Abstract
Finland provides unique opportunities to investigate population and medical genomics because of its adoption of unified national electronic health records, detailed historical and birth records, and serial population bottlenecks. We assembled a comprehensive view of recent population history (≤100 generations), the timespan during which most rare-disease-causing alleles arose, by comparing pairwise haplotype sharing from 43,254 Finns to that of 16,060 Swedes, Estonians, Russians, and Hungarians from geographically and linguistically adjacent countries with different population histories. We find much more extensive sharing in Finns, with at least one ≥ 5 cM tract on average between pairs of unrelated individuals. By coupling haplotype sharing with fine-scale birth records from more than 25,000 individuals, we find that although haplotype sharing broadly decays with geographical distance, there are pockets of excess haplotype sharing; individuals from northeast Finland typically share several-fold more of their genome in identity-by-descent segments than individuals from southwest regions. We estimate recent effective population-size changes through time across regions of Finland, and we find that there was more continuous gene flow as Finns migrated from southwest to northeast between the early- and late-settlement regions than was dichotomously described previously. Lastly, we show that haplotype sharing is locally enriched by an order of magnitude among pairs of individuals sharing rare alleles and especially among pairs sharing rare disease-causing variants. Our work provides a general framework for using haplotype sharing to reconstruct an integrative view of recent population history and gain insight into the evolutionary origins of rare variants contributing to disease.
Keywords: Finland, haplotypes, population genetics, rare variants, human history
Introduction
Most rare variants that play a critical role in diseases today arose during approximately the last 100 generations and provide signatures of population history.1 Recent large-scale DNA sequencing consortia efforts have demonstrated that one of the most predictive features of pathogenicity is allele frequency, given that most disease-causing variants are rare and thus relatively young.2, 3 These variants have not yet been fully exposed to the forces of natural selection that common, older variants have survived. However, aside from de novo variants in early-onset developmental phenotypes, the role of recently evolved, large-effect variants in common disease is largely uncharacterized. Stronger effects are most likely not confined to de novo variants but could persist for several generations; however, it has been difficult to identify this class of variation with single-variant analyses because such analyses have extremely limited power, especially for scenarios involving incomplete penetrance.4, 5 It is imperative that we better understand recent population genetic history because it bounds the ability of natural selection to purge deleterious variants during the most relevant period for producing disease-conferring variants subject to negative selection.6, 7 Furthermore, standard GWAS approaches typically include principal components to correct for population structure, but this is insufficient for rare variants.8 Haplotype-based methods have two major benefits over single-variant approaches for inferences into demographic history and disease association: (1) as opposed to commonly used site-frequency-based approaches,9 they are more informative of population history during the last tens to hundreds of generations, and (2) they can expose disease-causing rare variants at the population level without necessitating deep whole-genome sequencing. Rather, haplotype sharing can take advantage of massive, readily-available GWAS array data. Although these advantages have been theoretically recognized when sample sizes were relatively small,10, 11 they have been underutilized in the modern genomics era.
Finland provides a convenient example from which population history and rare disease associations can be inferred because of its unified electronic health records as well as the founder effect elicited by serial population bottlenecks. In addition to the out-of-Africa bottleneck experienced by all of Europe, Finland underwent multiple additional bottlenecks over the last few thousand years, and the Finnish founder population size is estimated to have included 3,000–24,000 individuals.12, 13, 14, 15, 16 Archaeological evidence indicates that Finland has been continuously but sparsely inhabited since the end of the last ice age ∼10.9 kya,14 when a small population of not more than a few thousand early hunter-gatherers first settled throughout Finland mostly from the south and to a lesser extent from the east and west.17 An ancient DNA study using samples dated to 6,000–9,500 years old across Sweden, Norway, and the Baltic Islands found evidence of multiple migration events into Scandinavia, where an east-west genetic gradient opposed the geographical gradient of modern populations.18 A cultural split in approximately 2,300 BC was hypothesized to separate the western and eastern areas of Finland, termed the early- and late-settlement regions (ESRs and LSRs), upon the arrival of the Corded Ware culture, which was primarily restricted to the southwestern and coastal regions of the country; this split has been supported by Y chromosome and mitochondrial DNA as well as historical data.17, 19, 20 During the last two millennia, a series of founding, extinction, and re-colonization events took place before continuous habitation coincident with agriculture.21 The ESR, encompassing the southern and western colonized regions of Finland, was more densely and permanently settled beginning ∼4,000 years ago, whereas the LSR, encompassing the northern and eastern regions of Finland, was more permanently inhabited beginning in the 1500s, pushing existing nomadic Sami people farther north into Lapland. Although Finland was a part of the Swedish Kingdom until 1809 and then became a semi-autonomous grand duchy controlled by tsarist Russia until it gained independence in 1917, immigration into western and especially eastern Finland was relatively low until after the collapse of the Soviet Union.22 Linguistically, the mother tongue of roughly 5% of the population is Swedish, and both Finnish and Swedish are taught at school. Bilingual Finns who speak Swedish as their mother tongue live mostly in the early-settlement region in restricted western and southern coastal regions.
Because of serial bottlenecks in Finland, the site-frequency spectrum is skewed toward more common variants than in other European populations, and deleterious alleles are more likely to be found in a homozygous state.16 The consequence of this is exemplified in the Finnish Disease Heritage (FinDis) database, which to date contains 36 monogenic diseases that are much more common in Finns than in any other population.23 Several complex diseases also show strong regional clines within Finland. For example, risk of schizophrenia and familial hypercholesterolemia is greatest in northeastern Finland.24, 25 Current Finnish demographic models are primarily based on single locus markers (i.e., the Y chromosome and mitochondria),12, 19, 20 and a few studies have recently expanded to incorporate autosomal data.22, 26, 27 Methods based on the site-frequency spectrum consider sites independently and are therefore optimized for inferring old demographic events (>100 generations ago); by contrast, haplotype-based demographic inference is optimized for detailing population history during the period most relevant for negatively selected traits (i.e., the last 100 generations).28, 29, 30, 31 Multiple lines of evidence indicate that recent history is particularly important for disadvantageous traits. For example, long runs of homozygosity (ROH), a special case of recent haplotype sharing, are enriched for deleterious variation,32 and increased ROH have been associated with decreased educational attainment as well as intellectual disability.33, 34 Furthermore, allele dating techniques indicate that pathogenic variants are on average considerably younger than neutral variants.3
Prior studies have used haplotype-based inference of pairwise sharing to query the recent demographic history of other populations, for example those with massive, densely connected genetic networks across the US.35 This type of inference has provided regional insights into how African American migration routes, which differ markedly from those of European Americans, have changed since the dawn of slavery in the US.36, 37 High levels of haplotype sharing have been observed in a number of other founder populations, including the Druze;38 Ashkenazi and other Jews;39, 40, 41 Indians;42 French Canadians;43 Hispanic/Latino and Native Americans;44, 45, 46 European isolates;47, 48, 49, 50, 51 and other European populations.30, 52, 53, 54 However, few studies have investigated recent demographic history in depth with pairwise haplotype sharing in Finns, who are among the best-studied population isolates.13, 22, 55
In this study, we combined biobank-scale genetic and detailed birth-record data to assemble a comprehensive inquiry into recent population history by employing genetic data from 43,254 Finnish individuals (∼0.8% of Finland’s total population) and 16,060 demographically distinct individuals from geographically or linguistically neighboring countries, including Sweden, Estonia, Russia, and Hungary. Although Finland is a poised example for population insights from haplotype sharing because of its serial population bottlenecks, our approach provides a general framework for using haplotype sharing to reconstruct an integrative view of recent population history within and across countries (e.g., elucidation of changes in migration, divergence, and population size over time). Through these analyses, we also demonstrate that elevated haplotype-sharing patterns resulting from multiple population bottlenecks provide insights into the origins of certain genetic diseases.
Material and Methods
Genotyping Datasets
Finnish samples were genotyped for various projects, all of which have been published previously and most of which were described in Surakka et al.56 In brief, study participants were as follows: European Network for Genetic and Genomic Epidemiology (ENGAGE) Consortium, Myocardial Infarction Genetics Exome Array Consortium,16 FINRISK (1992, 1997, 2002, and 2007) cohorts, Northern Finland Birth Cohort 1966, Corogene controls (which are also from FINRISK), Health 2000 samples from the GenMets study, the Helsinki Birth Cohort Study, the Cardiovascular Risk in Young Finns Study, and the Finnish Twin Cohort. All birth records are from the FINRISK study, which is a superset of several projects. The FINRISK 1997 cohort contains municipality-level birth records (n = 3,942), and the 2007 cohort contains region-level birth records (n = 5,448), which were genotyped across different projects and/or arrays (Table S1). Swedish samples used here were waves 5 and 6 (Sw5 and Sw6) and were genotyped as part of a schizophrenia study.57 Swedish genotype data are available upon application from the National Institute of Mental Health (NIMH) Genetics Repository (see Web Resources). Estonian samples are from the Estonian Genome Center, University of Tartu.58 Genotyping for individuals from St. Petersburg, Russia was performed as a part of a starvation study ongoing at the Broad Institute on a cohort previously described in Rotar et al.59 Hungarian samples included in the study were genotyped as part of the Hungarian Transdanubian Biobank.60 Genotyping details and sample sizes are shown in Table S2.
Exome Sequencing Data and Quality Control
Exome sequencing data from multiple studies of Finnish individuals were collected and harmonized as part of Sequencing Initiative Suomi (SISU) study (Table S3). The Finnish sequence data processing and variant calling is similar to that described previously.61 In brief, we filtered these data so that what remained were exomes with overlapping GWAS data from unrelated individuals in this study (n = 9,363), as described below in “Haplotypes Overlapping Exome Variants.” These individuals were primarily from the FINRISK study obtained through dbGaP.62 We performed sample and variant quality control after joint calling to assess the relationship between rare variation and pairwise haplotype sharing. We first removed any individuals with missingness > 10% at sites where allele frequency is greater than 0.001 and missingness is less than 10%. We then filtered to variants meeting genotype quality filters as follows: genotype quality ≥ 20, depth ≥ 10X, and allele balance > 0.2. We then extracted variants present at least twice, with a call rate > 0.8, and excluded variants that failed GATK VQSR quality.
Phasing and Imputation
All Finnish genotypes underwent quality control, phasing, and imputation, as described previously.56 Imputation was performed with the 1000 Genomes Project data.
Principal-Component Analysis
We combined best-guess genotypes for 43,254 Finnish individuals whose variants had been imputed with info score > 0.99 across all arrays, including the Affymetrix Genome-Wide Human SNP 6.0, Illumina Human 370k, 610k, 670k, Core Exome, and OmniExpress arrays. This resulted in ∼3.4 million accurately imputed common SNPs across all individuals. From these sites, we performed linkage-disequilibrium (LD) pruning by using PLINK v1.90b3f63 and keeping SNPs with minor-allele frequencey (MAF) > 0.05, missingness < 10%, and R2 ≤ 0.50 by using a window size of 50 SNPs and a 5 SNP overlap between windows. PCs were computed across 232,332 sites for all Finnish individuals with flashpca.64 We also generated a multi-population dataset of unrelated individuals with birth records, when available, from Finland; Sweden; Estonia; Hungary; and St. Petersburg, Russia. As before, we extracted best-guess Finnish imputed sites with info score > 0.99. We also filtered to individuals with ≤10% missingness, sites with ≤10% missingness, MAF ≥ 0.05, and LD R2 < 0.5. Because of array heterogeneity, we also filtered to sites on the Illumina Global Screening Array to avoid removing all Russian individuals because of high missingness. We then ran principal-component analysis (PCA) with 65,224 sites across n = 11,287 individuals.
Genetic Divergence
We computed FST among geographical regions by using PLINK v1.90b3f.63 For all analyses, we used the weighted Weir-Cockerham FST estimate.
Genetic Relatedness
We identified the maximal set of unrelated individuals separated by at least two degrees of relatedness by using KING v2.065 within each population. We identified a maximal unrelated set of 34,737 Finnish individuals, 7,863 Swedish individuals, 6,328 Estonian individuals, 294 Hungarian individuals, and 210 Russian individuals.
Haplotype Calling
We generated two sets of haplotypes: one by using IBDseq to assess effective population-size changes over time for Finland-only analyses,66 and another by using GERMLINE for all other analyses.67 We used IBDSeq rather than GERMLINE for the IBDNe analyses because of previous recommendations68 stating that switch errors in estimated haplotypes can cause erroneous haplotype breaks, resulting in spuriously recent inferences regarding the time to most recent common ancestor; IBDseq is less susceptible to these errors because it does not rely on phased data as input. We ran IBDseq on the maximal set of unrelated individuals with birth-record data (n = 9,008 individuals with 169,306 SNPs). To perform effective population-size inferences per region, we took the subset of haplotypes where both pairs of individuals were born in the same region.
For all other analyses, we used haplotypes called with GERMLINE. We first phased all genotype data together by using Eagle v2.3.2.69 We then generated haplotype calls by using GERMLINE (because of its computational tractability at large sample sizes) with the following parameters: -err_hom 0 -err_het 2 -bits 25 -h_extend -haploid. To investigate the decay of identity-by-descent (IBD) tract length, we used a minimum haplotype size of 1 cM (−min_m 1) within each population for unrelated samples with birth-record data and/or exome-sequencing data. When assessing haplotype sharing across the full set of unrelated genotyped Finns without respect to birth records, we set a minimum haplotype size (--min_m) of 3 cM for computational tractability and reasonable storage sizes. We removed haplotypes that fall partially or fully within centromeres, telomeres, acrocentric short chromosomal arms, heterochromatic regions, clones, and contigs identified in the UCSC hg19 genome “gaps” table.
Because we had used imputed sites to harmonize genotype data across arrays (albeit with very high fidelity with info score > 0.99), we assessed haplotype calling concordance from imputed data versus genotype-array data. As expected, we found high concordance between genotyped and high-quality imputed sites across datasets and geographical regions, especially for haplotypes longer than 2–3 cM (Figure S7).
Haplotype Calling for Effective Population-Size Analyses
Variants imputed with an info score > 0.99 that intersected across all six arrays on which Finnish samples were genotyped (Table S2) were included in the haplotype analyses, resulting in 3.4 million accurately imputed common SNPs across 43,254 individuals. High-imputation-quality best-guess genotypes were subsequently filtered to have MAF > 0.05, no indels, and LD R2 < 0.5. We ran IBDNe across regions of Finland by taking the subset of pairs of individuals who were both born in the same region. Demographic analyses included pairwise haplotypes for individuals from the FINRISK 1997 and 2007 cohorts, which contained the following number of individuals by region: 1,123 in region 1, 1,078 in region 2, 378 in region 4, 224 in region 5, 304 in region 6, 1,581 in region 7; 1,547 in region 8, 225 in region 9, 228 in region 10, 1,697 in region 11, and 184 in region 12 (region names are as in Table S4).
Mapping Cumulative Haplotype Sharing
Municipality-level maps of Finland, Sweden, and Estonia were downloaded in R SpatialPolygonsDataFrame (file format S4) format from the GADM database of Global Administrative Areas (see Web Resources) on 9/14/2015, 4/13/2017, and 7/24/2017, respectively. Pairwise sharing was computed for a maximal unrelated set of individuals (≥2nd degree relatives) with municipality- or region-level birth-record data (n = 8,630 individuals total: n = 5,020 with municipality-level data from FR97, and n = 3,610 with region-level data from FR07). From each city, all pairs where at least one individual had parents born within 80 km of each other and whose mean birth location was within 80 km of the city of interest were included. Municipalities are official and were numbered as described in the Web Resources, with three additional codes: 198 = no home in Finland, 199 = unknown, and 200 = abroad. To account for uncertainty when only region-level data were available, even weights were assigned to all municipalities within that region with the sum of the weights equal to 1; in contrast, a single municipality was given a weight of 1 in the municipality-level data.
Estimating Effective Migration Surfaces
We used estimating effective migration surfaces (EEMS) tool70 to estimate migration and diversity relative to geographic distance. We computed genetic dissimilarities for all unrelated pairwise individuals for whom municipality-level birth-record data were available and whose parents were both within 80 km; we used mean parental latitude and longitude when the parents’ data differed. We computed pairwise genetic dissimilarities by using the bed2diffs tool provided with EEMS on the intersected Finnish data, which included 232,332 SNPs for 2,706 individuals, as well as on the intersected Finnish, Swedish, Estonian, and Russian data, which included 88,080 genotyped SNPs across 10,993 individuals. We set the number of demes to 300 (but actually observed fewer than this) and adjusted the variances for all proposal distributions of migration, diversity, and degree-of-freedom parameters such that most were accepted 20%–30% of the time and all were accepted 10%–40% of the time, per recommendations in the manual. We increased the number of Markov chain Monte Carlo (MCMC) iterations, burn-in iterations, and thin iterations until the MCMC converged.
Whereas Finland birth records used in this analysis are at the municipality level, Swedish and Estonian birth records are at the region level. Because of differing birth-record densities and boundaries in Finland-only versus multi-country analyses, there are differing densities and numbers of observed demes. When setting nDemes = 300 across Finland, Sweden, Estonia, and St Petersburg, Russia, we observed 110 out of 274 demes. When setting nDemes = 300 across Finland alone, we observed 167 out of 266 demes.
Haplotypes Overlapping Exome Variants
All analyses of haplotypes paired with exome sequencing data were performed with Hail version 0.1. To map IDs between the genotype and exome sequencing data, we filtered genotype and exome data to variants with at least 1% frequency and less than 10% missingness in each dataset and subsequently removed individuals with greater than 10% missingness. We intersected these datasets, repeated the same filtering process, and identified 9,363 individuals with both data types by using the Hail IDB function, which is equivalent to plink --genome (minimum pi_hat = 0.95). Using haplotypes called with GERMLINE, we filtered out regions of the genome shared across all pairs at a rate greater than three times the standard deviation above the mean level of sharing (Figure S6) for improved computational tractability and to remove false positives that are common in regions of the genome with very high levels of sharing. We performed this filtering to make pairing of exome and haplotype pair data computationally tractable and remove false positives that are common in regions of the genome with very high levels of sharing. We overlaid the haplotype data with the exome data by using the annotate_variants_table function and calculated the number of pairs of individuals sharing haplotypes and genotypes for each variant (we excluded singletons and variants that failed VQSR filtering) by using a custom script in the Hail expression language. In brief, we determined the set of individuals carrying each genotype and then iterated over the pairs of individuals who share haplotypes; we counted cases where both members of the pair harbored the same genotype. The number of pairs that did not share a given genotype was simply computed as the number of pairs with the genotype minus the number of pairs that shared the genotype. We subsequently annotated variants with VEP version 85 by using transcripts from Gencode v19 and the LOFTEE plugin (Web Resources). We then computed a simple enrichment ratio for haplotype sharing at exome sequencing variants shared among heterozygous individuals (i.e., carriers) versus homozygous reference individuals, as follows: . We stratified haplotype enrichments across allele frequencies and predicted functional variant consequence as well as variants known to cause diseases in FinDis. Starting with a list of 50 FinDis annotated autosomal variants, we found that 40 exome sequencing variants were polymorphic and had overlapping haplotypes. Table S5 contains haplotype enrichments among carrier pairs relative to homozygous reference individuals.
Results
Population Substructure across Regions of Finland
To investigate fine-scale population structure within Finland, we assembled a panel of 43,254 Finnish individuals (Table S2, Material and Methods). We performed PCA on all individuals and used the subset of individuals with recorded birth record data to show that genetic variation in Finland broadly reflects geographical birthplace (Material and Methods, Figure 1A); we found highly significant correlations between PC1 and longitude (ρ = −0.72, p < 1 × 10−200) and between PC2 and latitude (ρ = −0.55, p < 1 × 10−200). The PCA and birth-record data also reflect variability in sampling and population density; high density in Helsinki and Turku contrasts with low density in the northernmost Lapland region (Figures S1A and S1B). Mean PC1 and PC2 across birth regions closely mirror geographical patterns, with the exception of southern Finland (region 1), which projects closer to central Finland than expected geographically; containing the capital city of Helsinki, southern Finland is the most populous region of Finland and consequently draws from across the country (Figures S1B and S1C). We also assessed genetic divergence across regions in Finland and identified relatively high levels of regional divergence in Finland compared to other European countries, e.g., the UK, Germany, Sweden, and Estonia;26, 71 mean FST between region pairs was 0.001 (Figure S1C). These results are consistent with an additional Finnish bottleneck with respect to nearby countries.
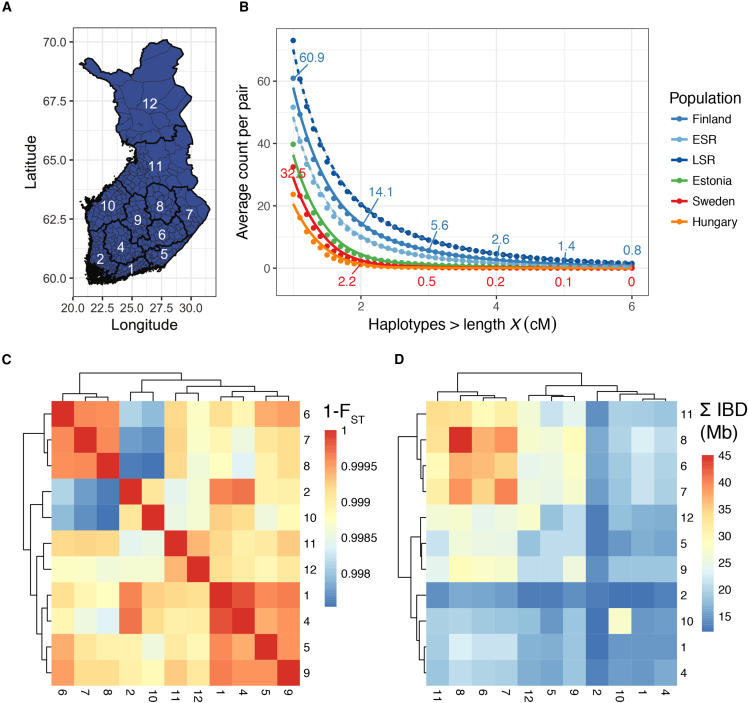
Figure 1.
Identity-by-Descent Haplotype Sharing and Genetic Divergence across Regions of Finland
(A) Regional map of Finland. Region names are shown in Table S4. Thin lines within regions represent municipality boundaries. Region 3 corresponds to the Åland Islands (not shown), a small Swedish-speaking archipelago in the Gulf of Bothnia.
(B) Distribution of average pairwise shared IBD segments in Finland (N = 7,669), specifically within two birth regions defined previously as having >95% posterior probability of clustering geographically in the early-settlement region (ESR; n = 428) and late-settlement region (LSR; n = 592),22 Estonia (n = 6,328), Sweden (n = 7,863), and Hungary (n = 294). All individuals included are unrelated and ancestrally representative of a given region or country. Numbers indicate average pairwise haplotypes shared at 1, 2, 3, 4, and 5 cM in Finland and Sweden.
(C) Hierarchical clustering of genetic similarity, as measured by 1 − FST across regions of Finland. Regions are numbered as in Table S4.
(D) Hierarchical clustering of cumulative IBD (minimum haplotype ≥ 3 cM) sharing across regions of Finland. Regions are numbered as in Table S4.
Regionally across Finland, we identified geographical clusters with high degrees of similarity. For example, Southern Savonia, Northern Karelia, and Northern Savonia (regions 6, 7, and 8, respectively) exhibit high degrees of genetic similarity (Figure 1C). We also identified genetic similarity clusters in the southern central regions of Southern Finland, Tavastia, Southern Karelia, and Central Finland (i.e., regions 1, 4, 5, and 9); western coastal regions of Southwest Finland and Ostrobothnia (2 and 10); and northern regions of Northern Ostrobothnia and Lapland (11 and 12). By comparing parent and offspring birthplaces, we show that within a single generation, offspring across Finland tend to move south, e.g., toward Helsinki (Kolmogorov-Smirnov two-sided test between and child’s and mean parents’ latitude: p = 8.7 × 10−3 (Figure S2).
Population Bottlenecks in Finland Are Reflected in Identity-by-Descent Sharing
To better understand the recent population history of Finland, we computed pairwise IBD sharing across all unrelated Finnish pairs of individuals (Material and Methods, Figures 1B and 1D). We performed hierarchical clustering of cumulative IBD sharing across pairs of individuals within and between regions of Finland, and we identified excess sharing in eastern Finland (regions 6, 7, and 8) compared with southwestern Finland (regions 1, 2, 4, and 10), where sharing was depleted (Figure 1D). Compared to genetic similarity from common variants (Figure 1C), haplotype-based clustering is more consistent with historical records that have documented the early- versus late-settlement regions in southwest and northeast Finland, respectively. Nonetheless, pairwise regional IBD and FST are highly correlated (Mantel test ρ = 0.89, p < 1 × 10−4 with 1,000 Monte Carlo repetitions). Previous work on serial founder effects showed that global genetic divergence increases with geographical distance,72 and we recapitulated this finding at the sub-country level within Finland (Figure S3A); we also identified decaying IBD sharing with increasing geographical distance within Finland (Figure S3B).
Because Finland historically has shared trade, language, and migration with neighboring countries and/or regions, including Sweden; Estonia; and St. Petersburg, Russia, we compared the relative level of allelic and haplotypic sharing within each population. We also compared these genetic data with individuals from Hungary because although it is geographically distal, it shares common linguistic roots; Finnish is a Uralic language that forms an outgroup to most European languages but is related to Estonian and Hungarian. Comparing pairwise IBD sharing within each of these countries, we found that cumulative IBD sharing between pairs of individuals is on average significantly greater across pairs of individuals in Finland than in Sweden, Estonia, Russia, and Hungary, which is expected from the Finnish population bottleneck (cumulative total of tracts ≥ 1 cM in length: = 22.9 cM versus = 107.0 cM, p < 1 × 10−50). Consistent with this observation, the average pair of Finns shares more haplotypes than the average pair in the other countries compared here, and these haplotypes are also longer: for example, 5.6 haplotypes ≥ 3 cM shared in Finland versus an order of magnitude fewer (0.5 haplotypes ≥ 3 cM) in Sweden (Figure 1B).
Recent Gene Flow and Migration Inference from IBD Sharing
We coupled haplotype sharing between pairs of individuals with municipality- and region-level birth-record data to determine relative rates of sharing among fine-scale locations in Finland. We used pairwise IBD to take the subset of individuals in which both parents were born within 80 km (∼50 miles) of each other. For each analysis, we further took the subset of pairs of individuals in which at least one individual had municipality-level birth records from within 80 km of a given city and assessed average pairwise IBD with other individuals across municipalities and regions of Finland. By comparing pairwise sharing from different Finnish cities, we found that IBD sharing is very uneven throughout the country, varying by several-fold, and that different geographical regions exhibit considerable substructure with differential IBD sharing patterns (Figure 2). This fine-scale structure is most likely driven by multiple bottlenecks, recent migration patterns, and variable population density (e.g., genetic diversity is higher, and thus IBD sharing is lower, in densely populated Helsinki than many rural areas because Helsinki ancestors have more diverse origins).
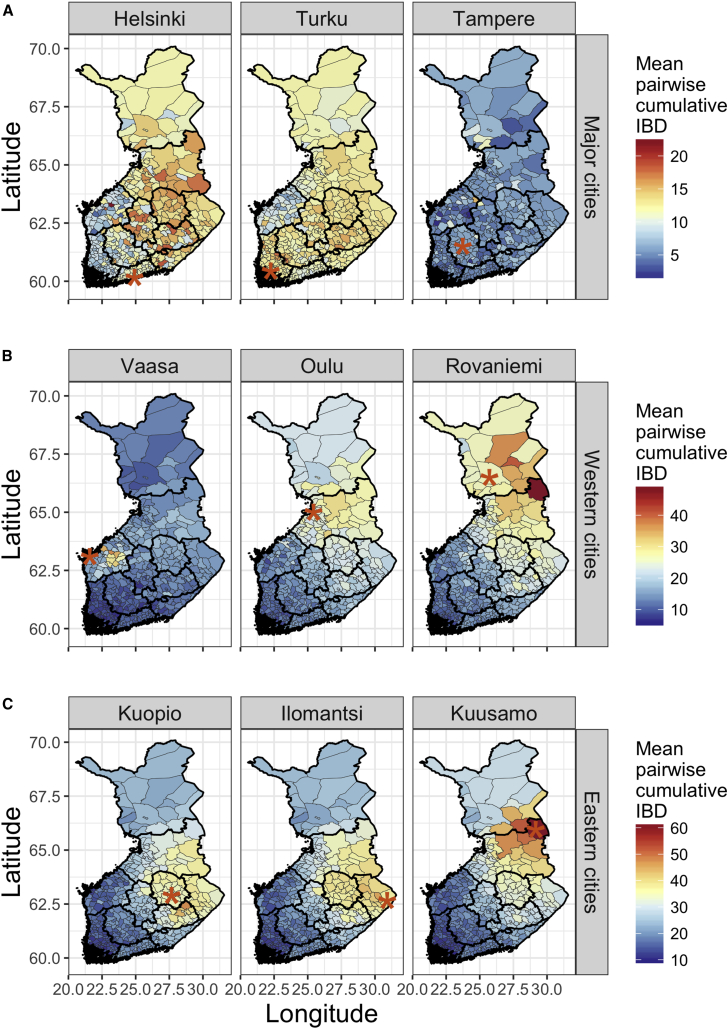
Figure 2.
Geographically Structured Haplotype Sharing between Pairs of Individuals across Finland
We used genetic data to take the subset of pairs of individuals whose birth records indicate that both parents were born within 80 km (∼50 miles) of each other. For each panel, we further took the subset of haplotypes from pairs of individuals in which at least one of the individual pairs lives within 80 km of cities indicated by red asterisks. Thinner lines outline municipalities, and thicker lines outline regions. The color shaded in each municipality indicates the weighted mean of cumulative IBD sharing for haplotypes ≥ 3 cM. Finland-wide city comparisons are grouped into three categories (note different scales for each): (A) major cities, (B) western cities, and (C) eastern cities.
For each city, the number of unique individuals whose parents are both from within an 80 km radius and total pairwise comparisons across Finland are as follows: n = 152 in Helsinki, 677,844 total pairwise comparisons; n = 227 in Turku, 1,003,794 total pairwise comparisons; n = 102 in Tampere, 457,419 total pairwise comparisons; n = 50 in Vaasa, 225,525 total pairwise comparisons; n = 185 in Oulu, 821,955 total pairwise comparisons; n = 13 in Rovaniemi, 58,877 total pairwise comparisons; n = 566 in Kuopio, 2,406,915 total pairwise comparisons; n = 363 in Ilomantsi, 1,580,502 total pairwise comparisons; and n = 25 in Kuusamo, 113,075 total pairwise comparisons.
Haplotype sharing is on average lowest when at least one individual lives in a major southern Finnish city (Figure 2A). Specifically, pairwise haplotype sharing is relatively low across Finland when at least one individual lives in Helsinki, Turku, and Tampere, which exhibit the lowest structure of cities compared here. Among individuals born in Helsinki, a relatively young capital (it became so in 1812), there is a subtle structure indicated by greater haplotype sharing with eastern Finland than western Finland on average; in contrast, individuals from the historical capital of Turku have more elevated haplotype sharing with nearby southwestern Finland (Figure 2). IBD sharing among western coastal cities (e.g., Vaasa, Oulu, and Rovaniemi) are intermediate and show varying patterns of regional haplotype sharing (Figure 2B). For example, Vaasa, a bilingual city with mostly Finnish and Swedish speakers surrounded by municipalities where people mostly speak Swedish, shows restricted patterns of elevated sharing specifically in Ostrobothnia (region 10). In contrast, Oulu and Rovaniemi in Northern Ostrobothnia and Lapland (regions 11 and 12) show broadly elevated patterns of sharing in the late-settlement region and depleted sharing in the early-settlement area. IBD sharing is generally highest among individuals living in northeastern cities in the late-settlement region (e.g., Kuopio, Ilomantsi, or Kuusamo); more structure is evident in the cosmopolitan cities, and greater sharing is evident in the late-settlement region (Figure 2C). Of all cities investigated, Kuusamo shows the most elevated IBD sharing: in haplotypes > 3 cM, ∼60 Mb on average is shared with nearby individuals, whereas ∼5–15 Mb is shared near Helsinki, Turku, and Tampere.
Fine-Scale Population Differentiation and Migration Rate Inference between Finland and Nearby Countries
We assessed how much sharing occurs within and between regions of Finland and neighboring countries and/or regions, including Sweden; Estonia; St. Petersburg, Russia; and Hungary (Figure 3A). PCA recapitulates geographic boundaries and Finnish bottlenecks: PC1 separates Finland from non-Finnish Europeans, and PC2 separates non-Finnish European populations along a cline (Figure 3B).58, 73 Birth regions also recapitulate expected trends; for example, southern Finns project closer in PCA space with northern Estonians than with individuals from other regions of either country (Figure 3B). Hierarchical clustering of genetic divergence (FST) within and between regions and countries demonstrates that divergence is typically smallest within countries, with the exception of Finland and the northernmost Swedish region, Norrbotten, which neighbors Finnish Lapland. These two areas cluster together, albeit with the greatest divergence within Finland plus Norrbotten (Figure S1D). Together with the migration-rate analysis, our results suggest that although Norrbotten is most genetically similar to Finnish Lapland, a migration barrier still separates these two counties. Individuals from the southwest coastal regions of Finland (regions 1, 2, 10, and 4; i.e., Southern Finland, Southwestern Finland, Ostrobothnia, and Tavastia) are more genetically similar to cosmopolitan Swedes than other Finns are (Figure S1D, Figure 3A). The divergence is greatest (FST ∼0.01) between eastern Finland (regions 6, 7, 8; i.e., Southern Savonia, North Karelia, and Northern Savonia) and the regions located within Hungary and southern Estonia (regions 30, 34, and 36) (Figure S1D). The elevated IBD sharing in Finland and the elevated divergence in relation to neighboring countries supports the utility of haplotypes for investigating recent population history as well as IBD mapping for identifying rare associations.74
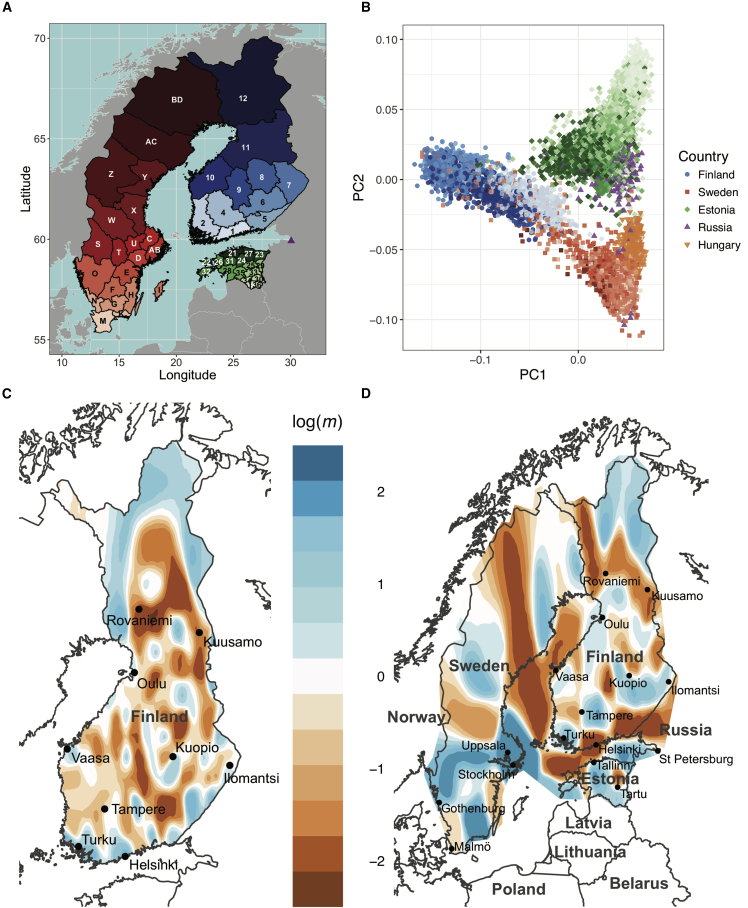
Figure 3.
Migration Rates and Haplotype Sharing within Finland and Between Neighboring Countries
(A) Map of regional Finnish, Swedish, and Estonian birthplaces. A purple triangle indicates St. Petersburg, Russia. Hungary is not shown. Finnish, Swedish, and Estonian region labels are shown in Table S4.
(B) PCA of unrelated individuals, colored by birth region (if available; otherwise, by country) as shown in (A).
(C and D) Migration rates inferred with EEMS. Values and colors indicate inferred rates, m: shades of blue indicate logarithmically higher migration at a given point on average (i.e., log(m) = 1 corresponds to effective migration that is 10-fold faster than the average), and shades of orange indicate migration barriers. (C) Migration rates among municipalities in Finland. (D) Migration rates within and between Finland; Sweden; Estonia; and St. Petersburg, Russia.
We also utilized the granular birth records to investigate geospatial migration rates (m) in Finland and among neighboring countries. We used a spatially explicit statistical model to estimate effective migration surfaces (via EEMS) by measuring effective migration rates from genetic differentiation (i.e., resistance distance) across neighboring demes.70 By measuring the genetic distance between evenly spaced demes relative to other pairs of demes across Finland and/or neighboring countries, we inferred locations where migration was uncommon (such locations are referred to as migration barriers and are depicted in dark orange) and where migration excesses occurred (these locations are depicted in blue) (Figures 3C and 3D). Across Finland, we found variable migration rates, many of which are consistent with known historical events (Figure 3C). For example, we identify migration barriers generally separating the early and late settlement area (i.e., between Tampere and Kuopio) as well as the northernmost Lapland region from the rest of Finland. In contrast, within Finland, there is increased migration in and directly surrounding several coastal cities, including Helsinki, Turku, Vaasa, and Oulu.
When one considers migration rates among individuals with birth records from Finland; Sweden; Estonia; and St. Petersburg, Russia (Figure 3D), the major migration routes within Finland remain broadly consistent. For example, a barrier to migration between the early and late settlement regions between Tampere and Kuopio remain, along with a barrier of migration into Lapland. The starkest difference is a barrier to migration along nearly the entire Finnish border (Figure 3D), most likely due to the absence of some neighboring comparison demes in Figure 3C (see also Figure S5), indicating little significant migration into Finland in the last 100 generations, consistent with the described patterns of low frequency variation presenting as a bottleneck or isolate. Apart from migration rate inferences along the border, subtle changes within Finland are most likely due to additional smoothing because of a larger area over which demes are spread (Material and Methods, Figure S5). Migration rates within Sweden are most elevated in southern regions near the largest cities, including Stockholm and Uppsala. As speculated previously,58 migration rates are generally elevated within Estonia but depleted along the west coast and between Tallinn and Tartu; rates are also depleted between the Estonia mainland and both Finland and Sweden. The strongest barriers to migration in and near Sweden are in the northwest as well as along the northwestern Finnish border separating Finnish Lapland and Sweden, although there are notably few individuals either sampled or living there, resulting in increased noise.
Regional Recent Effective Population Size Changes over Time
Haplotype sharing also enables an assessment of fluctuations in effective population size over time and across geographical regions. We inferred changes in effective population size over recent time across birth regions in Finland by using the haplotype-based IBDNe method.68 Across all birth regions, we identified a population expansion in the last 50 generations from around 103 to 105 and 106 (Figure 4 and Figure S4). The region with the largest current effective population size is Southern Finland (region 1, current Ne = 1.3 × 106, 95% CI = [5.5 × 105, 2.8 × 106]), which contains the capital city of Helsinki; these findings closely approximate current census data (current census population = ∼1.6 × 106). We inferred that Lapland (region 12), the northernmost and least populated region, had the least growth: current Ne = 6.9 × 104, 95% CI = [4.9 × 104, 8.8 × 104] (current census population = ∼1.8 × 105). The inferred effective population size is expected to be smaller than the census size because the census size includes multiple generations, variance in reproductive rates, and other factors.68
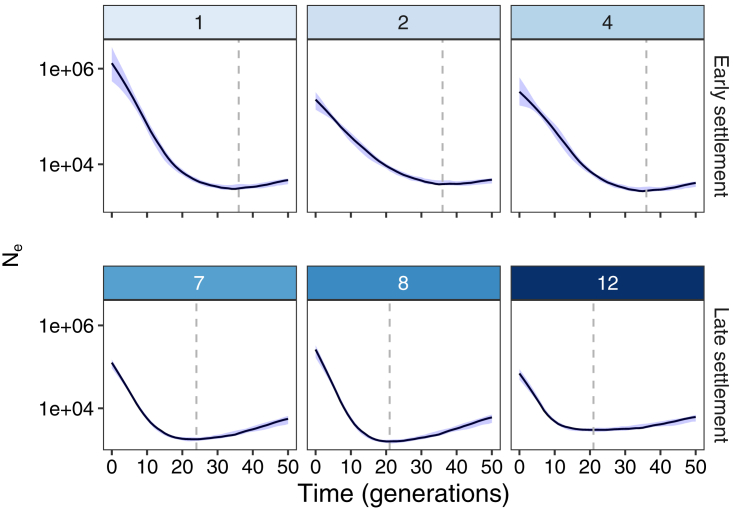
Figure 4.
Effective Population Size over Time by Birth Region in Finland
Representative regions within the early- and late-settlement regions are numbered as shown in Table S4. Dashed lines indicate the time at which the minimum Ne over the last 50 generations occurred in each region. The number of individuals in each region is shown in Figure S4. Error bars indicate 95% bootstrap confidence interval.
When comparing the early- and late-settlement regions, we found consistently earlier onset of population expansions in the early-settlement region. In the early-settlement region, for example, the population began expanding around 30–40 generations ago (circa 760–060 AD, if we assume a generation time of 30 years75). In contrast, the late-settlement region began expanding between approximately 15 and 25 generations ago (circa 1210–1510 AD) and had lower minimum effective population sizes (Figure 4). We also found significant evidence of a geographical cline, wherein populations began expanding earlier in regions farther south (ρ = 0.79, p = 4.2 × 10−3). For example, whereas Southern Finland and Southwestern Finland (regions 1 and 2) began growing ∼36 generations ago, the northernmost region of Lapland (region 12) only began growing ∼21 generations ago. We also infer larger current effective population sizes in the early- rather than late-settlement region, consistent with a higher population density for Finland in the early-settlement region. Together, the estimation of the regional expansion of the population in conjunction with IBD sharing within and between municipalities provides a clear picture of the population history as calculated entirely from genetic analysis of the modern Finnish population.
Haplotype Insights into Disease
To better understand the utility of IBD sharing for rare-variant interpretation, we coupled haplotype tracts with exome sequencing data (Material and Methods). A key consideration here is the quality and consistency of haplotype calls. To call the primary haplotypes used throughout this study, we used phased best-guess genotypes from very high-quality imputed sites (info score > 0.99) as input in GERMLINE to ensure overlap across several genotyping arrays. To look at an explicitly genotyped set, we compared haplotypes from the imputed set to haplotypes for overlapping samples genotyped as part of the ENGAGE consortium on the IlluminaCoreExome array because these had the largest number of individuals for whom birth records were available and who were also genotyped on a single array (Table S1 and Table S2). By phasing these overlapping samples and calling haplotypes separately, we could assess the fraction of haplotypes that overlapped, that were genotype specific, and that were imputation specific as a function of haplotype length and geography (Figure S7). As expected, shorter haplotypes (i.e., < 1.5 cM) were less concordant across call sets. In contrast, longer haplotypes were more concordant across call sets; a plateau began between 2 and 3 cM. We compared haplotype concordance across geographical regions, including the early- and late-settlement regions, and found highly consistent concordance, indicating that haplotype calls from high-quality imputed sites do not induce haplotype calling error rates that vary by geography specifically within Finland (Figure S7). Because haplotype sharing rates differ substantially across populations (Figure 1), this empirical relationship will most likely vary with population history (e.g., longer haplotype thresholds will most likely be useful in Sweden because the true rate of haplotype sharing is an order of magnitude lower for haplotypes > 3 cM). This empirical framework provides a valuable metric for assessing the appropriate threshold for reliably discovering high-fidelity IBD segments in different populations.
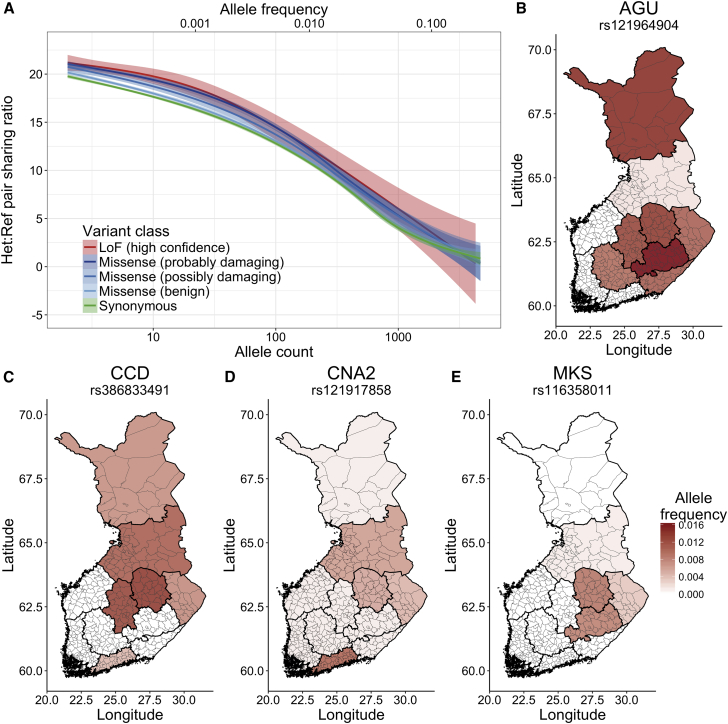
Because previous work in population genetics has suggested that haplotype lengths provide insight into the age of alleles76 and that younger alleles are more likely to be deleterious,3 we quantified the extent of haplotype sharing across predicted functional classes of variants and across genotype states. We found, as expected, that there is generally more haplotype sharing at the rare end of the allele-frequency spectrum (Figure 5A). Additionally, we identified greater haplotype sharing at the rarest allele frequencies in the predicted relative order of deleteriousness among missense variants (i.e., probably damaging > possibly damaging > benign), all of which exhibit greater haplotype sharing than synonymous variants. CpGs modestly disrupt haplotype patterns at the rarest allele frequencies (Figure 5A and Figure S8), which is most likely a product of mutational recurrence. Haplotype sharing rates are similar both in loss-of-function and missense-constrained genes (Figure S9), and these rates show similar signatures of mutational recurrence modestly disrupting haplotype sharing at CpG sites.
Figure 5.
Haplotype Sharing Enrichment across Variant Classes and in Finnish Heritage Diseases
(A) Haplotype sharing enrichment among pairs of individuals who are heterozygous versus homozygous reference (Material and Methods). Variant class curves differ most at the very low to low frequency ranges. The normal confidence intervals show the standard errors for each variant class.
(B)–(E) Allele frequency maps for known Finnish heritage disease variants. The same allele frequency scale is included for each of these plots and is shown on the bottom right. (B) AGU, Aspartylglucosaminuria. (C) CCD, congenital chloride diarrhea. (D) CNA2, cornea plana 2. (E) MKS, Meckel syndrome.
Additional haplotype summaries of these variants are shown in Table 1.
We also assessed the overlap of haplotypes for several known disease variants from the FinDis database (Material and Methods, Figures 5B and 5C). Across the genome, there is a 3% chance that two unselected Finns share a ≥1cM haplotype at any position. Considering a set of disease variants with 0.25%–1% frequency, we first confirmed that indeed homozygous reference individuals (non-carriers) share a haplotype spanning the mutation site at this same background rate. For pairs of individuals who are both carriers of a FinDis variant, however, the likelihood of sharing a haplotype ≥1 cM is an order of magnitude higher (∼30% or higher, Table 1). This enrichment of sharing among carriers belies the conceptual framework of IBD mapping, highlighting the power to detect rare, disease-associated loci. We focused on several validated causal variants known to confer disease (Table 1 and Table S5); such variants included alleles of the in-frame deletion of rs386833491 (ref = AACC and alt = A in Table 1), which is known to confer congenital chloride diarrhea. This allele is not imputable with the standard 1000 Genomes Project reference panel. We find a significant enrichment of haplotype lengths among pairs of individuals who are both heterozygous (mean length = 6.7 cM) versus both homozygous reference (mean length = 5.6 cM) for the rs386833491 deletion (t test p = 5.3 × 10−71, Figure S10). This deletion is most likely slightly more enriched for haplotype sharing beyond the other FinDis variants because of the regional specificity and origins in the late-settlement region (Figure 5C).
Table 1.
Enrichment of Haplotype Sharing that Overlaps FinDis Variants
| Disease Code | Gene | rsID | Chr | Pos | Ref | Alt | Freq | Reference Pair Ratio | Carrier Pair Ratio | Haplotype Enrichment |
|---|---|---|---|---|---|---|---|---|---|---|
| AGU | AGA | rs121964904 | 4 | 178359918 | C | G | 0.79% | 0.02 | 0.25 | 10.4 |
| CNA2 | KERA | rs121917858 | 12 | 91449319 | T | C | 0.52% | 0.03 | 0.82 | 23.5 |
| CCD | SLC26A3 | rs386833491 | 7 | 107427289 | AACC | A | 0.60% | 0.02 | 0.90 | 36.1 |
| MKS | CC2D2A | rs116358011 | 4 | 15538697 | C | T | 0.30% | 0.03 | 0.38 | 14.4 |
Haplotype enrichment is computed as in Figure 5 and the Material and Methods (in brief, the rate of haplotype sharing among pairs of heterozygous individuals per total number of heterozygous pairs relative to homozygous reference pairs). AGU, aspartylglucosaminuria; CNA2, cornea plana 2; CCD, congenital chloride diarrhea; MKS, Meckel syndrome.
See Table S5 for all reviewed FinDis variants.
Discussion
Bringing together genetic and birth-record data, we have constructed one of the most comprehensive genetic studies of population history to date. By coupling Finnish population history with spatial genetic analyses, we have inferred the timing of bottlenecks and expansions, deduced movement across Finland and neighboring countries, and assessed divergence and similarity during recent epochs. Our results demonstrate that prior dichotomous descriptions of the early- versus late-settlement regions correlate with our findings but are insufficient to explain the multi-generational continuous southwest-to-northeast migration trends that correspond with additional bottleneck signatures. We have recapitulated the observation that genes mirror geography at a broad scale,77 but we have shown that at a more granular level within this founder population, demographic fluctuations have been spatially structured by the local environment and other movement-inhibiting or movement-accelerating factors such as linguistic or cultural differences and forced migration events, e.g., by Swedish kings. This comprehensive study of Finnish population history was especially powerful for investigations of recent history over the last 100 generations through statistical analyses of pairwise genomic sharing via haplotypes among fine-scale regions.
The concept that haplotype tracts assessed from common-variant GWAS arrays can provide insight into both population history and rare disease without sequencing data harkens back to the International HapMap Project and earlier.10 Although these ideas have been around for decades, their implementation in biobank-scale data is now feasible and shows promise in isolated populations.78 Using data from Finland, we demonstrate that haplotypes provide insight into the evolutionary timeline and class of variants of greatest interest for this study: recent population history over the past 100 generations and rare, deleterious variants. Coupled with birth-record data, haplotype tracts allow deeper insight into fine-scale substructure, including differential sharing within and across coastal and inland municipalities in the early- and late-settlement regions of Finland, than common allele approaches alone.
Finland is particularly amenable for an investigation of recent population history because it has gone through multiple well-documented bottlenecks, has considerable population substructure compared to those of many other countries,22, 26, 27 and has a universal health care system with integrated registry information. The relatively high genetic divergence between the early- and late-settlement regions has been well documented in prior genetic analyses; we demonstrate much more granular resolution into differential rates of haplotypes across Finland at the level of municipality: for example, the several-fold differences in cumulative sharing across Finland between major urban southwest cities (e.g., Turku and Helsinki) and isolated late-settlement regions (e.g., Kuusamo).
The founder effects in Finland have resulted in a massive enrichment of longer haplotypes in Finns relative to non-Finnish European neighbors. Additionally, these effects have depleted genetic diversity overall and increased relatively common deleterious variants with respect to non-Finnish Europeans.16 A consequence of these bottleneck signatures is the utility of population-based linkage analysis for discovering deleterious variants at the rare end of the frequency spectrum. Many of the founder mutations contributing to the FinDis database were originally discovered through family-based linkage analysis.23 The emergence of biobank-scale genetic and clinical data allows researchers to use population-based linkage analysis to discover rare-variant associations with previously undiscovered diseases or in populations where risk was previously unrealized, such as in the case of a rare orthopedic collagen disorder that conferred extreme short stature and dysmorphic features in Puerto Ricans.78 Our work and previous studies suggest that coupling population-based linkage analysis with electronic health records provides a powerful tool for gaining rare-disease insights, particularly in populations that have gone through a historical bottleneck.29, 78, 79 Furthermore, researchers can query the role of these rare variants in complex disease by using GWAS arrays to construct kinship matrices from pairwise haplotype sharing to understand a more complete spectrum of allele frequencies in overall heritability.80, 81 This study demonstrates the utility of haplotype sharing for historical demographic inference and for identifying rare variants that confer risk of rare disorders in isolated populations, such as Finland, that have data from unified health care registries.
Acknowledgments
Thanks to the participants in the Finnish cohort studies. Thanks to the sequencing centers at Washington University, the Broad Institute, and the UK10K project for generation and deposition of exome sequencing data from FINRISK and other Finnish cohorts. We thank Eimear Kenny and Gillian Belbin for helpful discussions and Cotton Seed and Tim Poterba for helping to scale computational analyses. The Sweden Schizophrenia Study was supported by NIMH grant R01 MH077139. The Estonian Biobank was funded by an Estonian Research Council personal research grant (PUT1660). This research was supported by the Russian Science Foundation (17-15-01177); the National Human Genome Research Institute (5U54HG003079); the European Foundation for the Study of Diabetes (EFSD) New Horizons Programme (L.G. and L.K.); an EFSD/Novo Nordisk grant to R.B.P.; the Academy of Finland Center of Excellence in Complex Disease Genetics (grant 312063 to L.G., 312074 to A.P., and 312062 to S.R.); the Academy of Finland (grants 263401 and 267 882 to L.G.; 286500 to A.P.; 265240 and 263278 to J.K.; and 285380 to S.R.); the Sigrid Juselius Foundation ( A.P., L.G., and S.R.); the Finnish Foundation for Cardiovascular Research (A.P. and S.R.); the Nordic Information for Action eScience Center (62721 to A.P.); the 7th Research and Innovation Framework Programme (602633 to A.P.) (EUROHEADPAIN); and the Horizon 2020 Research and Innovation Programme (667301 to A.P. [COSYN] and 692145 to S.R. [ePerMed]); Biocentrum Helsinki (S.R.); University of Helsinki HiLIFE Fellow grant (S.R.); and the National Institutes of Health (R01HL113315-01 to A.P. and S.R.). M.J.D. is on the scientific advisory board of Ancestry DNA.
Published: April 26, 2018
Footnotes
Supplemental Data include 10 figures, five tables, and one Supplementary Note and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.003.
Contributor Information
Alicia R. Martin, Email: armartin@broadinstitute.org.
Mark J. Daly, Email: mjdaly@broadinstitute.org.
Web Resources
Database of Global Administrative Areas, http://www.gadm.org/
Hail, https://hail.is/
National Institute of Mental Health Genetics Repository, https://www.nimhgenetics.org/
GitHub (scripts from study), https://github.com/armartin/haplotypes
SISu Project, http://www.new.sisuproject.fi
Wikipedia.org (Finnish municipality codes), https://fi.wikipedia.org/wiki/Luettelo_Suomen_kuntanumeroista
Supplemental Data
FinDis consists of 36 monogenic diseases that are enriched in the Finnish bottleneck. Out of an initial list of 50 autosomal variants that are known to be major or minor causes of these diseases, 40 of these variants were polymorphic and in regions with high-quality haplotype calls. The reference pair ratio is (# hom ref pairs sharing a haplotype)/(total # hom ref pairs), and the carrier pair ratio is (# het pairs sharing a haplotype)/(total # het pairs). The haplotype enrichment is (carrier pair ratio)/(reference pair ratio).
References
- 1.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen M.D., Hubisz M.J., Gronau I., Siepel A. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 2014;10:e1004342. doi: 10.1371/journal.pgen.1004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiezun A., Pulit S.L., Francioli L.C., van Dijk F., Swertz M., Boomsma D.I., van Duijn C.M., Slagboom P.E., van Ommen G.J.B., Wijmenga C., Genome of the Netherlands Consortium Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLoS Genet. 2013;9:e1003301. doi: 10.1371/journal.pgen.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henn B.M., Botigué L.R., Peischl S., Dupanloup I., Lipatov M., Maples B.K., Martin A.R., Musharoff S., Cann H., Snyder M.P. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc. Natl. Acad. Sci. USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmueller K.E. The impact of population demography and selection on the genetic architecture of complex traits. PLoS Genet. 2014;10:e1004379. doi: 10.1371/journal.pgen.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathieson I., McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat. Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutenkunst R.N., Hernandez R.D., Williamson S.H., Bustamante C.D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 11.Bonnen P.E., Pe’er I., Plenge R.M., Salit J., Lowe J.K., Shapero M.H., Lifton R.P., Breslow J.L., Daly M.J., Reich D.E. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat. Genet. 2006;38:214–217. doi: 10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- 12.Sajantila A., Salem A.H., Savolainen P., Bauer K., Gierig C., Pääbo S. Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc. Natl. Acad. Sci. USA. 1996;93:12035–12039. doi: 10.1073/pnas.93.21.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peltonen L., Palotie A., Lange K. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 2000;1:182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 14.Palo J.U., Ulmanen I., Lukka M., Ellonen P., Sajantila A. Genetic markers and population history: Finland revisited. Eur. J. Hum. Genet. 2009;17:1336–1346. doi: 10.1038/ejhg.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S.R., Agarwala V., Flannick J., Chiang C.W.K., Altshuler D., Hirschhorn J.N., GoT2D Consortium Simulation of Finnish population history, guided by empirical genetic data, to assess power of rare-variant tests in Finland. Am. J. Hum. Genet. 2014;94:710–720. doi: 10.1016/j.ajhg.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim E.T., Würtz P., Havulinna A.S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Sequencing Initiative Suomi (SISu) Project Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmela, E. (2012). Genetic structure in Finland and Sweden: aspects of population history and gene mapping. PhD thesis (University of Helsinki).
- 18.Günther T., Malmström H., Svensson E.M., Omrak A., Sánchez-Quinto F., Kılınç G.M., Krzewińska M., Eriksson G., Fraser M., Edlund H. Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 2018;16:e2003703. doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poznik G.D., Xue Y., Mendez F.L., Willems T.F., Massaia A., Wilson Sayres M.A., Ayub Q., McCarthy S.A., Narechania A., Kashin S., 1000 Genomes Project Consortium Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat. Genet. 2016;48:593–599. doi: 10.1038/ng.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kittles R.A., Perola M., Peltonen L., Bergen A.W., Aragon R.A., Virkkunen M., Linnoila M., Goldman D., Long J.C. Dual origins of Finns revealed by Y chromosome haplotype variation. Am. J. Hum. Genet. 1998;62:1171–1179. doi: 10.1086/301831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tallavaara M., Pesonen P., Oinonen M. Prehistoric population history in eastern Fennoscandia. J. Archaeol. Sci. 2010;37:251–260. [Google Scholar]
- 22.Kerminen S., Havulinna A.S., Hellenthal G., Martin A.R., Sarin A.-P., Perola M., Palotie A., Salomaa V., Daly M.J., Ripatti S., Pirinen M. Fine-scale genetic structure in Finland. G3 (Bethesda) 2017;7:3459–3468. doi: 10.1534/g3.117.300217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peltonen L., Jalanko A., Varilo T. Molecular genetics of the Finnish disease heritage. Hum. Mol. Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 24.Stoll G., Pietiläinen O.P.H., Linder B., Suvisaari J., Brosi C., Hennah W., Leppä V., Torniainen M., Ripatti S., Ala-Mello S. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013;16:1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahtinen A.M., Havulinna A.S., Jula A., Salomaa V., Kontula K. Prevalence and clinical correlates of familial hypercholesterolemia founder mutations in the general population. Atherosclerosis. 2015;238:64–69. doi: 10.1016/j.atherosclerosis.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Salmela E., Lappalainen T., Fransson I., Andersen P.M., Dahlman-Wright K., Fiebig A., Sistonen P., Savontaus M.-L., Schreiber S., Kere J., Lahermo P. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in Northern Europe. PLoS ONE. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakkula E., Rehnström K., Varilo T., Pietiläinen O.P.H., Paunio T., Pedersen N.L., deFaire U., Järvelin M.-R., Saharinen J., Freimer N. The genome-wide patterns of variation expose significant substructure in a founder population. Am. J. Hum. Genet. 2008;83:787–794. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palamara P.F., Lencz T., Darvasi A., Pe’er I. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 2012;91:809–822. doi: 10.1016/j.ajhg.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning S.R., Thompson E.A. Detecting rare variant associations by identity-by-descent mapping in case-control studies. Genetics. 2012;190:1521–1531. doi: 10.1534/genetics.111.136937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralph P., Coop G. The geography of recent genetic ancestry across Europe. PLoS Biol. 2013;11:e1001555. doi: 10.1371/journal.pbio.1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szpiech Z.A., Xu J., Pemberton T.J., Peng W., Zöllner S., Rosenberg N.A., Li J.Z. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi P.K., Esko T., Mattsson H., Eklund N., Gandin I., Nutile T., Jackson A.U., Schurmann C., Smith A.V., Zhang W. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamsiz E.D., Viscidi E.W., Frederick A.M., Nagpal S., Sanders S.J., Murtha M.T., Schmidt M., Triche E.W., Geschwind D.H., State M.W., Simons Simplex Collection Genetics Consortium Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am. J. Hum. Genet. 2013;93:103–109. doi: 10.1016/j.ajhg.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han E., Carbonetto P., Curtis R.E., Wang Y., Granka J.M., Byrnes J., Noto K., Kermany A.R., Myres N.M., Barber M.J. Clustering of 770,000 genomes reveals post-colonial population structure of North America. Nat. Commun. 2017;8:14238. doi: 10.1038/ncomms14238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baharian S., Barakatt M., Gignoux C.R., Shringarpure S., Errington J., Blot W.J., Bustamante C.D., Kenny E.E., Williams S.M., Aldrich M.C., Gravel S. The Great Migration and African-American genomic diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathias R.A., Taub M.A., Gignoux C.R., Fu W., Musharoff S., O’Connor T.D., Vergara C., Torgerson D.G., Pino-Yanes M., Shringarpure S.S., CAAPA A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat. Commun. 2016;7:12522. doi: 10.1038/ncomms12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zidan J., Ben-Avraham D., Carmi S., Maray T., Friedman E., Atzmon G. Genotyping of geographically diverse Druze trios reveals substructure and a recent bottleneck. Eur. J. Hum. Genet. 2015;23:1093–1099. doi: 10.1038/ejhg.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atzmon G., Hao L., Pe’er I., Velez C., Pearlman A., Palamara P.F., Morrow B., Friedman E., Oddoux C., Burns E., Ostrer H. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet. 2010;86:850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behar D.M., Metspalu M., Baran Y., Kopelman N.M., Yunusbayev B., Gladstein A., Tzur S., Sahakyan H., Bahmanimehr A., Yepiskoposyan L. No evidence from genome-wide data of a Khazar origin for the Ashkenazi Jews. Hum. Biol. 2013;85:859–900. doi: 10.3378/027.085.0604. [DOI] [PubMed] [Google Scholar]
- 41.Campbell C.L., Palamara P.F., Dubrovsky M., Botigué L.R., Fellous M., Atzmon G., Oddoux C., Pearlman A., Hao L., Henn B.M. North African Jewish and non-Jewish populations form distinctive, orthogonal clusters. Proc. Natl. Acad. Sci. USA. 2012;109:13865–13870. doi: 10.1073/pnas.1204840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakatsuka N., Moorjani P., Rai N., Sarkar B., Tandon A., Patterson N., Bhavani G.S., Girisha K.M., Mustak M.S., Srinivasan S. The promise of discovering population-specific disease-associated genes in South Asia. Nat. Genet. 2017;49:1403–1407. doi: 10.1038/ng.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gauvin H., Moreau C., Lefebvre J.-F., Laprise C., Vézina H., Labuda D., Roy-Gagnon M.-H. Genome-wide patterns of identity-by-descent sharing in the French Canadian founder population. Eur. J. Hum. Genet. 2014;22:814–821. doi: 10.1038/ejhg.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno-Estrada A., Gravel S., Zakharia F., McCauley J.L., Byrnes J.K., Gignoux C.R., Ortiz-Tello P.A., Martínez R.J., Hedges D.J., Morris R.W. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gravel S., Zakharia F., Moreno-Estrada A., Byrnes J.K., Muzzio M., Rodriguez-Flores J.L., Kenny E.E., Gignoux C.R., Maples B.K., Guiblet W., 1000 Genomes Project Reconstructing Native American migrations from whole-genome and whole-exome data. PLoS Genet. 2013;9:e1004023. doi: 10.1371/journal.pgen.1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Homburger J.R., Moreno-Estrada A., Gignoux C.R., Nelson D., Sanchez E., Ortiz-Tello P., Pons-Estel B.A., Acevedo-Vasquez E., Miranda P., Langefeld C.D. Genomic insights into the ancestry and demographic history of South America. PLoS Genet. 2015;11:e1005602. doi: 10.1371/journal.pgen.1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong A., Masson G., Frigge M.L., Gylfason A., Zusmanovich P., Thorleifsson G., Olason P.I., Ingason A., Steinberg S., Rafnar T. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorjani P., Patterson N., Loh P.-R., Lipson M., Kisfali P., Melegh B.I., Bonin M., Kádaši L., Rieß O., Berger B. Reconstructing Roma history from genome-wide data. PLoS ONE. 2013;8:e58633. doi: 10.1371/journal.pone.0058633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panoutsopoulou K., Hatzikotoulas K., Xifara D.K., Colonna V., Farmaki A.-E., Ritchie G.R.S., Southam L., Gilly A., Tachmazidou I., Fatumo S. Genetic characterization of Greek population isolates reveals strong genetic drift at missense and trait-associated variants. Nat. Commun. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glodzik D., Navarro P., Vitart V., Hayward C., McQuillan R., Wild S.H., Dunlop M.G., Rudan I., Campbell H., Haley C. Inference of identity by descent in population isolates and optimal sequencing studies. Eur. J. Hum. Genet. 2013;21:1140–1145. doi: 10.1038/ejhg.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert E., Carmi S., Ennis S., Wilson J.F., Cavalleri G.L. Genomic insights into the population structure and history of the Irish Travellers. Sci. Rep. 2017;7:42187. doi: 10.1038/srep42187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Botigué L.R., Henn B.M., Gravel S., Maples B.K., Gignoux C.R., Corona E., Atzmon G., Burns E., Ostrer H., Flores C. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl. Acad. Sci. USA. 2013;110:11791–11796. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 54.Fiorito G., Di Gaetano C., Guarrera S., Rosa F., Feldman M.W., Piazza A., Matullo G. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 2016;24:1056–1062. doi: 10.1038/ejhg.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Browning S.R., Browning B.L. Identity-by-descent-based heritability analysis in the Northern Finland Birth Cohort. Hum. Genet. 2013;132:129–138. doi: 10.1007/s00439-012-1230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Surakka I., Horikoshi M., Mägi R., Sarin A.-P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., Timonen S., ENGAGE Consortium The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Multicenter Genetic Studies of Schizophrenia Consortium. Psychosis Endophenotypes International Consortium. Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haller T., Leitsalu L., Fischer K., Nuotio M.-L., Esko T., Boomsma D.I., Kyvik K.O., Spector T.D., Perola M., Metspalu A. MixFit: Methodology for computing ancestry-related genetic scores at the individual level and its application to the Estonian and Finnish population studies. PLoS ONE. 2017;12:e0170325. doi: 10.1371/journal.pone.0170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotar O., Moguchaia E., Boyarinova M., Kolesova E., Khromova N., Freylikhman O., Smolina N., Solntsev V., Kostareva A., Konradi A., Shlyakhto E. Seventy years after the siege of Leningrad: does early life famine still affect cardiovascular risk and aging? J. Hypertens. 2015;33:1772–1779. doi: 10.1097/HJH.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 60.Prasad R.B., Lessmark A., Almgren P., Kovacs G., Hansson O., Oskolkov N., Vitai M., Ladenvall C., Kovacs P., Fadista J. Excess maternal transmission of variants in the THADA gene to offspring with type 2 diabetes. Diabetologia. 2016;59:1702–1713. doi: 10.1007/s00125-016-3973-9. [DOI] [PubMed] [Google Scholar]
- 61.Rivas M.A., Graham D., Sulem P., Stevens C., Desch A.N., Goyette P., Gudbjartsson D., Jonsdottir I., Thorsteinsdottir U., Degenhardt F., UK IBD Genetics Consortium. NIDDK IBD Genetics Consortium A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat. Commun. 2016;7:12342. doi: 10.1038/ncomms12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borodulin K., Vartiainen E., Peltonen M., Jousilahti P., Juolevi A., Laatikainen T., Männistö S., Salomaa V., Sundvall J., Puska P. Forty-year trends in cardiovascular risk factors in Finland. Eur. J. Public Health. 2015;25:539–546. doi: 10.1093/eurpub/cku174. [DOI] [PubMed] [Google Scholar]
- 63.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abraham G., Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS ONE. 2014;9:e93766. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Browning B.L., Browning S.R. Detecting identity by descent and estimating genotype error rates in sequence data. Am. J. Hum. Genet. 2013;93:840–851. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe’er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Browning S.R., Browning B.L. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 2015;97:404–418. doi: 10.1016/j.ajhg.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loh P.-R., Palamara P.F., Price A.L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016;48:811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petkova D., Novembre J., Stephens M. Visualizing spatial population structure with estimated effective migration surfaces. Nat. Genet. 2016;48:94–100. doi: 10.1038/ng.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leslie S., Winney B., Hellenthal G., Davison D., Boumertit A., Day T., Hutnik K., Royrvik E.C., Cunliffe B., Lawson D.J., Wellcome Trust Case Control Consortium 2. International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nelis M., Esko T., Mägi R., Zimprich F., Zimprich A., Toncheva D., Karachanak S., Piskácková T., Balascák I., Peltonen L. Genetic structure of Europeans: a view from the North-East. PLoS ONE. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Q.S., Browning B.L., Browning S.R. Genome-wide haplotypic testing in a Finnish cohort identifies a novel association with low-density lipoprotein cholesterol. Eur. J. Hum. Genet. 2014;23:672–677. doi: 10.1038/ejhg.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tremblay M., Vézina H. New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am. J. Hum. Genet. 2000;66:651–658. doi: 10.1086/302770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sousa V., Hey J. Understanding the origin of species with genome-scale data: modelling gene flow. Nat. Rev. Genet. 2013;14:404–414. doi: 10.1038/nrg3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belbin G.M., Odgis J., Sorokin E.P., Yee M.-C., Kohli S., Glicksberg B.S., Gignoux C.R., Wojcik G.L., Van Vleck T., Jeff J.M. Genetic identification of a common collagen disease in puerto ricans via identity-by-descent mapping in a health system. eLife. 2017;6:68. doi: 10.7554/eLife.25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vacic V., Ozelius L.J., Clark L.N., Bar-Shira A., Gana-Weisz M., Gurevich T., Gusev A., Kedmi M., Kenny E.E., Liu X. Genome-wide mapping of IBD segments in an Ashkenazi PD cohort identifies associated haplotypes. Hum. Mol. Genet. 2014;23:4693–4702. doi: 10.1093/hmg/ddu158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zaitlen N., Kraft P., Patterson N., Pasaniuc B., Bhatia G., Pollack S., Price A.L. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9:e1003520. doi: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Evans L., Tahmasbi R., Jones M., Vrieze S., Abecasis G., Das S., Bjelland D., deCandia T., Yang J., Goddard M. Narrow-sense heritability estimation of complex traits using identity-by-descent information. bioRxiv. 2017 doi: 10.1101/164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FinDis consists of 36 monogenic diseases that are enriched in the Finnish bottleneck. Out of an initial list of 50 autosomal variants that are known to be major or minor causes of these diseases, 40 of these variants were polymorphic and in regions with high-quality haplotype calls. The reference pair ratio is (# hom ref pairs sharing a haplotype)/(total # hom ref pairs), and the carrier pair ratio is (# het pairs sharing a haplotype)/(total # het pairs). The haplotype enrichment is (carrier pair ratio)/(reference pair ratio).