Abstract
Ubiquitin-mediated control of protein stability is central to most aspects of plant hormone signaling. Attachment of ubiquitin to target proteins occurs via an enzymatic cascade with the final step being catalyzed by a family of enzymes known as E3 ubiquitin ligases, which have been classified based on their protein domains and structures. Although E3 ubiquitin ligases are conserved among eukaryotes, in plants they are well-known to fulfill unique roles as central regulators of phytohormone signaling, including hormone perception and regulation of hormone biosynthesis. This review will highlight up-to-date findings that have refined well-known E3 ligase-substrate interactions and defined novel E3 ligase substrates that mediate numerous hormone signaling pathways. Additionally, examples of how particular E3 ligases may mediate hormone crosstalk will be discussed as an emerging theme. Looking forward, promising experimental approaches and methods that will provide deeper mechanistic insight into the roles of E3 ubiquitin ligases in plants will be considered.
Roles of E3 ligases in phytohormone signaling
Ubiquitin E3 ligases are conserved among eukaryotes and fulfill a myriad of regulatory functions by facilitating the covalent attachment of ubiquitin to target proteins. Attachment typically occurs on lysine residues and can occur singly (monoubiquitination) or in chains (polyubiquitination). The types of attachments and degree of ubiquitination varies considerably among substrates and may be context dependent. This enzymatic reaction is carried out by a family of proteins called E3 ubiquitin ligases, which act at the end of a three-enzyme cascade to transfer ubiquitin from an E2 ubiquitin conjugating enzyme to a specific substrate(s). Ubiquitination of substrates is a tightly regulated process and can result in several functional outcomes, including protein degradation, changes in subcellular localization, and protein activation. In plants, numerous ubiquitin E3 ligases act as central regulators in phytohormone signaling pathways including auxin, brassinosteroid (BR), cytokinin (CK), ethylene, gibberellic acid (GA), jasmonate (JA), salicylic acid (SA), and strigolactone (SL) (for recent detailed reviews see (1–5)). This review will highlight up-to-date findings that have defined new E3 ligase-substrate interactions that mediate phytohormone signaling pathways, discuss examples of how some E3 ligases mediate hormone crosstalk, and touch on emerging approaches that will help us gain deeper mechanistic insight into these proteins.
There are over 1500 E3 ubiquitin ligase proteins encoded by the Arabidopsis genome which can be subdivided into different families (6). This includes the HECT (homologous to the E6AP carboxyl terminus) type, RING (really interesting new gene)1 family, Kelch-type and U-box containing ubiquitin protein ligases. The Cullin-RING ligase (CRL) family can be further subdivided into five subfamilies based on subunit organization and conserved domains: (1) SKP1-Cullin-F-box (SCF) type, (2) broad complex/tramtrack/bric-a-brac (BTB) type, (3) DDB1-binding/WD-40 domain containing proteins (DWD) type, (4) VON-HIPPEL LINDAU (VHL) type, and (5) SUPRESSOR OF CYTOKINE SIGNALING (SOCS) type (6). While CRLs have been called “molecular hubs” in plant hormone signaling pathways because of their central roles in hormone perception mechanisms and far-reaching cellular signaling effects, there are several examples of other types of E3 ubiquitin ligases playing roles in phytohormone signaling. Regardless of E3 ligase type, one common theme among these plant E3 proteins is that they interact with their substrates in a hormone-dependent manner (3). This is especially interesting given that both E3 ubiquitin ligases and plant hormones are diverse chemical structures with complex evolutionary histories.
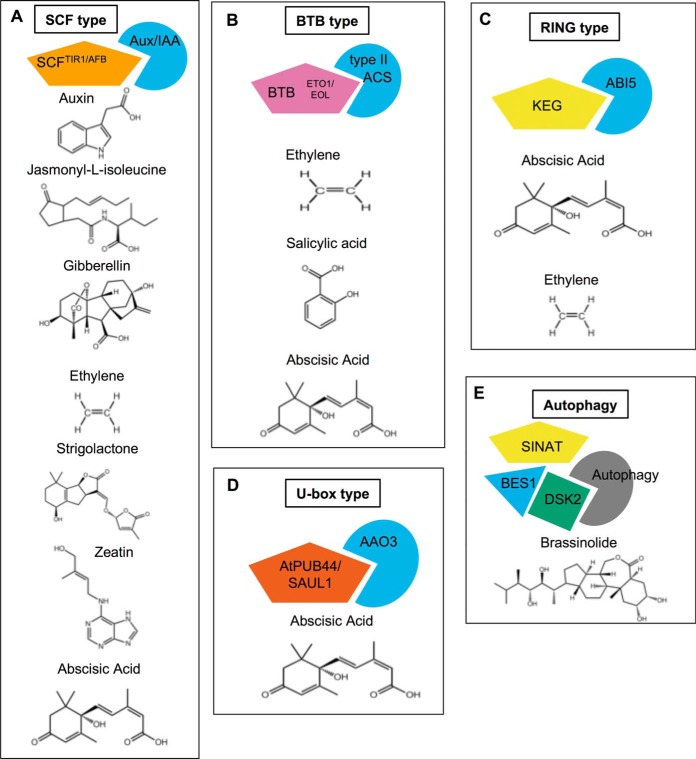
The exact roles of E3 ligases in hormone perception and/or signaling have been well established for most known cases (Fig. 1). The first hormone discovered to utilize ubiquitin E3 ligases as receptor molecules was auxin, which is perceived by SCFTIR1 and related Auxin F-box (AFB) family members (7, 8). Perception of auxin by SCFTIR1/AFB triggers rapid degradation of the Aux/IAA family of transcriptional repressors (9–13). Among all the various types of E3 ligases, the SCF type play prominent roles in perception of most phytohormones including jasmonic acid (14), gibberellin (15–17), ethylene (18), salicylic acid (SA) (19, 20), and strigolactones (21–25); also reviewed in (1). Other types of E3 ligases have also been linked to hormone signaling, including BTB type regulating ethylene biosynthesis (26–28) and CRL3-based E3 ligases modulating ABA signaling (Fig. 1B). RING type E3 ligases play key roles in ethylene biosynthesis (29) and several aspects of ABA pathways including biosynthesis, transcriptional regulation, and signaling (30–34) (Fig. 1C). U-box type E3s have been linked to ABA biosynthesis and downstream responses such as stomatal closure (35–37) (Fig. 1D). Finally, an E3 ligase and autophagy receptor protein have recently described to regulate BR signaling during stress (38, 39) (Fig. 1E). Overall, the number of E3 ligases linked to ABA signaling thus far is greater than for any other phytohormone, demonstrating the diversity and extent to which substrate ubiquitination can regulate ABA biosynthesis, signaling and downstream responses (1).
Fig. 1.
Several different types of E3 ubiquitin ligases have known roles in various aspects of phytohormone pathways. Illustrated here are representative E3-subtrate interactions from each class of E3 ligase with corresponding hormone(s) shown below. A, SCF type E3 ligases have been linked to several hormone pathways, including auxin, JA, GA, ethylene, strigolactones, cytokinins and ABA. B, BTB-type E3 ligases play roles in ethylene, SA and ABA pathways. C, RING-type E3 ligases are important regulators of ABA and ethylene signaling. D, One U-box type E3 ligase contributes to ABA biosynthesis. E, A novel mechanism for selective autophagy via the SINAT E3 ligases and ubiquitin receptor DSK2 has recently been linked to BR signaling.
Detailed biochemical studies have provided new insights into E3-substrate complex assembly and composition. For instance, protein structure studies on TIR1-auxin-AUX/IAA and JA-COI1-JAZ (JASMONATE ZIM-DOMAIN) complexes have revealed that small molecule co-factors are directly involved in SCFTIR1 and SCFCOI1 complexes (inositol hexakisphosphate (InsP6) and inositol pentakisphosphate (InsP5) respectively) (14, 40). Additionally, several of the key E3 ligases involved in auxin, JA and SL perception have been proposed to function as “co-receptor” complexes, whereby high-affinity hormone binding is facilitated by both the E3 ubiquitin ligase and substrate. This has been observed for the TIR1-auxin-AUX/IAA complex, the COI1-JA-Ile-JAZ complex, the ABI1 PUB12/13/U-box E3 ligase complex and the MAX2/D3-SL-D14 complex (11, 14, 41–45). These molecular interactions along with distinct naturally occurring forms of these hormones and co-receptor pairs could provide for increased complexity and specificity via varied combinatorial configurations, which may underlie downstream responses mediated by auxin, JA, ABI, and SL.
Substrate recognition has been mapped to minimal amino acid sequence motifs for several E3 targets related to auxin and JA signaling through careful biochemical studies; such sequences are not identifiable based solely on primary amino acid sequence and thus are not amenable to bioinformatics-based approaches. For instance, all Aux/IAAs contain a short consensus recognition motif, or “degron,” which directly engages with auxin-loaded TIR1 (13, 40, 46). However, regions outside the degron appear to contribute to differential hormone binding affinity among Aux/IAAs and F-box proteins (11, 46). JAZ proteins also contain a condensed degron sequence, which is a variable region in direct contact with the COI1-anchored JA-Ile molecule (14). Notably, this mechanism of interaction has been co-opted into a yeast assay for monitoring ubiquitin-mediated protein degradation in yeast called the auxin-inducible degradation (AID) system (46–49). This assay enables versatile conditional protein depletion and can be applied to various eukaryotic proteins that function as part of SCF complexes and/or substrates.
Novel E3 Ligase-substrate Interactions Involved in Hormone Signaling
Several new interactions have recently been described for several E3 ubiquitin ligases, which play various roles in hormone signaling. Several novel E3-substrate interactions have been described in BR pathways. One of the most surprising recent findings describes how BES1 (BRI1 EMS SUPPRESSOR 1) can be ubiquitinated by SINAT (SINA of Arabidopsis thaliana) E3 ligases, which promotes BES1-DSK2 (DOMINANT SUPPRESSOR OF KAR 2) interactions and subsequent degradation via selective autophagy (38, 39). Thus, both the proteasome and selective autophagy are involved in degrading BES1 while SINAT E3s ubiquitinate BES1 during starvation and light response. Although it is not clear why a single transcription factor would need to be degraded through several independent pathways, although it has been proposed that such multiple regulatory checkpoints could regulate BES1 levels to allow for integration of morphogenesis with distinct environmental cues such as light or stress. This work also points to an interesting possibility that other E3 ubiquitin ligase substrates could be degraded via selective autophagy mechanisms, but the signals that direct ubiquitinated proteins to autophagy versus proteasome mediated degradation need to be studied further. A novel F-box protein, KINK SUPPRESSED IN BZR1–1D (KIB1) was recently shown to mediate BR-induced ubiquitination and degradation of the glycogen synthase kinase-3 (GSK3)-like kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2) (50). Also, plasma membrane localization of the BRI1 receptor is regulated by ubiquitination but the E3 ligase(s) responsible are still unknown (51). Additionally, two different U-box type E3 ubiquitin ligases have been implicated as positive regulators of BR signaling in rice, ERECT LEAF 1 (ELF1) (52) and Taihu Dwarf (TUD1) (53) but further studies are required to identify the substrates of these ligases and their role(s) in BR-mediated plant growth.
Recent studies have continued to build on our understanding of how ABA pathways are regulated post-translationally. For example, the RING E3 SDIR targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate ABA signaling (54). Additionally, degradation of the ABA receptor ABI1 occurs by the PUB12/13 U-box E3 ligases (41). Perception of ABA by the pyrabactin resistance (PYR)/PYR1-like (PYL)/regulatory components of ABA receptor (RCAR) proteins with the co-receptor protein phosphatase type 2Cs facilitates activation of Snf1-related protein kinase 2 (SnRK2) kinases, which are in turn ubiquitinated and degraded by SNFAtPP2-B11 (55). AtPP2-B11 is an newly described F-box protein that functions as part of a canonical SCF E3 ligase complex to negatively regulate plant responses to ABA (55). Adding to the complexity of this pathway is ECERIFERUM9 (CER9), which encodes a putative RING domain-containing E3 ubiquitin ligase that is a novel negative regulator of ABA biosynthesis and ABA signaling during seed germination. CER9 is like Doa10 in S. cerevisiae, which targets substrates for degradation via the UPS. Further work on CER9 will be required to identify target protein(s) and their role in ABA signaling (56).
Other examples of recent ligase-substrate interactions include description of two novel E3 ligases involved in JA signaling, RING DOMAIN LIGASE 3 (RGLG3) and RGLG4 (57), and DEFECTIVE IN ANTHER DEHISCENCE1-Activating Factor (DAF) (58); the direct substrates of these ligases are not currently known and await further studies. Additionally, the plant U-box protein (PUB10) was recently found to regulate MYC2 degradation while de-ubiquitination enzymes UBP12 and UBP13 positively influence MYC2 stability (59, 60). Identification of UPB12 and UBP13 highlights our limited understanding of the roles of de-ubiquitination enzymes in plant hormone pathways and suggests that this aspect of ubiquitination could be better explored in plants. Finally, SA was one of the last major phytohormones without a known receptor. A recent study provides further evidence that the E3 ligase Nonexpressor of Pathogenesis-Related genes 1 (NPR1) gets degraded via interaction with the closely related NPR3/NPR4 substrate adaptor proteins (19, 61), thus NPR1 or NPR1-related proteins (NPR3 and NPR4) are the long-sought-after SA receptors (62).
New Insights into Ubiquitin Attachment to Substrates
Typically, ubiquitin is covalently attached to substrate proteins on lysine residues but there are several examples of non-canonical attachment mechanisms in other eukaryotes (63). With respect to E3 substrate proteins involved in phytohormone signaling the AuxIAA family is the best studied to date (13, 46, 47, 64, 65). A major outstanding question in the field was which residue(s) served as ubiquitin attachment sites and thus participate in auxin perception. Recent biochemical studies have provided surprising findings and underscore the need for further work related to ubiquitin attachment sites in plants. Specifically, biochemical and peptide mass spectrometry studies have recently demonstrated that conserved and variable lysine residues on AuxIAA proteins are ubiquitinated (65) but in the absence of lysine residues attachment can occur on serine and/or threonine positions (64). Taken together these studies demonstrate that ubiquitination of AuxIAA proteins can occur in several exposed flexible regions (i.e. “ubiquitination zones”) in a dose dependent manner. Additionally, several alternative linkage topologies may occur on AuxIAA proteins, including poly-mono-ubiquitination and/or multi-, poly-ubiquitination (65). By applying similar approaches to other well characterized substrates (Fig. 1) it will be possible to determine if such complex and/or flexible attachment properties are common or restricted to Aux/IAAs.
In other eukaryotes, post-translational modification (for e.g. phosphorylation) of substrate proteins is a common mechanism that regulates E3-substrate interactions, but the extent to which this occurs in plants is not clear. One unique example to date is the demonstration that s-nitrosylation of ABA INSENSITIVE 5 (ABI5) facilitates the subsequent degradation by the E3 ligase KEEP ON GOING (KEG) (66). Further studies focused on how E3 substrate proteins are modified (or not) will help address this discrepancy.
Making Connections: E3 Ligases and Targets Coordinate Phytohormone Crosstalk
Recently, several studies on E3 ligases in plants have demonstrated potential crosstalk mechanisms between various phytohormones. This finally moves hormone crosstalk out of the realm of transcriptional regulation and into the proteome, which potentially can impact hormone signaling in a more rapid and integrated fashion. For example, Dwarf and short grain 1 (DSG1) encodes U-box E3 that is collectively regulated by BR, ET, auxin and SA and functions to positively regulate cell division and elongation (67).
Another putative E3 ligase that integrates dueling hormone pathways to control photomorphogenesis is HIGH EXPRESSION OF OSMOSTICALLY RESPONSIVE GENES 1 (HOS1) (68, 69). HOS1 regulates hypocotyl expansion via auxin signaling and leaf expansion via ethylene, suggesting that this RING E3 ligase can regulate multiple hormone pathways in a tissue specific manner. Studies from the rice ortholog show that OsHOS1 directly regulates the stability of two ETHYLENE RESPONSE FACTOR transcription factors, rice ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN 1 (OsEREPB1), and OsEREBP2, which thereby regulates the JA mediated root curling response (70).
Hormone antagonism between ABA and SA also appears to involve E3 ligase activity, specifically via the regulation of NONEXPRESSOR OF PATHOGENESIS RELATED GENES 1 (NPR1). ABA and SA act antagonistically to influence NPR1 levels: whereas ABA promotes NPR1 degradation via the CUL3(NPR3/NPR4) complex, SA stabilizes NPR1 from ABA-promoted degradation through phosphorylation (71). The ABA pathway also integrates with JA signaling via the RING E3 ligase KEG modulating JASMONATE ZIM-DOMAIN (JAZ) protein stability (72).
A final example of potential hormone crosstalk involves targets of the SCFMAX2 E3 ubiquitin ligase, which is central to strigolactone (SL) signaling. Multiple proteins have been suggested as targets for SCFMAX2 including SUPPRRESSOR OF MORE AXILLARY GROWTH-LIKE (SMXL) family members, BRI1-EMS-SUPPRESSOR (BES1) and DELLAs (43, 73, 74). Thus, perception of SL could lead to alterations in BR and GA signaling that converge to regulate shoot development. Additionally, MAX2 can oppositely regulate GA and ABA biosynthesis to promote photomorphogenesis (75). Further work is required to determine the extent to which SL signaling directly regulates protein degradation in each of these individual hormone pathways, but SCFMAX2 holds a unique regulatory position among hormone crosstalk mechanisms.
Regulation of Hormone Related E3 Stability
Substrate stability has been an intense area of study, but E3 ligases have also been shown to have variable stability. A wealth of evidence demonstrates that various factors can contribute to E3 ligase stability, such as self-ubiquitination and protein-protein interactions. Considering the central roles F-box proteins play in phytohormone perception, it is of interest to know how these proteins themselves are regulated such that it impacts hormone perception and/or other downstream signaling events. Biochemical and genetic studies have recently explored stability of different types of E3 ligases involved in auxin, ABA and ethylene signaling. For example, the auxin receptor TIR1 has been recently found to undergo autocatalytic degradation based on key residues that affect binding to CUL1 that were identified from yeast two-hybrid assays (76). Furthermore, direct associations between SCFTIR1 and HEAT SHOCK PROTEIN 90 (HSP90) have been recently shown to regulate the stability of TIR1 in response to temperature, which contributes to auxin responsiveness (77). Additionally, oligomerization of TIR1 appears to contribute to SCFTIR1 function and auxin signaling (78), suggesting that the relative levels of TIR1 may influence auxin perception mechanisms as well. Another example comes from studies on ABA-regulated degradation of KEG; biochemical experiments have recently identified phosphorylation of KEG driven by Calcineurin B-like Interacting Protein Kinase 26 (CIPK26) as being important for KEG activity (79). Finally, the Ethylene Overproducer1 (ETO1)/ETO1-like family of E3 ligase proteins are negatively regulated by light such that ethylene biosynthesis is promoted during photomorphogenesis (80). Continued efforts in these areas to define protein-protein interactions, post-translational modifications and/or other contributing factors regulating stability of particular E3 ligases involved in phytohormone signaling should provide fresh insight into how particular complexes may be modified.
Looking Forward: Methods and Approaches to Gain Deeper Mechanistic Insights
To date, ∼100 proteins have been shown to be involved in hormone-mediated E3 ligase activity in model plant systems through detailed genetic and biochemical studies. There are still many outstanding questions in the field that could be addressed using peptide mass spectrometry-based approaches, including improved identification of plant ubiquitinated proteins. Various affinity purification techniques together with mass spectrometry have been fruitful in identifying ubiquitinated proteins in response to light and during seedling development (81, 82) but similar studies have yet to be performed following hormone treatments. Thus, we have only captured a portion of the plant ubiquitinome to date and new approaches and methods will need to be applied to go deeper. In other eukaryotic systems, the use of antibodies that recognize the Lys-E-Gly-Gly (diGLY) remnant that is generated following trypsin digestion of ubiquitinated proteins have been successful at quantitatively describing ubiquitination sites under various cellular conditions (83–85). This approach has been used in rice and wheat to describe ∼400 ubiquitinated in rice and ∼300 ubiquitinated proteins in wheat (86, 87), demonstrating that it could be effectively applied to other model plant species to perform comprehensive profiling of hormone-dependent ubiquitinated proteins.
Another successful approach to identify ubiquitinated proteins is the so-called StUbEx method which relies on replacing endogenous ubiquitin in human U2OS cell lines with a modified version that amenable to purification and identification via peptide mass spectrometry (88). Arabidopsis and other key plant species (Physcomitrella patens, Zea mays, Marchantia polymorpha) have >10 copies of ubiquitin genes which is a hurdle to reduction and replacement of this regulatory protein. However, Chlamydomonas reinhardtii is an algal model species that only contains four copies of ubiquitin and is amenable to targeted DNA replacement (89). Thus, application of the StUbEx approach to Chlamydomonas reinhardtii would be possible and perhaps allow deeper identification of ubiquitinated proteins related to plant hormone signaling. Altogether such high-throughput proteomics approaches could greatly expand our depth of understanding with respect to phytohormone signaling and aid in identification in exact sites of modification on substrates.
The recent identification of ubiquitination sites on Aux/IAA proteins (65) is a reminder that we do not know the exact sites of ubiquitination for most of the other known substrates in phytohormone signaling pathways. A sensor-based proteomic approach modeled after the Vx3K0 K63 polyubiquitin-specific sensor identified 107 proteins in juvenile Arabidopsis plants with K63 polyubiquitination events (90). This approach is based on three repetitions of ubiquitin interaction motifs from Saccharomyces cerevisiae VPS27 subunit of the Endosomal Sorting Complex Required for Transport (termed Vx3K0) combined with a helical linker that spaces the UIMs for selective binding to K63-linked polyubiquitin chains (90). Similar polyubiquitin-sensor methods have revealed localization a linkage type dependence of ubiquitin signaling events in mammalian cell lines (91, 92). With this method (90) now developed for Arabidopsis, further studies could be performed on various mutants and/or following hormone treatments to provide better resolution of known hormone regulated E3 substrates. Other types of linkages may require different approaches for identification in plants, but such data will greatly increase our understanding of ubiquitinated proteins.
An additional challenge in the field has been defining E3-target interactions. There are still hundreds of E3 ligases in Arabidopsis without known substrates and several known substrates without known ligases. Given the large number of E3 ligases with known roles in hormone pathways it would be reasonable to assume there are more UPL proteins involved in hormone signaling that have not been captured via traditional genetic or biochemical approaches. Because these enzymes are part of large gene families in plants with high degrees of functional overlap it can be challenging to perform functional studies; for instance, there are >700 F-box proteins encoded by the Arabidopsis genome. Additionally, interactions between E3 ligases and their substrates are often transient and rapid, which can make detection difficult. One approach to mitigate these issues involves expression of affinity-tagged dominant negative F-box proteins which allow in vivo substrate identification (93). A recent application of this method to generate a F-box “decoy” to trap and identify in planta substrates has been applied to circadian regulated F-box targets (94). In this method the authors removed the F-box domain from a small family of partially redundant paralogous F-box proteins to allow these proteins to interact with their cognate substrates without triggering ubiquitination. These mutant proteins were fused with a dual affinity tag (3xFLAG-6xHis) to allow for affinity-purification followed by mass spectrometry (AP-MS) to detect ligase-substrate interactions. This approach yielded several novel interacting proteins that were validated using heterologous systems including yeast two-hybrid and expression and co-immunoprecipitation in mammalian cell cultures. If this approach were applied to additional F-box proteins involved in phytohormones signaling further progress could be made toward substrate identification and mapping of ubiquitination sites. Additionally, the application of other successful AP-MS approaches (95, 96) to these studies may help to uncover substrate proteins for E3 ligases that have remained elusive.
Another strategy for mapping ligase-substrate interactions could involve high-throughput protein-protein interaction assays such as yeast two-hybrid screens. The excellent genome annotation in Arabidopsis and available rapid cloning techniques make such screens readily feasible. Given the large number of transcriptional regulators known to be E3 targets (1) it would be logical to screen existing transcription factor libraries (97) against a custom library of E3 ligases to identify potential ligase-substrate interactions; while some interactions may require additional molecules such as hormones, other protein-protein interactions will be detectable by such high-throughput screens. Additionally, researches can validate these omic results and overcome genetic redundancy amount E3 ligases by using multiplexed CRISPR/Cas9 techniques developed for Arabidopsis (98). Altogether the application of high-throughput omics approaches will deepen our understanding of E3 ligases involved in hormone signaling and provide novel insights to answer several outstanding questions.
Acknowledgments
Thank you to Trevor Nolan, Justin Walley, and two anonymous reviewers for critical comments of the manuscript.
Footnotes
* The author declares no conflict of interest.
1 The abbreviations used are:
- RING
- really interesting new gene
- CRL
- Cullin-RING ligase
- AFB
- auxin F-box
- AID
- auxin-inducible degradation
- NPR
- nonexpression of pathogenesis-related genes.
REFERENCES
- 1. Kelley D. R., and Estelle M. (2012) Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 160, 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guerra D. D., and Callis J. (2012) Ubiquitin on the Move: The Ubiquitin Modification System Plays Diverse Roles in the Regulation of Endoplasmic Reticulum- and Plasma Membrane-Localized Proteins. Plant Physiol. 160, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shabek N., and Zheng N. (2014) Plant ubiquitin ligases as signaling hubs. Nat. Struct. Mol. Biol. 21, 293–296 [DOI] [PubMed] [Google Scholar]
- 4. Vierstra R. D. (2012) The Expanding Universe of Ubiquitin and Ubiquitin-Like Modifiers. PLANT Physiol. 160, 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yan S., and Dong X. (2014) Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hua Z., and Vierstra R. D. (2011) The Cullin-RING Ubiquitin-Protein Ligases. Annu. Rev. Plant Biol. 62, 299–334 [DOI] [PubMed] [Google Scholar]
- 7. Dharmasiri N., Dharmasiri S., and Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- 8. Kepinski S., and Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 [DOI] [PubMed] [Google Scholar]
- 9. Santner A., Calderon-Villalobos L. I. A., and Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307 [DOI] [PubMed] [Google Scholar]
- 10. Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J. S., Jürgens G., and Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 11. Calderón Villalobos L. I. A., Lee S., De Oliveira C., Ivetac A., Brandt W., Armitage L., Sheard L. B., Tan X., Parry G., Mao H., Zheng N., Napier R., Kepinski S., and Estelle M. (2012) A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramos J. A., Zenser N., Leyser O., and Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dreher K. A. (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell Online 18, 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheard L. B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T. R., Kobayashi Y., Hsu F.-F., Sharon M., Browse J., He S. Y., Rizo J., Howe G. A., and Zheng N. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T. Y., Hsing Y. I. C., Kitano H., Yamaguchi I., and Matsuoka M. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 [DOI] [PubMed] [Google Scholar]
- 16. Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., Maeda T., Matsuoka M., and Yamaguchi I. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 [DOI] [PubMed] [Google Scholar]
- 17. Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M. N., Maier A., and Schwechheimer C. (2007) The DELLA Domain of GA INSENSITIVE Mediates the Interaction with the GA INSENSITIVE DWARF1A Gibberellin Receptor of Arabidopsis. Plant Cell Online 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qiao H., Chang K. N., Yazaki J., and Ecker J. R. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S. H., Tada Y., Zheng N., and Dong X. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L., Sonbol F. M., Huot B., Gu Y., Withers J., Mwimba M., Yao J., He S. Y., and Dong X. (2016) Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Chen L., Li Y., Yan C., Miao D., Sun Z., Yan J., Sun Y., Wang L., Chu J., Fan S., He W., Deng H., Nan F., Li J., Rao Z., Lou Z., and Xie D. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473 [DOI] [PubMed] [Google Scholar]
- 22. Zhao L. H., Zhou X. E., Yi W., Wu Z., Liu Y., Kang Y., Hou L., De Waal P. W., Li S., Jiang Y., Scaffidi A., Flematti G. R., Smith S. M., Lam V. Q., Griffin P. R., Wang Y., Li J., Melcher K., and Xu H. E. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chevalier F., Nieminen K., Sanchez-Ferrero J. C., Rodriguez M. L., Chagoyen M., Hardtke C. S., and Cubas P. (2014) Strigolactone Promotes Degradation of DWARF14, an / Hydrolase Essential for Strigolactone Signaling in Arabidopsis. Plant Cell 26, 1134–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stirnberg P., Furner I. J., and Ottoline Leyser H. M. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 [DOI] [PubMed] [Google Scholar]
- 25. Hamiaux C., Drummond R. S. M., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., and Snowden K. C. (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 [DOI] [PubMed] [Google Scholar]
- 26. Wang K. L. C., Yoshida H., Lurin C., and Ecker J. R. (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950 [DOI] [PubMed] [Google Scholar]
- 27. Yoshida H., Nagata M., Saito K., Kevin W. L. C., and Ecker J. R. (2005) Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christians M. J., Gingerich D. J., Hansen M., Binder B. M., Kieber J. J., and Vierstra R. D. (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lyzenga W. J., Booth J. K., and Stone S. L. (2012) The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 71, 23–34 [DOI] [PubMed] [Google Scholar]
- 30. Ko J. H., Yang S. H., and Han K. H. (2006) Upregulation of an arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y., Yang C., Li Y., Zheng N., Chen H., Zhao Q., Gao T., Guo H., and Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19, 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X., Garreton V., and Chua N. H. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stone S. L., Williams L. A., Farmer L. M., Vierstra R. D., and Callis J. (2006) KEEP ON GOING, a RING E3 Ligase Essential for Arabidopsis Growth and Development, Is Involved in Abscisic Acid Signaling. Plant Cell Online 18, 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Serrano M., Parra S., Alcaraz L. D., and Guzmán P. (2006) The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J. Mol. Evol. 62, 434–445 [DOI] [PubMed] [Google Scholar]
- 35. Raab S., Drechsel G., Zarepour M., Hartung W., Koshiba T., Bittner F., and Hoth S. (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 59, 39–51 [DOI] [PubMed] [Google Scholar]
- 36. Samuel M. A., Mudgil Y., Salt J. N., Delmas F., Ramachandran S., Chilelli A., and Goring D. R. (2008) Interactions between the S-Domain Receptor Kinases and AtPUB-ARM E3 Ubiquitin Ligases Suggest a Conserved Signaling Pathway in Arabidopsis. Plant Physiol. 147, 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo J., Shen G., Yan J., He C., and Zhang H. (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 46, 649–657 [DOI] [PubMed] [Google Scholar]
- 38. Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., and Wang X. (2017) SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev. Cell 41, 47–58.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nolan T. M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D. C., Walley J., and Yin Y. (2017) Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 41, 33–46.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tan X., Calderon-Villalobos L. I. A., Sharon M., Zheng C., Robinson C. V., Estelle M., and Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645 [DOI] [PubMed] [Google Scholar]
- 41. Kong L., Cheng J., Zhu Y., Ding Y., Meng J., Chen Z., Xie Q., Guo Y., Li J., Yang S., and Gong Z. (2015) Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6, 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., Yi W., Zhao L., Ma H., He Y., Wu Z., Melcher K., Qian Q., Xu H. E., Wang Y., and Li J. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., Sun S., Zhu W., Jia K., Yang H., and Wang X. (2013) Strigolactone/MAX2-Induced Degradation of Brassinosteroid Transcriptional Effector BES1 Regulates Shoot Branching. Dev. Cell 27, 681–688 [DOI] [PubMed] [Google Scholar]
- 44. Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., Ma W., Gao H., Chen J., Yang C., Wang D., Tan J., Zhang X., Guo X., Wang J., Jiang L., Liu X., Chen W., Chu J., Yan C., Ueno K., Ito S., Asami T., Cheng Z., Wang J., Lei C., Zhai H., Wu C., Wang H., Zheng N., and Wan J. (2013) D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stanga J. P., Smith S. M., Briggs W. R., and Nelson D. C. (2013) SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moss B. L., Mao H., Guseman J. M., Hinds T. R., Hellmuth A., Kovenock M., Noorassa A., Lanctot A., Villalobos L. I. A. C., Zheng N., and Nemhauser J. L. (2015) Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 169, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Havens K. A., Guseman J. M., Jang S. S., Pierre-Jerome E., Bolten N., Klavins E., and Nemhauser J. L. (2012) A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishimura K., and Kanemaki M. T. (2014) Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID). Curr. Protoc. Cell Biol. 2014, 20.9.1–20.9.16 [DOI] [PubMed] [Google Scholar]
- 49. Morawska M., and Ulrich H. D. (2013) An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu J. Y., Li Y., Cao D. M., Yang H., Oh E., Bi Y., Zhu S., and Wang Z. Y. (2017) The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 66, 648–657.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martins S., Dohmann E. M. N., Cayrel A., Johnson A., Fischer W., Pojer F., Satiat-Jeunemaître B., Jaillais Y., Chory J., Geldner N., and Vert G. (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakamoto T., Kitano H., and Fujioka S. (2013) An E3 ubiquitin ligase, ERECT LEAF1, functions in brassinosteroid signaling of rice. Plant Signal. Behav. 8, e27117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu X., Qian Q., Xu T., Zhang Y., Dong G., Gao T., Xie Q., and Xue Y. (2013) The U-Box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet. 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang H., Cui F., Wu Y., Lou L., Liu L., Tian M., Ning Y., Shu K., Tang S., and Xie Q. (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell Online 27, 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheng C., Wang Z., Ren Z., Zhi L., Yao B., Su C., Liu L., and Li X. (2017) SCFAtPP2-B11modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao H., Zhang H., Cui P., Ding F., Wang G., Li R., Jenks M. A., Lu S., and Xiong L. (2014) The putative E3 ubiquitin ligase ECERIFERUM9 regulates abscisic acid biosynthesis and response during seed germination and postgermination growth in Arabidopsis. Plant Physiol. 165, 1255–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang X., Wu Q., Ren J., Qian W., He S., Huang K., Yu X., Gao Y., Huang P., and An C. (2012) Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for Jasmonate-mediated responses in Arabidopsis. Plant Physiol. 160, 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Peng Y. J., Shih C. F., Yang J. Y., Tan C. M., Hsu W. H., Huang Y. P., Liao P. C., and Yang C. H. (2013) A RING-type E3 ligase controls anther dehiscence by activating the jasmonate biosynthetic pathway gene DEFECTIVE in ANTHER DEHISCENCE1 in Arabidopsis. Plant J. 74, 310–327 [DOI] [PubMed] [Google Scholar]
- 59. Jung C., Zhao P., Seo J. S., Mitsuda N., Deng S., and Chua N.-H. (2015) PLANT U-BOX PROTEIN10 Regulates MYC2 Stability in Arabidopsis. Plant Cell 27, 2016–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jeong J. S., Jung C., Seo J. S., Kim J.-K., and Chua N.-H. (2017) The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in Jasmonate responses. Plant Cell, tpc.00216.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kuai X., MacLeod B. J., and Després C. (2015) Integrating data on the Arabidopsis NPR1/NPR3/NPR4 salicylic acid receptors; a differentiating argument. Front. Plant Sci. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Attaran E., and He S. Y. (2012) The long-sought-after salicylic acid receptors. Mol. Plant 5, 971–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ciechanover A., and Ben-Saadon R. (2004) N-terminal ubiquitination: More protein substrates join in. Trends Cell Biol. 14, 103–106 [DOI] [PubMed] [Google Scholar]
- 64. Gilkerson J., Kelley D. R., Tam R., Estelle M., and Callis J. (2015) Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiol. 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Winkler M., Niemeyer M., Hellmuth A., Janitza P., Christ G., Samodelov S. L., Wilde V., Majovsky P., Trujillo M., Zurbriggen M. D., Hoehenwarter W., Quint M., and Calderón Villalobos L. I. A. (2017) Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 8, 15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Albertos P., Romero-Puertas M. C., Tatematsu K., Mateos I., Sánchez-Vicente I., Nambara E., and Lorenzo O. (2015) S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang N., Xing Y., Lou Q., Feng P., Liu S., Zhu M., Yin W., Fang S., Lin Y., Zhang T., Sang X., and He G. (2017) Dwarf and short grain 1, encoding a putative U-box protein regulates cell division and elongation in rice. J. Plant Physiol. 209, 84–94 [DOI] [PubMed] [Google Scholar]
- 68. Kim J. H., Lee H. J., and Park C. M. (2017) HOS1 acts as a key modulator of hypocotyl photomorphogenesis. Plant Signal. Behav. 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee K., and Seo P. J. (2015) The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal. Behav. 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lourenço T. F., Serra T. S., Cordeiro A. M., Swanson S. J., Gilroy S., Saibo N. J. M., and Oliveira M. M. (2015) The rice E3 ubiquitin ligase OsHOS1 modulates the expression of OsRMC, a gene involved in root mechano-sensing, through the interaction with two ERF transcription factors. Plant Physiol. 169, 2275–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ding Y., Dommel M., and Mou Z. (2016) Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J. 86, 20–34 [DOI] [PubMed] [Google Scholar]
- 72. Pauwels L., Ritter A., Goossens J., Durand A. N., Liu H., Gu Y., Geerinck J., Boter M., Vanden Bossche R., De Clercq R., Van Leene J., Gevaert K., De Jaeger G., Solano R., Stone S., Innes R. W., Callis J., and Goossens A. (2015) The RING E3 Ligase KEEP ON GOING Modulates JASMONATE ZIM-DOMAIN12 Stability. Plant Physiol. 169, 1405–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bennett T., Liang Y., Seale M., Ward S., Müller D., and Leyser O. (2016) Strigolactone regulates shoot development through a core signalling pathway. Biol. Open bio.021402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nakamura H., Xue Y.-L., Miyakawa T., Hou F., Qin H.-M., Fukui K., Shi X., Ito E., Ito S., Park S.-H., Miyauchi Y., Asano A., Totsuka N., Ueda T., Tanokura M., and Asami T. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4 [DOI] [PubMed] [Google Scholar]
- 75. Shen H., Luong P., and Huq E. (2007) The F-Box Protein MAX2 Functions as a Positive Regulator of Photomorphogenesis in Arabidopsis. PLANT Physiol. 145, 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu H., Zhang Y., Moss B. L., Bargmann B. O. R., Wang R., Prigge M., Nemhauser J. L., and Estelle M. (2015) Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 1, 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang R., Zhang Y., Kieffer M., Yu H., Kepinski S., and Estelle M. (2016) HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 7, 10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dezfulian M. H., Jalili E., Roberto D. K. A., Moss B. L., Khoo K., Nemhauser J. L., and Crosby W. L. (2016) Oligomerization of SCFTIR1 is essential for Aux/IAA degradation and auxin signaling in Arabidopsis. PLoS Genet. 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lyzenga W. J., Sullivan V., Liu H., and Stone S. L. (2017) The kinase activity of calcineurin B-like interacting protein kinase 26 (CIPK26) influences its own stability and that of the ABA-regulated ubiquitin ligase, Keep on Going (KEG). Front. Plant Sci. 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yoon G. M., and Kieber J. J. (2013) 14–3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Aguilar-Hernández V., Kim D. Y., Aguilar-Hernández V., Kim D. Y., Stankey R. J., Vierstra R. D., Scalf M., and Smith L. M. (2017) Mass spectrometric analyses reveal a central role for ubiquitylation in remodeling the Arabidopsis proteome during photomorphogenesis. Mol. Plant 10, 846–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kim D.-Y., Scalf M., Smith L. M., and Vierstra R. D. (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25, 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xu G., Paige J. S., and Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., and Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xie X., Kang H., Liu W., and Wang G. L. (2015) Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 14, 2017–2025 [DOI] [PubMed] [Google Scholar]
- 87. Zhang Y., Song L., Liang W., Mu P., Wang S., and Lin Q. (2016) Comprehensive profiling of lysine acetylproteome analysis reveals diverse functions of lysine acetylation in common wheat. Sci. Rep. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Akimov V., Olsen L. C. B., Hansen S. V. F., Barrio-Hernandez I., Puglia M., Jensen S. S., Solov'Yov I. A., Kratchmarova I., and Blagoev B. (2018) StUbEx PLUS - A modified stable tagged ubiquitin exchange system for peptide level purification and in-depth mapping of ubiquitination sites. J. Proteome Res. 17, 296–304 [DOI] [PubMed] [Google Scholar]
- 89. Ferenczi A., Pyott D. E., Xipnitou A., and Molnar A. (2017) Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Sci. 2017, 10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Johnson A., and Vert G. (2016) Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol. 171, 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sims J. J., Scavone F., Cooper E. M., Kane L. A., Youle R. J., Boeke J. D., and Cohen R. E. (2012) Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods 9, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van Wijk S. J. L., Fiskin E., Putyrski M., Pampaloni F., Hou J., Wild P., Kensche T., Grecco H. E., Bastiaens P., and Dikic I. (2012) Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell 47, 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sharma R., Williams P. J., Gupta A., McCluskey B., Bhaskaran S., Munoz S., and Oyajobi B. O. (2015) A dominant-negative F-box deleted mutant of E3 ubiquitin ligase, beta-TrCP1/FWD1, markedly reduces myeloma cell growth and survival in mice. Oncotarget 6, 21589–21602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lee C.-M., Feke A., Adamchek C., Webb K., Pruneda-Paz J., Bennett E. J., Kay S. A., and Gendron J. M. (2017) Decoys reveal the genetic and biochemical roles of redundant plant E3 ubiquitin ligases. bioRxiv [Google Scholar]
- 95. Lee C.-M., Adamchek C., Feke A., Nusinow D. A., and Gendron J. M. (2017) in Plant Genomics: Methods and Protocols, 231–249 [Google Scholar]
- 96. Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M. J., Evans B. S., Briggs S. P., Hicks L. M., Kay S. A., and Nusinow D. A. (2016) Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15, 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pruneda-Paz J. L., Breton G., Nagel D. H., Kang S. E., Bonaldi K., Doherty C. J., Ravelo S., Galli M., Ecker J. R., and Kay S. A. (2014) A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 8, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu Z., Chen Q., Chen W., Zhang X., Mei F., Zhang P., Zhao M., Wang X., Shi N., Jackson S., and Hong Y. (2017) Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in Arabidopsis. J. Integr. Plant Biol. [DOI] [PubMed] [Google Scholar]