Abstract
Hypoxia, a hallmark characteristic of glioblastoma (GBM) induces changes in the transcriptome and the proteome of tumor cells. We discovered that hypoxic stress produces significant qualitative and quantitative changes in the protein content of secreted exosomes from GBM cells. Among the proteins found to be selectively elevated in hypoxic exosomes were protein-lysine 6-oxidase (LOX), thrombospondin-1 (TSP1), vascular derived endothelial factor (VEGF) and a disintegrin and metalloproteinase with thrombospondin motifs 1 (ADAMTS1), well studied contributors to tumor progression, metastasis and angiogenesis. Our findings demonstrate that hypoxic exosomes induce differential gene expression in recipient glioma cells. Glioma cells stimulated with hypoxic exosomes showed a marked upregulation of small nucleolar RNA, C/D box 116–21 (SNORD116-21) transcript among others while significantly downregulated the potassium voltage-gated channel subfamily J member 3 (KCNJ3) message. This differential expression of certain genes is governed by the protein cargo being transferred via exosomes. Additionally, compared to normoxic exosomes, hypoxic exosomes increased various angiogenic related parameters vis-à-vis, overall tube length, branching intervals and length of isolated branches studied in tube formation assay with endothelial progenitor cells (EPCs). Thus, the intercellular communication facilitated via exosomes secreted from hypoxic GBM cells induce marked changes in the expression of genes in neighboring normoxic tumor cells and possibly in surrounding stromal cells, many of which are involved in cancer progression and treatment resistance mechanisms.
Keywords: Exosomes, Hypoxia, Glioblastoma, Transcriptome, Proteomics, Angiogenesis
Highlights
-
•
In GBM, hypoxic stress induces profound changes in the protein content of secreted exosomes.
-
•
Hypoxic exosomal contents induce angiogenesis and significant changes in recipient GBM cell transcriptome.
-
•
Hypoxic exosomes play a major role leading to tumor proliferation, tumor growth and cell survival.
1. Introduction
With a very poor survival of 14–16 months in newly diagnosed patients, GBM accounts for more than 50% of malignant gliomas [1], [2], [3]. Surgical resection of tumors with accompanying or concurrent radiation or chemotherapy, either alone or in combination, has not been an entirely successful curative approach for treating GBM. The tumors almost always recur either a short distance from the resected area or in some other part of the brain [4], [5], [6], [7]. In many cancers, including GBM, exosomes deliver potentially oncogenic cargo leading to modulation of tissue microenvironment and progression of the disease [8], [9], [10]. Endosomal in origin, exosomes constitute a set of vesicles, 30–100 nm in size, secreted by various cell types [11], [12], [13], [14]. They are involved in mediating intercellular communication by shuttling or transferring protein and genetic information [15], [16], [17]. We have previously reported that modulating a cell’s microenvironment with pro-inflammatory cytokines induces changes in the protein cargo of secreted exosomes, which in turn exert their effect on nearby or far away recipient cells [18]. In addition to inflammation, necrosis is another distinguishing feature of GBM tumors [19], [20]. Necrosis results from rapid growth of glioma cells resulting in competition of dividing cells for metabolic demands, from vascular thrombosis or from hypoxia in the central region of the tumors [21]. Along with necrosis, a hypoxic, yet viable, microenvironment is a prominently observed feature during GBM progression and in response to various treatment regimens employed to treat the disease [22], [23], [24]. These changes are linked to increased tumor growth rate in many instances. Although hypoxia, inflammation, and necrosis are hallmark characteristics of GBM tumors, and given their resistance to apoptosis, there is no comprehensive evidence, as of yet, on the effects of hypoxia on exosomal cargo and its ramification on recipient cells.
Hypoxic stress induces changes in the transcriptome and the proteome of GBM cells [25], [26], [27]. A recent report shed light on some proteins being ferried via exosomes reflecting the hypoxic status of secreting glioma cells [26]. Various angiogenic factors found in glioblastoma derived exosomes were shown to mediate stimulation of angiogenesis [10], [26]. Cells incubated with exosomes, irrespective of the origin of secreting cells, have also been shown to exhibit increased invasiveness in a Matrigel™ based invasion assay [28].
The primary aim of our study was to determine the effect of hypoxia on the protein content of exosomes and its effect on the global transcriptome of recipient cells that could go on to influence the characteristics of tumor progression. To understand the influence of hypoxia on tumor derived exosomes and the resulting effect of these exosomes on the surrounding tumor or stromal cells, the proteomic profile of exosomes secreted by hypoxic glioma cells and the transcriptome of cells treated with these exosomes was studied.
2. Methods and materials
2.1. Cell culture and reagents
Cell culture media – DMEM, phosphate buffered saline (PBS), penicillin/streptomycin were obtained from Corning(Thermo Fischer Scientific, Waltham, MA), fetal bovine serum (FBS) from Atlanta Biologicals Inc (Flowery Branch, GA). Pimonidazole was purchased from Hydroxyprobe Inc (Burlington, MA). MTS assay reagent (celltiter 96 aqueous one solution cell proliferation assay) was purchased from Promega Corporation (Madison, WI). The human glioma cell line, U87MG was purchased from ATCC. EPC cells were a gift from Joyce E. Bischoff, Ph.D. at Boston Children’s Hospital.
2.2. Cell culture and staining
U87MG cultures were maintained in complete DMEM media containing 10% FBS & 100 units/ml of penicillin/streptomycin at 37 °C in a 5% CO2 incubator. Overnight cell cultures grown to 80% confluence were used for experiments. For hypoxic conditions, cells cultured overnight were washed 3x with PBS and fresh serum free DMEM media was added and were then incubated for a further 18–24 h at 37 °C in an anaerobic chamber (Forma Scientific) with an atmospheric mixture of 5% CO2, 10% H2, 85% N2 producing oxygen concentrations below 0.5%. For control group, cells were cultured similarly, but in a normal 37 °C 5% CO2 incubator. In both groups cells were cultured in presence of 200 μM of the hypoxic marker, pimonidazole to allow assessment of adducts formed in hypoxic- derived exosomes.
Endothelial progenitor cells (EPCs) were cultured in complete EGM-2 BulletKit (Lonza, Morristown, NJ) medium containing 20% FBS and 1% penicillin streptomycin and L-glutamine.
2.3. Exosome isolation
Conditioned media from U87MG cultures was collected and exosomes isolated by sequential centrifugation as previously described [18], [29]. In brief the pooled conditioned media from normoxic or hypoxic cells was subjected to sequential centrifugation at 300g for 10 min and 10,000g for 30 min. The pellet formed was discarded and the supernatant was then subjected to ultracentrifugation at 100,000g for 2 h to isolate the exosomes. The supernatant was discarded, the formed exosome pellet was washed with PBS at 100,000g for 2 h and the washing discarded. The exosome pellet was then resuspended in 30–50 μl PBS dependent on the analysis technique or experimental requirements downstream.
2.4. Electron microscopy
Transmission electron images were obtained by adding 3 μl of exosome suspension onto 200 mesh formvar coated grids and allowed to dry at room temperature. The grids were washed with water, stained with 1% uranyl acetate solution for 5 min. After staining, the grids were washed once in 70% ethanol followed by 4× washes with molecular grade water. These grids were then loaded onto the sample holder of the electron microscope (JEOL 100cx) and exposed to 100 kV electron beam for capturing images.
2.5. Confocal microscopy
U87MG cells grown to 90% confluence in 4 chambered Lab-Tek Borosilicate Coverglass system were cultured under normoxic or hypoxic conditions in presence of 200 μM pimonidazole. After 24 h of culture under these conditions, cells were washed 3× with PBS and fixed with 4% paraformaldehyde for 20 min at room temp. The cells were further washed 3× with PBS and permeabilized and simultaneously blocked with 0.1% Triton-x 100 in 3% BSA containing PBS. Fixed cells were incubated with FITC conjugated primary antibody against pimonidazole (1:500 dilution, Hydroxyprobe Inc, Burlington, MA). Cells were washed and further incubated with antibodies against HIF-1α and galectin-1. The secondary antibodies used were Goat anti-mouse AF647 and Goat anti-rabbit AF488 (at 1:1000 dilution). The cells were counter stained with DAPI. Confocal images were acquired using the LSM 880 confocal microscope using the x20 lens.
2.6. In vitro angiogenesis (tube formation) assay
Overnight cell cultures of EPC cells grown to 80% confluence were used for experiments. Around 5000 cells were plated onto BD Matrigel coated 96 well plates in presence of either normoxic or hypoxic glioma exosomes. For control group, cells were cultured similarly, but without exosomes. 24 hrs later, images of the formed tubes in the wells were acquired. A total of 4 images (n = 4) for each sample were then processed through the FIJI imaging software using the plugin for angiogenesis analysis. The number of nodes, meshes, junctions and branching intervals were noted and tabulated.
2.7. Cell viability assay
Cell viability assay was performed according to the manufacturer’s instructions. In brief, U87MG glioma cells were seeded into 96 well plate at a density of 3000 cells per well and allowed to attach overnight (16–18 h). After a fresh media change to exosome free complete medium, the cells were then preincubated for 3 h with or without exosomes. To this was added 0.5 mM hydrogen peroxide (H2O2) to induce oxidative stress and cells were incubated for a further 24 h in a humidified, 5% CO2 atmosphere. To the cell culture medium in each well was added 20 μl of the MTS reagent, incubated for a further 1–4 h and the absorbance was read at 490 nm in a 96-well plate reader.
2.8. RNA sequencing
RNA was isolated from untreated and exosome treated U87MG cells 24 h post treatment using QIAGEN RNA-easy kit according to manufacturer’s specifications. RIN (RNA integrity number) 9 or higher for all samples used for sequencing. RNA was prepared for sequencing by use of the Illumina TruSeq mRNA Stranded Sample Preparation Kit (Illumina Inc, San Diego, CA). Briefly, messenger RNA (mRNA) was purified from 500 ng of total RNA using poly A selection. The resultant RNA was chemically fragmented and converted into single-stranded cDNA in a reverse transcription reaction using random hexamer priming. The second strand cDNA was generated, followed by end repair and the addition of a single ‘A’ base at each end of the molecule. Adapters that are necessary for attachment to the sequencer flow cell surface were ligated to each end of the cDNA. The adapters also contain unique sequences that serve as indices which allow samples to be pooled for multiplexed sequencing and later downstream analysis. The ligated cDNA served as the template for PCR amplification to enrich the final cDNA library for sequencing.
Sequencing cluster generation was accomplished on the Illumina NextSeq. 500 to bind denatured cDNA library molecules to the sequencing flow cell, followed by an isothermal reaction to amplify attached cDNAs, resulting in clonal clusters of approximately 1000 copies each.
The sequence of amplified cDNA clusters ligated to the flow cell was determined by sequencing by synthesis using Illumina TruSeq SBS reagents on the Illumina NextSeq. 500 instrument.
Data was then analyzed on the web based Advaitha pathway analysis software. Differentially expressed gene list were obtained using a threshold of 0.05 for statistical significance (p-value) and a log fold change of expression with absolute value of at least 0.6. These data were analyzed in the context of pathways obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Release 78.0 +/06-02, Jun 16) [30], [31], gene ontologies from the Gene Ontology Consortium database [32] (Gene Ontology Consortium, 2001), and diseases from the KEGG database [30], [31].
2.9. SDS-PAGE and western blotting
Around 7.5 μg or 10 μg of exosomal pellets and cell lysates, determined by BCA protein estimation, were boiled with 4× Laemmli’s sample buffer containing 10% beta-mercaptoethanol and run on a 4–20% SDS-PAGE denaturing gel (Bio-Rad Laboratories, Hercules, CA). The proteins were transferred to PVDF membranes using conventional transfer apparatus. The membranes were blocked with 5% Bovine serum albumin (BSA) dissolved in Tris-buffered saline (TBS) containing 0.01% tween-20 (TBS-t) for 1hr at room temp. Blots were probed sequentially with primary antibodies: rabbit anti-TSP-1 polyclonal antibody (1:1000), rabbit anti-LOX polyclonal antibody (1:1000), rabbit anti-ADAMTS1 polyclonal antibody (1:1000), rabbit polyclonal anti-thrombospondin-1, (all from ThermoFisher Scientific, Rockford, IL), rabbit polyclonal anti-CD63, (1:1000) (SBI Biosciences, Mountain View, CA) or mouse monoclonal anti-β-actin (1:5000) (Cell Signaling Technology, Beverly, MA)] diluted in blocking buffer for 1hr at room temp. Blots were washed 3× with TBS-t and then incubated with horse radish peroxide (HRP) conjugated anti-mouse or anti- rabbit antibody (1:10,000) (Santa Cruz Biotech, Dallas, TX) diluted in blocking buffer at room temperature for 1 h. Blots were washed thoroughly 3x with TBS-t and developed with a chemiluminescence developing reagent, SuperSignal® West Femto Maximum sensitivity substrate (ThermoFisher Scientific, Rockford, IL).
2.10. Mass spectrometry (MS)
Exosomes were lysed in RIPA buffer and the proteins were reduced, alkylated, and digested using filter-aided sample preparation [33]. Tryptic peptides were labeled using a tandem mass tag 6-plex isobaric label reagent set (Thermo) following the manufacturer’s instructions. Labeled peptides were separated into 36 fractions on a 100 × 1.0 mm Acquity BEH C18 column (Waters) using an UltiMate 3000 UHPLC system (Thermo) with a 40 min gradient from 99:1 to 60:40 buffer A:B ratio under basic pH conditions, and then consolidated into 12 super-fractions. Each super-fraction was then further separated by reverse phase Jupiter Proteo resin (Phenomenex) on an in-line 200 × 0.075 mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 60 min gradient from 97:3 to 67:33 buffer A:B ratio. Buffer A = 0.1% formic acid, 0.5% acetonitrile, Buffer B = 0.1% formic acid, 99.9% acetonitrile, both buffers adjusted to pH 10 with ammonium hydroxide for offline separation. Eluted peptides were ionized by electrospray (2.15 kV) followed by mass spectrometric analysis on an Orbitrap Fusion Tribrid mass spectrometer (Thermo) using multi-notch MS3 parameters. MS data were acquired using the FTMS analyzer in top-speed profile mode at a resolution of 240,000 over a range of 375–1500 m/z. Following CID activation with normalized collision energy of 35.0, MS/MS data were acquired using the ion trap analyzer in centroid mode over a range of 400–2000 m/z. Using synchronous precursor selection, up to 10 MS/MS precursors were selected for HCD activation with normalized collision energy of 65.0, followed by acquisition of MS3 reporter ion data using the FTMS analyzer in profile mode at a resolution of 30,000 over a range of 100–500 m/z. Proteins were identified and reporter ions quantified using MaxQuant (Max Planck Institute) with a parent ion tolerance of 3 ppm, a fragment ion tolerance of 0.5 Da, and a reporter ion tolerance of 0.01 Da. Scaffold Q+S (Proteome Software) was used to verify MS/MS based peptide and protein identifications (protein identifications were accepted if they could be established with less than 1.0% false discovery and contained at least 2 identified peptides; protein probabilities were assigned by the Protein Prophet algorithm [34] and to perform reporter ion-based statistical analysis. The network analyses were generated through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) [35].
2.11. Statistical analyses
For all results, statistical significance was calculated using GraphPad Prism’s one way ANOVA followed by Bonferroni’s multiple comparison test and results were expressed as mean ± SEM. Value of p < 0.05 was considered as statistically significant.
3. Results
3.1. Characterization of exosomes derived from hypoxic glioma cells
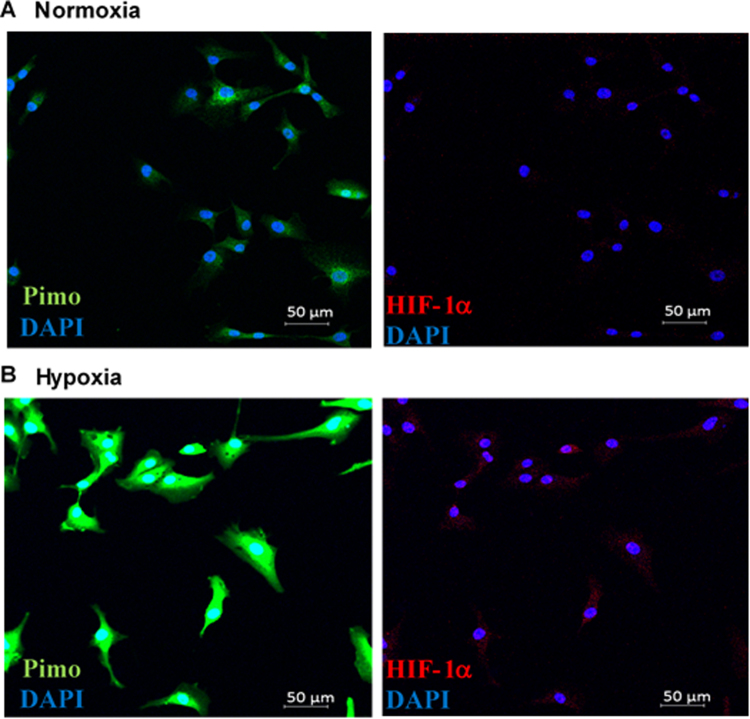
Confocal images of U87MG cells confirmed hypoxic stress in cells cultured in an anaerobic chamber showing increased intensity of pimonidazole staining (Sup Fig S1 A & B) as compared to cells cultured under normoxic conditions. Furthermore, increased staining for HIF-1α, upregulated under hypoxic conditions, also confirmed the hypoxic stress on U87MG cells. Exosomes were isolated from conditioned media of both groups of cells by sequential centrifugation and characterized by electron microscopy. Micropictograph analysis was used to confirm the morphology and determine the mean average diameters of the exosomes, with normoxic exosomes being around 71 ± 14 nm (Fig. 1A & C) in diameter while those derived from hypoxic cells were of a smaller diameter of 61 ± 13 nm (Fig. 1B & C). Western blot analysis shows enriched presence of exosomal marker CD63, a tetraspanin molecule commonly used as an exosomal marker protein, in exosomes compared to total U87MG cell lysates (Fig. 1D). These analyses confirm characterization of isolated vesicles as exosomes.
Fig. 1.
Characterization of exosomes. TEM of isolated vesicles: Electron micrographs display sets of distinct vesicles isolated from U87MG cells cultured under (A) normoxic conditions with average diameters of 71 ± 14 nm (C) while those derived from cells cultured under (B) hypoxic conditions had a slightly smaller average diameter of 61 ± 13 nm (C), (Scale bar: 100 nm). (D) Western blot analysis of equivalent amounts cell lysates and exosomes showed enrichment of CD63 in exosomes.
3.2. Quantitative and qualitative changes in exosomal proteome
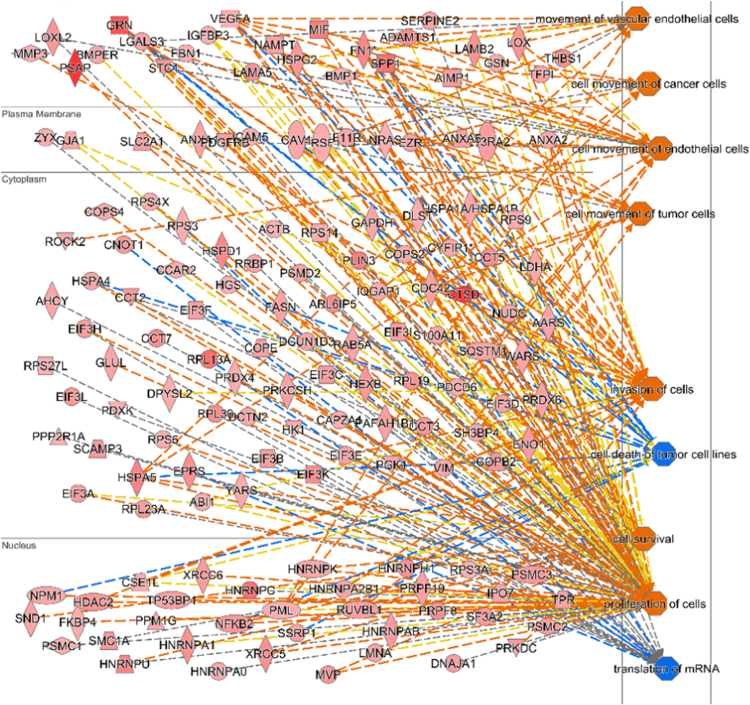
Our previous findings on cells’ microenvironment dictating the exosomal protein content [18], inspired us to investigate changes that are brought about in the exosomal cargo due to hypoxia. Exosomes isolated from glioma cells subjected to hypoxic and normoxic conditions were subjected to mass spectrometric analysis. Using the new TMT™ six-plex isobaric mass tagging MS analysis, a quantitative and comparative analysis of three independent sets of normoxic and hypoxic exosome was carried out. Mass spectra detected 136 different proteins (Supplementary info S1) which were differentially elevated (1.2 fold or more) in exosomes derived from cells exposed to hypoxia compared to those from normoxic cells. An Ingenuity pathway analysis (IPA) [35] of the hypoxic exosomal proteome showed that these differentially elevated proteins interact with proteins (Supplementary info S2) regulating various pathways such as upregulating the tumor growth, cell migration and tumor invasiveness pathways (orange nodes) while down regulating (blue nodes) the apoptosis and cell death pathways (Fig. 2).
Fig. 2.
MS and network analysis of hypoxic exosomal proteome. Ingenuity pathway analysis of 136 proteins that are elevated by 1.2 fold or greater in exosomes secreted by hypoxic U87MG cells. Network mapping of secreted proteins detected in hypoxic exosomes by MS (n = 3) shows their cellular localization, their interactive partners and the various biological functions that these proteins are potentially involved in. Nodes colored orange indicate upregulated pathways while nodes colored blue indicate downregulated pathways.
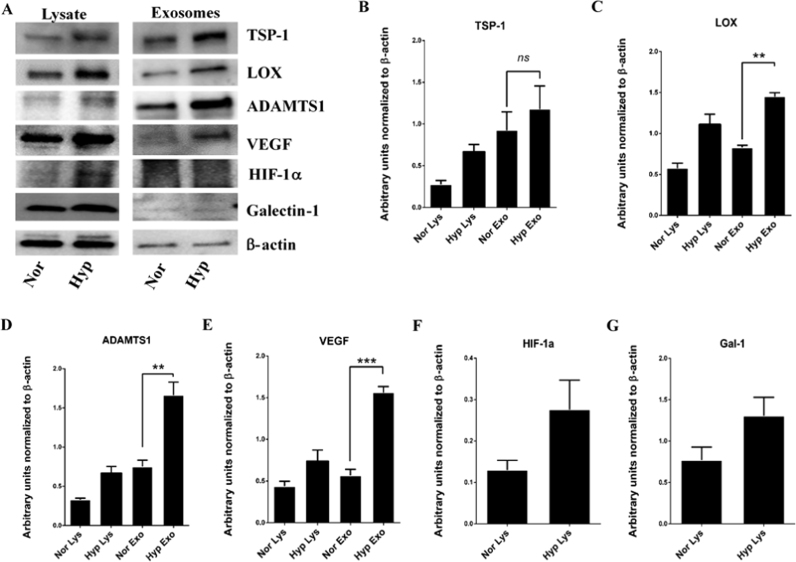
High grade GBM tumors are characterized by regions of intense neovascularization. MS analysis of the hypoxic exosomal proteome detected many of the major players which regulate angiogenesis such as TSP-1, LOX, VEGF and ADAMTS1. We further utilized the Western blot technique to validate and quantify these proteins (Fig. 3).
Fig. 3.
Elevated levels of proteins detected in hypoxic exosomes. (A) Western blot analysis of exosomes isolated from serum free, conditioned media of U87MG cells cultured under hypoxic conditions. Among the proteins detected in hypoxic exosomes were TSP-1, LOX, ADAMTS1, and VEGF. (B-E) Quantification of the western blots showed a significant increase in levels of secreted TSP-1, LOX, ADAMTS1, and VEGF in hypoxic exosomes compared to normoxic exosomes. Hif-1α (F) and Galectin-1 (G) were found to be elevated in hypoxic cell lysates although the increase was not significant. β-actin was used as loading control. Error bars, SEM (*p < 0.05, **p < 0.01, ***p < 0.001, ns=non significant).
TSP-1 and LOX have been reported to be involved in neovascularization of tumors while the ADAMTS1 has been shown to be involved in remodeling of ECM. MS analysis showed that these proteins were found to be significantly elevated in the exosomes secreted by cells exposed to hypoxia. The TSP-1 was found to be elevated 1.8 fold, LOX was elevated 2 fold, VEGF was elevated 2.5 fold while ADAMTS1 was also found to be elevated 1.8 fold in hypoxic exosomes. These results are in agreement with the presented MS quantification data.
3.3. Hypoxic exosomes induce angiogenesis and promote cell survival
Glioblastomas are characterized by increase in neovascularization to sustain growth, survival and proliferation of tumor cells [36], [37], [38]. Based on our previous MS and WB results showing elevated levels of some key players in angiogenesis, we were motivated to investigate the functional role of exosomes secreted by hypoxic glioma cells on the surrounding stromal cells.
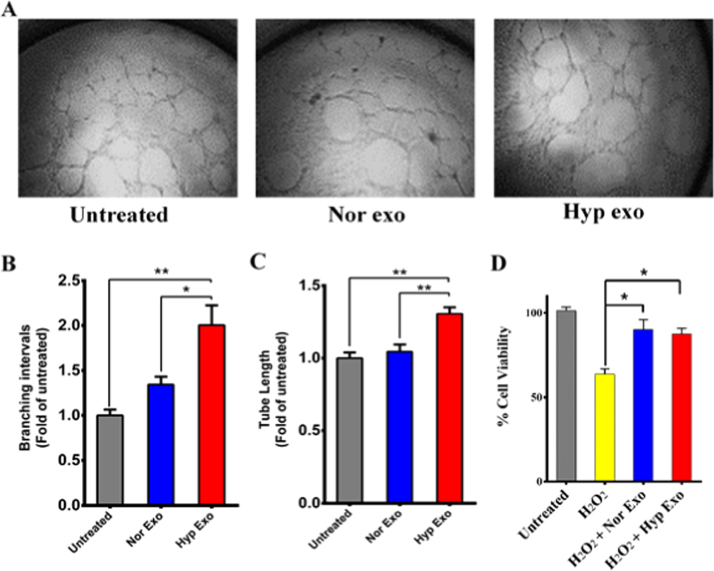
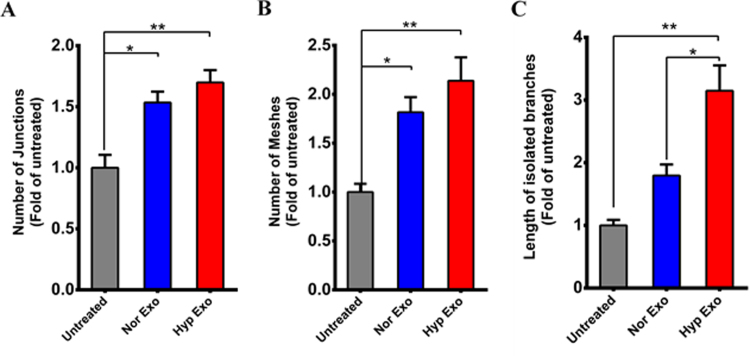
EPCs were treated with either normoxic or hypoxic exosomes and the effects on tube formation was measured. Treatment of EPCs with hypoxic exosomes stimulated tube forming ability in EPCs (Fig. 4A) by significantly increasing the formation of branching intervals (Fig. 4B) and tube length (Fig. 4C) over normoxic exosomes. Furthermore, although not significant, hypoxic exosomes induced a marked overall increase in the formation of junctions, meshes and isolated branch lengths in EPCs over normoxic exosomes (Sup Fig. 2 A, B and C).
Fig. 4.
Angiogenic effects of hypoxic exosomes. (A) In-vitro tube formation assay using EPC cells was performed in presence of either normoxic or hypoxic exosomes. A set of 4 different images was analyzed for each treatment and angiogenesis parameters were tabulated. Compared to untreated EPCs and those treated with normoxic exosomes, hypoxic exosome treatment produced a significant increase in the number of branching intervals, (B) and tube length, (C) in cultured EPCs. (D) Exosomes derived from both normoxic and hypoxic cells conferred significant cytoprotective effects to U87MG cells exposed to oxidative stress induced by 0.5 mM H2O2. Error bars, SEM (*p < 0.05, **p < 0.01).
Hypoxia induces oxidative stress in cells [39], [40], [41]. Exosomes provide substantial cytoprotective effects against oxidative stresses. U87MG glioma cells were exposed to hydrogen peroxide induced oxidative stress in the presence or absence of normoxic or hypoxic exosomes. Cells exposed to oxidative stress showed considerable decrease in cell viability. However, preincubation of glioma cells with either normoxic or hypoxic exosomes for 3–4 h, significantly diminished the effects of oxidative stress induced decrease in cell viability offering significant cytoprotective effects by 25–26% overall. The hypoxic exosomes however, did not offer any noticeable improvement over normoxic exosomes (Fig. 4D).
3.4. Exosome treatment induces significant changes in the transcriptome of recipient cells
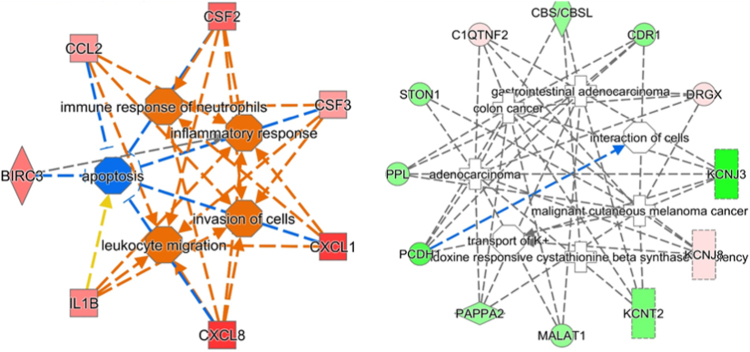
Intrigued by the response to exosomes by glioma cells under oxidative stress, we investigated the transcriptional changes in these cells as a consequence to exposure to either normoxic or hypoxic exosomes. U87MG cells grown in complete media supplemented with exosome free FBS were treated with hypoxic or normoxic exosomes. 24 h later, RNA was isolated from treated or untreated cells and the transcriptome of these cells was sequenced. Untreated cells were used as controls for RNA sequencing analysis. Compared to the transcriptome of untreated cells, in cells treated with normoxic exosomes, 29 differentially expressed (DE) genes were identified out of a total of 13102 genes detected (Supplementary info S3). Those genes which were found to be upregulated (Table 1 A – bold italics) were involved in apoptosis, interleukin signaling and cytokine mediated inflammation pathways (Fig. 5A). Downregulated genes (Table 1 A regular font) were found to be involved in TGF-beta, Wnt, Cadherin pathways.
Table 1.
Genes differentially expressed.
| A | |||
|---|---|---|---|
| symbol | entrez | logfc | adjpv |
| CXCL8 | 3576 | 1.29433 | 0.011312 |
| CXCL1 | 2919 | 1.24651 | 0.011312 |
| CSF2 | 1437 | 1.00003 | 0.011312 |
| TNFAIP3 | 7128 | 0.878881 | 0.011312 |
| GPX3 | 2878 | 0.877069 | 0.035951 |
| BIRC3 | 330 | 0.84613 | 0.011312 |
| KCCAT211 | 102724550 | 0.79094 | 0.011312 |
| RNF144B | 255488 | 0.776609 | 0.011312 |
| IL1B | 3553 | 0.771501 | 0.011312 |
| CCL2 | 6347 | 0.667585 | 0.011312 |
| SLC43A2 | 124935 | 0.655362 | 0.011312 |
| CSF3 | 1440 | 0.627342 | 0.011312 |
| SERPINB2 | 5055 | 0.623376 | 0.011312 |
| PPL | 5493 | − 0.605729 | 0.011312 |
| CLDN1 | 9076 | − 0.606167 | 0.035951 |
| IL20RB | 53833 | − 0.607983 | 0.011312 |
| STON1 | 11037 | − 0.624714 | 0.011312 |
| AKNA | 80709 | − 0.627865 | 0.011312 |
| DNAH10 | 196385 | − 0.633545 | 0.011312 |
| C6orf223 | 221416 | − 0.66998 | 0.035951 |
| PAPPA2 | 60676 | − 0.671331 | 0.011312 |
| SPRR2D | 6703 | − 0.675693 | 0.011312 |
| ANKRD22 | 118932 | − 0.678226 | 0.035951 |
| KCNT2 | 343450 | − 0.683391 | 0.011312 |
| INHBE | 83729 | − 0.694873 | 0.011312 |
| CDR1 | 1038 | − 0.697316 | 0.011312 |
| CBS | 875 | − 0.906225 | 0.011312 |
| MLXIPL | 51085 | − 0.949305 | 0.021512 |
| PCDH1 | 5097 | − 1.27402 | 0.031243 |
|
B | |||
|---|---|---|---|
| symbol | entrez | logfc | adjpv |
| SNORD116-21 | 100033432 | 10 | 0.013899 |
| KCNJ8 | 3764 | 1.08844 | 0.013899 |
| C1QTNF2 | 114898 | 0.998243 | 0.013899 |
| DRGX | 644168 | 0.915825 | 0.013899 |
| C4orf48 | 401115 | 0.882413 | 0.013899 |
| CBS | 875 | − 0.640255 | 0.013899 |
| PAPPA2 | 60676 | − 0.640677 | 0.013899 |
| MALAT1 | 378938 | − 0.646721 | 0.013899 |
| PPL | 5493 | − 0.691949 | 0.013899 |
| STON1 | 11037 | − 0.700129 | 0.013899 |
| CDR1 | 1038 | − 0.733006 | 0.013899 |
| KCNT2 | 343450 | − 0.834774 | 0.013899 |
| PCDH1 | 5097 | − 1.05263 | 0.013899 |
| KCNJ3 | 3760 | − 1.34574 | 0.013899 |
Genes significantly expressed in U87MG cells treated with normoxic exosomes (A) and cells treated with hypoxic exosomes (B). The genes listed in bold italics are significantly elevated and others significantly down regulated with logfc giving their fold change values along with their adjusted p values (adjpv) (n = 3).
Fig. 5.
Network analysis of differentially expressed genes in glioma cells exposed to exosomes. Ingenuity pathway analysis based on RNA seq results for differentially expressed genes in U87MG cells treated with normoxic exosomes (A) and hypoxic exosomes (B) shows regulation of varying pathways depending on the exosome source. Genes brighter in color are elevated while the fainter ones are downregulated.
Of the 6067 genes (Supplementary info S4) induced in cells exposed to hypoxic exosomes (Table 1 B – bold italics), five were significantly upregulated. The five genes are SNORD1, KCNJ8, C4orf48, C1QTNF2, and DRGX. Of the 7257 genes repressed (Table 1 B – regular font) in cells exposed to hypoxic exosomes, nine were significantly downregulated. These nine genes are CBS, PAPPA2, MALAT1, PPL, STON1, CDR1, KCNT2, PCDH1, and KCNJ3.
The list of differentially expressed genes was then analyzed for their pathways using the QIAGEN Integrated Pathways Analysis software [35] (Fig. 5A and B).
Analysis of the hypoxic exosome treated glioma cell transcriptome results showed that majority of these pathways are affected by a single downregulated gene, the KCNJ3 (potassium voltage gated channel subfamily J member 3). The significant changes observed in the transcriptome of hypoxic exosome treated glioma cells can thus be attributed to the functional exosomal cargo in ultimately regulating gene expression.
4. Discussion
A plethora of studies have shed light on how hypoxia plays an important role in tumor development, cell survival, proliferation, angiogenesis, and is involved in imparting intense resistance to multimodality anti-cancer treatments [36], [42], [43]. High grade gliomas are characterized by areas of hypoxia in the growing periphery, a necrotic core, regions of intense angiogenesis, inflammation, and resistance to apoptosis following chemo- and radiation therapy [44], [45], [46], [47]. Our investigations into hypoxia and its effects on the exosomes secreted by glioma cells point to some of the molecular mechanisms which may explain the stimulated angiogenesis and resistance to apoptosis observed in many pre-clinical and clinical studies on GBM [42], [46]. In general, the tumor microenvironment including the oxygen status surrounding tumor cells profoundly affects how cancer cells respond to chemo- and radiation therapies. Previous reports have shown that changes in the tissue microenvironment induce changes in the proteome and transcriptome of a cell [18]. Subsequently our study, in agreement with other recent reports [9], [18], [26], [27], [28], [48] have shown that these changes are reflected in the cargo of exosomes secreted by such cells. Glioma cells grown under hypoxic conditions show a marked increase in staining for pimonidazole adducts along with over expression of Hif-1α and galectin-1 (Fig. 3), both proteins usually over-expressed under hypoxic conditions.
Based on previous studies, we anticipated a change in the exosomal proteome under hypoxic stress [18], [26]. The physical, genetic and proteomic characteristics of secreted exosomes can vary due to the microenvironment they are produced from. Characterization of glioma exosomes revealed that the stress induced due to hypoxic conditions affects the exosome size distribution. Exosomes secreted by hypoxic cells were found to be of smaller size compared to normoxic cells. Our major effort in the work that followed was to understand the differences in the function of these hypoxic exosomes vs. exosomes secreted from normoxic cells. Prior studies have shown that exosomes in general impart significant cytoprotective effects against various stresses to cells [48], [49], [50], [51], [52]. Our results did agree with these observations. However, we did not find any significant difference in the cytoprotective effect against oxidative stress afforded by hypoxic exosomes compared to normoxic exosomes. We attribute this effect to the fact that even our ‘normoxic’ exosomes are derived from a glioma cell line and they all exhibit some level of cytoprotective effect when added to stressed cells, a result of carrying oncogenic cargo promoting cell survivability. However, the effect of these exosomes on EPC tube formation markedly differed. Hypoxic exosomes induced a clear increase in the node formation, mesh formation, number of branching intervals among other outputs in EPC cultured or treated with hypoxic exosomes. These results seem to implicate a pathway for the well-studied effect of hypoxia promoting increased neovascularization in the tumor microenvironment [27], [36], [37], [38], [44].
Although we found a general cytoprotective effect vis-à-vis normoxic or hypoxic exosomes, the effect of hypoxic exosomes on the transcriptome of glioma cells was significantly different compared to the one induced by normoxic exosomes. RNA seq analysis showed a number of differentially expressed genes and shown in Table 1. Although the number of genes differentially selectively expressed in cells treated with hypoxic exosomes was far lesser than the total genes induced by exosomes in general, there were major changes which help to explain how hypoxia might modulate the tumor environment and assist the tumor in progressing and resisting various therapeutic insults.
We were able to examine pathway analysis to identify a series of cellular functions likely to be induced in cells after exposure to hypoxic exosomes. Analysis of the hypoxia-exosome induced U87MG glial tumor cell transcriptome showed various metabolic pathway interruptions. Several of these interruptions were within amino acid metabolism and involved the biosynthesis of amino acids in general. Amino acid deficiency has been found to positively correlate with tumor angiogenesis that was induced with VEGF (vascular endothelial growth factor). Cystathionine-beta-synthase (CBS), which codes for the protein to catalyze the conversion of homocysteine to cystathionine, is the first step along the transsulfuration pathway, and a repression of CBS would inevitably retard amino acid biosynthesis which could activate the VEGF pathway in areas of hypoxia or those adjacent to hypoxic cells [53]. If these pathways are activated by a hypoxic tumor, it may be a link to neurological deficits associated with aggressive tumor growth, mediated by secreted exosomes from hypoxic regions.
Pathway analysis also identified possible interaction with the cGMP-PKG signaling pathway. Protein Kinase G is typically activated by cGMP and has downstream interactions that regulate a variety of physiological processes [54]. One of the downstream interactions is with mitochondrial ATP-sensitive potassium channels (mKATP) [55]. The specific mKATP is KCNJ8, which is a potassium ion channel [56]. Opening of the mKATP causes an increased concentration of reactive oxygen species (ROS). The ROS causes a feedback activation of the mKATP channel, leaving the channel open for a prolonged period of time, creating more ROS, which could lead to enhanced activity of the cell. For instance, the ROS may cause the intrinsic process of ischemic preconditioning (IPC) to occur. During IPC, short episodes of ischemia are repeatedly induced leading to an activated state of protective enzymatic pathways to reduce the impact/induce tolerance to subsequent ischemic incidents.
The down regulated genes were found to be involved in Wnt, GABA-BrII, and Cadherin mediated pathways. PCDH1, Stonin-1, and MALAT1 are all involved in intercellular communication. Stonin-1 is a non-coding RNA and MALAT1 is a non-protein coding gene [57]. PCDH1 is involved in both of the distinct categories. It is a membrane protein found at cell-to-cell boundaries [58], but it is also involved in neural cell adhesion [59].
The second and much more direct theme of these repressed genes is that of neurological functionality. Two genes (KCNT2 and KCNJ3) are voltage-gated potassium channels [60]. KCNT2 is an ATP-sensitive outward rectifying potassium channel. KCNJ3 is a G-protein controlled inward-rectifying potassium channel. CDR1 is a cerebellar degeneration related protein, and has been found in patients with paraneoplastic cerebellar degeneration [61]. PAPPA2 codes for a protein that cleaves insulin-like growth factor (IGF) which modulates neurological function. PPL codes for a protein that is suspected to be a contributor to intermediate filament, which provide cells with their structure. CBS codes for cystathionine-beta-synthase, which is a crucial intermediate in the transsulfuration pathway. The inhibition of CBS has been shown to effect a variety of biochemical processes, including the synthesis of glutathione [53]. One of the pathways that is affected by the downregulation of this particular gene is the oxytocin signaling pathway, the dysregulation of which negatively affects proliferation, differentiation and cellular migration.
There were also a number of significantly upregulated genes in glioma cells that were exposed to hypoxic exosomes and there were two prominent categories that emerged in our analysis. The first is that of intercellular communication. SNORD116-21 (HBII-85-21) [62], [63] is a small nucleolar RNA that could easily have been a component of the exosome communication, and C1QTNF1 which is a tumor necrosis factor related protein. Interestingly, the levels of small nucleolar RNA, SNORD116-21 was found to be increased 10-fold. In all, over half of the increased expression genes involve neuronal function and development. As mentioned, KCNJ8 is a voltage-gated potassium channel, critical to neurological membrane polarization. C4orf48 is a protein that has been shown to increase expression during neocortex and cerebellar development, and DRGX is the gene for the dorsal root ganglia homeobox, which is thought to relate to axon guidance.
The differential expression of genes in glioma cells exposed to hypoxic exosomes throws light on the paracrine signaling mechanisms which help the neighboring and exosome recipient tumor cells react to hypoxic microenvironment. Hypoxic exosomal proteins from glioma cells were found to carry a complement of proteins which were involved in proliferation of cells, proliferation of endothelial cells invasion of tumor, suppression of cell death, among other pathways (Fig. 4A and B). Many of the proteins identified are known to play an important role in tumor invasion and angiogenesis. Among the culprits identified were Protein-lysine 6-oxidase, Thrombospondin-1, Vascular endothelial growth factor and A disintegrin and metalloproteinase with thrombospondin motifs-1. Probing Western blots of hypoxic exosome proteins, we were able to confirm the elevated levels of LOX, ADAMTS1 and VEGF, known participants in hypoxia- induced angiogenesis and metastasis [64], [65], [66], [67], as well as Thrombospondin-1, known to be integrally related to vascular function/activation status and aberrant tumor vascularization [10], [18], [68], [69], [70] in hypoxic exosomes. The transformation observed in the EPCs and the differential expression of genes observed in the transcriptome of glioma cells treated with hypoxic exosomes can thus be attributed to the changes in the cargo of exosomes due to hypoxic stress.
The clinical manifestations in patients suffering from high grade gliomas such as cognitive disturbances, communication deficits, neurological deficits, seizures [71], [72] are due to the necrosis, inflammation, neovascularization and aggressive tumor growth. Many of these processes are modulated via intercellular communication through exosomes. The cargo of these exosomes changes depending on the microenvironment of the secreting cells. Hypoxic conditions, as seen in GBM tumor necrotic cores and their aggressively proliferating edges, induce changes in the cargo of exosomes secreted by glioma cells residing in these areas. The target cells of these exosomes react to the exosomal messages by differential expression of many genes. Based on our experimental results and pathway and network analysis of exosomal proteomic and target cell transcriptome results, the result of intercellular communication mediated by hypoxic exosomes, in part, explains why GBM tumors offer such intense resistance to aggressive multimodality anti-neoplastic treatments and such high rates of tumor recurrence. Our studies shed light on potential targets carried by exosomes and also provide information on the various genes that are differentially expressed which can potentially be targeted to halt the advance of proliferating cancer cells. By controlling the microenvironment of cells secreting exosomes, these vesicles can potentially be engineered to carry agents or drugs to target specific cancer cells thus avoiding or circumventing the dangerous side-effects of anti-neoplastic molecules.
Overall, our results suggest that glioblastoma tumor cells, growing in regions of hypoxia can secrete specific programs for changing neighboring or remote cell gene and protein expression, possibly contributing in a major way to progression and resistance of difficult to manage cancers such as GBM.
Acknowledgements
This study was supported by Center for Advanced Surface Engineering, under the United States National Science Foundation Grant No. IIA-1457888 and the Arkansas EPSCoR Program, ASSET III, Arkansas INBRE program grant (NIGMS) P20 GM103429 and J.D. Patterson Summer Undergraduate Research Fellowship. We would like to thank Dr. Susan Kadlubar, Division of Medical Genetics, UAMS, for her support in using the Qiagen Ingenuity Pathway Analysis software. We would like to acknowledge Jeffrey A. Kamykowski of UAMS Digital Microscopy Core for the confocal microscopy images.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.03.008.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Fig. S1.
Confocal images of U87 cells. U87MG cells grown under normoxic (A) and hypoxic (B) conditions in presence of pimonidazole. FITC conjugated anti-pimonidazole antibody was used to detect pimonidazole-induced adducts in cells. Cells were counter-stained for Hif-1α. Hypoxic cells showed increased binding of pimonidazole and increased fluorescence for Hif-1α relative to cells cultured in normoxic conditions.
Fig. S2.
Tube formation assay. In-vitro tube formation assay using EPC cells was performed in presence of either normoxic or hypoxic exosomes. Compared to untreated EPCs, those treated with either normoxic exosomes or hypoxic exosomes produced a significant increase in the number of (A) junctions and (B) meshes while length of isolated branches was significantly increased with hypoxic exosomes. Error bars, SEM (*p < 0.05, **p < 0.01).
References
- 1.Morgan L.L. The epidemiology of glioma in adults: a "state of the science" review. Neuro-oncology. 2015;17:623–624. doi: 10.1093/neuonc/nou358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diwanji T.P., Engelman A., Snider J.W., Mohindra P. Epidemiology, diagnosis, and optimal management of glioma in adolescents and young adults. Adolescent Health Med. Therap. 2017;8:99–113. doi: 10.2147/AHMT.S53391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia C.G., Kahn S.A., Geraldo L.H.M., Romano I., Domith I., Silva D., Dos Santos Assuncao F., Ferreira M.J., Portugal C.C., de Souza J.M., Romao L.F., Netto A.D.P., Lima F.R.S., Cossenza M. Combination therapy with sulfasalazine and valproic acid promotes human glioblastoma cell death through imbalance of the intracellular oxidative response. Mol. Neurobiol. 2018 doi: 10.1007/s12035-018-0895-1. [DOI] [PubMed] [Google Scholar]
- 4.Shirazi H.A., Grimm S., Raizer J., Mehta M.P. Combined modality approaches in the management of adult glioblastoma. Front. Oncol. 2011;1:36. doi: 10.3389/fonc.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson D.R., Chang S.M. Recent medical management of glioblastoma. Adv. Exp. Med. Biol. 2012;746:26–40. doi: 10.1007/978-1-4614-3146-6_3. [DOI] [PubMed] [Google Scholar]
- 6.Aoki T., Hashimoto N., Matsutani M. Management of glioblastoma. Expert Opin. Pharmacother. 2007;8:3133–3146. doi: 10.1517/14656566.8.18.3133. [DOI] [PubMed] [Google Scholar]
- 7.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A.A., Gijtenbeek J., Marosi C., Vecht C.J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J.G., Mirimanoff R.O., European Organisation for R., Treatment of Cancer Brain T., Radiation Oncology G., National Cancer Institute of Canada Clinical Trials G. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet. Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 8.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 9.Lotvall J., Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adhes. Migr. 2007;1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoorvogel W., Kleijmeer M.J., Geuze H.J., Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 12.Keller S., Sanderson M.P., Stoeck A., Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone R.M. Exosomes biological significance: a concise review. Blood Cells Mol. Dis. 2006;36:315–321. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 15.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J. Proteom. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Schartz N.E., Chaput N., Andre F., Zitvogel L. From the antigen-presenting cell to the antigen-presenting vesicle: the exosomes. Curr. Opin. Mol. Ther. 2002;4:372–381. [PubMed] [Google Scholar]
- 17.Fevrier B., Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Kore R.A., Abraham E.C. Inflammatory cytokines, interleukin-1 beta and tumor necrosis factor-alpha, upregulated in glioblastoma multiforme, raise the levels of CRYAB in exosomes secreted by U373 glioma cells. Biochem. Biophys. Res. Commun. 2014 doi: 10.1016/j.bbrc.2014.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siu A., Wind J.J., Iorgulescu J.B., Chan T.A., Yamada Y., Sherman J.H. Radiation necrosis following treatment of high grade glioma--a review of the literature and current understanding. Acta Neurochir. 2012;154:191–201. doi: 10.1007/s00701-011-1228-6. (discussion 201) [DOI] [PubMed] [Google Scholar]
- 20.Noch E., Khalili K. Molecular mechanisms of necrosis in glioblastoma: the role of glutamate excitotoxicity. Cancer Biol. Ther. 2009;8:1791–1797. doi: 10.4161/cbt.8.19.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis D.N. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 22.Mohyeldin A., Dalgard C.L., Lu H., McFate T., Tait A.S., Patel V.C., Wong K., Rushing E., Roy S., Acs G., Verma A. Survival and invasiveness of astrocytomas promoted by erythropoietin. J. Neurosurg. 2007;106:338–350. doi: 10.3171/jns.2007.106.2.338. [DOI] [PubMed] [Google Scholar]
- 23.Laurenti G., Benedetti E., D'Angelo B., Cristiano L., Cinque B., Raysi S., Alecci M., Ceru M.P., Cifone M.G., Galzio R., Giordano A., Cimini A. Hypoxia induces peroxisome proliferator-activated receptor alpha (PPARalpha) and lipid metabolism peroxisomal enzymes in human glioblastoma cells. J. Cell Biochem. 2011;112:3891–3901. doi: 10.1002/jcb.23323. [DOI] [PubMed] [Google Scholar]
- 24.Clarke R.H., Moosa S., Anzivino M., Wang Y., Floyd D.H., Purow B.W., Lee K.S. Sustained radiosensitization of hypoxic glioma cells after oxygen pretreatment in an animal model of glioblastoma and in vitro models of tumor hypoxia. PLoS One. 2014;9:e111199. doi: 10.1371/journal.pone.0111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deep G., Panigrahi G.K. Hypoxia-induced signaling promotes prostate cancer progression: exosomes role as messenger of hypoxic response in tumor microenvironment. Crit. Rev. Oncog. 2015;20:419–434. doi: 10.1615/CritRevOncog.v20.i5-6.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucharzewska P., Christianson H.C., Welch J.E., Svensson K.J., Fredlund E., Ringner M., Morgelin M., Bourseau-Guilmain E., Bengzon J., Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J.E., Tan H.S., Datta A., Lai R.C., Zhang H., Meng W., Lim S.K., Sze S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell Proteom. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho L., Guerrero P., Marchetti D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS One. 2013;8:e73790. doi: 10.1371/journal.pone.0073790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kore R.A., Abraham E.C. Phosphorylation negatively regulates exosome mediated secretion of cryAB in glioma cells. Biochim. Biophys. Acta. 2015 doi: 10.1016/j.bbamcr.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M., Goto S., Kawashima S., Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zagzag D., Amirnovin R., Greco M.A., Yee H., Holash J., Wiegand S.J., Zabski S., Yancopoulos G.D., Grumet M. Vascular apoptosis and involution in gliomas precede neovascularization: a novel concept for glioma growth and angiogenesis. Lab. Investig. 2000;80:837–849. doi: 10.1038/labinvest.3780088. [DOI] [PubMed] [Google Scholar]
- 37.Zagzag D., Friedlander D.R., Margolis B., Grumet M., Semenza G.L., Zhong H., Simons J.W., Holash J., Wiegand S.J., Yancopoulos G.D. Molecular events implicated in brain tumor angiogenesis and invasion. Pediatr. Neurosurg. 2000;33:49–55. doi: 10.1159/000028975. [DOI] [PubMed] [Google Scholar]
- 38.Zagzag D., Lukyanov Y., Lan L., Ali M.A., Esencay M., Mendez O., Yee H., Voura E.B., Newcomb E.W. Hypoxia-inducible factor 1 and VEGF upregulate CXCR4 in glioblastoma: implications for angiogenesis and glioma cell invasion. Lab. Investig. 2006;86:1221–1232. doi: 10.1038/labinvest.3700482. [DOI] [PubMed] [Google Scholar]
- 39.Giordano F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Investig. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solaini G., Baracca A., Lenaz G., Sgarbi G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta. 2010;1797:1171–1177. doi: 10.1016/j.bbabio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Cervellati F., Cervellati C., Romani A., Cremonini E., Sticozzi C., Belmonte G., Pessina F., Valacchi G. Hypoxia induces cell damage via oxidative stress in retinal epithelial cells. Free Radic. Res. 2014;48:303–312. doi: 10.3109/10715762.2013.867484. [DOI] [PubMed] [Google Scholar]
- 42.Mahase S., Rattenni R.N., Wesseling P., Leenders W., Baldotto C., Jain R., Zagzag D. Hypoxia-mediated mechanisms associated with antiangiogenic treatment resistance in glioblastomas. Am. J. Pathol. 2017;187:940–953. doi: 10.1016/j.ajpath.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao X.G., Wang C., Liu D.Y., Zhang X., Wang L., Yan M., Zhang W., Zhu J., Li Z.C., Mi C., Tian J.Y., Hou G.D., Miao S.Y., Song Z.X., Li J.C., Xue X.Y. Hypoxia upregulates HIG2 expression and contributes to bevacizumab resistance in glioblastoma. Oncotarget. 2016;7:47808–47820. doi: 10.18632/oncotarget.10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur B., Khwaja F.W., Severson E.A., Matheny S.L., Brat D.J., Van Meir E.G. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro-oncology. 2005;7:134–153. doi: 10.1215/S1152851704001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarassishin L., Casper D., Lee S.C. Aberrant expression of interleukin-1beta and inflammasome activation in human malignant gliomas. PLoS One. 2014;9:e103432. doi: 10.1371/journal.pone.0103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliver L., Olivier C., Marhuenda F.B., Campone M., Vallette F.M. Hypoxia and the malignant glioma microenvironment: regulation and implications for therapy. Curr. Mol. Pharmacol. 2009;2:263–284. doi: 10.2174/1874467210902030263. [DOI] [PubMed] [Google Scholar]
- 47.Sinha S., Koul N., Dixit D., Sharma V., Sen E. IGF-1 induced HIF-1alpha-TLR9 cross talk regulates inflammatory responses in glioma. Cell Signal. 2011;23:1869–1875. doi: 10.1016/j.cellsig.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 48.Eldh M., Ekstrom K., Valadi H., Sjostrand M., Olsson B., Jernas M., Lotvall J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One. 2010;5:e15353. doi: 10.1371/journal.pone.0015353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lv L.H., Wan Y.L., Lin Y., Zhang W., Yang M., Li G.L., Lin H.M., Shang C.Z., Chen Y.J., Min J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J. Biol Chem. 2012;287:15874–15885. doi: 10.1074/jbc.M112.340588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee C., Mitsialis S.A., Aslam M., Vitali S.H., Vergadi E., Konstantinou G., Sdrimas K., Fernandez-Gonzalez A., Kourembanas S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation. 2012;126:2601–2611. doi: 10.1161/CIRCULATIONAHA.112.114173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filipazzi P., Burdek M., Villa A., Rivoltini L., Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 2012;22:342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Corcoran C., Rani S., O'Brien K., O'Neill A., Prencipe M., Sheikh R., Webb G., McDermott R., Watson W., Crown J., O'Driscoll L. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One. 2012;7:e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawahara B., Moller T., Hu-Moore K., Carrington S.J., Faull K.F., Sen S., Mascharak P.K. Attenuation of antioxidant capacity in human breast cancer cells by carbon monoxide through inhibition of cystathionine beta-synthase activity: implications in chemotherapeutic drug sensitivity. J. Med. Chem. 2017 doi: 10.1021/acs.jmedchem.7b00476. [DOI] [PubMed] [Google Scholar]
- 54.Manivasagam S., Vellaichamy E. Suppression of Npr1, not Npr2 gene function induces hypertrophic growth in H9c2 cells in vitro. Biochem. Biophys. Res. Commun. 2017;491:250–256. doi: 10.1016/j.bbrc.2017.07.123. [DOI] [PubMed] [Google Scholar]
- 55.Peart J.N., Pepe S., Reichelt M.E., Beckett N., See Hoe L., Ozberk V., Niesman I.R., Patel H.H., Headrick J.P. Dysfunctional survival-signaling and stress-intolerance in aged murine and human myocardium. Exp. Gerontol. 2014;50:72–81. doi: 10.1016/j.exger.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper P.E., McClenaghan C., Chen X., Stary-Weinzinger A., Nichols C.G. Conserved functional consequences of disease-associated mutations in the slide-helix of Kir6.1 and Kir6.2 subunits of the ATP-sensitive potassium channel. J. Biol. Chem. 2017 doi: 10.1074/jbc.M117.804971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Z., Luo W., Wang J., Peng T., Sun G., Shi J., Li Z., Zhang B. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem. Biophys. Res. Commun. 2017;492:480–486. doi: 10.1016/j.bbrc.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 58.Faura Tellez G., Willemse B.W., Brouwer U., Nijboer-Brinksma S., Vandepoele K., Noordhoek J.A., Heijink I., de Vries M., Smithers N.P., Postma D.S., Timens W., Wiffen L., van Roy F., Holloway J.W., Lackie P.M., Nawijn M.C., Koppelman G.H. Protocadherin-1 localization and cell-adhesion function in airway epithelial cells in asthma. PLoS One. 2016;11:e0163967. doi: 10.1371/journal.pone.0163967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi S., Inoue Y., Kiyonari H., Abe T., Misaki K., Moriguchi H., Tanaka Y., Takeichi M. Protocadherin-17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev. Cell. 2016;38:331. doi: 10.1016/j.devcel.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 60.Blankenship M.L., Coyle D.E., Baccei M.L. Transcriptional expression of voltage-gated Na(+) and voltage-independent K(+) channels in the developing rat superficial dorsal horn. Neuroscience. 2013;231:305–314. doi: 10.1016/j.neuroscience.2012.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbagallo D., Condorelli A., Ragusa M., Salito L., Sammito M., Banelli B., Caltabiano R., Barbagallo G., Zappala A., Battaglia R., Cirnigliaro M., Lanzafame S., Vasquez E., Parenti R., Cicirata F., Di Pietro C., Romani M., Purrello M. Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou H., Zhao J., Yu C.H., Luo Q.J., Chen Y.Q., Xiao Y., Qu L.H. Identification of a novel box C/D snoRNA from mouse nucleolar cDNA library. Gene. 2004;327:99–105. doi: 10.1016/j.gene.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 64.Erler J.T., Bennewith K.L., Nicolau M., Dornhofer N., Kong C., Le Q.T., Chi J.T., Jeffrey S.S., Giaccia A.J. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 65.Erler J.T., Giaccia A.J. Lysyl oxidase mediates hypoxic control of metastasis. Cancer Res. 2006;66:10238–10241. doi: 10.1158/0008-5472.CAN-06-3197. [DOI] [PubMed] [Google Scholar]
- 66.Svensson K.J., Kucharzewska P., Christianson H.C., Skold S., Lofstedt T., Johansson M.C., Morgelin M., Bengzon J., Ruf W., Belting M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc. Natl. Acad. Sci. USA. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treps L., Perret R., Edmond S., Ricard D., Gavard J. Glioblastoma stem-like cells secrete the pro-angiogenic VEGF-A factor in extracellular vesicles. J. Extracell. Ves. 2017;6:1359479. doi: 10.1080/20013078.2017.1359479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Z., Kipnis J. Thrombospondin 1--a key astrocyte-derived neurogenic factor. FASEB J. 2010;24:1925–1934. doi: 10.1096/fj.09-150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma Y., Yabluchanskiy A., Lindsey M.L. Thrombospondin-1: the good, the bad, and the complicated. Circ. Res. 2013;113:1272–1274. doi: 10.1161/CIRCRESAHA.113.302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin Q., Qian J., Ge L., Shen L., Jia J., Jin J., Ge J. Effect and mechanism of thrombospondin-1 on the angiogenesis potential in human endothelial progenitor cells: an in vitro study. PLoS One. 2014;9:e88213. doi: 10.1371/journal.pone.0088213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sizoo E.M., Braam L., Postma T.J., Pasman H.R., Heimans J.J., Klein M., Reijneveld J.C., Taphoorn M.J. Symptoms and problems in the end-of-life phase of high-grade glioma patients. Neuro-oncology. 2010;12:1162–1166. doi: 10.1093/neuonc/nop045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sizoo E.M., Taphoorn M.J., Uitdehaag B., Heimans J.J., Deliens L., Reijneveld J.C., Pasman H.R. The end-of-life phase of high-grade glioma patients: dying with dignity? Oncologist. 2013;18:198–203. doi: 10.1634/theoncologist.2012-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material