Abstract
Ssy5 is a signaling endoprotease that plays a key role in regulating central metabolism, cellular aging, and morphological transitions important for growth and survival of yeast (Saccharomyces cerevisiae) cells. In response to extracellular amino acids, Ssy5 proteolytically activates the transcription factors Stp1 and Stp2, leading to enhanced Ssy1–Ptr3–Ssy5 (SPS) sensor–regulated gene expression. Ssy5 comprises a catalytic (Cat) domain and an extensive regulatory prodomain. Ssy5 is refractory to both broad-spectrum and serine protease–specific inhibitors, confounding its classification as a protease, and no information about Ssy5's cleavage-site preferences and its mechanism of substrate selection is available. Here, using mutational and inhibition experiments, we investigated the biogenesis and catalytic properties of Ssy5 and conclusively show that it is a serine protease. Atypical for the majority of serine proteases, Ssy5's prodomain was obligatorily required in cis during biogenesis for the maturation of the proteolytic activity of the Cat domain. Autolysis and Stp1 and Stp2 cleavage occurred between a cysteine (at the P1 site) and a serine or alanine (at the P′1 site) and required residues with short side chains at the P1 site. Substitutions in the Cat domain affecting substrate specificity revealed that residues Phe-634, His-661, and Gly-671 in the S1-binding pocket of this domain are important for Ssy5 catalytic function. This study confirms that the signaling protease Ssy5 is a serine protease and provides a detailed understanding of the biogenesis and intrinsic properties of this key enzyme in yeast.
Keywords: Saccharomyces cerevisiae, serine protease, signal transduction, substrate specificity, yeast, enzyme structure, environmental sensing, receptor activated proteolysis, signaling protease, SPS sensor, zymogen, signaling protease
Introduction
Proteases are instrumental in the control of many biological processes, including the cell cycle, growth, differentiation, apoptosis, metabolic homeostasis, and signaling (1, 2). Understanding the complexity of protease-regulated processes and the intrinsic activity of this class of enzymes is thus important (3). Consistent with their central regulatory roles and druggable catalytic activities, proteases constitute an important focus of research in medicine and biotechnology.
Protease-dependent signal transduction depends on cleavage of substrates to regulate their activation, inactivation, or functional modification of signaling components. Protease signaling is inherently irreversible and thus differs from other types of signaling events in cells. Consequently, the regulatory mechanisms controlling protease activity require tight coordination to avoid deleterious effects (3, 4). Unwanted protease activity may contribute to the development and progression of disease, such as Alzheimer disease and cancer (5–9). In plants and fungi, proteases are instrumental in the processing, maturation, or destruction of specific sets of proteins in response to developmental cues or to variations in environmental conditions (5).
The Ssy5 signaling endoprotease is a core component of the nutrient-induced SPS4-sensing pathway of Saccharomyces cerevisiae. This signaling pathway enables yeast cells to detect the presence of extracellular amino acids and initiate a response that induces their uptake (10–15). The binding of extracellular amino acids to the SPS sensor, a plasma membrane-localized protein complex, results in the endoproteolytic activation of the homologous effector transcription factors Stp1 and Stp2. Stp1 and Stp2 are synthesized as latent precursors excluded from the nucleus by negative regulatory activities intrinsic to their N-terminal domains (10). Upon amino acid induction, Ssy5 cleaves off the N-terminal domains, enabling the processed forms of Stp1 and Stp2 to efficiently target the nucleus and to induce the expression of genes encoding broad-specificity amino acid permeases (10–15).
Ssy5 is expressed as an inactive zymogen that during biogenesis is subject to an autocatalytic event that cleaves the N-terminal prodomain from the C-terminal Cat domain (16, 17). For most proteases, autolytic processing is directly involved in catalytic activation; however, autolysis does not suffice for Ssy5 activation. Following autolysis, the prodomain remains associated with the Cat domain and prevents substrate processing (16, 17). The prodomain functions as the inhibitory subunit of the SPS sensor. At this stage Ssy5 constitutes a primed protease able to bind but not proteolytically process Stp1 and Stp2, the only known substrates. Although uncommon, similar inhibition mechanisms can be found in secreted prokaryotic bacterial proteases, including the paradigmatic α-lytic protease. Similar to Ssy5, α-lytic protease is expressed with an N-terminal prodomain essential for the catalytic domain to reach a correctly folded and active state (18–20). Following autolysis, the α-lytic protease prodomain remains strongly associated with the catalytic domain, sterically blocking the active site until secreted, at which point the inhibitory prodomain is degraded by extracellular proteases (21).
The mechanisms regulating Ssy5 activation have recently been elucidated and shown to be more complex than for α-lytic protease activation (22, 23). The primary amino acid-induced signal is initiated by the binding of amino acids to Ssy1, which stabilizes a signaling conformation (24). As a consequence, and in a strictly Ptr3-dependent manner, a phosphodegron in the N-terminal Ssy5 prodomain is brought in close spatial proximity to the plasma membrane-anchored yeast casein kinases, Yck1/2, thus facilitating its hyperphosphorylation. Phosphorylation serves as a switch-like trigger to recruit the ubiquitin E3 ligase complex Skp1–Cullin–F-boxGrr1, which mediates the ubiquitylation and proteasomal degradation of the hyperphosphorylated phosphodegron. As an outcome the liberated Cat domain processes Stp1 and Stp2.
Currently, little is known regarding the catalytic properties of Ssy5. The Cat domain contains a conserved and predicted catalytic triad (His-465, Asp-545, and Ser-640) suggesting that Ssy5 is a serine protease. Even though mutation of the predicted nucleophilic Ser-640 results in complete loss of signaling (25), Ssy5 has been refractory to both broad-spectrum and serine protease-specific inhibitors, e.g. diisopropyl fluorophosphate (26), which has confounded its classification as a serine protease. Moreover, no information regarding cleavage-site preferences and the mechanism of substrate selection is available.
Here, we have investigated the biogenesis and catalytic properties of Ssy5. The data provide a detailed understanding of the intrinsic sequence requirements within the catalytic site that are important for autolysis during biogenesis and processing of Stp1 and Stp2. We report under sensitive assay conditions that the catalytic activity of Ssy5 is responsive to the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF), a finding that conclusively confirms that Ssy5 is a serine protease. Moreover, we have identified the scissile bonds in Stp1 and Stp2 and established the rules for their recognition as substrates, and we analyzed the residues in the substrate-binding pocket involved in substrate recognition.
Results
Ssy5 is a serine protease
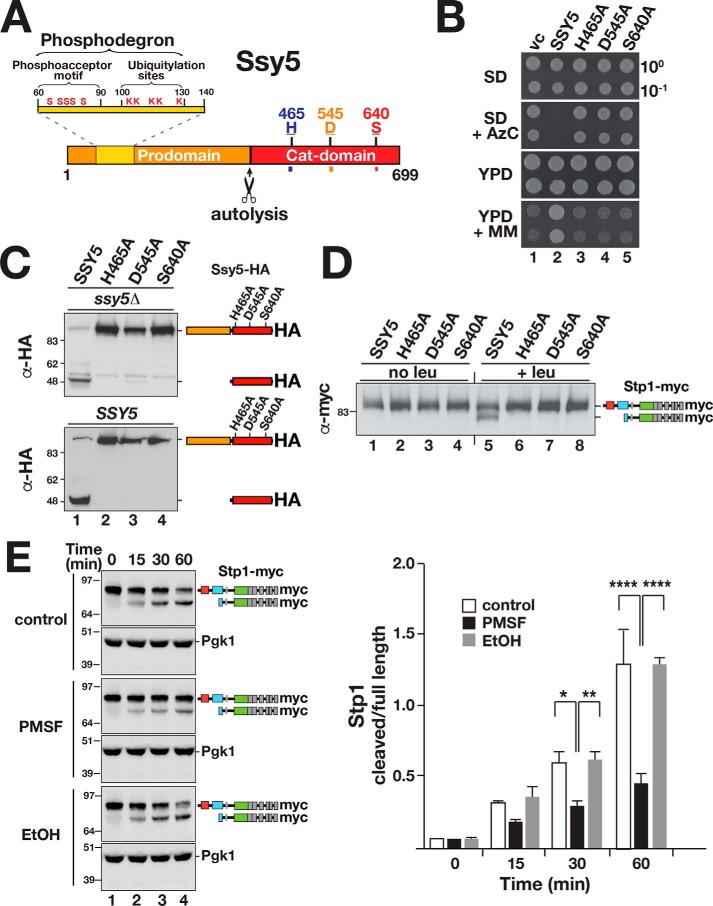
The Ssy5 Cat domain contains three residues, His-465, Asp-545, and Ser-640, that appear in a sequence context similar to the catalytic triad of well-characterized serine proteases (Fig. 1A). The replacement of the putative nucleophilic serine 640 with alanine results in the loss of amino acid-induced SPS sensor signaling (25, 27). To test the requirement of the catalytic triad, we constructed alleles with alanine substitutions at the three critical residues, His-465, Asp-545, and Ser-640, and assessed the ability of these mutant alleles to complement ssy5Δ in growth-based assays. Azetidine-2-carboxylic acid (AzC) is a toxic proline analog and 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino-]-sulfonyl}-benzoic acid (MM) is an inhibitor of branched-chain amino acid biosynthesis. When grown on YPD media, cells lacking a functional Ssy5 are resistant to AzC and sensitive to MM; mutant cells do not take up the toxic proline analog and are unable to take up sufficient branched-chain amino acids to support growth. The mutant alleles failed to complement ssy5Δ indicating that they encoded nonfunctional proteins (Fig. 1B, dilutions 3–5). Notably, the mutant proteins did not exhibit autolytic processing (Fig. 1C, upper panel, lanes 2–4) and were incapable of proteolytically processing Stp1 (Fig. 1D). Next, we examined whether autolytic processing of Ssy5 is a strictly intramolecular event. The cleavage of the mutant Ssy5 proteins was assessed in WT cells with an intact and functional SSY5 locus. No processing was observed (Fig. 1C, lower panel, lanes 2–4), indicating that Ssy5 cleavage occurs via an intramolecular event and cannot be catalyzed in trans.
Figure 1.
Ssy5 is a serine protease that undergoes autolytic processing. A, schematic diagram of the Ssy5. Scissors indicate the autolytic cleavage site. The prodomain (residues 1–381) includes a phosphodegron (residues 60–130) containing the phosphoacceptor motif (conserved serine residues and ubiquitylation sites, Ser and Lys, respectively) (22). The Cat domain (residues 382–699) presents a chymotrypsin-like catalytic triad composed of residues His-465, Asp-545, and the nucleophilic Ser-640. B, growth of ssy5Δ strain CAY265 transformed with pRS316 (vc), pCA177 (SSY5), pCA215 (ssy5-His-465A), pCA216 (ssy5-Asp-545A), or pCA217 (ssy5-Ser-640A). Dilutions of cultures grown in SD were spotted onto SD, SD+AzC, YPD, and YPD+MM, and the plates were incubated at 30 °C. C, autolytic processing of Ssy5 requires a functional catalytic triad. Immunoblot analysis of extracts prepared from HKY77 (ssy5Δ; upper panel) and CAY29 (SSY5; lower panel) carrying pCA177 (SSY5), pCA215 (ssy5-His-465A), pCA216 (ssy5-Asp-545A), or pCA217 (ssy5-Ser-640A). Cells were grown in SD in the absence of inducing amino acids. The immunoreactive forms of the HA-tagged constructs are schematically depicted at their corresponding positions of migration. D, Stp1 cleavage by Ssy5 requires a functional catalytic triad. Immunoblot analysis of protein extracts from HKY77 (ssy5Δ) carrying plasmids as in C grown in SD (no leu) and 30 min after the induction by leucine (+leu). The immunoreactive forms of Stp1 are schematically depicted. E, serine protease inhibitor PMSF inhibits Ssy5. Immunoblot analysis of in vitro Stp1 cleavage reactions. Lysates of strain AMY001 (ssy5Δstp1Δstp2Δprb1Δ) carrying pCA177 (Ssy5-HA) were prepared 30 min after induction with 1.3 mm leucine and mixed with lysates of AMY001 expressing Stp1-myc (pCA211) in the absence or presence of either 5 mm PMSF or an equal volume of ethanol 99.5% (EtOH). The immunoreactive forms of myc-tagged Stp1 are schematically represented at their corresponding positions of migration (left panel). The signal intensities of the cleaved and full-length forms of Stp1 were normalized to loading control Pgk1 (α-Pgk1 antibody) and quantified. The ratios between the values of cleaved and full-length forms of Stp1 were determined at the different time points and the mean values plotted (right panel). Error bars show standard deviation (n = 3). Two-way ANOVA followed by Tukey's multiple comparison test was performed using GraphPad Prism version 7.0. Significance values are indicated by the following: *, p = 0.013; **, p = 0.005; ****, p < 0.001. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
Ssy5 protease activity can be assayed in cell-free lysates in a manner that faithfully reproduces SPS sensor signaling (16). Briefly, this is carried out by combining protein lysates from ssy5Δ stp1Δ stp2Δ cells that express either Stp1-myc or Ssy5-HA. Stp1 cleavage is observed only if the Ssy5-HA expressing strain has been induced with leucine prior to lysate preparation. By quantifying the levels of processed and full-length forms of Stp1, and plotting their ratio over a period of 60 min, we noted a clear reduction in Ssy5-dependent processing activity in the presence of 5 mm PMSF (Fig. 1E). The solvent alone (EtOH) did not affect processing. The mutational and chemical inhibition data provide conclusive evidence that Ssy5 is a serine protease.
Prodomain is required as a translational fusion for Cat domain folding and substrate recognition
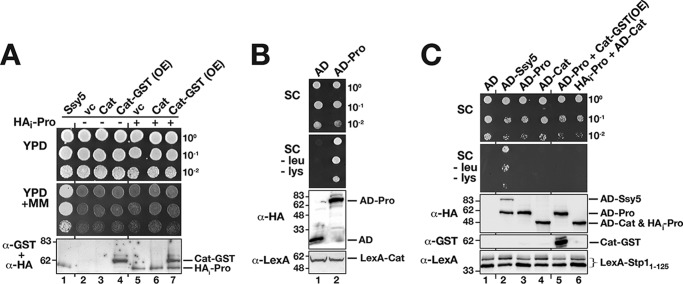
Co-expression of pro- and Cat domains, individually placed under the control of the weak endogenous SSY5 promoter (PSSY5), does not reconstitute a functional protease (16). This finding, consistent with the results obtained in Fig. 1, C and D, suggested that the functional association between the pro- and Cat domains is established in a co-translational manner. We tested this notion by overexpressing the Cat domain from the strong TDH3 promoter on a multicopy plasmid in combination with a single-copy vector expressing the prodomain (HAi-Pro) (Fig. 2A, lane 7) and did not detect any complementation of the ssy5Δ phenotype (Fig. 2A, dilution 7). Thus, the Cat domain does not fold into an active conformation even in the presence of high levels of co-expressed prodomain.
Figure 2.
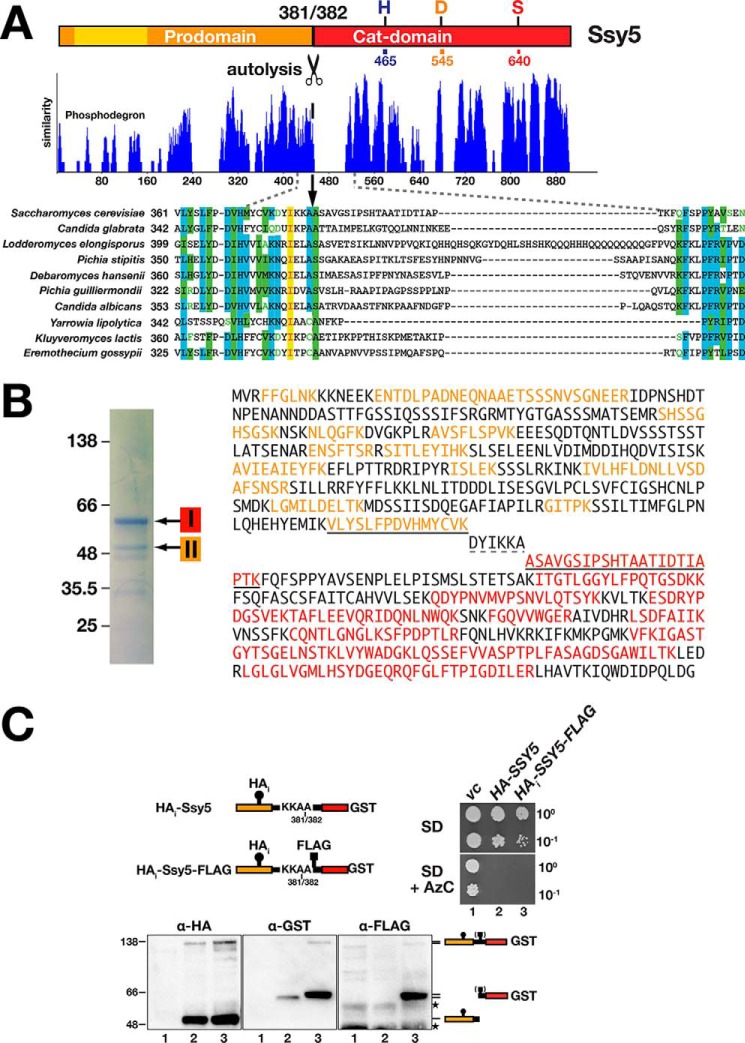
Prodomain is required in cis for Cat domain function and substrate interaction. A, co-expressed pro- and Cat domains of Ssy5 do not form a catalytically active protease. Growth of CAY309 (ssy5Δ) carrying plasmids pSH120 (SSY5), pRS316 (vc), pSH078 (PSSY5-Cat), or pSH098 (PTDH3-Cat-GST, overexpression construct, OE) and pRS314 (−) or pSH043 (HAi-Pro, +) as indicated. 10-Fold dilution series of cell suspensions were spotted onto YPD and YPD+MM. Immunoblot analysis of cell lysates (α-HA and α-GST antibodies) is shown; the immunoreactive forms of the HAi-Pro and Cat-GST are indicated at their corresponding positions of migration. B, co-expressed pro- and Cat domains of Ssy5 interact. Plasmids directing the expression of prey proteins, either the activation domain alone (AD, pJG4–5) or AD fused to the prodomain (AD-Pro, pSH096), were introduced into the two-hybrid reporter strain SHY016 expressing the LexA-Cat bait construct (pSH095). Dilutions of cultures pregrown in SC (containing galactose) were spotted onto galactose containing SC and SC minus leucine and lysine as indicated. Immunoblot analysis of bait and prey constructs in cell lysates is shown; the immunoreactive forms of the prey and bait constructs are indicated at their corresponding positions of migration. C, Stp1 interacts with Ssy5 in a conformationally-dependent manner. Dilutions of the yeast two-hybrid reporter strain SHY016 carrying bait construct LexA-Stp11-125 (pCA147) together with prey plasmids AD (pJG4-5), AD-Ssy5 (pSH043), AD-Pro (pSH096), AD-Cat (pSH125), AD-Pro (pSH096), and overexpressed Cat (pSH076, Cat-GST, and OE), or with HAi-Pro (pSH076) and AD-Cat (pSH125). Immunoblot analysis of bait and prey constructs in cell lysates; the immunoreactive forms of expressed proteins are indicated at their corresponding position of migration. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
Next, we used a directed two-hybrid approach to investigate whether the lack of Cat domain function was due to its inability to interact with the co-expressed prodomain. We constructed a bait plasmid composed of the Cat domain fused to the LexA DNA-binding domain, and a prey composed of the prodomain fused to the B42 transcriptional activator (AD), and we tested whether this interaction pair activated LexA-operated LYS2 and LEU2 reporters. Co-expression resulted in growth on the reporter-selective media (Fig. 2B). Thus, individually expressed pro- and Cat domains do interact but not in a manner that facilitates Cat domain folding (Fig. 2B).
We exploited our previous finding that Stp1 interacts with Ssy5 in a two-hybrid setup to assess the folding status of individually and co-expressed domains. Three prey proteins, i.e. full-length Ssy5 (AD-Ssy5), prodomain (AD-Pro), and Cat domain (AD-Cat), were fused to the B42 transcriptional activator and tested for their ability to interact with the first 125 N-terminal residues of Stp1 fused to LexA. As reported previously, the N terminus of Stp1 interacts with full-length Ssy5 (Fig. 2C, dilution 2) (16). In contrast, no interaction between Stp1 and individually expressed pro- and Cat domains was observed (Fig. 2C, dilutions and lanes 3 and 4). Other Ssy5 fragments, including regions 220–699, 388–699, 1–448, and 91–220, also failed to interact with Stp1 (data not shown). Finally, we assessed whether co-expression of the Cat domains (Cat-GST) or prodomains (HAi -Pro) with the AD-Pro and AD-Cat constructs, respectively, would enable Stp1 binding; no interaction was detected (Fig. 2C, dilutions and lanes 5 and 6). Taken together, our data indicate that the pro- and Cat domains of Ssy5 must be expressed as a single peptide to enable the Cat domain to fold into a catalytically active protease capable of substrate recognition.
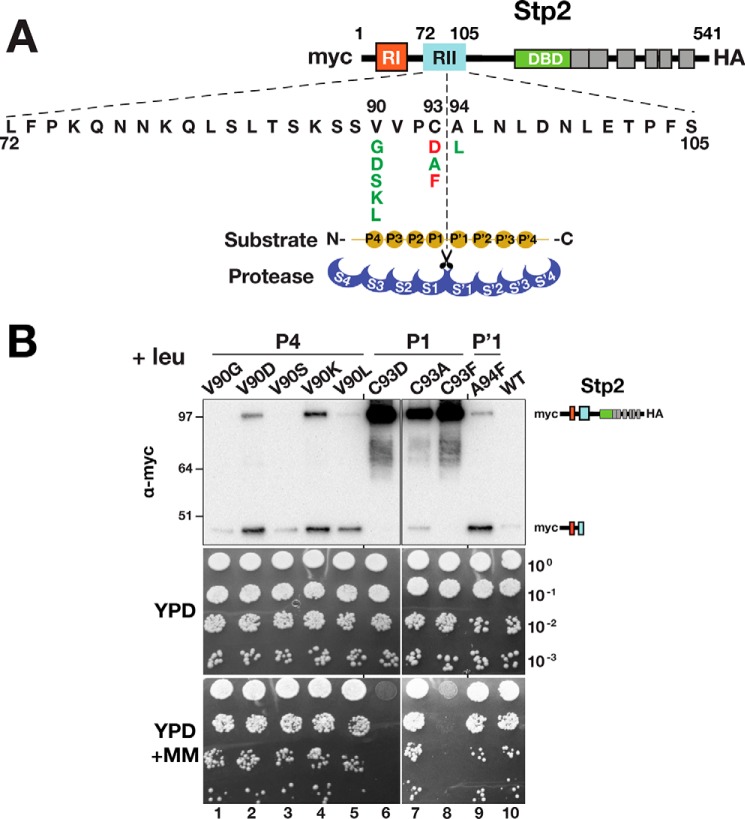
Ssy5 cleaves its substrates between a cysteine and a serine (Stp1) or alanine (Stp2)
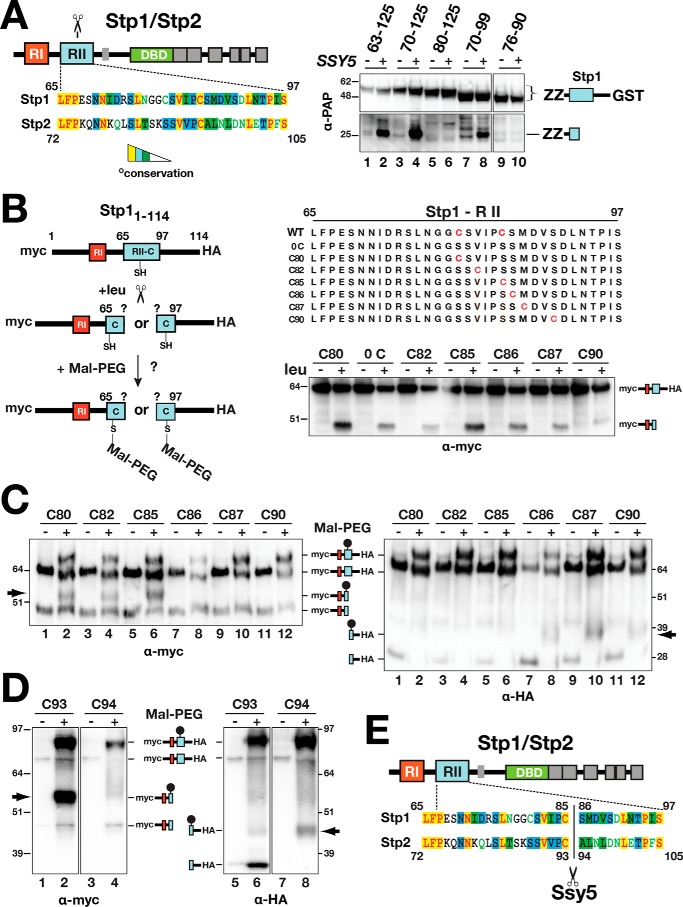
To better understand the catalytic properties of Ssy5, we determined the precise cleavage site in its only known substrates Stp1 and Stp2. Based on mutational analysis, the conserved N-terminal regulatory motif region II (RII) is predicted to encompass the Ssy5 cleavage site (11). Consistent with this notion, a minimal fragment of Stp1, including RII, amino acid residues 70–99, is cleaved in an SPS sensor-dependent manner (Fig. 3A, right panel, lane 8). Truncated versions of Stp1 (residues 1–114) and Stp2 (residues 1–102) carrying N-terminal 13xmyc- and C-terminal 6xHA-tags, without cysteine (0C; serine substituted at positions Cys-80 and Cys-85) and single cysteine residues at distinct positions (Cys-80, Cys-82, Cys-85, Cys-86, Cys-87, and Cys-80) were created (Fig. 3B). These constructs were tested as substrates; cleavage was monitored by immunoblot analysis, and all were processed in an SPS sensor-dependent manner (Fig. 3B, right panel). Detection of cysteine residues present in the cleavage products of the Stp1/Stp2 derivatives was achieved through labeling with the thiol-reactive reagent PEG-maleimide (Fig. 3B, left panel) and assessing the altered electrophoretic mobility by immunoblot analysis (Fig. 3C).
Figure 3.
Determination of the Ssy5 cleavage site in Stp1 and Stp2. A, schematic representation of Stp1 and Stp2 indicating the location of regulatory regions I (RI) and II (RII) and the DNA-binding domains (DBD) (left panel). The Ssy5 cleavage site (scissors) is within region II. Sequence alignment of region II from Stp1 and Stp2 (AlignX, Vector NTI); identical (yellow), conservative (blue), and similar (green) residues are highlighted, and residues with weak (green text) or no similarity (black) are indicated. Strain CAY328 (ssy5Δ) carrying pRS316 (−) or pSH120 (SSY5, +) and plasmids expressing PGAL1-promoted Stp1 fragments, aa 63–125 (pCA221), aa 70–125 (pCA222), aa 80–125 (pCA237), aa 70–99 (pCA246), or aa76–90 (pCA254) in the context of a fusion between a dual immunoglobulin-binding Z domain (ZZ) and GST, was grown overnight in YP with 2% EtOH and subsequently suspended at an OD600 of 1 in YP with 2% galactose for 2 h to induce expression of the Stp1 fragments. Extracts were analyzed by immunoblot using PAP (1:5000), a polyclonal horseradish peroxidase-conjugated immunocomplex that binds the ZZ tag (right panel). The immunoreactive forms of the Stp1 constructs are depicted at their corresponding positions of migration. B, schematic depiction of the maleimide-PEG labeling strategy to map the scissile bonds in Stp1 cleaved by Ssy5 (left panel). WT 13xMYC-STP11–114-ZZ-6xHA and engineered constructs (right panel) containing no cysteine or a single cysteine residue (red) were individually introduced into CAY123 (stp1Δstp2Δ). Extracts, prepared from the strains grown in SD (−leu) and 30 min after induction by leucine (+leu), were analyzed by immunoblotting with α-myc antibodies; the immunoreactive forms of the myc-tagged constructs are depicted at their corresponding positions of migration (right panel). C, Stp1 cleavage. Strain CAY123 (stp1Δstp2Δ) carrying pAM014 (C80), pAM018 (C82), pAM017 (C85), pAM020 (C86), pAM021 (C87), or pAM022 (C90) was grown in SD, and extracts, prepared 30 min after induction by 1.3 mm leucine, were treated with maleimide-PEG as indicated. Depending on the site of cleavage and the localization of the cysteine residue, the maleimide-PEG label will increase the molecular mass of either the N- or C-terminal fragment by 5 kDa (arrows). Immunoblots were developed with α-myc (left panel) or α-HA (right panel) antibodies. The immunoreactive forms of myc-tagged N- and C-terminal HA-tagged constructs are depicted at their corresponding positions of migration. D, Stp2 cleavage. Immunoblot analysis of protein extracts from strain CAY123 (stp1Δstp2Δ) carrying pAM080 (C93) or pAM081 (C94). Cells were grown, and extracts were treated with maleimide-PEG as in C. The immunoreactive forms of myc-tagged N- and C-terminal HA-tagged constructs are depicted at their corresponding positions of migration. E, summary diagram of the scissile bonds in Stp1 (aa 85/86) and Stp2 (93/94) cleaved by Ssy5. Molecular markers (kDa) are indicated at the position of migration (left and right of immunoblots).
Upon leucine induction and PEG-maleimide treatment, a molecular weight shift in the 13myc-tagged N-terminal cleavage product was observed when cysteine occupied amino acid positions 80, 82, and 85 (Fig. 3C, left panel, lanes 2, 4, and 6, arrow) but not when placed at positions 86, 87, and 90 (Fig. 3C, left panel, lanes 8, 10, and 12). These results indicate that Cys-85 is the C-terminal amino acid of the N-terminal cleavage fragment. A molecular weight shift of the C-terminal 6xHA-tagged cleavage fragment was observed when cysteine was placed at positions 86, 87, and 90 (Fig. 3C, right panel, lanes 8, 10, and 12, arrow) but not when present at positions 80, 82, and 85 (Fig. 3C, right panel, lanes 2, 4, and 6). Together, these findings demonstrate that Ssy5 catalyzes the cleavage of Stp1 between cysteine 85 and serine 86. Using a similar approach, we unambiguously identified the Ssy5 cleavage site in Stp2 and found that cleavage occurs between cysteine 93 and alanine 94 within a conserved sequence motif that aligns well with Stp1 (Fig. 3, A, D and E). This was accomplished by analyzing a Stp2 fragment encompassing residues 1–102. Together, the data establish the precise and conserved cleavage sites in both Stp1 and Stp2 (Fig. 3E).
Ssy5 requires a small amino acid residue at the P1 site
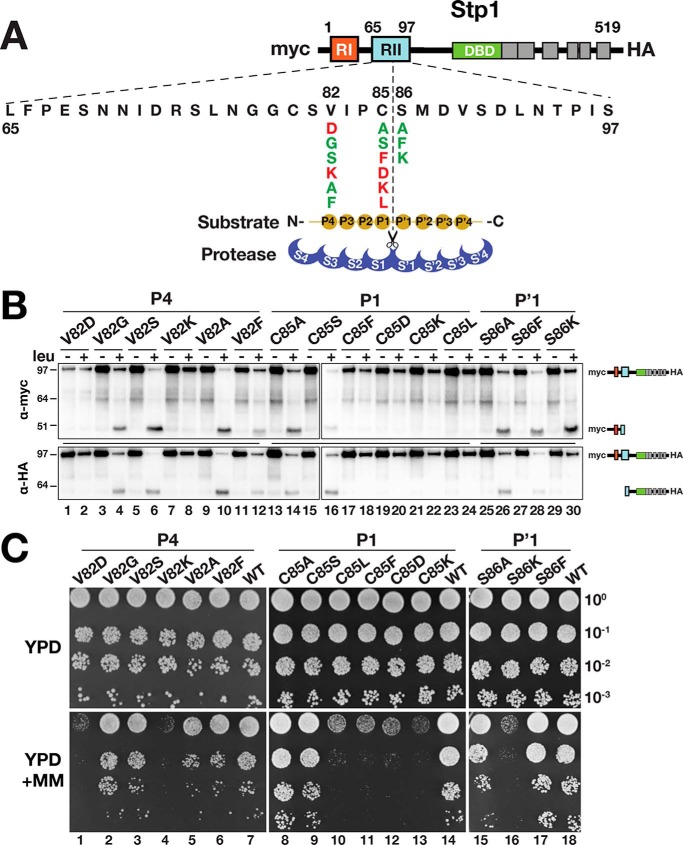
Knowledge regarding the precise cleavage site in Stp1 and Stp2 provided the basis to scrutinize the substrate sequence requirements. According to the Schechter and Berger nomenclature (28), the active site of a protease is composed of several subsites, S pockets, that accommodate consecutive amino acids of the substrate (called P sites). The substrate specificity of chymotrypsin-like serine proteases is usually classified in terms of the P1–S1 interaction (29). We constructed mutant Stp1 variants carrying amino acid substitutions at the P1 and P′1 positions as well as at the P4 position. Three categories of amino acids were used to replace the native residues at the desired positions: charged (aspartate and lysine); bulky hydrophobic (leucine and phenylalanine); and small (serine, alanine, and glycine) (Fig. 4A). Immunoblot analysis revealed that charged and hydrophobic residues at the P1 position render Stp1 noncleavable (Fig. 4B, lanes 17–24). Accordingly, these variants failed to complement the stp1Δ stp2Δ growth phenotype on MM (Fig. 4C, dilutions 10–13). The effect of residue replacements at the P4 and P′1 position (Fig. 4A) revealed that the conserved valine at the P4 position is important for Stp1 cleavage. When charged amino acids were placed in the P4 position, Stp1 was not cleaved (Fig. 4B, lanes 1, 2, 7, and 8) and did not support growth on MM (Fig. 4C, dilutions 1 and 4). Substitutions with alanine and phenylalanine at the P′1 position did not impair cleavage (Fig. 4B, lanes 25–30). However, lysine at this position rendered Stp1 noncleavable and unable to support growth on MM (Fig. 4C, dilution 16). Further investigation of the S86K variant revealed that the C-terminal cleavage fragment was unstable; the cleavage product could not be detected by immunoblot analysis (Fig. 4B, lane 30). An N-terminal lysine is known to be destabilizing in the context of N-end rule substrates (30, 31), and thus it is likely that lack of growth on MM is due to the inherent instability of the modified C-terminal fragment.
Figure 4.
Ssy5 exhibits a strong preference for substrates with a small amino acid at the P1 position. A, schematic presentation of Stp1 with expanded RII; the Ssy5 cleavage site, substrate P and P′ sites, as well as the corresponding S and S′ protease subsites are depicted according to Ref. 28. Amino acid substitutions at the P4, P1, and P′1 positions and summary of their effect on Ssy5 cleavage and growth is as shown in B and C: red, impaired; green, neutral. B, Ssy5 cannot cleave substrates with either bulky or charged amino acid residues at the P1 position or charged amino acid residues at the P4 position. Immunoblot analysis of protein extracts from strain CAY123 (stp1Δstp2Δ) carrying plasmids pAM029 (V82D), pAM030 (V82G), pAM046 (V82S), pAM047 (V82K), pAM048 (V82A), pAM049 (V82F), pAM050 (C85A), pAM051 (C85S), pAM053 (C85F), pAM054 (C85D), pAM055 (C85K), pAM052 (C85L), pAM056 (S86A), pAM057 (S86F), or pAM058 (S86K). Immunoblot developed with α-myc (upper panel) or α-HA (lower panel). The immunoreactive forms of N-terminal myc-tagged and C-terminal HA-tagged Stp1 fragments are schematically depicted at their corresponding positions of migration. C, growth of strain CAY123 (stp1Δstp2Δ) carrying plasmids as in B, including pMB31 (WT). 10-Fold dilutions of cultures grown in SD were spotted onto YPD and YPD+MM, and plates were incubated at 30 °C for 2 days and photographed. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
To test whether these sequence requirements are specific to Stp1, we performed the same range of amino acid substitutions in Stp2. As observed for Stp1, charged and large hydrophobic residues at the P1 position rendered Stp2 noncleavable and unable to complement stp1Δ stp2Δ phenotypes (Fig. 5B, lanes and dilutions 6 and 8). Interestingly and contrary to Stp1, a charged amino acid at the P4 position did not impair Stp2 cleavage or its ability to complement stp1Δ stp2Δ (Fig. 5B, lanes and dilutions 2 and 4). A less strict P4 requirement for Stp2 cleavage could be the consequence of the nonconserved residues spanning the P4–P′4 region, positively affecting Ssy5 subsite cooperativities. Alternatively, differences at more remote sites of Stp2 could contribute to an enhanced affinity for Ssy5.
Figure 5.
Ssy5 cleavage of Stp2 is not impaired by charged amino acids at the P4 position. A, schematic presentation of Stp2 with expanded RII; the Ssy5 cleavage site, substrate P and P′ sites, as well as the corresponding S and S′ protease subsites are depicted (28). Amino acid substitutions at the P4, P1, and P′1 positions and summary of their effect on Ssy5 cleavage and growth as in B: red, impaired; green, neutral. B, Ssy5 cleavage of Stp2 (upper panels). Immunoblot analysis of protein extracts from strain CAY123 (stp1Δstp2Δ) carrying plasmids pAM059 (V90G), pAM060 (V90D), pAM063 (V90S), pAM064 (V90K), pAM061 (C93A), pAM062 (C93D), pAM069 (C93F), pAM068 (A94F), or pMB30 (WT). Immunoblots were developed with α-myc; the immunoreactive forms of N-terminal myc-tagged Stp2 are schematically depicted at their corresponding positions of migration. Growth of strain CAY123 (stp1Δstp2Δ) carrying plasmids as in the upper panel. 10-Fold dilutions of cultures grown in SD were spotted onto YPD and YPD+MM; plates were incubated at 30 °C for 2 days and photographed. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
Autolytic cleavage in Ssy5 occurs between Ala-381 and Ala-382
It has previously been proposed, based on sequence homology between Stp1 and Ssy5 and the availability of mutations that abolish autolysis, that autolytic processing of Ssy5 occurs somewhere between acids 409 and 432 (25). By contrast, N-terminal sequence analysis of purified Cat domain identified the N-terminal amino acid as Ala-382, suggesting that autolysis occurs between amino acids 381 and 382 (17). Strikingly, Ala-382 and the region immediately preceding this residue are highly conserved in all Ssy5 orthologs; by contrast, the succeeding sequences are highly variable (Fig. 6A). We sought to independently define the site of autolysis. First, we purified Ssy5 (HAi-Ssy5-GST) and resolved the Cat domain (band I) and prodomain (band II) by SDS-PAGE (Fig. 6B, left panel). The bands were subjected to in-gel trypsin digestion, and the extracted peptides were identified by mass spectrometry (MS) (Fig. 6B, right panel). One of the peptides recovered from band 1 had a mass corresponding to the peptide ASAVGSIPSHTAATIDTIAPTK (aa 382–403). Sequencing of this peptide by MS/MS confirmed its identity (data not shown). This peptide is the most N-terminal in our band 1 data set, and its N terminus could not have been generated by trypsin digestion of Ssy5. We conclude that this peptide defines the N terminus of the purified Cat domain, a result that is consistent with cleavage occurring between amino acids 381 and 382 (17).
Figure 6.
Biochemical and in vivo determination of the Ssy5 autolytic cleavage site. A, Ssy5 from S. cerevisiae and orthologs from the related fungi were compared using sequence alignment algorithm AlignX (Vector NTI). The level of similarity of the aligned proteins is plotted; numbers correspond to amino acid residues of the longest ortholog (Lodderromyces elongisporus). The sequence alignment of the region containing the putative autoprocessing site (17) (scissors; between residues 381 and 382 in S. cerevisiae) is shown expanded below the plot. Identical (yellow), conservative (blue), and similar (green) residues are highlighted; residues with weak (green text) or no similarity (black) are indicated. B, mass spectrometric analysis of the Ssy5 pro- and Cat domain. Ssy5 was purified from lysates from CAY324 (ssy5Δ) carrying pCA260 (HAi-SSY5) grown in YPD. HAi-Ssy5 has a 3xHA tag (HAi) inserted internally in the prodomain and has GST fused to the C terminus of the Cat domain. Coomassie staining revealed two prominent species corresponding in size to the Cat domain (band I) and the prodomain (band II). The bands were subjected to trypsin digestion, and resulting peptides were analyzed by MS. The Ssy5 protein sequence with peptides recovered from band I and band II are highlighted in red and orange, respectively. The diagnostic peptides identified, i.e. the most C-terminal peptide of the prodomain and the most N-terminal peptide of the Cat domain are underlined (solid line). Amino acids DYIKKA (dashed underline) were not identified. C, in vivo analysis of Ssy5 autolysis. Schematic presentation of Ssy5 constructs; the positions of the 3xHA tag (HAi), autolytic processing site (between Ala-381 and Ala-382), FLAG-tag (inserted after Ala-382), and C-terminal GST fusion are indicated. Growth of strain PLY1351 (ssy5Δ) transformed with pRS316 (vc), pCA260 (HAi-Ssy5), or pTP103 (HAi -Ssy5-FLAG) were spotted on SD and SD medium containing AzC (right panel). Immunoblot analysis of extracts from the strains (lane 1, vc; lane 2, HAi-Ssy5; lane 3, HAi-Ssy5-FLAG) grown in SD (lower panel). The immunoreactive forms of Ssy5 are represented at their position of migration; the parentheses around the FLAG-tag symbol indicates that it is uniquely present in extracts in lanes 3. The position of unrelated cross-reacting proteins is marked with stars. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
To further investigate the cleavage site, we inserted a FLAG tag immediately after Ala-382 in a doubly tagged HAi-Ssy5-GST construct (Fig. 6C). The resulting HAi-SSY5-FLAG382 allele complements ssy5Δ phenotypes and therefore encodes a functional protein (Fig. 6C, right panel). Immunoblot analysis revealed that the internal FLAG tag co-migrates with the GST-tagged Cat domain of autoprocessed Ssy5 (Fig. 6C, lower panel) indicating that cleavage occurs N-terminal of alanine 382. Taken together, our data conclusively demonstrate that autolytic processing occurs between Ala-381 (P1) and Ala-382 (P′1) residues.
Ssy5 autolysis exhibits a less stringent sequence requirement
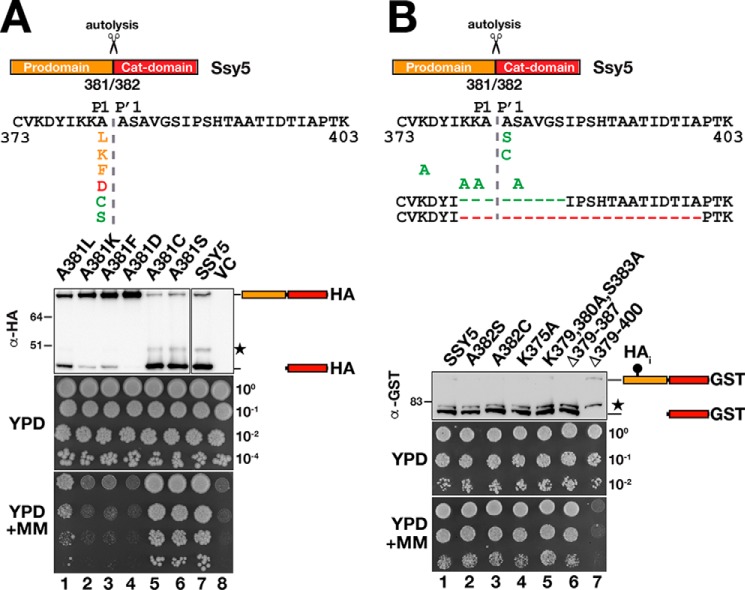
Autolysis occurs during biogenesis as the catalytic cleft attains a functional conformation. We tested the possibility that as an intramolecular event, autolysis may exhibit a less stringent sequence requirement. We replaced the P1 alanine (Ala-381) with a set of amino acids as indicated in Fig. 7A. Autolysis and complementation of ssy5Δ phenotypes were blocked or impaired when a charged (Lys, Asp) or bulkier hydrophobic (Leu, Phe) residue was introduced at the P1 position (Fig. 7A, lanes and dilutions 1–4). By contrast, replacing Ala-381 with C or S did not impair autolysis or growth (Fig. 7A, lanes and dilutions 5 and 6; compare with lane and dilution 7). These results show that autolysis presents similar sequence specificity requirements as Stp1 and Stp2 cleavage, at the P1 site.
Figure 7.
Ssy5 autolysis exhibits a less strict sequence requirement. Mutational analysis of the Ssy5 autoprocessing site was performed. Shown is a schematic presentation of Ssy5 expanding the sequence between amino acids 373 and 403 and including single amino acid substitutions at the P1 and P′1 positions, at other positions and deletions, as indicated. Mutations that block (red), impair (orange), or do not affect autolysis (green) are indicated. A, immunoblot analysis of extracts and growth of HKY77 (ssy5Δ) carrying plasmids pAM105 (A381L), pAM106 (A381K), pAM107 (A381F), pAM108 (A381D), pAM109 (A381C), pAM110 (A381S), pCA177 (SSY5), or pRS316 (VC). Immunoreactive forms of HA-tagged Ssy5 are indicated at their corresponding position of migration. The position of an unrelated cross-reacting protein is marked with a star. Ssy5 function was assessed by a growth-based assay, and 10-fold dilutions of cultures grown in SD were spotted onto YPD and YPD+MM. B, immunoblot analysis of extracts and growth of the CAY265 (ssy5Δ) carrying plasmids pSH120 (SSY5), pSH091 (A382S), pSH093 (A382C), pSH127 (K375A), pSH117 (K379A, K380A, and S383A), pSH128 (Δ379–387), or pSH129 (Δ379–400). Immunoreactive forms of Ssy5-GST are indicated at their corresponding position of migration. The position of an unrelated cross-reacting protein is marked with a star. Ssy5 function was assessed by growth on YPD and YPD+MM as in A. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
Next, we created Ssy5 mutant variants carrying substitutions at different residues surrounding the cleavage site, including the P′1 site (Fig. 7B). None of the mutant proteins, even the construct carrying a stretch of six continuous alanine residues (K379A, K380A, Ala-382, S383A, Ala-384), affected autolysis (Fig. 7B). Strikingly, a deletion encompassing nine residues (Δ379–387) spanning the autolytic cleavage site also did not impair cleavage or functionality of Ssy5 (Fig. 7B, lane and dilution 6); apparently, an autolytic cleavage event takes place at an alternative site in this mutant protein. By contrast, a more extensive deletion of 21 residues (Δ379–400), extending further into the nonconserved region upstream of the cleavage site, resulted in a nonfunctional ssy5 allele that weakly expressed a full-length protein devoid of autolytic activity (Fig. 7B, lane and dilution 7). The data indicate that the autolytic activity of Ssy5 exhibits similar but not as strict sequence specificity as observed for Stp1 and Stp2 cleavage. The P1 site residue appears to be the most important cleavage determinant.
Three residues in the Ssy5 catalytic cleft determine substrate specificity
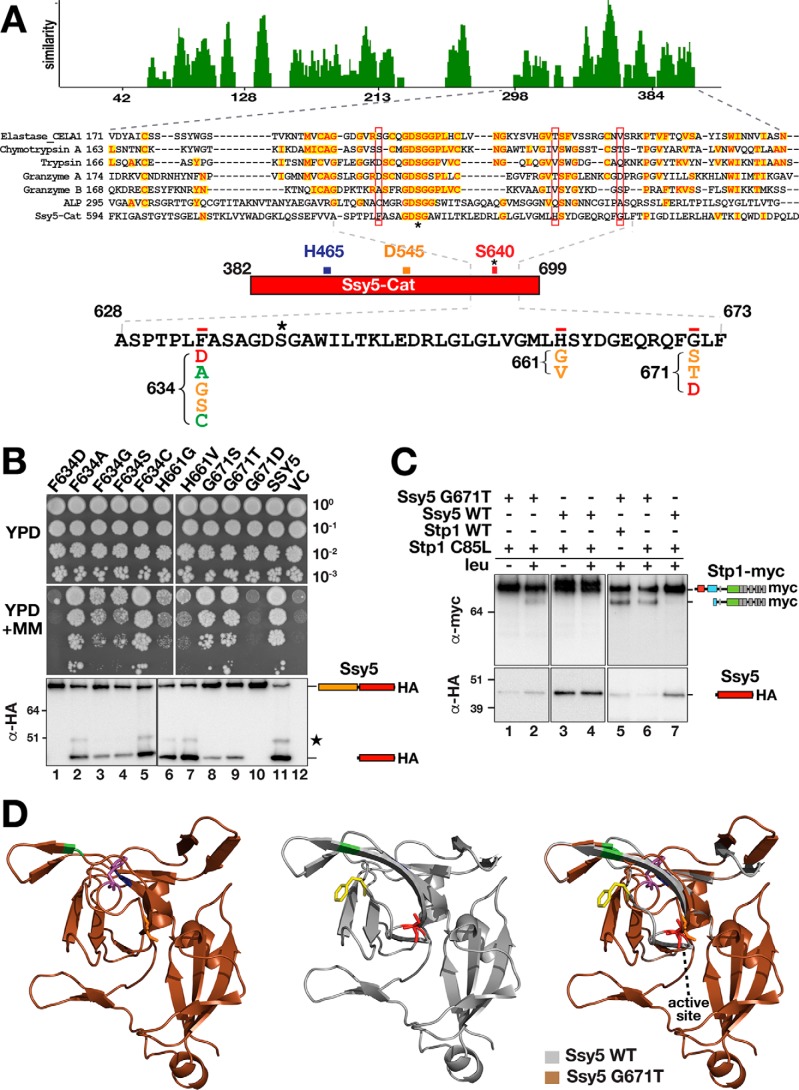
The findings indicating that a small amino acid residue at the P1 position is important for Ssy5 substrates suggested the existence of a shallow S1 subsite. The S1 site in chymotrypsin-like serine proteases is a pocket adjacent to the nucleophilic Ser-195, formed by residues 189–192, 214–216, and 224–228 (chymotrypsin numbering). The residues at positions 189, 216, and 226 usually determine specificity (29, 32). By comparing the Cat domain of Ssy5 with well-characterized serine proteases, we identified only a few conserved residues flanking the active site Ser-640 (Fig. 8A). Based on the amino acid sequence alignment with chymotrypsin A, we tested Ssy5 residues Phe-634, His-661, and Gly-671 for their ability to influence substrate specificity by replacing them with residues found in other serine proteases at the same position (Fig. 8A). We assessed autolysis and the ability of the mutant variants to complement the ssy5Δ growth phenotype (Fig. 8B). The results indicate that the mutant proteins could be classified into three categories as follows: category 1, inactive proteins, F634D and G671D, which did not exhibit autolytic cleavage and failed to complement ssy5Δ (Fig. 8B, dilutions and lanes 1 and 10); category 2, partially active proteins, F634G, F634S, H661G, H661V, G671S, and G671T, which exhibited reduced autolytic activities and/or partial complementation of ssy5Δ (Fig. 8B, dilutions and lanes 3, 4, and 6–9); and category 3, fully active proteins, F634A and F634C, which exhibited autolytic processing and full complementation of ssy5Δ (Fig. 8B, dilutions and lanes 2 and 5). These results suggest that the three residues selected are indeed important for Ssy5 catalytic activity.
Figure 8.
Mutational analysis of the Ssy5 catalytic cleft. A, sequences of the Ssy5 Cat domain and well-described serine proteases, elastase, chymotrypsin, trypsin, granzyme A, granzyme B, and α-lytic protease (ALP), were aligned according to the AlignX (Vector NTI) algorithm. The level of similarity is plotted using the residue numbers corresponding to α-lytic protease sequence. The alignment of the region containing the predicted active site, between residues 594 and 694 in Ssy5-Cat, is shown expanded below the plot. Identical (red, yellow highlight) as well as residues with no similarity (black) are indicated. The active-site serine is indicated by an asterisk. Residues 634, 661, and 671 in the Ssy5 Cat domain, predicted to affect specificity, are boxed in red. The Ssy5 sequence between 628 and 673 is expanded, and single amino acid substitutions at the 634, 661, and 671 positions are shown. Substitutions that block (red), impair (orange), or do not affect autolysis or growth (green) are indicated. B, growth and immunoblot analysis of extracts from HKY77 (ssy5Δ) carrying plasmids pAM111 (F634D), pAM112 (F634A), pAM113 (F634G), pAM114 (F634S), pAM115 (F634C), pAM116 (H661G), pAM117 (H661V), pAM118 (G671S), pAM119 (G671T), pAM120 (G671D), pCA177 (SSY5), or pRS316 (VC). 10-Fold dilutions of cultures grown in SD were spotted onto YPD and YPD+MM, and the immunoreactive forms of HA-tagged Ssy5 are indicated at their corresponding position of migration. C, Ssy5 G671T substitution exhibits altered substrate specificity. Immunoblot analysis of extracts from HKY77 (ssy5Δ) expressing Ssy5 and Stp1 proteins are as indicated: Ssy5-G671T (pAM119); stp1-C85L (pAM130); Ssy5 (pCA177); and Stp1 (pCA204). Extracts were prepared from uninduced (−) or leucine (1.3 mm, 30 min)-induced (+) cells. Immunoreactive forms of HA-tagged Ssy5 and myc-tagged Stp1 are indicated at their corresponding position of migration. D, molecular modeling of Ssy5 (middle, gray) and Ssy5 G671T (left, brown) were obtained using PHYRE2 and visualized with PyMOLTM. The differences between both structures are shown as color mismatch (right). The specific location of Phe-634, His-661, and Gly-671 are indicated in yellow, green, and blue, respectively. The active site Ser-640 is colored in red. Alterations in the active site and Phe-640 position predicted to occur as a consequence of the G671T substitution are indicated in orange and magenta highlights, respectively. Molecular markers (kDa) are indicated at the position of migration (left of immunoblots).
Next, we considered the possibility that the partially active category 2 mutant proteins may exhibit altered substrate specificities. We tested this by assessing whether these mutant proteins could cleave any of the P1 substitution mutations of Stp1 that were refractory to cleavage by WT Ssy5, i.e. C85F, C85D, C85K, and C85L (see Fig. 4). Protein extracts were prepared from strains individually expressing all possible combinations of category 2 Ssy5 mutants and the Stp1-P1 mutants. Immunoblot analysis showed that the Ssy5-G671T mutant exhibited an altered substrate affinity. In addition to processing Stp1 (Fig. 8C, lane 5), the Ssy5-G671T mutant also processed Stp1-C85L (Fig. 8C, lanes 2 and 6). Consequently, Ssy5-G671T accommodated the Stp1 mutant with a bulkier lateral chain at the P1 position and could catalyze its endoproteolytic processing.
To gain a better appreciation of the structural consequences of introducing the G671T mutation, we modeled the Cat domain of Ssy5 and its G671T mutant using PHYRE2 (33). The Ssy5 sequence fitted best to the α-lytic protease homolog of the thermophilic bacteria Thermobifida fusca (34). The structural models include the majority of the Cat domain, i.e. residues 456–681 (Fig. 8D), and have confidence levels of 98.4% for Ssy5 (middle panel) and 98.6% for Ssy5-G671T (left panel). By comparing the models, the Ssy5-G671T mutation is predicted to alter the structure close to the active site and in a more distal loop (Fig. 8D, right panel, brown and gray mismatch). Also, compared with native Ssy5, the nucleophilic Ser-640 attains a different orientation in Ssy5-G671T (Fig. 8D, red-Ssy5; orange-Ssy5-G671T). Strikingly, in the Ssy5-G671T mutant, residue Phe-634 attains an orientation that opens up the S1 subsite pocket (Fig. 8D compare yellow-Ssy5 with magenta-Ssy5-G671T). The orientation of the other residues tested, His-661 and Gly-671, are not predicted to significantly alter the Ssy5 structure (Fig. 8D, green and blue, respectively). The structural changes predicted by modeling are fully consistent with the ability of the G671T mutation to accommodate a bulkier R-group at the P1 position.
Discussion
Here, we report that Ssy5-dependent cleavage of Stp1 is inhibited by PMSF, a finding that provides the first experimental confirmation that Ssy5 is a serine protease. Previously, based on sequence homology, Ssy5 had been classified as a serine protease (MEROPS accession MER0043119), the single member of family S64 belonging to the Clan PA (35). The Cat domain of Ssy5 contains three residues (His-465, Asp-545, and Ser-640) predicted to constitute a chymotrypsin-like catalytic triad (25). Consistent with PMSF inhibition, we found that all three residues of the catalytic triad are critical for proteolytic activity.
The only known physiological substrates for Ssy5 are the transcription factors Stp1 and Stp2. To find the recognition sequence for Ssy5, we characterized the cleavage site and identified the scissile bonds. Sequence alignments show that the cysteine residue preceding the scissile bond (P1 site) is conserved in Stp1 and Stp2 orthologs. The proteolytic activity of Ssy5 requires small amino acid residues (serine, alanine, and cysteine) to be in the P1 position. The placement of charged or bulky hydrophobic residues at this position resulted in mutants Stp1 and Stp2 that were not cleaved and consistently unable to complement stp1Δstp2Δ phenotype.
Interestingly, in Stp1, we identified an additional more remote sequence determinant at the P4 position. This is unusual because few serine proteases are known to display selectivity for P4 residues (36). Specificity at the P1 and P4 residues is characteristic of the family of eukaryotic pro-protein–processing proteases for which the S. cerevisiae Kex2 protease is the prototype and of subtilisins (37, 38). Subtilisins are known for possessing an extended binding cleft comprising a minimum of eight subsites (38). Searching the MEROPS database reveals that only three serine proteases are known to cleave P4 valine- and P1 cysteine-containing substrates (35), indicating that the Ssy5 recognition sequence is uncommon.
Curiously, the P4 substitutions impairing Stp1 cleavage did not affect Ssy5's ability to process Stp2. This could be due to different residues, spanning the P4–P′4 region of Stp1 and Stp2, inducing a different Ssy5 subsite cooperativity. It is known that binding of a particular substrate residue at a protease subsite can have either a positive or negative influence on the binding of particular residues at other subsites (2). Subsite cooperativity has been observed in a wide range of proteases, often between nonadjacent subsites (2). The existence of different, more remote, binding sites facilitating a more stable attachment of Stp2 to Ssy5 substrate-binding site could also explain a less strict P4 requirement.
As observed for Stp1 and Stp2, autolytic processing of Ssy5 displays similar amino acid requirements at the P1 position. However, it is important to note that the severity of such mutations on autolysis is lower when compared with the effects on Stp1 and Stp2 cleavage. Furthermore, alanine substitutions around the autolysis site and even the deletion of nine residues spanning the active site did not affect autolysis. This suggests that compared with Stp1 and Stp2 cleavage, autolytic processing exhibits a less strict sequence requirement. This may be the consequence of autolysis occurring before the active site attains its final conformation. Likewise, autolytic processing of α-lytic protease precedes and is necessary for folding and catalytic maturation of the C-terminal domain (19, 39). Interestingly, as observed for Stp1, the P4 position in Ssy5 appears to be important for autolysis; we have reported that several substitutions of the conserved isoleucine 378 impair function as assessed by growth-based assays (40).
Together, the results indicate that Ssy5 substrate specificity is primarily determined by the nature of the amino acid residue present at the P1 position, a feature typical of the chymotrypsin family of serine proteases (32). Serine proteases have substrate-binding pockets that are tailored to bind the side chain of the residue preceding the scissile bond (P1) (41). Our finding that only small amino acids are accepted at the P1 position suggested Ssy5 possesses a shallow S1 subsite that cannot accommodate large amino acid side chains. Three residues in the putative Ssy5 active site, Phe-634, His-661, and Gly-671, were found to influence Ssy5 autolytic processing and activity. Interestingly, the mutation G671T renders an Ssy5 mutant variant with altered substrate specificity. This mutant retained the ability to cleave Stp1 but gained the capacity to cleave a mutant Stp1 variant (C85L) not processed by WT Ssy5. Apparently, residue Gly-671 is a major determinant of the Ssy5 S1 subsite architecture. Consistent with this notion, structural modeling predicted that the G671T mutation would alter the active site by affecting the orientation of Ser-640 and Phe-634. The predicted structural changes appear to explain the impaired autolytic activity and altered specificity demonstrated by the G671T mutant.
With regard to Ssy5 biogenesis, we found that autolytic processing is a strictly intramolecular event. This differentiates Ssy5 from classical serine proteases that usually are activated by intermolecular proteolytic processing of an inactive zymogen precursor (42–45). Intriguingly, we found that the individually expressed Cat domain and prodomain physically interact; however, this interaction does not suffice to enable the Cat domain to attain a folding status compatible with substrate binding. These results align well with our previous observations (16) and suggest that the functional association between the pro- and Cat domains is required early during Ssy5 biogenesis.
The prodomain likely functions as a co-translational chaperone, or a folding scaffold, for the Cat domain. Similarly, the prodomain of the well-characterized secreted bacterial α-lytic proteases act as folding catalysts essential for their Cat domains to achieve functional native states within a biologically relevant time scale (18, 20, 46). The unusually large size of the α-lytic protease prodomain (166 amino acids) has been postulated to overcome the folding barrier that results in the high kinetic stability of the Cat domain (34). Consistently, correctly folded α-lytic Cat domain exhibits an equally large unfolding barrier (20, 47, 48). The fact that Ssy5 also has an extremely large prodomain (381 amino acids), suggests that the Ssy5 Cat domain may also exhibit high kinetic stability.
Modeling the Ssy5 sequence fitted best with the structure of the T. fusca protease A (TFPA), a thermophilic homolog of the α-lytic protease (34). TFPA exhibits enhanced stability at high temperatures, which has been attributed to a specific structural element, i.e. the domain bridge, that links the N- and C-terminal domains of the protease (34). The α-lytic protease subfamily contains some of the best-studied kinetically stable proteins, and the high free-energy barrier (ΔG) associated with folding and unfolding is central to their biological function in the harsh extracellular environment (34, 49). Consistent with the exceptional fit to the structure of TFPA, the Stp1-processing activity of the Cat domain is stable for a period exceeding 1 month in extracts stored on ice, as assessed by in vitro cleavage assays (data not shown).
It is interesting to consider the consequences of a protease with high-kinetic stability functioning in the framework of an intracellular signaling pathway. A major conundrum arises because within the context of SPS sensor signaling, amino acid induction irreversibly activates the Stp1- and Stp2-processing activity of Ssy5 Cat domain. An enhanced kinetic stability may be required to spare the Cat domain from degradation by the proteasome while it is still attached to a phosphorylated and polyubiquitylated prodomain (22, 50). However, this raises the question of how cells turn off the proteolytic activity of a kinetically stable intracellular protease once the inhibitory prodomain is degraded. Attempts to answer this question will open a new window to further understand the unusual catalytic properties and biological role of Ssy5. Finally, future work has the potential to facilitate the pharmacological targeting of Ssy5 as a means to control the growth of fungi, in which the SPS sensing pathway plays a central role in regulating metabolism, e.g. in the human pathogen Candida albicans (51, 52).
Experimental procedures
Media
Standard media, including yeast extract/peptone/dextrose (YPD) medium, ammonia-based synthetic minimal dextrose (SD) medium, supplemented as required to enable growth of auxotrophic strains, and ammonia-based synthetic complete dextrose (SC) were prepared as described previously (10). When needed, l-leucine was added at a concentration of 1.3 mm to induce SPS sensor signaling. Sensitivity to MM on complex medium is described elsewhere (11). Sensitivity to AzC was tested at a final concentration of 1 mm on solid leucine-supplemented SD medium.
Strains and plasmids
The construction of all S. cerevisiae strains, except AMY001 has been documented elsewhere, as indicated (Table 1). AMY001 was isolated as a G418-resistant clone obtained by transformation of MBY93 with a PCR product amplified from CAY324 that spans the PRB1 locus. With the exception of the two-hybrid strain SHY016, all strains are isogenic descendants of the S288C-derived strain AA255/PLY115 (53). The plasmids used in this work are listed in Table 2. The sequences of mutagenic oligonucleotides and PCR primers for homologous recombination are available upon request.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Ref. or source |
|---|---|---|
| AMY001 | MATa ura3-52 ssy5Δ1::hisG stp1Δ51::Agleu2 stp2Δ50::hphMX4 prb1Δ::kanMX | This work |
| CAY29 | MATa ura3-52 | 10 |
| CAY123 | MATa ura3-52 stp1Δ51::Agleu2 stp2Δ50::hphMX4 | 10 |
| CAY265 | MATa ura3-52 ssy5Δ2::hisG gap1Δ::PAGP1-lacZ | 16 |
| CAY309 | MATa lys2Δ201 trp1ΔFA ura3-52 ssy5Δ2::hisG | 16 |
| CAY324 | MATa ura3-52 ssy5Δ2::hisG prb1Δ::kanMX | 16 |
| CAY328 | MATa ura3-52 trp1Δ101::loxP ssy5Δ2::hisG | 16 |
| HKY77 | MATa lys2Δ201 ura3-52 ssy5Δ2::hisG | 14 |
| HKY84 | MATa lys2Δ201 ura3-52 ssy1Δ13::hisG ssy5Δ2::hisG | 14 |
| HKY85 | MATa lys2Δ201 ura3-52 ptr3Δ15::hisG ssy5Δ2::hisG | 14 |
| MBY93 | MATa ura3-52 ssy5Δ1:: hisG stp1Δ51::Agleu2 stp2Δ50::hphMX4 | 58 |
| PLY126 | MATa lys2Δ201 ura3-52 | 59 |
| SHY016 | MATα his3 trp1–100 ura3-52 leu2::6×lexAOp-LEU2 lys2Δ::8×lexAOp-LYS2 asi1Δ8::loxP-kanMX-loxP GAL2 | 16 |
Table 2.
Plasmids used in this study
| Plasmid | Description | Ref. |
|---|---|---|
| pAM014 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C85S)-ZZ-6xHA | This work |
| pAM016 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S C85S)-ZZ-6xHA | This work |
| pAM017 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S)-ZZ-6xHA | This work |
| pAM018 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S C85S V82C)-ZZ-6xHA | This work |
| pAM020 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S C85S S86C)-ZZ-6xHA | This work |
| pAM021 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S C85S M87C)-ZZ-6xHA | This work |
| pAM022 | pRS202 (URA3) containing 13xMYC-STP1 (1–114) (C44S C80S C85S S90C)-ZZ-6xHA | This work |
| pAM029 | pMB31 containing 13xMYC-STP1 (V82G)-6xHA | This work |
| pAM030 | pMB31 containing 13xMYC-STP1 (V82D)-6xHA | This work |
| pAM046 | pMB31 containing 13xMYC-STP1 (V82S)-6xHA | This work |
| pAM047 | pMB31 containing 13xMYC-STP1 (V82K)-6xHA | This work |
| pAM048 | pMB31 containing 13xMYC-STP1 (V82A)-6xHA | This work |
| pAM049 | pMB31 containing 13xMYC-STP1 (V82F)-6xHA | This work |
| pAM050 | pMB31 containing 13xMYC-STP1 (C85A)-6xHA | This work |
| pAM051 | pMB31 containing 13xMYC-STP1 (C85S)-6xHA | This work |
| pAM052 | pMB31 containing 13xMYC-STP1 (C85L)-6xHA | This work |
| pAM053 | pMB31 containing 13xMYC-STP1 (C85F)-6xHA | This work |
| pAM054 | pMB31 containing 13xMYC-STP1 (C85D)-6xHA | This work |
| pAM055 | pMB31 containing 13xMYC-STP1 (C85K)-6xHA | This work |
| pAM056 | pMB31 containing 13xMYC-STP1 (S86A)-6xHA | This work |
| pAM057 | pMB31 containing 13xMYC-STP1 (S86F)-6xHA | This work |
| pAM058 | pMB31 containing 13xMYC-STP1 (S86K)-6xHA | This work |
| pAM059 | pMB30 containing 13xMYC-STP2 (V90G)-6xHA | This work |
| pAM060 | pMB30 containing 13xMYC-STP2 (V90D)-6xHA | This work |
| pAM061 | pMB30 containing 13xMYC-STP2 (C93A)-6xHA | This work |
| pAM062 | pMB30 containing 13xMYC-STP2 (C93D)-6xHA | This work |
| pAM063 | pMB30 containing 13xMYC-STP2 (V90S)-6xHA | This work |
| pAM064 | pMB30 containing 13xMYC-STP2 (V90K)-6xHA | This work |
| pAM067 | pMB30 containing 13xMYC-STP2 (V90L)-6xHA | This work |
| pAM068 | pMB30 containing 13xMYC-STP2 (A94F)-6xHA | This work |
| pAM069 | pMB30 containing 13xMYC-STP2 (C93F)-6xHA | This work |
| pAM080 | pRS202 (URA3) containing 13xMYC-STP2 (1–102) (C59S C60S)-ZZ-6xHA) | This work |
| pAM081 | pRS202 (URA3) containing 13xMYC-STP2 (1–102) (C59S C60S C93S A94C)-ZZ-6xHA | This work |
| pAM105 | pCA177 containing SSY5 (A381L)-6xHA-KlTRP1 | This work |
| pAM106 | pCA177 containing SSY5 (A381K)-6xHA-KlTRP1 | This work |
| pAM107 | pCA177 containing SSY5 (A381F)-6xHA-KlTRP1 | This work |
| pAM108 | pCA177 containing SSY5 (A381D)-6xHA-KlTRP1 | This work |
| pAM109 | pCA177 containing SSY5 (A381C)-6xHA-KlTRP1 | This work |
| pAM110 | pCA177 containing SSY5 (A381S)-6xHA-KlTRP1 | This work |
| pAM111 | pCA177 containing SSY5 (F634D)-6xHA-KlTRP1 | This work |
| pAM112 | pCA177 containing SSY5 (F634A)-6xHA-KlTRP1 | This work |
| pAM113 | pCA177 containing SSY5 (F634G)-6xHA-KlTRP1 | This work |
| pAM114 | pCA177 containing SSY5 (F634S)-6xHA-KlTRP1 | This work |
| pAM115 | pCA177 containing SSY5 (F634C)-6xHA-KlTRP1 | This work |
| pAM116 | pCA177 containing SSY5 (H661G)-6xHA-KlTRP1 | This work |
| pAM117 | pCA177 containing SSY5 (H661V)-6xHA-KlTRP1 | This work |
| pAM118 | pCA177 containing SSY5 (G671S)-6xHA-KlTRP1 | This work |
| pAM119 | pCA177 containing SSY5 (G671T)-6xHA-KlTRP1 | This work |
| pAM120 | pCA177 containing SSY5 (G671D)-6xHA-KlTRP1 | This work |
| pAM130 | pCA204 containing STP1 (C85L)-13xMYC-kanMX | This work |
| pCA147 | pEG202 (HIS3) KmR G418R containing PADH1-STP (1–124)-LexA | This work |
| pCA177 | pRS316 (URA3) containing SSY5-6xHA-KlTRP1 | This work |
| pCA204 | pRS317 (LYS2) containing STP1-13xMYC-kanMX | This work |
| pCA215 | pRS316 (URA3) containing SSY5 (H465A)-6xHA-KlTRP1 | This work |
| pCA216 | pRS316 (URA3) containing SSY5 (D545A)-6xHA-KlTRP1 | This work |
| pCA217 | pRS316 (URA3) containing SSY5 (S640A)-6xHA-KlTRP1 | This work |
| pCA218 | pRS316 (URA3) containing PTDH3-SSY5–6xHA-KlTRP1 | This work |
| pCA221 | pEG-KT (leu2-d) KlTRP1-loxP-GAL1-loxP-ZZ-TEV-STP1 (63-125)-GST | 16 |
| pCA222 | pEG-KT (leu2-d) KlTRP1-loxP-GAL1-loxP-ZZ-TEV-STP1 (70–125)-GST | This work |
| pCA237 | pEG-KT (leu2-d)KlTRP1-loxP-GAL1-loxP-ZZ-TEV-STP1 (80–125)-GST | This work |
| pCA246 | pEG-KT (leu2-d) KlTRP1-loxP-GAL1-loxP-ZZ-TEV-STP1 (70–99)-GST | This work |
| pCA254 | pEG-KT (leu2-d) KlTRP1-loxP-GAL1-loxP-ZZ-TEV-STP1 (76–90)-GST | This work |
| pCA260 | pRS202 (URA3) containing PTDH3-HAi-SSY5-GST-SBP-kanMX | 16 |
| pMB31 | pRS202 (URA3) containing 13xMYC-STP1–6xHA | 58 |
| pMB30 | pRS202 (URA3) containing 13xMYC-STP2–6xHA | 60 |
| pSH043 | pJG4-5 (TRP1) containing PGAL1-SSY5 (1–699) | 16 |
| pSH076 | pRS316 (URA3) containing SSY5 (1–381)-GST | 16 |
| pSH078 | pRS316 (URA3) containing SSY5 (382–699)-GST | This work |
| pSH091 | pRS316 (URA3) containing SSY5 (A382S)-GST | This work |
| pSH093 | pRS316 (URA3) containing SSY5 (A382C)-GST | This work |
| pSH095 | pEG202 (HIS3) KmR G418R containing PADH1--LexA-SSY5 (382–699) | This work |
| pSH096 | pJG4-5 (TRP1) containing B42-HA-SSY5 (1–381) | This work |
| pSH097 | pJG4-5 (TRP1) containing PGAL1-HAi-SSY5 (1–381) | This work |
| pSH098 | pRS202 (URA3) containing PTDH3-SSY5 (382–699)-GST | This work |
| pSH117 | pRS316 (URA3) containing SSY5 (K379A K380A S383A)-GST | This work |
| pSH120 | pRS316 (URA3) containing HAi-SSY5 (1–699)-GST | 50 |
| pSH125 | pJG4-5 (TRP1) containing B42-HA-SSY5 (382–699) | This work |
| pSH127 | pRS316 (URA3) containing SSY5 (K375A)-GST | This work |
| pSH128 | pRS316 (URA3) containing SSY5 (Δ379–387)-GST | This work |
Directed yeast two-hybrid assay
Two-hybrid interactions between a LexA-fused Cat domain (pSH095) and an AD-fused Ssy5 pro-domain (pSH096) were tested in strain SHY016, which harbors an asi1 null allele to allow nuclear targeting of the LexA-Stp1(1–125) (16). Direct interactions, between the bait plasmid pCA147, expressing LexA-Stp1(1–125), and an AD-fused Ssy5 prodomain (pSH096) and Cat domain (pSH125) were tested in the same strain. Interactions were assayed for growth on SC plates lacking uracil, tryptophan, histidine, leucine, and lysine containing 4% galactose and 0.1% glucose as carbon sources.
Immunoblot analysis
Whole-cell extracts were prepared under denaturing conditions as described previously (54). Primary antibodies were diluted as follows: 12CA5 ascites fluid (anti-hemagglutinin (HA) mAb (55), 1:5000; purified goat polyclonal anti-GSH S-transferase (GST) antibody (ab6613, Abcam), 1:5000; anti-myc 9E10 mAb (Roche Applied Science), 1:5000; 3F10 anti-HA–horseradish peroxidase (HRP) (Roche Applied Science), 1:5000; anti-GST 3–4C mAb (Zymed Laboratories Inc.), 1:500; anti-myc-HRP 9E10 mAb (Roche Applied Science), 1:5000; anti-FLAG polyclonal (Sigma), 1:5000; anti-Pgk1 (Molecular Probes), 1:10,000. Dual immunoglobulin-binding Z domain (ZZ)-tagged proteins were detected using anti-HRP polyclonal antibody horseradish peroxidase immunocomplex (PAP, DakoCytomation), 1:5000. Immunoreactive bands were visualized by chemiluminescence detection (SuperSignal West Dura Extended-Duration Substrate; Thermo Fisher Scientific) of horseradish peroxidase using an LAS1000 system (Fuji Photo Film Co., Ltd.) or by near-IR fluorescence detection (Odyssey Fc; LI-COR Biosciences, Lincoln, NE). Quantification was performed using Image Studio Lite software (LI-COR Biosciences) by normalizing background-corrected signals to the Pgk1 loading control.
Maleimide-PEG labeling assay
Labeling of protein extracts with maleimide-PEG was performed following the procedure described in Ref. 56, with minor adjustments. Briefly, after TCA precipitation and washing, oxidized thiol groups present in the protein extracts were reduced by addition of 50 mm Bond-BreakerTM tris(2-carboxyethyl)phosphine solution (Thermo Fisher Scientific Inc.) in denaturing buffer consisting of 6 m urea, 200 mm Tris-HCl (pH 8.5), 10 mm EDTA, and 0.5% (w/v) SDS. After TCA precipitation, reduced cysteines were subsequently alkylated by addition of 20 mm methoxypolyethylene glycol maleimide (maleimide-PEG) (Sigma) in denaturing buffer. Samples were mixed with nonreducing SDS sample buffer and loaded onto 4–12% SDS-polyacrylamide gels.
Mass spectrometry
Ssy5 was purified from a 3-liter culture of ssy5Δ strain CAY324 carrying pCA260 (HAi-Ssy5) grown under inducing conditions (SC −ura) to optimize cell yield as described (16). After separation on SDS-PAGE, the bands corresponding to the prodomain and Cat domain were excised from the gel and subjected to in-gel trypsin digestion, essentially as described (57), however, with the following minor modifications. The destaining was performed with 35% acetonitrile (ACN) in 50 mm NH4HCO3. Prior to LC-MS analysis, peptides extracted from the gel were solubilized in 10 μl 5% ACN, 0.1% formic acid (FA).
Peptide samples were injected (4.9 μl) using partial loop mode onto a nanoAcquity UPLC system (Waters Corp.), plumbed for single pump trapping using a C18 trap column (5 μm, 2 cm) and a BEH C18 analytical column (1.7 μm, 10 cm) (Waters Corp.). Trapping was performed for 2 min at 15 μl min−1 using solvent A (0.1% FA). Subsequently, an analytical separation was performed at 400 nl min−1 with the following gradient: 1% B (100% ACN, 0.1% FA) (0–1 min), 1–40% B (1–55 min), 40–85% B (55–56 min), 85% B (56–60 min), 85–1% B (60–61 min), and 1% B (61.90 min).
The gradient flow from the nanoAcquity was delivered at 400 nl min−1 to a Q-TofTM II mass spectrometer (Micromass, Waters Corp.) with an ESI interface consisting of the Z spray source fitted with a nanoelectrospray probe (source voltage 2.9 kV, source temperature 80 °C, cone voltage 35 V, cone gas flow 50 liters/h). Data were acquired in data-dependent mode with one full scan (400–1700 m/z) followed by maximum four MS/MS scans (50–1800 m/z). External TOF mass calibration was performed prior to the UPLC-MS analysis. This was obtained by direct infusion of a solution containing [Glu1]fibrinopeptide B (1 μm) in 50% ACN, 0.1% FA, data acquisition in TOF MS/MS mode over the m/z range 50–2000, the collision energy set to 30 V, and mass set to 785.84 m/z ([Glu1]fibrinopeptide B, [M + 2H]2+).
Raw data files were converted to PKL files (background subtracted, deisotoped, and centroided) using the Protein Lynx Global Server (PLGS) software package (Waters Corp.) and searched against the S. cerevisiae entries in the National Center for Biotechnology Information nonredundant protein database (NCBInr) using Mascot (Matrix Science, London, UK) with the following search parameters: enzyme, trypsin or none; fixed modifications, carbamidomethylcysteine; variable modification, oxidized methionine; peptide tolerance, 100 ppm; peptide charge, +1: number of missed cleavage sites, up to 2: MS/MS tolerance, 0.1 Da.
In vitro protease assay
Yeast cells were grown in SD to an OD600 of 1. The cultures expressing Ssy5 were induced for 30 min with 1.3 mm leucine. Cells were disrupted by bead beating (three times for 20 s) in lysis buffer (50 mm HEPES-KOH, 100 mm NaCl, 5 mm DTT at pH 8.0), and nonlysed cells were removed by low-speed centrifugation (1200 × g) for 15 min. Protease reactions were prepared on ice in lysis buffer (pH 7.4) by mixing equal amounts of Stp1- and Ssy5-containing lysate. Reactions were incubated at 30 °C for 60 min, stopped by the addition of 2× sample buffer, boiled, and separated on SDS-PAGE. The ability to inhibit Stp1 cleavage was tested using a final concentration of 5 mm PMSF or an equal volume of 99.5% EtOH, added 30 min prior to substrate addition.
Author contributions
A. M., T. P., S. H., C. A., and P. O. L. conceptualization; A. M., T. P., S. H., G. S., C. A., and P. O. L. data curation; A. M., T. P., S. H., G. S., C. A., and P. O. L. formal analysis; A. M., T. P., S. H., G. S., C. A., and P. O. L. investigation; A. M., T. P., S. H., G. S., V. B., and C. A. methodology; A. M. and P. O. L. writing-original draft; A. M., T. P., S. H., V. B., C. A., and P. O. L. writing-review and editing; V. B. and P. O. L. supervision; V. B. and P. O. L. funding acquisition; P. O. L. visualization; P. O. L. project administration.
Acknowledgments
We thank members of the Ljungdahl laboratory for constructive comments throughout the course of this work.
This work was supported by Swedish Research Council Grants 2007-3894, 2011-5925, and 2015-04202 (to P. O. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- SPS
- Ssy1–Ptr3–Ssy5
- ACN
- acetonitrile
- AD
- activation domain
- AzC
- azetidine-2-carboxylic acid
- Cat
- catalytic
- EtOH
- ethanol
- FA
- formic acid
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- MM
- 2-{[({[(4-methoxy-6-methyl)-1,3,5-triazin-2-yl]-amino}carbonyl)amino]-sulfonyl}benzoic acid
- PMSF
- phenylmethylsulfonyl fluoride
- SC
- synthetic complete dextrose
- SD
- synthetic minimal dextrose
- vc
- vector control
- YPD
- yeast extract/peptone/dextrose
- ZZ
- dual immunoglobulin-binding Z domain
- aa
- amino acid
- TFPA
- T. fusca protease A
- PEG
- polyethylene glycol
- ACN
- acetonitrile
- FA
- formic acid.
References
- 1. Hooper N. M. (2002) Proteases: a primer. Essays Biochem. 38, 1–8 10.1042/bse0380001 [DOI] [PubMed] [Google Scholar]
- 2. Ng N. M., Pike R. N., and Boyd S. E. (2009) Subsite cooperativity in protease specificity. Biol. Chem. 390, 401–407 [DOI] [PubMed] [Google Scholar]
- 3. Turk B. (2006) Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799 10.1038/nrd2092 [DOI] [PubMed] [Google Scholar]
- 4. Turk B., Turk D., and Turk V. (2012) Protease signalling: the cutting edge. EMBO J. 31, 1630–1643 10.1038/emboj.2012.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. López-Otín C., and Bond J. S. (2008) Proteases: multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–30437 10.1074/jbc.R800035200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dean R. A., Butler G. S., Hamma-Kourbali Y., Delbé J., Brigstock D. R., Courty J., and Overall C. M. (2007) Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Mol. Cell. Biol. 27, 8454–8465 10.1128/MCB.00821-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. auf dem Keller U., Prudova A., Gioia M., Butler G. S., and Overall C. M. (2010) A statistics-based platform for quantitative N-terminome analysis and identification of protease cleavage products. Mol. Cell. Proteomics 9, 912–927 10.1074/mcp.M000032-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole S. L., and Vassar R. (2008) The role of amyloid precursor protein processing by BACE1, the β-secretase, in Alzheimer disease pathophysiology. J. Biol. Chem. 283, 29621–29625 10.1074/jbc.R800015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Egeblad M., and Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- 10. Andréasson C., and Ljungdahl P. O. (2002) Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16, 3158–3172 10.1101/gad.239202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andréasson C., and Ljungdahl P. O. (2004) The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24, 7503–7513 10.1128/MCB.24.17.7503-7513.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wielemans K., Jean C., Vissers S., and André B. (2010) Amino acid signaling in yeast: post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J. Biol. Chem. 285, 855–865 10.1074/jbc.M109.015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tumusiime S., Zhang C., Overstreet M. S., and Liu Z. (2011) Differential regulation of transcription factors Stp1 and Stp2 in the Ssy1-Ptr3-Ssy5 amino acid sensing pathway. J. Biol. Chem. 286, 4620–4631 10.1074/jbc.M110.195313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forsberg H., and Ljungdahl P. O. (2001) Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21, 814–826 10.1128/MCB.21.3.814-826.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ljungdahl P. O., and Daignan-Fornier B. (2012) Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190, 885–929 10.1534/genetics.111.133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andréasson C., Heessen S., and Ljungdahl P. O. (2006) Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20, 1563–1568 10.1101/gad.374206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poulsen P., Lo Leggio L., and Kielland-Brandt M. C. (2006) Mapping of an internal protease cleavage site in the Ssy5p component of the amino acid sensor of Saccharomyces cerevisiae and functional characterization of the resulting pro- and protease domains by gain-of-function genetics. Eukaryot. Cell 5, 601–608 10.1128/EC.5.3.601-608.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silen J. L., and Agard D. A. (1989) The α-lytic protease pro-region does not require a physical linkage to activate the protease domain in vivo. Nature 341, 462–464 10.1038/341462a0 [DOI] [PubMed] [Google Scholar]
- 19. Peters R. J., Shiau A. K., Sohl J. L., Anderson D. E., Tang G., Silen J. L., and Agard D. A. (1998) Pro region C terminus: protease active site interactions are critical in catalyzing the folding of α-lytic protease. Biochemistry 37, 12058–12067 10.1021/bi980883v [DOI] [PubMed] [Google Scholar]
- 20. Cunningham E. L., and Agard D. A. (2003) Interdependent folding of the N- and C-terminal domains defines the cooperative folding of α-lytic protease. Biochemistry 42, 13212–13219 10.1021/bi035409q [DOI] [PubMed] [Google Scholar]
- 21. Cunningham E. L., and Agard D. A. (2004) Disabling the folding catalyst is the last critical step in α-lytic protease folding. Protein Sci. 13, 325–331 10.1110/ps.03389704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Omnus D. J., Pfirrmann T., Andréasson C., and Ljungdahl P. O. (2011) A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol. Biol. Cell 22, 2754–2765 10.1091/mbc.E11-04-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Omnus D. J., and Ljungdahl P. O. (2013) Rts1-protein phosphatase 2A antagonizes Ptr3-mediated activation of the signaling protease Ssy5 by casein kinase I. Mol. Biol. Cell 24, 1480–1492 10.1091/mbc.E13-01-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu B., Ottow K., Poulsen P., Gaber R. F., Albers E., and Kielland-Brandt M. C. (2006) Competitive intra- and extracellular nutrient sensing by the transporter homolog Ssy1p. J. Cell Biol. 173, 327–331 10.1083/jcb.200602089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abdel-Sater F., El Bakkoury M., Urrestarazu A., Vissers S., and André B. (2004) Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24, 9771–9785 10.1128/MCB.24.22.9771-9785.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfirrmann T., and Ljungdahl P. O. (2012) in Handbook of Proteolytic Enzymes (Rawlings N. D., and Salvesen G. S., eds) pp. 3103–3110, Academic Press, Elsevier, Oxford, UK [Google Scholar]
- 27. Andréasson C. (2004) Ligand-activated Proteolysis in Nutrient Signaling. Ph.D. thesis, Karolinska Institutet [Google Scholar]
- 28. Schechter I., and Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 10.1016/S0006-291X(67)80055-X [DOI] [PubMed] [Google Scholar]
- 29. Hedstrom L. (2002) Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 10.1021/cr000033x [DOI] [PubMed] [Google Scholar]
- 30. Bartel B., Wünning I., and Varshavsky A. (1990) The recognition component of the N-end rule pathway. EMBO J. 9, 3179–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U.S.A. 93, 12142–12149 10.1073/pnas.93.22.12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perona J. J., and Craik C. S. (1995) Structural basis of substrate specificity in the serine proteases. Protein Sci. 4, 337–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelley L. A., and Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 34. Kelch B. A., and Agard D. A. (2007) Mesophile versus thermophile: insights into the structural mechanisms of kinetic stability. J. Mol. Biol. 370, 784–795 10.1016/j.jmb.2007.04.078 [DOI] [PubMed] [Google Scholar]
- 35. Rawlings N. D., Waller M., Barrett A. J., and Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 10.1093/nar/gkt953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Page M. J., Macgillivray R. T., and Di Cera E. (2005) Determinants of specificity in coagulation proteases. J. Thromb. Haemost. 3, 2401–2408 10.1111/j.1538-7836.2005.01456.x [DOI] [PubMed] [Google Scholar]
- 37. Rockwell N. C., and Fuller R. S. (1998) Interplay between S1 and S4 subsites in Kex2 protease: Kex2 exhibits dual specificity for the P4 side chain. Biochemistry 37, 3386–3391 10.1021/bi972534r [DOI] [PubMed] [Google Scholar]
- 38. Grøn H., Meldal M., and Breddam K. (1992) Extensive comparison of the substrate preferences of two subtilisins as determined with peptide substrates which are based on the principle of intramolecular quenching. Biochemistry 31, 6011–6018 10.1021/bi00141a008 [DOI] [PubMed] [Google Scholar]
- 39. Sauter N. K., Mau T., Rader S. D., and Agard D. A. (1998) Structure of α-lytic protease complexed with its pro region. Nat. Struct. Biol. 5, 945–950 10.1038/2919 [DOI] [PubMed] [Google Scholar]
- 40. Pfirrmann T., Lokapally A., Andréasson C., Ljungdahl P., and Hollemann T. (2013) SOMA: a single oligonucleotide mutagenesis and cloning approach. PLoS ONE 8, e64870 10.1371/journal.pone.0064870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pizzi E., Tramontano A., Tomei L., La Monica N., Failla C., Sardana M., Wood T., and De Francesco R. (1994) Molecular model of the specificity pocket of the hepatitis C virus protease: implications for substrate recognition. Proc. Natl. Acad. Sci. U.S.A. 91, 888–892 10.1073/pnas.91.3.888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eder J., and Fersht A. R. (1995) Pro-sequence-assisted protein folding. Mol. Microbiol. 16, 609–614 10.1111/j.1365-2958.1995.tb02423.x [DOI] [PubMed] [Google Scholar]
- 43. Stroud R. M., Kossiakoff A. A., and Chambers J. L. (1977) Mechanisms of zymogen activation. Annu. Rev. Biophys. Bioeng. 6, 177–193 10.1146/annurev.bb.06.060177.001141 [DOI] [PubMed] [Google Scholar]
- 44. Whitcomb D. C., and Lowe M. E. (2007) Human pancreatic digestive enzymes. Dig. Dis. Sci. 52, 1–17 10.1007/s10620-006-9589-z [DOI] [PubMed] [Google Scholar]
- 45. Neurath H., and Dixon G. H. (1957) Structure and activation of trypsinogen and chymotrypsinogen. Fed. Proc. 16, 791–801 [PubMed] [Google Scholar]
- 46. Fujishige A., Smith K. R., Silen J. L., and Agard D. A. (1992) Correct folding of α-lytic protease is required for its extracellular secretion from Escherichia coli. J. Cell Biol. 118, 33–42 10.1083/jcb.118.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sohl J. L., Jaswal S. S., and Agard D. A. (1998) Unfolded conformations of α-lytic protease are more stable than its native state. Nature 395, 817–819 10.1038/27470 [DOI] [PubMed] [Google Scholar]
- 48. Baker D., Sohl J. L., and Agard D. A. (1992) A protein-folding reaction under kinetic control. Nature 356, 263–265 10.1038/356263a0 [DOI] [PubMed] [Google Scholar]
- 49. Jaswal S. S., Sohl J. L., Davis J. H., and Agard D. A. (2002) Energetic landscape of α-lytic protease optimizes longevity through kinetic stability. Nature 415, 343–346 10.1038/415343a [DOI] [PubMed] [Google Scholar]
- 50. Pfirrmann T., Heessen S., Omnus D. J., Andréasson C., and Ljungdahl P. O. (2010) The prodomain of Ssy5 protease controls receptor-activated proteolysis of transcription factor Stp1. Mol. Cell Biol. 30, 3299–3309 10.1128/MCB.00323-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martínez P., and Ljungdahl P. O. (2005) Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25, 9435–9446 10.1128/MCB.25.21.9435-9446.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miramón P., and Lorenz M. C. (2016) The SPS amino acid sensor mediates nutrient acquisition and immune evasion in Candida albicans. Cell. Microbiol. 18, 1611–1624 10.1111/cmi.12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Antebi A., and Fink G. R. (1992) The yeast Ca2+-ATPase homolog, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3, 633–654 10.1091/mbc.3.6.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silve S., Volland C., Garnier C., Jund R., Chevallier M. R., and Haguenauer-Tsapis R. (1991) Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 11, 1114–1124 10.1128/MCB.11.2.1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., and Lerner R. A. (1984) The structure of an antigenic determinant in a protein. Cell. 37, 767–778 10.1016/0092-8674(84)90412-4 [DOI] [PubMed] [Google Scholar]
- 56. Tetsch L., Koller C., Dönhöfer A., and Jung K. (2011) Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 11, 74 10.1186/1471-2180-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kinter M., and Sherman N. E. (2005) in Protein Sequencing and Identification Using Tandem Mass Spectrometry, pp. 147–165, John Wiley & Sons, Inc., New York [Google Scholar]
- 58. Boban M., and Ljungdahl P. O. (2007) Dal81 enhances Stp1- and Stp2-dependent transcription necessitating negative modulation by inner nuclear membrane protein Asi1 in Saccharomyces cerevisiae. Genetics 176, 2087–2097 10.1534/genetics.107.075077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Klasson H., Fink G. R., and Ljungdahl P. O. (1999) Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416 10.1128/MCB.19.8.5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boban M., Zargari A., Andréasson C., Heessen S., Thyberg J., and Ljungdahl P. O. (2006) Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Biol. 173, 695–707 10.1083/jcb.200601011 [DOI] [PMC free article] [PubMed] [Google Scholar]