Abstract
In higher plants, two nuclear gene families, FtsZ1 and FtsZ2, encode homologs of the bacterial protein FtsZ, a key component of the prokaryotic cell division machinery. We previously demonstrated that members of both gene families are essential for plastid division, but are functionally distinct. To further explore differences between FtsZ1 and FtsZ2 proteins we investigated the phenotypes of transgenic plants overexpressing AtFtsZ1-1 or AtFtsZ2-1, Arabidopsis members of the FtsZ1 and FtsZ2 families, respectively. Increasing the level of AtFtsZ1-1 protein as little as 3-fold inhibited chloroplast division. Plants with the most severe plastid division defects had 13- to 26-fold increases in AtFtsZ1-1 levels over wild type, and some of these also exhibited a novel chloroplast morphology. Quantitative immunoblotting revealed a correlation between the degree of plastid division inhibition and the extent to which the AtFtsZ1-1 protein level was elevated. In contrast, expression of an AtFtsZ2-1 sense transgene had no obvious effect on plastid division or morphology, though AtFtsZ2-1 protein levels were elevated only slightly over wild-type levels. This may indicate that AtFtsZ2-1 accumulation is more tightly regulated than that of AtFtsZ1-1. Plants expressing the AtFtsZ2-1 transgene did accumulate a form of the protein smaller than those detected in wild-type plants. AtFtsZ2-1 levels were unaffected by increased or decreased accumulation of AtFtsZ1-1 and vice versa, suggesting that the levels of these two plastid division proteins are regulated independently. Taken together, our results provide additional evidence for the functional divergence of the FtsZ1 and FtsZ2 plant gene families.
The first identified proteins of the chloroplast division machinery were homologs of the essential bacterial cell division protein FtsZ (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998). In contrast with most bacterial genomes that contain only a single gene encoding FtsZ, the nuclear genome of Arabidopsis contains at least three FtsZ genes encoding members of two distinct protein families, FtsZ1 and FtsZ2. AtFtsZ1-1 and AtFtsZ2-1, members of the FtsZ1 and FtsZ2 families, respectively, have been shown to be essential for chloroplast division. When the level of either gene is diminished, chloroplast division is inhibited, yielding cells with as few as one very large chloroplast. One important difference between AtFtsZ1-1 and AtFtsZ2-1 is their predicted localization. AtFtsZ1-1 is targeted to the chloroplast, as is a closely related FtsZ protein from pea (Gaikwad et al., 2000), and is thought to be a component of a division ring that forms on the stromal side of the inner envelope membrane. AtFtsZ2-1, in contrast, lacks a chloroplast transit peptide, is not targeted to the chloroplast in vitro, and is hypothesized to be a constituent of a division ring that assembles on the cytoplasmic surface of the outer envelope membrane. The two rings together are postulated to coordinate chloroplast division (Osteryoung et al., 1998).
Bacterial FtsZ is a self-associating cytoskeletal GTPase evolutionarily and structurally related to the eukaryotic tubulins (de Boer et al., 1992; RayChaudhuri and Park, 1992; Mukherjee and Lutkenhaus, 1994; Erickson, 1997; Yu and Margolin, 1997; Löwe and Amos, 1998; Nogales et al., 1998a, 1998b). Early in the bacterial division cycle, prior to the onset of cytokinesis, FtsZ assembles into a ring at the division plane that encircles the cell on the inner surface of the cytoplasmic membrane. At least eight other essential proteins are then recruited to the division site and function to complete cytokinesis. Throughout this process, the FtsZ ring remains localized at the leading edge of the division septum (Bi and Lutkenhaus, 1991; Bramhill, 1997; Lutkenhaus and Addinall, 1997; Pogliano et al., 1997; Rothfield and Justice, 1997; Nanninga, 1998; Rothfield et al., 1999).
Overexpression of FtsZ in bacteria has yielded important insights into the properties of this protein. First, FtsZ appears to be rate limiting in the division process since slight overproduction of FtsZ increases cell division (Ward Jr. and Lutkenhaus, 1985). This increased division results in the formation of small, inviable “minicells” that lack chromosomes due to the occurrence of divisions not only at midcell, but also near the cell poles. Second, high overexpression of FtsZ inhibits cell division and results in the formation of bacterial filaments (Ward Jr. and Lutkenhaus, 1985). This appears to be due in part to a stoichiometric imbalance between FtsZ and other division proteins because the filamentation phenotype can be relieved by simultaneous overexpression of FtsA or ZipA, two other bacterial cell division proteins that interact with FtsZ directly and colocalize with FtsZ to the midcell (Ward Jr. and Lutkenhaus, 1985; Wang and Gayda, 1990; Dai and Lutkenhaus, 1992; Hale and de Boer, 1997, 1999; Wang et al., 1997; Mosyak et al., 2000).
Although chloroplast division involves some proteins homologous to components of the bacterial division machinery, division of higher plant chloroplasts differs from bacterial cell division since it requires the coordinated activities of at least two FtsZ proteins and does not involve cell wall ingrowth at the division site (Osteryoung and Pyke, 1998; Osteryoung et al., 1998). In the studies described here we sought to further explore the functional differences between FtsZ1 and FtsZ2 proteins by analyzing the phenotypes of Arabidopsis plants overexpressing AtFtsZ1-1 or AtFtsZ2-1. The results reveal additional parallels between chloroplast and bacterial cell division with regard to the behavior of FtsZ, but support a difference in the roles played by FtsZ1 and FtsZ2 in chloroplast division. The data also suggest that the levels of AtFtsZ1-1 and AtFtsZ2-1 are regulated independently in Arabidopsis, and that FtsZ1 may have an additional function inside the organelle in regulating chloroplast morphology.
RESULTS
Production of Antibodies Specific for Recognition of AtFtsZ1-1 or AtFtsZ2-1
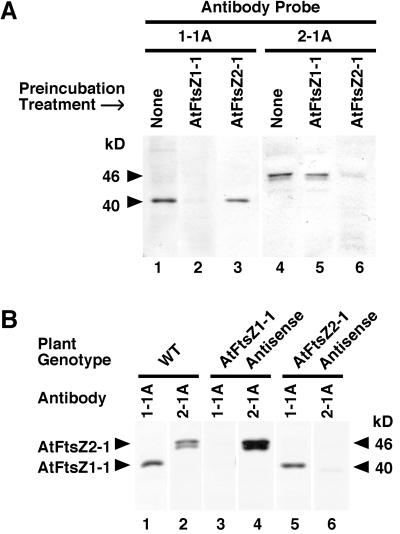
In preparation for investigating AtFtsZ1-1 and AtFtsZ2-1 protein levels in this and other studies we produced antipeptide antibodies specific for detection of these two polypeptides on immunoblots. The specificities and reactivities of the affinity-purified antibodies, designated 1-1A and 2-1A, respectively, were analyzed in a series of immunoblotting and competition binding assays (Fig. 1). Each antibody was highly selective for its target protein. In Arabidopsis leaf extracts the l-1A antibodies reacted with a single polypeptide of 40 kD (Fig. 1, A, lanes 1 and 3 and B, lanes 1 and 5), whereas the 2-1A antibodies reacted primarily with a polypeptide of 46 kD, though one of 45 kD was also detected (Fig. 1, A, lanes 4 and 5 and B, lanes 2 and 4). In competition binding assays, immunoreactivity of the 1-1A antibody with the 40-kD protein was prevented when the antibodies were preincubated with recombinant AtFtsZ1-1 protein (Fig. 1A, lane 2), but not with recombinant AtFtsZ2-1 protein (Fig. 1A, lane 3). Likewise, immunoreactivity of the 2-1A antibodies with the 46- and 45-kD polypeptides was blocked when the antibodies were preincubated with recombinant AtFtsZ2-1 protein (Fig. 1A, lane 6), but not with AtFtsZ1-1 protein (Fig. 1A, lane 5). Furthermore, the 1-1A antibodies detected the 40-kD polypeptide in extracts from wild-type plants and transgenic plants expressing an AtFtsZ2-1 antisense construct (Fig. 1B, lanes 1 and 5), but not in plants expressing an antisense AtFtsZ1-1 construct (Fig. 1B, lane 3). Conversely, the 2-1A antibodies detected the 46- and 45-kD polypeptides in extracts from wild-type plants and transgenic plants expressing an AtFtsZ1-1 antisense construct (Fig. 1B, lanes 2 and 4), but not in plants expressing an antisense AtFtsZ2-1 construct (Fig. 1B, lane 6). Neither antibody cross-reacted with prokaryotic FtsZ proteins in bacterial extracts (not shown). From these results, we conclude that the 1-1A antibodies specifically recognize AtFtsZ1-1, which migrates at 40 kD, whereas the 2-1A antibodies specifically recognize either two distinct forms of AtFtsZ2-1, which migrate at 46 and 45 kD, or AtFtsZ2-1 and a closely related polypeptide. The absence of both bands in the AtFtsZ2-1 antisense lines (Fig. 1B, lane 6) is most consistent with the former possibility.
Figure 1.
Specificity of AtFtsZ antipeptide antibodies. A, Immunoblots of proteins isolated from wild-type Arabidopsis leaf extracts were probed with 1-1A (lanes 1–3) or 2-1A (lanes 4–6) antibodies raised against peptide from AtFtsZ1-1 or AtFtsZ2-1, respectively. Antibodies were preincubated with purified, recombinant AtFtsZ1-1 protein (lanes 2 and 5) or AtFtsZ2-1 protein (lanes 3 and 6). B, Immunoblot of proteins isolated from leaf extracts of wild-type (lanes 1 and 2), AtFtsZ1-1 antisense (lanes 3 and 4), or AtFtsZ2-1 antisense (lanes 5 and 6) plants. Blots were probed with either 1-1A (lanes 1, 3, and 5) or 2-1A (lanes 2, 4, and 6) antibodies. The 46- and 40-kD polypeptides are indicated by markers. Equivalent volumes of plant extracts were loaded in each lane.
Overproduction of AtFtsZ1-1 Inhibits Chloroplast Division in Transgenic Arabidopsis
AtFtsZ1-1 was overexpressed in Arabidopsis under the control of the cauliflower mosaic virus 35S (35S) promoter in the vector pART27 (Gleave, 1992). The kanamycin-resistant (kanr) transgenic plant lines used in this report were confirmed to be independent transformants by Southern-blot analysis (data not shown). With the exception of a slight twist in some of the leaves, all kanr plants exhibited normal growth and development. However, microscopic examination of mesophyll cells revealed distinct phenotypes in plants overexpressing AtFtsZ1-1 when compared with wild-type plants of the Columbia ecotype, which typically contain 80 to 100 chloroplasts in fully expanded mesophyll cells (Osteryoung et al., 1998), or to control plants transformed with the empty pART27 vector (Fig. 2A). The phenotypes of the transgenic plants consistently fell into three categories defined by chloroplast number and size. Transgenic plants were classified as “wild-type-like” if the size and number of chloroplasts in mesophyll cells were similar to those in cells from wild-type or vector controls. Plants with 15 to 30 enlarged chloroplasts per cell were classified as “intermediate” (Fig. 2B), and those with five or fewer very large chloroplasts per cell were termed “severe” (Fig. 2C). Most plants with severe phenotypes contained only one or two chloroplasts per cell and these organelles usually appeared flattened, filling the thin layer of cytoplasm surrounding the vacuole. Despite the enlarged size, chloroplast ultrastructure consistently appeared normal (data not shown). Similar “severe” phenotypes have been described for the Arabidopsis plastid division mutant arc6 (Pyke and Leech, 1994), for plants transformed with antisense AtFtsZ1-1 or AtFtsZ2-1 transgenes (Osteryoung et al., 1998), and for FtsZ knockout mutants in the moss Physcomitrella patens (Strepp et al., 1998). The proportion of T1 individuals exhibiting wild-type-like, intermediate, and severe phenotypes were 19%, 39%, and 42%, respectively (Table I). Therefore, over 80% of the kanr plants expressing the AtFtsZ1-1 sense transgene displayed defects in chloroplast division.
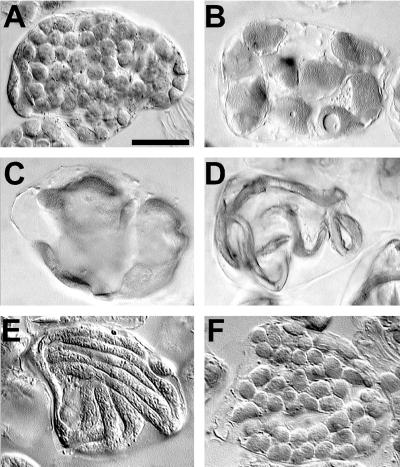
Figure 2.
Phenotypes of transgenic plants overexpressing AtFtsZ1-1 or AtFtsZ2-1. Mesophyll cells are shown from the first leaves of 23-d-old plants transformed with the empty pART27 vector (A), the AtFtsZ1-1 sense transgene (B–E), or the AtFtsZ2-1 sense transgene (F). Tissue was prepared for imaging with differential interference contrast optics using methods described previously (Pyke and Leech, 1991). Bar = 25 μm in all figures. A three-dimensional rotating reconstruction of cells with phenotypes similar to those shown in C and D can be found in a video supplement at www.plantphysiol.org.
Table I.
Phenotypic distribution of transgenic T1 plants
| Phenotype | Percentage of Transgenic
Plantsa
|
|
|---|---|---|
| AtFtsZ1-1 | AtFtsZ2-1 | |
| Wild type-like | 19 | 72 |
| Intermediate | 39 | 14 |
| Severe | 42 | 14 |
Total no. of plants analyzed: AtFtsZ1-1, 129; AtFtsZ2-1, 127.
The Severity of Chloroplast Division Inhibition Is Proportional to AtFtsZ1-1 Protein Level
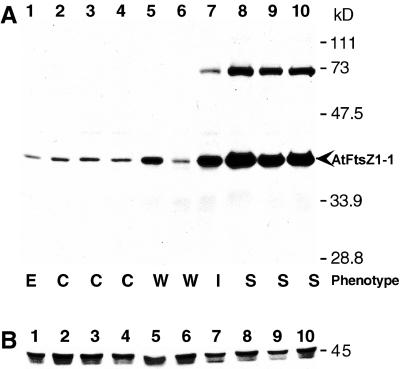
AtFtsZ1-1 protein levels in T3 kanr plants representing wild-type-like, intermediate, and severe phenotypes were investigated by immunoblot analysis. Proteins in leaf homogenates were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the affinity-purified AtFtsZ1-1 antipeptide antibodies. An immunoreactive 40-kD polypeptide was detected that varied in amount among different transgenic lines (Fig. 3A, lanes 5–10), but comigrated with authentic AtFtsZ1-1 from wild-type and empty-vector control extracts (Fig. 3A, lanes 1–4). These results indicate that most of the AtFtsZ1-1 protein in the overexpression lines was properly targeted to the chloroplast and processed. However, a slower-migrating immunoreactive polypeptide of 72 kD was also detected in plants with increased levels of the 40-kD polypeptide (Fig. 3A, lanes 7–10). Competition binding assays have shown that this polypeptide does contain AtFtsZ1-1 (not shown), and may represent a non-dissociated form of the protein, possibly a dimer. A similarly migrating polypeptide is occasionally observed in wild-type plants as well (data not shown). The increased levels of the 72-kD polypeptide in overexpression lines with high AtFtsZ1-1 levels may be related to the ability of FtsZ proteins to form dimers and multimers in a concentration-dependent fashion (Di Lallo et al., 1999; Sossong et al., 1999; Rivas et al., 2000; White et al., 2000). An FtsZ1 homolog from pea has also been shown to form multimers in vitro (Gaikwad et al., 2000).
Figure 3.
Immunoblot analysis of plant extracts overexpressing AtFtsZ1-1. Proteins in extracts from 23-d-old transgenic plants were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antipeptide antibodies raised against AtFtsZ1-1 (A) or AtFtsZ2-1 (B). Lane 1, Empty vector control (E); lanes 2 through 4, wild type (C); lanes 5 through 10, transgenic plants with wild-type-like (W, lanes 5 and 6), intermediate (I, lane 7), or severe (S, lanes 8–10) phenotypes. Equal loading of all samples was confirmed by staining the membranes with ponceau S (data not shown).
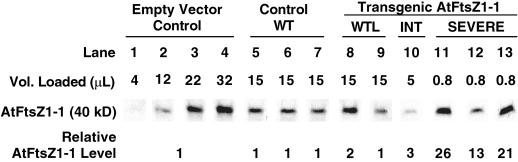
Visual inspection of immunoblots from the AtFtsZ1-1 overexpression lines suggested a correlation between the level of AtFtsZ1-1 accumulation and the severity of chloroplast division defect. In extracts from plants with wild-type-like (Fig. 3A, lanes 5 and 6) and intermediate (Fig. 3A, lane 7) phenotypes, the levels of the 40-kD AtFtsZ1-1 polypeptide were similar to, or slightly higher than, those seen in extracts from the vector controls (Fig. 3A, lane 1) and non-transformed wild-type plants (Fig. 3A, lanes 2–4), whereas in plants with severe phenotypes (Fig. 3A, lanes 8–10), AtFtsZ1-1 protein levels were noticeably elevated. To further analyze this relationship, the level of AtFtsZ1-1 in the transgenic plants relative to that in control plants was quantified by immunoblotting (Fig. 4). For this purpose, a calibration curve was constructed from densitometric analysis of the 40-kD polypeptide in four lanes loaded with increasing volumes of whole leaf extract from control plants transformed with the empty vector (Fig. 4, lanes 1–4). For transgenic extracts, the volumes analyzed were adjusted to maintain protein levels within the linear range of the standard curve (Fig. 4, lanes 5–13). The relative level of the 40-kD AtFtsZ1-1 polypeptide in each sample was then calculated from the standard curve based on the densitometry readings and sample volume. The calculated protein levels in extracts from three separate non-transformed wild-type plants (Fig. 4, lanes 5–7) were comparable with those from empty vector control plants, indicating that the increased protein levels observed in the transgenic plants were due to expression of the AtFtsZ1-1 transgene and not to the vector. In transgenic plants classified as wild-type-like, the relative level of AtFtsZ1-1 protein was similar to the control level or elevated no more than 2-fold (Fig. 4, lanes 8 and 9). Protein levels between 3-fold (Fig. 4, lane 10) and 6-fold (not shown) above control levels were measured in plants with an intermediate chloroplast phenotype. Although data from only a single intermediate line are shown in Figures 3 and 4, two additional lines with intermediate phenotypes also had approximately 3-fold more AtFtsZ1-1 than controls, whereas one line had 6-fold more (not shown). Plants exhibiting the most severe phenotypes had AtFtsZ1-1 levels ranging from 13- to 26-fold over control levels (Figs. 3A, lanes 8–10, and 4, lanes 11–13). Our data indicate a correlation between the level of AtFtsZ1-1 and the severity of chloroplast division inhibition. However, we cannot rule out the possibility that the observed division defects resulted from accumulation of the 72-kD form of AtFtsZ1-1 rather than from overproduction of the protein per se. This 72-kD band was not quantified, but its levels in the transgenic lines appeared to correlate with those of the 40-kD polypeptide (Fig. 3, lanes 7–10, and data not shown).
Figure 4.
Relative levels of the 40-kD AtFtsZ1-1 polypeptide in plants expressing the AtFtsZ1-1 transgene. Densitometry readings from an immunoblot loaded with increasing amounts of extract from an empty vector control plant (lanes 1–4) were used to construct a standard concentration curve for AtFtsZ1-1. AtFtsZ1-1 levels in the other plant extracts (lanes 5–13), all loaded so that the densitometry reading produced by the 40-kD AtFtsZ1-1 polypeptide on immunoblots fell within the linear range of the standard curve, were then calculated, taking into account the volume loaded. The volume loaded, signal produced on immunoblots, and calculated level of AtFtsZ1-1 relative to that in the empty-vector controls are shown for three Columbia wild type plants (lanes 5–7), and transgenic plants with wild-type-like (WTL, lanes 8 and 9), intermediate (INT, lane 10), or severe (SEVERE, lanes 11–13) phenotypes.
AtFtsZ1-1 Overexpression Produces a Novel Chloroplast Morphology in Some Transgenic Plants
In addition to the phenotypes described above, an interesting and unusual phenotype was encountered in three independent AtFtsZ1-1 overexpression lines with severe phenotypes. A small number of cells in these plants contained chloroplasts that appeared long and narrow, giving the impression of worms or noodles (Fig. 2, D and E). Two of these plant lines had only a few noodle-like chloroplasts per cell (Fig. 2D), whereas one line had about 15 (Fig. 2E). The diameter of these chloroplasts varied somewhat, but was comparable with that of wild-type chloroplasts. The length, however, was many times longer than in wild type, and the chloroplasts meandered in unique patterns around the cytoplasm of the cell. Only a small proportion of the cells in these plants displayed the noodle-like phenotype; the vast majority of cells exhibited a typically severe chloroplast morphology. The noodle-shaped chloroplasts were only observed in transgenic plants with high levels of AtFtsZ1-1 protein, including those represented in lanes 8 through 10 of Figure 3, but have not been found in all such lines.
Slight Overexpression of AtFtsZ2-1 Does Not Disrupt Chloroplast Division
The effect of AtFtsZ2-1 overexpression in transgenic plants was also investigated. The T1 generation of kanr plants transformed with the AtFtsZ2-1 transgene contained cells with wild-type-like, intermediate, and severe chloroplast phenotypes, similar to those described for the AtFtsZ1-1 transgenic plants, and had no obvious abnormalities in growth or development. However, in contrast to the AtFtsZ1-1 overexpression lines, 72% of the AtFtsZ2-1 kanr T1 individuals exhibited a wild-type-like phenotype (Fig. 2F), whereas plants with intermediate and severe phenotypes each constituted only 14% of the total (Table I). Furthermore, in subsequent generations the lines with intermediate and severe phenotypes reverted to the wild-type-like phenotype. The extent of this reversion was such that only one transgenic line retained reduced chloroplast numbers by the T3 generation. Because of this trend, T1 and T3 plants were studied further.
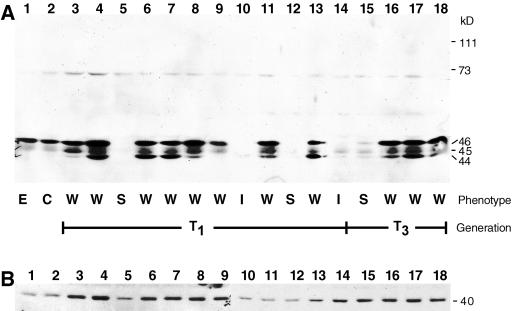
AtFtsZ2-1 protein levels were analyzed in whole-leaf extracts from T1 and T3 plants by immunoblotting. In extracts from transgenic plants exhibiting wild-type-like phenotypes (Fig. 5A, lanes 2, 3, 6–9, 11, 13, and 16–18), the same 46- and 45-kD polypeptides present in wild-type and empty-vector control plants (Fig. 5A, lanes 1 and 2) were detected. The levels of the 46-kD polypeptide in these lines were comparable with control levels, although some extracts exhibited a slight increase (Fig. 5A, lanes 4 and 8). We have observed that the intensity of the 45-kD band varies considerably among individuals in wild type (not shown), and the levels of the 45-kD protein in the transgenic plants did not appear to vary outside this range. The more obvious result of AtFtsZ2-1 overexpression was the accumulation of a 44-kD polypeptide, detected in all kanr plants with a wild-type-like phenotype, but not detected in any of the controls. Accumulation of this polypeptide was not correlated with any noticeable plastid division defect. In contrast, all transgenic lines with intermediate or severe phenotypes (Fig. 5A, lanes 5, 10, 12, and 14), including the line retaining a severe phenotype into the T3 generation (lane 15), had reduced levels of the 46- and 45-kD species present in controls and did not accumulate the 44-kD polypeptide present in lines with the wild-type-like phenotypes. These data suggest that inhibition of chloroplast division in the AtFtsZ2-1 transgenic plants resulted from cosuppression rather than overexpression of AtFtsZ2-1.
Figure 5.
Immunoblot analysis of transgenic plant extracts expressing the AtFtsZ2-1 transgene. Proteins in extracts from 23-d-old transgenic plants were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antipeptide antibodies raised against AtFtsZ2-1 (A) or AtFtsZ1-1 (B). Lane 1, Empty vector control (E); lane 2, wild type (C); lanes 3 through 18, T1 or T3 transgenic plants with wild-type-like (W, lanes, 3, 4, 6–9, 11, 13, and 16–18), intermediate (I, lanes 10 and 14), or severe (S, lanes 5 and 15) phenotypes. Equal loading of all samples was confirmed by staining the membranes with ponceau S (data not shown).
AtFtsZ1-1 and AtFtsZ2-1 Accumulation Are Regulated Independently of One Another
To learn whether overproduction of AtFtsZ1-1 was accompanied by a change in AtFtsZ2-1 levels or vice versa, the levels of both proteins in extracts from each set of transgenic plants were analyzed on duplicate immunoblots. In all AtFtsZ1-1 overexpression lines analyzed, AtFtsZ2-1 protein remained at wild-type levels (Fig. 3B). In a converse manner, the level of AtFtsZ1-1 protein in the AtFtsZ2-1 transgenic lines was unaffected by the level of AtFtsZ2-1 protein (Fig. 5B). Further, antisense repression of AtFtsZ1-1, though reducing AtFtsZ1-1 protein to nearly undetectable levels (Fig. 1B, lane 3) and severely inhibiting chloroplast division (Osteryoung et al., 1998), had no affect on accumulation of AtFtsZ2-1 (Fig. 1B, lane 4) or vice versa (Fig. 1B, lanes 5 and 6). Therefore, we conclude that the phenotypes associated with manipulation of AtFtsZ1-1 or AtFtsZ2-1 expression levels, whether from an increase or decrease, result from altered accumulation of only one and not both proteins. In addition, although AtFtsZ1-1 and AtFtsZ2-1 are co-expressed in wild-type plants (Fig. 1B, lanes 1 and 2; Osteryoung et al., 1998), the collective results of overexpression and antisense experiments suggest that FtsZ1 and FtsZ2 protein levels are regulated independently in Arabidopsis.
DISCUSSION
Correlation between AtFtsZ1-1 Accumulation and Plastid Number
Plants producing AtFtsZ1-1 at levels ranging from 13- to as high as 26-fold over wild-type levels exhibited drastically reduced numbers of enlarged chloroplasts, indicating a severe inhibition of chloroplast division. This phenotype is comparable with the filamentation phenotype observed in Escherichia coli cells expressing FtsZ at high levels. However, when bacterial FtsZ levels were only slightly elevated, extra divisions were induced near the cell poles, resulting in the formation of minicells. Furthermore, when levels of overexpression were below those resulting in filamentation, the minicell phenotype was proportional to the degree of FtsZ overexpression (Ward Jr. and Lutkenhaus, 1985). Based on these data we anticipated that slightly elevated AtFtsZ1-1 protein levels might increase the frequency of chloroplast division, yielding plants with cells containing smaller, more numerous chloroplasts. Instead, AtFtsZ1-1 levels as little as 3-fold over wild-type levels inhibited, rather than increased, chloroplast division. However, we have observed a few transgenic lines with more than 150 tiny chloroplasts in mesophyll cells (data not shown), but these plants were chlorotic and died as seedlings, preventing further analysis of their phenotypes or AtFtsZ1-1 expression levels. Nevertheless, these observations suggest that elevated AtFtsZ1-1 levels could, under a limited set of circumstances, increase the frequency of chloroplast division. Based on the observation that plants with 2-fold more AtFtsZ1-1 than wild type had wild-type numbers of chloroplasts, whereas plants with 3-fold more protein had intermediate numbers, indicating partial inhibition of division, we would predict that AtFtsZ1-1 accumulation only within this narrow range would lead to increased numbers of chloroplasts, and that levels above this are inhibitory for plastid division. This idea is consistent with the behavior of FtsZ in bacteria and with the possibility that the plastid division defect resulting from AtFtsZ1-1 overproduction reflects a stoichiometric imbalance in plastid division components. It is possible that the use of a weak promoter instead of the strong 35S promoter might allow identification of more plants with only slightly elevated levels of AtFtsZ1-1, and perhaps increased chloroplast numbers, for further study. It is also possible that simultaneous overexpression of additional chloroplast division proteins (including AtFtsZ2-1) might be required to permit an increase in the plastid division frequency. This is suggested by the finding that the increased frequency of cell divisions observed when FtsZ is overexpressed in E. coli only occurs when FtsA is also overexpressed at similar levels (Begg et al., 1998). Plant survival alternatively may be compromised by increased chloroplast numbers, which could account for the small number of plants identified with this phenotype. This conjecture is supported in part by the phenotype of the Arabidopsis arc1 chloroplast division mutant, which is characterized by slightly increased numbers of small chloroplasts (Pyke and Leech, 1992; Marrison et al., 1999). arc1 grows more slowly and is pale early in its development compared with either wild type or other arc mutants that have reduced numbers of enlarged chloroplasts.
Expression of the AtFtsZ2-1 Sense Transgene Does Not Produce a Plastid Division Phenotype
In contrast to the dramatic phenotypes associated with AtFtsZ1-1 overexpression, more than 70% of the plants transformed with the AtFtsZ2-1 sense transgene displayed a wild-type-like phenotype. In fact, immunoblotting results (Fig. 5A, lanes 5, 10, 12, 14, and 15) indicated that all plastid division defects observed among these transgenic lines were due to cosuppression of endogenous AtFtsZ2-1 expression rather than to overexpression, similar to the plastid division defects observed in plants expressing an AtFtsZ2-1 antisense transgene (Fig. 1B, lane 6; Osteryoung et al., 1998). However, authentic AtFtsZ2-1 protein did not accumulate substantially over wild-type levels in the overexpression experiments (Fig. 5A), which may indicate that higher levels are lethal or that AtFtsZ2-1 accumulation is more tightly regulated than that of AtFtsZ1-1. The only phenotype associated with AtFtsZ2-1 transgene expressioin (when it did not result in cosuppression) was the presence of a 44-kD form of AtFtsZ2-1 not detected in wild-type plants. Because this polypeptide was smaller than the one produced by in vitro translation of the predicted AtFtsZ2-1 open reading frame (Osteryoung et al., 1998; R.S. McAndrew, S. Vitha and K.W. Osteryoung, unpublished data), it may represent a degradation product or an aberrantly processed form of AtFtsZ2-1. Accumulation of this polypeptide at the observed levels had no effect on plastid division, however. Overall, the differences in the phenotypes of plants expressing AtFtsZ1-1 and AtFtsZ2-1 transgenes further support a difference in the functions of FtsZ1 and FtsZ2 proteins.
Aberrant Chloroplast Morphology Associated with High Levels of AtFtsZ1-1 Protein
The long, narrow chloroplasts observed in some of the AtFtsZ1-1 overexpression lines occurred only in transgenic lines exhibiting severe plastid division defects and high AtFtsZ1-1 protein levels. Although at present we cannot be certain of the biological relevance of this unique phenotype, it could suggest an additional role for chloroplast-localized FtsZ1 proteins in the control of chloroplast shape. Immunofluorescence data indicating the presence of longitudinally oriented AtFtsZ1-1-containing filaments in the noodle-shaped chloroplasts (S. Vitha and R. McAndrew, unpublished observations) are consistent with this idea. The assembly of many such filaments in plastids with high AtFtsZ1-1 levels could restrict their ability to expand laterally, allowing plastid expansion to occur only longitudinally to produce narrow, elongated chloroplasts. The noodle phenotype alternatively could reflect an abnormality in the formation of stromules, narrow tubular connections between plastids through which protein molecules can pass (Kohler et al., 1997). At present there is no direct evidence that plant FtsZ proteins participate in chloroplast shape determination or stromule biogenesis, but the notion that FtsZs function in multiple processes, like their tubulin structural homologs, is not incompatible with their cytoskeletal properties.
MATERIALS AND METHODS
Construction of Sense Transgenes and Plant Transformation
Full-length cDNAs for AtFtsZ1-1 (accession no. U39877) and AtFtsZ2-1 (accession no. AF089738) in the plasmid vector pZL1 (Gibco-BRL, Cleveland) were obtained as described previously (Osteryoung et al., 1998). A gel-purified SmaI-ClaI fragment containing the complete AtFtsZ1-1 cDNA sequence was ligated directionally behind the 35S promoter in pART7 (Gleave, 1992) digested with the same enzymes. The resulting plasmid was digested with NotI and the fragment containing the AtFtsZ1-1 insert was ligated into NotI-digested pART27 (Gleave, 1992) to create the plasmid pAP202 containing the AtFtsZ1-1 transgene. The AtFtsZ2-1 transgene was constructed by digesting the pART27 derivative pSN506 (Norris et al., 1998) with EcoRI and HindIII, and replacing the insert with an EcoRI-HindIII fragment containing the complete AtFtsZ2-1 cDNA to create pRF501. pAP202 and pRF501 were purified from Escherichia coli and transferred to Agrobacterium tumefaciens GV3101 (Koncz and Schell, 1986). Restriction analysis confirmed that no rearrangements occurred in the transfer to Agrobacterium. Arabidopsis ecotype Columbia was transformed by vacuum infiltration (Bechtold et al., 1993; Bent et al., 1994) with pAP202, and by floral dip (Clough and Bent, 1998) with pRF501 or the pART27 empty vector.
Selection and Propagation of Transgenic Plants
T1 seed from the inoculated plants were collected, sown on plates containing nutrient medium (4.3 g/L Murashige and Skoog salts, 1% [w/v] Suc, B5 vitamins, and 0.8% [w/v] Phytagar [Gibco-BRL]) and 100 mg/L kanamycin (Fisher Scientific, Hampton, NH), incubated at 4°C for 2 d, and moved to controlled environment chambers for germination. Chambers were set at a relative humidity of 40%, with 16 h of light daily (125 μmol m−2 s−1) at 20°C, and 8 h of darkness at 18°C. The age of the plants was taken from the date of seed transfer to the growth chamber following cold treatment. After 10 d in the growth chamber, kanr plants were transferred to a mixture of Supersoil potting mix (Rod McLellan Co., San Francisco) and vermiculite (4:1).
T2 and T3 seeds were sown on Rockwool (GrodanHP; Agro Dynamics, East Brunswick, NJ) saturated with Hoagland nutrient solution containing 100 mg/L kanamycin (Gibeaut et al., 1997). Seeds were covered with plastic and incubated at 4°C for 2 d, then transferred to growth chambers for germination. After 14 d, kanr plants were transferred to soil and grown as described above.
Microscopic Analysis
When plants were 18 d post-germination, the first leaf was removed with a razor blade and prepared for microscopic analysis as described (Pyke and Leech, 1991). Samples were then viewed with differential interference contrast optics using a BH-2 microscope (Olympus America, Melville, NY). Images were captured by computer using a DEI-750 digital charge-coupled device camera (Optronics, Goleta, CA) and Adobe Photoshop (Adobe Systems, San Jose, CA) software.
Generation of Antipeptide Antibodies
Peptides corresponding to regions of AtFtsZ1-1 and AtFtsZ2-1 predicted to constitute highly accessible and immunogenic epitopes were designed using the crystal structure of Methanococcus janaschii (Löwe and Amos, 1998), epitope mapping data for monoclonal antibodies against E. coli FtsZ (Voskuil et al., 1994), and molecular modeling programs. Peptides corresponding to residues 201 through 215 in AtFtsZ1-1 (EGRKRSLZALEAIEK) and residues 168 through 184 in AtFtsZ2-1 (RRRTVQAQEGLASLRD) were synthesized, purified by HPLC, and coupled to keyhole limpet hemocyanin (Pierce, Rockford, IL). These peptides, designated 1-1A and 2-1A, respectively, were injected into rabbits (nos. 4164 and 4166, respectively) for the production of polyclonal antibodies (Alpha Diagnostics, San Antonio, TX). The antisera obtained were partially purified by ammonium sulfate precipitation (50% final saturation) followed by dialysis against phosphate-buffered saline (140 mm NaCl, 3 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4, pH 7.4), as described (Harlow and Lane, 1988). Antibodies were further purified on affinity columns prepared by immobilizing the peptide antigens to SulfoLink Coupling Gel (Pierce) according to the manufacturer's standard protocol, yielding final protein concentrations of 0.7 and 0.9 mg ml−1 for antibodies 1-1A and 2-1A, respectively, determined using the Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA). These preparations were diluted for immunoblotting as described below.
Immunoblotting Procedures
Tissue for immunoblot analysis was collected from leaves of 21-d-old plants with microscopically verified mesophyll cell phenotypes. Tissue was homogenized in a microcentrifuge tube with 10 μL of grinding buffer {60 mm Tris [tris(hydroxymethyl)-aminomethane]-HCl, pH 8.0, 100 mm dithiothreitol, 2% [w/v] SDS, 15% [w/v] Suc, 5 mm ε-amino-N-caproic acid, 1 mm benzamidine HCl, and 0.01% [w/v] bromphenol blue} per milligram of leaf tissue plus a few grains of sterilized sand. Homogenized tissue was heated for 15 min at 70°C and stored at −20°C until use. Prior to electrophoresis, samples were reheated to 70°C for 5 min and centrifuged (3 min, 14,000g) to remove particulates. Proteins were separated by standard SDS-PAGE on 11% (w/v) polyacrylamide gels (Bio-Rad, Richmond, CA) and transferred to nitrocellulose membranes (0.45 μm, Micron Separations, Westborough, MA). Except where indicated, sample volumes loaded on gels were equivalent. Membranes were blocked for 30 min in TBST (50 mm Tris-HCl, pH 7.4, 200 mm NaCl, and 0.2% [v/v] Tween 20) containing 2% (w/v) Carnation non-fat dry milk (Nestle Food Company, Glendale, CA), then incubated in TBST plus 2% (w/v) nonfat dry milk (TBST-milk) containing affinity-purified AtFtsZ1-1 or AtFtsZ2-1 antibodies at dilutions of 1:1,500 and 1:3,000, respectively. Incubations with primary antibody were carried out in Seal-a-Meal bags (Dazey Corporation, Century, KS) shaken vigorously overnight at room temperature. After four 10-min washes in TBST, membranes were incubated with horseradish peroxidase-conjugated goat-anti-rabbit secondary antibodies (Fisher, Pittsburgh) for 15 min at 1:4,000 dilution in TBST-milk. Following four 10-min washes in TBST, membranes were developed using Renaissance Western Blot Chemiluminescence Reagent (NEN Life Science Products, Boston) and the signal was recorded on X-OMAT 1s film (Kodak, Rochester, NY).
Competition Binding Assays
To determine antibody specificity, immunoblotting experiments were performed as described above, except that prior to probing membranes, each diluted antibody was preincubated for 2 to 4 h in TBST-milk with an equimolar amount of purified, recombinant AtFtsZ1-1 (residues 41–269) or AtFtsZ2-1 (residues 92–282) on a rocking platform. These truncated versions of AtFtsZ1-1 and AtFtsZ2-1 were obtained by expressing the corresponding cDNA fragments in the expression vector pJC40 (Clos and Brandau, 1994) as 10-histidine-tagged FtsZ fusion proteins in E. coli BL21(λDE3)/plysS cells, following induction with isopropylthio-β-galactoside (1 mm) at 37°C. The recombinant proteins were purified from inclusion bodies in cell lysates using metal chelation chromatography (His-Bind Resin, Novagen, Madison, WI) according to the manufacturer's protocol for denaturing conditions. Protein concentrations of truncated AtFtsZ1-1 or AtFtsZ2-1 following chromatography, determined using the Bio-Rad Protein Assay reagent (Bio-Rad), were 1.75 and 1.24 mg ml−1, respectively.
AtFtsZ1-1 Quantification
Quantification of FtsZ protein levels was performed by scanning the film used for chemiluminescent detection of signals on immunoblots into the computer using a Mirage IIse imager (UMAX Technologies, Fremont, CA) and Binuscan software (Binuscan, New York). Densitometry measurements were made from the scanned films using Molecular Analyst software (Bio-Rad). Densitomery measurements were used to quantify the levels of AtFtsZ1-1 from a standard curve prepared by evaluating the intensities of the 40-kD polypeptide in four lanes loaded with increasing amounts of an AtFtsZ1-1-containing plant extract prepared from a control plant transformed with the empty pART27 vector. A best-fit curve calculated from the data had an R2 value of 0.98. This curve was used to calculate the relative amount of protein in the other samples, which were loaded so that the signal generated on the immunoblot was in the linear range of the standard curve. The level of AtFtsZ1-1 protein in each extract was calculated relative to the control sample.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge Travis Gallagher for excellent care and feeding of plants.
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. MCB–9604412) and by the Nevada Agricultural Experiment Station.
Indicates Web-only data.
LITERATURE CITED
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie Paris. 1993;316:1194–1199. [Google Scholar]
- Begg K, Nikolaichik Y, Crossland N, Donachie WD. Roles of FtsA and FtsZ in activation of division sites. J Bacteriol. 1998;180:881–884. doi: 10.1128/jb.180.4.881-884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- Clos J, Brandau S. pJC20 and pJC40: two high-copy-number vectors for T7 RNA polymerase-dependent expression of recombinant genes in Escherichia coli. Protein Expr Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Di Lallo G, Anderluzzi D, Ghelardini P, Paolozzi L. FtsZ dimerization in vivo. Mol Microbiol. 1999;32:265–274. doi: 10.1046/j.1365-2958.1999.01344.x. [DOI] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- Gaikwad A, Babbarwal V, Pant V, Mukherjee SK. Pea chloroplast FtsZ can form multimers and correct the thermosensitive defect of an Escherichia coli ftsZ mutant. Mol Gen Genet. 2000;263:213–221. doi: 10.1007/s004380051162. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 1997;115:317–319. doi: 10.1104/pp.115.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PAJ. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 298–299. [Google Scholar]
- Kohler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. Exchange of protein molecules through connections between higher plant plastids. Science. 1997;276:2039–2042. doi: 10.1126/science.276.5321.2039. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Addinall SG. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- Marrison JL, Rutherford SM, Robertson EJ, Lister C, Dean C, Leech RM. The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 1999;18:651–662. doi: 10.1046/j.1365-313x.1999.00500.x. [DOI] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Downing KH, Amos LA, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998a;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alphabeta tubulin dimer by electron crystallography. Nature. 1998b;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Norris SR, Shen X, DellaPenna D. Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 1998;117:1317–1323. doi: 10.1104/pp.117.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Pyke KA. Plastid division: evidence for a prokaryotically derived mechanism. Curr Opin Plant Biol. 1998;1:475–479. doi: 10.1016/s1369-5266(98)80038-1. [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial FtsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1991;96:1193–1195. doi: 10.1104/pp.96.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 1992;99:1005–1008. doi: 10.1104/pp.99.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM. A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 1994;104:201–207. doi: 10.1104/pp.104.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D, Park JT. Escherichia coli cell-division gene FtsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Rivas G, Lopez A, Mingorance J, Ferrandiz MJ, Zorrilla S, Minton AP, Vicente M, Andreu JM. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer: the primary steps for FtsZ assembly. J Biol Chem. 2000;275:11740–11749. doi: 10.1074/jbc.275.16.11740. [DOI] [PubMed] [Google Scholar]
- Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Rothfield LI, Justice SS. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- Sossong TM, Jr, Brigham-Burke MR, Hensley P, Pearce KH., Jr Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil JL, Westerbeek CA, Wu C, Kolk AH, Nanninga N. Epitope mapping of Escherichia coli cell division protein FtsZ with monoclonal antibodies. J Bacteriol. 1994;176:1886–1893. doi: 10.1128/jb.176.7.1886-1893.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Gayda RC. High-level expression of the FtsA protein inhibits cell septation in Escherichia coli K-12. J Bacteriol. 1990;172:4736–4740. doi: 10.1128/jb.172.8.4736-4740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–4034. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.