Abstract
N-alpha-acetylation is a common co-translational protein modification that is essential for normal cell function in humans. We previously identified the genetic basis of an X-linked infantile lethal Mendelian disorder involving a c.109T>C (p.Ser37Pro) missense variant in NAA10, which encodes the catalytic subunit of the N-terminal acetyltransferase A (NatA) complex. The auxiliary subunit of the NatA complex, NAA15, is the dimeric binding partner for NAA10. Through a genotype-first approach with whole-exome or genome sequencing (WES/WGS) and targeted sequencing analysis, we identified and phenotypically characterized 38 individuals from 33 unrelated families with 25 different de novo or inherited, dominantly acting likely gene disrupting (LGD) variants in NAA15. Clinical features of affected individuals with LGD variants in NAA15 include variable levels of intellectual disability, delayed speech and motor milestones, and autism spectrum disorder. Additionally, mild craniofacial dysmorphology, congenital cardiac anomalies, and seizures are present in some subjects. RNA analysis in cell lines from two individuals showed degradation of the transcripts with LGD variants, probably as a result of nonsense-mediated decay. Functional assays in yeast confirmed a deleterious effect for two of the LGD variants in NAA15. Further supporting a mechanism of haploinsufficiency, individuals with copy-number variant (CNV) deletions involving NAA15 and surrounding genes can present with mild intellectual disability, mild dysmorphic features, motor delays, and decreased growth. We propose that defects in NatA-mediated N-terminal acetylation (NTA) lead to variable levels of neurodevelopmental disorders in humans, supporting the importance of the NatA complex in normal human development.
Keywords: NAA15, NAA10, NatA complex, N-terminal acetylation (NTA), N-terminal acetyltransferases (NATs), neurodevelopmental disorder, Ogden syndrome, intellectual disability, autism, congenital heart defects
Main Text
Advances in sequencing technologies such as whole-exome or genome sequencing (WES/WGS) have led to disease-gene association discoveries, functional annotation of the human genome, and improved diagnostic rates in individuals with suspected genetic disorders refractory to conventional diagnostic testing. An estimated diagnostic rate that often exceeds 25% can be achieved when WES/WGS is applied to otherwise undiagnosed complex cases.1, 2, 3, 4, 5 NAA15 (N-alpha-acetyltransferase 15, MIM: 608000) was previously characterized as one of fifty-two risk genes for neurodevelopmental disorders by targeted sequencing of a large autism spectrum and intellectual disability (ASID) cohort.6 In another study of de novo changes in severe congenital heart disease (CHD), likely gene disrupting (LGD) variants in NAA15 were identified in two affected individuals in a cohort of 362 severe CHD cases; one of these individuals was known to have additional neurodevelopmental defects.7 In an effort to further characterize the clinical and molecular spectrum associated with genetic defects in NAA15, we ascertained, from 33 unrelated families, 38 individuals with truncating, presumably LGD (nonsense, frameshifting and splice) variants in NAA15 via a collaborative world-wide effort among multiple institutions. As a result of comprehensive clinical evaluation and molecular analyses in all individuals, we propose that deleterious variants in NAA15 are associated with variable levels of intellectual disability, developmental delay, autism spectrum disorder, dysmorphic features, and congenital cardiac anomalies.
This study was performed in accordance with protocols approved by the institutional review boards of the participating institutions (see Supplemental Data). Three affected individuals were recruited from the UK Deciphering Developmental Disorders (DDD) project (families 20, 27, and 30). Written informed consent was obtained from all study participants. The key clinical features of our cohorts are summarized in Table 1. Detailed clinical summaries for each subject are provided in the Supplemental Data. The use of GeneMatcher, a web-based tool for connecting researchers with an interest in the same gene,8 facilitated contact between international collaborators.
Table 1.
Summary of Phenotypes
| Phenotype | Number of individuals with phenotype | Number of individuals with relevant data | Percentage |
|---|---|---|---|
| Brain Structure and Function | |||
| Intellectual disability (ID)a | 23 | 23 | 100 |
| ASD, ADHD, or behavioral issues | 30 | 33 | 91 |
| Abnormal brain MRI | 2 | 11 | 18 |
| Speech delay | 32 | 33 | 97 |
| Seizures | 6 | 26 | 23 |
| Motor Impairments | |||
| Motor delay and related abnormalities | 31 | 32 | 97 |
| Muscle tone issues | 7 | 18 | 39 |
| Feeding difficulties | 8 | 14 | 57 |
| Cardiovascular | |||
| Congenital cardiac defects | 4 | 19 | 21 |
| Major vessel anomalies | 2 | 19 | 11 |
| Arrhythmias | 1 | 19 | 5 |
| Hypertension | 1 | 19 | 5 |
| Other | |||
| Mild dysmorphism | 18 | 28 | 64 |
| Skeletal or connective-tissue defects | 8 | 20 | 40 |
In individuals > 5 years of age, when IQ testing or other cognitive testing was performed.
All subjects have variable degrees of neurodevelopmental disabilities, including impaired motor abilities (HP: 0001270), intellectual disability (HP: 0001249), impaired verbal abilities (HP: 0000750), and autism spectrum disorder (HP: 0000729) (Table 1, Table S1, and Table S2). Many subjects have impaired motor function, including fine-motor difficulties (n = 5, or 12%), mild ataxia (n = 1), abnormality of movement (n = 1), motor delay (n = 22, or 60%), and hypotonia (n = 5, or 14%). Various levels of intellectual disability are reported in almost all study subjects with available data; such disability includes mild, moderate, or severe intellectual disability and learning difficulties with or without behavioral issues (Table 1, Table S1, and Table S2). The majority of affected individuals have verbal issues, including complete absence of speech, delayed language development, the need for sign language, or other speech difficulties. Most subjects also present with autism spectrum disorder (ASD) and/or other behavioral abnormalities. Individual 11 was noted to have marked hypersomnolence in early years, in apparent similarity to what was recently reported in a girl with a missense variant in NAA10.9 Minor facial dysmorphology was reported in some individuals (Table 1), but there were no consistent features noted nor a recognizable pattern of facial dysmorphology (Figure 1 and Figure S1). The birth weight and length were low in some individuals; the most notable feature was a birth weight ≤ 1st percentile in 7 out of 25 (28%) individuals with available information (Table S3). Some of the individuals remain small throughout life, whereas others are of normal stature and a few are above average height (Table S3).
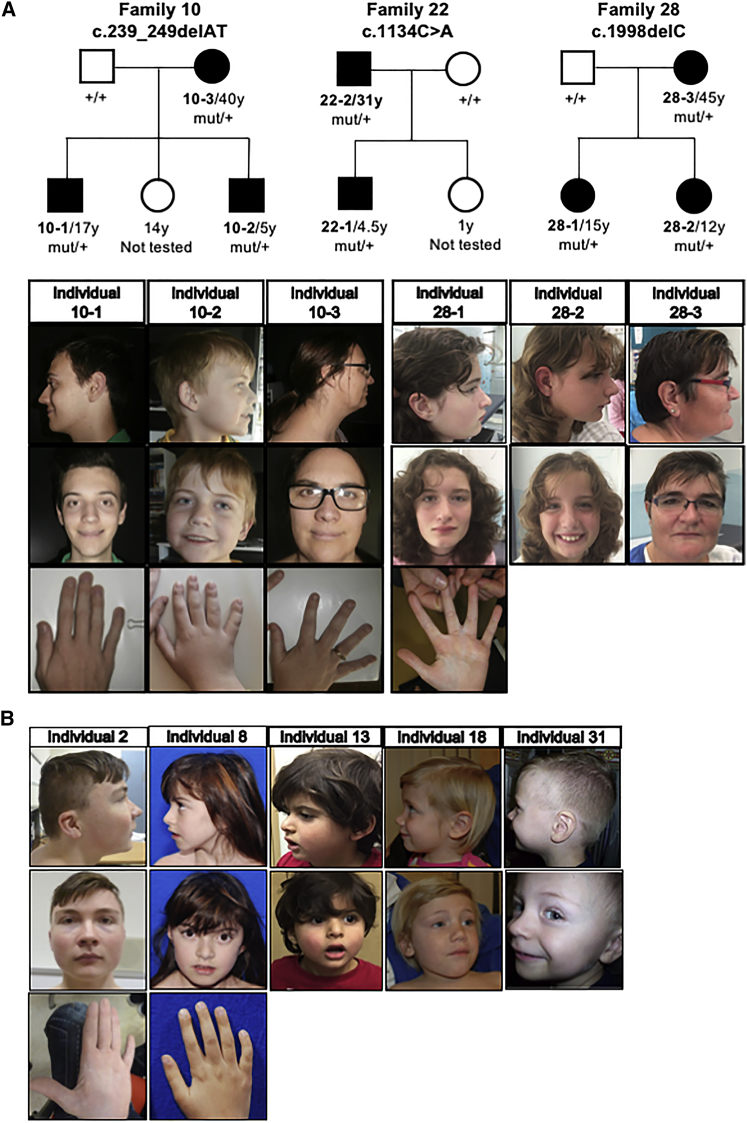
Figure 1.
Pedigrees, Mild Facial Dysmorphology, and Hands of Individuals with Familial or de novo NAA15 LGD Variants
(A) Pedigrees are shown for the three families with inherited variants. Family 10, Individual 10-1: at age 17 years and 6 months, with prominent eyebrows, broad nose, and prominent chin. Hand appears normal. Individual 10-2: at 6 years and 6 months, with very well-developed philtral pillars. Hand appears normal. Individual 10-3: mother, with long mentum of the chin and relatively thick alae nasi. Hand appears normal. Family 28, Individual 28-1: at age 15 years, partial syndactyly in one hand, but otherwise not with particularly notable dysmorphology. Individual 28-2: sister, at age 12 years, who was not noted to have any obvious dysmorphology. Individual 28-3: Mother at age 45 years, with broad nose but otherwise not with notable dysmorphology.
(B) Minor facial dysmorphology was noted in some probands, but there were no reliably consistent features shared among them. Individual 2: at 17 years old, noted to have brachycephaly, appearance of ocular hypertelorism with short palpebral fissures, prominent nose tip with a longer columella of the nose, trapezoidal philtrum, and micrognathia without retrognathia. Also noted are small low-set, posteriorly rotated ears, with thickened and overfolded helix; hypoplastic distal phalanges on digits 2, 3, and 4; 5th finger with brachyclinodactyly; and persistence of fetal finger pads on the 3rd and 4th digit. Individual 8: at the age 8 years 9 months, noted to have thin philtrum, bulbous nasal tip, and 5th finger with brachyclinodactyly. Individual 13: at 4 years old, no facial dysmorphism noted. Individual 18: at 4 years and 3 months, with bulbous nose tip, thick alae nasi and anteverted nares, prominent cupid’s bow and philtrum, long mentum of the chin, and simple ears. Individual 31: with epicanthus inversus, smooth philtrum, thin vermilion border of the upper lip, and sparse lateral eyebrows.
Almost all individuals have normal or uncharacterized cardiac function (Table 1), with four exceptions. Individual 2 (Figure 1) has atrial ectopic (multifocal) tachycardia (HP: 0011701), treated with verapamil, and hypertension (HP: 0000822). Individual 3 had a ventricular septal defect (VSD), repaired surgically during infancy. Individual 17 has a Marfanoid habitus, with an aortic root at the upper limit of normal. By far the most severely affected, individual 19 has heterotaxy syndrome associated with a complex cardiac diagnosis of dextrocardia involving left superior and inferior venae cavae, total anomalous pulmonary venous return to the innominate vein, tricuspid atresia, hypoplastic right ventricle, double-outlet right ventricle, and transposed great arteries with severe pulmonary stenosis. The variant in this individual (c.1009_1012delGAAA) was previously reported in a cohort of 1,213 subjects with CHD and an increased prevalence of extracardiac congenital anomalies (CAs) and risk of neurodevelopmental disabilities (NDDs).7, 10 Another LGD variant, c.2282C>A (p.Ser761∗) in NAA15, was first reported in an individual with pulmonary stenosis, single left coronary artery, and tetralogy of Fallot (in the context of no reported neurodevelopmental disability), although we have been unable to obtain additional information on this individual.7 A more recent analysis of this now expanded cohort of 2,871 CHD probands, including 2,645 parent-offspring trios, did not find any new variants in NAA15.11 Given the low prevalence of CHD in our own cohort of 38 individuals, one caveat is that the expression of severe congenital heart disease could be due to variation at a second locus, a noncoding mutation outside of the exome, or some other additional variation undetected thus far.
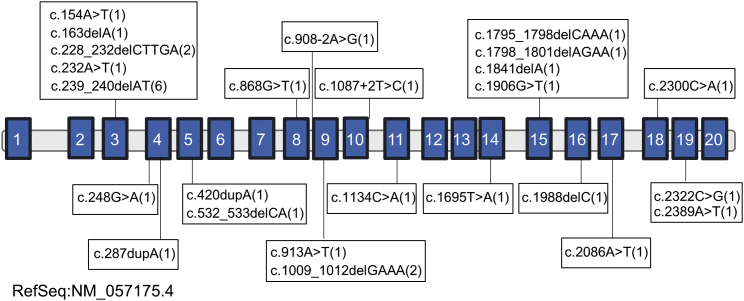
A total of 25 presumably LGD variants contained in 12 of the 20 exons and two intron-exon boundaries of NAA15 were identified from 33 unrelated families (Figure 2, Table S1, Table S2); these included nonsense variants (n = 13), canonical splice-site variants (n = 2), and frameshift variants (n = 10). The inheritance pattern of the variants was determined to be de novo for most subjects (22 families) through testing of parental samples. Familial inheritance was observed in three families (families 10, 22, and 28), and the corresponding NAA15 LGD variant segregated with the neurocognitive phenotypes, including in one mildly affected parent in each family and in affected siblings in families 10 and 28. For Family 10, the read count data did not demonstrate any somatic mosaicism in the blood sample from the mother. Among the 25 variants identified, there were three recurrent variants, including c.228_232delCTTGA (p.Asp76Glufs∗20) (families 3 and 4, de novo), c.239_240delAT (p.His80Argfs∗17) (families 6-11B, de novo in families 6 and 7, familial in family 10, and unknown inheritance in the rest of the families), and c.1009_1012delGAAA (p.Glu337Argfs∗5) (families 19 and 20, de novo). We examined genomic context around the three recurrent loci to look for micro-homology that might increase the propensity for recurrent mutations and found that the most recurrent mutation, c.239_240delAT, occurs in the middle of one of 20 reported mutation hotspots, CATGT.12 In addition, this recurrent variant, and another one, c.228_232delCTTGA, are close to each other in exon 3 in an area that is computationally predicted13 to form a quasipalindromic structure (lying distal to an even larger quasipalindromic structure), and the third recurrent mutation c.1009_1012delGAAA in exon 9 lies just distal to a quasipalindromic structure (Figure S2).14
Figure 2.
Exonic Localization of NAA15 LGD Variants Identified in Subjects in This Study
Schematic representation of the genomic structure of human NAA15. Solid blue rectangles indicate exons, and the horizontal bars represent introns. NAA15 variants with their relative positions in the gene are shown, and the number of affected individuals with the specific variants is shown in parentheses.
Data from the Exome Aggregation Consortium (ExAC) study of 60,706 control individuals show that NAA15 is likely intolerant to LGD variants (pLI = 1.00),15 that the residual variation intolerance score (RVIS) = −0.89 (among the 10.2% most LGD intolerant of human genes), and that LoF-FDR[ExAC] = 0.000224349.16 Excluding small cohorts (<100 probands, Table S4), we in total identified fourteen de novo variants in NAA15 from six independent rare disease cohorts with a total sample size of ∼36,731. Ten out of 14 cases are reported in detail here; the remaining four lack sufficient phenotype information. Our aggregate frequency of de novo LGD variants in affected individuals (∼4.0 per 10,000) is significantly higher than the background rates estimated by Samocha et al.17 for LGD mutations (expected ∼0.038 per 10,000; p < 2.2 × 10−16). If we further restrict the analysis to the three largest cohorts, each of which included more than 5,000 probands, the observed enrichment remains highly significant (nine de novo LGD variants among 33,831 total probands; p = 2.48 × 10−14). We acknowledge that there are limitations to comparing results from ExAC to a clinically ascertained cohort, particularly when one undertakes a genotype-first approach by actively searching for singleton cases with variants in NAA15 by using different sequencing platforms and coverage levels.18 However, the average coverage for NAA15 in ExAC and gnomAD databases is 47× versus approximately 20× coverage levels provided by clinically offered exome tests, suggesting that the increased number of LGD variants in the current study is not due to higher exon coverage levels in clinical sequencing. Only six LGD variants in NAA15 are reported in ExAC (Table S5), and 11 NAA15 LGD variants are reported in the Genome Aggregation Database (gnomAD) (Table S6). Two of the variants that are recurrent and de novo in our research cohort (c.239_240delAT [p.His80Argfs∗17] and c.228_232delCTTGA [p.Asp76Glufs∗20]) are present one time each in ExAC (and also duplicated in gnomAD, given that gnomAD includes many variants from ExAC). It should be noted that phenotypic information as well as the variant inheritance are not available on these individuals in ExAC or gnomAD. Given that the three parents in the inherited families (families 10, 22, and 28) were only mildly affected, it is possible that such individuals could be found in cohorts such as ExAC or gnomAD. A recent study showed that ∼2.8% of the ExAC population is associated with possible disease-associated genotypes,19 and it is well-known that genetic background can influence the expressivity of any given variant.
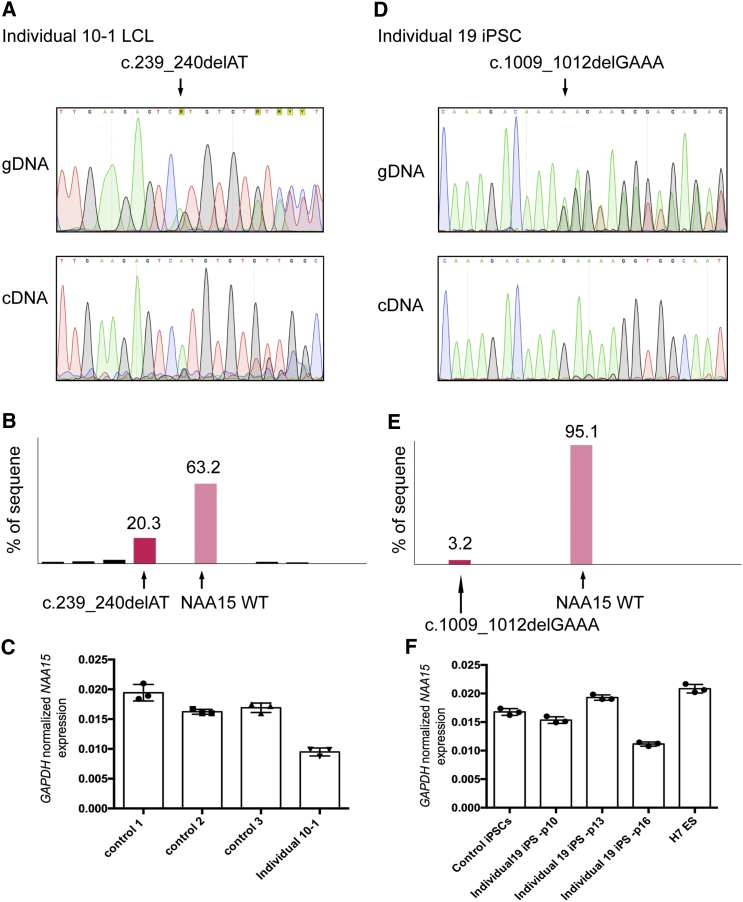
We sought to confirm whether any of the LGD variants might trigger nonsense-mediated decay (NMD) of the respective mutant RNA. For this, we made use of two research-subject-derived cell lines, including one lymphoblastoid cell line (LCL) from individual 10-1 (c.239_240delAT) (Figures 3A–3C and Supplemental Methods) and one induced pluripotent stem cell (iPS) line from individual 19 (c.1009_1012delGAAA) (Figures 3D–3F). Quantitative RT-PCR with primers 3′ to the mutation demonstrated approximately 50% decreased total RNA in one cell passage from the LCLs from individual 10-1 in comparison to three control LCLs (Figure 3C, left panel), whereas the same assay (Figure 3F, right panel) and an additional Taqman assay (Figure S3) showed more variability in total RNA isolated from three different passages of the iPS line from individual 19 than from one control iPS line and a control human embryonic stem cell (hESC) line. Nonetheless, RT-PCR with primers spanning the mutation sites, followed by Sanger sequencing, did demonstrate substantially reduced mutant transcript in the LCL from individual 10-1 (Figure 3B) and almost complete absence of the mutant transcript in three different passages of the iPS line from individual 19 (a representative result from passage 16 is shown in Figure 3E). This reduction most likely occurs because the variant transcript is targeted for degradation via the nonsense-mediated decay (NMD) pathway.20
Figure 3.
Expression Analysis of NAA15 in Research-Subject-Derived Cell Lines
(A and D) Sanger sequencing of genomic DNA (top panel) and reverse-transcribed cDNA (bottom panel) isolated from a lymphoblastoid cell line (LCL) of individual 10-1 (c.239_240delAT) (A) and an induced pluripotent stem cell (iPS) line (passage 16) of individual 19 (c.1009_1012delGAAA) (D).
(B and E) Quantification of different cDNA species from cDNA Sanger sequencing showing the relative ratio of WT NAA15 versus c.239_240delAT (LCL line) (B) and (c.1009_1012delGAAA) (passage 16 iPS cell line) (E).
(C and F) NAA15 mRNA expression level analyzed by qPCR in research subject-derived cell lines (at passage numbers p10, p13, and p16), as compared to control cell lines (at passage 16). Error bars are standard deviation (SD), and the assay was performed three times per sample.
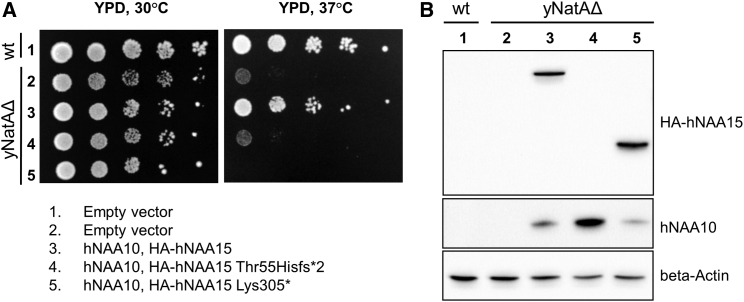
We further explored the functional effects for two of the other LGDs in a yeast assay in which the human NatA complex can functionally replace yeast NatA, as shown by complementation of growth phenotypes21, 22 and partial rescue of the NatA-specific Nt-acetylome.23 Mutant NAA15 (p.Thr55Hisfs∗2 [c.163delA] from family 2 and p.Lys305∗ [c.913A>T] from family 18) failed to rescue the temperature-sensitive growth phenotype of yNatAΔ (Figure 4A and Tables S7, S8, and S9), suggesting that the two variants lead to reduced or abolished NatA activity, at least as assessed in this heterologous system. We further verified human NatA expression in the yNatA deletion strain by immunoblotting (Figure 4B) against the HA epitope that was incorporated N-terminal to NAA15. In the context of overexpression from a plasmid, we detected both full-length HA-NAA15 and HA-NAA15 p.Lys305∗, but not HA-NAA15 p.Thr55Hisfs∗2, suggesting that the mRNA for HA-NAA15 p.Thr55Hisfs∗2 is most likely undergoing complete NMD and/or that this truncated mini-protein is unstable, whereas truncated mini-protein HA-NAA15 p.Lys305∗ can be expressed in this system but nonetheless does not provide functional rescue.
Figure 4.
Truncation Mutations of Human NAA15 Impair NatA Function and Yeast Viability
(A) Serial dilution spot assay depicting the sensitivity of human NAA15 Thr55Hisfs∗2 and Lys305∗ mutants to increased temperature in a ynaa10Δ, ynaa15Δ double-deletion background (yNatAΔ).
(B) Confirmation of human NatA expression by immunoblot analysis with anti-hNAA10 and anti-HA (for HA-hNAA15 detection) along with anti-beta Actin as a loading control.
Distributed throughout the entire gene of NAA15, the 25 LGD variants we reported here are predicted to undergo NMD, leading to degradation of the mutant mRNA and thus loss of the aberrant protein product. Expression analysis from research-subject-derived lymphoblast cells or IPSCs confirmed under-representation of the mutant transcript in cDNA. In addition, the functional deficiency of human mutated NAA15 was further supported by the growth rescue experiment in the yeast NatA-deficient strain, in which mutant human NAA15 failed to restore the growth-deficiency phenotype. In light of these results, we propose haploinsufficiency of NAA15 as the most likely mechanism for this newly recognized disease, although we readily acknowledge that some of the LGDs might not trigger complete NMD or might do so differentially in different tissues, leaving open the possibility for expression of a truncated NAA15 protein, which could possibly act via a dominant-negative or gain-of-function mechanism in some individuals. De novo missense variants (c.1014G>T [p.Lys338Asn] and c.841G>C [p.Glu281Gln]) have been previously reported in two individuals with autism and intellectual disability, respectively;24, 25 however, the deleterious effect of these missense variants has not been established and so will also require further functional studies, segregation in families, and/or proof of recurrence in multiple affected individuals. Further supporting our postulated mechanism of haploinsufficiency, when we searched the DECIPHER database26 and our clinical cohorts for individuals with small microdeletions involving NAA15, the smallest deletion we could find is in a 31-year-old man carrying a de novo 2.73 Mb deletion, including NAA15 and 17 other predicted genes. This man was noted as having mild intellectual disability, mild dysmorphic features, motor delays in childhood, a low birth weight (−2SD), and adult height, weight, and head circumference all at the 10th centile (Figure S4). He has poor vision as a result of cortical visual impairment (CVI), which was not reported (but also not formally screened for) in any of the above reported individuals but which was found in some of the individuals with NAA10 mutations.27 It is also possible that his CVI could be due to some other missing gene in the CNV interval. There are currently 18 large heterozygous CNV deletions, including NAA15 in the DECIPHER database;26 these deletions range in size from 3.27 Mb to 24.30 Mb, and many are noted to be associated with global developmental delay or intellectual disability, supporting the case for haploinsufficiency of at least some of the genes in these CNV intervals. One individual with a de novo 5.2 Mb deletion died from a sudden cardiac event at the age of 35 (see Supplemental Case Reports).
Human NAA15 encodes an 866 amino acid (∼105 kDa) protein, NAA15, containing tetratricopeptide repeat domains and a putative bipartite nuclear localization signal.28 Many studies have shown that NAA15 acts as the auxiliary subunit binding with the catalytic subunit NAA10 and localizes it to the ribosome, where this complex (named the NatA complex) serves as an N-terminal acetyltransferase (NAT).29 This complex is evolutionarily conserved from yeast to vertebrates,23 and the X-ray crystal structure of the 100 kDa holo-NatA complex from Schizosaccharomyces pombe shows that the NatA-NAA15 auxiliary subunit contains 13 tetratricopeptide motifs and adopts a ring-like topology that wraps around the NatA-NAA10 subunit, an interaction that alters the NAA10 active site for substrate-specific acetylation.30 Mutation or loss of the NatA subunits in yeast (Saccharomyces cerevisiae) or human HeLa cells results in inhibited cell growth, cell apoptosis, and failure to enter the G0 phase in the cell cycle.31, 32 Nat1 (ortholog of NAA15) knock-down flies have impaired locomotor activity and early adult lethality.6 NAA10 and NAA15 are both highly expressed in regions of cell division and migration during brain development and are downregulated as neurons differentiate in early postnatal development.33, 34 NAA15 has been shown to be expressed at low levels in most adult tissues (e.g., nervous system, heart, and reproductive system) (see GTEx Portal). However, RNA-seq data from human brain tissue suggests that upregulation of NAA15 occurs in utero at eight weeks after conception and is developmentally downregulated thereafter, the highest expression being in the occipital neocortex and anterior cingulate (medial prefrontal) cortex (Figures S5A and S5B), supporting a role for NAA15 in development of the nervous system. Similarly, in mice, upregulated expression of NAA15 has been shown in regions of neuronal migration, and proliferation in the neonatal mouse brain has been shown along with reduced expression as neurons differentiate during early postnatal development.33, 34
Genetic defects in NAA10, which is X-linked and encodes another member of the NatA complex, are associated with Ogden syndrome (MIM: 300855), Lenz microphthalmia (MIM: 309800), and intellectual disability (with variable cardiac involvement).27, 35, 36, 37, 38, 39 In the case of Ogden syndrome, a total of eight boys from two families had a distinct combination of dysmorphology, hypotonia, global developmental delays, cardiac anomalies, cardiac arrhythmias and cardiomegaly, and the identical missense mutation segregated in multiple affected individuals in two unrelated families.40 Different variants in NAA10 have been reported, sometimes with only a mild intellectual-disability phenotype in heterozygous females, but also sometimes with hydrocephaly, supernumerary vertebrae, congenital heart defects, and arrhythmias, which are always more severe in the males.9, 27, 35, 36, 37 Although developmental delay and/or intellectual disability might be the only presenting feature, the additional cardiac, growth, dysmorphic features and other findings vary in type and severity. For the one family in which affected members had Lenz microphthalmia syndrome and a splice-site variant in NAA10, and in which proband-derived fibroblasts lacked expression of full-length NAA10 and displayed a cell-proliferation defect,41 it is not known why this family alone has such a dramatic ocular phenotype, although it is worth noting that 9/13 (69%) of the female subjects reported with missense variants in NAA10 had some milder form of eye anomalies, including astigmatism, hyperopia and/or myopia.27 Most studies have reported that missense mutations in NAA10 decrease the enzymatic function of NAA10 and/or decrease its binding to NAA15.21, 22, 27, 35, 39, 40
In total, the presentations involving NAA10 and NAA15 appear to have phenotypic overlap but variability, and as such should be referred to more broadly as “NAA10-related syndrome” and “NAA15-related syndrome.” The extensive phenotypic variability is most likely related to genetic background differences and also to the spatial and temporal tissue-specific acetylation of a few N-terminal acetylation substrates by the NatA complex, although there are also suggested N-terminal acetylation (NTA)-independent functions for NAA10.38, 42 In the past few years, the first instance of NTA with relevance to cardiac function was reported and involved NTA of the cardiac voltage-gated sodium channel, Nav1.5, in tissues from individuals with end-stage heart failure.43, 44 Indeed, protein quality control is of major relevance in heart failure.45 Also, a 2015 study linked NTA and N-end-rule degradation to blood pressure regulation46, 47 and demonstrated that N-terminal mutants of Rgs2, a key G-protein regulator, are differentially processed by NATs and the two branches of the N-end-rule pathway, leading to an imbalance in the signaling governing blood pressure. In regard to more common diseases and basic biology, there is emerging evidence that NTA of proteins are overexpressed or otherwise dysregulated in a variety of cancers, including lung, prostate, and liver cancers.48, 49, 50, 51, 52, 53, 54 NTA has been linked to neurodegenerative diseases such as Parkinson, Alzheimer, and Huntington disease, and NatA/NAA10 has been shown to contribute to the regulation of amyloid β-protein generation, to modulate the stabilization of Sup35 amyloid formation, and to prevent aggregation of Htt,55, 56, 57, 58, 59, 60 supporting the importance of NTA in the progression of these diseases. Current findings link NTA to degradation of some proteins via Ac/N-degron-mediated recruitment of specific ubiquitin ligases.47, 61, 62, 63, 64 NTA might also influence protein-complex formation, as exemplified by the NEDD8 ligation enzymes,65 along with prion formation.60 Also, protein-specific targeting to membranes of the nucleus,66 Golgi67, 68 and lysosomes69 was shown to require NTA, but a general role in targeting is not supported. 39, 40, 70
In conclusion, we propose that disruption of NatA complex functionality can cause developmental disorders with variable expressivity. Future identification of additional affected individuals and studies in model organisms will be required if we are to continue to refine the clinical phenotype and determine the underlying mechanism whereby reduced expression or perturbed function of NAA15 results in these phenotypes.
Declaration of Interests
G.J.L serves on advisory boards for GenePeeks and Seven Bridges Genomics. The Department of Molecular and Human Genetics at BCM derives revenue from molecular testing offered at Baylor Genetics Laboratories. J.R.L has stock ownership in 23 and Me, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. E.E.E. is on the scientific advisory board of DNAnexus. W.K.C. is on the scientific advisory board of the Regeneron Genetics Center. Richard Person and Rebecca Willaert are employees of GeneDx, a wholly owned subsidiary of OPKO Health.
Acknowledgments
T.A. thanks Nina Glomnes for technical support. We appreciate the study participants and their families, without whom this work would not have been possible. Megan Cho at GeneDx facilitated the collaborations with the clinicians who submitted samples to GeneDx. The Baylor-Hopkins Center for Mendelian Genomics (BHCMG) has been funded by the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) grant number UM1HG006542. The DDD Study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant WT098051). H.A.F.S. is supported by funds from the LB692 Nebraska Tobacco Settlement Biomedical Research Development Program. J.E.P. was supported by NHGRI grant K08 HG008986. This research was further supported in part by the following to E.E.E.: the Simons Foundation Autism Research Initiative (SFARI 303241 and 337701) and National Institutes of Health (R01MH101221). E.E.E. is an investigator of the Howard Hughes Medical Institute. The collection of the proband from HudsonAlpha was supported by Clinical Sequencing Evidence-Generating Research (CSER) study grant number (NIH/NHGRI 4UM1HG007301-04). S.V. and T.A. were funded by the Research Council of Norway (249843), the Norwegian Cancer Society (PR-2009-0222), and the Norwegian Health Authorities of Western Norway (912176). G.J.L. is supported by funds from the Stanley Institute for Cognitive Genomics at Cold Spring Harbor Laboratory. Additional acknowledgments can be found in the Supplemental Data.
Published: April 12, 2018
Footnotes
Supplemental Data include Supplemental Case reports, Figures S1–S4, a Supplemental Note, Supplemental Materials and Methods, Figures S1–S5, Tables S1–S9, Supplemental References, and Supplemental Acknowledgments and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.004.
Contributor Information
Linyan Meng, Email: lmeng@bcm.edu.
Gholson J. Lyon, Email: gholsonjlyon@gmail.com.
Web Resources
BrainSpan: Atlas of the Developing Brain, http://www.brainspan.org/ (accessed 09/22/17)
GTEx, https://www.gtexportal.org/ (accessed 10/26/17)
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 4.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 6.Stessman H.A., Xiong B., Coe B.P., Wang T., Hoekzema K., Fenckova M., Kvarnung M., Gerdts J., Trinh S., Cosemans N. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017;49:515–526. doi: 10.1038/ng.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sidhu M., Brady L., Tarnopolsky M., Ronen G.M. Clinical manifestations associated with the N-terminal-acetyltransferase NAA10 gene mutation in a girl: Ogden Syndrome. Pediatr. Neurol. 2017;76:82–85. doi: 10.1016/j.pediatrneurol.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R., Zeng X., Qi H., Chang W., Sierant M.C. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Růžička M., Kulhánek P., Radová L., Čechová A., Špačková N., Fajkusová L., Réblová K. DNA mutation motifs in the genes associated with inherited diseases. PLoS ONE. 2017;12:e0182377. doi: 10.1371/journal.pone.0182377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ripley L.S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc. Natl. Acad. Sci. USA. 1982;79:4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett J.C., Buxbaum J., Cutler D., Daly M., Devlin B., Gratten J., Hurles M.E., Kosmicki J.A., Lander E.S., MacArthur D.G. New mutations, old statistical challenges. bioRxiv. 2017 [Google Scholar]
- 19.Tarailo-Graovac M., Zhu J.Y.A., Matthews A., van Karnebeek C.D.M., Wasserman W.W. Assessment of the ExAC data set for the presence of individuals with pathogenic genotypes implicated in severe Mendelian pediatric disorders. Genet. Med. 2017;19:1300–1308. doi: 10.1038/gim.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 21.Van Damme P., Støve S.I., Glomnes N., Gevaert K., Arnesen T. A Saccharomyces cerevisiae model reveals in vivo functional impairment of the Ogden syndrome N-terminal acetyltransferase NAA10 Ser37Pro mutant. Mol. Cell. Proteomics. 2014;13:2031–2041. doi: 10.1074/mcp.M113.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dörfel M.J., Fang H., Crain J., Klingener M., Weiser J., Lyon G.J. Proteomic and genomic characterization of a yeast model for Ogden syndrome. Yeast. 2017;34:19–37. doi: 10.1002/yea.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J.E., Vandekerckhove J., Lillehaug J.R., Sherman F., Gevaert K. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J.J., Halvardson J., Zander C.S., Zaghlool A., Georgii-Hemming P., Mansson E., Brandberg G., Savmarker H.E., Frykholm C., Kuchinskaya E. Exome sequencing reveals NAA15 and PUF60 as candidate genes associated with intellectual disability. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2018;177:10–20. doi: 10.1002/ajmg.b.32574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bragin E., Chatzimichali E.A., Wright C.F., Hurles M.E., Firth H.V., Bevan A.P., Swaminathan G.J. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42:D993–D1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saunier C., Støve S.I., Popp B., Gérard B., Blenski M., AhMew N., de Bie C., Goldenberg P., Isidor B., Keren B. Expanding the phenotype associated with NAA10-related N-terminal acetylation deficiency. Hum. Mutat. 2016;37:755–764. doi: 10.1002/humu.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fluge Ø., Bruland O., Akslen L.A., Varhaug J.E., Lillehaug J.R. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene. 2002;21:5056–5068. doi: 10.1038/sj.onc.1205687. [DOI] [PubMed] [Google Scholar]
- 29.Arnesen T., Anderson D., Baldersheim C., Lanotte M., Varhaug J.E., Lillehaug J.R. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 2005;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liszczak G., Goldberg J.M., Foyn H., Petersson E.J., Arnesen T., Marmorstein R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat. Struct. Mol. Biol. 2013;20:1098–1105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen J.R., Kayne P.S., Moerschell R.P., Tsunasawa S., Gribskov M., Colavito-Shepanski M., Grunstein M., Sherman F., Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnesen T., Gromyko D., Pendino F., Ryningen A., Varhaug J.E., Lillehaug J.R. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25:4350–4360. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 33.Sugiura N., Adams S.M., Corriveau R.A. An evolutionarily conserved N-terminal acetyltransferase complex associated with neuronal development. J. Biol. Chem. 2003;278:40113–40120. doi: 10.1074/jbc.M301218200. [DOI] [PubMed] [Google Scholar]
- 34.Sugiura N., Patel R.G., Corriveau R.A. N-methyl-D-aspartate receptors regulate a group of transiently expressed genes in the developing brain. J. Biol. Chem. 2001;276:14257–14263. doi: 10.1074/jbc.M100011200. [DOI] [PubMed] [Google Scholar]
- 35.Popp B., Støve S.I., Endele S., Myklebust L.M., Hoyer J., Sticht H., Azzarello-Burri S., Rauch A., Arnesen T., Reis A. De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur. J. Hum. Genet. 2015;23:602–609. doi: 10.1038/ejhg.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casey J.P., Støve S.I., McGorrian C., Galvin J., Blenski M., Dunne A., Ennis S., Brett F., King M.D., Arnesen T., Lynch S.A. NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Sci. Rep. 2015;5:16022. doi: 10.1038/srep16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 38.Dörfel M.J., Lyon G.J. The biological functions of Naa10 - From amino-terminal acetylation to human disease. Gene. 2015;567:103–131. doi: 10.1016/j.gene.2015.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myklebust L.M., Van Damme P., Støve S.I., Dörfel M.J., Abboud A., Kalvik T.V., Grauffel C., Jonckheere V., Wu Y., Swensen J. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum. Mol. Genet. 2015;24:1956–1976. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rope A.F., Wang K., Evjenth R., Xing J., Johnston J.J., Swensen J.J., Johnson W.E., Moore B., Huff C.D., Bird L.M. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esmailpour T., Riazifar H., Liu L., Donkervoort S., Huang V.H., Madaan S., Shoucri B.M., Busch A., Wu J., Towbin A. A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet. 2014;51:185–196. doi: 10.1136/jmedgenet-2013-101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C.C., Peng S.H., Shen L., Lee C.F., Du T.H., Kang M.L., Xu G.L., Upadhyay A.K., Cheng X., Yan Y.T. The role of N-alpha-acetyltransferase 10 protein in DNA methylation and genomic imprinting. Mol. Cell. 2017;68:89–103.e7. doi: 10.1016/j.molcel.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltran-Alvarez P., Tarradas A., Chiva C., Pérez-Serra A., Batlle M., Pérez-Villa F., Schulte U., Sabidó E., Brugada R., Pagans S. Identification of N-terminal protein acetylation and arginine methylation of the voltage-gated sodium channel in end-stage heart failure human heart. J. Mol. Cell. Cardiol. 2014;76:126–129. doi: 10.1016/j.yjmcc.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 44.Marionneau C., Abriel H. Regulation of the cardiac Na+ channel NaV1.5 by post-translational modifications. J. Mol. Cell. Cardiol. 2015;82:36–47. doi: 10.1016/j.yjmcc.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira J.C., Boer B.N., Grinberg M., Brum P.C., Mochly-Rosen D. Protein quality control disruption by PKCβII in heart failure; rescue by the selective PKCβII inhibitor, βIIV5-3. PLoS ONE. 2012;7:e33175. doi: 10.1371/journal.pone.0033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aksnes H., Drazic A., Arnesen T. (Hyper)tension release by N-terminal acetylation. Trends Biochem. Sci. 2015;40:422–424. doi: 10.1016/j.tibs.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Park S.E., Kim J.M., Seok O.H., Cho H., Wadas B., Kim S.Y., Varshavsky A., Hwang C.S. Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science. 2015;347:1249–1252. doi: 10.1126/science.aaa3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min L., Xu H., Wang J., Qu L., Jiang B., Zeng Y., Meng L., Jin H., Shou C. N-α-acetyltransferase 10 protein is a negative regulator of 28S proteasome through interaction with PA28β. FEBS Lett. 2013;587:1630–1637. doi: 10.1016/j.febslet.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 49.Shim J.H., Chung Y.H., Kim J.A., Lee D., Kim K.M., Lim Y.S., Lee H.C., Lee Y.S., Yu E., Lee Y.J. Clinical implications of arrest-defective protein 1 expression in hepatocellular carcinoma: A novel predictor of microvascular invasion. Dig. Dis. 2012;30:603–608. doi: 10.1159/000343090. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z., Wang Z., Guo J., Li Y., Bavarva J.H., Qian C., Brahimi-Horn M.C., Tan D., Liu W. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proc. Natl. Acad. Sci. USA. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z.H., Gong J.L., Yu M., Yang H., Lai J.H., Ma M.X., Wu H., Li L., Tan D.Y. Up-regulation of human arrest-defective 1 protein is correlated with metastatic phenotype and poor prognosis in breast cancer. Asian Pac. J. Cancer Prev. 2011;12:1973–1977. [PubMed] [Google Scholar]
- 52.Hua K.T., Tan C.T., Johansson G., Lee J.M., Yang P.W., Lu H.Y., Chen C.K., Su J.L., Chen P.B., Wu Y.L. N-α-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218–231. doi: 10.1016/j.ccr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Lee C.-F., Ou D.S.C., Lee S.-B., Chang L.-H., Lin R.-K., Li Y.-S., Upadhyay A.K., Cheng X., Wang Y.-C., Hsu H.-S. hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalvik T.V., Arnesen T. Protein N-terminal acetyltransferases in cancer. Oncogene. 2013;32:269–276. doi: 10.1038/onc.2012.82. [DOI] [PubMed] [Google Scholar]
- 55.Asaumi M., Iijima K., Sumioka A., Iijima-Ando K., Kirino Y., Nakaya T., Suzuki T. Interaction of N-terminal acetyltransferase with the cytoplasmic domain of beta-amyloid precursor protein and its effect on A beta secretion. J. Biochem. 2005;137:147–155. doi: 10.1093/jb/mvi014. [DOI] [PubMed] [Google Scholar]
- 56.Pezza J.A., Villali J., Sindi S.S., Serio T.R. Amyloid-associated activity contributes to the severity and toxicity of a prion phenotype. Nat. Commun. 2014;5:4384. doi: 10.1038/ncomms5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raychaudhuri S., Sinha M., Mukhopadhyay D., Bhattacharyya N.P. HYPK, a Huntingtin interacting protein, reduces aggregates and apoptosis induced by N-terminal Huntingtin with 40 glutamines in Neuro2a cells and exhibits chaperone-like activity. Hum. Mol. Genet. 2008;17:240–255. doi: 10.1093/hmg/ddm301. [DOI] [PubMed] [Google Scholar]
- 58.Arnesen T., Starheim K.K., Van Damme P., Evjenth R., Dinh H., Betts M.J., Ryningen A., Vandekerckhove J., Gevaert K., Anderson D. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol. Cell. Biol. 2010;30:1898–1909. doi: 10.1128/MCB.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes W.M., Klaips C.L., Serio T.R. Defining the limits: Protein aggregation and toxicity in vivo. Crit. Rev. Biochem. Mol. Biol. 2014;49:294–303. doi: 10.3109/10409238.2014.914151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmes W.M., Mannakee B.K., Gutenkunst R.N., Serio T.R. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat. Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hwang C.-S., Shemorry A., Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang C.S., Shemorry A., Auerbach D., Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shemorry A., Hwang C.-S., Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim H.K., Kim R.R., Oh J.H., Cho H., Varshavsky A., Hwang C.S. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156:158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott D.C., Monda J.K., Bennett E.J., Harper J.W., Schulman B.A. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murthi A., Hopper A.K. Genome-wide screen for inner nuclear membrane protein targeting in Saccharomyces cerevisiae: roles for N-acetylation and an integral membrane protein. Genetics. 2005;170:1553–1560. doi: 10.1534/genetics.105.043620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Behnia R., Panic B., Whyte J.R.C., Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 68.Setty S.R.G., Strochlic T.I., Tong A.H.Y., Boone C., Burd C.G. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 69.Hofmann I., Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- 70.Aksnes H., Osberg C., Arnesen T. N-terminal acetylation by NatC is not a general determinant for substrate subcellular localization in Saccharomyces cerevisiae. PLoS ONE. 2013;8:e61012. doi: 10.1371/journal.pone.0061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.