Abstract
Platelets are recruited to sites of vascular injury, where they are activated and aggregate to form a hemostatic plug. This process requires the activation of the small GTPase Rap1B by its cognate guanine nucleotide exchange factor CalDAG-GEFI. Studies on platelet function suggest that CalDAG-GEFI activity is regulated by changes in cytosolic calcium, but the exact molecular mechanism is poorly understood. Here we show that purified CalDAG-GEFI is autoinhibited and directly regulated by calcium. Substitutions of putative calcium-binding residues within the canonical EF hands of CalDAG-GEFI diminish its capacity to activate Rap1B. Structural differences between active (WT) and inactive (EF hand variant) CalDAG-GEFI protein were determined by hydrogen–deuterium exchange MS. The highest differential rates of deuterium uptake in WT over EF hand variant CalDAG-GEFI were observed in regions within the catalytic Cdc25 domain and a putative autoinhibitory linker connecting the Cdc25 and EF hand domains. Exchange activity in the EF hand variant was fully restored by an additional substitution, valine 406 to glutamate, which is thought to disrupt the interface between the autoinhibitory linker and the Cdc25 domain. Overall, our results suggest a model for how CalDAG-GEFI remains in an autoinhibited state when levels of cytosolic calcium in resting platelets are low. In response to cellular stimulation, calcium mobilization and binding to the EF hands causes conformational rearrangements within CalDAG-GEFI, including the autoinhibitory linker that frees the catalytic surface of CalDAG-GEFI to engage and activate Rap1B. The data from this study are the first evidence linking CalDAG-GEFI activity directly to calcium.
Keywords: calcium, guanine nucleotide exchange factor (GEF), GTPase, platelet, structural model, autoinhibition, conformational dynamics, EF hand
Introduction
Human survival depends on our ability to prevent blood loss at sites of vascular injury. Upon damage to the endothelial lining, blood platelets detect exposed extracellular matrix and locally produced thrombin, become activated, and form a hemostatic plug. Critical to plug formation is the engagement of αIIbβ3 integrins on the cell surface, a process that depends on the inside-out signaling to these receptors (1).
Given the unique high-shear environment found in blood vessels, signal transduction in platelets leading to integrin inside-out activation has been optimized for sensitivity and speed. Upon receptor stimulation, the second messenger calcium is rapidly released from the dense tubule system. Calcium also enters through channels in the plasma membrane, effectively increasing intracellular calcium concentrations 50-fold. Calcium plays an important role in the activation of αIIbβ3. Integrin activation also depends strongly on the focal adhesion proteins talin-1 and kindlin-3 as well as the small GTPase Ras-related protein (Rap) 1 (1, 2). The Rap1 isozymes Rap1A and Rap1B are guanine nucleotide binding proteins and members of the large superfamily of Ras small GTPases. Small GTPases act like molecular switches, cycling between an inactive GDP-bound and an active GTP-bound state. The biological role of Rap1B in platelets has been well studied. Genetically modified mice lacking the Rap1B isozyme have a number of platelet defects, including a marked reduction in integrin activation in response to agonist stimulation (3).
Small GTPase activity is modulated by guanine nucleotide exchange factors (GEFs)2 and GTPase-activating proteins (GAPs). The most abundant Rap-GEF and Rap-GAP in platelets are calcium- and diacylglycerol-regulated guanine nucleotide exchange factor I (CalDAG-GEFI, RasGRP2) and Ras p21 protein activator 3 (RASA3, GAP1IP4bp), respectively (4, 5). CalDAG-GEFI contains a Ras exchange motif (REM) domain with no known function, a catalytic Cdc25 domain, a pair of calcium-binding EF hands, and an atypical C1 domain with no known function. Studies in mice, dogs, and humans lacking functional CalDAG-GEFI demonstrate that CalDAG-GEFI/Rap1B signaling is crucial for rapid activation of αIIbβ3, required for platelet adhesion at sites of vascular injury (5–8).
EF hands are composed of pairs of helices that typically bind calcium using acid residues within the intervening loop. These residues are defined by their relative positions (1, 3, 5, and 12), and position 12 in particular is a highly conserved glutamate that coordinates calcium through bidentate interactions. Mutating this glutamate cripples calcium binding (9, 10). Calcium binding generally leads to major conformational changes in the EF hands and other regions of the protein (11).
Our functional studies in platelets suggested that calcium is involved the regulation of CalDAG-GEFI activity (12). In this work, we present biochemical evidence that both EF hands contribute to CalDAG-GEFI activation and that this requires calcium. Hydrogen–deuterium exchange MS studies further suggest that calcium binding induces global conformational changes within CalDAG-GEFI, most prominently in the EF hands and a putative autoinhibitory linker sequence connecting the EF hands and Cdc25 domain. Analysis of different mutant forms of CalDAG-GEFI confirms that the linker is important for autoinhibiting CalDAG-GEFI in the absence of calcium. Thus, our work provides the first evidence that CalDAG-GEFI activity is directly regulated by calcium and that release of autoinhibition is the molecular mechanism underlying CalDAG-GEFI activation.
Results
CalDAG-GEFI activation is calcium-dependent
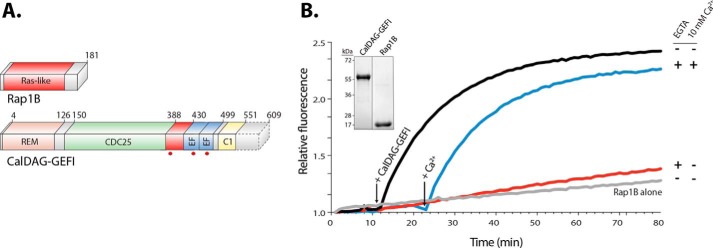
Previous studies using isothermal titration calorimetry determined that the isolated EF hands of CalDAG-GEFI (amino acid residues 417–495) bind calcium with very high affinity (Kd ∼80 nm) (13). To determine whether calcium affects catalytic activity in full-length CalDAG-GEFI, we established a cell-free nucleotide exchange assay using purified human CalDAG-GEFI and Rap1B proteins. Purified CalDAG-GEFI (amino acid residues 1–551, WT) was slightly truncated at the C terminus to increase protein stability. Rap1B (amino acid residues 1–181) was purified with a substitution, cysteine 181 to serine, to increase solubility (Fig. 1A). Purification was performed using affinity and size exclusion chromatography. Proteins were >95% pure as confirmed by SDS-PAGE (Fig. 1B, inset). Proteins were stable at 4 °C for up to 15 h. Compared with Rap1B alone, exchange activity increased by ∼16-fold in samples containing Rap1B and CalDAG-GEFI. Of note, exchange activity was not altered by addition of free calcium (data not shown). However, treatment of CalDAG-GEFI with 20 mm EGTA, a calcium-selective chelator, dramatically reduced catalytic activity. Importantly, exchange activity was restored upon addition of 10 mm free calcium, demonstrating that the loss of activity in EGTA-treated samples was not due to protein instability (Fig. 1B). These studies suggested that CalDAG-GEFI was purified in a calcium-bound state and that chelation of calcium crippled its capacity for nucleotide exchange.
Figure 1.
CalDAG-GEFI requires calcium to activate Rap1B. A, domain architecture. Rap1B is primarily composed of a Ras-like domain (red), whereas CalDAG-GEFI is a multidomain protein consisting of a REM domain (salmon), a catalytic Cdc25 domain (green), a putative autoinhibitory linker (red), two calcium-binding EF hands (blue), and an atypical C1 domain (yellow). CalDAG-GEFI was truncated (dotted lines) at residue 551 for purification. Truncated residues are not conserved and do not impact the capacity of CalDAG-GEFI to activate Rap1B. Substitutions in CalDAG-GEFI used in this work are marked (red circles) below its domain architecture. B, activation of Rap1B by CalDAG-GEFI monitored by the increased fluorescence of BODIPY FL GDP loaded onto Rap1B. Nucleotide (100 nm) and GTPase (1 μm) were incubated in four wells monitored simultaneously (λex/em = 480/520). Select reactions also included 10 mm EGTA as indicated. At 12 min (left arrow), 400 nm CalDAG-GEFI was added to all reactions except the one marked Rap1B alone. Addition of 10 mm free Ca2+ (right arrow) reconstituted the exchange activity. Inset, purified CalDAG-GEFI and Rap1B (2 μg) used in nucleotide exchange reactions; stained gel after SDS-PAGE.
Both EF hands are critical for regulating CalDAG-GEFI exchange activity
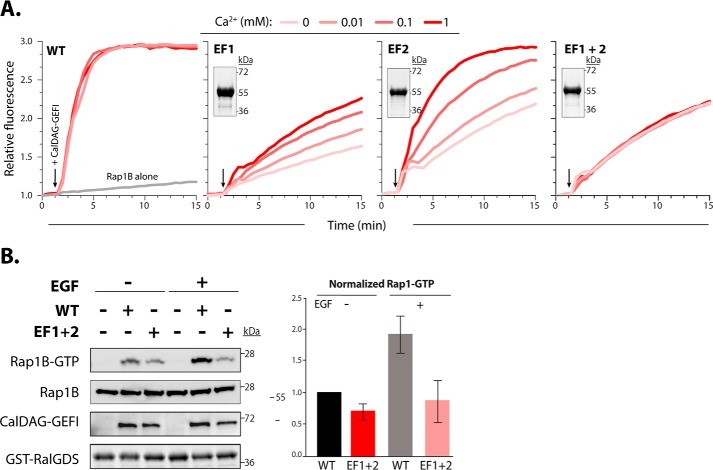
To determine whether and how the individual EF hands regulate CalDAG-GEFI activity, we mutated a key residue in the calcium binding loops (Fig. 2A). Substitutions of glutamic acid at position 12 in the calcium binding loop of EF hands has been shown to reduce the binding affinity for calcium by over 100-fold (14, 15). Glutamic acid substitution for alanine at position 450 in the N-terminal EF hand (EF1) or position 479 in the C-terminal EF hand (EF2) markedly decreased nucleotide exchange toward Rap1B. Catalytic activity was restored by adding increasing concentrations of calcium in EF2, whereas only partial recovery was observed in EF1. Catalytic activity in the double EF hand mutant (EF1+2) could not be restored (Fig. 2A). These data provide strong evidence that calcium binding to EF1 is essential for exchange activity in CalDAG-GEFI, whereas EF2 is important but not essential in this process. To validate our in vitro findings in a cellular context, we studied agonist-induced Rap1 activation in human embryonic kidney 293T cells expressing WT or EF1+2 CalDAG-GEFI (Fig. 2B). Cellular stimulation with epidermal growth factor (EGF) led to a significant increase in Rap1-GTP levels in cells expressing WT CalDAG-GEFI. In contrast, Rap1 activation was not observed in EGF-stimulated cells expressing EF1+2 CalDAG-GEFI.
Figure 2.
Mutations in the calcium-binding EF hands reduce the capacity of CalDAG-GEFI to active Rap1B. A, nucleotide exchange was monitored as described in Fig. 1. EF1 and EF2 indicate mutant forms of CalDAG-GEFI substituted (E450A and E479A) at equivalent positions within the N- and C-terminal EF hands, respectively. EF1+2 indicates the double mutant. Insets, stained gels of purified mutants of CalDAG-GEFI (2 μg) after SDS-PAGE. B, left panel, stained gel for GST-RBD pulldown of active Rap1B from human embryonic kidney 293T cells expressing WT or EF1+2 proteins after SDS-PAGE and Western blot analysis (representative of three independent experiments). Cells were incubated for 5 min in the presence and absence of 100 ng/ml EGF before lysis. Right panel, quantification of Rap1-GTP levels (mean ± S.D.).
Calcium binding to CalDAG-GEFI induces conformational rearrangements required for its activity
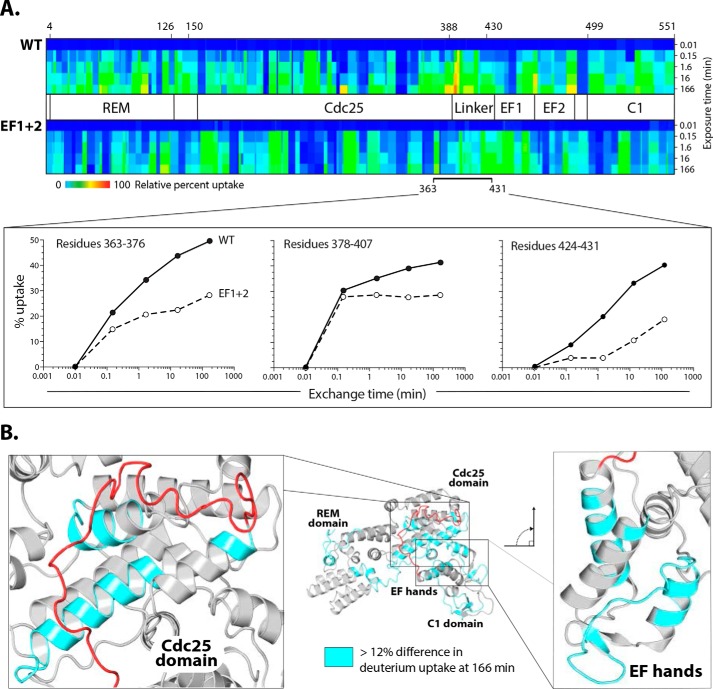
We next performed hydrogen–deuterium exchange MS experiments (HDX-MS) to determine the differences in deuterium uptake between WT and EF1+2 proteins. Measuring the rate of deuterium uptake defines the stability of hydrogen bond networks between residues stabilizing secondary structures in proteins as well as residues that are more solvent-accessible (16, 17). The most stable hydrogen bonds have the slowest exchange, whereas more dynamic regions exchange faster. WT and EF1+2 proteins were exposed to deuterated water for a designated amount of time and then quenched and digested for mass spectrometric analysis. We recovered 326 peptides from the WT sample and 329 from the EF1+2 sample, with complete coverage of both samples.
The relative difference in deuterium uptake was represented in a heat map to highlight the regions with slowest (Fig. 3A, dark blue) and fastest (Fig. 3A, red) deuterium exchange. WT and EF1+2 showed unique deuterium exchange signatures, suggesting conformational differences between the active and the apo form of the protein. A more than 5% difference in uptake between two samples is considered significant (17). Deuterium uptake in a putative autoinhibitory region linking the Cdc25 domain and EF1 was markedly higher in the WT sample (23 and 25%, respectively) (Fig. 3A). Different regions had more than 12% higher rates of exchange in WT compared with EF1+2 (Fig. 3B), including a small portion of the REM domain (19%), the Cdc25-Rap1B interface (20%), and the EF hands (17%). Displacement of the linker in response to calcium binding would provide a likely explanation for the very fast deuterium exchange rates measured for the residues comprising the Cdc25 domain-Rap1B interface in the WT protein.
Figure 3.
Differential hydrogen–deuterium exchange between CalDAG-GEFI WT and EF1+2. A, Heat maps of deuterium uptake for CalDAG-GEFI and EF1+2. Proteins were incubated in deuterated water for the indicated times prior to measurements of deuterium uptake using MS. The largest differences in uptake between the two proteins span the putative autoinhibitory linker and EF hands. Heat maps for individual peptides in the linker region are shown below. B, homology model of CalDAG-GEFI. Light blue map enhanced deuterium uptake (>12%, excluding the linker) of CalDAG-GEFI relative to EF1+2. The majority of these regions cluster within the binding site for Rap1B and EF hands shown in expanded views.
Valine 406 contributes to maintain CalDAG-GEFI in an autoinhibited state
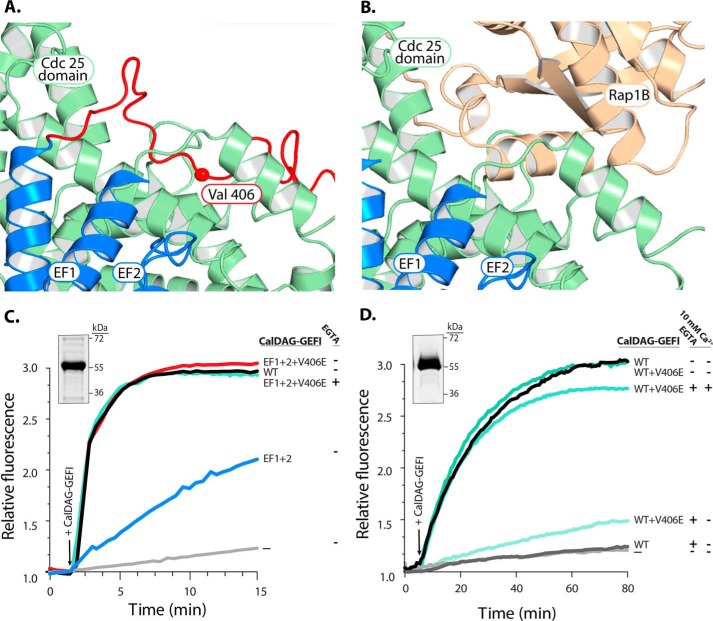
We next determined critical residues within the putative autoinhibitory linker region using a homology model of inactive CalDAG-GEFI, based on predictions by the Iterative Threading Assembly Refinement (I-TASSER) server (for more details, see “Experimental Procedures”). We identified amino acids 406–410 (VLEEW) as the residues that insert directly into the Rap1B binding groove (Fig. 4, A and B). This region of the linker is fully conserved in all mammals. To test whether displacement of this linker is sufficient to activate CalDAG-GEFI, we purified both WT and EF1+2 protein with an additional substitution, a valine 406 for glutamic acid, and tested the capacity of these proteins for nucleotide exchange toward Rap1B. We selected to substitute valine 406, as our homology model suggested a key role for this residue in anchoring the autoinhibitory linker in the Rap1 binding groove. Consistent with this conclusion, the V406E mutation completely restored catalytic activity in EF1+2 CalDAG-GEFI, suggesting that the autoinhibitory linker was displaced in this mutant in absence of bound calcium (Fig. 4C). The V406E mutation had a weaker effect in the WT protein, where nucleotide exchange activity was still markedly impaired in the presence of EGTA (Fig. 4D). These studies suggest that the interaction of V406 with the Cdc25 domain contributes to the autoinhibited state of CalDAG-GEFI.
Figure 4.
Substitution of a conserved valine fully restores the exchange activity of EF1+2. A, homology model of CalDAG-GEFI with the putative autoinhibitory linker highlighted in red. B, equivalent homology model with the linker removed and Rap1B docked onto the structure. C, substitution of valine 406 (V406E) fully restores the exchange capacity of EF1+2. Inset, stained gel of purified EF1+2+V406E after SDS-PAGE. D, exchange activity of CalDAG-GEFI WT+V406E both in the presence and absence of EGTA and exogenous calcium. Inset, stained gel of purified WT+V406E after SDS-PAGE.
Discussion
The four members of the CalDAG-GEF (RasGRP) family are critical for the proper function of different blood cell types (18). They all possess a characteristic domain structure with an N-terminal REM/Cdc25 catalytic domain and a C-terminal regulatory domain consisting of a pair of EF hands and a C1 domain. In this work, we investigated the mechanistic details by which calcium affects nucleotide exchange activity in CalDAG-GEFI, a key regulator of Rap1 signaling in platelets. Compared with CalDAG-GEFII (RasGRP1), the best studied family member, CalDAG-GEFI shows significant differences in the Cdc25 catalytic domain and the EF hand and C1 regulatory domains. CalDAG-GEFII is a RasGEF that exists as a dimer in solution. Binding of diacylglycerol (DAG) to its C1 domain is critical for dimer release and, thus, CalDAG-GEFII function. In contrast, CalDAG-GEFI is primarily a Rap-GEF, does not dimerize, and does not contain a typical DAG-binding C1 domain. There are also significant differences with regard to the EF hand regulatory domain; while CalDAG-GEFII contains only one active, low-affinity (Kd > 1 μM) EF hand, CalDAG-GEFI contains two fully functional EF hands with high affinity for calcium (Kd < 100 nm) (13). The high affinity for calcium is consistent with the documented role of CalDAG-GEFI in the rapid, calcium-dependent activation of Rap1 and integrin αIIbβ3 that is required for platelet adhesion under shear stress conditions (12). Using biochemical and biophysical approaches, we demonstrate that both EF hands are critical for CalDAG-GEFI function, and we provide evidence that calcium binding induces global conformational changes in CalDAG-GEFI, most prominently in an autoinhibitory linker region that prevents Rap1 binding to the Cdc25 domain in the absence of calcium.
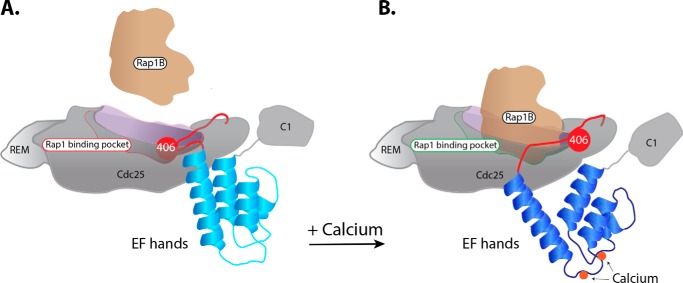
Our data suggest a straightforward model for the regulation of CalDAG-GEFI exchange activity (Fig. 5). In circulating platelets with low intracellular levels of calcium, CalDAG-GEFI is in an autoinhibited state, stabilized by the linker region between the Cdc25 domain and the EF hands. This linker blocks the catalytic surface of the Cdc25 domain so that it cannot engage Rap1B. Upon an external stimulus that raises intracellular levels of calcium, the EF hands bind calcium and change conformation. These conformational changes are coupled to movement of the autoinhibitory linker that reveals the catalytic surface needed to engage Rap1B. After Rap1B binding and GTP-for-GDP exchange, the complex dissociates freeing GTP-bound Rap1B to engage downstream effectors.
Figure 5.
Model for calcium-dependent rearrangements within CalDAG-GEFI required for its engagement and activation of Rap1B. A and B, at low concentrations of intracellular calcium (A), the linker encompassing valine 406 blocks the surface of the Cdc25 domain that engages Rap1B. As calcium levels rise (B), the EF hands bind calcium and change conformation. This rearrangement moves the linker, liberating the surface of CalDAG-GEFI needed to bind Rap1B.
Based on isothermal titration calorimetry experiments, the individual EF hands of CalDAG-GEFI bind one calcium ion with high affinity (Kd ∼80 nm) (13). This result strongly suggests that the two EF hand domains may act in concert to activate CalDAG-GEFI. Indeed, disabling either EF hand markedly impaired the exchange activity. Addition of free calcium in a dose-dependent manner restored exchange activity in the EF2 mutant. In contrast, only a partial recovery of function was observed for the EF1 mutant. Disabling both EF hands in CalDAG-GEFI rendered CalDAG-GEFI unresponsive to calcium. These studies suggest that EF2 is critical for high-affinity calcium binding, whereas EF1 is important for inducing conformational changes required for CalDAG-GEFI catalytic activity.
Regulating activity via high-affinity binding to calcium explains perfectly why CalDAG-GEFI plays such a crucial role in platelet function. Platelets circulate at high velocity in the blood stream, patrolling the vasculature for breaches in the endothelial lining. To fulfill their hemostatic function, platelets need to be able to sense minute changes in their environment and to rapidly change from an anti-adhesive to a pro-adhesive state. Even weak platelet agonists such as ADP cause rapid and significant changes in cytoplasmic calcium. CalDAG-GEFI can quickly integrate this calcium signal and mediate a near-immediate activation of Rap1 and αIIbβ3 integrin.
However, such high sensitivity to calcium also poses a challenge, as unwanted CalDAG-GEFI activation could lead to premature platelet activation and thrombosis. In fact, one would expect that, if CalDAG-GEFI binds calcium at such high affinity, then a large pool of CalDAG-GEFI would be activated in resting, circulating platelets. Thus, additional regulatory mechanisms must be in play to prevent unwanted platelet activation. One important mechanism that would offset CalDAG-GEFI signals is negative regulation by RASA3, a Rap-GAP highly expressed in the platelet membrane. In mice, loss of RASA3 function leads to platelet preactivation, rapid platelet clearance, and severe thrombocytopenia because of unbalanced CalDAG-GEFI signaling (4).
It is also possible that CalDAG-GEFI function depends on more than just calcium binding to its EF hands. For example, the C1-like domain may provide additional regulatory activity. Within the CalDAG-GEF family, CalDAG-GEFI is the only member that contains an atypical C1 domain with very low affinity for diacylglycerol (Kd ∼ 2 μm versus Kd ∼ 5 nm for the other family members) (19). Structural studies by Iwig et al. (13) demonstrated that DAG binding to the C1 domain in CalDAG-GEFII is important to release C1 domain dimerization, causing protein translocation to the membrane. In contrast, CalDAG-GEFI is a monomer, and platelets lacking CalDAG-GEFI aggregate normally in response to stimulation with phorbol ester (5), a DAG mimetic that engages typical C1 domains. However, platelets expressing a truncated CalDAG-GEFI lacking the C1 domain are defective in their integrin activation and aggregation responses (20). Thus, the C1 domain clearly plays a role in CalDAG-GEFI function, but further studies will be required to unravel the underlying molecular mechanism(s).
The comparison of hydrogen–deuterium exchange profiles for active (WT) and inactive (EF1+2) CalDAG-GEFI provides important structural insights into how GEF activity is regulated by binding of calcium. Central to this regulation are the autoinhibitory linker and the EF hands. The autoinhibitory linker is bracketed by residues with some of the largest deuterium uptake differentials between the active and crippled forms of CalDAG-GEFI. Furthermore, the regions of the Cdc25 domain critical for GEF activity, based on the homology model, also showed markedly increased deuterium uptake in the active protein compared with the EF1+2 mutant. These differences support the contention that the autoinhibitory linker moves away from the catalytic surface of the Cdc25 domain upon engagement of the EF hands by calcium. The fact that substitution of valine 406, centered within the autoinhibitory linker, is sufficient to fully rescue the exchange capacity of the EF1+2 mutant strongly supports our interpretation of the HDX data. Our studies with the WT V406E mutant further suggest that residues in the vicinity of valine 406 contribute to stabilizing the autoinhibitory linker in the Rap1 binding groove. At this point, we can only speculate why the EF1+2 mutant is more sensitive to introduction of the V406E mutant. We observed that, in the presence of EGTA, the EF1+2 mutant has a higher exchange activity relative to the WT protein (data not shown). This finding may suggest that the point mutations in the EF hands have a minimal effect on the structure of the protein, slightly increasing exchange activity. Introduction of the V406E mutation then shifts the protein into a conformation that allows full activity in our cell-free assay. In the WT protein, introduction of the V406E mutation has a milder effect.
It is interesting that additional regions of CalDAG-GEFI also exhibit differential hydrogen–deuterium exchange. These regions include portions of the REM domain, the region between REM and Cdc25 domains, and portions of the C1 domain. Presumably, the calcium-dependent activation of CalDAG-GEFI is initiated by structural alterations within the EF hands, but these alterations consequently propagate throughout the protein. The functional effect of these additional alterations should be determined in future work, as there is precedent in Sos1 that the REM and Cdc25 domains cooperate to activate Ras (21). Intriguingly, the autoinhibitory linker might be expected to contain the highest differences in hydrogen–deuterium exchange based on the proposed model. However, this is not the case, suggesting that it is not completely disordered upon calcium-dependent activation. Instead, it seems likely that this region adopts an alternative, stable conformation upon calcium binding, perhaps interacting with the EF hands themselves.
Taken together, our data provide mechanistic insight into the functional consequences of calcium binding to the EF hands of CalDAG-GEFI and the structural rearrangements required for GEF activity to occur. These findings will be important in our understanding of how mutations in patients impair CalDAG-GEFI function and in efforts to design inhibitors of CalDAG-GEFI signaling.
Experimental procedures
Reagents
Horseradish peroxidase–conjugated secondary antibodies and HisTrap HP affinity and size exclusion columns were purchased from GE Healthcare (Marlborough, MA). FLAG M2 (catalog no. F3165-2MG) mAb was purchased from Sigma-Aldrich (St. Louis, MO). EGF (catalog no. E9644), GDP-BODIPY (catalog no. G22360), and EGTA (catalog no. E1219) were purchased from Gibco/Thermo Fisher Scientific (Waltham, MA).
Protein purification
A plasmid containing the gene coding for human CalDAG-GEFI was purchased from Harvard University's Human ORFeome v5.1. The gene was PCR-amplified using primers introducing a stop at position 1653 in the nucleotide sequence. The complementary DNA was subcloned into a p15LIC2 bacterial expression vector. The expressed protein, CalDAG-GEFI (1–551), contained all functional domains plus an N-terminal His6 tag required for purification. Cloning was performed according to a ligation-independent cloning protocol. CalDAG-GEFI (1–551) EF hand mutant EF1 was made by substituting glutamic acid to alanine at position 450. EF hand mutant EF2 was made by substituting glutamic acid to alanine at position 479. The EF hand double mutant EF1+2 was made by substituting glutamic acid to alanine at positions 450 and 479. The EF hand linker mutant EF1+2+V406 was made by substituting valine to glutamic acid at position 406 using EF1+2 complementary DNA for the PCR template. All mutations were carried out using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol.
CalDAG-GEFI proteins were expressed in a Rosetta strain of Escherichia coli (Novagen). Cell cultures were grown at 37 °C in lysogeny broth, 0.5 μm ZnSO4, 50 μg/ml ampicillin, and chloramphenicol. Protein expression was induced by addition of 500 μm isopropyl β-d-thiogalactopyranoside and cells were incubated for 15 h at 20 °C. Cell pellets were resuspended in 20 mm PIPES (pH 6.8), 300 mm NaCl, 0.5 μm ZnSO4, 10 mm β-mercaptoethanol, 10 mm imidazole, 5% glycerol, and 1× protease inhibitors (Roche). Cell pellets were lysed using an Emulsiflex C5 cell homogenizer (Avestin) and clarified by ultracentrifugation at 45,000 rpm for 45 min at 4 °C. Filtered supernatant was applied to a HisTrap HP affinity column (GE Healthcare) equilibrated with buffer A (20 mm PIPES (pH 6.8), 300 mm NaCl, 8 mm imidazole, and 5% glycerol). Protein was eluted from the column with 400 mm imidazole using buffer B (20 mm PIPES (pH 6.8), 300 mm NaCl, 1 m imidazole, and 5% glycerol). Peak fractions were collected, treated with tobacco etch virus protease overnight at 4 °C and dialyzed in buffer A without imidazole. A second pass over the HisTrap HP affinity column removed the cleaved His tag and tobacco etch virus protease from the sample. Flow-through fractions were concentrated using a Vivaspin column (Sartoris) with a 10,000 molecular weight cutoff filter and then loaded onto a Superdex 200 10/300 size exclusion column (GE Healthcare) equilibrated with S200 size exclusion buffer containing 20 mm PIPES (pH 6.8), 300 mm NaCl, and 4 mm DTT. The final protein product was 95% pure, monomeric, and concentrated to 20 mg/ml. Aliquots were flash-frozen in liquid nitrogen and stored at −80 °C.
Human Rap1B (1–181) C181S was cloned into the pProEXHtb vector and purified from E. coli. 500 μm isopropyl β-d-thiogalactopyranoside was used to induce protein expression, and cultures were incubated for 8 h at 20 °C. Protein purification was done using the same chromatography steps as for CalDAG-GEFI proteins listed above with buffer modifications, substituting the PIPES buffer with Tris-HCl (pH 7.0), 1 μm ZnSO4 with 5 mm MgCl2, and adding 50 μm GDP to protein lysis buffer.
Nucleotide exchange assay
Nucleotide exchange on Rap1B was measured using a fluorescence-based assay as described previously (22). The reaction volume was 100 μl containing 1 μm Rap1B, between 300–500 nm CalDAG-GEFI, and 100 nm BODIPY-GDP (Life Technologies). The reaction buffer contained 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 5% glycerol, and 0.004% NP-40. Reactions were performed in a black-bottom 96-well plate (Corning). The fluorescent signal was measured using a Pherastar microplate reader (BMG Labtech) (wavelength excitation/emission = 480/520 nm and 1-nm slits). For calcium chelation experiments, 100 μm CalDAG-GEFI was treated with 10 mm EGTA for 2 h at 4 °C. The sample was spun at 4,000 rpm for 30 s and then transferred to a new tube containing reaction buffer, diluting calcium-free CalDAG-GEFI to 400 nm. Samples were treated with 10 mm calcium where indicated.
Rap1B pulldown assays
Rap1B pulldown experiments were performed as described recently (12).
HDX-MS
HDX-MS experiments on WT CalDAG-GEFI and EF1+2 were performed using an Ultra Performance Liquid Chromatography (UPLC) HDX system coupled with a Q-Tof Premier mass spectrometer (Waters Corp., Milford, MA) as described previously (23). Briefly, each protein sample was buffer-exchanged into PBS and then diluted to 1.5 mg/ml. 1 μl of sample was diluted 1:7 (v/v) in 10 mm phosphate, 99.9% D2O, pD 7.0 at 20 °C using a robotic autosampler to initiate hydrogen–deuterium exchange. Exchange reactions were quenched after 0.01, 0.15, 1.6, 16, and 166 min by placing them at 1 °C and adding an equal volume of precooled quenching buffer (100 mm phosphate, 0.5 m tris(2-carboxyethyl)phosphine, 0.8% formic acid, and 2% acetonitrile (pH 2.5)). Each sample and each time point were done in replicates of six. Samples from each time point were fragmented through a Waters Enzymate BEH pepsin column, and the peptic fragments were separated on an in-line C18 HPLC column and then analyzed by MS. Mass assignment for each peptide without HDX was inspected manually; any assignment with a mass deviation >0.2 Da was removed. The extent of deuterium incorporation in each peptide was calculated and tabulated using the Waters software. The relative fractional deuterium uptake is represented as a heat map and was measured for each peptide by calculating the number of residues that exchange an amide proton for deuterium over the total number of protons that are able to exchange (peptide length −1) and averaged for each time point.
Homology model
The homology model of CalDAG-GEFI was built using the I-TASSER server available to academic researchers through the Zhang laboratory at the University of Michigan. I-TASSER generated our model by comparing the protein sequence of CalDAG-GEFI with protein sequences of all proteins that have crystal or cryo-EM structures deposited in the Protein Data Bank. These structures were used as templates to predict secondary structures in CalDAG-GEFI based on sequence homology to the template sequence. In addition, the sequence in CalDAG-GEFI lacking a template, typically disordered loop regions, was modeled ab initio as secondary structures having the lowest free energy. I-TASSER predicted five homology models of CalDAG-GEFI, ranked in order from highest to lowest probability; we chose the highest probable model.
Author contributions
A. A. C., J. S., and W. B. conceptualization; A. A. C., W. D., J. R., and W. B. data curation; A. A. C., W. D., J. R., R. L., J. S., and W. B. formal analysis; A. A. C. and W. B. validation; A. A. C., J. R., R. L., J. S., and W. B. investigation; A. A.C., W. D., J. R., J. S., and W. B. methodology; A. A. C., J. S., and W. B. writing-original draft; A. A. C., J. S., and W. B. writing-review and editing; W. D. software; R. L., J. S., and W. B. supervision; J. S. and W. B. funding acquisition; J. S. and W. B. project administration; W. B. resources.
Acknowledgments
We thank the members of the W. B. and J. S. laboratories for stimulating discussions and technical assistance. We also thank Leslie Parise, Keith Burridge, and Victoria Bautch for insights and suggestions.
This work was supported by NHLBI, National Institutes of Health Grants R01 HL130404 and R01 HL121650 and NIGMS, National Institutes of Health Grant P01GM103723. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- REM
- Ras exchange motif
- EGF
- epidermal growth factor
- HDX
- hydrogen–deuterium exchange
- DAG
- diacylglycerol.
References
- 1. Shattil S. J., Kim C., and Ginsberg M. H. (2010) The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 10.1038/nrm2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stefanini L., and Bergmeier W. (2016) RAP1-GTPase signaling and platelet function. J. Mol. Med. 94, 13–19 10.1007/s00109-015-1346-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chrzanowska-Wodnicka M., Smyth S. S., Schoenwaelder S. M., Fischer T. H., and White G. C. 2nd (2005) Rap1b is required for normal platelet function and hemostasis in mice. J. Clin. Invest. 115, 680–687 10.1172/JCI22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stefanini L., Paul D. S., Robledo R. F., Chan E. R., Getz T. M., Campbell R. A., Kechele D. O., Casari C., Piatt R., Caron K. M., Mackman N., Weyrich A. S., Parrott M. C., Boulaftali Y., Adams M. D., et al. (2015) RASA3 is a critical inhibitor of RAP1-dependent platelet activation. J. Clin. Invest. 125, 1419–1432 10.1172/JCI77993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crittenden J. R., Bergmeier W., Zhang Y., Piffath C. L., Liang Y., Wagner D. D., Housman D. E., and Graybiel A. M. (2004) CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat. Med. 10, 982–986 10.1038/nm1098 [DOI] [PubMed] [Google Scholar]
- 6. Westbury S. K., Canault M., Greene D., Bermejo E., Hanlon K., Lambert M. P., Millar C. M., Nurden P., Obaji S. G., Revel-Vilk S., Van Geet C., Downes K., Papadia S., Tuna S., Watt C., et al. (2017) Expanded repertoire of RASGRP2 variants responsible for platelet dysfunction and severe bleeding. Blood 130, 1026–1030 10.1182/blood-2017-03-776773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boudreaux M. K., Catalfamo J. L., and Klok M. (2007) Calcium-diacylglycerol guanine nucleotide exchange factor I gene mutations associated with loss of function in canine platelets. Transl. Res. 150, 81–92 10.1016/j.trsl.2007.03.006 [DOI] [PubMed] [Google Scholar]
- 8. Lozano M. L., Cook A., Bastida J. M., Paul D. S., Iruin G., Cid A. R., Adan-Pedroso R., Ramón González-Porras J., Hernández-Rivas J. M., Fletcher S. J., Johnson B., Morgan N., Ferrer-Marin F., Vicente V., Sondek J., et al. (2016) Novel mutations in RASGRP2, which encodes CalDAG-GEFI, abrogate Rap1 activation, causing platelet dysfunction. Blood 128, 1282–1289 10.1182/blood-2015-11-683102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Declercq J.-P., Evrard C., Lamzin V., and Parello J. (1999) Crystal structure of the EF-hand parvalbumin at atomic resolution (0.91 Å) and at low temperature (100 K). Evidence for conformational multistates within the hydrophobic core. Protein Sci. 8, 2194–2204 10.1110/ps.8.10.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar S., Ahmad E., Kumar S., Khan R. H., and Gourinath S. (2012) Flexibility of EF-hand motifs: structural and thermodynamic studies of calcium binding protein-1 from Entamoeba histolytica with Pb2+, Ba2+, and Sr2+. BMC Biophys. 5, 15 10.1186/2046-1682-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewit-Bentley A., and Réty S. (2000) EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 10, 637–643 10.1016/S0959-440X(00)00142-1 [DOI] [PubMed] [Google Scholar]
- 12. Stefanini L., Roden R. C., and Bergmeier W. (2009) CalDAG-GEFI is at the nexus of calcium-dependent platelet activation. Blood 114, 2506–2514 10.1182/blood-2009-04-218768 [DOI] [PubMed] [Google Scholar]
- 13. Iwig J. S., Vercoulen Y., Das R., Barros T., Limnander A., Che Y., Pelton J. G., Wemmer D. E., Roose J. P., and Kuriyan J. (2013) Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. eLife 2, e00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cates M. S., Berry M. B., Ho E. L., Li Q., Potter J. D., and Phillips G. N. (1999) Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin. Structure/Folding and Design 7, 1269–1278 [DOI] [PubMed] [Google Scholar]
- 15. Busch E., Hohenester E., Timpl R., Paulsson M., and Maurer P. (2000) Calcium affinity, cooperativity, and domain interactions of extracellular EF-hands present in BM-40. J. Biol. Chem. 275, 25508–25515 10.1074/jbc.M001770200 [DOI] [PubMed] [Google Scholar]
- 16. Konermann L., Pan J., and Liu Y.-H. (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 10.1039/C0CS00113A [DOI] [PubMed] [Google Scholar]
- 17. Morgan C. R., and Engen J. R. (2010) Investigating Solution-Phase Protein Structure and Dynamics by Hydrogen Exchange Mass Spectrometry. Curr. Protoc. Protein Sci. 10.1002/0471140864.ps1706s58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stone J. C. (2011) Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer 2, 320–334 10.1177/1947601911408082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czikora A., Lundberg D. J., Abramovitz A., Lewin N. E., Kedei N., Peach M. L., Zhou X., Merritt R. C. Jr., Craft E. A., Braun D. C., and Blumberg P. M. (2016) Structural basis for the failure of the C1 domain of Ras guanine nucleotide releasing protein 2 (RasGRP2) to bind phorbol ester with high affinity. J. Biol. Chem. 291, 11133–11147 10.1074/jbc.M116.725333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stolla M., Stefanini L., Roden R. C., Chavez M., Hirsch J., Greene T., Ouellette T. D., Maloney S. F., Diamond S. L., Poncz M., Woulfe D. S., and Bergmeier W. (2011) The kinetics of αIIbβ3 activation determines the size and stability of thrombi in mice: implications for antiplatelet therapy. Blood 117, 1005–1013 10.1182/blood-2010-07-297713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margarit S. M., Sondermann H., Hall B. E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., and Kuriyan J. (2003) Structural evidence for feedback activation by Ras·GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 10.1016/S0092-8674(03)00149-1 [DOI] [PubMed] [Google Scholar]
- 22. Ren J., Cook A. A., Bergmeier W., and Sondek J. (2016) A negative-feedback loop regulating ERK1/2 activation and mediated by RasGPR2 phosphorylation. Biochem. Biophys. Res. Commun. 474, 193–198 10.1016/j.bbrc.2016.04.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng W., Wang Y., Druzak S. A., Healey J. F., Syed A. K., Lollar P., and Li R. (2017) A discontinuous autoinhibitory module masks the A1 domain of von Willebrand factor. J. Thromb. Haemost. 15, 1867–1877 10.1111/jth.13775 [DOI] [PMC free article] [PubMed] [Google Scholar]