Abstract
In this investigation, a gene (CDF_Amyl) encoding extracellular α-amylase in Aspergillus niger strain CSA35 associated with cassava spoilage was amplified using specific primers and characterized in silico. The gene had a partial nucleotide sequence of 968 bp and encoded a protein of 222 aa residues with a molecular weight and isoelectric point of 25.13 kDa and 4.17, respectively. Its catalytic site was located in the active site domain. BLASTp analysis showed that the protein primary sequence of the α-amylase gene had 98% and 99% homologies with the α-amylase of A. niger and A. oryzae RIB40, respectively. The gene is more closely related to α-amylase genes from fungi than to bacterial, plant, or animal α-amylase genes. Restriction mapping of the gene showed it can be digested with restriction enzymes like NcoI, PstI, SmaI, and BcLI among others but not with EcoRI and EcoRV. Its protein product had a hydrophobicity score of − 0.43 but no transmembrane helix. The CDF_Amyl protein was subcellularly localized in the secretory pathway, an indication of its release into extracellular space after secretion. Also, the 3D structure of the CDF-Amyl protein was barrel-shaped with domains characteristic of α-amylases. The encoded α-amylase Vmax is 6.90 U/mg protein and Km is 6.70 mg/ml. It was concluded that the unique characteristics of the CDF_Amyl gene and its deduced protein could find applications in biotechnological, food and pharmaceutical industries where cloning and further modification of this gene would be required for product development and improvement.
Keywords: Aspergillus niger; Alpha-amylase gene; Bioinformatics characterisation; Restriction enzymes; Phylogenetics, evolution
Highlights
-
•
The study amplified α-amylase gene from a new strain of Aspergillus niger CSA35.
-
•
Restriction enzymes like SmaI and BcLI can cut CDF_Amyl but not EcoRI and EcoRV.
-
•
Its protein product had a hydrophobicity score of - 0.43; no transmembrane helix.
-
•
The CDF_Amyl protein is subcellularly localized in the fungi secretory pathway.
-
•
CDF_Amyl and its protein product could be applied in biotechnological industries.
1. Introduction
α-Amylases (EC 3.2.1.1.) hydrolyse polysaccharides such as starch, glycogen etc via random breakdown of their internal α − 1, 4-glucosidic bonds to produce oligosaccharides of various sizes [16], [31]. They are among the most sought-after enzymes because of their high relevance in present-day biotechnology. α-Amylases have wide applications in starch processing, brewing industries, detergent, textile, pharmaceutical industries etc [31], [41]. Although they can be obtained from almost all living organisms, the enzymes from microbial sources (basically bacteria and fungi) tend to be more suitable for industrial applications.
α-Amylases are grouped as family 13 of the glycosyl hydrolases. They have similar structures and catalytic sites and do not vary in their catalytic mechanism. A typical α-amylase has a three dimensional (3D) structure resembling a barrel shape with three characteristic domains –A, B and C [14], [27]. Situated in loops at the C-termini of β-strands in the A domain are highly conserved residues of amino acid which partake in catalysis and binding of substrates [27]. The B domain is involved in binding of Ca2+ and also participates in binding of substrate. The C-domain's role, a second protrusion in front of or behind the A-domain, however, is not fully known; but a study by Holm et al. [21] on genetic aberrations in the C-domain of Bacillus stearothermophilus α-amylase suggests that the domain plays a role in enzyme activity.
Amylases vary in molecular weights; usually from about 10–210 kDa [40]. The presence of carbohydrate moieties in some organisms’ amylases tends to raise their molecular weights; for example, glycoproteins have been observed in Aspergillus oryzae, Lipomyces kononenkoae, Bacillus stearothermophilus and Bacillus subtilis strains. The isoelectric point (pI) of amylases ranges from 3.25 to 10.1 [12]. The pI refers to the pH at which an enzyme's overall electric charge is zero [10]. Knowledge of enzymes’ pI values is used for electro focussing.
Over the years, a number of application softwares or programmes have been developed and are being used for easy study of gene and protein sequences in a new field of molecular biology presently referred to as bioinformatics or computational biology ([46]; [47]). These bioinformatics tools make characterisation of such nucleotide or protein sequences less stressful, time conservative and cost effective aside their highly precise or accurate outcomes. Some of these computational tools are utilized in an array of in silico analyses ranging from determination of evolutionary relatedness of genes or enzyme proteins (phylogeny), molecular weights, isoelectric points (pI), transmembrane helices, hydrophobicity, subcellular localization of proteins, three-dimensional (3D) structure of proteins, conserved domains, detection of introns, to in silico determination of restriction enzyme cleavage sites in gene sequences ([25], [7], [8]; Blum et al. [9]; [33], [24]).
Food spoilage generally has been closely linked with the action of some microorganisms capable of degrading some nutrient materials in food substances [1], [39]. Cassava (Manihot esculenta) and its products, being notable rich sources of carbohydrate, water and some minerals [6], support the growth of a number of microbes, including fungi [3], [42]. Hence, the spoilage of cassava tubers or their processed products after a long period of exposure to air or dirts, are often times associated with visible growth of Aspergillus species on or around them with characteristic colour(s) and odour [4]. These saprophytic fungi are believed to grow and survive on the cassava tubers by secreting starch hydrolyzing enzymes such as amylases, amidst others, thereby breaking down cassava starch to simple sugars for nutrition [28], [40].
The present study amplified and characterized the α-amylase gene of Aspergillus niger strain CSA35 using in silico analyses. The fungi, which grow on decaying cassava tubers and peels, had been identified in a previous study. The characterisation of the fungal cassava starch-degrading α-amylase gene and its deduced protein sequence would facilitate the potential industrial applications of the α-amylase gene and its protein product.
2. Material and methods
2.1. Preparation of cassava flour
Peeled cassava (M. esculenta) tubers were processed into flour according to the method described by Oyewole and Sanni [34].
2.2. YPD agar preparation
Preparation of yeast extract-peptone dextrose (YPD) agar was carried out as described by Tonukari et al. [44]. Thereafter, it was slightly cooled and poured into plates to solidify.
2.3. Growth of fungi
Aspergillus niger CSA35, previously identified using 18S rRNA sequence and reported to be associated with cassava spoilage in Nigeria [4], was inoculated into a sterile YPD agar in a Petri dish. It was allowed to grow and sporulate in the dark for one week and then sub-cultured in sterilized Cassava Starch Agar (CSA) which contained cassava flour (2%) as the main carbon source, NaNO2 (1%) and agar powder (1.5%), for additional 7 days. To avoid growth of bacteria, 20% ampicillin (5 µl) was included in the subcultures. Thereafter, the hyphae were harvested.
2.4. Genomic DNA isolation
Genomic DNA was extracted from the fungi using a modified cetyl trimethyl ammonium bromide (CTAB) method [49] and purified using ethanol precipitation. The presence and quality of the isolated genomic DNA was ascertained via agarose gel electrophoresis using 0.8% agarose gel stained with ethidium bromide (5 µg/µl). Images of bands obtained in the gel were viewed and captured using a UV-light automated Gel Documentation machine (Enduro, USA).
2.5. PCR amplification of α-amylase gene
Alpha amylase gene was amplified by polymerase chain reaction (PCR) method as described by Zidani et al. [49] using the specific forward and reverse primers (AmyFP 5′-CATCTGGATCACCCCCGTTA-3′ and AmyRP 5′-AGACTTACGAAGCGAACCGT-3′, respectively) and the isolated fungal genomic DNA as template. The PCR mixture (25 µl) contained, 1.25 µl each of forward and reverse primers (5 mM each), 1.25 µl of 2.5 mM dNTP mix (dATP, dGTP, dTTP, dCTP) (Promega), 10 × NH4 buffer (1.25 µl), 1.0 µl of 50 mM MgCl2 (Promega), 1.0 µl of 5 unit/µl of Taq DNA polymerase, 7.25 µl of dd·H2O and 7.25 µl of 10 ng/µl of genomic DNA. The PCR program was performed on a thermal cycler (Biometra) system as follow: Stage 1 (x1): 94 °C for 5 min to achieve the initial DNA denaturation; Stage 2 (x 35): 94 °C for 30 s, for denaturation; 60 °C for 30 s, for annealing; and 72 °C for 5 min, for extension. Stage 3 (x1): 72 °C for 5 min and 10 °C for infinity (∞). The presence of amplified α-amylase genes was established by agarose gel electrophoresis using 1.0% agarose stained with 3 µl of ethidium bromide (5 µg/µl) and viewed with Enduro Gel Doc machine.
2.6. Gene sequencing
The purified α-amylase amplicons were sequenced using an automated Applied Biosystem 3130xl Genetic Analyzer (ABI Sequencer, USA). The sequencing was carried out at the Bioscience Center, International Institute for Tropical Agriculture (IITA), Ibadan, Nigeria.
2.7. Bioinformatics characterisation of putative α-amylase gene
The nucleotide sequence of α-amylase obtained was translated to its corresponding protein primary sequence using the European Biotechnology Information (EBI) EMBOSS translation tool and National Center for Biotechnology Information (NCBI) BLASTX tool; the most suitable reading frame was selected and exons identified. Identifications of closely matched α-amylase sequences and conserved domains were carried out with the aid of NCBI BLASTp and CD tools available online at http://blast.ncbi.nlm.nih.gov/Blast.cgi and https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi respectively [30], while multiple sequence alignment and construction of phylogeny tree were done using EMBL EBI ClustalW2 and Phylogeny tools available at http://www.ebi.ac.uk/Tools. Restriction mapping was carried out on the cassava-degrading fungal amylase gene sequence (CDF_Amyl) using a suitable bioinformatics tool available online (http://www.bioinformatics.nl/tools/tacg4/form4.html). Prediction of CDF_Amyl molecular weight (Mw) and isoelectric point (pI) was done using ExPASy - Compute pI/Mw tool available online at http://web.expasy.org/compute_pi/, while the three-dimensional (3D) structure was determined using Phyre2 software available at http://www.sbg.bio.ic.ac.uk/phyre2/ [25]. The subcellular localization of gene protein product was predicted using MultiLoc2 software available at http://abi.inf.uni-tuebingen.de/Services/MultiLoc2 (Blum et al., 2009). In silico determinations of hydrophobic/hydrophilic nature and transmembrane helices of the deduced protein were carried out using the translated amino acid sequence of the gene as input data in specialized bioinformatics applications available at http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html [20], [32], [33] and http://www.cbs.dtu.dk/services/TMHMM-2.0/ [24], respectively.
3. Results and discussion
Genes in their native forms generally comprise protein-coding nucleotide sequences called exons and non protein-coding sequences referred to as introns (de Conti et al., 2013). During posttranslational modifications, however, introns are spliced leaving exons that encode synthesis of functional proteins (de Conti et al. [13]). In this study, we observed the presence of four (4) introns and five (5) exons in the Aspergillus niger CSA35 α-amylase gene (CDF_Amyl) (Fig. 1). The gene has a partial nucleotide sequence of 926 bp and a deduced protein sequence of 222 amino acids (Fig. 1). Most fungi have been reported to harbor at least one fungal-type α-amylase gene that is evolutionarily intron-rich (da Lage et al. [11]).
Fig. 1.
Partial nucleotide sequence and deduced amino acid sequence of the cassava degrading fungi amylase gene (CDF_Amyl). Four introns (non coding regions) were found in the CDF_Amyl gene sequence. The introns were denoted in lower case letters (italicized) while the exons (5) in upper case letters.
The protein-protein BLAST (BLASTp) result obtained showed that the cassava starch-degrading partial α-amylase gene shares 98–99% identity with α-amylase proteins from Aspergillus oryzae RIB40, A. niger, A. flavus NRRL3357, A. kawachii (Table 1) and several other Aspergillus species and strains already deposited in NCBI Genbank. This is an indication that the amplified alpha amylase gene from Aspergillus niger CSA35 actually encodes α-amylase protein. Preliminary biochemical analysis of the encoded α-amylase protein secreted by the fungus showed that it has a maximum specific activity (Vmax) of 6.90 U/mg protein and a Michaelis constant (Km) of 6.70 mg/ml using soluble starch as substrate [5].
Table 1.
BLASTp result of CDF_Amyl protein-protein matches and source organisms.
| Matched protein/organism | Max score | Total score | Query cover | E-value | Identity | Accession number |
|---|---|---|---|---|---|---|
| Alpha-amylase A type− 1/2 [Aspergillus oryzae RIB40] | 462 | 462 | 100% | 4e− 159 | 99% | XP_003189619.1 |
| Chain A, Orthorhombic Crystal Structure Of Aspergillus niger Alpha-Amylase | 461 | 461 | 100% | 7e− 159 | 98% | 2GUY_A |
| Alpha-amylase, putative [Aspergillus flavus NRRL3357] | 461 | 461 | 100% | 1e− 158 | 98% | XP_002374124.1 |
| Alpha-amylase [Aspergillus kawachii] | 461 | 461 | 100% | 1e− 158 | 98% | BAD01051.1 |
| Taka-amylase A (Taa-G1) precursor [Aspergillus oryzae] | 460 | 460 | 100% | 3e− 158 | 98% | AAA32708.1 |
| Alpha-amylase [Aspergillus awamori] | 460 | 460 | 100% | 3e− 158 | 98% | BAD06002.1 |
| Alpha amylase Precursor [Aspergillus awamori] | 459 | 459 | 100% | 4e− 158 | 98% | Q02905.1 |
| Alpha amylase Precursor [Aspergillus shirousami] | 455 | 455 | 100% | 3e− 157 | 98% | P30292.1 |
| Taka-amylase A precursor [Aspergillus oryzae] | 450 | 450 | 100% | 2e− 154 | 97% | BAA00336.1 |
| Alpha-amylase [Aspergillus sojae] | 499 | 499 | 100% | 7e− 154 | 95% | BAM28635.1 |
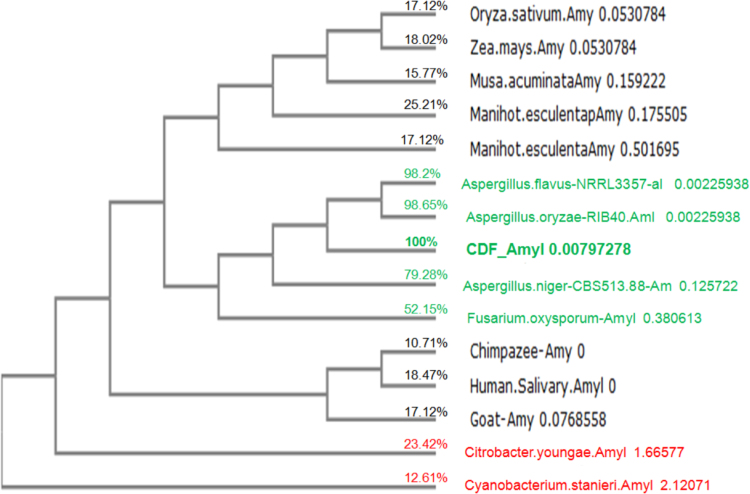
The evolutionary relationship (phylogeny) between the cassava starch-degrading fungal partial α-amylase gene (CDF_Amyl) and α-amylase genes in plants (rice, maize, banana, and cassava plant), other fungi, bacteria, animals (Chimpazee, goat) and human saliva is as shown in Fig. 2. Phylogenies help to provide necessary information on biological diversity, for structuring classifications as well as for proper understanding of events that occurred during evolution [7], [8]. The α-amylase gene amplified from the Aspergillus niger CSA35, associated with cassava spoilage in Nigeria, is more closely related to α-amylase genes from fungi than to bacterial, plant, or animal α-amylase genes (Fig. 2), as expected in the absence of horizontal gene transfers [23].
Fig. 2.
Evolutionary relationship between cassava-degrading fungal amylase (CDF_Amyl) and those in plants, fungi, bacteria, lower animals and human in NCBI GenBank.
The amino acid residues present in the conserved domain of the fungal partial amylase gene (CDF_Amyl) are as shown in Fig. 3a. α-Amylase genes generally possess catalytic regions that comprise aspartate (D), glutamate (E) and histidine (H) residues that are involved in the enzymes’ mechanism of starch hydrolysis [26]. In the present research work, the amino acid residues present in the conserved domain of the CDF_Amyl gene include histidine (H), arginine (R), aspartate (D), and glutamate (E) amongst others (Fig. 3a). The catalytic site is present in the active site region (Fig. 3b). These findings agree with the report of Liu et al. [26] and affirm that the amylase gene (CDF-Amyl) belonged to the superfamily of α-amylases.
Fig. 3.
a. Conserved domain amino acids of cassava-degrading fungi amylase (CDF_Amyl). #Hash marks above the aligned sequences show the location of conserved feature residues. The amino acid residues present in the conserved domain of CDF_Amyl gene include 46 His (H), 128 Arg (R), 130 Gln (D), 154 Glu (E), and 220 His (H) amongst others. 7TAA_A denotes Taka Amylase from Aspergillus oryzae (GenBank Accession no. 7TAA_A); query = CDF_Amyl gene (from this study); 2AAA_A = alpha-amylases from Aspergillus niger (Accession: 2AAA_A); gi 27373051) = alpha-amylase from Lipomyces kononenkoae (GenBank: AAO12212.1); gi 50260808 = hypothetical amylase protein from Cryptococcus neoformans var. neoformans B-3501A (GenBank: EAL23458.1); gi 74582599 = alpha-amylase from Schizosaccharomyces pombe 972 h (UniProtKB/Swiss-Prot: O74922.1); gi 145510486 = hypothetical amylase protein from Paramecium tetraurelia strain d4-2 (NCBI Reference Sequence: XP_001441176.1); gi 167515550 = hypothetical amylase protein from Monosiga brevicollis MX1 (NCBI Reference Sequence: XP_001742116.1); gi 300104136 = glycoside hydrolase family 13 protein from Schizophyllum commune H4–8 (GenBank: EFI95542.1); gi 340501008 = hypothetical amylase protein from Ichthyophthirius multifiliis (GenBank: EGR27831.1). All amylases compared for conserve domain amino acids are from eukaryotic organisms. b. Active and catalytic site regions of cassava-degrading fungi amylase (CDF_Amyl). The CDF_Amyl belonged to the superfamily of amylases. The catalytic site region of the amylase is within the active site region.
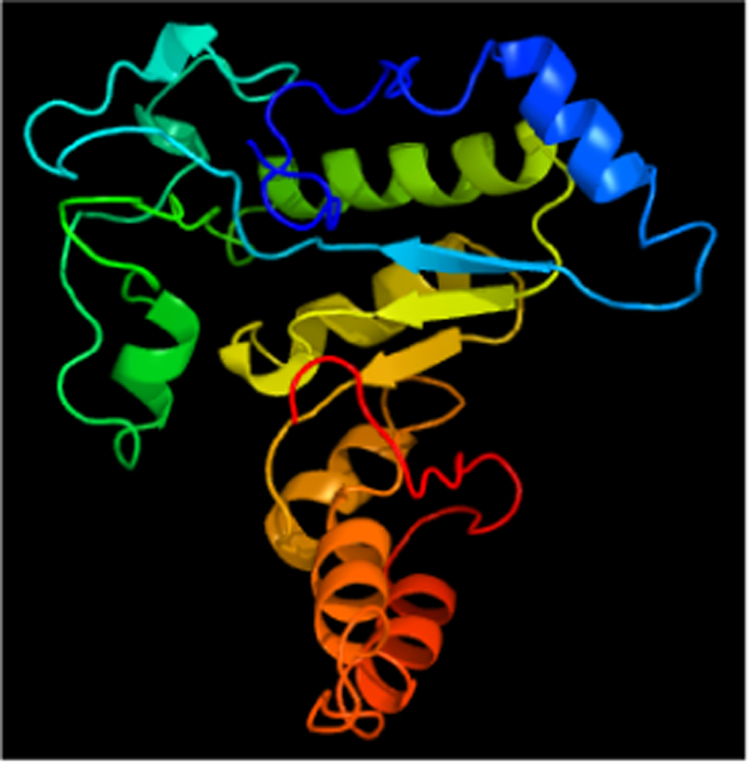
A typical structural characteristic of α-amylase proteins is their barrel-shape appearance [27]. The barrel shape is made up of curly polar α-helixes in the outer parts of the protein and β-sheets close to the middle of the protein. The 3D-protein structure of the cassava starch-degrading fungal amylase (CDF_Amyl) reported in this study (Fig. 4) is identical to that of α-amylase isolated from Aspergillus oryzae [27] and also agrees with the typical α-amylase structure described by de Souza and Magalhães [14] and a number of others already deposited in NCBI genbank.
Fig. 4.
3D-Protein structure of Aspergillus sp. CSA35 α-amylase. Image coloured by rainbow, N to C terminus.
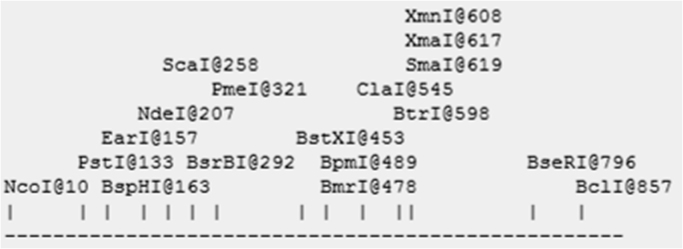
As clearly depicted by the endonuclease restriction linear map shown in Fig. 5, a large array of restriction enzymes (such as PstI, SmaI, NcoI, BcLI etc) can be used to cut the cassava-degrading fungal α-amylase gene (CDF_Amyl, 926 bp) when considering cloning and gene expression experiments. However, restriction enzymes like EcoRI and EcoRV will be unable to cut the CDF_Amyl gene (Fig. 5). These findings are in agreement with those of Yu [48] who observed that PstI and SmaI can cut alpha-amylase gene (αAmy3 gene enhancer) isolated from rice (Oryza sativa). They are, however, in contrast to the report of Puspasari et al. [36] who digested raw starch–degrading α-amylase from Bacillus aquimaris MKSC6.2 using EcoRI and EcoRV during a gene expression study. The choice of restriction enzyme to use when considering digestion of the partial amylase gene (CDF_Amyl) in molecular biology studies, however, will not only depend on the choice of restriction enzyme but also on a number of other factors such as the size of gene fragment required, the ability of the choice restriction enzyme to cut both the potential vector of interest and the amylase gene (CDF_Amyl), as well as the goal or nature of the research being considered, etc [18], [29], [43].
Fig. 5.
Endonuclease restriction map of the cassava degrading fungi amylase gene (CDF_Amyl).
The bioinformatics analysis also showed that the extracellular enzyme (alpha amylase) secreted by Aspergillus niger CSA35 has an isoelectric point (pI) of 4.17 and a molecular weight (Mw) of 25.13 kDa (Table 2). Molecular weights of amylases vary from about 10–210 kDa but most of these enzymes are in the range of 50–60 kDa. Amylases from Bacillus caldolyticus and Chloroflexus aurantiacus have the lowest (10 kDa) and highest (210 kDa) molecular weights, respectively [19], [40]. The pI values are quite useful for understanding of enzyme behaviour against different purification procedures at different pH values. Amylase isoelectric points are generally in the range of 3.25–10.1 [12]. The pI value obtained in this study is also in accord with the report of Ahmad et al. [2] who opined that alpha amylase from Aspergillus oryzae is an acidic enzyme with optimum pH of 4.5 and low isoelectric point (pI = 4.2). The knowledge of the isoelectric point and the Mw would be of great advantage to laboratories or industries when considering small or large scale extraction of the CDF_Amyl protein.
Table 2.
Isoelectric point and molecular weight of CDF_Amyl.
| Amylase | Isoelectric point (pI) | Molecular weight (Dalton) | Number of amino acid residues |
|---|---|---|---|
| CDF_Amyl | 4.17 | 25,128.71 | 222 |
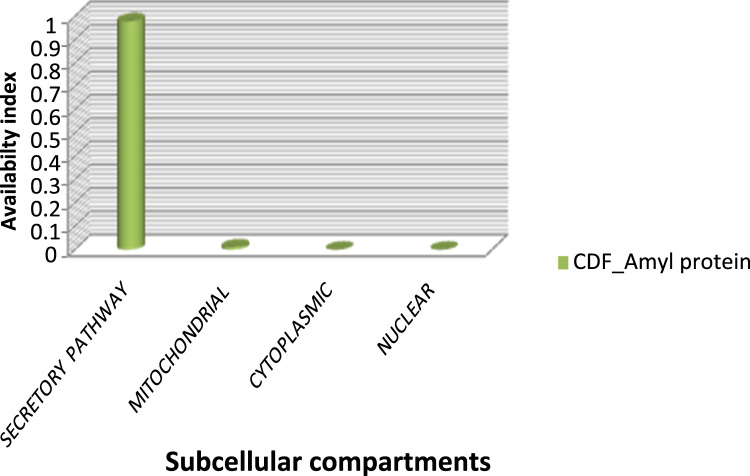
In silico analysis of the subcellular localization of the α-amylase protein in A. niger CSA35 revealed that the enzyme (CDF_Amyl) is predominantly found in the secretory pathway within the cassava degrading fungi (Fig. 6). Soluble proteins are synthesized in the secretory pathway and thereafter delivered into the extracellular space where they are utilized for cellular functions [17]. Our report is in agreement with the reports of de Souza and Magalhães [14] and Deb et al. [15] that α-amylases are mainly extracellularly localized.
Fig. 6.
Subcellular localization of CDF_Amyl in Aspergillus sp. CSA35.
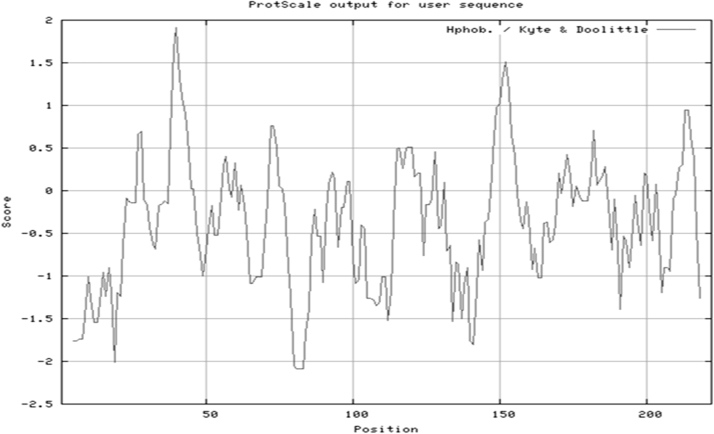
Many studies have shown that the amino acid sequence of proteins play a crucial role in determining solubility or hydrophobicity of expressed proteins [22], [32], [33]. In this study, it was observed that the CDF_Amyl protein had a hydrophobicity score of − 0.43 which indicates that the amino acid sequence is of a soluble protein (Fig. 7). Our present finding that the cassava starch-degrading fungal α-amylase (CDF_Amyl) is a soluble protein also agrees with the fact that the enzyme is localized in the secretary pathway (Fig. 6), since proteins secreted into extracellular space are mostly hydrophilic in nature [38].
Fig. 7.
Hydrophobicity prediction of CDF_Amyl. The overall average hydrophobicity score is − 0.43 (below zero) which indicated that the protein is a soluble protein.
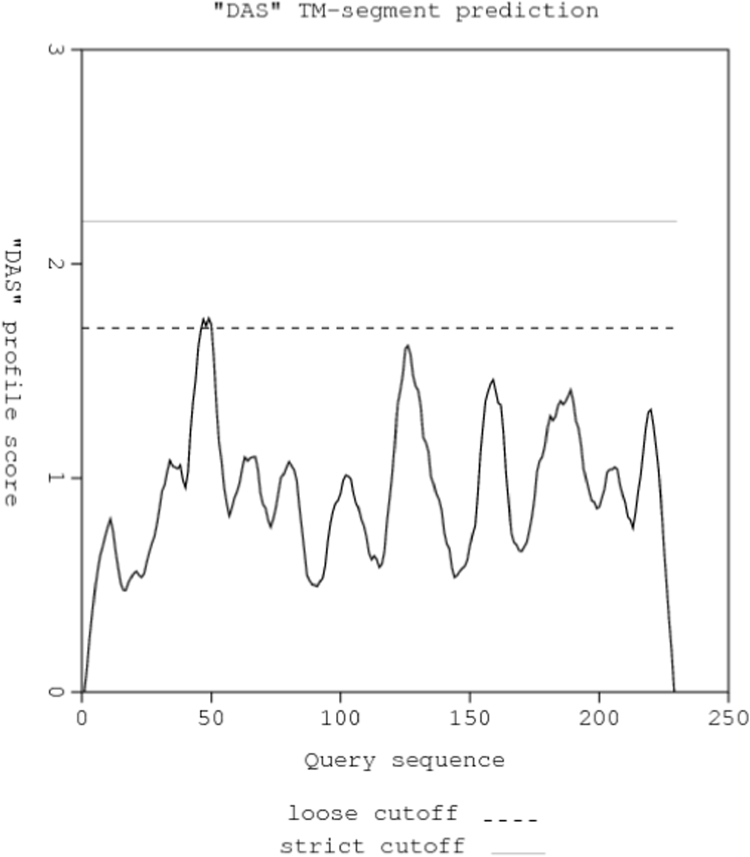
The results of the in silico analysis also indicate that the amylase protein (CDF_Amyl) under study is not a transmembrane protein (Fig. 8). This is also in agreement with the previous findings of this study that the alpha amylase (CDF_Amyl) from A. niger CSA35 is not hydrophobic but a secreted, soluble protein. According to Phillips et al. [35], transmembrane proteins are typically hydrophobic in nature since they span through the non polar phospholipid bilayers of membranes and a large proportion of their structures are embedded in the lipids. Separation of such proteins from biomembranes is usually difficult except with use of nonpolar solvents, detergents, and/ or denaturing agents [37]. However, the CDF_Amyl protein, being a non-transmembrane and hydrophilic protein, is not bound to the fungal cell membrane but rather secreted into the fungi growth medium. This characteristic can be exploited during industrial isolation and purification of the cassava starch-degrading fungal α-amylase to obtain the functional protein in its intact or undenatured form [45].
Fig. 8.
Prediction of transmembrane segments in CDF_Amyl. No transmembrane helix was found in the CDF_Amyl protein which showed that the protein is a non TM-protein. DAS, dense alignment surface.
4. Conclusion
The present study findings strongly affirmed that the amplified amylase gene from A. niger CSA35 is a member of the α-amylase super-family. The unique characteristics of the CDF_Amyl gene and its deduced protein could find applications in biotechnological, food and pharmaceutical industries where cloning and further modification of this gene would be required for product development and improvement.
Acknowledgements
The authors greatly appreciate all research personnels at the African Research Laboratories (ARL), Otorho-Agbon, Delta State and Bioscience Center, IITA, Ibadan, Nigeria for their kind assistance during the research.
Acknowledgments
Conflict of interests
The authors do not declare any conflict of interests.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.03.006.
Appendix A. Transparency document
Supplementary material
References
- 1.Adebayo C.O., Aderiye B.I., Akpor O.B. Assessment of bacterial and fungal spoilage of some Nigerian fermented and unfermented foods. Afri. J. Food Sci. 2014;8(3):140–147. [Google Scholar]
- 2.Ahmad R., Mohsin M., Ahmad T., Sardar M. Alpha amylase assisted synthesis of TiO2 nanoparticles: structural characterization and application as antibacterial agents. J. Hazardous Mater. 2015;283:171–177. doi: 10.1016/j.jhazmat.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 3.Andersen B., Frisvad J.C., Søndergaard I., Rasmussen I.S., Larsen L.S. Associations between fungal species and water-damaged building materials. Appl. Environ. Microbiol. 2011;77(12):4180–4188. doi: 10.1128/AEM.02513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avwioroko O.J., Tonukari N.J. Isolation and molecular identification of Aspergillus species associated with the spoilage of cassava in Nigeria. Nig. J. Sci. Environ. 2014;13(1):55–68. [Google Scholar]
- 5.Avwioroko O.J., Tonukari N.J., Asagba S.O. Biochemical characterization of crude α-amylase of Aspergillus spp. associated with the spoilage of cassava (Manihot esculenta) tubers and processed products in Nigeria. Adv. Biochem. 2015;3(1):15–23. [Google Scholar]
- 6.Bamidele O.P., Ogundele F.G., Ojubanire B.A., Fasogbon M.B., Bello O.W. Nutritional composition of “gari” analog produced from cassava (Manihot esculenta) and cocoyam (Colocasia esculenta) tuber. Food Sci. Nutr. 2014;2(6):706–711. doi: 10.1002/fsn3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baum D.A. Reading a phylogenetic tree: the meaning of monophyletic groups. Nature Educ. 2008;1(1):190. [Google Scholar]
- 8.Baum D.A., Offner S. Phylogenies and tree thinking. Am. Biol. Teacher. 2008;70:222–229. [Google Scholar]
- 9.Blum T., Briesemeister S., Kohlbacher O. MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics. 2009;10:274. doi: 10.1186/1471-2105-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chingin K., Astorga-Wells J., Pirmoradian Najafabadi M., Lavold T., Zubarev R.A. Separation of polypeptides by isoelectric point focusing in electrospray-friendly solution using a multiple-junction capillary fractionator. Anal. Chem. 2012;84(15):6856–6862. doi: 10.1021/ac3013016. [DOI] [PubMed] [Google Scholar]
- 11.da Lage J.L., Binder M., Hua-Van A., Janeček Š., Casane D. Gene make-up: rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evolut. Biol. 2013;13(1):40. doi: 10.1186/1471-2148-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva T.M. Fungal amylases: applications and functional properties. In: Maria de Lourdes T.M.P., Rai M., editors. 'Fungal Enzymes'. CRC Press, Taylor & Francis Group; New York: 2013. pp. 152–172. [Google Scholar]
- 13.De Conti L., Baralle M., Buratti E. Exon and intron definition in pre‐mRNA splicing. Wiley Interdisciplinary Rev. 2013;4(1):49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 14.de Souza P.M., Magalhães P.O. Application of microbial α-amylase in industry – a review. Braz. J. Microbiol. 2010;41:850–861. doi: 10.1590/S1517-83822010000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deb P., Talukdar S.A., Mohsina K., Sarker P.K., Sayem S.M.A. Production and partial characterization of extracellular amylase enzyme from Bacillus amyloliquefaciens P-001. SpringerPlus. 2013;2:154–164. doi: 10.1186/2193-1801-2-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delkash-Roudsaria S., Zibaee A., Mozhdehi M.R.A. Digestive α-amylase of Bacterocera oleae Gmelin (Diptera: J. King Saud Univ. Sci. 2014;26(1):53–58. [Google Scholar]
- 17.Farhan H., Rabouille C. Signalling to and from the secretory pathway. J. Cell Sci. 2011;124:171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 18.Galka P., Jamez E., Joachim G., Soumillion P. QuickLib, a method for building fully synthetic plasmid libraries by seamless cloning of degenerate oligonucleotides. PLOS One. 2017;12(4):e0175146. doi: 10.1371/journal.pone.0175146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta R., Gigras P., Goswami V.K., Chauhan B. Microbial amylase: a biotechnological perspective. Process Biochem. 2003;38:1599–1616. [Google Scholar]
- 20.Hirokawa T., Boon-Chieng S., Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 21.Holm L., Koivula A.K., Lehtovaara P.M., Hemminki A., Knowles J.K.C. Random mutagenesis used to probe the structure and function of Bacillus stearothermophilus alpha-amylase. Protein Engr. 1990;3:181–191. doi: 10.1093/protein/3.3.181. [DOI] [PubMed] [Google Scholar]
- 22.Huang H.L., Charoenkwan P., Kao K.F., Lee H.C., Chang F.L., Huang W.L., Ho S.J., Shu L.S., Chen W.L., Ho S.Y. Prediction and analysis of protein solubility using a novel scoring card method with dipeptide composition. BMC Bioinform. 2012;13(Suppl 17):S3. doi: 10.1186/1471-2105-13-S17-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husnik F., McCutcheon J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018;16(2):67. doi: 10.1038/nrmicro.2017.137. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda M., Arai M., Lao D.M., Shimizu T. Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 2002;2(1):19–33. [PubMed] [Google Scholar]
- 25.Kelley L.A., Sternberg M.J.E. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y., Lei Y., Zhang X., Gao Y., Xiao Y., Peng H. Identification and phylogenetic characterization of a new subfamily of α-amylase enzymes from marine microorganisms. Mar. Biotechnol. 2012;14(3):253–260. doi: 10.1007/s10126-011-9414-3. [DOI] [PubMed] [Google Scholar]
- 27.Maarel M.J.E.C., Veen B., Uitdehaag J.C.M., Leemhuis H., Dijkhuizen L. Properties and applications of starch-converting enzymes of the α-amylase family. J. Biotechnol. 2002;94:137–155. doi: 10.1016/s0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 28.Maheshwari R., Bharadwaj G., Bhat M.K. Thermophilic fungi: their physiology and enzymes. Microbiol. Mol. Biol. Rev. 2000;64(3):461–488. doi: 10.1128/mmbr.64.3.461-488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manna S., Harman A., Accari J., Barth C. Altering the selection capabilities of common cloning vectors via restriction enzyme mediated gene disruption. BMC Res. Notes. 2013;6(1):85. doi: 10.1186/1756-0500-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucl. Acids Res. 2017;45(D1):D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metin K., Koc O., Ateşlier B.B., Biyik H.H. Purification and characterization of α-amylase produced by Penicillium citrinum HBF62. Afr. J. Biotechnol. 2010;9(45):7692–7701. [Google Scholar]
- 32.Mitaku S., Hirokawa T. Physicochemical factors for discriminating between soluble and membrane proteins: hydrophobicity of helical segments and protein length. Protein Eng. 1999;12(11):953–957. doi: 10.1093/protein/12.11.953. [DOI] [PubMed] [Google Scholar]
- 33.Mitaku S., Hirokawa T., Tsuji T. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics. 2002;18:608–616. doi: 10.1093/bioinformatics/18.4.608. [DOI] [PubMed] [Google Scholar]
- 34.O.B. Oyewole, L.O. Sanni. Constraints in traditional cassava processing: A case of ‘fufu’ production. in: Transformation Alimentaire Du Manioc (Cassava Food Processing). Editors: Tom Agbor Egbe, AlainBrauman, Dany Griffon, Serge Treche. Institut Francais de Recherche Scientifique Pour le Development en Cooperation (ORSTOM), Paris, France. ISBN 2-7099-1279-1. Pp. 523–529.
- 35.Phillips R., Ursell T., Wiggins P., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459(7245):379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puspasari F., Radjasa O.K., Noer A.S., Nurachman Z., Syah Y.M., Maarel M., Dijkhuizen L., Janeček Š., Natalia D. Raw starch–degrading α‐amylase from Bacillus aquimaris MKSC 6.2: isolation and expression of the gene, bioinformatics and biochemical characterization of the recombinant enzyme. J. Appl. Microbial. 2013;114(1):108–120. doi: 10.1111/jam.12025. [DOI] [PubMed] [Google Scholar]
- 37.Rabilloud T. Humana Press; Chicago: 2009. Solubilization of Proteins in 2DE: An Outline. In Two-Dimensional Electrophoresis Protocols; pp. 19–30. [DOI] [PubMed] [Google Scholar]
- 38.Rabouille C. Pathways of unconventional protein secretion. Trends in cell biology. 2017;27(3):230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Ray B., Bhunia A. CRC press; Florida: 2013. Fundamental Food Microbiology. [Google Scholar]
- 40.Saranraj P., Stella D. Fungal amylase: a review. Int. J. Microbiol. Res. 2013;4(2):203–211. [Google Scholar]
- 41.Siddique F., Hussain I., Mahmood M.S., Ahmed S.I., Iqbal A. Isolation and characterization of a highly thermostable αlpha-amylase enzyme produced by Bacillus licheniformis. Pak. J. Agri. Sci. 2014;51(2):309–314. [Google Scholar]
- 42.Sussman A.S. Longevity and survivability of fungi. The fungi. 2013;3:447–486. [Google Scholar]
- 43.Taheri A., Robinson S.J., Parkin I., Gruber M.Y. Revised selection criteria for candidate restriction enzymes in genome walking. PLOS One. 2012;7(4):e35117. doi: 10.1371/journal.pone.0035117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tonukari N.J., Oliseneku E.E., Avwioroko O.J., Aganbi E., Orororo O.C., Anigboro A.A. A novel pig feed formulation containing Aspergillus niger CSA35 pretreated-cassava peels and its effect on growth and selected biochemical parameters of pigs. Afr. J. Biotechnol. 2016;15(19):776–785. [Google Scholar]
- 45.Vit O., Petrak J. Integral membrane proteins in proteomics. How to break open the black box? J. Proteom. 2017;153:8–20. doi: 10.1016/j.jprot.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Voit E.O. Cambridge University Press; United Kingdom: 2000. Computational Analysis of Biochemical Systems: A Practical Guide for Biochemists and Molecular Biologists. [Google Scholar]
- 47.Wetie A.G.N., Sokolowska I., Woods A.G., Roy U., Deinhardt K., Darie C.C. Protein–protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches. Cell. Mol. Life Sci. 2014;71(2):205–228. doi: 10.1007/s00018-013-1333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S.M. Yu, Rice α-amylase transcriptional enhancers. U.S. Patent No. 7,094,958. Washington, DC: U.S. Patent and Trademark Office, 2006.
- 49.Zidani S., Ferchichi A., Chaieb M. Genomic DNA extraction method from pearl millet (Pennisetum glaucum) leaves. Afr. J. Biotechnol. 2005;4(8):862–866. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material