Abstract
Salinity causes osmotic stress to crops and limits their productivity. To understand the mechanism underlying soybean salt tolerance, proteomics approach was used to identify phosphoproteins altered by NaCl treatment. Results revealed that 412 of the 4698 quantitatively analyzed phosphopeptides were significantly up-regulated on salt treatment, including a phosphopeptide covering the serine 59 in the transcription factor GmMYB173. Our data showed that GmMYB173 is one of the three MYB proteins differentially phosphorylated on salt treatment, and a substrate of the casein kinase-II. MYB recognition sites exist in the promoter of flavonoid synthase gene GmCHS5 and one was found to mediate its recognition by GmMYB173, an event facilitated by phosphorylation. Because GmCHS5 catalyzes the synthesis of chalcone, flavonoids derived from chalcone were monitored using metabolomics approach. Results revealed that 24 flavonoids of 6745 metabolites were significantly up-regulated after salt treatment. We further compared the salt tolerance and flavonoid accumulation in soybean transgenic roots expressing the 35S promoter driven cds and RNAi constructs of GmMYB173 and GmCHS5, as well as phospho-mimic (GmMYB173S59D) and phospho-ablative (GmMYB173S59A) mutants of GmMYB173. Overexpression of GmMYB173S59D and GmCHS5 resulted in the highest increase in salt tolerance and accumulation of cyaniding-3-arabinoside chloride, a dihydroxy B-ring flavonoid. The dihydroxy B-ring flavonoids are more effective as anti-oxidative agents when compared with monohydroxy B-ring flavonoids, such as formononetin. Hence the salt-triggered phosphorylation of GmMYB173, subsequent increase in its affinity to GmCHS5 promoter and the elevated transcription of GmCHS5 likely contribute to soybean salt tolerance by enhancing the accumulation of dihydroxy B-ring flavonoids.
Keywords: iTRAQ, Metabolomics, Oxidative stress, Phosphoproteome, Protein Modification*, Stress response, Flavonoid Metabolism, GmMYB173, Metabonomics, Phosphoproteomics, Soybean
Salinity is one of the main abiotic stresses affecting the growth and productivity of crops (1). In saline soils, the sodium ion causes most of the damages to plant cells at the molecular level through ionic toxicity and homeostasis disruption (2–4). Furthermore, the salt stress interferes with the photosynthesis in chloroplasts and the electron transport in mitochondria and accelerates the production of reactive oxygen species (ROS) and simultaneously causes oxidative stress (5, 6). Plants have evolved two main strategies to eliminate these excessive ROS, the enzymatic degradation, for example elimination of ROS by superoxide dismutase or ascorbate peroxidase (7), and deoxidization by small molecules such as ascorbic acid, mannitol, and flavonoids (8, 9).
Compared with that of ascorbic acid and mannitol, the role of the flavonoid family in plant tolerance to salinity is just beginning to unfold (9–11), although more than 10,000 species of flavonoids have been isolated from plants thus far (12, 13). It is well known that all flavonoids are derivatives of chalcone, an intermediate that is directly synthesized from p-coumaroyl-CoA and alonyl-CoA to 15-carbon skeletons at a 3:1 ratio (12). Derived from chalcone, various categories of flavonoids can be synthesized, including flavanones (e.g. naringenin), isoflavone (e.g. formononetin), flavonols (e.g. quercetin) and others, based on their structural variation in the C-ring and the functional groups at C-3 and C-4 (supplemental Fig. S1) (12). Most of the natural flavonoids are conjugated with glycosides (11), such as naringenine-7-rhamnosidoglucoside (naringin) and daidzein-7-O-β-d-glucopyranoside (daidzin). Flavonoids have been found to have a wide array of physiological functions in plant tolerant responses to abiotic stresses, including protection against harmful UV light (14–16) and hydrogen peroxide scavenging (5, 17–19). The hydroxyl groups of flavonoids play a critical role during the ROS scavenging processes (20–23).
Three categories of enzymes, including the chalcone synthase (CHS)1, chalcone isomerase (CHI), and cytochrome P450 monooxygenase (CPM), play critical roles in flavonoid synthesis. CHS can catalyze the conversion of 4-coumaroyl-CoA and malonyl-CoA to chalcone. The chalcone could be directly converted to flavanone by CHI. Then, CPM catalyzes the conversion of flavanones (e.g. naringenin) to isoflavones (e.g. formononetin). Interestingly, CHS, CHI, and CPM usually play key roles in plant response to salt stress (24–26). Piao et al. demonstrated that AtCHS expression was significantly increased concomitantly with the accumulation of anthocyanins in Arabidopsis plants subjected to salt stress (27). Martens Yan et al. found that the RNA interference of GmFNSII (coding for CYP93B, an enzyme in cytochrome P450 monooxygenase subfamily) significantly reduced the tolerance of soybean roots to salinity because of the decreased flavone level and higher H2O2 accumulation (9). Our recent study documented that salt tolerance of Arabidopsis and soybean were positively regulated by CHS and negatively regulated by CHI and CPM (28). It is noteworthy that CHS is a single copy gene in the Arabidopsis genome (29), and our results confirmed that the AtCHS knockout mutant is extremely sensitive to salt stress, demonstrating that chalcone and its downstream derivatives play a key role in protecting plants from the damage caused by saline stresses (28). CHS genes in leguminous plants is comprised of a multiple gene family, for instance there are at least nine CHS genes in soybean (29). Expressions of different CHS family members exhibit distinct temporal and spatial patterns in response to developmental and environmental cues (30), implying CHS members in soybean might be involved in different physiological processes. Identifying the specific roles of a CHS gene in soybean is a challenge to traditional genetic and physiological studies.
Genes encoding CHS, CHI, and CPM that are involved in different flavonoid biosynthetic steps, are usually regulated by MYB transcription factors (TFs) (31–34). The MYBs can be grouped into four types, MYB1R, R2R3-MYB, R1R2R3-MYB, and 4RMYB, based on the variation in the repeat numbers of the MYB DNA-binding domains (32). Among them, the R2R3-MYB subfamily is well known to regulate flavonoid metabolism (35). In Arabidopsis, AtMYB75, AtMYB90, MYB112, MYB113, and MYB114 have been shown to play important roles in activating the transcription of genes in the anthocyanin biosynthetic pathway (34, 36, 37). In Scutellaria baicalensis, SbMYB8 was reported to improve drought tolerance by regulating flavonoid biosynthesis (35). Soybean GmMYBJ2 was found to notably improve the seed germination rate of transgenic Arabidopsis undergoing water-deficit stress (38). Usually, the MYB-type TFs interact with other transcription factors such as bHLH (37) and WD40 proteins to regulate the production of anthocyanins (25, 39).
The physiological responses to salinity rely on diverse interplays, such as those between kinases and protein phosphorylation (28), between MYB-type TFs and salt-responsive enzymes (e.g. CHS, CHI, and CPM) (32, 40), and those between enzymes and metabolites (24). All these form complicated networks to help plants adapt to the ever-changing environments (41). Hence, it is essential to gain an in-depth and integrated knowledge for understanding the nature of these complicated responses. Omics studies, including the proteomics and metabolomics, allow researchers to obtain comprehensive insight and to connect the nodes of the molecular networks underlying plants' abiotic tolerances (41). Interestingly, the quantitative analyses of omics provide us precise network structure and kinetic data to simulate the dynamics of a biological process (42).
Using phosphoproteomic and proteomic approaches, our previous studies revealed that several key proteins (such as MYB TFs, CHS, CHI and CPM) are involved in soybean tolerance to salinity (28). We also proposed a salt tolerance pathway involving flavonoid metabolism, mostly mediated by phosphorylated MYB TFs. In this study, we are introducing a mechanism by which the phosphorylation of GmMYB173 regulates flavonoid syntheses via expressional control of GmCHS5 and enhances the tolerance of soybean to salt stress.
EXPERIMENTAL PROCEDURES
Plant Materials, Growth Conditions, and Salt Treatments
Seeds of Glycine max cultivar Union85140 were surface sterilized and germinated in wet filter papers as previously described (28). Two days after germination, the seedlings were transplanted in pearlite and sphagnum peat soil (v:v = 1:3). After transferring to the growth chamber, the light intensity was set at 200 μmol × m−2 × s−1 with a photo period of 18 h per day. The temperature was adjusted to 25/18 °C for day/night cycle. The seedlings were watered with 1/4 fold Fahräeus medium every 4 days and deionized water was also used to irrigate every 2 days after application of abovementioned medium. At the three-trifoliate stage, the seedlings were treated with 200 mm NaCl for 24 h. Then the samples were collected and immediately stored at −80 °C until further use.
Protein Extraction and Digestion with FASP
The TCA/Acetone extraction method was used to isolate total proteins from soybean roots as previously described (28). Five grams of root tissue for each sample was ground into fine powder in liquid nitrogen. The powder was thoroughly suspended in 45 ml of pre-cooled TCA/Acetone (v:v = 1:9) and kept at −20 °C overnight for protein extraction. The homogenate was centrifuged at 7000 × g for 20 min and the pellet was washed three times with 40 ml acetone. Then the residual acetone was removed by applying vacuum and 50 mg white powder of each sample was resuspended in 1 ml SDT lysis buffer (4% SDS, 100 mm Tris-HCl, 1 mm DTT, 1 mm PMSF, pH7.6, including 1 × PhosSTOP phosphatase inhibitor mixture from Roche, Mannheim, Germany). The solution was boiled for 15 min in a water bath, sonicated for 100 s, centrifuged at 13,400 × g for 15 min. The protein concentration in supernatant was quantified via BCA (bicinchoninic acid) method and 20 μg was used to run a SDS-PAGE gel for QC test.
The protein digestion procedure was performed as previously described (28). Before digestion, 200 μg of protein for each sample were processed to remove residual SDS in the samples by filter aided sample preparation (FASP) method. Next, the concentrated proteins were digested with 8 μg of trypsin at 37 °C for 16–18 h. After digestion, the peptide solution was passed through a Microcon filtration device (MWCO 10 kd) and the peptide concentration was quantified by measuring OD280. All the procedures were carried out at 4 °C unless exceptions were stated.
Eight-plex iTRAQ Labeling and Phosphopeptide Enrichment
One hundred micrograms of digested peptide for each sample was subjected to AB Sciex iTRAQ labeling. The eight-plex iTRAQ labeling was performed according to the manufacturer's instructions (28). Each sample was analyzed with four biological replicates. The average value of 4 replicates serves as normalizing reference (REF) for the peptide content calculation.
For phosphopeptides enrichment, TiO2 beads were used as previously described (28). The labeled peptides were acidified with 50 μl DHB buffer (3% 2, 5-dihydroxybenzoic acid, 80% acetonitrile and 0.1% TFA), and then incubated with 25 μg of TiO2 beads (10 μ in diameter, Sangon Biotech) for 40 min at room temperature. After the incubation, the TiO2 beads were spun down and the pellets were packed into plastic tips (fit to 10 μl pipette). The peptide-TiO2 beads were washed with 20 μl of wash solution I (20% acetic acid, 300 mm octanesulfonic acid sodium salt and 20 mg/ml DHB) three times, then washed with 20 μl wash solution II (70% water; 30% acetonitrile) three times. The phosphopeptides were then eluted using freshly prepared ABC buffer (50 mm ammonium phosphate, pH 10.5). The enriched phosphopeptide solution was lyophilized and re-dissolved in 20 μl 0.1% TFA solution for nano-RPLC-MS/MS analysis.
Nano-RPLC-MS/MS Analysis of Phosphorylated Peptides
For Nano-RPLC-MS/MS analysis, 5 μg (≤ 10 μl) phosphopeptide solution was loaded onto a two dimensional EASY-nLC1000 system coupled to a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific). In the nanoLC separation system, mobile phase A solution containing 0.1% formic acid in water, and mobile phase B solution containing 84% acetonitrile and 0.1% formic acid. Before peptide loading, the Thermo EASY SC200 trap column (RP-C18, 3 μm, 100 mm × 75 μm) was pre-equilibrated with 95% mobile phase A for 30 min. The phosphopeptides were first transferred to the Thermo scientific EASY column (2 cm × 100 μm 5 μm-C18) and then separated via the trap column using a gradient of 0–55% mobile phase B for 220 min. Then, the columns were rinsed with 100% mobile phase B for 8 min and re-equilibrated to the initial conditions for 12 min. The flow rate of the above procedures was 0.25 μl per min.
Experimental Design and Statistical Rationale for Mass Spectrometric Data Analyses
In this research, four biological replicas of control (C1∼C4) and NaCl treated (T1∼T4) groups of soybean roots were analyzed in one eight-plex iTRAQ set (supplemental Fig. S2). In addition, each biological replicate of these labeled peptides was measured three times. The MS data, which ranged from 350 to 1800 m/z, was acquired at the resolution of 70,000. To obtain MS/MS spectra, the 10 most abundant ions from each MS scan were subsequently dissociated by higher energy collisional dissociation (HCD) in alternating data-dependent mode with a resolution no less than 17500 (supplemental Table S1).
The raw data were extracted by Mascot2.2, and analyzed by Proteome Discoverer1.4 (Thermo Scientific). To identify the phosphopeptides, the mascot data was searched against the 74305 entries curated in the peptide database of soybean (uniprot_Glycine_74305_20140429.fasta, http://www.uniprot.org/, on April 29, 2014).
The PhosphoRS algorithm (from the Proteome Discoverer1.4) was used for confidence assessments on the localizations of phosphorylation in peptide sequences (42). According to the phosphoRS, a peptide that passed both phosphoRS probabilities > 0.75 and phosphoRS score > 0.5 was considered as a truly phosphopeptide. For each spectra of a specific sample, the median of its intensity values was used for quantification analysis. The average value of 8 median of a specific spectra was used as reference (supplemental Table S1) for data normalization in an Eight-plex iTRAQ Labeled sample group. A Student's t test was performed using the standard deviation to assess the changes between two samples Phosphopeptides with FDR (False Discovery Rate) < 0.05 and p value < 0.05 were considered as significantly changed.
Metabolomic Analysis
The metabolomics analysis was conducted as previously described (44), with a few minor modifications. Five grams of root tissue for each sample was homogenized in liquid nitrogen. For metabolites extraction, 50 mg homogenate was suspended in 1 ml of pre-cooled methanol: acetonitrile: water (2:2:1, v/v/v) solution. The mixtures were briefly vortexed, and then sonicated for 30 min, this vortex-sonication step was repeated twice. After 60 min incubation at −20 °C, the extract was centrifuged at 14,000 × g for 15 min.
Two μl of supernatant was injected into the Agilent 1290 Infinity LC system coupled to a Triple TOF 6600 Mass Spectrometer (AB SCIEX). In the UHPLC separation system, mobile phase A solution contains 25 mm ammonium acetate and 25 mm ammonia in water, and mobile phase B solution contains 100% acetonitrile. The ACQUITY UPLC BEH Amide separation column (Waters, 1.7 μm, 2.1 mm × 100 mm) was pre-equilibrated with 85% mobile phase B at 4 °C before loading the metabolites. The metabolites were then separated using a gradient of 85–65% mobile phase B for 12 min. The mobile phase B was then kept at 40% for 3 min followed by 5 min rinse with 85% of mobile phase B. The flow rate of the total mobile phase was set as 300 μl per min. Each sample was analyzed with 10 biological replicates. The equipment was calibrated with standardized as suggested by the manufacturer.
The separated metabolites were ionized with ESI positive and negative mode, respectively. The parameters of MS analysis are listed in supplemental Table S2. The raw data was converted into mzML format using the ProteoWizard software, and then the XCMS program was used to calibrate the retention-time, extract peak-integration and normalization of the data from different eight-plexes. The identity of each ion was searched against the METLIN database (https://metlin.scripps.edu/index.php) for molecular annotation.
Bioinformatic Analysis
Peptide motifs were extracted using the motif-X algorithm (45). The width of the generated motifs was set as seven amino acids and S or T was selected as the central amino acid.
Chromatin Immunoprecipitation-quantitative PCR
35S:GmMYB173-HA transgenic lines were used for ChIP-qPCR as described by Schmidt et al. with minor modification. ChIP analysis with the Merck Milipore ChIP kit according to the manufacturer's instructions. Soybean roots were cross-linked with 1% (v/v) formaldehyde for 15 min. A rabbit anti-HA polyclonal antibody was used to precipitate the GmMYB173-HA: DNA complexes. The immune-complex was then resuspended in 30 μl elution buffer and boiled for 3 h in water bath, and then treated with phenol-chloroform mixture to remove the protein in the DNA-containing solution. For each qPCR reaction, 1 μl of the ChIPed DNA was used. Enrichments were calculated by first normalizing the obtained CT values with the input CT and subsequent computation of ΔΔCT (CT(IgG)-CT(anti-HA)). Primers used are listed in supplemental Table S3.
Electrophoresis Mobility Shift Assay (EMSA)
DNA fragment encoding the N-terminal 167 aa containing the DNA-binding domain (aa 74–130) of GmMYB173 were amplified using PCR and with primer pair listed in supplemental Table S3. The amplified fragment was cloned in pEASY-blunt (Transgene, Tianjin, China). Site-directed mutagenesis was used to generate the S59A and S59D mutants. All the constructs in pEASY-blunt were sequenced and sub-cloned into pET32a in the reading frame to express the 6 x His-tags at both N- and C-ends of the 167 aa GmMYB173 truncated protein or its mutants. These expression constructs were transformed into E. coli strain BL21 (DE3) pLysS, and the WT or mutated versions of recombinant GmMYB173 protein was purified through nickel-affinity chromatography (Ni-NTA, Qiagene). Probe labeling and EMSA were carried out as previously described (46). Briefly oligo primers contain a 30 bp tested promoter sequence were synthesized and four 4 hot A were added at the end of both strands using Klenow fragment of DNA polymer I). 32P-labeled CHS5-P3 probe (see supplemental Table S3, 104 cpm per sample) was incubated with 2 μg of tested protein at room temperature for 30 min. The reaction mixture was separated on 8% of polyacrylamide gel in 0.5 x TBE buffer, and the dried gel was exposed to X-ray film for 48 to 96 h.
In Vitro Kinase Activity Assays
The full-length cDNA of soybean casein kinase-II (GmCK2) was generated with PCR using a pair of primers with EcoRI and XhoI added to the 5′- and 3′-ends (supplemental Table S3). The fragment was cloned in pEASY-blunt (Transgene), sequenced and sub-cloned into pET32a to express the 6 × His-tags at both N- and C-ends. The purified GmCK2α was used to test whether recombinant GmMYB173 (aa 1–167, see the previous section) could be phosphorylated by GmCK2α. The wild type and S59A mutant of GmMYB173 was tested as substrate. The substrate phosphorylation assay was performed in 20 μl of kinase buffer containing HEPES (25 mm), MgCl2 (5 mm), 50 μm ATP, 1 μCi of [γ-32P] ATP, 0.4 μg GmCK2α and 0.4 μg GmMYB173-WT or GmMYB173-S59A for 30 min at 25 °C. The reactions were stopped with SDS loading buffer and electrophoresed on a 12% SDS-PAGE. The dried gel was exposed X-ray film for 72 h.
RNA Extraction and Quantitative RT-PCR
RNA extraction, mRNA reverse transcription and quantitative real-time PCR were performed as previously described (28). Briefly, 1 g of root sample was ground to a fine powder in liquid nitrogen and then the total RNA was isolated according to the manufacturer's instructions (RNApure Plant Kit, CWBiotech, Beijing, China) and subjected to DNA digestion with 20 units of DNase I for 15 min at 25 °C. The isolated total RNA visualized using electrophoresis on 1.5% (w/v) agarose gels for quality assurance.
About 3 μg of RNA template was used to synthesize cDNAs by HiFiScript 1st Strand cDNA Synthesis Kit (CWBiotech) following the manufacturer's instructions. UltraSYBR Mixture (CWBiotech) was used for qRT-PCR on a Bio-Rad CFX96 PCR detector. The primers (supplemental Table S3) were selected using the Primer Blast online software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) against the Glycine max (taxid:3847) database. Each reaction mixture consisted of 10 ng cDNA template, 1 μl each forward and reverse primers (100 mm stocks), and 10 μl SYBR green in a final volume of 20 μl. PCR was run with an initial denaturing temperature of 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 32 s. Melting curve analysis was also performed at the end of the run to verify that only one product was amplified. PCR efficiencies and expression levels were calculated for target and reference genes from a serial dilution of the cDNA template. Target gene transcript levels were normalized to the transcript level of gene GmActin11 (28).
Plant Expression Constructs
Primer sequences are listed in supplemental Table S3 and all constructs were verified by sequencing. Full-length cDNAs for soybean GmMYB173 (Glyma17G094400) and GmCHS5 (Glyma01g43880.1) were downloaded from the Phytozome database (http://www.phytozome.net/soybean).
For gene overexpression and ChIP analysis, the full-length CDS of target genes from Union85140, were cloned into the pDL28-HA/Ubq10::dsRed vector, a derivate of pDL28 carrying an HA-tag and a RFP-tag (46), between SacI and KpnI (GmMYB173) sites or BamH I single site (GmCHS5), downstream of the 35S promoter. The original pDL28-HA/Ubq10::dsRed vector was used as a negative control. All these constructs were transformed into the Union85140 via Agrobacterium rhizogenes strain K599. The composites were treated with 200 mm NaCl in 1/4 fold Fahräeus medium for 24 h. The transgenic roots were verified by PCR.
For subcellular localization analysis, the full-length CDS of GmMYB173 was cloned into the pDL28 vector between SacI and KpnI sites, downstream to the 35S promoter (defined as pDL28-GmMYB173). Then, the RFP-tag was inserted into the pDL28-GmMYB173 construct between KpnI and SalI, downstream of the GmMYB173 (defined as pDL28-GmMYB173-RFP). The pDL28-GmMYB173-RFP was transformed into the Union85140 via A. rhizogenes strain K599.
For RNAi assays, the Oligo Engine 2.0 software was used to select an approximate 300 bp (300∼800) length fragment from the coding region of the target gene. The fragment was amplified from cDNA library and cloned into the pHANNIBAL vector in opposite orientations on either side of a PDK intron for constructing an invert repeat (13). The RNAi construct was cloned into the pDL28-HA/Ubq10::dsRed vector between SalI and SpeI (GmMYB173 and GmCHS5) sites downstream of the 35S promoter. The pDL28-Gene RNAi vector was inserted into A. rhizogenes strain K599 for transformation.
For site mutation, the primers (supplemental Table S3) were generated by QuikChange Primer Design software (http://www.genomics.agilent.com/primerDesignProgram.jsp). Full-length cDNA of the target gene was inserted into pEASY®-Blunt Zero Cloning Vector (TRANS) and mutated by PCR. The PCR product was added with 1 U DpnI enzyme (Thermo Scientific) and incubated in metal bath at 37 °C for 1 h. The mutated cDNA was then inserted into pDL28 vector between SacI and KpnI sites.
Subcellular Localization
Soybean roots were mounted between glass slides and cover slips, and then imaged using an inverted Carl Zeiss LSM 710 laser scanning microscope (47).
HPLC Analysis
HPLC method for flavone quantification was operated as described previously with minor modifications (13). Briefly, 25 mg roots were ground in liquid nitrogen, and extracted with 1.25 ml of pre-cooled methanol: acetic acid: water (9 : 1 : 10, v/v/v) at room temperature for 30 min. After 5 min of incubation, the supernatant of extract was filtered through a 0.22 μm membrane. Then, 10 μl of the purified extract was injected into a Waters HPLC system and separated by a 150 mm × 4.6 mm × 5 μm RP-Spherisorb-ODS2-C18 analytical column. The flavone compounds were monitored with a 600 Controller and a PDA Detector between the wavelengths of 200 and 600 nm. The mobile phase A contained 10% acetonitrile and 0.1% trifluoroacetic acid in water (v/v/v), and mobile phase B contained 90% acetonitrile and 0.1% trifluoroacetic acid in water (v/v/v). The separation gradient was set as 10–40% mobile phase B for 30 min at a flow rate of 0.5 ml per min, followed by 40–100% mobile phase B for 5 min. Then the column was rinsed with 100% of mobile phase B for 2 min. Peak identification was confirmed by both retention time against standards and UV absorption spectra. The flavonoid cyanidin 3-arabinoside chloride (CAS: 111613-04-8) was purchased from Miragen.
Scavenging Activity of the Superoxide Anion (O2·y-) Assay
To access the root's scavenging activity to the superoxide anion (O2ÿ-), the guaiacol method was operated as previously described (28). 50 mg of root homogenate was dissolved in 1 ml of 0.05 m phosphate buffer (pH 5.5) for extraction of antioxidant enzymes. After 10 min incubation, the extract was centrifuged at 12,000 × g and 4 °C for 10 min. Then, 0.5 ml supernatant (crude enzyme extract) was added into 2 ml of the reaction buffer, which contained 0.5 ml 0.05 m guaiacol (substrate, overdose), 1 ml 0.05 m phosphate buffer and 0.5 ml 2% hydrogen peroxide (H2O2). The increased OD470 value because of the enzyme-dependent guaiacol oxidation was recorded every 30 s until the reaction time reached 4 min.
Free Radical Scavenging Activity on ABTSÿ+
For analyzing the scavenging activity of soybean roots on cationic radical, the ABTSÿ+ decolorization assay was conducted as previously described (28). Briefly, 10 μl extracts were mixed with 1.0 ml of ABTSÿ+ working solution containing 2.45 mm potassium persulphate and 7 mm ABTS (OD734 = 0.70 ± 0.02). After 30 min incubation at 30 °C, the OD734 of reaction mixture was measured.
Rapid Function Test of GmMYB173S59p in Soybean Hairy Root System
The pDL28-GmMYB173S59A–RFP (OE-S59A), pDL28-GmMYB173S59D-RFP (OE-S59D) and pDL28-GmMYB173-RFP (OE-WT) constructs were transformed into the cotyledonary node of Union85140 via A. rhizogenes strain K599 as previously described (28). The pDL28-RFP empty vector (EV) was used as a negative control. After transgenic roots were verified by fluorescent stereo microscope (Nikon SMZ1500), the composites were transplanted in pearlite and sphagnum peat soil (v/v = 1:3). A week after transplanting, the composites were treated with 200 mm NaCl in 1/4 fold Fahräeus medium every 4 days and deionized water was also irrigated every 2 days after application of abovementioned salt medium. The roots were harvested 4 weeks after transplanting.
RESULTS
Phosphoprotein Profiles of Soybean Root Reveal Prominent Changes of Transcription Factors
Protein phosphorylation in cells is one of the most active posttranslational modifications in response to various stresses; and phosphoproteomics provides the most effective and comprehensive approach to study the phosphorylation status of all the proteins in plants subject to a particular treatment (28). In this research, quantitative phosphoproteomic approaches were used to find the key signaling components in soybean roots under saline treatment. In total, 4697 phosphorylated sites from 2239 phosphoproteins were quantitatively analyzed (supplemental Table S4 and S5). The software MEME Suite and motif-X were used to extract overrepresented motifs from the 656 quantitatively analyzed phosphorylated peptides with significant changes between the treated and control groups (supplemental Table S4). The intensities of phosphopeptides from treatment (IpT) were compared with those from control (IpC) and the ratio (IpT : IpC) with significant differences (p value < 0.05) were divided into two groups. The up-regulated group (Up) includes phosphopeptides with IpT > IpC and down-regulated group (Down) includes phosphopeptides with IpT < IpC. Nine phosphorylation motifs in both Up and Down groups were significantly enriched (Fig. 1A, 2B). Interestingly, we were able to observe conserved phosphopeptides only when Ser was set as the central phosphorylated residue in both groups. In both Up and Down groups, the amino acid closely neighboring the phosphorylated Ser was mainly Pro or Asp or Glu (Fig. 1). There were four phosphorylation motifs ([sP], [sDxD], [sPxR] and [sxD]) enriched from both Up and Down groups. Five motifs ([sDxE], [Ds], [sxs], [sxE] and [sxP]) were only found in the Up group and five motifs ([sDD], [DsxE], [sDx], [ss] and [sE]) were only detected in the Down group. These differentially phosphorylated motifs were then searched against relevant databases to identify kinases which could target them as substrates, and our results indicate that [sP], [sDxE], [sxs], [sxE] are potential substrates of plant ERK1 and ERK2 kinases, [sxD] is recognized by casein kinase-II (CK2), sPxR is a substrate of CDK1/2/4/6 kinase, growth associated histone HI kinase, and Cdc2 kinase (48).
Fig. 1.
Phosphorylation motifs deduced from peptides with different modification levels following salt stress and verification of GmMYB173 as a substrate of GmCK2α. A, Phosphorylation motifs expressed as positional weight matrices extracted from the phosphopeptides in the Up group using motif-X software. B, Phosphorylation motifs extracted from the phosphopeptides in the Down group. C, GmMYB173 could be a substrate of GmCK2α. Phosphorylation assay was carried out as described in experimental procedure section. S59A and WT: recombinant proteins of the N-terminal 167 aa of GmMYB173-S59A and GmMYB173-WT fused to a Trx- and a 6 × His-tags at its N-terminal end, and a 6 × His-tag at its C-terminal end. Data indicated that the Ser59 is the unique site in GmMYB173.
Fig. 2.
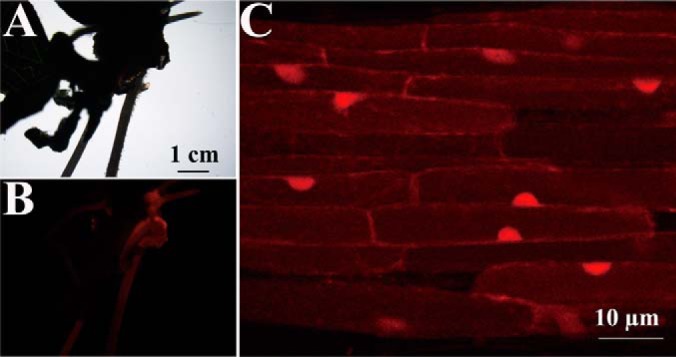
GmMYB173-RFP is localized in the nucleus of soybean root cells. The transgenic roots with red fluorescence were selected by Nikon SMZ1500 fluorescent stereo microscope (A, B), and then used for subcellular localization analysis with Zeiss LSM710 confocal microscope (C).
Several transcription factors, including MYB, bZIP, pH-response transcription factors, calmodulin-binding transcription activators (CAMTA), ethylene-responsive factors (ERF), WRKY, and GTE were found with fluctuating phosphorylation modifications (supplemental Table S4 and S5). Furthermore, multiple transcription factors including MYB, WRKY and bZIP, have been quantitatively analyzed with one or more phosphorylated peptides. Interestingly, the phosphorylation intensities of peptides DAVAAGYASADDAAPQNSGR in GmMYB176 (D8L1Z5), DALAAGYAsADDAAPQNSGR in GmMYB173 (Q0PJH9) and DDAAGYAsADDAAPINSDK in GmMYB183 (Q0PJI2) increased significantly under salt stress (supplemental Table S4 and S5). In addition, the phosphorylated peptide QGsLTLPR in GmbZIPs (I1KM05) increased significantly after salt treatment, whereas another phosphorylated peptide SNNVDNsPISPHYVINR in this protein declined significantly. Moreover, the ADAsPRTDISTDVDTDDKNPR in GmWRKY (A0A0B2RZI4) was observed with significant decline.
The MYB type TFs were reported to play pivotal roles in regulating the expression of enzymes involved in flavonoids syntheses (32). Consistently we observed that all the salt responsive GmCHS, GmCHI and GmCPM contained several MYB-TF binding motifs in their promoters (28). Hence, the phosphorylation of TFs GmMYB173, GmMYB176 and GmMYB183 could be important nodes connecting the upstream stress signals to the transcriptional control of the salt responsive genes. GmMYB176 was published before (49) and GmMYB183 will be covered in a separate study, GmMYB173 will be the focus of the current study. Because the only phosphorylation peptide found in GmMYB173 contains a CK2 recognition motif (sxD), we expressed and purified the GmCK2α and confirmed that the GmMYB173 protein is a substrate of the soybean kinase GmCK2α (Fig. 1C).
Metabolite Profiles of Soybean Root Reveal Prominent Changes in Flavonoids
The metabolites were ionized by negative- or positive-charged mode, and then analyzed using tandem MS. Ten biological replicates of each treatment were used to estimate the fold changes of metabolites in salt treated and control groups (supplemental Table S6 and S7) in the label-free metabolomics study as previously reported (50). In total, we were able to identify 2077 and 4668 metabolites in the negatively- or positively-charged mode respectively; of which 41 and 66 were found to be flavonoids (supplemental Table S6 and S7).
The flavonoids with significant and reproducible differences (p < 0.05) are listed in Table I. Salt treatment was found to stimulate the accumulation of 24 dihydroxy B-ring flavonoids including luteolin 3′-methyl ether 7-glucuronosyl-(1->2)- glucuronide, quercetin 3,7,3′-tri-O-sulfate, cyanidin 3-(6″-succinyl-glucoside) and cyanidin 3-arabinoside chloride (Table I). On the other hand, the concentrations of 40 monohydroxy B-ring flavonoids were found to be significantly decreased following salt treatment; and formononetin, genistein, daidzin, daidzein, naringenin, malonyldaidzin, malonylglycitin, and formononetin 7-O-(6″-acetylglcoside) are some of the examples found in the down regulated flavonoids. These results imply that branching into different classes of flavonoids during their metabolism process could have significant impacts on the performance of soybean plants grown under salt stress. As the precursor of all flavonoids, the accumulation of chalcone is essential for the syntheses of other flavonoids, which are believed to be positively correlated with soybean tolerance to salt stress, such as luteolin 3′-methyl ether 7-glucuronosyl-(1->2)-glucuronide, quercetin 3,7,3′-tri-O-sulfate, cyanidin 3-(6″-succinyl-glucoside) and cyanidin 3-arabinoside chloride. Hence through the regulation of flavonoid syntheses, CHS acts as a hub in the complicated network leading to the establishment of soybean tolerance to salinity. The homologs of CHI and CPM that catalyze the conversion of chalcone to derivatives with different modifications (for example, the positive regulator quercetin or the negative effector formononetin) could play distinct roles in soybean response to salinity.
Table I. Flavonoid contents with significant changes under salt stress.
| Name | Fold change | p value |
|---|---|---|
| Neoraunone | 35.5518 | 0.0000 |
| Robustone | 32.0782 | 0.0000 |
| Repenol | 20.2519 | 0.0000 |
| Kaempferol 3-sulfate-7-alpha-arabinopyranoside | 5.9515 | 0.0000 |
| Rotenonone | 5.9184 | 0.0000 |
| 6-Hydroxykaempferol 3,5,7,4′-tetramethyl ether 6-rhamnoside | 5.7315 | 0.0000 |
| Echinoisosophoranone | 5.2733 | 0.0017 |
| Luteolin 3′-methyl ether 7-glucuronosyl-(1->2)-glucuronide | 4.7629 | 0.0000 |
| Atovaquone | 3.9426 | 0.0000 |
| Corylin | 3.3739 | 0.0971 |
| Isochamaejasmin | 2.8030 | 0.0000 |
| Naringin | 2.7699 | 0.0000 |
| Medicagol | 2.4899 | 0.0000 |
| Dalpanin | 2.4896 | 0.0002 |
| Quercimeritrin | 2.3617 | 0.0000 |
| Quercetin 3,7,3′-tri-O-sulfate | 2.2331 | 0.0000 |
| Calopogoniumisoflavone B | 1.9353 | 0.0016 |
| Cyanidin 3-(6′′-succinyl-glucoside) | 1.7405 | 0.0053 |
| Karanjin | 1.5249 | 0.0000 |
| Glabrone | 1.4590 | 0.0151 |
| Lilaline | 1.4260 | 0.0058 |
| Pongamoside A | 1.4181 | 0.0007 |
| Irisflorentin | 1.3598 | 0.0067 |
| Cyanidin-3-arabinoside | 1.1340 | 0.0141 |
| Flavaprenin 7,4′-diglucoside | 0.9019 | 0.0377 |
| Kaempferol 3-(4′′-(E)-p-coumarylrobinobioside)-7-rhamnoside | 0.8417 | 0.0126 |
| Silybin | 0.8271 | 0.0220 |
| 6-C-Glucopyranosylepicatechin | 0.8199 | 0.0019 |
| Formononetin | 0.8045 | 0.0224 |
| Neodulin | 0.7874 | 0.0439 |
| Syringetin 3-glucuronide | 0.7454 | 0.0001 |
| Kaempferol 3-(2′′-hydroxypropionylglucoside)-4′-glucoside | 0.7187 | 0.0000 |
| Pyranodelphinin B | 0.7136 | 0.0294 |
| Ikarisoside A | 0.6994 | 0.0011 |
| Quercetin-7-(6 -acetylglucoside) | 0.6934 | 0.0011 |
| Genistein | 0.6816 | 0.0001 |
| Malonylgenistin | 0.6774 | 0.0320 |
| Patuletin 3-(6′′-(E)-feruloylglucoside) | 0.6268 | 0.0001 |
| 6-Methoxykaempferol 3-rhamnoside-7-(4′′′-acetylrhamnoside) | 0.6121 | 0.0000 |
| Evodiamine | 0.5602 | 0.0000 |
| Mirificin | 0.5558 | 0.0084 |
| Delphinidin 3-(6′′-malonylglucoside) | 0.5177 | 0.0020 |
| Cyanidin 3-(6′′-dioxalylglucoside) | 0.5110 | 0.0001 |
| Davallioside A | 0.4955 | 0.0000 |
| Frutinone A | 0.4625 | 0.0000 |
| Formononetin 7-O-(6′′-acetylglcoside) | 0.4621 | 0.0000 |
| Daidzin | 0.4509 | 0.0000 |
| Isoliquiritigenin 2′-glucosyl-(1->4)-rhamnoside | 0.4224 | 0.0000 |
| Quercetin 3-(6′′-malonylglucoside)-7-glucoside | 0.4188 | 0.0000 |
| Pelargonidin 3-O-[2-O-(beta-D-xylopyranosyl)-6-O-(methyl-malonyl)-bata-D-galactopyranoside] | 0.4034 | 0.0085 |
| Lupalbigenin | 0.3966 | 0.0000 |
| Glycitin | 0.3597 | 0.0044 |
| Isorhamnetin 3-rhamnoside | 0.3424 | 0.0000 |
| Clitoriacetal | 0.3262 | 0.0000 |
| Cladrin 7-O-glucoside | 0.3014 | 0.0001 |
| Phenindione | 0.2664 | 0.0000 |
| Apigenin 7-(2′′-acetyl-6′′-methylglucuronide) | 0.2375 | 0.0000 |
| RHOIFOLIN | 0.2308 | 0.0000 |
| Daidzein | 0.2203 | 0.0000 |
| Malonyldaidzin | 0.1301 | 0.0001 |
| Macrocarposide | 0.1231 | 0.0000 |
| Malonylglycitin | 0.0992 | 0.0000 |
| Malvidin 3-glucoside-pyruvate | 0.0925 | 0.0000 |
| Naringenin | 0.0153 | 0.0000 |
GmMYB173 Is a Direct Regulator of GmCHS5 in Response to Salinity
Although the protein coded by Glyma17G094400 (GmMYB173) was categorized into the MYB type transcription factor family (51), empirical data about the putative role of GmMYB173 as a transcription factor, as well as its physiological function, are rarely reported. In this study, GmMYB173 was tagged with RFP at its C-terminal end and expressed in roots of soybean via A. rhizogenes-mediated transformation (Fig. 2A, 2B). Consistent with its role as a transcription factor, our results indicated that the RFP-tagged GmMYB173 is localized to the nucleus in the root cells of soybean (Fig. 2C).
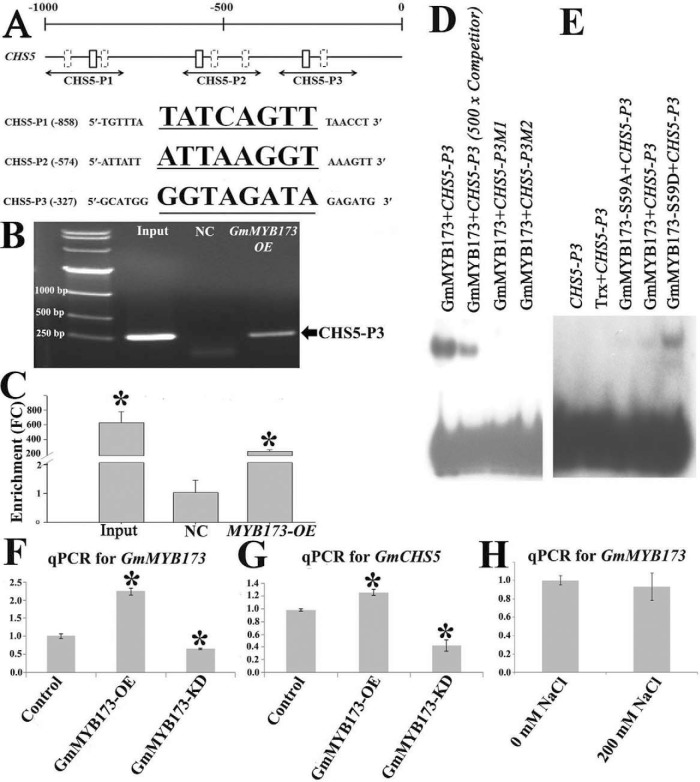
We further searched the MYB TF binding motif (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A) (48) in the −1 kb promoter regions of GmCHS5 (Glyma01g43880.1), a salt-responsive gene we recently identified (28), and eight copies of the MYB-binding cis-element were found in its promoter.
To test whether GmMYB173 interact with the promoter of GmCHS5 in soybean cells, we carried out chromatin immunoprecipitation assays coupled to quantitative PCR (ChIP-qPCR). The sheared chromatin prepared from the transgenic soybean roots was precipitated using a rabbit polyclonal antibody against HA-tag. A 234-fold enrichment was observed in qRT-PCR amplification of ChIPed DNA from soybean roots expressing 35S::GmMYB173–2xHA/Ubq10::RFP using primer pairs spanning the MYB-binding site CHS5-P3 in the GmCHS5 promoter as compared with ChIPed DNA from soybean roots carrying the empty vector (Fig. 3B, 3C). No enrichment was observed in qRT-PCR amplification using primer pairs spanning CHS5-P1 and P2 sites (Fig. 3A).
Fig. 3.
GmMYB173 binds to the promoter of GmCHS5 via an MYB-binding element and regulates its expression. A, Position of MYB TF binding motifs, (G/A/T)(G/A/T)T(C/A)(A/G)(A/G)(G/T)(T/A), boxes in the promoter of GmCHS5. Fragments selected for amplification in ChIP-qPCR assay are presented proportionally underneath the promoter; sequences for probes in EMSA (solid boxes) are listed. B, C, Chromatin immuno-precipitation assay verified the GmMYB173 binding to promoter of GmCHS5 in vivo. ChIP assay reveals that GmMYB173 binds the GmCHS5 promoter (B); ChIP-qPCR-based in vivo binding assay with promoter fragments spanning the GmCHS5-P3 fragment (C, NC: Negative control, root sample transformed with the empty vector, FC: fold change). EMSA assay showed that GmMYB173 specially binds to the GmCHS5-P3 fragment from the GmCHS5 promoter (D), and Phosphorylation at S59 of GmMYB173 enhances this interaction (E). (GmCHS5-P3, ttttGTTGATGCATGGGGTAGATAGAGATGCATA) or two mutated version with its GGTAGATA core sequence changed to GCCAGATA (M1) or GGCCGATA (M2) was labeled as probe. EMSAs were carried as described in the section of Experimental Procedures. pET32a-Trx tag: the 184 aa Trx protein with two his-tags at its C terminus expressed from the empty pET32b vector. Recombinant proteins of the N-terminal 167 aa of GmMYB173, GmMYB173-S59A, and GmMYB173-S59D are fused to a Trx- and a 6 × His-tags at its N-terminal end, and a 6 × His-tag at its C-terminal end. Competitor is 5-hundred folds of unlabeled double strand GmCHS5-P3 fragment. F, G, Transcription of GmMYB173 (F) and GmCHS5 (G) in untransformed control, GmMYB173-OE (overexpression) and GmMYB173-KD (knock-down) roots; expression of GmCHS5 was found to be correlated with expression of GmMYB173. H, Transcription of GmMYB173 in soybean roots treated with 200 mm NaCl. Data represent mean values ± S.E., each sample was analyzed with three biological replicates. An asterisk indicates a significant difference based on P (≤ 0.05) of the Student's t test.
To ascertain the interaction between GmMYB173 and the promoter of GmCHS5, we performed EMSAs using double-stranded GmCHS5 probes containing a MYB motif (GmCHS5-P3: GGTAGATA) and two mutants (GmCHS5-P3M1: GCCAGATA; GmCHS5-P3M2: GGCGGATA) as described in the experimental procedure. The result showed that GmMYB173 binds to the CHS5-P3 probe in a sequence dependent manner (Fig. 3D). We also tested GmCHS5-P1 and GmCHS5-P2 probes with EMSA, consistently not interactions were detected between them and the DNA binding domain of GmMYB173. Because the S59 phosphorylation site is close to the DNA-binding domain (aa 74–130), we compared interaction between GmCHS5-P3 probe and the WT, phospho-ablative (S59A) mutant of GmMYB173, our results supported that phosphorylation at S59 significantly facilitate the binding of GmMYB173 to the promoter of GmCHS5 via the GmCHS5-P3 MYB-binding element (Fig. 3E).
To study whether endogenous level of GmMYB173 affects the transcription of GmCHS5, we generated the transgenic roots of soybean where the expression of GmMYB173 was elevated by overexpression (GmMYB173-OE) or knocked down through RNAi (GmMYB173-KD). We analyzed the transcriptional levels of GmCHS5 in GmMYB173-OE and GmMYB173-KD roots (Fig. 3F). Roots that carry the empty vector (pDL28-HA) were used as a control. Over expression of GmMYB173 resulted in the elevated expression of GmCHS5 and knockdown of GmMYB173 resulted in the decreased expression of GmCHS5. The differences in both cases are statistically significant, and moreover an approximate 2-fold increase in the transcription of GmCHS5 was observed in GmMYB173-OE roots when compared with that in GmMYB173-KD roots (Fig. 3G). These results suggested that GmMYB173 positively correlates with the transcription of GmCHS5, supporting a regulatory role for GmMYB173. We then compared the transcription of GmMYB173 before and after salt treatment, no significant difference was detected (Fig. 3H). Mass spec data generated in the current study revealed a significantly increase in phosphorylation of GmMYB173 at S59 following salt treatment (supplemental Table S4). Together our data support that the salt triggered increase in GmCHS5 expression is very likely realized though the phosphorylation facilitated binding of GmMYB173 to the promoter of GmCHS5.
Endogenous GmMYB173 Level and Phosphorylation Status Affect Flavonoid Metabolism
Because GmCHS5 encodes a chalcone synthase that plays a critical role in the accumulation of chalcone and downstream flavonoids and the establishment of soybean tolerance to salt stress (13). To further validate that both GmMYB173 and GmCHS5 enhance soybean salt tolerance through the metabolism of flavonoids, we analyzed the endogenous flavonoids in composite seedlings (Fig. 4, Table II, and supplemental Table S7) whose roots were transformed with constructs aimed at overexpressing or knocking down of GmMYB173 and GmCHS5.
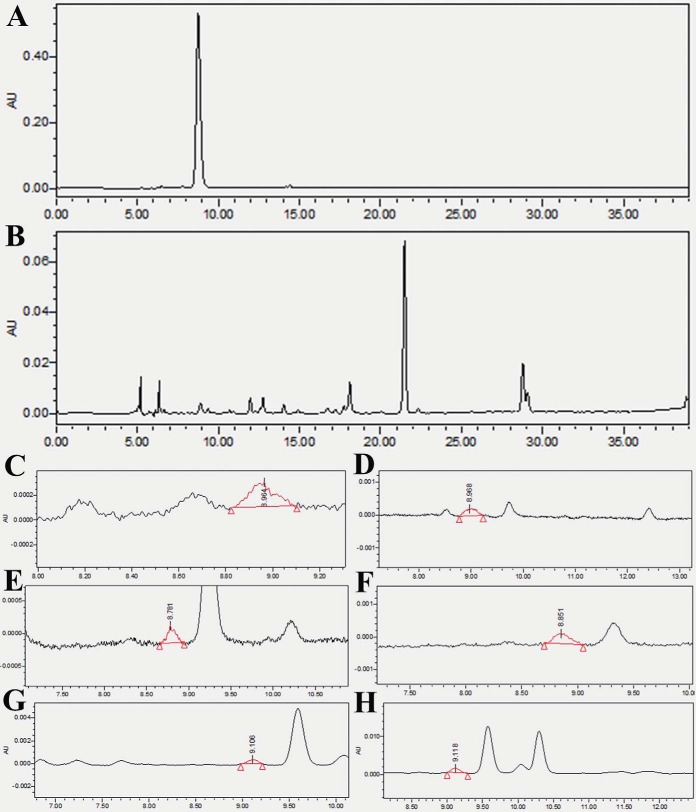
Fig. 4.
HPLC-based analysis of cyanidin 3-arabinoside chloride (C3A) contents in transgenic roots of soybean. A, The chromatographic spectrum observed from C3A standard separation. B, The chromatographic spectrum observed from HPLC separation of transgenic GmMYB173-OE roots without salt treatment. C–H The chromatographic spectrum observed from HPLC separation of transgenic GmCHS5-OE control (C), GmCHS5-OE salt stress (D), GmMYB173-OE control (E), GmMYB173-OE salt stress (F), GmMYB173S59D control (G), GmMYB173S59D salt stress (H) roots.
Table II. Flavonoid contents in OE or KD roots in response to salt stress-salinity-2.
| Control | NaCl | |
|---|---|---|
| Cyanidin-3-arabinoside (μg/g fresh weight) | ||
| Empty vector | ND | ND |
| GmPCHS5-OE | 0.0266 ± 0.0007 | 0.0654 ± 0.0006 |
| GmPCHS5-KD | ND | ND |
| GmMYB173-OE | 0.0346 ± 0.0045 | 0.0668 ± 0.0073 |
| GmMYB173S59D | 0.0533 ± 0.0024 | 0.1760 ± 0.0047 |
| GmMYB173S59A | ND | ND |
| GmMYB173-KD | ND | ND |
Cyanidin 3-arabinoside chloride (C3A) was proposed to be a positive effector to soybean tolerance and is also one of the most easily detected flavonoids (supplemental Table S6 and S7). Hence, C3A in the composite soybean roots is selected for analysis. Introduction of GmCHS5-KD construct into the roots of the composite soybean plants rendered the C3A undetectable in both salt treatment and untreated samples. In contrast, the content of C3A in soybean roots transformed with GmCHS5-OE construct was measured to be 0.0266 ± 0.0007 without treatment (control) and 0.0654 ± 0.0006 μg/g of fresh roots when treated with salt (Fig. 4, Table II).
It is well-known that the function of transcription factors is usually modulated through phosphorylation (52–56). To test the relevance of phosphorylation at S59 of GmMYB173 on its function and subsequent flavonoid metabolism, we generated phospho-mimic (GmMYB173S59D) and phospho-ablative (GmMYB173S59A) mutants, and the 35S promoter-driven mutants were expressed in the roots of composite soybean plants as performed with the wild-type GmMYB173. While monitoring the endogenous C3A level, we found that the accumulation of C3A in roots expressing phospho-mimic mutant GmMYB173S59D reached the highest levels and was almost undetectable in the roots expressing phospho-ablative mutant GmMYB173S59A (Fig. 4, Table II). These data suggest that phosphorylation at S59 is required for the activation of GmMYB173 and the metabolism of flavonoids.
GmCHS5 and GmMYB173 Pathway Mediate ROS Elimination in Soybean Roots under Salt Stress
The antioxidant capacity in plant tissue is generally accepted to be positively correlated with plant tolerance to salinity. The antioxidant properties in soybean root were analyzed using H2O2 guaiacol and ABTS·+ (2, 2′-azinobis (3-ethylbenzothiazoline 6-sulfonate)) radical scavenging capacity assays as previously described (28). To test the relevance of ROS elimination capacity with flavonoid metabolism, all the above mentioned transgenic roots (for flavonoid content evaluation) were also analyzed for their ROS elimination capacity.
Consistent with cyanidin 3-arabinoside chloride contents, the ROS elimination capacities showed similar trends in the tested transgenic roots (Table III). Generally, the scavenging capacities of both the H2O2 and ABTS·+ in all transgenic roots were found to be stimulated by salt stress. In addition, both GmMYB173 and GmCHS5 were shown to act as positive regulators for boosting the ROS scavenging capacities in soybean roots. For example, under salt stress the H2O2elimination capacities were measured as 1.6467 ± 0.0094 U × g−1 × min−1 in transgenic roots expressing GmCHS-KD and 2.6500 ± 0.0424 U × g−1 × min−1 in soybean roots expressing GmCHS-OE (Table III). In addition, expressing the GmMYB173S59D construct in soybean roots resulted in the highest capacities for eliminating both H2O2 and ABTS·+; whereas the expressed GmMYB173-KD construct coincided with the lowest ROS scavenging capacities (Table III). To summarize, both the overexpression of GmMYB173 and GmCHS5 enhanced the ROS elimination ability of soybean roots. Moreover, the phosphorylation of GmMYB173 could play a positive role in enhancing soybean's ability in ROS scavenging.
Table III. ROS elimination capacity of OE or KD roots.
| R(−)Na(−) | R(−)Na(+) | ||
|---|---|---|---|
| H2O2 (U × g−1 × min−1) | |||
| GmCHS5 | OE | 1.7800 ± 0.0990 | 2.6500 ± 0.0424 |
| KD | 1.1913 ± 0.0265 | 1.6467 ± 0.0094 | |
| GmMYB173 | OE-WT | 1.7925 ± 0.0672 | 2.0100 ± 0.0085 |
| OE-S59D | 1.9625 ± 0.0318 | 2.3100 ± 0.0707 | |
| OE-S59A | 1.1025 ± 0.0318 | 1.2967 ± 0.0613 | |
| KD | 0.5800 ± 0.0177 | 1.0488 ± 0.0018 | |
| ABTS·+ (10 μmol × g−1) | |||
| GmCHS5 | OE | 0.5091 ± 0.0155 | 0.5651 ± 0.0150 |
| KD | 0.3336 ± 0.0191 | 0.3699 ± 0.0082 | |
| GmMYB173 | OE-WT | 0.3237 ± 0.0076 | 0.4225 ± 0.0041 |
| OE-S59D | 0.4038 ± 0.0037 | 0.5084 ± 0.0383 | |
| OE-S59A | 0.2772 ± 0.0034 | 0.3157 ± 0.0012 | |
| KD | 0.1913 ± 0.0060 | 0.2518 ± 0.0367 |
Phosphorylation of GmMYB173S59 Improve Salt Tolerance of Transgenic Soybean Roots
We compared the salt tolerance of transgenic roots, our results showed that roots expressing GmMYB173S59D (OE-S59D) showed higher tolerance than those transformed with GmMYB173S59A (OE-S59A) and GmMYB173 (OE-WT) constructs (Fig. 5). Furthermore, the transgenic roots whose GmMYB173 expression is knocked-down (KD) displayed the lowest tolerance to salt stress and even turned brown at the end of salt treatment. These results indicate that GmMYB173 is a positive regulatory factor in soybean salt tolerance and the phosphorylation at S59 plays a pivotal role in activating GmMYB173 function in vivo.
Fig. 5.
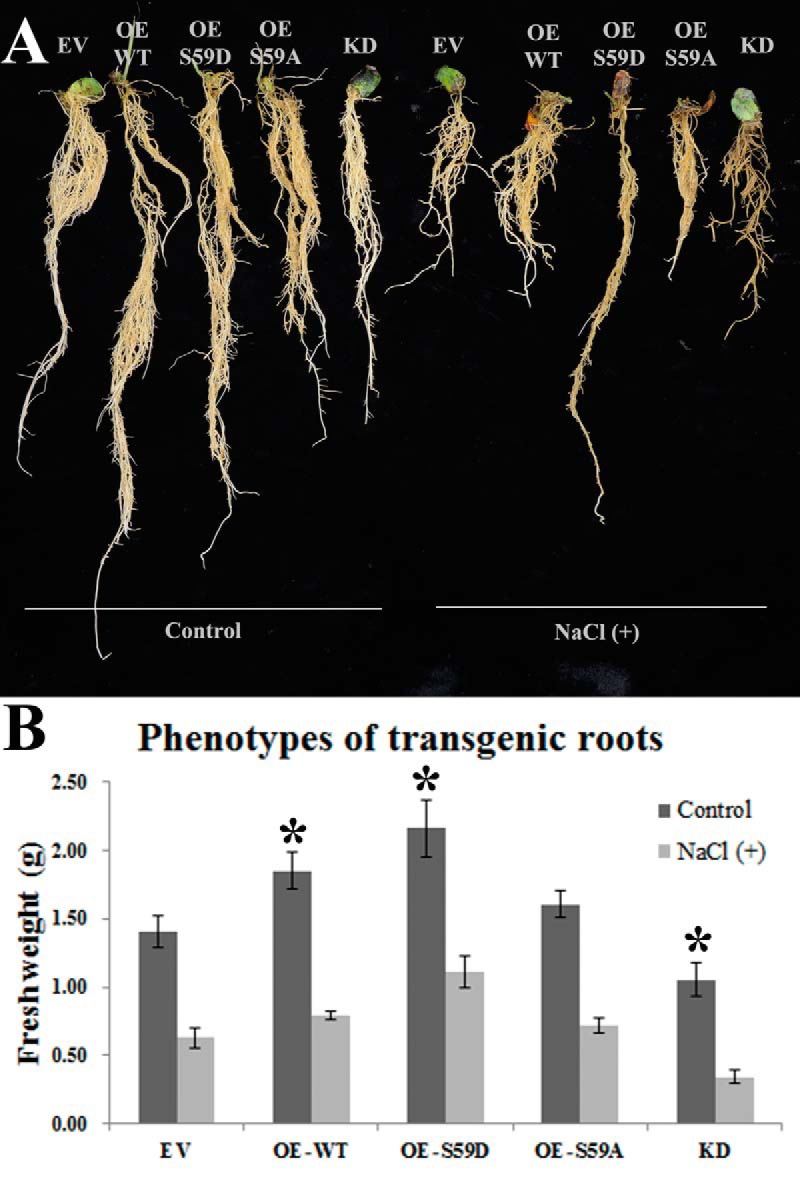
Effects of salt treatment on transgenic roots of composite soybean plants. Composite soybean plants were generated by infection with A. rhizogenes strain K599 carrying empty vector (EV), overexpression constructs of GmMYB173 (OE-WT), GmMYB173S59D (OE-S59D), GmMYB173S59A (OE-S59A) and RNAi-mediated knock down construct of GmMYB173 (KD). Transgenic roots were confirmed by DsRed fluorescence as described in materials and methods section. One-week old composite plants were treated in 1/4 fold Fahräeus medium without (control) or with 200 mm NaCl [NaCl (+)] for 4 weeks. A, Plants are typical representatives of three repeats of salt treatments. B, Fresh weights of transgenic roots: data expressed are means ± s.d. of three repeats; an asterisk indicates a significant difference from the control (EV) based on p ≤ 0.05 of student's t test.
DISCUSSION
Salt stress usually enhances the production of reactive oxygen species in the stressed plants and results in oxidative stress (19, 57). The excess ROS initiates MAPK cascades which modify several types of transcription factors, such as MYB (58), ERF (ethylene-responsive) and WRKY via phosphorylation. The genes encoding for superoxide dismutase and ascorbate peroxidase are among those transcriptionally activated to eliminate the excessive ROS (59–61). It is known that severe salt stress could inactivate these antioxidant enzymes while it up-regulates the biosynthesis of flavonoids (62).
The anti-oxidative function of flavonoids is not new; and the chemical basis of the anti-oxidative potential of flavonoids has been ascribed to the hydroxy groups present in their structures (10). Brown et al. demonstrated that the presence of ortho 3′, 4′-dihydroxy in the B ring was crucial for Cu2+-quercetin chelate formation by enhancing its antioxidant activity; and quercetin is oxidized by H2O2 much faster than kaempferol (10, 63). The flavonoid derivatives, primarily produced through ortho-dihydroxy B-ring glycosylation, have also been proven to donate electrons or hydrogen atoms in the anti-oxidative reactions (64); and accumulation of quercetin 3-O-glycosides in Ligustrum vulgare root was reported to be significantly increased on salt stress (65). Quercetin derivatives were also documented to play significant roles in reducing singlet oxygen (1O2) in Phillyrea latifolia leaves (66).
It is now generally accepted that the process leading to the production of dihydroxy B-ring-substituted flavonoids and their derivatives plays positive roles in soybean salt tolerance, whereas the formation of monohydroxy B-ring/-substituted flavonoids and their derivatives have negative contributions to soybean resistance to oxidative stresses (67–69). Consistently we found at the metabolome level, the contents of endogenous dihydroxy B-ring-substituted flavonoids, such as cyanidin 3-arabinoside chloride, cyanidin 3-(6″-succinyl-glucoside), quercetin 3-(6″-methylglucuronide), luteolin 3′-methyl ether 7-glucuronosyl-(1->2)-glucuronide and quercetin 3, 7, 3′-tri-O-sulfate, were found to be significantly up-regulated; the monohydroxy B-ring/-substituted flavonoids, such as daidzin, genistein and formononetin, were found to be down-regulated in soybean roots in response to salt stress (Table I). Apparently, when stressed by salinity, excess ROS will be induced, and the soybean root needs the flavonoids that have higher radical-scavenging capacity to get better protection.
Flavonoids are synthesized via the shikimate-phenylpropanoid pathway. In Arabidopsis thaliana, over 30 enzymes, such as chalcone synthase (CHS), chalcone flavonone isomerase (CHI) and cytochrome P450 monooxygenase (CPM), have been characterized for their roles involved in flavonoid synthesis (69). The physiological functions of CHS, CHI and CPM attracted intensive attention of scientists interested in understanding the legume's response to stresses, but the regulation of flavonoid biosynthesis in response to environmental cues remained largely elusive until recently.
CHSs form a multiple gene family in legumes in sharp contrast to Arabidopsis which contains only one CHS gene in its genome (29). Differential expression of the CHS family members in soybean in different developmental stages, among various organs, and in response to multiple biological and environmental stimuli has been documented (30), indicating that synthesis of flavonoids is affected in response to developmental and environmental cues.
The transcriptional controls of flavonoid synthesis genes are almost dictated by MYB-type TFs (34, 58, 70). There are 156 MYB members in soybean, and expression of about one third of them was affected by salt or drought treatment (32). Among these stress-responding MYBs, GmMYB76, GmMYB92, and GmMYB177 could function as negative regulators of ABA signaling (32). Previous studies have also shown that overexpression of some MYB transcription factor genes improves stress tolerance. Overexpression of Craterostigma plantagineum CpMYB10 in Arabidopsis can enhance osmotic stress tolerance of the transgenic seedlings germinated in Petri dishes and improve tolerance to desiccation and salt in adult plants grown in soil (71). Overexpression of rice OsMYB4 in Arabidopsis increases plant tolerance to chilling and freezing (72).
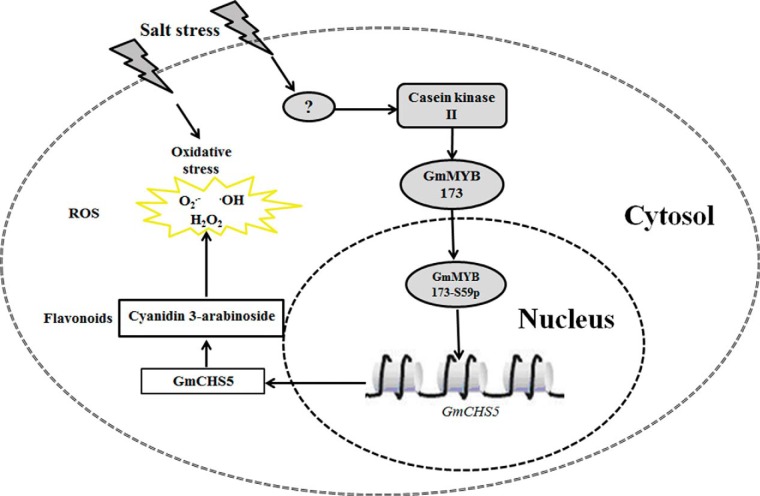
In the present study, the phosphorylation of GmMYB173 at Ser59, likely catalyzed by the soybean kinase GmCK2α, was shown to be induced by salt stress (Fig. 1C), and positively correlated with soybean tolerance to salt stress. GmMYB173 was confirmed to target the promoter of GmCHS5 encoding for a chalcone synthase in vivo; and the expression of GmCHS5 was shown to be up-regulated in GmMYB173-OE roots especially when it is phospho-mimically mutated, leading to enhanced accumulation of dihydroxy B-ring flavonoids, such as quercetin derivatives. Hence, GmMYB173 may contribute to salt tolerance at least via activation of GmCHS5 and preferred accumulation of dihydroxy B-ring flavonoids, but how this preference over monoxy B-ring flavonoids is realized remains to be addressed.
It is noteworthy to mention that the unique phosphorylation site (Ser-59) in GmMYB173 is conserved among orthologs in different plants, including Glycine max, Phaseolus vulgaris, Cajanus cajan, Vigna angularis, Arachis hypogaes, Medicago truncatula, and Citrus arietinum (Fig. 6), indicating the phosphorylation is a conserved regulatory even, which were shown to enhance the affinity of these MYB factors to their target CHS genes. In the currently study, we found the transcription of GmCHS5 is positively correlated with the endogenous level of GmMYB173 and its transcription activating capability which is positively regulated via phosphorylation at Ser59. In addition, we found that the expression of GmMYB173 is not induced by salt treatment. Hence the stress signals must be transduced into the expression of GmCHS5 and the subsequently preferred accumulation of dihydroxy B-ring flavonoids via the phosphorylation of GmMYB173, a post-translational regulation believed to be more expeditious than a regulation through a transcriptional process (Fig. 7).
Fig. 6.

The phosphorylated peptide GYAsADDA in GmMYB173 protein is highly conserved in several plants. The peptide GYAsADDA was found in homologs of GmMYB173 in several plants, including Glycine max, Phaseolus vulgaris, Cajanus cajan, Vigna angularis, Arachis hypogaes, Medicago truncatula and Citrus arietinum.
Fig. 7.
A hypothetical model illustrating how the transcription factor GmMYB173 regulates the expression of target gene GmCHS5 during soybean response to salinity. After the perception of salinity signal, the GmMYB173 is phosphorylated by casein kinase II and then activates the transcription of downstream target genes including GmCHS5. These enzymes catalyze the production of dihydroxy B-ring flavonoids (such as cyanidin 3-arabinoside chloride), and the accumulation of these flavonoids help reduce the ROS and the establishment of soybean tolerance to salinity.
DATA AVAILABILITY
The mass spectrometry raw data are available in the PRIDE Archive (https://www.ebi.ac.uk/pride/archive/) via identifier PXD006599. GenBank accession numbers for the proteins mentioned in this article are: GmMYB173, Glyma17G094400; GmCHS5, Glyma01g43880.1.
Supplementary Material
Acknowledgments
We thank Miss Zhifang Jiang (from Hangzhou Normal University) for her kind assistance during the HPLC analysis. We are grateful to Dr. Rui Wang and Dr. Jing Yu from Shanghai Applied Protein Technology Co. Ltd. for their kind help during bioinformatics analysis.
Footnotes
* This work was supported by the grants 31301053 and U1130304 from the National Science Foundation of China, LY17C020004 from the Natural Science Foundation of Zhejiang Province, 20170432B01 from the Agricultural Self Declaration Project of Hangzhou Science and Technology Bureau, PF14002004014, PD11002002018001, 2016XJSGWXM27 and 2016XJSGWXM32 from Hangzhou Normal University.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CHS
- chalcone synthase
- CHI
- chalcone isomerase
- CPM
- cytochrome P450 monooxygenase
- TF
- transcription factor.
REFERENCES
- 1. Sobhanian H., Aghaei K., and Komatsu S. (2011) Changes in the plant proteome resulting from salt stress: Toward the creation of salt-tolerant crops? J. Proteomics 74, 1323–1337 [DOI] [PubMed] [Google Scholar]
- 2. Zhu J. K. (2011) Plant salt tolerance. Trends Plant Sci. 6, 66–71 [DOI] [PubMed] [Google Scholar]
- 3. Katiyar-Agarwal S. Zhu J, Kim K., Agarwal M., Fu X., Huang A., and Zhu J. K. (2006) The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qu L. Q., Huang Y. Y., Zhu C. M., Zeng H. Q., Shen C. J., Liu C., Zhao Y., and Pi E. X. (2016) Rhizobia-inoculation enhances the soybean's tolerance to salt stress. Plant Soil 400, 209–222 [Google Scholar]
- 5. Fahnenstich H., Scarpeci T. E., Valle E. M., Flugge U. I., and Maurino V. G. (2008) Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 148, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Affenzeller M. J., Darehshouri A., Andosch A., Lutz C., and Lutz-Meindl U. (2009) Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 60, 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu T., Pi E. X., Tsai S. N., Lam H. M., Sun S. M., Kwan Y. W., and Ngai S. M. (2011) GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biol. 11, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Y. H., Lam H. M., Pi E. X., Zhan Q. L., Tsai S. N., Wang C. M., Kwan Y. W., and Ngai S. M. (2013) Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J. Agr. Food Chem. 61, 8711–8721 [DOI] [PubMed] [Google Scholar]
- 9. MartensYan J. H., Wang B. A., Jiang Y. N., Cheng L. J., and Wu T. L. (2014) GmFNSII-controlled soybean flavone metabolism responds to abiotic stresses and regulates plant salt tolerance. Plant Cell Physiol. 55, 74–86 [DOI] [PubMed] [Google Scholar]
- 10. Yamasaki H., Sakihama K., and Ikehara N. (1997) Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. Bioch. 115, 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y., Lin F. Z., Yang H., Yue L., Hu F., Wang J. L., Luo Y. Y., and Cao F. L. (2014) Effect of varying NaCl doses on flavonoid production in suspension cells of Ginkgo biloba: relationship to chlorophyll fluorescence, ion homeostasis, antioxidant system and ultrastructure. Acta Physiol. Plant 36, 3173–3187 [Google Scholar]
- 12. Aoki T., Akashi T., and Ayabe S-i. (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J. Plant Res. 113, 475–488 [Google Scholar]
- 13. Jiang Y. N., Wang B. A., Li H., Yao L. M., and Wu T. L. (2010) Flavonoid production is effectively regulated by RNA1 interference of two flavone synthase genes from Glycine max. J. Plant Biol. 53, 425–432 [Google Scholar]
- 14. Shen X., Zhou Y., Duan L., Li Z., Eneji A. E., and Li J. (2010) Silicon effects on photosynthesis and antioxidant parameters of soybean seedlings under drought and ultraviolet-B radiation. J. Plant Physiol. 167, 1248–1252 [DOI] [PubMed] [Google Scholar]
- 15. Bandurska H., Niedziela J., and Chadzinikolau T. (2013) Separate and combined responses to water deficit and UV-B radiation. Plant Sci. 213, 98–105 [DOI] [PubMed] [Google Scholar]
- 16. Soledad O. N., Florencia M. M., Laura F. M., Raul D. G., Balbina A. A., and Pia O. F. (2015) Potassium phosphite increases tolerance to UV-B in potato. Plant Physiol. Bioch. 88, 1–8 [DOI] [PubMed] [Google Scholar]
- 17. Graham T. L., Graham M. Y., Subramanian S., and Yu O. (2007) RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144, 728–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia L. G., Sheng Z. W., Xu W. F., Li Y. X., Liu Y. G., Xia Y. J., and Zhang J. H. (2012) Modulation of anti-oxidation ability by proanthocyanidins during germination of Arabidopsis thaliana seeds. Mol. Plant 5, 472–481 [DOI] [PubMed] [Google Scholar]
- 19. Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., Michael A. J., Tohge T., Yamazaki M., and Saito K. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chobot V., Huber C., Trettenhahn G., and Hadacek F. (2009) (±)-catechin: chemical weapon, antioxidant, or stress regulator? J. Chem. Ecol. 35, 980–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakao K., Fujii M., and Hou D. X. (2009) Acetyl derivate of quercetin increases the sensitivity of human leukemia cells toward apoptosis. BioFactors 35, 399–405 [DOI] [PubMed] [Google Scholar]
- 22. Kim H. J., Suh H. J., Kim J. H., Park S., Joo Y. C., and Kim J. S. (2010) Antioxidant activity of glyceollins derived from soybean elicited with Aspergillus sojae. J. Agric. Food Chem. 58, 11633–11638 [DOI] [PubMed] [Google Scholar]
- 23. Yang X., Kang S. M., Jeon B. T., Kim Y. D., Ha J. H., Kim Y. T., and Jeon Y. J. (2011) Isolation and identification of an antioxidant flavonoid compound from citrus-processing by-product. J. Sci. Food Agric. 91, 1925–1927 [DOI] [PubMed] [Google Scholar]
- 24. Koes R., Verweij W., and Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242 [DOI] [PubMed] [Google Scholar]
- 25. Ferreyra M. L. F., Rius S. P., and Casati P. (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishida J. K., Wakatake T., Yoshida S., Takebayashi Y., Kasahara H., Wafula E., dePamphilis C. W., Namba S., and Shirasu K. (2016) Local auxin biosynthesis mediated by a YUCCA flavin monooxygenase regulates Haustorium development in the Parasitic plant Phtheirospermum japonicum. Plant Cell 28, 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piao H. L., Lim J. H., Kim S. J., Cheong G. W., and Hwang I. (2001) Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 27, 305–314 [DOI] [PubMed] [Google Scholar]
- 28. Pi E. X., Qu L. Q., Hu J. W., Huang Y. Y., Qiu L. J., Lu H., Jiang B., Liu C., Peng T. T., Zhao Y., Wang H. Z., Tsai S. N., Ngai S. M., and Du L. Q. (2016) Mechanisms of soybean roots' tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteomics 15, 266–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmutz J., Cannon S. B., Schlueter J., Ma J. X., Mitros T., Nelson W., Hyten D. L., Song Q. J., Thelen J. J., Cheng J. L., Xu D., Hellsten U., May G. D., Yu Y., Sakurai T., Umezawa T., Bhattacharyya M. K., Sandhu D., Valliyodan B., Lindquist E., Peto M., Grant D., Shu S. Q., Goodstein D., Barry K., Futrell-Griggs M., Abernathy B., Du J. C., Tian Z. X., Zhu L. C., Gill N., Joshi T., Libault M., Sethuraman A., Zhang X. C., Shinozaki K., Nguyen H. T., Wing R. A., Cregan P., Specht J., Grimwood J., Rokhsar D., Stacey G., Shoemaker R. C., and Jackson S. A. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 [DOI] [PubMed] [Google Scholar]
- 30. Wingender R., Rohrig H., Horicke C., Wing D., and Schell J. (1989) Differential regulation of soybean chalcone synthase genes in plant defense, symbiosis and upon environmental stimuli. Mol. Gen. Genet. 218, 315–322 [DOI] [PubMed] [Google Scholar]
- 31. Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8, 272–279 [DOI] [PubMed] [Google Scholar]
- 32. Liao Y., Zou H. F., Wang H. W., Zhang W. K., Ma B., Zhang J. S., and Chen S. Y. (2008) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047–1060 [DOI] [PubMed] [Google Scholar]
- 33. Schwinn K. E., Boase M. R., Bradley J. M., Lewis D. H., Deroles S. C., Martin C. R., and Davies K. M. (2014) MYB and bHLH transcription factor transgenes increase anthocyanin pigmentation in petunia and lisianthus plants, and the petunia phenotypes are strongly enhanced under field conditions. Front. Plant Sci. 5, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lotkowska M. E., Tohge T., Fernie A. R., Xue G. P., Balazadeh S., and Mueller-Roeber B. (2015) The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yuan Y., Qi L. J., Yang J., Wu C., Liu Y. J., and Huang L. Q. (2015) A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tiss. Org. 120, 961–972 [Google Scholar]
- 36. Borevitz J. O., Xia Y., Blount J., Dixon R. A., and Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gonzalez A., Zhao M., Leavitt J. M., and Lloyd A. M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 [DOI] [PubMed] [Google Scholar]
- 38. Su L. T., Wang Y., Liu D. Q., Li X. W., Zhai Y., Sun X., Li X. Y., Liu Y. J., Li J. W., and Wang Q. Y. (2015) The soybean gene, GmMYBJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana. Acta Physiol. Plant 37, 138 [Google Scholar]
- 39. Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. M., Srinivasan N., Blundell T. L., Esch J. J., Marks M. D., and Gray J. C. (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Z. Y., Liu X., Wang X. D., Zhou M. P., Zhou X. Y., Ye X. G., and Wei X. N. (2012) An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New Phytol. 196, 1155–1170 [DOI] [PubMed] [Google Scholar]
- 41. Bassel G. W., Gaudinier A., Brady S. M., Hennig L., Rhee S. Y., and De Smet I. (2012) Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24, 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Albert R. (2007) Network inference, analysis, and modeling in systems biology. Plant Cell 19, 3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 44. Niehaus T. D., Nguyen T. N., Gidda S. K., ElBadawi-Sidhu M., Lambrecht J. A., McCarty D. R., Downs D. M., Cooper A. J., Fiehn O., Mullen R. T., and Hanson A. D. (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26, 3010–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 46. Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., and Poovaiah B. W. (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158 [DOI] [PubMed] [Google Scholar]
- 47. Chen F., Dong G., Wu L., Wang F., Yang X., Ma X., Wang H., Wu J., Zhang Y., Wang H., Qian Q., and Yu Y. (2016) A nucleus-encoded chloroplast protein YL1 is involved in chloroplast development and efficient biogenesis of chloroplast ATP synthase in rice. Sci. Rep. 6, 32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prasad T. S. K., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., Balakrishnan L., Marimuthu A., Banerjee S., Somanathan D. S., Sebastian A., Rani S., Ray S., Kishore C. J. H., Kanth S., Ahmed M., Kashyap M. K., Mohmood R., Ramachandra Y. L., Krishna V., Rahiman B. A., Mohan S., Ranganathan P., Ramabadran S., Chaerkady R., and Pandey A. (2009) Human protein reference database-2009 update. Nucleic Acids Res. 37, D767–D772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yi J. X., Derynck M. R., Li X. Y., Telmer P., Marsolais F., and Dhaubhadel S. (2010) A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62, 1019–1034 [DOI] [PubMed] [Google Scholar]
- 50. Masclaux-Daubresse C., Clement G., Anne P., Routaboul J. M., Guiboileau A., Soulay F., Shirasu K., and Yoshimoto K. (2014) Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26, 1857–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Du H., Yang S. S., Liang Z., Feng B. R., Liu L., Huang Y. B., and Tang Y. X. (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mao G. H., Meng X. Z., Liu Y. D., Zheng Z. Y., Chen Z. X., and Zhang S. Q. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meng X. Z., Xu J., He Y. X., Yang K. Y., Mordorski B., Liu Y. D., and Zhang S. Q. (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ni W. M., Xu S. L., Chalkley R. J., Pham T. N. D., Guan S. H., Maltby D. A., Burlingame A. L., Wang Z. Y., and Quail P. H. (2013) Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25, 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhai Q. Z., Yan L. H., Tan D., Chen R., Sun J. Q., Gao L. Y., Dong M. Q., Wang Y. C., and Li C. Y. (2013) Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. Plos Genet. 9, e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou X. M., Wang H., Burg M. B., and Ferraris J. D. (2013) Inhibitory phosphorylation of GSK-3 beta by AKT, PKA, and PI3K contributes to high NaCl-induced activation of the transcription factor NFAT5 (TonEBP/OREBP). Am. J. Physiol. Renal. 304, F908–F917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schmidt R., Mieulet D., Hubberten H. M., Obata T., Hoefgen R., Fernie A. R., Fisahn J., Segundo B. S., Guiderdoni E., Schippers J. H. M., and Mueller-Roeber B. (2013) SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morse A. M., Whetten R. W., Dubos C., and Campbell M. M. (2009) Post-translational modification of an R2R3-MYB transcription factor by a MAP kinase during xylem development. New Phytol. 183, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 59. Levee V., Major I., Levasseur C., Tremblay L., MacKay J., and Seguin A. (2009) Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 184, 48–70 [DOI] [PubMed] [Google Scholar]
- 60. Fukao T., Yeung E., and Bailey-Serres J. (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. El-kereamy A, Bi Y. M., Ranathunge K., Beatty P. H., Good A. G., and Rothstein S. J. (2012) The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. Plos One 7, e52030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Agati G., Azzarello E., Pollastri S., and Tattini M. (2012) Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 196, 67–76 [DOI] [PubMed] [Google Scholar]
- 63. Brown J. E., Khodr H., Hider R. C., and Rice-Evans C. A. (1998) Structural dependence of flavonoid interactions with Cu2+ ions: implications for their antioxidant properties. Biochem. J. 330(Pt 3), 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hernandez I., Alegre L., Van Breusegem F., and Munne-Bosch S. (2009) How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 14, 125–132 [DOI] [PubMed] [Google Scholar]
- 65. Agati G., Biricolti S., Guidi L., Ferrini F., Fini A., and Tattini M. (2011) The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 168, 204–212 [DOI] [PubMed] [Google Scholar]
- 66. Agati G., Matteini P., Goti A., and Tattini M. (2007) Chloroplast-located flavonoids can scavenge singlet oxygen. New Phytol. 174, 77–89 [DOI] [PubMed] [Google Scholar]
- 67. Zhang J., Subramanian S., Stacey G., and Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57, 171–183 [DOI] [PubMed] [Google Scholar]
- 68. Uchida K., Akashi T., and Aoki T. (2015) Functional expression of cytochrome P450 in Escherichia coli: An approach to functional analysis of uncharacterized enzymes for flavonoid biosynthesis. Plant Biotechnol. 32, 205–213 [Google Scholar]
- 69. Fatima T., Kesari V., Watt I., Wishart D., Todd J. F., Schroeder W. R., Paliyath G., and Krishna P. (2015) Metabolite profiling and expression analysis of flavonoid, vitamin C and tocopherol biosynthesis genes in the antioxidant-rich sea buckthorn (Hippophae rhamnoides L.). Phytochemistry 118, 181–191 [DOI] [PubMed] [Google Scholar]
- 70. Zhang J. A., Subramanian S., Zhang Y. S., and Yu O. (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 144, 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villalobos M. A., Bartels D., and Iturriaga G. (2004) Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol. 135, 309–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vannini C., Locatelli F., Bracale M., Magnani E., Marsoni M., Osnato M., Mattana M., Baldoni E., and Coraggio I. (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 37, 115–127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry raw data are available in the PRIDE Archive (https://www.ebi.ac.uk/pride/archive/) via identifier PXD006599. GenBank accession numbers for the proteins mentioned in this article are: GmMYB173, Glyma17G094400; GmCHS5, Glyma01g43880.1.