Abstract
Invasive infections by the human pathogenic fungus Aspergillus fumigatus start with the outgrowth of asexual, airborne spores (conidia) into the lung tissue of immunocompromised patients. The resident alveolar macrophages phagocytose conidia, which end up in phagolysosomes. However, A. fumigatus conidia resist phagocytic degradation to a certain degree. This is mainly attributable to the pigment 1,8-dihydroxynaphthalene (DHN) melanin located in the cell wall of conidia, which manipulates the phagolysosomal maturation and prevents their intracellular killing. To get insight in the underlying molecular mechanisms, we comparatively analyzed proteins of mouse macrophage phagolysosomes containing melanized wild-type (wt) or nonmelanized pksP mutant conidia. For this purpose, a protocol to isolate conidia-containing phagolysosomes was established and a reference protein map of phagolysosomes was generated. We identified 637 host and 22 A. fumigatus proteins that were differentially abundant in the phagolysosome. 472 of the host proteins were overrepresented in the pksP mutant and 165 in the wt conidia-containing phagolysosome. Eight of the fungal proteins were produced only in pksP mutant and 14 proteins in wt conidia-containing phagolysosomes. Bioinformatical analysis compiled a regulatory module, which indicates host processes affected by the fungus. These processes include vATPase-driven phagolysosomal acidification, Rab5 and Vamp8-dependent endocytic trafficking, signaling pathways, as well as recruitment of the Lamp1 phagolysosomal maturation marker and the lysosomal cysteine protease cathepsin Z. Western blotting and immunofluorescence analyses confirmed the proteome data and moreover showed differential abundance of the major metabolic regulator mTOR. Taken together, with the help of a protocol optimized to isolate A. fumigatus conidia-containing phagolysosomes and a potent bioinformatics algorithm, we were able to confirm A. fumigatus conidia-dependent modification of phagolysosomal processes that have been described before and beyond that, identify pathways that have not been implicated in A. fumigatus evasion strategy, yet.
Mass spectrometry proteomics data are available via ProteomeXchange with identifiers PXD005724 and PXD006134.
Keywords: Infectious disease, Cell-cell interactions, Host-Pathogen Interaction, Microbes, Pathogens, Subcellular Separation, Virulence
Infections with the opportunistic pathogen Aspergillus fumigatus pose a major threat to human health. It is estimated that there are worldwide more than 200,000 cases of invasive aspergillosis per year affecting patients with a severe underlying immune suppression because of hematologic diseases, genetic immune deficiencies or solid organ or hematopoietic stem cell transplantation (1, 2). A. fumigatus produces asexual spores (conidia) that are distributed via the air. On inhalation conidia reach the lower airway tract of human individuals. In immunocompromised hosts, the impaired immune cell function allows fungal colonization of the lung tissue and the establishment of a life-threatening infection (3).
Resident alveolar macrophages belong to the first line of immune defense (4). They are activated by fungal surface structures such as the β-1,3-glucans of the conidial cell wall which bind to the C-type lectin receptor Dectin-1 and thereby inducing phagocytosis. Activated macrophages engulf conidia in a phagosome, which gradually acquires biocidal properties from fusion with lysosomes to generate a mature phagolysosome with acidic luminal pH because of vacuolar ATPase (vATPase)1 activity (4, 5). Small Rab GTPases govern the dynamic fusion and fission processes required for generation of mature phagolysosomes and thus belong to the targets of intracellular pathogens to interfere with the maturation process (6–8). For example, Mycobacterium tuberculosis prevents the recruitment of Rab14 to the phagolysosome and thereby arrests its maturation (7).
The gray-green pigment of A. fumigatus conidia consists of 1,8-dihydroxynaphthalene (DHN) melanin that represents an important virulence determinant (9). Because of melanin wt conidia can reduce the production of and quench reactive oxygen species (ROS) and thus lower the amounts of ROS in the phagolysosome. Furthermore, wt conidia inhibit apoptosis, survive in host cells, germinate and cause damage to host cells (10–13). A mutant strain producing white conidia was shown to be defective in the polyketide synthase gene pksP and, hence, unable to produce DHN melanin (12). Inside the phagolysosome pksP conidia are faster degraded than wt conidia by luminal acidification and activity of lytic enzymes (14). DHN melanin is the crucial and sufficient factor to block acidification as demonstrated in experiments with melanin particles, i.e. “melanin ghosts” (14). However, the molecular mechanisms of the interference of the wt conidium with the phagosomal maturation are still not well understood. Previous work of our group showed that A. fumigatus wt conidia-containing phagolysosomes fused with vesicles of the endocytic compartment to a certain extent, but showed reduced acidification required to establish the fungicidal environment for degradation of conidia (13). The activity of the vATPase is essential to drive the acidification of conidia-containing phagolysosomes (14).
For several pathogenic microorganisms, proteomic studies unraveled the specific protein composition of phagolysosomes for a better understanding of the phagosomal maturation process and its modulation by pathogens (15–23). These studies demonstrated that the protein composition of the phagosome is highly dynamic, specific for the ingested particle, depends on the stage of phagosomal maturation, the type of cell line and the activation status of the phagocyte. Therefore, we set out to analyze the phagolysosomal proteome after phagocytosis of wt and pksP mutant conidia to shed light on processes inhibited by wt conidia during the maturation of phagosomes. In this study, a magnetic label-based purification protocol for conidia-containing phagolysosomes and the application of a label-free protein quantification method was developed. Bioinformatics was employed to determine the regulatory modules of differentially abundant proteins of the macrophage phagolysosome. We confirmed a differential regulation of vATPase subunits and apoptosis induction. These processes were already identified by us in previous studies as targets of A. fumigatus for immune evasion. Moreover, the combination of quantitative proteomics and bioinformatics led to the identification of A. fumigatus-modulated processes and of macrophage intracellular functions. These processes include vesicle trafficking and endocytosis, degradative and immune functions of the phagolysosome, NADPH oxidase activity to produce ROS, MAPK and mTOR signaling as well as metabolic reprogramming of the macrophage.
EXPERIMENTAL PROCEDURES
Materials and Chemicals
Primary antibodies were purchased from Abcam (Germany) (Ndufb9 ab200198; Vamp8 ab76021), Santa Cruz Biotechnology (Germany) (Lamp1 sc-19992, Cathepsin D sc-10725), Cell Signaling Technology (Germany) (mTOR #2983; Rab5 #3547) and Thermo Fisher Scientific (Germany) (Cathepsin Z PA5–47048). Secondary antibodies for Western blotting analyses were goat anti-rabbit IgG-HRP (sc-2030), goat anti-rat IgG-HRP (sc-2031) from Santa Cruz Biotechnology and rabbit anti-goat IgG-HRP (#31402) from Thermo Fisher Scientific. Secondary antibody for immunofluorescence was goat anti-rabbit IgG-DyLight 633 (#35562, Thermo Fisher Scientific).
Cells and A. fumigatus Strains
Mouse RAW 264.7 macrophages (ATCC TIB-71) were cultured in DMEM (Biozyme, Germany) with FBS (GE Healthcare, Germany), ultraglutamine (Biozyme) and gentamycin at 37 °C with 5% (v/v) CO2. A. fumigatus wt strain ATCC 46645 and pksP mutant (11, 12) strain were cultured on Aspergillus minimal medium (AMM) agar plates. After 5 days of incubation at 37 °C, conidia were harvested in 10 ml of 0.9% (w/v) NaCl/0.01% (v/v) Tween 80 (Roth, Germany) using a cell scraper and filtered through a 40 μm cell strainer (BD, Germany) to remove mycelia. The number of conidia was counted using a hemocytometer.
Magnetic Labeling of Conidia and Purification of Phagolysosomes
The protocol for isolation of conidia-containing phagolysosomes by magnetic separation was adapted from Steinhäuser et al. (24). 2 × 108 wt conidia were labeled with 10 mg/ml EZ link sulfo-NHS-LC biotin (Thermo Fisher Scientific) in 50 mm Na2CO3. Initial experiments demonstrated that for the same amount of pksP mutant conidia 10 μg/ml NHS-biotin linker yielded a comparable efficiency of magnetic loading. Cells were incubated with the linker for 2 h at 4 °C on a rotator and then washed. Fifty microliters of the streptavidin-coupled magnetic beads (Miltenyi, Germany) in the concentration provided by the supplier (Thermo Fisher Scientific) were added in labeling buffer (PBS with 2 mm EDTA) to the conidia suspension and incubated for 15 min at 4 °C on a rotator. Coincubation was performed in 4-well plates. Each well contained 4 × 106 RAW 264.7 macrophage cells.
For infection, an MOI of 5 was used, which corresponds to 2 × 107 magnetically labeled conidia. An MOI of 5 yielded the required amount of protein needed for the analysis of the subcellular proteome. Coincubation was stopped after 2 h. The cells were washed with PBS, scratched off and collected in homogenization buffer (3 mm imidazole, 250 mm sucrose, pH 7.4 and proteinase inhibitors (cOmplete, Roche, Switzerland) and benzonase nuclease (Merck Millipore, Germany)). Cell lysis was achieved by pressing the cell suspension 60 times through a needle (27G) and was monitored microscopically. DNase (Epicenter, Germany) was added and the lysate was incubated for 5 min at 37 °C. For sampling of the whole cell proteome, proteins of the lysate were precipitated and processed as described below. To purify the phagolysosomal fraction the lysate was loaded onto a QuadroMACS separator (Miltenyi). Phagolysosomes with magnetically labeled conidia were retained on the stand by magnetic force and proteins were directly eluted from the column with 98 °C heated elution buffer (2% (w/v) SDS, 100 mm TEAB, 10% (v/v) glycerol, 1 mm TCEP).
Processing of Proteins
The eluted protein solution was reduced by evaporation in a SpeedVac at 60 °C to a volume of 100 μl. Proteins were precipitated with methanol and chloroform as described elsewhere (25) and protein concentrations determined using the Direct Detect Infrared Spectrometer (Merck Millipore). One hundred micrograms of the precipitated protein were resuspended in 100 mm TEAB buffer, proteins were reduced with 200 mm TCEP, alkylated with 375 mm iodoacetamide and digested with 4 μg trypsin (SERVA) over night at 37 °C. The reaction was stopped with formic acid and the sample was resuspended in acetonitrile/TFA for LC-MS/MS analysis.
Western Blotting
Sixteen and 8 μg of protein were loaded on a 4–12% (w/v) Bis-Tris Protein Gel (NuPAGE, Thermo Fisher Scientific) and separated by SDS-PAGE in 1× MES running buffer (Thermo Fisher Scientific). The protein was blotted with a semi-dry blot device (Bio-Rad, Germany) onto a low fluorescent PVDF membrane (GE Healthcare Life Sciences) using a Tris/glycine transfer buffer (25 mm Tris-HCl, 192 mm glycine, 20% (v/v) methanol). The blot was incubated with specific primary antibodies as specified and horse reddish peroxidase-coupled secondary antibodies were used for detection. The reaction mix of the Smart Protein Layer Kit (NH DyeAGNOSTICS, Germany) was added to the sample prior to the experiment for a fluorescent labeling of the total amount of proteins, which was used as a loading control (26). Fluorescence and chemiluminescence were detected with the Fusion FX7 system (Vilber Lourmat, Germany) and signal intensities and quantifications were determined with the Bio-1D analysis software (Vilber Lourmat) as described in the manufacturer's instructions. Western blots were conducted for each of the three replicates of conidia-containing phagolysosome purifications.
Experimental Design and Statistical Rational
By using a quantitative label-free proteomics approach, we compared the subproteome of enriched phagolysosomes containing melanized wt with nonmelanized pksP mutant conidia. Three biological replicates of wild-type and pksP conidia-containing phagolysosomes were measured each in three analytical replicates. Analytical replicates were treated as fractions (merged together for database searches) whereas biological replicates were analyzed independently of each other. Mean values and standard deviations for biological replicates of each comparison group were calculated. Differentially abundant proteins were determined using a log2 fold-change cut-off threshold of 1. By the identification of regulatory modules an additional level of information was provided. A regulatory module denotes a region in the organism-specific protein-protein interaction network, which is significantly enriched with differentially abundant proteins. Proteins that are differentially abundant but not within the module are disregarded because they are not well connected with other differentially abundant proteins. Thus, confidence in the reliability of the measured fold-change is gained by additional topological information from the protein-protein interaction network. For evaluation of the enrichment of the phagolysosomal fraction, we further compared the phagolysosome-enriched subcellular proteome with the whole cell proteome of macrophages, which had ingested either wt or pksP conidia of A. fumigatus. The whole cell sample was generated once and analyzed in three technical replicates.
LC-MS/MS Analysis of Protein Samples
Two different methods were applied. For the sub-proteome analysis of the conidia-containing phagolysosomal fraction an analytical method designated as method M#1 was used. For comparison of the phagolysosomal sub-proteome with the whole cell proteome method M#2 was applied.
LC-MS/MS analysis was carried out on an Ultimate 3000 RSLC nano system coupled to a QExactive Plus mass spectrometer (both Thermo Fisher Scientific). Peptides were enriched on a nano-trap column (Acclaim PepMap 100, length 20 mm, diameter 75 μm, particle size 3 μm) at a flow rate of 5 μl/min. Trapped peptides were separated after a valve switch on Acclaim PepMap RSLC nano columns with 150 mm (M#1) or 500 mm (M#2) length (Thermo Fisher Scientific). The mobile phase consisted of eluent (A) 0.1% (v/v) formic acid in H2O and eluent (B) 0.1% (v/v) formic acid in 90/10 ACN/H2O. We applied either 135 min (M#1) or 360 min (M#2) gradient elution using the following gradients:
M#1: 0–5 min at 4% B, 10 min at 6.5% B, 15 min at 7.5% B, 20 min at 8% B, 25 min at 8.6% B, 30 min at 9.2% B, 35 min at 9.9% B, 40 min at 10.6% B, 45 min at 11.3% B, 50 min at 12.2% B, 55 min at 13.4% B, 60 min at 14.9% B, 65 min at 17% B, 70 min at 19.1% B, 75 min at 22.4% B, 80 min at 26% B, 88 min at 32% B, 94 min at 40% B, 100 min at 52% B, 103 min at 68% B, 106–114 min at 96% B, 115–135 min at 4% B.
M#2: 0–4 min at 4% B, 90 min at 9% B, 130 min at 12.5% B, 180 min at 17% B, 200 min at 20% B, 220 min at 24% B, 250 min at 35% B, 260 min at 44% B, 265 min at 50% B, 270 min at 55% B, 275 min at 70% B, 280–290 min at 96% B, 291–360 min at 4% B.
Positively charged ions were generated with a stainless-steel emitter at 2.2 kV spray voltage using a Nanospray Flex Ion Source (Thermo Fisher Scientific). The MS instrument was operated in Full MS/dd MS2 (TopN) mode. Precursor ions were measured in full scan mode within a mass range of either m/z 300–1600 (M#1) or m/z 300–1500 (M#2) at a resolution of 70k/140k FWHM (M#1/M#2) using a maximum injection time of 120 ms and an AGC (automatic gain control) target of 106. Up to 10 of the most abundant precursor ions per scan cycle with an assigned charge state of z = 2–6 were selected for data-dependent acquisition using an isolation width of m/z 2.0. HCD fragmentation was conducted at a normalized collision energy of 30 V using N2. Dynamic exclusion of precursor ions was 35 s (M#1) or 40 s (M#2). Fragment ions were monitored at a resolution of 17.5k (FWHM) using a maximum injection time of 120 ms and an AGC target of 2 × 105. The fixed first mass was set to m/z 120. The LC-MS/MS instrument was operated by means of the Thermo/Dionex Chromeleon Xpress 6.8 software and the Thermo QExactive Plus Tune/Xcalibur 3.0.63 software.
Protein Database Search and Label-free Quantitation
Thermo raw files were processed by the Proteome Discoverer (PD) software v1.4 (M#1) and v2.1 (M#2) (Thermo Fisher Scientific). Tandem mass spectra were searched against databases (download date 21/09/2016) of Mus musculus (UniProt) and A. fumigatus (Aspergillus Genome Database, AspGD). It was searched against 66,868 database entries (protein sequences) using the algorithms of Mascot v2.4.1 (Matrix Science, UK), Sequest HT (as integral part of the Thermo software suite Proteome Discoverer 1.4.0.288 (M#1) and 2.1.0.81 (M#2)) and MS Amanda (release version 1.0.0.4756). Two missed cleavages were allowed for tryptic peptides. The precursor mass tolerance was 10 ppm and the fragment mass tolerance was 0.02 Da. Dynamic modification was oxidation of methionine. Static modification was cysteine carbamidomethylation. Percolator node and a reverse decoy database was used for q-value validation of the peptide spectral matches (PSMs) using a strict target false discovery (FDR) rate of < 1%. At least 2 peptides per protein were required for positive protein hits. Label-free quantification was performed with the precursor ions area - Top3 method included in PD. It compares the 3 most abundant peptides of each protein by using the peak area of the respective precursor ion (27). The mass tolerance was set to 2 ppm and the signal-to-noise ratio should be > 3. The abundance values were normalized based on the total peptide amount. Only unique peptides were considered for quantification. The significance threshold for differential protein regulation was set to factor ≥ 2.0 (up- or down-regulation). The mass spectrometry proteomics data were deposited at the ProteomeXchange Consortium via the PRIDE (28) partner repository.
Identification of the Regulatory Module from LC-MS/MS Data
Identification of the murine regulatory module was performed using ModuleDiscoverer (29) like the approach described in (30). In brief, differentially abundant proteins were mapped onto the murine protein-protein interaction network obtained from the STRING database. Identification of the regulatory module was then performed by the extraction of network regions that are significantly enriched with differentially abundant proteins. These sub-networks were analyzed regarding their biological function using the GOstats package for R (31). Mapping of different protein identifiers was achieved by applying the org.Mm.eg.db annotation package for R (32) as well as the Ensembl BioMarts resource (33) using the biomaRt package for R (34). A detailed description is provided in the supplement.
Prediction of Host-Pathogen Interactions
To predict the protein interactions of the M. musculus macrophages and A. fumigatus conidia within the phagolysosome, the regulatory module and the detected fungal proteins were mapped to an existing database of predicted host-pathogen interactions (HPI). The data set HPI was calculated by using experimental verified interactions from model organisms. These sources for interactions deliver the backbone to predict interfacial interaction of host and pathogen (35). By using traits that characterize the source interacting proteins, such as amino acid sequence, GO annotation, pathway membership, expression level, or domain information, pairs of interacting proteins of host and pathogen could be set up, filtered and refined. The establishment of protein pairs, as well as the filtering was exhibited like Remmele et al. (35), whereas the refinement steps involved a scoring system based on experimental exchange of orthologs and analog prediction methods. Details about the prediction and scoring method are given in the supplementary information.
Immunofluorescence
Conidia were stained with 0.1 mg/ml calcofluor white (CFW, Fluorescence Brightener 26, Sigma Aldrich, Germany) for 25 min at room temperature or with 0.1 mg/ml fluorescein isothiocyanate (FITC, Sigma-Aldrich) in 100 mm Na2CO3 for 30 min at 37 °C. Cells were infected with conidia at an MOI of 2 and coincubated for 2 h at 37 °C with 5% (v/v) CO2. After coincubation, cells were fixed with 3.7% (v/v) formaldehyde for 10 min at room temperature. For immunofluorescence detection, cells were treated with 0.25% (v/v) Triton X-100 (VWR, Germany), blocked with 3% (w/v) bovine serum albumin (Sigma-Aldrich) and 3% (v/v) goat serum (Santa Cruz Biotechnology) and then incubated overnight at 4 °C with the primary antibody. Secondary antibody was added for 1 h at room temperature. All images were acquired on a Zeiss LSM 780 confocal microscope with a Zeiss Plan-appochromat 63×/1.4 oil objective. Three technical and two biological replicates were analyzed for each strain. Six images were taken per technical replicate, which resulted in 36 images in total per strain. The total number of ingested conidia per image was determined. The conidia-containing phagolysosomes with a specific fluorescence from the target molecule, as indicated by a circular fluorescence signal around the conidium, were considered positive for the desired marker. With the number of positive phagolysosomes and the total number of ingested conidia the ratio of target-positive phagolysosomes was determined.
RESULTS
Development of a Protocol for Isolation of Conidia-containing Phagolysosomes
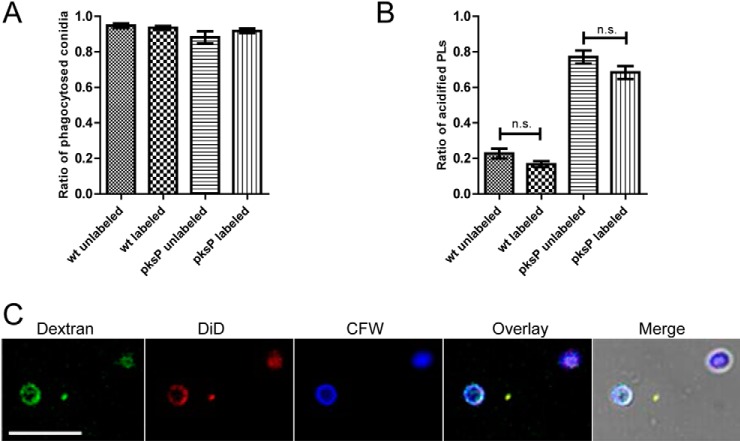
To address the question, whether the manipulation of the phagosome maturation by wt conidia is reflected in the phagolysosomal proteome, proteins were extracted from phagolysosomes containing melanized wt and nonmelanized pksP mutant conidia based on a protocol for the isolation of Mycobacteria-containing phagolysosomes (24). Magnetic labeling of conidia enabled the isolation of the phagolysosomal fraction after lysis of infected macrophages (Fig. 1). The concentration of NHS-biotin linker was adjusted to the different surface properties (11) of the two types of conidia. Therefore, linker-binding capacities of conidia of the two strains were monitored microscopically by the addition of streptavidin-coupled Alexa-fluorophore (data not shown). Magnetic beads were bound to the conidia by a streptavidin tag. To exclude effects of labeling on phagocytosis and intracellular processing of the conidia, phagocytosis ratio and the ratio of acidified phagolysosomes of labeled and unlabeled wt and pksP mutant conidia were determined. After 2 h of coincubation of conidia with macrophages, no differences in phagocytosis of labeled and unlabeled conidia were detected (Fig. 2A). Furthermore, 16.8% and 20.8% of labeled and unlabeled wt conidia-containing phagolysosomes, respectively, and 68.4% and 77.1% of labeled and unlabeled pksP mutant conidia-containing phagolysosomes, respectively, were acidified (Fig. 2B). Thus, the label on the conidial surface had no significant effect on the acidification of the phagolysosome.
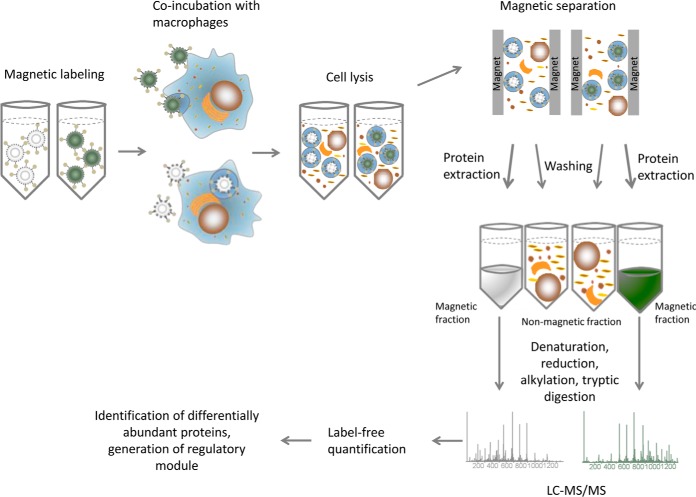
Fig. 1.
Flowchart for the purification of conidia-containing phagolysosomes, proteome detection and data analysis. The procedure starts with magnetic labeling of conidia that are then incubated with macrophages for 2 h. The cell lysate is loaded onto a magnetic column to separate the magnetic fraction from the cell debris. The protein is extracted on-column, then precipitated and concentrated for LC-MS/MS measurement. After label-free quantification the data was analyzed to identify regulated proteins and modules.
Fig. 2.
Effect of magnetic labeling on characteristics of conidia. Magnetic labeling of conidia has no effect on phagocytosis and acidification of phagolysosomes (PLs). A, The phagocytosis ratio was determined after 2 h coincubation of macrophages with magnetically labeled or unlabeled conidia of the wt and the pksP mutant. B, The ratio of acidified phagolysosomes was determined 2 h post-infection of macrophages with conidia. C, Integrity of the phagolysosomal membrane is not impaired by isolation of conidia-containing phagolysosomes. Cells were loaded overnight with fluorescent dextran beads. Conidia were labeled with CFW and added to the cells for 2 h. Cell lysis was performed by shearing the cell suspension and the lysate stained with DiD, specific for eukaryotic cell membranes. Images show a triple staining of particles with dextran, DiD and CFW indicating an intact phagolysosomal compartment.
Wt and pksP conidia-containing phagolysosomes were isolated two hours after infection of RAW 264.7 macrophages, because at this time point acidified phagolysosomes had been formed (36). After coincubation of conidia and macrophages, shearing of cells provided the best method to ensure a thorough cell lysis without affecting the integrity of the phagolysosomes. To determine the integrity and quality of the isolated phagolysosomes, membranes were stained with the lipophilic dye DiD. The localization of fluorescent microbeads (3000 MW Dextran beads, Thermo Fisher Scientific), which are taken up by pinocytosis and transferred via lysosomes to the phagolysosome, was monitored microscopically. A clear fluorescence signal of DiD and dextran beads surrounded the conidia indicating the presence of a continuous and intact phagolysosomal membrane (Fig. 2C). The complete lysate was loaded onto a magnetic column and washed extensively to remove contaminating cell debris. The proteins of retained phagolysosomes were extracted on the column, and the flow-through was collected, concentrated and processed for LC-MS/MS measurements (Fig. 1).
Identification of Differentially Abundant Proteins from LC-MS/MS Data
LC-MS/MS measurement and protein identification resulted in a dual proteome set consisting of 2431 murine phagolysosomal and 65 A. fumigatus proteins (supplemental Table S1). 95% of the proteins were detected in both wt und pksP mutant conidia-containing phagolysosomes, suggesting that quantitative differences predominate. To identify the differences, a label-free quantification was performed using the “Top Three” method (27) and the relative abundance of proteins in pksP mutant versus wt conidia-containing phagolysosomes was calculated. Based on a log2 fold-change cut-off threshold of 1, we identified 637 differentially abundant proteins in the phagolysosome and 22 differentially abundant proteins from A. fumigatus. Of the 637 regulated proteins of the phagolysosome, 472 were overrepresented in the pksP mutant conidia-containing phagolysosome and 165 in the wt conidia-containing phagolysosome. On the fungal side, 8 proteins were found to be specific for pksP mutant conidia and 14 proteins for wt conidia.
For a quality control of the purification protocol the phagolysosomal proteome was compared with the whole cell proteome, which was obtained by extraction of proteins directly from the lysate of macrophages infected with wt or pksP mutant conidia. The whole cell proteome comprised 1986 murine proteins (supplemental Table S2). 53% of total phagolysosomal proteins and 39% of differentially abundant proteins in the phagolysosome were also detected in the whole cell proteome. Thus, a proportion of 47% of total phagolysosomal proteins and 61% of differentially produced proteins was only found in the purified sample, indicating, that the purification protocol of phagolysosomes allowed for enrichment of these organelles. This assumption was further substantiated because proteins assigned to the gene ontology (GO) “lysosome” (count = 69, p-value = 5.3 × 10−6) and “lysosomal membrane” (count = 63, p-value = 3.2 × 10−10) showed a strong enrichment in the phagolysosomal fraction compared with the whole cell proteome (count = 41, p-value 0.06 and count = 40, p-value = 1.8 × 10−4). Further, the p-value for the GO-term “cytosol” is larger in the phagolysosomal fraction (p-value = 8.5 × 10−21, count = 362) compared with the p-value in the whole cell sample (p-value = 4.2 × 10−41, count = 359) indicating that the phagolysosomal fraction contains less signal from the cytosol compared with the whole cell samples. Based on this evaluation, the purification protocol clearly accomplished a concentration of the phagolysosomal fraction.
Regulatory Module Implies Interaction of Regulated Proteins
To deduce regulated processes from the data set of differentially abundant proteins, we used a bioinformatics approach that identified regulatory modules within the protein-protein interaction (PPI) network provided by the STRING database (37). This analysis was based on the projection of experimentally identified differentially abundant proteins. As a result, a host regulatory module was defined that contained the regulated proteins but also nonregulated and nondetected proteins depicted as knots and their connections visualized as edges. Within the network, submodules represent groups of proteins with a higher connectivity based on STRING that are functionally or structurally related.
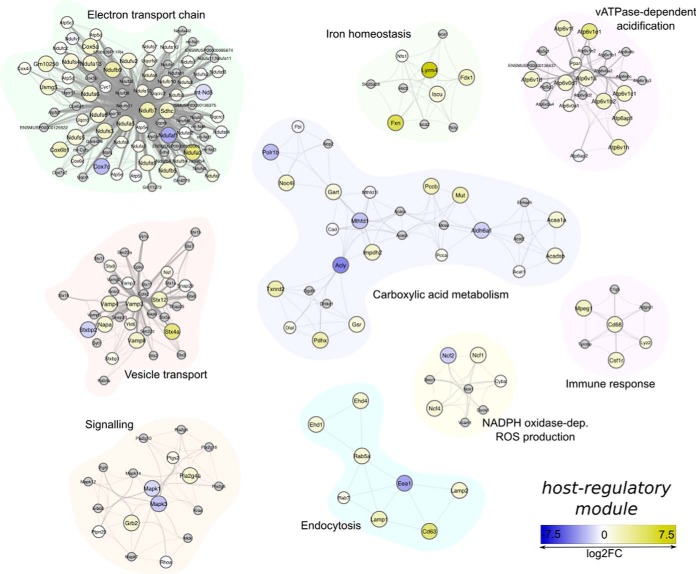
The assembled host regulatory module is composed of 302 proteins connected by 3448 edges. One hundred seventy-eight of the 302 proteins were detected by LC-MS/MS. One hundred nine of these 178 proteins were identified as differentially abundant. Seventy-nine proteins were enriched in the pksP mutant and 30 in the wt conidia-containing phagolysosome. In the host regulatory module 17 submodules were classified. Fourteen of these submodules comprising five or more proteins were considered for further discussion and used in enrichment analysis of GO and KEGG pathway terms (supplemental Table S3). The submodules representing vATPase-dependent vacuolar acidification, signaling, endocytosis and vesicle transport, immune response, generation of ROS via NADPH oxidase and electron transport chain were the focus for further validation experiments and discussion (Fig. 3). In detail, these submodules include proteins with annotated functions in MAPK signaling pathway, actin polymerization, SNARE-mediated membrane trafficking and Rab GTPase-regulated vesicle fusion, hydrolytic activities and NADPH oxidase-dependent ROS production.
Fig. 3.
Host regulatory module. A total of 302 proteins are connected by 3448 edges in the displayed module. 178 proteins were detected in the LC-MS/MS analysis and 109 classified as differentially regulated. The graph displays regions of the protein-protein interactome that are significantly enriched and shown here to be differently regulated. The color scale from blue to yellow represents the log2 fold change of protein abundances, proteins in gray were not detected by mass spectrometry, but complete the respective submodule because of their close connectivity to the neighboring partner. The edge size is indicative of the confidence score of the interaction.
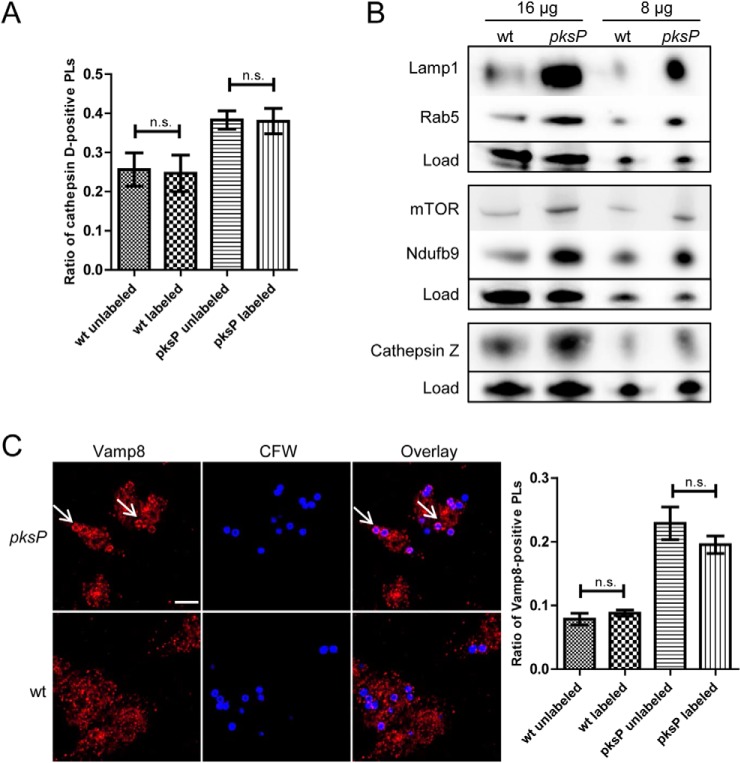
To verify the data obtained by LC-MS/MS analysis, label-free quantification and the regulatory module analysis, the abundances of several representative proteins were quantified by Western blotting analyses or immunofluorescence (Fig. 4). First, to exclude that magnetic labeling interferes with recruitment of phagolysosomal proteins the localization of cathepsin D was monitored by immunofluorescence. Both, labeled and unlabeled wt conidia reduced recruitment of cathepsin D to the phagolysosomal membrane, whereas pksP conidia-containing phagolysosomes showed a typical pattern of cathepsin D recruitment, independent of magnetic labeling (Fig. 4A). Representative proteins included proteins enriched in pksP conidia-containing phagolysosomes: Cathepsin Z, a lysosomal protease, which was 2-fold higher abundant; Lamp1, a phagolysosomal marker protein represented in the “endocytosis” module with a 3.8-fold higher abundance; Nduf9, a component of the NADH dehydrogenase complex represented in the module “electron transport chain” with 5.9-fold higher abundance. The higher abundance of selected proteins in pksP conidia-containing phagolysosomes was confirmed by Western blot analysis (Fig. 4B). Because we found LAMTOR, a regulator of the mTOR signaling pathway, enriched in pksP conidia-containing phagolysosomes, and, second, the regulatory module indicated a regulation of carboxylic acid metabolism and cellular respiration, we analyzed the abundance of the kinase mTOR. Both processes are potentially controlled by mTOR. Although not detected by LC-MS/MS, we could confirm a differential regulation of mTOR in conidia-containing phagolysosomes by Western blotting (Fig. 4B). The vesicle trafficking protein 8 (Vamp8), which was assigned to the “vesicle trafficking” module and which was 2.8-fold enriched in pksP conidia-containing phagolysosomes, was analyzed for its localization by immunofluorescence. A differential recruitment was clearly confirmed (Fig. 4C).
Fig. 4.
Recruitment of proteins to the phagolysosomal membrane. A, The effect of magnetic labeling on recruitment of cathepsin D to the phagolysosomal membrane 2 h post infection was determined by immunofluorescence. Columns represent mean values of the proportion of cathepsin D-positive phagolysosomes (PLs) ± SDs. B, Western blotting analyses were performed to determine abundances of selected proteins in the phagolysosomal fraction. 16 or 8 μg of protein extract from purified wt or pksP mutant conidia-containing phagolysosomes were separated by SDS-PAGE. Antibodies against selected candidate proteins from the proteome data were applied. Because of the lack of suitable house-keeping proteins the samples were mixed with the Smart Protein Layer kit that enabled a normalization of the bands to the total loaded protein. C, wt conidia-containing phagolysosomes fail to recruit Vamp8. Images are representative of Vamp8 immunofluorescence staining of macrophages 2 h post infection with wt or pksP mutant conidia at an MOI = 2. Scale bars refer to 10 μm and arrows indicate Vamp8-positive phagolysosomes. Columns represent the mean values of the proportion of Vamp8-positive phagolysosomes ± SDs.
Fungal Proteins in the Phagolysosome
The host regulatory module provided information about processes that are regulated in the macrophage on phagocytosis of A. fumigatus conidia. The dual proteome analysis allowed us to investigate differentially abundant fungal proteins as potential effectors interfering with the host endocytic pathway. It is likely that fungal proteins in the phagolysosome are either actively secreted or released from the fungal cell surface. However, it needs to be considered that the two strains face different conditions: Although pksP mutant conidia are exposed to an acidic environment and tackled by phagolysosomal degrading enzymes 2 h after phagocytosis, the wt conidia reside in more favorable conditions in a phagolysosome with neutral pH. Accordingly, the fungal proteome enriched from pksP mutant conidia-containing phagolysosomes was composed of proteins induced on oxidative stress or on encounter with immune cells, e.g. a GTPase regulating vesicular transport, an RNA helicase, alcohol dehydrogenases and a transaldolase of the pentose phosphate pathway (supplemental Table S4).
Proteins enriched in wt conidia-containing phagolysosomes included a catalase, drug response and mitochondrial unfolded protein response elements and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Further proteins were histone H2A1, a component of ribosome biogenesis, transcription and mRNA processing, the signaling protein 14-3-3 and a high abundant conidial protein with unknown function that was found by Asif et al. (38) to be present on conidia (supplemental Table S5).
DISCUSSION
Here, we set out to characterize the phagolysosomal proteome and to identify phagolysosomal proteins and processes that are altered after phagocytosis of melanized A. fumigatus conidia to allow their survival inside the macrophage. The phagolysosome is the place of the direct interaction between conidia and the host, hence, it can be expected that modifications in its protein composition influence its fungicidal activity. On the level of the whole cell, changes in abundance of proteins in specific organelles like the phagolysosome might be masked, e.g., by highly abundant cytosolic proteins. Therefore, we analyzed the proteomes of both whole macrophage cells and phagolysosomes. Because previously we showed that DHN melanin of the conidia interferes with the maturation of phagolysosomes (14) we compared the phagolysosomal proteomes of macrophages, which were infected either with wt or nonpigmented pksP mutant conidia of A. fumigatus. For this purpose, we developed a method to purify conidia-containing phagolysosomes and analyzed their proteomes. By bioinformatical analyses of differentially regulated phagolysosomal proteins we compiled a regulatory module that represents the regulated proteins. By combining the different methods, we deduced a model in which wt conidia modulate endosomal trafficking and fusion processes, intracellular signaling and metabolic pathways to prevent acidification of the phagolysosome, delivery of degradative enzymes, induction of autophagy, apoptosis and proinflammatory immune response to survive within the phagolysosome.
Our analysis required the isolation of conidia-containing phagolysosomes. A common protocol to isolate phagolysosomes makes use of the sedimentation properties of the ingested latex beads and was first described by Wetzel et al. (39). However, because of the sedimentation properties of conidia-containing phagolysosomes their separation on a sucrose gradient is difficult. Alternatively, Steinhäuser et al. described the purification of Mycobacterium tuberculosisbacteria-containing phagolysosomes based on magnetic separation (24). The cell lysis was optimized such that the cytoplasmic membrane was disrupted and at the same time the integrity of the phagolysosomal membrane was maintained. A strategy of magnetic separation, protein extraction and concentration was developed to yield the conidia-containing phagolysosomal proteome that could be measured by LC-MS/MS.
The outer phagolysosomal membrane is surrounded by a mesh of sticky actin fibers that trap organelles in its proximity during the isolation procedure (40). The occurrence of proteins typically present in nonphagosomal cell compartments such as the nucleus, ribosomes or mitochondria has been reported earlier (15). Their frequency was especially high in cells with a high autophagocytic activity. This can be explained by the dynamic interaction and exchange between those two organelles (21, 41). Additionally, mitochondria have been implicated in a broad number of cellular functions, including TLR-dependent ROS production in response to intracellular pathogens (42) and induction of apoptosis via intrinsic and extrinsic pathways (43).
The comparison of the protein abundances in the wt and pksP mutant conidia-containing phagolysosomes is a challenging endeavor, because of the complexity of the experimental set-up and high variations of the biological replicates. The results of studies on the phagolysosomal proteome vary depending on different factors: (1) the usage of cell lines or primary cells (23), (2) the ingested particles, e.g., latex beads, apoptotic cells, pathogen-associated structures or pathogens (16–18, 22), (3) the age of the phagolysosome (15, 17), (4) the analyzed fraction, e.g., entire phagolysosome or membrane domains (detergent-resistant or detergent-soluble membranes) (15), and (5) the activation status of the cells, e.g., stimulation with cytokines prior to the infection (19).
With the help of a label-free quantification, around 30% of the identified proteins were classified as significantly regulated. The differential regulation of cathepsin Z, Lamp1, Rab5 and NADH dehydrogenase was verified by Western blotting and the differential recruitment of Vamp8 was confirmed by immunofluorescence. Interestingly, the regulation of mTOR, which was not detected by the LC-MS/MS measurement, but was predicted from the interaction network, was found to be differentially regulated by Western blot analysis and thus confirmed the bioinformatics approach. The putative low abundance of mTOR might explain why this protein was only detected by sensitive Western blotting but not by LC-MS/MS. The comprehensive LC-MS/MS analysis was based on a bottom-up data-dependent shotgun approach, e.g., not only the general detection sensitivity of the tryptic peptides of a certain protein is decisive. Much more important for highly complex peptide samples is the dynamic range of the analysis. e.g., relative abundance of the tryptic peptides of mTOR in relation to all other peptides, because the 10 most abundant precursor ions per data-dependent scan cycle are selected for fragmentation. Because murine mTOR is a large protein of about 289 kDa, which implicates numerous potentially detectable peptides, it can be assumed that mTOR is of relative low abundance. At least the abundance of mTOR is lower than for all proteins identified in our study.
Taken together, the bioinformatics compilation of the differentially abundant proteins into the regulatory module allowed for an identification of processes that are modified in the phagolysosome after ingestion of either wt or pksP conidia.
vATPase-dependent Acidification
Acidification of the phagolysosome is prerequisite for the degradation of phagocytosed material and is driven by the vATPase, the vacuolar ATP-dependent proton pump (44). Activity of the enzyme complex is regulated via assembly and disassembly of the membrane bound V0 and cytosolic V1 domain (45). In a previous study, we showed that activity of the vATPase mainly drives the acidification of phagolysosomes containing pksP conidia (14). Here, the proteomic data confirmed the increased abundance of V1 subunits in pksP conidia-containing phagolysosomes.
Lysosome-Endosomal Trafficking
The endocytic trafficking and transport system is also modulated in the wt conidia-containing phagolysosome. We detected several proteins of the SNARE, SNAP and syntaxin family, which mediate intracellular trafficking, docking and fusion processes (46). They were mainly down-regulated in macrophages infected with wt conidia. Also, the small GTPase Rab5 and early endosomal antigen 1 (EEA1) were regulated. Rab5 and its effector EEA1 mediate homotypic fusion of the early phagosome with early endosomes and are regarded as markers for an early stage of phagolysosomal maturation (47, 48). Rab5 had a lower abundance in wt conidia-containing phagolysosomes, whereas EEA1 was more abundant. Likewise, EH domain-containing proteins, which control endocytic fusion and colocalize with Rab5 and EEA1 were less abundant in wt conidia-containing phagolysosomes. This implies that wt conidia-containing phagosomes are less fusogenic and arrest the maturation of the phagolysosome at an early stage. This finding is in line with earlier observations when we determined the fusion of A. fumigatus conidia-containing phagosomes with lysosomes by measuring the localization of the phagolysosomal markers cathepsin D and Lamp1 (13). The reduced acquisitions of those markers by wt conidia-containing phagosomes prompted us to speculate that A. fumigatus wt conidia interfere with the endocytic pathway by preventing fusions of the phagosome with lysosomes. However, when we analyzed the fusion rate of dextran beads-containing lysosomes with phagosomes (14), only a slight delay in the delivery of the beads to wt conidia-containing phagosomes was observed. Thus, it is conceivable that A. fumigatus conidia do not generally block endocytic fusions, but rather selectively interfere with the delivery of proteins and enzymes required for the maturation of the phagolysosome into a biocidal compartment. An interference with endosomal trafficking and fusions by targeting small Rab GTPases has been reported for many pathogens such as Candida albicans, Mycobacterium tuberculosis, Coxiella burnetii, Helicobacter pylori, Salmonella enterica, Chlamydia species and others (reviewed in (49)).
Generation of Energy
Components of the oxidative phosphorylation (OXPHOS) system, i.e. protein complexes of the mitochondrial electron transport chain, are enriched in the pksP conidia-containing phagolysosomal sample. The occurrence of those components, although they are of nonphagolysosomal origin, might represent a specific effect of mitochondrial involvement in phagolysosomal processes. We found subunits of all four complexes of the electron transport chain as well as 3-ketoacyl-CoA thiolase and acyl-CoA dehydrogenase of the fatty acid β-oxidation enriched in the pksP mutant conidia-containing phagolysosome, indicating that wt conidia influence the host cell energy metabolism. In line with this assumption, we found mTOR, a regulator of cellular metabolism (50), enriched in phagolysosomes of macrophages that were challenged with pksP conidia. Furthermore, mitochondrial ROS, which are generated by the electron transport chain, were described to contribute to the killing of intracellular pathogens (42, 51).
Intracellular Signaling
The proteomic data indicate a reduced presence of components of the Akt-mTOR pathway in the wt conidia-containing phagolysosomal proteome. This signaling axis plays a major role in inducing autophagy under starvation conditions to recycle nutrients from the phagosome. By targeting the autophagy-activating pathways ingested wt conidia interfere with the generation of the biocidal autophagosome (52). In line with this hypothesis, we found components of the autophagy machinery such as Vamp8, Snap29 and Vti1b of the autophagosomal SNARE complex (53) and Rubicon (Run domain Beclin-1-interacting and cysteine-rich domain-containing protein) (54) in the regulated modules confirming a role of this specific intracellular degradation process in the clearance of A. fumigatus conidia. Rubicon has recently been identified as the central switch from autophagy to Microtubule-Associated Protein 1 Light Chain 3 (LC3)-associated phagocytosis (LAP), an Atg5-dependent autophagy mechanism, where the LC3BI protein binds phosphatidylethanolamine (PE) to form the active, lipidated LC3BII protein that binds to phagolysosomes (55, 56). Rubicon recruits the NADPH oxidase (NOX) and activates its ROS producing activity, which in turn is required for LC3B lipidation (54). In line, Chamilos et al. (52, 57) demonstrated that DHN melanin of A. fumigatus conidia blocks LAP activation in macrophages.
Previous work of our lab showed that wt conidia of A. fumigatus block apoptosis of macrophages by hijacking the PI3K/Akt signaling pathway (58). The sustained activation of Akt promotes cell survival and inhibits induction of the apoptosis process and likely depicts a strategy of the pathogen to hide inside the macrophage and evade further immune responses (58, 59). Among the differential abundant proteins were elements of the apoptosis or cell survival regulation pathways: MAPK, AKT, mTOR and Rac signaling pathway, the ubiquitin-proteasome system, members of the 14-3-3 family proteins, prostaglandin E synthase among others. Pro-apoptotic proteins such as Bak1 (60), Aifm2 and Praf2 were higher abundant in pksP mutant conidia-containing phagolysosomes, whereas the anti-apoptotic regulator Bcl2-like 13 protein was enriched in wt conidia-containing phagolysosomes.
Degradation and Antigen Processing Capacity
Different studies demonstrated the requirement for ROS for an efficient fungal clearance (61, 62). ROS are generated by the NOX complex which consists of five subunits. Our data suggest impaired NOX assembly and, thus, reduced production of ROS after ingestion of wt conidia, because one of the five NOX subunits, p47phox (Ncf1) was less abundant in wt conidia-containing phagolysosomes. Also, upstream elements of the Rac GTP-binding protein family, which regulate NOX assembly, such as guanine nucleotide exchange factors (GEFs) ArhGEFs and Rac GTPase activating protein (RacGAP1) showed low levels in the wt sample. Langfelder et al. showed a 10-fold increase in ROS production of PMNs that were challenged with A. fumigatus pksP mutant conidia compared with wt conidia (12). Jahn et al. demonstrated that this increase can be attributed at least in part to the fact that pksP mutant conidia lack ROS quenching by DHN melanin (63). Besides its fungicidal activity, signaling function was also ascribed to ROS in mediating the induction of the autophagy machinery (64) and pro-apoptotic signaling (58, 65). Consistently, Volling et al. hypothesized that DHN melanin inhibits apoptosis by quenching ROS (58, 63).
Cathepsins, lysosomal peptidases and enzymes to degrade glycans and glycolipids were enriched in the proteome of pksP mutant conidia-containing phagolysosomes indicating a reduced degradation capacity of wt conidia-containing phagolysosomes. In line with this finding is the reduced antigen processing and presentation machinery of those phagolysosomes. For example, phagolysosomes containing wt conidia had lower levels of the macrophage activation marker CD68 or macrophage colony-stimulating factor receptor 1 (CSFR1).
The interference of A. fumigatus with the described intracellular processes of macrophages is conceivably because of an interaction of fungal proteins with host proteins. We performed an initial bioinformatics analysis to predict potential interaction candidates on the basis of already described host-pathogen protein interactions. This analysis suggests fungal heat shock proteins Hsp70 and 90, the translation elongation factor Tef1 and a 14-3-3 protein ArtA to interact with a range of host proteins involved in processes such as vATPase activity, endocytic trafficking and signaling (supplemental Fig. S1). Experiments evaluating the prediction of these interactions, e.g., on a biochemical level or using immune cells with a knock down of the genes of interest, are required to identify the role of protein-protein interactions for the immune modulation by A. fumigatus.
Altogether, this study provides a detailed map of proteins and a comprehensive overview of macrophage intracellular processes that are regulated on ingestion of A. fumigatus conidia. It delivers a protocol to obtain conidia-containing phagolysosomes, a bioinformatics method for integrating quantitative LC-MS/MS data into the context of the entire protein-protein interaction network to identify significantly regulated processes and confirms previous findings about the A. fumigatus immune evasion strategy, e.g., inhibition of vATPase-dependent phagolysosomal acidification and the prevention of apoptosis induction. Intriguingly, our data suggest an interference of wt conidia with endocytic vesicle trafficking, MAPK and mTOR signaling, ROS production via NADPH oxidase, and degradative functions of the phagolysosome. Further, the reprogramming of energy metabolism and activation of macrophage immune responses, has not been reported before. These findings lay the basis for mechanistic studies that will help to unravel the complexity of immune evasion strategies of A. fumigatus.
DATA AVAILABILITY
The mass spectrometry proteomics data were deposited at the ProteomeXchange Consortium via the PRIDE (28) partner repository with the data set identifiers PXD005724 (https://www.ebi.ac.uk/pride/archive/projects/PXD005724) and PXD006134 (https://www.ebi.ac.uk/pride/archive/projects/PXD006134).
Supplementary Material
Acknowledgments
We thank Andreas Thywissen for initial experiments and Silke Steinbach for excellent technical assistance.
Footnotes
* This work was supported by the excellence graduate school Jena School for Microbial Communication JSMC and the CRC/Transregio 124 Human-pathogenic fungi and their human host - Networks of interaction - FungiNet, funded by the Deutsche Forschungsgemeinschaft (projects A1, B2 and Z2) as well as the InfectControl-project FINAR funded by the German Ministry for Education and Research (BMBF) and the Leibniz Science Campus InfectoOptics (project HotAim).
 This article contains supplemental material.
This article contains supplemental material.
Hella Schmidt, hella.schmidt@leibniz-hki.de; Sebastian Vlaic, sebastian.vlaic@leibniz-hki.de; Thomas Krüger, thomas.krueger@leibniz-hki.de; Franziska Schmidt, franziska.schmidt@leibniz-hki.de; Johannes Balkenhol, johannes.balkenhol@uni-wuerzburg.de; Thomas Dandekar, dandekar@biozentrum.uni-wuerzburg.de; Reinhard Guthke, reinhard.guthke@leibniz-hki.de; Olaf Kniemeyer, olaf.kniemeyer@leibniz-hki.de; Thorsten Heinekamp, thorsten.heinekamp@leibniz-hki.de.
1 The abbreviations used are:
- vATPase
- vacuolar ATPase
- 1,8-DHN
- 1,8-dihydroxynaphthalene
- A. fumigatus
- Aspergillus fumigatus
- AGC
- automatic gain control
- AMM
- Aspergillus minimal medium
- Atg
- autophagy-related genes
- CD
- cluster of differentiation
- CFW
- calcofluor white
- CoA
- coenzyme A
- CSFR
- colony-stimulating factor receptor
- DMEM
- Dulbecco's modified eagle medium
- EEA
- early endosomal antigen
- FBS
- fetal bovine serum
- FcR
- fragment crystallizable receptor
- FDR
- false discovery rate
- FWHM
- full width of half maximum
- GAP
- GTPase-activating protein
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GEF
- guanine nucleotide exchange factor
- GO
- gene ontology
- HCD
- higher-energy collisional dissociation
- HPI
- host-pathogen interaction
- HRP
- horse-reddish peroxidase
- Hsp
- heat shock protein
- IgG
- immunoglobulin G
- KEGG
- Kyoto encyclopedia of genes and genomes
- Lamp
- lysosome-associated membrane protein
- LAP
- LC3-associated phagocytosis
- LC3
- microtubule-associated protein 1 light chain 3
- MAPK
- mitogen-activated protein kinase
- MOI
- multiplicity of infection
- mRNA
- messenger ribonucleic acid
- mTOR
- molecular target of rapamycin
- Nduf
- NADH dehydrogenase
- NOX
- NADPH oxidase
- OXPHOS
- oxidative phosphorylation
- PD
- Proteome Discoverer
- PE
- phosphatidylethanolamine
- PI3K
- phosphoinositide-3-kinase
- pks
- polyketide synthase
- PMN
- polymorphonuclear
- PPI
- protein-protein interaction
- PSM
- peptide spectrum matches
- ROS
- reactive oxygen species
- Rubicon
- Run domain beclin-1-interacting and cysteine-rich domain-containing protein
- SNAP
- synaptosomal-associated protein
- SNARE
- soluble N-ethylmaleimide-sensitive-factor attachment receptor
- TCEP
- tris-(2-carboxyethyl)-phosphine
- TEAB
- tetraethylammonium bicarbonate
- Tef
- translation elongation factor
- TLR
- toll-like receptor
- Vamp
- vesicle-associated membrane protein
- Vti
- vesicle transport through interaction with t-SNAREs
- wt
- wild type.
REFERENCES
- 1. Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., and White T. C. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. [DOI] [PubMed] [Google Scholar]
- 2. Kontoyiannis D. P., Marr K. A., Park B. J., Alexander B. D., Anaissie E. J., Walsh T. J., Ito J., Andes D. R., Baddley J. W., Brown J. M., Brumble L. M., Freifeld A. G., Hadley S., Herwaldt L. A., Kauffman C. A., Knapp K., Lyon G. M., Morrison V. A., Papanicolaou G., Patterson T. F., Perl T. M., Schuster M. G., Walker R., Wannemuehler K. A., Wingard J. R., Chiller T. M., and Pappas P. G. (2010) Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 50, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 3. Heinekamp T., Schmidt H., Lapp K., Pahtz V., Shopova I., Koster-Eiserfunke N., Kruger T., Kniemeyer O., and Brakhage A. A. (2015) Interference of Aspergillus fumigatus with the immune response. Semin. Immunopathol. 37, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dagenais T. R., and Keller N. P. (2009) Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22, 447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park S. J., and Mehrad B. (2009) Innate immunity to Aspergillus species. Clin. Microbiol. Rev. 22, 535–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thi E. P., Lambertz U., and Reiner N. E. (2012) Sleeping with the enemy: how intracellular pathogens cope with a macrophage lifestyle. PLoS Pathog. 8, e1002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koul A., Herget T., Klebl B., and Ullrich A. (2004) Interplay between Mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2, 189–202 [DOI] [PubMed] [Google Scholar]
- 8. Meresse S., Steele-Mortimer O., Moreno E., Desjardins M., Finlay B., and Gorvel J. P. (1999) Controlling the maturation of pathogen-containing vacuoles: a matter of life and death. Nat. Cell. Biol. 1, E183–E188 [DOI] [PubMed] [Google Scholar]
- 9. Heinekamp T., Thywissen A., Macheleidt J., Keller S., Valiante V., and Brakhage A. A. (2012) Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 3, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slesiona S., Gressler M., Mihlan M., Zaehle C., Schaller M., Barz D., Hube B., Jacobsen I. D., and Brock M. (2012) Persistence versus escape: Aspergillus terreus and Aspergillus fumigatus employ different strategies during interactions with macrophages. PLoS ONE 7, e31223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jahn B., Koch A., Schmidt A., Wanner G., Gehringer H., Bhakdi S., and Brakhage A. A. (1997) Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect. Immun. 65, 5110–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langfelder K., Jahn B., Gehringer H., Schmidt A., Wanner G., and Brakhage A. A. (1998) Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187, 79–89 [DOI] [PubMed] [Google Scholar]
- 13. Jahn B., Langfelder K., Schneider U., Schindel C., and Brakhage A. A. (2002) PKSP-dependent reduction of phagolysosome fusion and intracellular kill of Aspergillus fumigatus conidia by human monocyte-derived macrophages. Cell Microbiol. 4, 793–803 [DOI] [PubMed] [Google Scholar]
- 14. Thywissen A., Heinekamp T., Dahse H. M., Schmaler-Ripcke J., Nietzsche S., Zipfel P. F., and Brakhage A. A. (2011) Conidial dihydroxynaphthalene melanin of the human pathogenic fungus Aspergillus fumigatus interferes with the host endocytosis pathway. Front. Microbiol. 2, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goyette G., Boulais J., Carruthers N. J., Landry C. R., Jutras I., Duclos S., Dermine J. F., Michnick S. W., LaBoissiere S., Lajoie G., Barreiro L., Thibault P., and Desjardins M. (2012) Proteomic characterization of phagosomal membrane microdomains during phagolysosome biogenesis and evolution. Mol. Cell. Proteomics 11, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rogers L. D., and Foster L. J. (2007) The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 104, 18520–18525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dill B. D., Gierlinski M., Hartlova A., Arandilla A. G., Guo M., Clarke R. G., and Trost M. (2015) Quantitative proteome analysis of temporally resolved phagosomes following uptake via key phagocytic receptors. Mol. Cell. Proteomics 14, 1334–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee B. Y., Jethwaney D., Schilling B., Clemens D. L., Gibson B. W., and Horwitz M. A. (2010) The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol. Cell. Proteomics 9, 32–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trost M., English L., Lemieux S., Courcelles M., Desjardins M., and Thibault P. (2009) The phagosomal proteome in interferon-γ-activated macrophages. Immunity 30, 143–154 [DOI] [PubMed] [Google Scholar]
- 20. Shui W., Gilmore S. A., Sheu L., Liu J., Keasling J. D., and Bertozzi C. R. (2009) Quantitative proteomic profiling of host-pathogen interactions: the macrophage response to Mycobacterium tuberculosis lipids. J. Proteome Res. 8, 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shui W., Sheu L., Liu J., Smart B., Petzold C. J., Hsieh T. Y., Pitcher A., Keasling J. D., and Bertozzi C. R. (2008) Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl. Acad. Sci. U.S.A. 105, 16952–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shui W., Petzold C. J., Redding A., Liu J., Pitcher A., Sheu L., Hsieh T., Keasling J. D., and Bertozzi C. R. (2011) Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J. Proteome Res. 339, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo M., Hartlova A., Dill B. D., Prescott A. R., Gierlinski M., and Trost M. (2015) High-resolution quantitative proteome analysis reveals substantial differences between phagosomes of RAW 264.7 and bone marrow derived macrophages. Proteomics 15, 3169–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steinhauser C., Heigl U., Tchikov V., Schwarz J., Gutsmann T., Seeger K., Brandenburg J., Fritsch J., Schroeder J., Wiesmuller K. H., Rosenkrands I., Walther P., Pott J., Krause E., Ehlers S., Schneider-Brachert W., Schutze S., and Reiling N. (2013) Lipid-labeling facilitates a novel magnetic isolation procedure to characterize pathogen-containing phagosomes. Traffic 14, 321–336 [DOI] [PubMed] [Google Scholar]
- 25. Wessel D., and Flugge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 26. Faden F., Eschen-Lippold L., and Dissmeyer N. (2016) Normalized quantitative Western blotting based on standardized fluorescent labeling. Methods Mol. Biol. 1450, 247–258 [DOI] [PubMed] [Google Scholar]
- 27. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 28. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vlaic S., Conrad T., Tokarski-Schnelle C., Gustafsson M., Dahmen U., Guthke R., and Schuster S. (2018) ModuleDiscoverer: Identification of regulatory modules in protein-protein interaction networks. Sci. Rep. 8, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barrenas F., Chavali S., Alves A. C., Coin L., Jarvelin M. R., Jornsten R., Langston M. A., Ramasamy A., Rogers G., Wang H., and Benson M. (2012) Highly interconnected genes in disease-specific networks are enriched for disease-associated polymorphisms. Genome Biol. 13, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Falcon S., and Gentleman R. (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 [DOI] [PubMed] [Google Scholar]
- 32.Carlson M. org.Mm.eg.db: Genome wide annotation for Mouse. R package version 3.2.3, https://bioconductor.org/
- 33. Kinsella R. J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., and Flicek P. (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database Vol. 2011, Article ID bar030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durinck S., Spellman P. T., Birney E., and Huber W. (2009) Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Remmele C. W., Luther C. H., Balkenhol J., Dandekar T., Muller T., and Dittrich M. T. (2015) Integrated inference and evaluation of host-fungi interaction networks. Front. Microbiol. 6, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Desjardins M., Huber L. A., Parton R. G., and Griffiths G. (1994) Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124, 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asif A. R., Oellerich M., Amstrong V. W., Riemenschneider B., Monod M., and Reichard U. (2006) Proteome of conidial surface associated proteins of Aspergillus fumigatus reflecting potential vaccine candidates and allergens. J. Proteome Res. 5, 954–962 [DOI] [PubMed] [Google Scholar]
- 39. Wetzel M. G., and Korn E. D. (1969) Phagocytosis of latex beads by Acahamoeba castellanii (Neff). 3. Isolation of the phagocytic vesicles and their membranes. J. Cell Biol. 43, 90–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garin J., Diez R., Kieffer S., Dermine J. F., Duclos S., Gagnon E., Sadoul R., Rondeau C., and Desjardins M. (2001) The phagosome proteome: insight into phagosome functions. J. Cell Biol. 152, 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., and Green D. R. (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 [DOI] [PubMed] [Google Scholar]
- 42. West A. P., Brodsky I. E., Rahner C., Woo D. K., Erdjument-Bromage H., Tempst P., Walsh M. C., Choi Y., Shadel G. S., and Ghosh S. (2011) TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang C., and Youle R. J. (2009) The role of mitochondria in apoptosis. Annu. Rev. Genet. 43, 95–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haas A. (2007) The phagosome: compartment with a license to kill. Traffic 8, 311–330 [DOI] [PubMed] [Google Scholar]
- 45. Lafourcade C., Sobo K., Kieffer-Jaquinod S., Garin J., and van der Goot F. G. (2008) Regulation of the V-ATPase along the endocytic pathway occurs through reversible subunit association and membrane localization. PLoS ONE 3, e2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waters M. G., and Hughson F. M. (2000) Membrane tethering and fusion in the secretory and endocytic pathways. Traffic 1, 588–597 [DOI] [PubMed] [Google Scholar]
- 47. Christoforidis S., McBride H. M., Burgoyne R. D., and Zerial M. (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625 [DOI] [PubMed] [Google Scholar]
- 48. Simonsen A., Lippe R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., and Stenmark H. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
- 49. Brumell J. H., and Scidmore M. A. (2007) Manipulation of Rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71, 636–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Byles V., Covarrubias A. J., Ben-Sahra I., Lamming D. W., Sabatini D. M., Manning B. D., and Horng T. (2013) The TSC-mTOR pathway regulates macrophage polarization. Nat. Commun. 4, 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roca F. J., and Ramakrishnan L. (2013) TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153, 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chamilos G., Akoumianaki T., Kyrmizi I., Brakhage A. A., Beauvais A., and Latge J. P. (2016) Melanin targets LC3-associated phagocytosis (LAP): a novel pathogenetic mechanism in fungal disease. Autophagy 12, 888–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L. M., Li J., Vivona S., Pfuetzner R. A., Brunger A. T., and Zhong Q. (2015) ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., and Yue Z. (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boyle K. B., and Randow F. (2015) Rubicon swaps autophagy for LAP. Nat Cell Biol. 17, 843–845 [DOI] [PubMed] [Google Scholar]
- 56. Martinez J., Malireddi R. K., Lu Q., Dias Cunha L., Pelletier S., Gingras S., Orchard R., Guan J. L., Tan H., Peng J., Kanneganti T. D., Virgin H. W., and Green D. R. (2015) Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Akoumianaki T., Kyrmizi I., Valsecchi I., Gresnigt M. S., Samonis G., Drakos E., Boumpas D., Muszkieta L., Prevost M. C., Kontoyiannis D. P., Chavakis T., Netea M. G., van de Veerdonk F. L., Brakhage A. A., El-Benna J., Beauvais A., Latge J. P., and Chamilos G. (2016) Aspergillus cell wall melanin blocks LC3-associated phagocytosis to promote pathogenicity. Cell Host Microbe 19, 79–90 [DOI] [PubMed] [Google Scholar]
- 58. Volling K., Thywissen A., Brakhage A. A., and Saluz H. P. (2011) Phagocytosis of melanized Aspergillus conidia by macrophages exerts cytoprotective effects by sustained PI3K/Akt signalling. Cell Microbiol. 13, 1130–1148 [DOI] [PubMed] [Google Scholar]
- 59. Volling K., Brakhage A. A., and Saluz H. P. (2007) Apoptosis inhibition of alveolar macrophages upon interaction with conidia of Aspergillus fumigatus. FEMS Microbiol. Lett. 275, 250–254 [DOI] [PubMed] [Google Scholar]
- 60. Pardo J., Urban C., Galvez E. M., Ekert P. G., Muller U., Kwon-Chung J., Lobigs M., Mullbacher A., Wallich R., Borner C., and Simon M. M. (2006) The mitochondrial protein Bak is pivotal for gliotoxin-induced apoptosis and a critical host factor of Aspergillus fumigatus virulence in mice. J. Cell Biol. 174, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Philippe B., Ibrahim-Granet O., Prevost M. C., Gougerot-Pocidalo M. A., Sanchez Perez M., Van der Meeren A., and Latge J. P. (2003) Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 71, 3034–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grimm M. J., Vethanayagam R. R., Almyroudis N. G., Dennis C. G., Khan A. N., D'Auria A. C., Singel K. L., Davidson B. A., Knight P. R., Blackwell T. S., Hohl T. M., Mansour M. K., Vyas J. M., Rohm M., Urban C. F., Kelkka T., Holmdahl R., and Segal B. H. (2013) Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice. J. Immunol. 190, 4175–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jahn B., Boukhallouk F., Lotz J., Langfelder K., Wanner G., and Brakhage A. A. (2000) Interaction of human phagocytes with pigmentless Aspergillus conidia. Infect. Immun. 68, 3736–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang J., Canadien V., Lam G. Y., Steinberg B. E., Dinauer M. C., Magalhaes M. A., Glogauer M., Grinstein S., and Brumell J. H. (2009) Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. U.S.A. 106, 6226–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jacobson M. D. (1996) Reactive oxygen species and programmed cell death. Trends Biochem. Sci. 21, 83–86 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data were deposited at the ProteomeXchange Consortium via the PRIDE (28) partner repository with the data set identifiers PXD005724 (https://www.ebi.ac.uk/pride/archive/projects/PXD005724) and PXD006134 (https://www.ebi.ac.uk/pride/archive/projects/PXD006134).