Abstract
High-mobility group box 1 (HMGB1) is a chromatin-associated protein that, in response to stress or injury, translocates from the nucleus to the extracellular milieu, where it functions as an alarmin. HMGB1's function is in part determined by the complexes (HMGB1c) it forms with other molecules. However, structural modifications in the HMGB1 polypeptide that may regulate HMGB1c formation have not been previously described. In this report, we observed high-molecular weight, denaturing-resistant HMGB1c in the plasma and peripheral blood mononuclear cells of individuals with systemic lupus erythematosus (SLE) and, to a much lesser extent, in healthy subjects. Differential HMGB1c levels were also detected in mouse tissues and cultured cells, in which these complexes were induced by endotoxin or the immunological adjuvant alum. Of note, we found that HMGB1c formation is catalyzed by the protein–cross-linking enzyme transglutaminase-2 (TG2). Cross-link site mapping and MS analysis revealed that HMGB1 can be cross-linked to TG2 as well as a number of additional proteins, including human autoantigens. These findings have significant functional implications for studies of cellular stress responses and innate immunity in SLE and other autoimmune disease.
Keywords: protein cross-linking, transglutaminase, lipopolysaccharide (LPS), calcium, autoimmune disease, autoimmunity, high-mobility group box 1, systemic lupus erythematosus
Introduction
HMGB1 is a nonhistone chromatin-associated protein that undergoes a number of post-translational modifications that determine cellular localization and functional properties (reviewed in Refs. 1, 2). Under basal conditions, HMGB1 resides in the nucleus of most cell types as a DNA-binding protein important for the regulation of chromatin architecture (3). In response to stress, HMGB1 translocates to the cytosol, where it plays a role in innate immunity as a cytosolic sensor and is secreted as an endogenous danger signal (4, 5). The diverse functions of extranuclear HMGB1 are in part dependent on the complexes it forms with other molecules (6). HMGB1 has a remarkable ability to form complexes with proinflammatory molecules, including LPS4 (7), bacterial DNA, (8), and endogenous factors such as IL-1β (9) and nucleosomes (10). This is underscored by the emerging role of HMGB1 in rheumatic diseases such as systemic lupus erythematosus (SLE), where the presence of HMGB1 in complex with endogenous nuclear components such as dsDNA or nucleosomes has been shown to play an important role in breaking immune tolerance against nuclear antigens (10). Although noncovalent interaction of HMGB1 with other molecules to form complexes is well-established, structural modifications in the HMGB1 polypeptide that may regulate such interaction have not been described.
Transglutaminase 2 (TG2) is a member of the protein glutamine γ-glutamyltransferase enzyme family (EC 2.3.2.13) that catalyzes the calcium-dependent cross-linking of protein targets. TG2-mediated cross-linking occurs via a transamidation reaction between γ-carboxamide groups of peptide-bound glutamines (glutamine donors) and ϵ-amino groups of peptide-bound lysines (lysine donors), resulting in the formation of ϵ-(γ-glutamyl) lysine isopeptide bonds (11–13). Because TG2 is a prominent stress response enzyme and the primary autoantigen in celiac disease (14), TG2-mediated protein complex formation has been investigated as a possible mechanism by which immune tolerance can be broken to self-molecules.
Here we note the presence of high-molecular weight HMGB1 protein complexes (designated HMGB1c hereafter) that were enriched in plasma and PBMCs from SLE patients. These complexes were partially resistant to reducing agents, which was suggestive of covalent HMGB1 binding that was independent of disulfide bonding. Subsequent studies revealed that these HMGB1c form via TG2 transamidation activity, demonstrating that HMGB1 is a TG2–cross-linking substrate. Moreover, TG2 mediated HMGB1c formation in response to stimulation with LPS or the adjuvant alum. Some cross-linking sites are mapped to nuclear localization signal (NLS) regions of the HMGB1 polypeptide, and constituents of in vitro cross-linked HMGB1c included a number of autoantigens. These results identify and characterize, for the first time, HMGB1-TG2 interaction as well as the presence of TG2-dependent HMGB1c, with significant implications for cellular stress responses, innate immunity, and autoimmune disease.
Results
High-molecular weight HMGB1 complexes are present in plasma from systemic lupus erythematosus patients
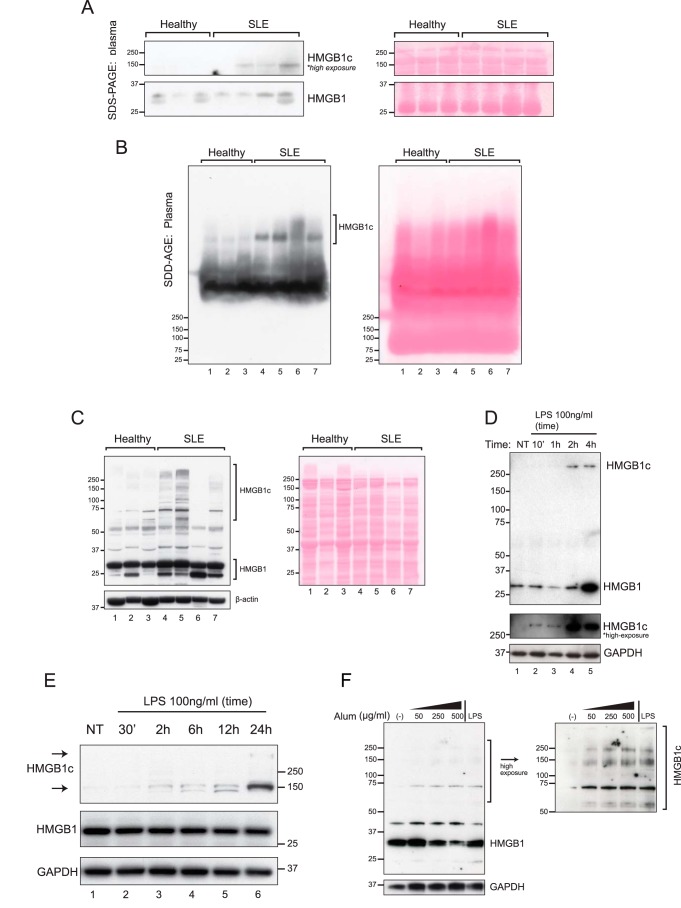
Elevated levels of HMGB1 have been described in SLE (15). In this study, we further demonstrated the presence of HMGB1 in large protein complexes in SLE patients. In standard SDS-PAGE followed by Western blot analysis, the plasma levels of the prototypical 29-kDa HMGB1 varied slightly among individual samples, but there were no significant differences observed between SLE patients and healthy controls (Fig. 1A, bottom panels). However, an additional and larger HMGB1 signal of ∼150 kDa was observed in three of four SLE patients but not in all three controls (Fig. 1A, top panels, and Fig. S1). To further characterize the larger HMGB1 species, next we performed semidenaturing detergent–agarose gel electrophoresis (SDD-AGE), a well-established technique for separating large, SDS-denaturing resistant protein complexes (16, 17). SDD-AGE followed by Western blotting resolved two distinct patterns of the high-molecular weight HMGB1 complex: a faster-migrating complex that was uniformly observed in all plasma samples and a lupus-associated slower-migrating complex (Fig. 1B). Notably, the 29-kDa and the 150-kDa HMGB1 signals seen with SDS-PAGE were not detected by SDD-AGE. Because staining of the membrane showed a lack of proteins in the immediate regions of 25 kDa, the 29-kDa HMGB1 species could have been lost by diffusion. Alternately, under the less stringent condition of SDD-AGE, the partially unfolded 29-kDa HMGB1 could migrate aberrantly or, more likely, remain associated with its interacting proteins and yielded signals with retarded gel mobility. Both SDS-PAGE and SDD-AGE analysis show that HMGB1 circulates in the blood as large protein complexes and that the levels of such complexes are significantly higher in SLE patients than in healthy subjects. A similar pattern of HMGB1 expression was observed in the corresponding PBMCs: near-uniform expression of the 29-kDa HMGB1 in all samples but an enrichment of slower-migrating HMGB1c bands spanning from 75 to 250 kDa in three of four SLE patients (Fig. 1C). Although SDD-AGE proved useful for separating large complexes that are likely too large to enter SDS-PAGE gels, HMGB1c detected under the conditions of SDD-AGE could involve covalent as well as noncovalent linkages. Because we were interested in mechanisms that could catalyze the novel, covalently linked form of HMGB1 complexes, standard SDS-PAGE (that is, the presence of a reducing agent and heating of the samples before gel loading) was employed for subsequent experiments.
Figure 1.
High-molecular weight HMGB1 complexes are present in plasma and PBMCs from SLE patients. A, SDS-PAGE followed by immunoblotting with HMGB1 antibodies compared HMGB1 in plasma samples of healthy individuals (n = 3) and SLE patients (n = 4). HMGB1 (29 kDa) and high-molecular weight HMGB1 complexes (HMGB1c) are depicted as lower and higher exposures from the same blot (full blot exposures are shown in Fig. S1). The protein membrane was initially stained with Ponceau S to show protein loading (right panel). B, the plasma samples in A separated on SDD-AGE gels followed by HMGB1 immunoblot showed SLE-associated HMGB1c. The protein membrane stained with Ponceau S showed comparable protein loading (right panel). C, immunoblot with SDS-PAGE showed that HMGB1c was enriched in PBMCs from three of four SLE patients compared with healthy controls. Incubating the membrane with β-actin and initially staining with Ponceau S showed comparable protein loading. D, induction of HMGB1 and HMGB1c by LPS in PBMCs. Shown is an immunoblot of protein lysates of human PBMCs stimulated with vehicle (control) or with LPS (100 ng/ml) at the time interval indicated at the top of each lane and probed with HMGB1 antibody (top panel). A longer exposure of the HMGB1c region of the gel is depicted in the center panel, with GAPDH in the bottom panel. E, HMGB1c is induced by LPS stimulation in RAW 264.7 cells. Cells were stimulated with vehicle (NT) or LPS (100 ng/ml) for the times indicated and lysed in a hypotonic buffer to obtain cytosolic extracts. Immunoblots of protein lysates reveal the time-dependent increase in cytosolic HMGB1c, with the appearance of an additional, slower-migrating HMGB1c at the top of the gel (lane 6). F, alum induces HMGB1c in a concentration-dependent manner in the mouse macrophage-like cell line RAW 264.7. Shown is an immunoblot analysis of total cell lysates incubated with different concentrations of alum, shown on the top of each lane, for 6 h. As a positive control, cells were stimulated with LPS (100 ng/ml) for 24 h. Data shown in D--F are representative of three to four independent experiments with similar results.
Immunoblotting of unstimulated PBMC protein lysates from healthy subjects revealed the presence of a single 29-kDa HMGB1 band (Fig. 1D, lane 1). In contrast, LPS increased p29 HMGB1 levels after 4 h and also induced the formation of an HMGB1c migrating in excess of the 250-kDa size marker at 10 min and peaking at 2 h (Fig. 1D, lanes 2–5). Next we stimulated RAW264.7 cells, a mouse macrophage-like cell line, with LPS and found that HMGB1c is also induced in this cell line in a time-dependent manner. Low levels of a 150-kDa HMGB1c were present in control cells (Fig. 1E, lane 1) but increased over time, resulting in a substantial increase the p150 HMGB1c and appearance of a slower-migrating band near the top of the gel (Fig. 1E, lane 6).
LPS is a well-known activator of the NLRP3 inflammasome through TLR4 ligation, which also is induced through an reactive oxygen species–dependent mechanism (18, 19). Remarkably the NLRP-3 agonist alum (20) induced the formation of HMGB1c in a concentration-dependent manner in RAW 264.7 cells (Fig. 1F). Formation of HMGB1c despite exposure to heat and reducing agents was suggestive of disulfide-independent covalent bonding. Moreover, induction of HMGB1c described above by LPS and alum in cells suggested that the formation of these putative covalent linkages is induced in response to an inflammatory state.
LPS induces HMGB1c in vivo
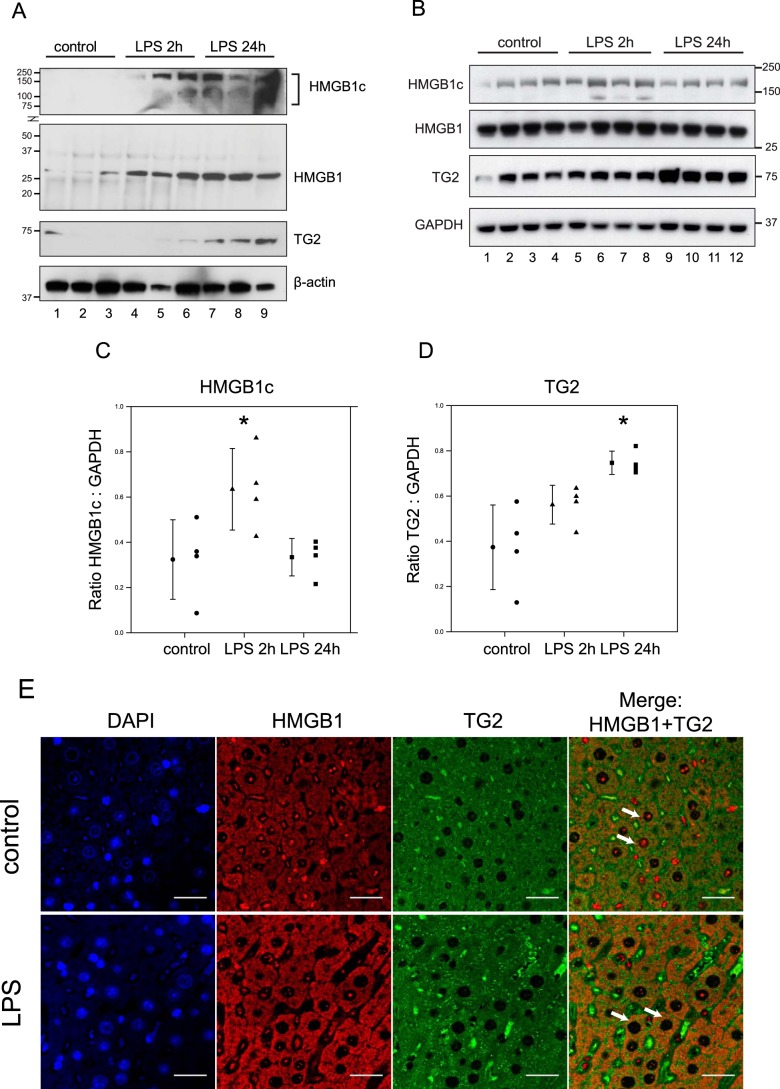
Given that LPS induced the formation of HMGB1c and is known to increase calcium flux (21) and raise intracellular reactive oxygen species levels (22), we hypothesized that HMGB1c formation may be driven by the reactive oxygen species- and calcium-activated protein–cross-linking enzyme transglutaminase 2 (TG2). TG2 mediates cross-linking via a protein transamidation reaction between an ϵ-amino group of a lysine residue (designated as Lys donor hereafter) and a γ-carboxamide group of glutamine residue (Gln donor). Because TG2 expression and cross-linking activity have been shown to be induced by LPS in rodent models (23), we first examined HMGB1 and TG2 expression in spleens and livers at 2 h and 24 h after intraperitoneal injection of LPS (2 mg/kg) into mice (n = 3–4) by Western blot analysis. Minimal amounts of HMGB1, HMGB1c, and TG2 were detected in spleens of unstimulated mice (Fig. 2A, lanes 1–3). However, LPS significantly increased the expression of HMGB1 and HMGB1c at 2 h and 24 h post-injection (Fig. 2A, lanes 4–9). In the liver, in addition to the 29-kDa HMGB1 band, modest amounts of HMGB1c were observed in unstimulated mice (Fig. 2B, lanes 1–4). The expression of HMGB1c was significantly induced by LPS at 2 h but returned to near basal levels at 24 h (Fig. 2, B, lanes 5-12, and C). Consistent with a previous report (23), TG2 levels in the liver were increased 24 h after LPS administration (Fig. 2, B and D). Similarly, immunohistochemistry followed by confocal microscopy showed that stimulation with LPS for 2 h led to translocation of HMGB1 from the nucleus to the cytosol, where it exhibited increased co-localization with TG2 (Fig. 2E). These observations are in agreement with previous reports showing that LPS induces the translocation of HMGB1 from the nucleus to the cytosol (24).
Figure 2.
Induction of HMGB1c and TG2 by LPS in mice and HMGB1c by alum in monocytes. LPS (2 mg/kg) was injected intraperitoneally into BALB/c mice (n = 3–4/group). A–D, tissues, spleens (A), and livers (B–D) were harvested at 0, 2, and 24 h after LPS injection, lysed, and subjected to immunoblot analysis using the indicated antibodies. C–E, the HMGB1c signal intensities in B were normalized to GAPDH and are depicted as scatterplots alongside sample mean with standard deviation error bars (*, p = 0.02 for C; *, p = 0.004 for D). E, immunohistochemistry followed by confocal microscopy of mouse liver after 2-h stimulation showed LPS-induced HMGB1 nuclear-to-cytosolic translocation, where it showed apparent colocalization with TG2 (scale bars = 30 μm). DAPI, 4′,6-diamidino-2-phenylindole.
HMGB1c formation is mediated by TG2 transamidation activity
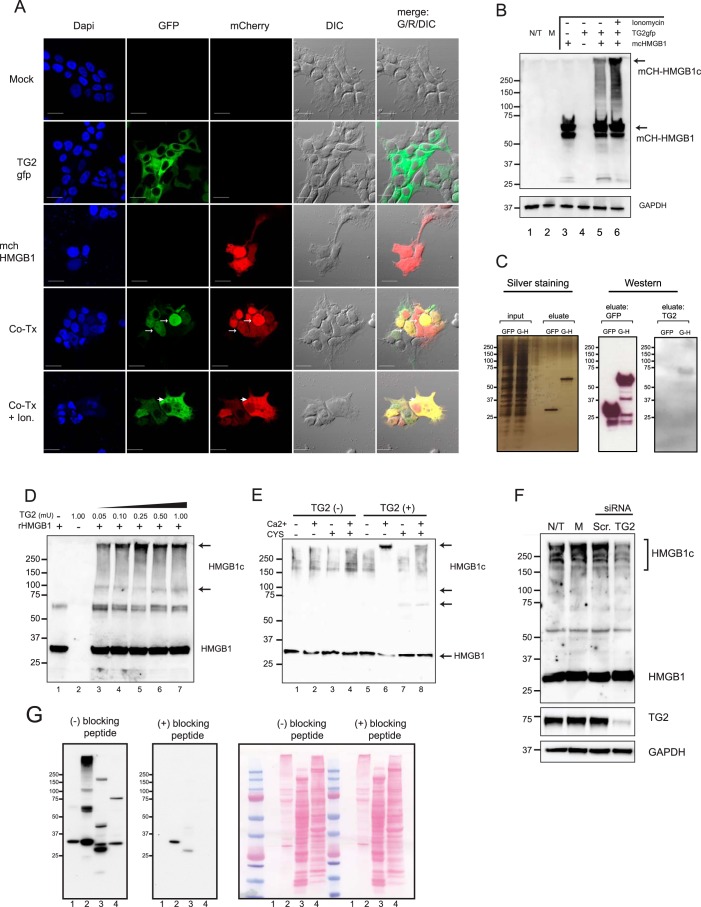
To test the hypothesis that the formation of HMGB1c is mediated by TG2, we first transfected human embryonic kidney 293T cells with expression vectors for HMGB1, TG2, or both. For the ease of monitoring by microscopy, these recombinant proteins were tagged with a fluorochrome: mCherry for HMGB1 (mch-HMGB1, red fluorescence) and GFP for TG2 (TG2-GFP, green fluorescence). Twenty-four hours after transfection, imaging with confocal microscopy revealed that, in individual transfections, mch-HMGB1 was mainly localized in the nucleus, whereas TG2-GFP was in the cytosol (Fig. 3A). In co-transfection experiments, however, TG2-GFP was also observed in the nucleus and co-localized with mch-HGBM1 (Fig. 3A, arrows). Furthermore, when co-transfected cells were stimulated with ionomycin, an agent that increases intracellular Ca2+ concentrations and activates TG2, mch-HMGB1 was observed in the cytosol, co-localizing with TG2-GFP (Fig. 3A, arrowheads). These data suggest that HMGB1 and TG2 are interacting, as they can affect the subcellular localization of each other in transient transfection experiments. Formation of HMGB1c in these transfection experiments was demonstrated by Western blot analysis. Using total cell lysates, mCherry antibodies yielded a signal of ∼58 kDa from mch-HMGB1 singly transfected cells, the expected size of mCherry fused to HMGB1 (Fig. 3B, lane 3). A slightly faster-migrating species, most likely a partial degradation product of mch-HMGB1, was also observed. However, no signals with slower gel mobility were observed. In contrast, co-transfecting mch-HMGB1 with TG2-GFP yielded slower-migrating mch-HMGB1c, whose formation was further induced by ionomycin (Fig. 3B). We conclude that TG2 was sufficient to induce the formation of HMGB1c, which was further stimulated by ionomycin.
Figure 3.
TG2 is associated with HMGB1 and catalyzes the formation of HMGB1c. A and B, human kidney 293T cells were transfected with plasmid constructs for mCherry-HMGB1 (mch-HMGB1), TG2-GFP, or both (Co-Tx) and then analyzed by confocal microscopy at 24 h (A) and by Western blotting of total cell lysates (B). In some samples, transfected cells were incubated with ionomycin (Ion, 5 μm for 90 min) before analysis. R, red; G, green; DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole. N/T, not transfected. C, co-immunoprecipitation experiments showed interaction of HMGB1 and TG2. 293T cells were transfected with a construct for GFP-HMGB1 (G-H) or vector control (GFP only) and harvested at 24 h. Total cell lysates were incubated with GFP antibodies coupled to magnetic agarose beads for 30 min and washed. Left panel, silver staining of input and eluates resolved by SDS-PAGE. Immunoblots were performed with eluates using GFP antibodies (center panel) and TG2 antibodies (right panel). D, recombinant HMGB1 is a TG2 substrate. rHMGB1, a His6-HMGB1 fusion protein produced in E. coli, was incubated with varying concentrations of porcine TG2 for 60 min at 37 °C in reaction buffers supplemented with Ca2+ (5 mm). The reactions were terminated by addition of SDS loading buffer and subjected to Western blotting with HMGB1 antibodies. E, endogenous HMGB1 is a TG2 substrate. Total RAW 264.7 cell lysates were incubated with TG2 (0.2 milliunits/μl) with or without Ca2+ (5 mm) and cystamine (10 mm), as indicated, for 1 h and then subjected to immunoblotting with HMGB1 antibodies. Western blot analysis showed that the activation of TG2 by Ca2+ induced, whereas inhibition of TG2 by cystamine inhibited, the formation of HMGB1c. F, knockdown of TG2 by siRNA inhibited the formation of HMGB1c. Human pulmonary fibroblasts were transfected with TG2-targeted siRNA (TG2, 25 nm) or scramble control (Scr, 100 nm). After 72 h, these cells, together with untransfected or mock-transfected (M) cells exposed to the transfection reagent only, were harvested and analyzed by Western blotting using HMGB1 antibodies. G, confirmation of HMGB1 antibody specificity for HMGB1c with a blocking peptide. HMGB1 immunoblots were incubated in the absence (left panel) or presence (right panel) of a synthetic peptide used to raise the HMGB1 antibody. (lane 1, recombinant HMGB1; lane 2, HMGB1c catalyzed by the addition of recombinant HMGB1 to a cell lysate; lanes 3 and 4, endogenous HMGB1 and HMGB1c from mouse liver (lane 3) and human pulmonary fibroblast cell lysates (lane 4). Data shown are representative of three or more independent experiments.
Next we demonstrated that HMGB1 interacted with TG2 by co-immunoprecipitation experiment. Constructs for enhanced GFP-HMGB1 or the vector control (expressing GFP only) were transfected into 293T cells. Subsequently, total lysates were subjected to immunoprecipitation using GFP-Trap coupled to magnetic agarose beads. Silver staining of the protein gel showed that GFP was 27 kDa, whereas GFP-HMGB1 was 53 kDa (Fig. 3C). In Western blotting, both recombinant proteins reacted with GFP antibodies. However, only the eluate from the GFP-HMGB1 sample but not the GFP sample yielded an appropriate TG2 signal of 75 kDa with TG2 antibodies. Hence, the result of the immunoprecipitation experiment showed that HMGB1 interacts with TG2.
To demonstrate that TG2 mediates the formation of HMGB1c, we incubated recombinant HMGB1 (rHMGB1, His6 tag fused to human recombinant HMGB1 produced in Escherichia coli) with porcine liver TG2 protein in a buffer supplemented with Ca2+ (5 mm). Western blotting of the reaction mixtures demonstrated that TG2 was sufficient for the formation of HMGB1c, which formed in a TG2 concentration–dependent manner up to a TG2 enzyme concentration of 0.25 milliunits/μl (Fig. 3D, lane 5). Reaction products with more TG2 were less reactive to the anti-HMGB1 antibody used to process the Western blots in Fig. 3D, lanes 6 and 7, but may have been too large to enter the SDS-PAGE gels. Next we incubated TG2 with total RAW 264.7 cell lysates to determine whether TG2 mediates the formation of HMGB1c with endogenous HMGB1 as a substrate. Western blot analysis showed that, in RAW 264.7 cells, HMGB1 exists as a 29-kDa band as well as HMGB1c variants with sizes ranging from ∼150 kDa to in excess of 250 kDa (Fig. 3E, lane 1). Activation of TG2 with Ca2+ culminated in a decrease of HMGB1 p29 and the formation of HMGB1c accumulating near the top of the gel (Fig. 3E, lane 6, arrow). These complexes migrated with an apparent narrower size range but may also have been too large to enter the gel. Significantly, cystamine, a TG2 transamidation inhibitor, suppressed the formation of HMGB1c while inducing a smaller HMGB1c of 60 kDa and 100 kDa (Fig. 3E, lanes 7 and 8), suggesting that the latter could be intermediates. Because HMGB1 is 29 kDa, the 60-kDa HMGB1c could represent HMGB1 homodimers or HMGB1 associated with another protein of similar size. We also examined the effect of TG2 loss of function on HMGB1c. Knockdown of TG2 expression with TG2-targeting siRNAs in primary human pulmonary fibroblasts (hPFBs), a cell line shown previously to have high basal levels of TG2 expression and transamidation activity (25) and reduced levels of HMGB1c (Fig. 3F). To confirm the specificity of the anti-HMGB1 antibody used to detect HMGB1c so far, a peptide-blocking experiment was performed (Fig. 3G), where HMGB1 immunoblots were incubated with or without a synthetic peptide used to raise the HMGB1 antibody. Notably, the blocking peptide suppressed the signals from recombinant HMGB1 (Fig. 1G, lane 1), HMGB1c catalyzed by the addition of TG2 and recombinant HMGB1 to a cell lysate (Fig. 3G, lane 2), as well as endogenous HMGB1 and HMGB1c from mouse liver and hPFB cell lysates (Fig. 3G, lanes 3 and 4, respectively). Suppression of the multiple HMGB1c forms in the in vitro cross-link reaction as well as endogenous forms in cell and tissue lysates confirms the specificity of the HMGB1 antibody for HMGB1c. The data suggest, for the first time, that, in addition to the well-characterized p29 form, HMGB1 exists as covalent, cross-linked protein complexes in various cells and tissues.
HMGB1 contains multiple K donors and is covalently bound to TG2 via isopeptide bonds
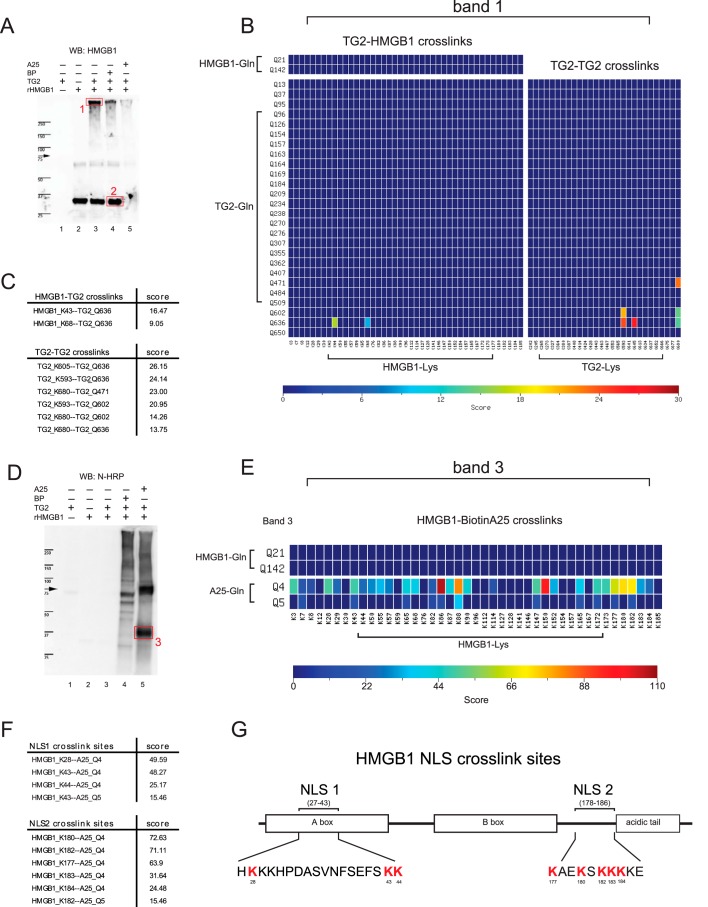
TG2 has multiple functions, including GTPase-, protein kinase-, protein disulfide isomerase-, and Ca2+-dependent transamidation activity (reviewed in Ref. 26). So far, our data demonstrate Ca2+ dependence and TG2 inhibitor antagonism of the HMGB1c formation, which supports the notion that TG2 catalyzes the formation of HMGB1c via its transamidation activity. Therefore, the side chains of one or more glutamyl (Gln) and lysinyl (Lys) residues in HMGB1 should serve as TG2 substrates, and, potentially, HMGB1c could consist of HMGB1 forming intra- and/or intermolecular isopeptide bonds. To determine whether HMGB1 harbors such Gln and/or Lys donors, we first performed substrate incorporation assays using two well-characterized biotinylated TG2 substrates as affinity probes. These probes were 5-(biotinamido) pentylamine (BP), a TG2 K donor substrate, and the synthetic biotinylated peptide TVQQEL (A25), a Gln donor substrate (27). rHMGB1 was incubated with TG2 in the presence of either BP or A25. Subsequently, the cross-linking reactions were stopped by addition of SDS loading buffer and subjected to Western blotting with anti-HMGB1 antibodies to detect HMGB1 (Fig. 4A). A duplicated blot was incubated with NeutrAvidin-HRP (N-HRP) to determine whether one or both biotinylated peptides were incorporated into rHMGB1 (Fig. 4D). For the BP incorporation assay, anti-HMGB1 yielded a signal of 29 kDa (Fig. 4A, lane 4, band 2), but no signal at the corresponding region using N-HRP was seen (Fig. 4D, lane 4). Hence, these results suggest that the p29 HMGB1 isoform does not form isopeptide bonds with BP and lacks Gln donors. Weak signals from larger species ranging from 50 kDa and distributed to the top of the gel were also observed. Those signals could be derived from contamination in the TG2 preparation or some from TG2. For A25, the HMGB1 antibodies did not yield a signal from p29 (Fig. 4A, lane 5), whereas N-HRP yielded two prominent signals of 75 kDa and 37 kDa (Fig. 4D, lane 5). The former species likely was TG2 cross-linked to A25, and the latter was HMGB1 cross-linked to multiple A25. Because A25 was ∼0.9 kDa in size, the data suggest that multiple A25 Gln donors were incorporated into the HMGB1 polypeptide, resulting in an approximate size shift from 29 to 37 kDa in SDS-PAGE. In addition, the data suggest that HMGB1 may not contain any Gln donor targeted by TG2 in substrate incorporation assays.
Figure 4.
Identification and mapping of HMGB1 cross-linking sites by substrate incorporation assays. Substrate incorporation assays were performed, using as affinity probes two well-characterized biotinylated TG2 substrates: BP and A25. A and D, rHMGB1 was incubated in the presence or absence of TG2 and BP or A25 peptide for 1 h, size-fractionated on SDS-PAGE gels, and analyzed by immunoblot using HMGB1 antibodies (A) or N-HRP (D). WB, Western blot. B and C, a protein region (band 1, indicated by a box in A, lane 3) was excised from the gel for tandem mass spectrometry analysis and subsequent cross-link mapping using the MassMatrix database search software. B, cross-links detected by the database search are depicted as probability maps, where each cell represents a potential ϵ-(γ-glutamyl) lysine isopeptide bond. Band 1, which reacted with HMGB1 antibodies, contained TG2-HMGB1 cross-links, where HMGB1 Lys-44 and Lys-68 formed isopeptide bonds with Gln-636 of TG2. C, summary table of cross-linking mapping shown in B. In addition to the HMGB1 and TG2 cross-links, TG2-TG2 isopeptide bonds were detected in band 1, indicating that TG2 formed cross-links with itself. E and G, HMGB1 is a TG2 lysine donor substrate and cross-linked to A25 at multiple sites. Band 3 from D, lane 5, was excised from the gel and (E) cross-link mapping revealed that A25 was cross-linked to a number of lysines, some of which reside within HMGB1 NLS regions. F and G, Lys donors present in NLS1 or NLS2 of HMGB1 are summarized in F, and in the HMGB1 protein is schematically shown in G. The blots depicted in A and D are representative of data obtained from three independent experiments.
Formation of HMGB1c was also was observed when TG2 was incubated with rHMGB1 (Fig. 4A, lane 3). Notably, HMGB1c formation was partially diminished by BP (lane 4) and almost completely suppressed by A25 (lane 5). These observations were in agreement with the substrate incorporation assay, which showed that A25 was efficiently incorporated into HMGB1 whereas BP was not.
To identify the cross-linking sites in the substrate incorporation assays, three protein regions, including bands 1 and 2 (Fig. 4A) and band 3 (Fig. 4D), were excised from the gels and subjected to tandem LC-MS analysis followed by cross-link mapping using the MassMatrix database search software (28). Because mass loss of -NH3+, a unique characteristic of cross-linking via transamidation, was included in the search criteria for analysis of the mass spectra, all cross-links identified from the excised gel bands were TG2-mediated. The identified cross-links are depicted in Fig. 4, B and E, as probability maps, where each cell represents a potential ϵ-(γ-glutamyl) lysine isopeptide bond formed between the Gln donors and Lys donors. For band 1, two TG2-HMGB1 cross-links were identified in which HMGB1 Lys-44 and Lys-68 individually cross-linked to Gln-636 of TG2. Significantly, the results show that HMGB1 as a substrate was covalently cross-linked to its enzyme TG2 in HMGB1c. In addition, multiple TG2-TG2 isopeptide bonds were detected (Fig. 4, B and C), indicating that TG2 also cross-linked itself, as reported previously (29, 30). However, no HMGB1-HMGB1 cross-links were detected. This observation also agrees with the results of the substrate incorporation assay, which showed the lack of a Gln donor in HMGB1. For band 3, cross-link mapping revealed that, of the 43 HMGB1 lysines, 38 formed isopeptide bonds with either Gln-4 or, to a lesser extent, Gln-5 of the A25 peptide, indicating that HMGB1 contains multiple Lys donors (Fig. 4E and Table S1). This finding also agrees with the gel retardation observed for the A25-incorporated HMGB1 (Fig. 4D, lane 5). Notably, many Lys donors identified lie within NLS1 or NLS2 of HMGB1 (Fig. 4, F and G). In agreement with the substrate incorporation assay, no cross-linking of BP to HMGB1 was detected in band 2 (data not shown), further suggesting that HMGB1 lacks Gln donor sites. Taken together, we demonstrate that HMGB1 is a TG2 transamidation substrate and that HMGB1 can cross-link to the C terminus of TG2 via two isopeptide bonds.
HMGB1 is cross-linked to several autoantigens
Next we examined the TG2-catalyzed HMGB1 transamidation reaction kinetics and studied the formation of HMGB1c at various time points by Western blotting. Two major HMGB1c formed with different kinetics were detected (Fig. 5A). The fast-migrating HMGB1c migrating at ∼60 kDa was detected as early as 5 min, whereas the slower-migrating complex migrating near the top of the gel was detected after 30 min. The kinetics of their appearance suggest that the faster-migrating HMGB1c was most likely an intermediate of the slower-migrating HMGB1c. Next we studied the effect of cell extracts on HMGB1c formation by supplementing the kinetic assay with NIH-3T3 cell lysate. This cell line was chosen for its characteristically low TG2 expression and concordant absence of detectable HMGB1c (data not shown). The pattern of HMGB1c formation was largely similar to that without cell extracts, except that the amount of the larger HMGB1c was significantly increased by addition of the extracts, particularly at the 45-min and 60-min time points (Fig. 5B). Components in the cell extracts could contribute to the formation of HMGB1c by favoring the cross-links between HMGB1 and TG2 or by providing additional endogenous transamidation substrates that were cross-linked to HMGB1. Given the lack of detection of HMGB1 Gln donor sites by cross-link mapping, the latter scenario suggests that cell lysate may have enhanced HMGB1c formation by providing additional endogenous Gln donors that were cross-linked to HMGB1 Lys donor sites, resulting in increased HMGB1c formation. This latter scenario further suggests that HMGB1c may be heteromeric in nature and formed by the cross-linking of HMGB1 to other proteins.
Figure 5.
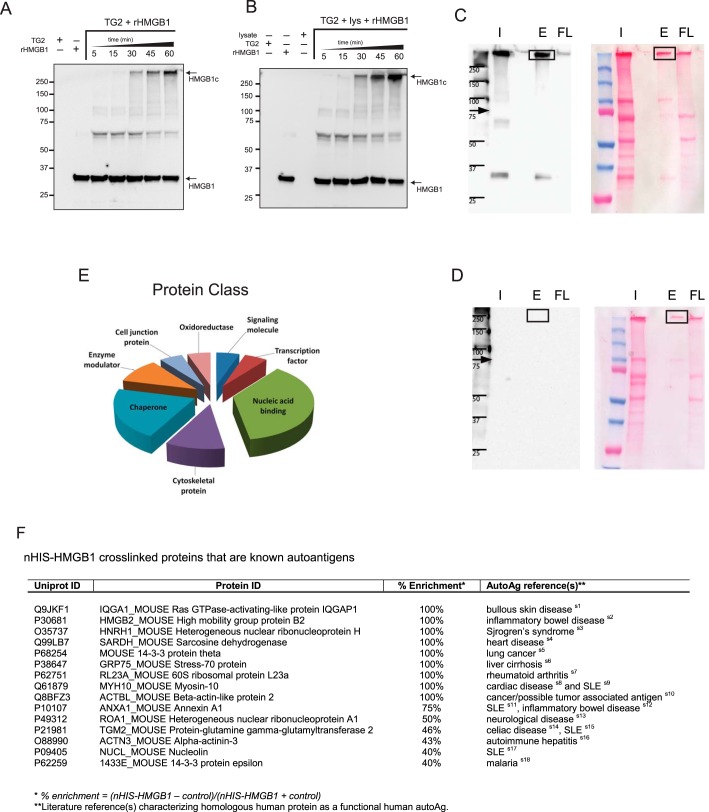
Isolation and identification of TG2-mediated HMGB1 cross-linked proteins by mass spectrometry. A and B, NIH-3T3 cell lysates enhanced TG2-mediated HMGB1 cross-linking. Porcine TG2 was incubated with rHMGB1 (His6-HMGB1) supplemented without (A) or with NIH3T3 cell lysates (B). The reaction mixtures were sampled over a 60-min interval, size-fractionated on SDS-PAGE, and analyzed by immunoblotting using anti-His antibodies. C and D, consequently, TG2 was incubated with NIH-3T3 cell lysate and with (C) and without (D) rHMGB1for 1 h. The reaction mixtures were purified on cobalt columns to isolate HMGB1– and HMGB1–cross-linked proteins. Samples were resolved by SDS-PAGE and subjected to immunoblot analysis with HMGB1 antibodies (left panel) and staining with Ponceau S (right panel). I, input; E, eluates; FL, flow-through. E, the functional classification of the proteins cross-linked to rHMGB1 as identified by mass spectrometry and subjected to gene ontology analysis using the PANTHER gene ontology search tool. F, HMGB1-associated proteins that are autoantigens. Data depicted are representative of four (A and B) or three (C–F) independent experiments.
Finally, to identify putative proteins that can form isopeptide bonds with HMGB1, rHMGB1 was used as bait to isolate HMGB1c after incubation with TG2 and NIH-3T3 cell lysates in a calcium reaction buffer. Because the His tag in rHMGB1 binds to divalent metal ions, rHMGB1c was purified from the reaction mixtures using cobalt columns, and then the eluate was size-fractionated on reducing SDS-PAGE gels. In Western blot analysis, HMGB1c yielded a strong signal with anti-HMGB1 antibodies, which migrated as a discrete species, as revealed by protein staining of a duplicated gel (highlighted with boxes in Fig. 5C). Consequently, that region of the gel was excised and subjected to LC-MS/MS analysis. As a control, the corresponding gel region was isolated from a control binding reaction that lacked rHMGB1 (Fig. 5D). Additional control reactions with rHMGB1 and cell lysate in the absence of TG2 did not induce HMGB1c formation and, thus, were not subjected to MS analysis (Fig. S3). To account for the possibility that some proteins may have been cross-linked to HMGB1 while also binding to the column nonspecifically, an “enrichment score” was calculated from the spectral counts to compare proteins identified in the rHMGB1 reaction with the control reaction: % enrichment = (rHMGB1 – control)/(rHMGB1 + control).
Using this formula, an enrichment score of +100% represents protein hits unique to the rHMGB1 cross-linked reaction in HMGB1c. A score of 0% represents spectral counts for a given identified protein equal in rHMGB1 and control reactions, and a −100% score represents nonspecific binding only. In that experiment, a total of 66 proteins have a positive score. In addition to HMGB1 itself, HMGB1c included 17 unique proteins (scores of 100%), and 10 other proteins have scores higher than 40%, including TG2 (Table S2). Classification of the proteins with unique hits using the PANTHER gene ontology search tool indicated that a majority of those proteins consisted of nucleic acid binding proteins (41.2%), cytoskeletal proteins (11.8%), or chaperones (23.5%) (Fig. 5E). Although collectively diverse in structure and function, cytoskeletal proteins (31), chaperones (32), and proteins involved in the synthesis and processing of DNA and RNA (33) tend to be prone to forming autoantigens. Remarkably, many of the HMGB1c-associated proteins identified by this study are known autoantigens (Fig. 5F).
TG2 transamidation activity, described above, is typically quenched before analysis by heating in SDS loading buffer or by addition of the alkylating agent iodoacetamide (IAA). Because the presence of SDS and excess reducing agents was incompatible with downstream purification on the Co2+ column, we instead used IAA. Although cysteine alkylation is the predominant reaction under limiting quantities of IAA reagent, excess IAA can also alkylate imidazoles such as histidine residues. This could interfere with binding of the His-tagged rHMGB1 to the column. To address this issue, we performed the analysis depicted in Fig. 5, C–F, and Table S2 under four different conditions. Under condition 1, reaction mixtures were immediately purified on the column after the incubation period without addition of IAA. Under condition 2, TG2 enzyme activity was quenched with IAA prior to column purification. Because the incorporation of rHMGB1 into large protein complexes could also block interaction of the His tag to the column through steric interference, two additional cross-link reactions were performed where HMGB1 was prebound to the column before adding TG2 and lysate. These reactions were subsequently purified without quenching TG2 enzymatic activity (condition 3) or after quenching with IAA (condition 4). Only conditions 1–3 yielded sufficient peptide mass for analysis after gel extraction and digestion. Protein identification by LC-MS/MS analysis for conditions 2 and 3 were representative of group 1 (as depicted in Fig. 5, C–F, and Table S2). Protein identification by LC-MS/MS analysis for Conditions 2 and 3 is representative of Group 1 (as depicted in Fig. 5 (C–F) and Table S2, data not shown).
Discussion
Here we report, for the first time, TG2–HMGB1 interaction and the existence of heat- and reducing agent–resistant HMGB1 protein complexes that are formed via TG2-mediated transamidation. Unlike the prototypical 29-kDa HMGB1 isoform (HMGB1 p29) that is ubiquitously expressed, the abundance of HMGB1c varies in a cell- and tissue-specific manner. For example, in unstimulated mice, although only minimal levels of HMGB1c were found in the spleen (Fig. 2A), substantially more HMGB1c was observed in the liver as a slower-migrating band of ∼150 kDa (Fig. 2, B and C). Although HMGB1c was induced by LPS in both tissues, the kinetics and extent of induction were different. Similar variations in HMGB1c levels were observed in human and mouse cells, with HMGB1c high in human primary pulmonary fibroblasts (Fig. 3F) but low in PBMCs (Fig. 1C) and RAW 264.7 cells (Fig. 1D). Moreover, TG2 cross-links HMGB1 in vitro into complexes that include TG2, the related high-mobility group box family member HMGB2, as well as a number of additional cellular proteins, some of which are known autoantigens. These results suggest that HMGB1c is regulated by TG2 via different mechanisms, depending on tissue or cell type and associated physiological conditions.
Previous studies have shown that TG2 is distributed in multiple subcellular compartments, including the extracellular matrix, plasma membrane, cytosol, mitochondria, and nucleus (26). Furthermore, the activity of TG2 is positively regulated by Ca2+ and negatively regulated by GTP, and hence TG2 likely would perform distinct functions in different subcellular locations and body fluid compartments because of the differences in the ionic compositions of these microenvironments. In most cell types, TG2 is located mainly in the cytosol, with only a minimal amount present in the nucleus. The carboxyl end of TG2, called the β-barrel 2 region, contains an NLS (KQKRK, residues 598 to 602) and a nuclear export signal (residues 657 to 664). Together, the balance of these two signals has been proposed to determine the nuclear abundance of TG2 (34). In this study, we showed that Gln-636 of TG2, a residue located in between these two regions, could be cross-linked to Lys-43 and Lys-68 of HMGB1 (Fig. 4, B and C) and that more nuclear TG2 was observed in HMGB1-overexpressing cells (Fig. 3A). The mechanism by which HMGB1 and TG2 affect the subcellular localization of each other remains to be elucidated. Because HMGB1 can be cross-linked to TG2 by isopeptide bonds, it is plausible that such cross-linking could shift the dynamics of import and export of TG2 to nuclear retention. The interaction of this enzyme and substrate pair should have significant physiological consequences because nuclear TG2 has been shown to be involved in several pathological conditions, including neurodegenerative diseases, liver diseases, and cancers, by affecting cellular processes in cell growth or survival, differentiation and apoptosis via post-translational modification, and/or interaction with transcriptional factors and chromatin-associated proteins such as histones (34). Alternatively, HMGB1 association with TG2 in the nucleus could reduce cytosolic TG2.
Given the prominent role of TG2 in the stress response and regulation of tissue homeostasis, it is not surprising that both gain and loss of TG2 function have been attributed to the development of inflammatory disorders and autoimmunity. TG2 knockout mice have compromised anti-inflammatory responses and develop an autoimmune phenotype that has been attributed to impaired clearance of apoptotic cells (35). Rapid and efficient clearance of apoptotic cell debris by phagocytes plays a key role in preventing the exposure of toxic, immunogenic materials from dying cells to the immune system (reviewed in Ref. 36). Moreover, increased calcium flux during apoptosis is known to induce substantial TG2 transamidation activity (37), where protein cross-linking may stabilize dying cells prior to clearance, inhibiting the leakage of cellular components that could result in the inappropriate extracellular presence of self-antigens (38). Thus, deficient cross-linking of immunogenic self-proteins could also contribute to autoimmune phenomena in the TG2−/− mouse model, which may exacerbate autoimmunity in the context of impaired apoptotic cell clearance.
In contrast, the pro-inflammatory nature of TG2 is exemplified by reduced sensitivity of TG2−/− mice to endotoxic shock (39). Moreover, inappropriate gain of TG2 function via increased transamidation activity could have pathological consequences. An example is celiac disease, where TG2 modification of gluten peptides triggers an autoimmune reaction leading to the formation of anti-TG2 antibodies and destruction of the intestinal mucosa (14). Although the ability of disparate TG2 functional states to similarly induce inflammatory and autoimmune disorders is counterintuitive at face value, our discovery of HMGB1–TG2 interactions in this study may help shed light on the complex role of TG2 in tissue homeostasis, inflammation, and autoimmunity. The presence of HMGB1 in complexes with other partner molecules may enhance the ability of the associated partners to signal through their cognate receptors (41, 42). Circulating levels of HMGB1 have been shown to increase in patients with SLE, which often correlates with disease activity (43–45). Moreover, the presence of HMGB1 in blood-borne immune complexes in patients with SLE has been shown to be necessary for instigating an immune response (8). Proteins in HMGB1c are covalently linked via isopeptide bonds, which are particularly resistant to protease activity. Thus, HMGB1c may have enhanced stability, resulting in sustained signaling of constituent partner pro-inflammatory ligands. In this capacity, HMGB1c could contribute to regulating the local inflammatory milieu.
TG2-mediated cross-linking of HMGB1 to autoantigens may shed light on a possible novel mechanism for targeting of self-proteins during autoimmune responses. HMGB1c may facilitate the adjuvant function that has been attributed to HMGB1 in vaccine development (46–48). Interestingly, alum, a frequently used adjuvant, induced the formation of HMGB1c (Fig. 1F). Moreover, our data show that LPS is an inducer of HMGB1c, which may implicate HMGB1c as an in vivo adjuvant that facilitates a robust response to infectious insults in a normal host. However, HMGB1 and associated complexes may also play a central role in the breaking of immune tolerance. Because only an estimated 1–2% of proteins are targets of autoimmunity, and the molecular characteristics that incline self-proteins to become autoantigens is currently not completely understood (49), the observed high incidence of autoantigens (Fig. 5F) among proteins cross-linked to HMGB1 is remarkable. Furthermore, it has been shown that HMGB1 bound to nucleosomes can induce anti-histone and anti-dsDNA antibodies in nonautoimmune mice in a TLR2-dependent manner (10). Taken together, the presence of HMGB1c in SLE plasma suggests that HMGB1c may be an indicator for and possible driver of auto-inflammatory processes.
A third potential role for TG2-mediated HMGB1 modification is in the regulation of HMGB1 intra-cellular trafficking and secretion. Translocation of HMGB1 from the nucleus to the cytosol is known to be induced by stressful stimuli such as oxidative insult, starvation, and endotoxin (24, 50, 51). These stressors activate signaling cascades that culminate in the modification of HMGB1 within two NLSs, inducing its cytosolic translocation (24, 52, 53). HMGB1 NLS1 spans amino acids 27–43, whereas NLS2 is located at residues 178–186. Both regions contain lysine clusters at residues 28, 29, and 30 (NLS1) and 180 and 182–184 (NLS2) that, upon hyperacetylation, trigger cytosolic translocation (24). Remarkably, cross-linking to A25 peptide was detected at lysines 28, 43, and 44, which reside in or immediately adjacent to HMGB1 NLS1. Similarly, HMGB1 was cross-linked to the A25 probe at lysines 177, 180, 182, and 183, which reside in or immediately adjacent to HMGB1 NLS2 (Fig. 4, F and G). The presence of TG2 lysine cross-linking sites within the HMGB1 NLS, shown previously to be important for acetylation-induced cytosolic translocation, is suggestive of a role for TG2-mediated HMGB1 transamidation in the regulation of trafficking during cellular stress responses. HMGB1c formation possibly functions in part as a homeostatic response at odds with the classical acetylated, monomeric, and pro-inflammatory forms of HMGB1 that are released in response to stress. A recent report suggested that LPS-induced HMGB1 secretion may be inhibited by the deacetylase sirtuin 1 (SIRT1) (54). Thus, it is tempting to speculate that the availability of certain HMGB1 lysines for transamidation may be in part regulated by the balance of acetyltransferase/deacetylase activity in the cell under different conditions. Ultimately, TG2-mediated transamidation may drive HMGB1 toward a cytosolic and/or extracellular fate, the function of which may be determined by the identity of HMGB1–cross-linked constituents.
Extracellular HMGB1 is a mediator of inflammation during infection or tissue injury. Immune cells actively release HMGB1 in response to infection, which, in turn, orchestrates innate and adaptive immune responses. However, whether the quantity of HMGB1 in the blood may serve as a biomarker for systemic inflammatory disorders remains controversial. Cumulative evidence suggests that the biological effect of HMGB1 depends not only on its quantity but also on its many post-translational modifications. Based on our data and those of others, we propose that HMGB1c have biological functions distinct from the prototypical p29 form. Furthermore the structure of HMGB1c, as imparted by the identity and configuration of constituent cross-linked-molecules, may also have important functional implications. The serum or plasma levels of HMGB1 are determined by ELISA more often than immunoblotting because of the higher throughput and more quantitative nature of the former technique. However, discrepancies between the two quantitation methods have been reported (55). This would necessitate that a quantitation of HMGB1 levels needs to be carried out, using both methods for accuracy. Moreover, the biological activity of extracellular HMGB1 depends on its binding partners and post-translational modifications. For example, the redox states of Cys-23, Cys-45, and Cys-106 determine the biological activity of the extracellular HMGB1 (56). The completely reduced form binds the chemokine CXC12 and signal through the CXCR4 receptor to induce chemotaxis. The form with a disulfide bond between Cys-23, Cys-45, and Cys-106 in the thiol form can induce cytokine production by signaling through TLR4. Lastly, the completely oxidized form with cysteines in the form of sulfonates is unable to stimulate cytokines or induce chemotaxis but can induce tolerance. Similarly, TG2-mediated HMGB1 transamidation may add to the functional diversity of HMGB1 via HMGB1c formation. Although the composition and function of HMGB1c in vivo remain to be established, when taken together, our data suggest that this novel transglutaminase-dependent cross-linking has important functional implications for HMGB1 biology in the context of cellular stress responses, innate immunity, and autoimmune disease.
Experimental procedures
Antibodies and reagents
Antibodies for TG2 (catalog no. 3557), GAPDH-HRP (catalog no. 8884), and β-actin (catalog no. 4970) were obtained from Cell Signaling Technology. HMGB1 (catalog no. 18256), mCherry (catalog no. 167453), and TG2 (catalog no. 2386) were obtained from Abcam. GFP (catalog no. TA150041) and His (catalog no. TA150088) were obtained from Origene. HMGB1 (catalog no. H9664) was obtained from Sigma-Aldrich. Cystamine (catalog no. C8707), ionomycin (catalog no. I3909), and LPS (catalog no. L2880) were purchased from Sigma. Imject Alum (catalog no. 77161) and a silver staining kit (catalog no. 24612) were purchased from Thermo Scientific. The various Alexa fluorophore–conjugated secondary antibodies used for immunofluorescent staining were purchased from Cell Signaling Technology. TG2 from guinea pig liver (Sigma-Aldrich, catalog no. T5398) and recombinant HMGB1 with an N-terminal His6 tag produced in E. coli (Genway Biotech Inc., catalog no. GWB-37D275) were purchased. A25 (GenScript), BP (Thermo Scientific), and HMGB1-blocking peptide (Abcam, catalog no. ab18650) were purchased.
Plasmid constructs
A plasmid for expressing human TG2 with a C-terminal GFP tag was purchased from Origene (catalog no. RG217353). A plasmid construct with human HMGB1 complementary DNA inserted after the C terminus of the mCherry tag in mCherry2-C1 was prepared by standard molecular cloning procedures. Human HMGB1 cloned into plasmid mEGF-C1 to produce GFP-HMGB1 fusion proteins was also prepared. mCherry2-C1 and mEGFP-C1 were gifts from Michael Davidson (Addgene plasmids 54563 and 54759).
Mice
BALB/c mice purchased from The Jackson Laboratory (Bar Harbor, ME) were housed at the OSU animal facility. All animal maintenance and protocols were specifically approved for this study by the Institutional Animal Care and Use Committee through the University Laboratory Animal Resources. Mice were stimulated with LPS (2 mg/kg) via intraperitoneal injection, and tissues were harvested 2 or 24 h afterward. Tissues from unstimulated mice served as controls.
Cell lines and human peripheral blood mononuclear cells
hPFBs, kindly provided by Dr. Daren L. Knoell (Ohio State University, Columbus, OH), were described previously (25). RAW 264.7 cells (TIB-71), 293T cells (CRL-3216), and NIH-3T3 fibroblasts (CRL-1658) were purchased from the ATCC. Human peripheral blood mononuclear cells (PBMCs) were obtained from SLE patients meeting the revised criteria of the American College of Rheumatology (57). Patients were recruited for this study from the OSU Wexner Medical Center clinics. Healthy volunteers were recruited from the American Red Cross and local communities using the Research Match program through the Center for Clinical and Translational Science at Ohio State University. Participation was in accordance with an approved institutional review board protocol at OSU Wexner Medical Center and abided by the Declaration of Helsinki principles. Samples obtained for processing were either whole blood collected into heparinized tubes or filtered blood samples. PBMCs were isolated using a Ficoll-Paque Plus density gradient and cultivated as described previously (58).
Protein gel electrophoresis, immunoblotting, and co-immunoprecipitation
SDD-AGE using 1.5% agarose gel was performed according to Ref. 17, except that gels were run at 4 °C. Proteins were transferred from SDD-AGE gels onto nitrocellulose membranes via a reverse capillary method as described in Ref. 16. Standard SDS-PAGE included incubating samples in loading buffer with 100 mm DTT at 95 °C for 5 min and resolving protein samples on BoltTM 4–12% BisTris Plus gels, 10-well (Thermo Scientific, catalog no. NW04120BOX). Immunoblot using chemiluminescent HRP substrate (Thermo Scientific SuperSignalTM West Dura Extended Duration Substrate, catalog no. 34075) was performed as described previously (25). To maximize transfer of HMGB1c for SDS-PAGE, methods for high-efficiency transfer of high-molecular weight protein complexes, which included a single-use transfer buffer (48 mm Tris, 390 mm glycine, 0.05% SDS, and 20% methanol) and two-stage electrophoretic transfer process (60 mA for 16 h followed by 450 mA for an additional 3 h), were performed as described previously (25). Signals were detected on film or using a ChemiDoc XRS charge-coupled device imaging camera (Bio-Rad). Chemiluminescence exposure times to film/CCD camera were varied over a period of 5–500 s to better resolve HMGB1c signals. In experiments in which the expression of more than one protein was examined, duplicated gels were used. Subsequently, all filters were incubated with the loading controls GAPDH or β-actin. In figures where multiple proteins from duplicate blots are depicted in separate panels, one sample control panel is shown to indicate equal expression of GAPDH or β-actin in that group of samples. Signal intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD; version 1.50i) and normalized to the loading control (GAPDH). The data were reported as scatterplots alongside sample means with standard deviation error bars. To show the specificity of the HMGB1 antibodies, duplicated SDS-PAGE protein blots were incubated with HMGB1 antibodies with or without a 5-fold excess of HMGB1 blocking peptide. Immunoprecipitation was performed with total cell lysates prepared from 293T cells transfected with GFP-HMGB1 or GFP alone using a GFP-Trap®_MA kit according to the manufacturer's protocol (Chromotek, catalog no. gtmakl-20).
TG2 cross-linking reactions and substrate incorporation assays
Total cell lysates (0.25 μg/μl) prepared in a modified radioimmune precipitation assay lysis buffer (50 mm MOPS (pH 7.5), 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 150 mm NaCl, 1 mm DTT, 0.2 mm PMSF, and protease inhibitor mixture) were incubated with TG2 (1.0 milliunits/μl) in cross-linking reaction buffer (50 mm MOPS (pH 7.5), 1 mm DTT, 0.2 mm PMSF, and protease inhibitor mixture) and with or without CaCl2 (5 mm). For Ca2+-free reactions, buffers were also supplemented with 2 mm EGTA. Some reactions also contained a TG2 inhibitor, cystamine (10 mm). For recombinant HMGB1, rHMGB1 (20 ng/μl) was incubated with TG2 at varying concentrations (0.05–1.0 milliunits/μl) as described in the text. In substrate incorporation assays, the cross-linking reaction contained rHMGB1 (20 ng/μl) and TG2 (1.0 milliunits/μl) with or without 2 mm BP or A25. All cross-linking reactions were incubated for 60 min at 37 °C, and reaction products were size-fractionated on 4–12% polyacrylamide gels, transferred to nitrocellulose membranes, and processed for immunoblot analysis using HMGB1 antibodies or NeutrAvidin-HRP.
TG2 siRNA knockdown
To inhibit endogenous TG2 protein expression, a duplex siRNA sequence (target sequence, 5′-AAGGGCGAACCACCTGAACAA-3′) was synthesized (Qiagen) and used in human pulmonary fibroblast transfection studies to specifically target the coding sequence of human TG2 mRNA (60). A scrambled-sequence siRNA (ON-TARGET Plus control siRNA, nontargeting siRNA 1, catalog no. D-0018100-01-05, Dharmacon) was used as a negative control. Transfection was performed using 25 nm siRNA and RNAimax transfection reagent (Invitrogen) according to the manufacturer's protocol. Cells were harvested and analyzed 72 h after transfection.
Immunohistochemical staining and confocal microscopy
Liver histological sections (5 μm) from mice stimulated with LPS for 2 h were prepared by the OSU Comprehensive Cancer Center (CCC) Comparative Pathology and Mouse Phenotyping core and used for immunohistochemical staining as described previously (59) Primary antibodies were anti-HMGB1 (rabbit monoclonal, 1:1000 dilution) and anti-TG2 (Abcam, mouse monoclonal, 1:50 dilution), and the secondary antibodies were Alexa 555–conjugated anti-rabbit IgG (1:500 dilution) and Alexa 488–conjugated anti-mouse IgG (1:500 dilution) (Cell Signaling Technology), respectively. To visualize proteins with fluorescent tags, 293T cells were grown to 50% confluence on 18-mm 1.5 poly-d-lysine–coated coverslips (Neuvitro) in a 12-well tissue culture plate and transfected with plasmids (1 μg of each) encoding TG2-GFP, mCherry-HMGB1, or both using Lipofectamine 3000 (Thermo Scientific, catalog no. L3000008) according to the manufacturer's protocol. Twenty-four hours later, cells were fixed by adding 16% formaldehyde directly to the culture medium to a final concentration of 4% and processed for confocal microscopy. DNA was stained with 4′,6-diamidino-2-phenylindole. All confocal imaging was performed at the OSU Campus Microscopy and Imaging Facility on the Olympus FV1000 Spectral scanning laser confocal system using a ×40 oil objective (UPLFLN, numerical aperture 1.3) for liver histological sections or a ×60 oil objective (PLAPON, numerical aperture 1.4) for 293T cells. Confocal images were processed on Olympus Fluoview software v4.2.
Purification of rHMGB1 cross-linked protein complexes and MS analysis
Cross-link reactions were constituted as described above with NIH-3T3 cell lysates in the presence or absence of rHIS-HMGB1 and incubated with 200 μl of Talon metal affinity resin (Clontech) for 2.5 h at 4 °C with gentle agitation. Resin-bound proteins were sedimented at 700 × g for 5 min and washed four times in wash buffer (50 mm sodium phosphate (pH 7.4), 300 mm NaCl, 5 mm β-mercaptoethanol, 0.2 mm PMSF, and protease inhibitor mixture), followed by elution in 200 μl of 2× SDS loading buffer and heating at 95 °C for 5 min. The eluted proteins were separated on 4–12% SDS-PAGE gels, transferred onto nitrocellulose, stained with Ponceau S, and then subjected to immunoblot analysis with HMGB1 antibodies. Duplicated gels were stained with Coomassie stain (Bio-Rad, catalog no. 161-0786). The bands of interest were excised from the gel, destained in water, dehydrated with acetonitrile, rehydrated with 100 mm ammonium bicarbonate, digested with trypsin (Promega, catalog no. V5111) in 50 mm ammonium bicarbonate digestion buffer overnight, and then extracted using a 45% HPLC water/50% acetonitrile/5% formic acid solution.
Protein concentration in the samples was determined using Nanodrop. Samples were run on a Thermo Scientific LTQ Orbitrap XL mass spectrometer coupled to a Dionex Ultimate 3000 HPLC system. The HPLC stationary phase was a reversed phase C18 column (Michrom Bioresources Magic C18AQ, 200 μm × 150 mm, 3 μm, 200 Å). The mobile phases for this study were phase A (0.1% formic acid) and phase B (0.1% formic acid in acetonitrile) with a gradient of increasing percentage of phase B (from 2% to 40% over 40 min) at a flow rate of 2 μl/min. The MS RAW files were converted to mzxml and searched using the in-house search engine MassMatrix (40) with the UniProt SwissProt Mouse Database (retrieved February 20, 2015) to obtain protein identification. Cross-linking analysis was performed using the XMapper software integrated into MassMatrix.
Author contributions
W. L. W., S. Agarwal, M. A. F., L.-C. W., and W. N. J. conceptualization; W. L. W. and M. A. F. data curation; W. L. W., L. W., M. A. F., and L.-C. W. formal analysis; W. L. W. and W. N. J. supervision; W. L. W. and W. N. J. funding acquisition; W. L. W. validation; W. L. W., L. W., T. T. W., M. G., O. A., J. H., G. V., N. Y., S. Ardoin, S. Agarwal, M. A. F., and L.-C. W. investigation; W. L. W., L. W., T. T. W., and M. A. F. visualization; W. L. W., T. T. W., M. A. F., and L.-C. W. methodology; W. L. W. and L.-C. W. writing-original draft; W. L. W. project administration; W. L. W., L. W., M. G., O. A., S. Agarwal, M. A. F., L.-C. W., and W. N. J. writing-review and editing; M. A. F., L.-C. W., and W. N. J. resources; M. A. F. software.
Supplementary Material
Acknowledgments
We are grateful for the participation of our patient and healthy volunteers as well as the Clinical Research Center and the Research Match Program through CCTS at OSUWMC. Funding is provided through OSUWMC and the CCTS is supported by UL1TR001070 from the National Center for Advancing Translational Science. The lupus research program at The Ohio State University is supported in part by a grant from the Lupus Research Alliance.
This work was supported in part by National Institutes of Health Grant T32CA090223 (to W. L. W.) and P30CA016058 (to L.-C. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3, Tables S1 and S2, and references.
- LPS
- lipopolysaccharide
- SLE
- systemic lupus erythematosus
- NLS
- nuclear localization signal
- SDD-AGE
- semidenaturing detergent–agarose gel electrophoresis
- PBMC
- peripheral blood mononuclear cell
- hPFB
- human pulmonary fibroblast
- BP
- 5-(biotinamido) pentylamine
- N-HRP
- NeutrAvidin–horseradish peroxidase
- IAA
- iodoacetamide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- OSU
- Ohio State University
- PMSF
- phenylmethylsulfonyl fluoride.
References
- 1. Yang H., Antoine D. J., Andersson U., and Tracey K. J. (2013) The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukocyte Biol. 93, 865–873 10.1189/jlb.1212662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harris H. E., Andersson U., and Pisetsky D. S. (2012) HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat. Rev. Rheumatol. 8, 195–202 10.1038/nrrheum.2011.222 [DOI] [PubMed] [Google Scholar]
- 3. Ueda T., and Yoshida M. (2010) HMGB proteins and transcriptional regulation. Biochim. Biophys. Acta 1799, 114–118 10.1016/j.bbagrm.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 4. Lotze M. T., and Tracey K. J. (2005) High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 5, 331–342 10.1038/nri1594 [DOI] [PubMed] [Google Scholar]
- 5. Yanai H., Ban T., Wang Z., Choi M. K., Kawamura T., Negishi H., Nakasato M., Lu Y., Hangai S., Koshiba R., Savitsky D., Ronfani L., Akira S., Bianchi M. E., Honda K., et al. (2009) HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103 10.1038/nature08512 [DOI] [PubMed] [Google Scholar]
- 6. Bianchi M. E. (2009) HMGB1 loves company. J. Leukocyte Biol. 86, 573–576 10.1189/jlb.1008585 [DOI] [PubMed] [Google Scholar]
- 7. Youn J. H., Oh Y. J., Kim E. S., Choi J. E., and Shin J. S. (2008) High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-α production in human monocytes. J. Immunol. 180, 5067–5074 10.4049/jimmunol.180.7.5067 [DOI] [PubMed] [Google Scholar]
- 8. Tian J., Avalos A. M., Mao S. Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., Hua J., An L. L., Audoly L., La Rosa G., Bierhaus A., et al. (2007) Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 8, 487–496 10.1038/ni1457 [DOI] [PubMed] [Google Scholar]
- 9. Sha Y., Zmijewski J., Xu Z., and Abraham E. (2008) HMGB1 develops enhanced proinflammatory activity by binding to cytokines. J. Immunol. 180, 2531–2537 10.4049/jimmunol.180.4.2531 [DOI] [PubMed] [Google Scholar]
- 10. Urbonaviciute V., Fürnrohr B. G., Meister S., Munoz L., Heyder P., De M. F., Bianchi M. E., Kirschning C., Wagner H., Manfredi A. A., Kalden J. R., Schett G., Rovere-Querini P., Herrmann M., and Voll R. E. (2008) Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 205, 3007–3018 10.1084/jem.20081165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folk J. E., and Finlayson J. S. (1977) The ϵ-(γ-glutamyl)lysine crosslink and the catalytic role of transglutaminases. Adv. Protein Chem. 31, 1–133 10.1016/S0065-3233(08)60217-X [DOI] [PubMed] [Google Scholar]
- 12. Stamnaes J., Fleckenstein B., and Sollid L. M. (2008) The propensity for deamidation and transamidation of peptides by transglutaminase 2 is dependent on substrate affinity and reaction conditions. Biochim. Biophys. Acta 1784, 1804–1811 10.1016/j.bbapap.2008.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stamnaes J., Pinkas D. M., Fleckenstein B., Khosla C., and Sollid L. M. (2010) Redox regulation of transglutaminase 2 activity. J. Biol. Chem. 285, 25402–25409 10.1074/jbc.M109.097162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E. O., and Schuppan D. (1997) Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 10.1038/nm0797-797 [DOI] [PubMed] [Google Scholar]
- 15. Abdulahad D. A., Westra J., Limburg P. C., Kallenberg C. G., and Bijl M. (2010) HMGB1 in systemic lupus erythematosus: its role in cutaneous lesion development. Autoimmun. Rev. 9, 661–665 10.1016/j.autrev.2010.05.015 [DOI] [PubMed] [Google Scholar]
- 16. Halfmann R., and Lindquist S. (2008) Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. e838 10.3791/838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bagriantsev S. N., Kushnirov V. V., and Liebman S. W. (2006) Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 412, 33–48 10.1016/S0076-6879(06)12003-0 [DOI] [PubMed] [Google Scholar]
- 18. Martinon F., Agostini L., Meylan E., and Tschopp J. (2004) Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14, 1929–1934 10.1016/j.cub.2004.10.027 [DOI] [PubMed] [Google Scholar]
- 19. Bronner D. N., Abuaita B. H., Chen X., Fitzgerald K. A., Nuñez G., He Y., Yin X. M., and O'Riordan M. X. (2015) Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity 43, 451–462 10.1016/j.immuni.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenbarth S. C., Colegio O. R., O'Connor W., Sutterwala F. S., and Flavell R. A. (2008) Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453, 1122–1126 10.1038/nature06939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watanabe N., Suzuki J., and Kobayashi Y. (1996) Role of calcium in tumor necrosis factor-α production by activated macrophages. J. Biochem. 120, 1190–1195 10.1093/oxfordjournals.jbchem.a021540 [DOI] [PubMed] [Google Scholar]
- 22. Gandhirajan R. K., Meng S., Chandramoorthy H. C., Mallilankaraman K., Mancarella S., Gao H., Razmpour R., Yang X. F., Houser S. R., Chen J., Koch W. J., Wang H., Soboloff J., Gill D. L., and Madesh M. (2013) Blockade of NOX2 and STIM1 signaling limits lipopolysaccharide-induced vascular inflammation. J. Clin. Invest. 123, 887–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowness J. M., and Tarr A. H. (1997) Increase in transglutaminase and its extracellular products in response to an inflammatory stimulus by lipopolysaccharide. Mol. Cell Biochem. 169, 157–163 10.1023/A:1006846400478 [DOI] [PubMed] [Google Scholar]
- 24. Bonaldi T., Talamo F., Scaffidi P., Ferrera D., Porto A., Bachi A., Rubartelli A., Agresti A., and Bianchi M. E. (2003) Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 22, 5551–5560 10.1093/emboj/cdg516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Willis W. L., Hariharan S., David J. J., and Strauch A. R. (2013) Transglutaminase-2 mediates calcium-regulated crosslinking of the Y-box 1 (YB-1) translation-regulatory protein in TGFβ1-activated myofibroblasts. J. Cell. Biochem. 114, 2753–2769 10.1002/jcb.24624 [DOI] [PubMed] [Google Scholar]
- 26. Gundemir S., Colak G., Tucholski J., and Johnson G. V. (2012) Transglutaminase 2: a molecular Swiss army knife. Biochim. Biophys. Acta 1823, 406–419 10.1016/j.bbamcr.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruoppolo M., Orrù S., D'Amato A., Francese S., Rovero P., Marino G., and Esposito C. (2003) Analysis of transglutaminase protein substrates by functional proteomics. Protein Sci. 12, 1290–1297 10.1110/ps.0239103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H., Hsu P. H., Zhang L., Tsai M. D., and Freitas M. A. (2010) Database search algorithm for identification of intact cross-links in proteins and peptides using tandem mass spectrometry. J. Proteome. Res. 9, 3384–3393 10.1021/pr100369y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fleckenstein B., Qiao S. W., Larsen M. R., Jung G., Roepstorff P., and Sollid L. M. (2004) Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J Biol Chem. 279, 17607–17616 10.1074/jbc.M310198200 [DOI] [PubMed] [Google Scholar]
- 30. Stamnaes J., Iversen R., du Pré M. F., Chen X., and Sollid L. M. (2015) Enhanced B-cell receptor recognition of the autoantigen transglutaminase 2 by efficient catalytic self-multimerization. PLoS ONE 10, e0134922 10.1371/journal.pone.0134922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shrivastav M., Mittal B., Aggarwal A., and Misra R. (2002) Autoantibodies against cytoskeletal proteins in rheumatoid arthritis. Clin. Rheumatol. 21, 505–510 10.1007/s100670200124 [DOI] [PubMed] [Google Scholar]
- 32. Routsias J. G., and Tzioufas A. G. (2006) The role of chaperone proteins in autoimmunity. Ann. N.Y. Acad. Sci. 1088, 52–64 10.1196/annals.1366.029 [DOI] [PubMed] [Google Scholar]
- 33. Gilbert D., Brard F., Jovelin F., and Tron F. (1996) Do naturally occurring autoantibodies participate in the constitution of the pathological B-cell repertoire in systemic lupus erythematosus? J. Autoimmun. 9, 247–257 10.1006/jaut.1996.0031 [DOI] [PubMed] [Google Scholar]
- 34. Kuo T. F., Tatsukawa H., and Kojima S. (2011) New insights into the functions and localization of nuclear transglutaminase 2. FEBS J. 278, 4756–4767 10.1111/j.1742-4658.2011.08409.x [DOI] [PubMed] [Google Scholar]
- 35. Szondy Z., Sarang Z., Molnar P., Nemeth T., Piacentini M., Mastroberardino P. G., Falasca L., Aeschlimann D., Kovacs J., Kiss I., Szegezdi E., Lakos G., Rajnavolgyi E., Birckbichler P. J., Melino G., and Fesus L. (2003) Transglutaminase 2−/− mice reveal a phagocytosis-associated crosstalk between macrophages and apoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 100, 7812–7817 10.1073/pnas.0832466100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanaka M., and Miyake Y. (2007) Apoptotic cell clearance and autoimmune disorder. Curr. Med. Chem. 14, 2892–2897 10.2174/092986707782360006 [DOI] [PubMed] [Google Scholar]
- 37. Fesus L., Thomazy V., Autuori F., Ceru M. P., Tarcsa E., and Piacentini M. (1989) Apoptotic hepatocytes become insoluble in detergents and chaotropic agents as a result of transglutaminase action. FEBS Lett. 245, 150–154 10.1016/0014-5793(89)80210-8 [DOI] [PubMed] [Google Scholar]
- 38. Piredda L., Amendola A., Colizzi V., Davies P. J., Farrace M. G., Fraziano M., Gentile V., Uray I., Piacentini M., and Fesus L. (1997) Lack of “tissue” transglutaminase protein cross-linking leads to leakage of macromolecules from dying cells: relationship to development of autoimmunity in MRLIpr/Ipr mice. Cell Death Differ. 4, 463–472 10.1038/sj.cdd.4400267 [DOI] [PubMed] [Google Scholar]
- 39. Falasca L., Farrace M. G., Rinaldi A., Tuosto L., Melino G., and Piacentini M. (2008) Transglutaminase type II is involved in the pathogenesis of endotoxic shock. J. Immunol. 180, 2616–2624 10.4049/jimmunol.180.4.2616 [DOI] [PubMed] [Google Scholar]
- 40. Xu H., and Freitas M. A. (2009) MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 10.1002/pmic.200700322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wähämaa H., Schierbeck H., Hreggvidsdottir H. S., Palmblad K., Aveberger A. C., Andersson U., and Harris H. E. (2011) High mobility group box protein 1 in complex with lipopolysaccharide or IL-1 promotes an increased inflammatory phenotype in synovial fibroblasts. Arthritis Res. Ther. 13, R136 10.1186/ar3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hreggvidsdóttir H. S., Lundberg A. M., Aveberger A. C., Klevenvall L., Andersson U., and Harris H. E. (2012) High mobility group box protein 1 (HMGB1)-partner molecule complexes enhance cytokine production by signaling through the partner molecule receptor. Mol. Med. 18, 224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang W., and Pisetsky D. S. (2008) Expression of high mobility group protein 1 in the sera of patients and mice with systemic lupus erythematosus. Ann. Rheum. Dis. 67, 727–728 [DOI] [PubMed] [Google Scholar]
- 44. Ma C. Y., Jiao Y. L., Zhang J., Yang Q. R., Zhang Z. F., Shen Y. J., Chen Z. J., and Zhao Y. R. (2012) Elevated plasma level of HMGB1 is associated with disease activity and combined alterations with IFN-α and TNF-α in systemic lupus erythematosus. Rheumatol. Int. 32, 395–402 10.1007/s00296-010-1636-6 [DOI] [PubMed] [Google Scholar]
- 45. Li J., Xie H., Wen T., Liu H., Zhu W., and Chen X. (2010) Expression of high mobility group box chromosomal protein 1 and its modulating effects on downstream cytokines in systemic lupus erythematosus. J. Rheumatol. 37, 766–775 10.3899/jrheum.090663 [DOI] [PubMed] [Google Scholar]
- 46. Muthumani G., Laddy D. J., Sundaram S. G., Fagone P., Shedlock D. J., Kannan S., Wu L., Chung C. W., Lankaraman K. M., Burns J., Muthumani K., and Weiner D. B. (2009) Co-immunization with an optimized plasmid-encoded immune stimulatory interleukin, high-mobility group box 1 protein, results in enhanced interferon-γ secretion by antigen-specific CD8 T cells. Immunology 128, e612–e620 10.1111/j.1365-2567.2009.03044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fagone P., Shedlock D. J., Bao H., Kawalekar O. U., Yan J., Gupta D., Morrow M. P., Patel A., Kobinger G. P., Muthumani K., and Weiner D. B. (2011) Molecular adjuvant HMGB1 enhances anti-influenza immunity during DNA vaccination. Gene Ther. 18, 1070–1077 10.1038/gt.2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rovere-Querini P., Capobianco A., Scaffidi P., Valentinis B., Catalanotti F., Giazzon M., Dumitriu I. E., Müller S., Iannacone M., Traversari C., Bianchi M. E., and Manfredi A. A. (2004) HMGB1 is an endogenous immune adjuvant released by necrotic cells. EMBO Rep. 5, 825–830 10.1038/sj.embor.7400205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Plotz P. H. (2003) The autoantibody repertoire: searching for order. Nat. Rev. Immunol. 3, 73–78 10.1038/nri976 [DOI] [PubMed] [Google Scholar]
- 50. Tang D., Kang R., Livesey K. M., Cheh C. W., Farkas A., Loughran P., Hoppe G., Bianchi M. E., Tracey K. J., Zeh H. J. 3rd, and Lotze M. T. (2010) Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190, 881–892 10.1083/jcb.200911078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang D., Shi Y., Kang R., Li T., Xiao W., Wang H., and Xiao X. (2007) Hydrogen peroxide stimulates macrophages and monocytes to actively release HMGB1. J. Leukocyte Biol. 81, 741–747 10.1189/jlb.0806540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Evankovich J., Cho S. W., Zhang R., Cardinal J., Dhupar R., Zhang L., Klune J. R., Zlotnicki J., Billiar T., and Tsung A. (2010) High mobility group box 1 release from hepatocytes during ischemia and reperfusion injury is mediated by decreased histone deacetylase activity. J. Biol. Chem. 285, 39888–39897 10.1074/jbc.M110.128348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Youn J. H., and Shin J. S. (2006) Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 177, 7889–7897 10.4049/jimmunol.177.11.7889 [DOI] [PubMed] [Google Scholar]
- 54. Kim Y. M., Park E. J., Kim J. H., Park S. W., Kim H. J., and Chang K. C. (2016) Ethyl pyruvate inhibits the acetylation and release of HMGB1 via effects on SIRT1/STAT signaling in LPS-activated RAW264.7 cells and peritoneal macrophages. Int. Immunopharmacol. 41, 98–105 10.1016/j.intimp.2016.11.002 [DOI] [PubMed] [Google Scholar]
- 55. Urbonaviciute V., Fürnrohr B. G., Weber C., Haslbeck M., Wilhelm S., Herrmann M., and Voll R. E. (2007) Factors masking HMGB1 in human serum and plasma. J. Leukocyte Biol. 81, 67–74 10.1189/jlb.0306196 [DOI] [PubMed] [Google Scholar]
- 56. Tang D., Kang R., Zeh H. J. 3rd, and Lotze M. T. (2011) High-mobility group box 1, oxidative stress, and disease. Antioxid. Redox Signal. 14, 1315–1335 10.1089/ars.2010.3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hochberg M. C. (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40, 1725 10.1002/art.1780400928 [DOI] [PubMed] [Google Scholar]
- 58. Young N. A., Friedman A. K., Kaffenberger B., Rajaram M. V., Birmingham D. J., Rovin B. H., Hebert L. A., Schlesinger L. S., Wu L. C., and Jarjour W. N. (2013) Novel estrogen target gene ZAS3 is overexpressed in systemic lupus erythematosus. Mol. Immunol. 54, 23–31 10.1016/j.molimm.2012.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu L. C., Goettl V. M., Madiai F., Hackshaw K. V., and Hussain S. R. (2006) Reciprocal regulation of nuclear factor κB and its inhibitor ZAS3 after peripheral nerve injury. BMC. Neurosci. 7, 4 10.1186/1471-2202-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mann A. P., Verma A., Sethi G., Manavathi B., Wang H., Fok J. Y., Kunnumakkara A. B., Kumar R., Aggarwal B. B., and Mehta K. (2006) Overexpression of tissue transglutaminase leads to constitutive activation of nuclear factor-κB in cancer cells: delineation of a novel pathway. Cancer Res. 66, 8788–8795 10.1158/0008-5472.CAN-06-1457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.