexpressing HeLa cells by using anti-His6 tag or anti-actin antibodies. The sizes in kilodaltons of molecular markers are indicated at the left. D, [3H]Trp uptake into HeLa cells, in which human FL TrpRS or IDO1 was overexpressed. Data are expressed as [3H]Trp uptake relative to that of the empty vector control. All data are expressed as mean ± S.D. of at least four independent experiments. **, p < 0.01 versus empty control. E, effects of a competitor (3 μm) on [3H]Trp uptake into HeLa cells, in which human FL TrpRS was overexpressed. Results are expressed as a percentage of the value of the control without any competitor. All data are expressed as mean ± S.D. of at least three independent experiments. **, p < 0.01 versus the none control.
Because His-346 residue of human IDO1 is the proximal heme ligand and H346A IDO1 mutant cannot bind heme and catalyze the first step in Trp catabolism (8, 9), the effect of overexpressing this mutant on Trp uptake was investigated. Indeed, unlike WT IDO1, this mutant did not stimulate Trp uptake into HeLa cells (Fig. 2D), suggesting that heme binding to IDO1 is crucial for its function. These results are consistent with a previous study showing that the expression of human IDO in HeLa cells induces expression of a high-affinity Trp transport system (16).
Next, the potent and selective System L inhibitor BCH was employed to determine whether Trp transport was mediated solely by the ubiquitous System L transporter or together with another transport system. Competition studies for Trp uptake into HeLa cells, in which human FL TrpRS was overexpressed, in the absence or presence of 3 μm competitor were performed. BCH did not influence the amount of 150 nm [3H]Trp taken up into cells (Fig. 2E), suggesting that at low Trp concentrations, a novel transport system with high affinity, but not System L, was induced.
TrpRS catalyzes the condensation of Trp with ATP to give Trp-AMP. Although the adenylate is highly labile, it is sequestered in the active site to protect it from hydrolysis before it is condensed with the 3′ end of tRNA to yield the aminoacyl-tRNA. The nonhydrolyzable Trp-AMP analogue Trp-SA is a TrpRS-specific inhibitor that binds tightly to the active site. Interestingly, Trp uptake was reduced in the presence of Trp-SA or unlabeled Trp (Fig. 2E), confirming that Trp uptake is mediated by TrpRS.
Identification of FL TrpRS amino acid residues crucial for Trp uptake into cells by site-directed mutagenesis
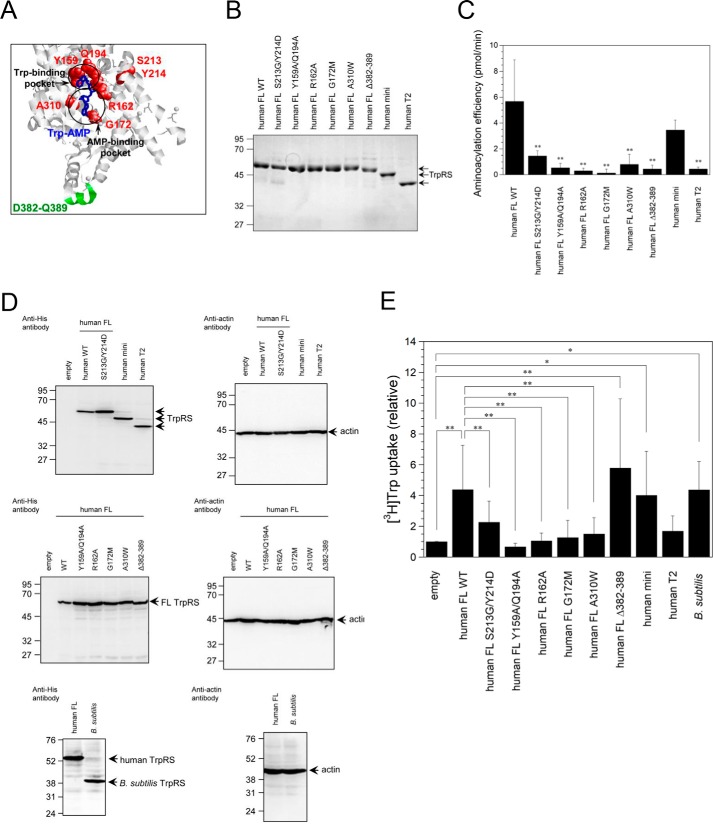
To identify residues crucial for Trp uptake activity of TrpRS, several site-directed human FL TrpRS mutants whose structure and function have previously been well-characterized were produced (33, 42–44). Tertiary structural position of amino acid residues crucial for Trp-, ATP-, and tRNA-binding in TrpRS is shown in Fig. 3A. It has been reported that although deletion of TrpRS amino acid residues 382–389 (Δ382–389 mutant) crucial for the interaction with the anticodon of tRNA does not have aminoacylation activity, the adenylate formation step was unaffected (43). Furthermore, previous reports showed that the potential polymorphic variant of human TrpRS S213G/Y214D TrpRS double mutant or R162A TrpRS mutant cannot bind ATP as tightly as FL WT TrpRS (42, 44). The apparent Km of ATP for S213G/Y214D TrpRS mutant is about 10-fold higher than that for WT TrpRS, whereas both Km values for Trp are almost the same (42, 45). Arg-162 is modeled to form salt bridges with both α- and β-phosphates of ATP (44). The R162A mutation decreased the ATP-binding affinity by about 60-fold (44). Two more single mutants, A310W and G172M in which the AMP pocket is blocked, were prepared. Both mutations did not affect Trp binding but weakened the binding of TrpRS to the AMP moiety (33). Furthermore, the Y159A/Q194A TrpRS double mutant was created to disrupt Trp-binding pocket. Indeed, previous studies showed that Y159A/Q194A TrpRS cannot bind to Trp (33).
Figure 3.
[3H]Trp uptake into site-directed generated mutant TrpRS-overexpressing HeLa cells. A, tertiary structural position of amino acid residues crucial for Trp-, ATP-, or tRNA-binding in human FL TrpRS (PDB ID: 1R6T). Residues crucial for Trp- or ATP-binding in human TrpRS are highlighted in red. Trp-AMP and 382–389 resides crucial for tRNA binding in human TrpRS are indicated in blue and green, respectively. B, SDS-PAGE analysis of purified recombinant TrpRS proteins. The gel was stained with Coomassie Brilliant Blue. The sizes in kilodaltons of molecular markers are indicated at the left. C, aminoacylation activities of WT and TrpRS mutants toward yeast tRNATrp. Aminoacylation efficiencies were determined from initial rates and calculated as picomole/min of aminoacylated tRNATrp synthesized during a 1-min incubation. All data are expressed as mean ± S.D. of at least three independent experiments, each carried out in duplicate. **, p < 0.01 versus full-length WT TrpRS. D, Western blot analyses of TrpRS-overexpressing HeLa cells. The sizes in kilodaltons of molecular markers are indicated at the left. E, Initial rates of [3H]Trp uptake into TrpRS-overexpressing HeLa cells. Data are expressed as [3H]Trp uptake relative to that of the empty vector control. All data are expressed as mean ± S.D. of at least four independent experiments. **, p < 0.01; *, p < 0.05.
Recombinant His6-tagged human WT TrpRS and TrpRS mutants were purified following expression in Escherichia coli and their purity was confirmed by SDS-PAGE. A band corresponding to His6-tagged human FL (54 kDa), mini (49 kDa), or T2 TrpRS (44 kDa) was observed (Fig. 3B).
Because previous data showed that eukaryotic TrpRS can aminoacylate yeast tRNATrp (46–51), we investigated the aminoacylation of yeast tRNATrp by recombinant TrpRS. Aminoacylation activity of human FL S213G/Y214D, Y159A/Q194A, R162A, G172M, A310W, or Δ382–389 TrpRS mutant was significantly lower than that of FL WT TrpRS (Fig. 3C). In addition, human mini TrpRS, but not T2 TrpRS, exhibited similar aminoacylation activity as human FL TrpRS (Fig. 3C).
[3H]Trp uptake rates into HeLa cells overexpressing site-directed TrpRS mutants were measured. Expression levels of these TrpRS mutants in HeLa cells were confirmed by Western blot analyses (Fig. 3D). As expected, bands corresponding to His6-tagged human FL TrpRS (56 kDa), mini (51 kDa), and T2 TrpRS (46 kDa) were detected (Fig. 3D). Overexpression of the S213G/Y214D, Y159A/Q194A, R162A, G172M, or A310W mutants of TrpRS Trp- or ATP-binding site did not stimulate Trp uptake rates into cells significantly (Fig. 3E). In contrast, overexpression of the tRNA-binding site Δ382–389 mutant, human mini TrpRS, and human FL TrpRS enhanced Trp uptake into cells (Fig. 3E). Finally, human T2 TrpRS that cannot bind ATP as tightly as FL or mini TrpRS (44) had no significant effect on Trp uptake into the cells (Fig. 3E).
Because it has been reported that human, but not Bacillus subtilis, TrpRS can charge human tRNATrp (52), we next investigated the effect of B. subtilis TrpRS on Trp uptake into HeLa cells. As shown in Fig. 3, D and E, overexpression of B. subtilis TrpRS (40 kDa) significantly stimulated Trp uptake into the cells.
Addition of purified TrpRS protein into cell culture enhances Trp uptake into HeLa cells
The effect of adding purified TrpRS protein to cell culture medium on [3H]Trp uptake into HeLa cells was investigated. Human FL WT, FL S213G/Y214D, FL A310W, FL Δ382–389, mini and T2 TrpRSs, and B. subtilis TrpRS were purified following expression in E. coli. Their purity was confirmed by SDS-PAGE (Figs. 3B and 4B). The addition of human FL WT, human FL Δ382–389, human mini, or B. subtilis TrpRS into the cell culture stimulated Trp uptake into the cells whereas human FL S213G/Y214D, FL A310W, or T2 TrpRS did not (Fig. 4A). These results are consistent with those of TrpRS-overexpressing cells.
Figure 4.
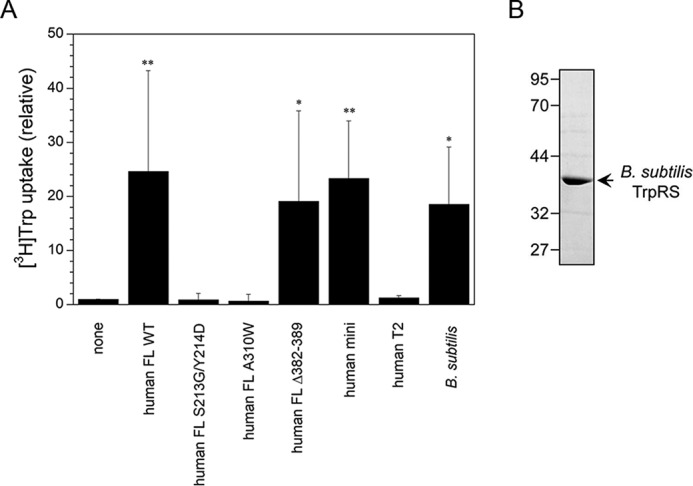
Analysis of the effect of adding TrpRS to the cell culture medium on [3H]Trp uptake into HeLa cells. A, [3H]Trp uptake into HeLa cells. Data are expressed as [3H]Trp uptake relative to that of the none control. All data are expressed as mean ± S.D. of at least four independent experiments. **, p < 0.01; *, p < 0.05 versus the none control. B, SDS-PAGE of purified His6-tagged B. subtilis TrpRS protein (the predicted molecular size 38 kDa). The sample was analyzed on 12.0% SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue. The sizes in kilodaltons of molecular markers are indicated at the left.
Tyrosine uptake into HeLa cells
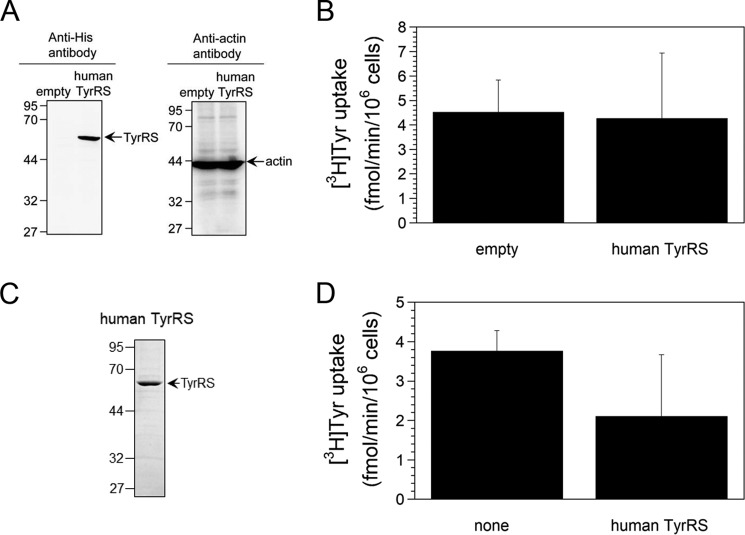
Finally, to test the specificity of human TrpRS we investigated whether human TyrRS can regulate Tyr uptake into cells by overexpressing His6-tagged human TyrRS (62 kDa) in HeLa cells. Overexpression of this protein was confirmed by Western blot analyses (Fig. 5A). It had no effect on [3H]Tyr uptake into HeLa cells (Fig. 5B). Furthermore, His6-tagged human TyrRS protein (60 kDa) was purified following expression in E. coli and its purity was confirmed by SDS-PAGE (Fig. 5C). Addition of the purified human TyrRS into the cell culture medium did not influence [3H]Tyr uptake into cells (Fig. 5D). Taken together, these results demonstrate that human TyrRS cannot regulate Tyr uptake into cells.
Figure 5.
[3H]Tyr uptake into HeLa cells. A, Western blot analyses of His6-tagged human TyrRS-overexpressing HeLa cells. The sizes in kilodaltons of molecular markers are indicated at the left. B, [3H]Tyr uptake into His6-tagged human TyrRS-overexpressing HeLa cells. Data are expressed as mean ± S.D. of five independent experiments. C, SDS-PAGE of purified human TyrRS protein. The gel was stained with Coomassie Brilliant Blue. The sizes in kilodaltons of molecular markers are indicated at the left. D, analysis of the effect of TyrRS protein addition to the cell culture medium on [3H]Tyr uptake into HeLa cells. All data are expressed as mean ± S.D. of at least three independent experiments.
Discussion
In the present study, we demonstrated that Trp uptake into IFN-γ–treated cells is mediated by a novel Trp-selective transport system with a few hundred-fold higher affinity for Trp than the known System L transporter. In particular, we showed that the uptake of low Trp concentrations (150 nm) is only dependent on the novel Trp-selective transport system and is regulated by TrpRS. Competition studies showed that the high-affinity system did not correspond to any known transporter activity and displayed a marked selectivity for Trp over other amino acids. The present results show that TrpRS can regulate Trp uptake into the cells.
Moreover, we demonstrated that Trp- and ATP-binding sites, but not the tRNA-binding site, of human TrpRS play important roles in the regulation of Trp uptake into the cells. Furthermore, we showed that B. subtilis TrpRS stimulated Trp uptake into HeLa cells as did human TrpRS. It has been reported that prokaryote and eukaryote cytoplasmic TrpRSs cannot cross-aminoacylate their respective tRNATrp (52). For example, only human TrpRS charges human tRNATrp and B. subtilis TrpRS charges B. subtilis tRNATrp but they cannot cross-charge each other (52). Taken together, these results are consistent with the conclusion that tRNA-binding site of TrpRS is not crucial for Trp uptake into the cells.
TrpRS and TyrRS amino acid sequences and their tertiary and quaternary structure of the catalytic domains are similar (53–58). Human mini TrpRS and mini TyrRS belong to a family of regulators of angiogenesis and have opposing cell-signaling activities (30, 31, 59). Human mini TrpRS acts as an angiostatic factor, whereas human mini TyrRS functions as an angiogenic factor (30, 31, 59). In the present study, we demonstrated that human TrpRS regulates Trp uptake into cells, whereas human TyrRS did not regulate Tyr uptake into the cells. This functional difference may be related to the characteristics that the expression of human TrpRS, but not human TyrRS, is regulated by IFN-γ.
The present study clarified that both TrpRS and IDO1 play important roles in Trp uptake. What is the relationship of TrpRS and IDO1 for Trp uptake? IDO1 catalyzes the degradation of Trp into kynurenine (7–9). Bhutia et al. (17) clarified that the treatment of cells with kynurenine, which is a physiological agonist for the aryl hydrocarbon receptor (AhR), increased Trp uptake into cells at low concentration of Trp (100 nm). Moreover, they showed that activation of AhR by various pharmacological AhR agonists also increased Trp uptake at the low Trp concentration and proposed that IDO1 may induce expression of a novel Trp-selective transporter by the activation of AhR by kynurenine (17). On the other hand, because the first step of aminoacylation reaction by TrpRS is reversible, TrpRS can play roles in transport of Trp into cells. A possible mechanism of how TrpRS regulates Trp uptake is summarized in Fig. 6. At first, IFN-γ stimulates TrpRS expression in human cells. A part of TrpRS may then be secreted from the cells. Indeed, previous studies demonstrated that TrpRS is secreted into the extracellular space (60) and that extracellular TrpRS exists in cell culture medium (61, 62). Moreover, human TrpRS was recently reported to be rapidly secreted from cells upon pathogen infection and act as a primary defense system against infection (63). After it is secreted, extracellular TrpRS regulates Trp uptake into human cells. It should also be noted that Km value of Trp for TrpRS in tRNATrp aminoacylation reaction is ∼2 μm (41, 42) and is significantly higher than that for the novel Trp transport system. Further studies are necessary to identify the cell-surface molecule that binds to extracellular TrpRS for Trp uptake and to determine whether the novel Trp transport system with high affinity and selectivity toward Trp is TrpRS itself or a novel Trp transporter that can be regulated by TrpRS.
Figure 6.
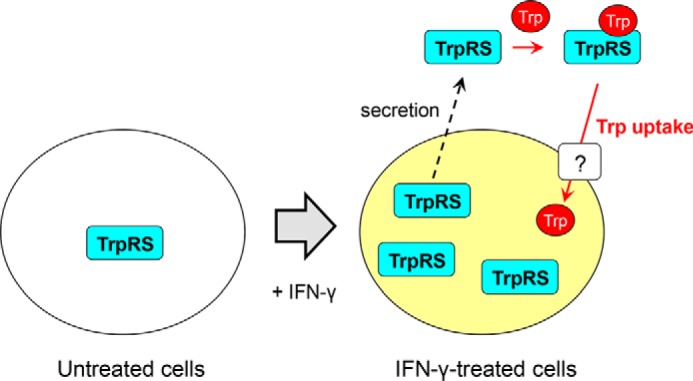
A schematic model of Trp uptake regulatory mechanism by TrpRS into IFN-γ–treated human cells.
Experimental procedures
Reagents
Cys, Asp, Glu, Phe, His, Ile, Lys, Leu, Met, Pro, Gln, Arg, Ser, Thr, Val, Tyr, tryptamine, and BCH, an inhibitor of System L amino acid transporters, were obtained from Sigma-Aldrich. Ala, Gly, Asn, and Trp were purchased from Wako Pure Chemical Industries (Osaka, Japan). Brewer's yeast tRNA and human IFN-γ were purchased from Roche Diagnostics. l-[5-3H]Trp ([3H]Trp) and l-[ring 3,5-3H]Tyr ([3H]Tyr) were purchased from PerkinElmer Life Sciences and American Radiolabeled Chemicals, Inc. (St. Louis, MO), respectively.
Cell lines
The human acute monocytic leukemia cell line THP-1 (ATCC TIB-202) was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in RPMI 1640 medium containing 10% (v/v) FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm glutamine (all from Invitrogen), and 0.05 mm 2-mercaptoethanol (Wako Pure Chemical Industries). HeLa cells (RCB0007), derived from a human epithelial fatal cervical carcinoma, were obtained from the RIKEN Cell Bank (Tsukuba, Japan). This cell line was maintained in culture in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/liter of glucose, 10% (v/v) FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine (all from Invitrogen). Both THP-1 and HeLa cell lines were cultured in a humidified atmosphere containing 5% CO2 at 37 °C.
Trp or Tyr uptake measurements
THP-1 or HeLa cells were washed twice in PBS, pH 7.4, and resuspended in PBS at 1 × 106 cells/ml. [3H]Trp (final concentration 150 nm) or [3H]Tyr (final concentration 150 nm) in the absence or presence of competitor (final concentration 3 μm) or protein (final concentration 500 nm) was then added to a tube containing 300 μl of resuspended cells. Trp or Tyr uptake was measured at 0, 2, 4, 6, and 8 min at room temperature (∼20 °C) by harvesting 50 μl aliquots by rapid filtration through Whatman® GF/C glass microfiber filters (GE Healthcare) on a vacuum filtration manifold (Merck Millipore). Filters were washed five times with 5 ml PBS, air dried over 2 h, and then subjected to scintillation counting (LS 6500 Multi-Purpose Scintillation Counter, Beckman Coulter). In all assays, initial uptake rate (v) was expressed as femtomole min−1 (1 × 106 cells)−1.
Treatment of THP-1 or HeLa cells with IFN-γ
THP-1 or HeLa cells were seeded at 1.5 × 105 cells/ml on CellBIND® Surface 35-mm cell culture dishes (Corning) or Falcon® Tissue Culture–treated 60-mm dishes (Corning) the day before experiments. The cells were treated with IFN-γ (100 units/ml) for 24 h at 37 °C.
Transfection of siRNA into HeLa cells and treatment of HeLa cells with IFN-γ
HeLa cells were seeded at a density of 7 × 104 cells/ml on Falcon® Tissue Culture–treated 60-mm dishes (Corning) the day before experiments. The siGENOME Non-targeting siRNA (Dharmacon, Lafayette, CO), siGENOME human TrpRS siRNA smart pool (Dharmacon), or siGENOME human IDO1 siRNA smart pool (Dharmacon) was transfected using Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instructions. The cells were incubated for 48 h after transfection prior to treatment with IFN-γ (100 units/ml) for 24 h at 37 °C.
Plasmid preparation
Human IDO1 cDNA clone was obtained from RIKEN BioResource Center (Ibaraki, Japan). B. subtilis TrpRS cDNA was synthesized using GeneArt® StringsTM DNA fragments service (Invitrogen). The nucleotide sequence of the cDNA coding for human FL TrpRS (amino acids 1–471), human mini TrpRS (amino acids 48–471), human T2 TrpRS (amino acids 94–471), B. subtilis TrpRS (amino acids 1–330), human IDO1 (amino acids 1–403), or human TyrRS (amino acids 1–528) was cloned into the eukaryotic pcDNA3.1/myc-His(−) B expression vector, which gives a gene product with a C-terminal tag of His6 residues. Moreover, cDNA fragments of human FL, mini, T2 TrpRS, and human TyrRS were separately cloned into the prokaryotic expression vector pET20b (Novagen, Madison, WI) so as to generate a gene product with a C-terminal tag of His6 residues (47, 48). A cDNA fragment of B. subtilis TrpRS was also cloned into the pET20b expression vector. Quik Change Site-Directed Mutagenesis system (Stratagene, La Jolla, CA) was used to introduce substitutions at specific sites. The identity of all final constructs were confirmed by DNA sequencing (Fasmac Co. Ltd., DNA sequencing services, Atsugi, Japan) to ensure no mistakes had been introduced during amplification.
Plasmid transfection into HeLa cells
HeLa cells were seeded at a density of 1.5 × 105 cells/ml on Falcon® Tissue Culture–treated 60-mm dishes (Corning) the day before experiments. The pcDNA3.1/myc-His(−) B expression vector or control vector (empty vector) was transfected using Lipofectamine® 2000 (Invitrogen) according to the manufacturer's instructions. The cells were collected 24 h after transfection.
Western blot analyses
The samples were separated through 12.0% SDS-polyacrylamide gels. Proteins were transferred onto Hybond-P PVDF membranes (GE Healthcare), which were then blocked with PBS containing 5% skim milk (Wako Pure Chemical Industries). The membranes were incubated for 1 h in PBS containing rabbit polyclonal antibodies against human mini TrpRS, which were prepared by custom polyclonal antibody production services (Takara Bio Inc., Shiga, Japan); the mouse mAb directed against His6 residues (Invitrogen); rabbit anti-IDO1 polyclonal antibodies (GeneTex, Irvine, CA); or mouse anti–β-actin mAb (Sigma-Aldrich). After washing, the membranes were incubated with an HRP-linked F(ab′)2 fragment of donkey anti-rabbit IgG or an HRP-linked whole antibody of sheep anti-mouse IgG (GE Healthcare Biosciences) for 1 h, respectively. The membranes were again washed three times with the buffer, and the proteins were visualized using ECLTM Western Blotting Detection Reagents (GE Healthcare Biosciences). Chemiluminescent signals were detected using a LAS-4000 mini luminescent image analyzer (GE Healthcare Biosciences).
Preparation of proteins
The pET20b expression constructs for His6-tagged human FL, mini, T2 TrpRS, B. subtilis TrpRS, and human TyrRS were introduced into E. coli BL21(DE3) (Novagen). The cells were grown at 37 °C to an A600 of ∼0.8 and then heterologous gene expression was induced by the addition of 0.4 mm isopropyl β-d-thiogalactopyranoside. Cells were harvested 4 h after induction. According to the procedures described by Novagen, the recombinant His6-tagged proteins were purified on a nickel affinity column (His·Bind® resin, Novagen) from the supernatant of lysed cells. Endotoxin was removed from the protein solutions by phase separation using Triton X-114 (64, 65). Protein concentration of human TrpRS, B. subtilis TrpRS, or human TyrRS was determined by using Bradford protein assay reagent (Bio-Rad Laboratories) and BSA (Sigma-Aldrich) as the standard. The protein concentration of human IDO1 was determined from the reported absorption coefficient of the Soret band at 404 nm (ϵ404 = 172 mm−1 cm−1) (66).
Aminoacylation assays
Aminoacylation activity was assayed at room temperature (∼20 °C) in buffer containing the following: 150 mm Tris-HCl, pH 7.5, 150 mm KCl, 10 mm MgCl2, 4 mm ATP, and 20 μm Trp (1 μm [3H]Trp) in the absence or presence of a competitor (200 μm). Prior to assays, Brewer's yeast tRNA was heated at 70 °C for 2 min and reannealed at the ambient temperature for 30 min. The reactions were initiated by adding the purified samples (100 nm) to the buffer that included Brewer's yeast tRNA (500 μm). At 0, 2, 4, and 6 min, reaction samples were removed and spotted onto Whatman No. 3MM paper filters. The filter discs were added to cold 5% TCA that included 2 mm Trp. The filters were washed three times in cold 5% TCA and 2 mm Trp, once in ethanol, and dried. The washed filters were then counted in a liquid scintillation counter (LS6500, Beckman Coulter).
Statistical analyses
Data were analyzed by one-way analysis of variance (ANOVA) followed by Tukey-Kramer post hoc tests.
Author contributions
M. M. and K. W. data curation; M. M., T. Y., and K. W. formal analysis; M. M. and K. W. funding acquisition; M. M., T. Y., and K. W. validation; M. M., T. Y., and K. W. investigation; M. M. and K. W. visualization; M. M., T. Y., and K. W. methodology; M. M., T. Y., and K. W. writing-review and editing; K. W. conceptualization; K. W. resources; K. W. software; K. W. supervision; K. W. writing-original draft; K. W. project administration.
Acknowledgments
The TrpRS-specific inhibitor, Trp-SA, was a gift from Drs. Tatsuo Yanagisawa and Shigeyuki Yokoyama from RIKEN Structural Biology Laboratory, Japan.
This work was supported in part by the Kurata Memorial Hitachi Science and Technology Foundation Kurata Grant (to K. W.), the Smoking Research Foundation research grant (to K. W.), and Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 21570129, 26440047, and 17K07329 (to K. W.) and 16J09853 (to M. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- MDM
- monocyte-derived macrophage
- DC
- dendritic cell
- FL
- full-length
- BCH
- 2-aminobicyclo-[2.2.1]heptane-2-carboxylic acid
- Trp-SA
- 5′-O-[N-(l-tryptophanyl)sulfamoyl]adenosine
- AhR
- aryl hydrocarbon receptor
- His6
- six histidine.
References
- 1. Munn D. H., Zhou M., Attwood J. T., Bondarev I., Conway S. J., Marshall B., Brown C., and Mellor A. L. (1998) Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281, 1191–1193 10.1126/science.281.5380.1191 [DOI] [PubMed] [Google Scholar]
- 2. Munn D. H., Shafizadeh E., Attwood J. T., Bondarev I., Pashine A., and Mellor A. L. (1999) Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 189, 1363–1372 10.1084/jem.189.9.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terness P., Bauer T. M., Röse L., Dufter C., Watzlik A., Simon H., and Opelz G. (2002) Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: Mediation of suppression by tryptophan metabolites. J. Exp. Med. 196, 447–457 10.1084/jem.20020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uyttenhove C., Pilotte L., Théate I., Stroobant V., Colau D., Parmentier N., Boon T., and Van den Eynde B. J. (2003) Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 9, 1269–1274 10.1038/nm934 [DOI] [PubMed] [Google Scholar]
- 5. Nathan C. F., Murray H. W., Wiebe M. E., and Rubin B. Y. (1983) Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158, 670–689 10.1084/jem.158.3.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor M. W., and Feng G. (1991) Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 5, 2516–2522 10.1096/fasebj.5.11.1907934 [DOI] [PubMed] [Google Scholar]
- 7. Shimizu T., Nomiyama S., Hirata F., and Hayaishi O. (1978) Indoleamine 2,3-dioxygenase. Purification and some properties. J. Biol. Chem. 253, 4700–4706 [PubMed] [Google Scholar]
- 8. Littlejohn T. K., Takikawa O., Truscott R. J., and Walker M. J. (2003) Asp274 and His346 are essential for heme binding and catalytic function of human indoleamine 2,3-dioxygenase. J. Biol. Chem. 278, 29525–29531 10.1074/jbc.M301700200 [DOI] [PubMed] [Google Scholar]
- 9. Sugimoto H., Oda S., Otsuki T., Hino T., Yoshida T., and Shiro Y. (2006) Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 103, 2611–2616 10.1073/pnas.0508996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwidzinski E., Bunse J., Aktas O., Richter D., Mutlu L., Zipp F., Nitsch R., and Bechmann I. (2005) Indolamine 2,3-dioxygenase is expressed in the CNS and down-regulates autoimmune inflammation. FASEB J. 19, 1347–1349 10.1096/fj.04-3228fje [DOI] [PubMed] [Google Scholar]
- 11. Segawa H., Fukasawa Y., Miyamoto K., Takeda E., Endou H., and Kanai Y. (1999) Identification and functional characterization of a Na+-independent neutral amino acid transporter with broad substrate selectivity. J. Biol. Chem. 274, 19745–19751 10.1074/jbc.274.28.19745 [DOI] [PubMed] [Google Scholar]
- 12. Yanagida O., Kanai Y., Chairoungdua A., Kim D. K., Segawa H., Nii T., Cha S. H., Matsuo H., Fukushima J., Fukasawa Y., Tani Y., Taketani Y., Uchino H., Kim J. Y., Inatomi J., Okayasu I., Miyamoto K., Takeda E., Goya T., and Endou H. (2001) Human L-type amino acid transporter 1 (LAT1): Characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta 1514, 291–302 10.1016/S0005-2736(01)00384-4 [DOI] [PubMed] [Google Scholar]
- 13. del Amo E. M., Urtti A., and Yliperttula M. (2008) Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 35, 161–174 10.1016/j.ejps.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 14. Kudo Y., Boyd C. A. R., Sargent I. L., and Redman C. W. G. (2003) Decreased tryptophan catabolism by placental indoleamine 2,3-dioxygenase in preeclampsia. Am. J. Obstet. Gynecol. 188, 719–726 10.1067/mob.2003.156 [DOI] [PubMed] [Google Scholar]
- 15. Seymour R. L., Ganapathy V., Mellor A. L., and Munn D. H. (2006) A high-affinity, tryptophan-selective amino acid transport system in human macrophages. J. Leukoc. Biol. 80, 1320–1327 10.1189/jlb.1205727 [DOI] [PubMed] [Google Scholar]
- 16. Silk J. D., Lakhal S., Laynes R., Vallius L., Karydis I., Marcea C., Boyd C. A., and Cerundolo V. (2011) IDO induces expression of a novel tryptophan transporter in mouse and human tumor cells. J. Immunol. 187, 1617–1625 10.4049/jimmunol.1000815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhutia Y. D., Babu E., and Ganapathy V. (2015) Interferon-γ induces a tryptophan-selective amino acid transporter in human colonic epithelial cells and mouse dendritic cells. Biochim. Biophys. Acta 1848, 453–462 10.1016/j.bbamem.2014.10.021 [DOI] [PubMed] [Google Scholar]
- 18. Kudo Y., Boyd C. A. R., Sargent I. L., and Redman C. W. G. (2001) Tryptophan degradation by human placental indoleamine 2,3-dioxygenase regulates lymphocyte proliferation. J. Physiol. 535, 207–215 10.1111/j.1469-7793.2001.00207.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fleckner J., Rasmussen H. H., and Justesen J. (1991) Human interferon γ potently induces the synthesis of a 55-kDa protein (γ2) highly homologous to rabbit peptide chain release factor and bovine tryptophanyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 88, 11520–11524 10.1073/pnas.88.24.11520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubin B. Y., Anderson S. L., Xing L., Powell R. J., and Tate W. P. (1991) Interferon induces tryptophanyl-tRNA synthetase expression in human fibroblasts. J. Biol. Chem. 266, 24245–24248 [PubMed] [Google Scholar]
- 21. Kisselev L., Frolova L., and Haenni A. L. (1993) Interferon inducibility of mammalian tryptophanyl-tRNA synthetase: New perspectives. Trends Biochem. Sci. 18, 263–267 10.1016/0968-0004(93)90178-P [DOI] [PubMed] [Google Scholar]
- 22. Reano A., Richard M. H., Denoroy L., Viac J., Benedetto J. P., and Schmitt D. (1993) Gamma interferon potently induces tryptophanyl-tRNA synthetase expression in human keratinocytes. J. Invest. Dermatol. 100, 775–779 10.1111/1523-1747.ep12476463 [DOI] [PubMed] [Google Scholar]
- 23. Fleckner J., Martensen P. M., Tolstrup A. B., Kjeldgaard N. O., and Justesen J. (1995) Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine 7, 70–77 10.1006/cyto.1995.1009 [DOI] [PubMed] [Google Scholar]
- 24. Shaw A. C., Røssel Larsen M., Roepstorff P., Justesen J., Christiansen G., and Birkelund S. (1999) Mapping and identification of interferon gamma-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis 20, 984–993 10.1002/(SICI)1522-2683(19990101)20:4/5%3C984::AID-ELPS984%3E3.0.CO%3B2-R [DOI] [PubMed] [Google Scholar]
- 25. Liu J., Shue E., Ewalt K. L., and Schimmel P. (2004) A new γ-interferon-inducible promoter and splice variants of an anti-angiogenic human tRNA synthetase. Nucleic Acids Res. 32, 719–727 10.1093/nar/gkh240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schimmel P. (1987) Aminoacyl-tRNA synthetases: General scheme of structure-functional relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 56, 125–158 10.1146/annurev.bi.56.070187.001013 [DOI] [PubMed] [Google Scholar]
- 27. Schimmel P. R., and Söll D. (1979) Aminoacyl-tRNA synthetases: General features and recognition of transfer RNAs. Annu. Rev. Biochem. 48, 601–648 10.1146/annurev.bi.48.070179.003125 [DOI] [PubMed] [Google Scholar]
- 28. Wakasugi K., and Schimmel P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 10.1126/science.284.5411.147 [DOI] [PubMed] [Google Scholar]
- 29. Wakasugi K., and Schimmel P. (1999) Highly differentiated motifs responsible for two cytokine activities of a split human tRNA synthetase. J. Biol. Chem. 274, 23155–23159 10.1074/jbc.274.33.23155 [DOI] [PubMed] [Google Scholar]
- 30. Wakasugi K., Slike B. M., Hood J., Ewalt K. L., Cheresh D. A., and Schimmel P. (2002) Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J. Biol. Chem. 277, 20124–20126 10.1074/jbc.C200126200 [DOI] [PubMed] [Google Scholar]
- 31. Wakasugi K., Slike B. M., Hood J., Otani A., Ewalt K. L., Friedlander M., Cheresh D. A., and Schimmel P. (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 173–177 10.1073/pnas.012602099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X. L., Schimmel P., and Ewalt K. L. (2004) Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem. Sci. 29, 250–256 10.1016/j.tibs.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 33. Zhou Q., Kapoor M., Guo M., Belani R., Xu X., Kiosses W. B., Hanan M., Park C., Armour E., Do M. H., Nangle L. A., Schimmel P., and Yang X. L. (2010) Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 17, 57–61 10.1038/nsmb.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sajish M., Zhou Q., Kishi S., Valdez D. M. Jr., Kapoor M., Guo M., Lee S., Kim S., Yang X. L., and Schimmel P. (2012) Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 8, 547–554 10.1038/nchembio.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krause S. W., Rehli M., Kreutz M., Schwarzfischer L., Paulauskis J. D., and Andreesen R. (1996) Differential screening identifies genetic markers of monocyte to macrophage maturation. J. Leukoc. Biol. 60, 540–545 10.1002/jlb.60.4.540 [DOI] [PubMed] [Google Scholar]
- 36. Matsunaga T., Ishida T., Takekawa M., Nishimura S., Adachi M., and Imai K. (2002) Analysis of gene expression during maturation of immature dendritic cells derived from peripheral blood monocytes. Scand. J. Immunol. 56, 593–601 10.1046/j.1365-3083.2002.01179.x [DOI] [PubMed] [Google Scholar]
- 37. Tolstrup A. B., Bejder A., Fleckner J., and Justesen J. (1995) Transcriptional regulation of the interferon-γ-inducible tryptophanyl-tRNA synthetase includes alternative splicing. J. Biol. Chem. 270, 397–403 10.1074/jbc.270.1.397 [DOI] [PubMed] [Google Scholar]
- 38. Turpaev K. T., Zakhariev V. M., Sokolova I. V., Narovlyansky A. N., Amchenkova A. M., Justesen J., and Frolova L. Y. (1996) Alternative processing of the tryptophanyl-tRNA synthetase mRNA from interferon-treated human cells. Eur. J. Biochem. 240, 732–737 10.1111/j.1432-1033.1996.0732h.x [DOI] [PubMed] [Google Scholar]
- 39. Miyanokoshi M., Tanaka T., Tamai M., Tagawa Y., and Wakasugi K. (2013) Expression of the rodent-specific alternative splice variant of tryptophanyl-tRNA synthetase in murine tissues and cells. Sci. Rep. 3, 3477 10.1038/srep03477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Otani A., Slike B. M., Dorrell M. I., Hood J., Kinder K., Ewalt K. L., Cheresh D., Schimmel P., and Friedlander M. (2002) A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 178–183 10.1073/pnas.012601899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kisselev L. L., Favorova O. O., and Kovaleva G. K. (1979) Tryptophanyl-tRNA synthetase from beef pancreas. Methods Enzymol. 59, 234–257 10.1016/0076-6879(79)59087-9 [DOI] [PubMed] [Google Scholar]
- 42. Ewalt K. L., Yang X. L., Otero F. J., Liu J., Slike B., and Schimmel P. (2005) Variant of human enzyme sequesters reactive intermediate. Biochemistry 44, 4216–4221 10.1021/bi048116l [DOI] [PubMed] [Google Scholar]
- 43. Yang X. L., Otero F. J., Ewalt K. L., Liu J., Swairjo M. A., Köhrer C., RajBhandary U. L., Skene R. J., McRee D. E., and Schimmel P. (2006) Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 25, 2919–2929 10.1038/sj.emboj.7601154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X. L., Guo M., Kapoor M., Ewalt K. L., Otero F. J., Skene R. J., McRee D. E., and Schimmel P. (2007) Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure 15, 793–805 10.1016/j.str.2007.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frolova L. Y., Sudomoina M. A., Grigorieva A. Y., Zinovieva O. L., and Kisselev L. L. (1991) Cloning and nucleotide sequence of the structural gene encoding for human tryptophanyl-tRNA synthetase. Gene 109, 291–296 10.1016/0378-1119(91)90624-K [DOI] [PubMed] [Google Scholar]
- 46. Johnson J. D., Spellman J. M., White K. H., Barr K. K., and John T. R. (2002) Human tryptophanyl-tRNA synthetase can efficiently complement the Saccharomyces cerevisiae homologue, Wrs1P. FEMS Microbiol. Lett. 216, 111–115 10.1111/j.1574-6968.2002.tb11423.x [DOI] [PubMed] [Google Scholar]
- 47. Wakasugi K., Nakano T., and Morishima I. (2005) Oxidative stress-responsive intracellular regulation specific for the angiostatic form of human tryptophanyl-tRNA synthetase. Biochemistry 44, 225–232 10.1021/bi048313k [DOI] [PubMed] [Google Scholar]
- 48. Wakasugi K. (2007) Human tryptophanyl-tRNA synthetase binds with heme to enhance its aminoacylation activity. Biochemistry 46, 11291–11298 10.1021/bi7012068 [DOI] [PubMed] [Google Scholar]
- 49. Wakasugi K. (2010) An exposed cysteine residue of human angiostatic mini tryptophanyl-tRNA synthetase. Biochemistry 49, 3156–3160 10.1021/bi1000239 [DOI] [PubMed] [Google Scholar]
- 50. Wakasugi K. (2010) Species-specific differences in the regulation of the aminoacylation activity of mammalian tryptophanyl-tRNA synthetases. FEBS Lett. 584, 229–232 10.1016/j.febslet.2009.11.073 [DOI] [PubMed] [Google Scholar]
- 51. Nakamoto T., Miyanokoshi M., Tanaka T., and Wakasugi K. (2016) Identification of a residue crucial for the angiostatic activity of human mini tryptophanyl-tRNA synthetase by focusing on its molecular evolution. Sci. Rep. 6, 24750 10.1038/srep24750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu F., Chen X., Xin L., Chen L., Jin Y., and Wang D. (2001) Species-specific differences in the operational RNA code for aminoacylation of tRNATrp. Nucleic Acids Res. 29, 4125–4133 10.1093/nar/29.20.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doublié S., Bricogne G., Gilmore C., and Carter C. W. Jr. (1995) Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure 3, 17–31 10.1016/S0969-2126(01)00132-0 [DOI] [PubMed] [Google Scholar]
- 54. Ribas de Pouplana L., Frugier M., Quinn C. L., and Schimmel P. (1996) Evidence that two present-day components needed for the genetic code appeared after nucleated cells separated from eubacteria. Proc. Natl. Acad. Sci. U.S.A. 93, 166–170 10.1073/pnas.93.1.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brown J. R., Robb F. T., Weiss R., and Doolittle W. F. (1997) Evidence for the early divergence of tryptophanyl- and tyrosyl-tRNA synthetase. J. Mol. Evol. 45, 9–16 10.1007/PL00006206 [DOI] [PubMed] [Google Scholar]
- 56. Yang X. L., Skene R. J., McRee D. E., and Schimmel P. (2002) Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl. Acad. Sci. U.S.A. 99, 15369–15374 10.1073/pnas.242611799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang X. L., Otero F. J., Skene R. J., McRee D. E., Schimmel P., and Ribas de Pouplana L. (2003) Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl. Acad. Sci. U.S.A. 100, 15376–15380 10.1073/pnas.2136794100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kise Y., Lee S. W., Park S. G., Fukai S., Sengoku T., Ishii R., Yokoyama S., Kim S., and Nureki O. (2004) A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat. Struct. Mol. Biol. 11, 149–156 10.1038/nsmb722 [DOI] [PubMed] [Google Scholar]
- 59. Ewalt K. L., and Schimmel P. (2002) Activation of angiogenic signaling pathways by two human tRNA synthetases. Biochemistry 41, 13344–13349 10.1021/bi020537k [DOI] [PubMed] [Google Scholar]
- 60. Paley E. L. (2011) Tryptamine-induced tryptophanyl-tRNAtrp deficiency in neurodifferentiation and neurodegeneration interplay: Progenitor activation with neurite growth terminated in Alzheimer's disease neuronal vesicularization and fragmentation. J. Alzheimers Dis. 26, 263–298 10.3233/JAD-2011-110176 [DOI] [PubMed] [Google Scholar]
- 61. Kapoor M., Zhou Q., Otero F., Myers C. A., Bates A., Belani R., Liu J., Luo J. K., Tzima E., Zhang D. E., Yang X. L., and Schimmel P. (2008) Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 283, 2070–2077 10.1074/jbc.M706028200 [DOI] [PubMed] [Google Scholar]
- 62. Lee C. W., Chang K. P., Chen Y. Y., Liang Y., Hsueh C., Yu J. S., Chang Y. S., and Yu C. J. (2015) Overexpressed tryptophanyl-tRNA synthetase, an angiostatic protein, enhances oral cancer cell invasiveness. Oncotarget 6, 21979–21992 10.18632/oncotarget.4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ahn Y. H., Park S., Choi J. J., Park B. K., Rhee K. H., Kang E., Ahn S., Lee C. H., Lee J. S., Inn K. S., Cho M. L., Park S. H., Park K., Park H. J., Lee J. H., et al. (2016) Secreted tryptophanyl-tRNA synthetase as a primary defense system against infection. Nat. Microbiol. 2, 16191 10.1038/nmicrobiol.2016.191 [DOI] [PubMed] [Google Scholar]
- 64. Aida Y., and Pabst M. J. (1990) Removal of endotoxin from protein solutions by phase separation using Triton X-114. J. Immunol. Methods 132, 191–195 10.1016/0022-1759(90)90029-U [DOI] [PubMed] [Google Scholar]
- 65. Liu S., Tobias R., McClure S., Styba G., Shi Q., and Jackowski G. (1997) Removal of endotoxin from recombinant protein preparations. Clin. Biochem. 30, 455–463 10.1016/S0009-9120(97)00049-0 [DOI] [PubMed] [Google Scholar]
- 66. Papadopoulou N. D., Mewies M., McLean K. J., Seward H. E., Svistunenko D. A., Munro A. W., and Raven E. L. (2005) Redox and spectroscopic properties of human indoleamine 2,3-dioxygenase and a His303Ala variant: Implications for catalysis. Biochemistry 44, 14318–14328 10.1021/bi0513958 [DOI] [PubMed] [Google Scholar]