Abstract
Primary ciliary dyskinesia (PCD) is a genetically and phenotypically heterogeneous disorder characterized by destructive respiratory disease and laterality abnormalities due to randomized left-right body asymmetry. PCD is mostly caused by mutations affecting the core axoneme structure of motile cilia that is essential for movement. Genes that cause PCD when mutated include a group that encode proteins essential for the assembly of the ciliary dynein motors and the active transport process that delivers them from their cytoplasmic assembly site into the axoneme. We screened a cohort of affected individuals for disease-causing mutations using a targeted next generation sequencing panel and identified two unrelated families (three affected children) with mutations in the uncharacterized C11orf70 gene (official gene name CFAP300). The affected children share a consistent PCD phenotype from early life with laterality defects and immotile respiratory cilia displaying combined loss of inner and outer dynein arms (IDA+ODA). Phylogenetic analysis shows C11orf70 is highly conserved, distributed across species similarly to proteins involved in the intraflagellar transport (IFT)-dependant assembly of axonemal dyneins. Paramecium C11orf70 RNAi knockdown led to combined loss of ciliary IDA+ODA with reduced cilia beating and swim velocity. Tagged C11orf70 in Paramecium and Chlamydomonas localizes mainly in the cytoplasm with a small amount in the ciliary component. IFT139/TTC21B (IFT-A protein) and FLA10 (IFT kinesin) depletion experiments show that its transport within cilia is IFT dependent. During ciliogenesis, C11orf70 accumulates at the ciliary tips in a similar distribution to the IFT-B protein IFT46. In summary, C11orf70 is essential for assembly of dynein arms and C11orf70 mutations cause defective cilia motility and PCD.
Keywords: primary ciliary dyskinesia, Kartagener syndrome, mutation, intraflagellar transport, dynein, Paramecium
Introduction
Cilia are complex organelles that form a distinct cellular compartment projecting from the surface of non-dividing cells. They have a microtubular-based axoneme core structure, contained within a specialized membrane that extends from the plasma membrane. This basic structure is highly conserved across species to serve various physiological functions.1 Motile and non-motile types of cilia in the human body have differences in their axoneme structural arrangement and motilities.2, 3 Non-motile primary cilia present on most cells contain nine pairs of microtubules forming a circular scaffold of nine-fold symmetry termed 9+0. Nodal cilia of the embryonic left-right organizer (node) have a similar structure but with dynein “arm” motor subunits attached to the peripheral microtubules that provide motility machinery for ciliary beating.4 Motile cilia of the respiratory epithelium, fallopian tubes, and brain ependyma (which lines the ventricular system) have a 9+2 arrangement, where the peripheral microtubules attached to dynein arm motors surround a central pair of microtubules. The sperm flagellum has the same structure as motile cilia with subtle differences in the distribution of dynein arms along the axoneme.5 Lastly, kinocilia of the inner ear hair cells have a 9+2 organization but lack beating motility, containing outer but not inner dynein arms.
The main function of cilia/flagella motility is in generating fluid flow or movement within fluids. Defective cilia motility leads to primary ciliary dyskinesia (PCD [MIM: 244400]), a genetic disorder in which left-right positioning of body organs is randomized such that laterality problems affect about half of affected individuals. Such laterality defects may in some cases be associated with cardiac, spleen, and other organ problems.6 Children with PCD suffer from chronic, widespread upper and lower airway infections and congestion, progressing toward bronchiectasis. Symptoms often present early in life with neonatal respiratory distress syndrome. Affected individuals can have hydrocephalus, hearing defects, retinal dystrophy, and infertility in adults especially in males due to sperm dysmotility.7, 8
PCD is inherited as an autosomal-recessive or X-linked disease. The genetic cause of PCD, where known, has mainly been mutations affecting one of the hundreds of genes encoding essential components of the complex motile ciliary machinery. PCD is dominated by mutations affecting the axonemal dynein arm motors, for example in subunits of the outer dynein arm (ODA) such as DNAH5 (MIM: 603335)9 or in proteins involved in ODA targeting and anchoring such as CCDC103 (MIM: 614677), ARMC4 (MIM: 615408), and ODA docking complex (ODA-DC) components such as CCDC151 (MIM: 615956).10, 11, 12, 13, 14, 15, 16
Perhaps more surprising than the finding that mutations in cilia structural components are a frequent cause of PCD has been a growing number of PCD loci that do not encode parts of the cilium per se but rather encode proteins that act in the apparently complicated process of the assembly and transport of ciliary dynein motors.17, 18 Many such assembly factors work strictly in the cytoplasm in a chaperoning step and cause PCD when mutated, such as DNAAF1 (MIM: 613190),19, 20, 21 DNAAF2 (MIM: 612517),22 DNAAF3 (MIM: 614566),23 DYX1C1 (DNAAF4 [MIM: 608706]),24, 25 HEATR2 (DNAAF5 [MIM: 61486]),26, 27 LRRC6 (MIM: 614930),28, 29, 30 and PIH1D3 (MIM: 300933).31, 32 Other factors, identified in both the human population through analysis of individuals with PCD and in genetic screens for cilia motility defects in model organisms, resemble chaperones in that they affect the assembly of multiple ciliary dynein isoforms but also resemble transport factors based on their presence in a detergent-soluble ciliary compartment. These include ZMYND10 (MIM: 607070)33, 34 and C21orf59 (MIM: 615494).35, 36
A number of additional proteins involved in dynein assembly have been identified from non-human studies including ODA8, ODA5, and ODA10 of the flagellated model organism Chlamydomonas reinhardtii. ODA5 and ODA10 directly interact as a complex required for formation of an assembly-competent outer dynein arm in the cytoplasm, participating along with ODA8 in the later stages of cytoplasmic ODA assembly.37, 38 ODAs are then trafficked into cilia for assembly onto the ODA-DC using the internal cargo transport system, intraflagellar transport (IFT), and subsequent steps involve proteins thought to be involved in an IFT-dependent trafficking step, such as ODA16 (DAW1 or WDR69 in human), which has so far not been linked to disease in humans.39, 40, 41 The anterograde or base-tip direction of cilia cargo transport involves members of the IFT-B subcomplex, one of which (IFT46) interacts with ODA16. ODA16 therefore seems to function as an adaptor between IFT-B and the ODA, required for the IFT-based transport of ODAs into the cilia/flagella compartment.40
To reach a genetic diagnosis of PCD, bi-allelic mutations in autosomal-recessive or hemizygous state in an X-linked gene should be identified. Reported mutations in known genes account for only about 70% of PCD cases, so additional genes are still to be identified.17 Here we describe mutations in a gene, C11orf70, that cause PCD and demonstrate that C11orf70 encodes a protein needed for assembly of both outer and inner arm axonemal dyneins in respiratory cilia, a hallmark of the axonemal dynein assembly co-chaperones. Using model organisms, we show that C11orf70 orthologs perform a conserved dynein assembly function and are present in both the cytoplasm and in cilia. Furthermore, ciliary abundance of this protein is detergent sensitive and dependent on continued IFT trafficking, as expected for a factor important for late steps in ciliary dynein assembly.
Material and Methods
Subjects and Families
The study was ethically approved by the London Bloomsbury Research Ethics Committee (08/H0713/82). Written informed consent was obtained from all participants or their guardians prior to enrollment in the study. Affected individuals were diagnosed with PCD through standard diagnostic screening by the Royal Brompton Hospital PCD diagnostic service, according to European Respiratory Society diagnostic guidelines.42
Next Generation Sequencing
Genomic DNA was extracted from whole blood samples using a standard phenol-chloroform protocol. Genetic screening was done as previously described using a targeted next generation sequencing panel including all genes that are recognized to cause isolated heterotaxy or PCD when mutated and other candidate genes that are predicted to play a role in cilia motility.31 The panel was designed using the Agilent SureDesign tool (Agilent Technologies) to capture all coding regions of the included genes and 25 bp at the exon-intron boundaries. Library preparation was done using the SureSelectQXT Target Enrichment Kit (Agilent Technologies). Paired end sequencing (2 × 75 cycles) was performed using the NextSeq 500/550 High Output v2 kit and NextSeq platform (Illumina). A multiplex of 48 samples from affected individuals with PCD were sequenced per flow cell in each run using barcoding indices. Sequencing data were subjected to quality control and further analysis using an in-house pipeline as previously described.43 Alignment of Fastq sequence files with the human genome reference (GRCh37/hg19) used BWA (0.6.1-r104),44 with variant calling performed using Freebayes45 and variant annotation using Alamut Batch (Interactive Biosoftware) softwares. VCF files were then converted into Excel format to enable manual variant filtration and prioritization. Only coding nonsynonymous and splicing variants (±5 bp from exon-intron boundary) were retained for further analysis. Common sequence variants were then filtered out based on their minor-allele frequency (MAF) to exclude all variants with MAF ≥ 0.01 in the dbSNP build 141, ExAC, Exome Variant Server, and 1000 Genomes databases. Retained variants were evaluated and prioritized based on their predicted impact at the protein level and predicted pathogenicity scores using a number of in silico softwares: Human Splicing Finder, SIFT, PolyPhen-2, Mutation Taster, Combined Annotation Dependent Depletion (CADD) score, and Multivariate Analysis of Protein Polymorphism (MAPP). Conservation across species was studied using PhastCons, PhyloP, and the BLOSUM62 matrix within BLAST.
Sanger Sequencing
Prioritized variants in C11orf70 were further confirmed in the probands and segregated within other family members by Sanger sequencing. Primers flanking the exon harboring the mutations were designed using the NCBI Primer-BLAST tool with genomic sequence amplification performed using standard PCR conditions and annealing temperatures based on the primer Tm temperature. Primers are listed in Table S1. Analysis of the sequencing data used SnapGene software (GSL Biotech).
Air-Liquid Interface (ALI) Culture
BMI1 transformed normal human bronchial epithelial cells (BMI1 NHBE) were cultured at air liquid interface on collagen-coated transwell inserts (passage 7) for 4 weeks as previously described.46, 47
Quantitative RT-PCR
Total RNA was extracted using Trizol Reagent (QIAGEN). cDNA was produced using Omniscript RT kit (QIAGEN). Real-time qPCR reactions were done in triplicate using the CFX96 touch real-time PCR system and iQ SYBR green Supermix and data analysis using Bio-Rad CFX Manager 3.1 software (Bio-Rad Labs). Primers used for qPCR in human and Paramecium cells are listed in Table S1.
Transmission Electron Microscopy (TEM) and Immunofluorescence in Human Subjects
Nasal brushings from affected individuals were fixed in 4% glutaraldehyde for transmission electron microscopy. Processing and quantification was as previously described.48 Briefly, samples were bound in agar, post fixed in osmium tetroxide, dehydrated, and embedded in araldite before being cut into thin sections for analysis. Sections were stained with 2% methanolic uranyl acetate and lead citrate to provide contrast and assessed on a Jeol 1400+ transmission electron microscope. Images of cilia cross-sections were acquired with a digital camera (AMT 16X, Deben UK). To calculate the percentage loss of dynein arms, we divided the observed number of cilia cross sections with absent arms on all nine microtubular doublets by the total number of cilia examined. Immunofluorescence for PCD diagnosis was conducted as previously described.49 All antibodies used in this study are listed in Tables S2 and S3.
High-Speed Video Microscopy Analysis (HSVM)
Nasal epithelial cells were re-suspended in Medium 199 (pH7.2, Sigma-Aldrich Co.) and assessed by light microscopy at 37°C in a 1 mm chamber. Well-ciliated continuous epithelial strips were selected for assessment by high-speed video. Cilia were recorded using a 63× oil immersion lens on an upright DM60 Leica microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 500 frames per second (Troubleshooter, Fastec Imaging). Ciliary beat pattern was assessed and ciliary beat frequency calculated according to the number of frames required to complete 10 full beats.50
Paramecium Gene Silencing
We identified the Paramecium C11orf70 ortholog as GSPATG00011350001 (GenBank: XM_001443078.1) using public databases CILDB and ParameciumDB. A 561 bp long DNA segment corresponding to the N terminus of the gene was cloned in L4440 plasmid between the two convergent T7 promotors.51 Possible RNAi off-target effects were assessed using the ParameciumDB RNAi off-target tool. E. coli HT115 bacteria (lacking RNAase III activity) were transformed with this silencing vector and grown in Lysogeny broth (LB) medium. Paramecia (Paramecium tetraurelia d4-2 strain) were fed with the transformed bacteria over 3 days as previously described.52 The RNAi experiment was repeated several times to ensure reproducibility of the results (n > 10). RNAi of the ND7 gene,53 which affects trichocyst exocytosis without affecting cilia motility, was used as a negative control. Knockdown of ND7 was attested by trichocyst retention after picric acid treatment, as described previously53 and the RNAi conditions that we used for ND7 ablation were published previously.53, 54 To assess the degree of gene knockdown at the RNA level, a mass culture protocol was used to get sufficient quantities of paramecia for RNA extraction as previously described.55 Total RNA was extracted using the RNAeasy Micro kit (QIAGEN). cDNA was produced using 1 μg RNA, Superscript IIII (Invitrogen), and random hexanucleotides (Invitrogen). qPCR was performed to assess the degree of knockdown at the RNA level (primers listed in Table S1).
Paramecium Cilia Function Tests
Paramecium velocity was tested using 3–5 paramecia per test transferred into a drop of conditioned BHB solution and tracked for 10 s every 0.3 s to assess the swimming pattern under dark field microscopy, using MetaVue software. ImageJ software was used for image analysis and measurement of the swimming velocity.56 For Paramecium cilia beating analysis, knockdown cells were adhered to a slide covered with Cell-TakTM (BD Bioscience) after silencing for 72 hr as previously described.57 Cilia beating was recorded using a 63× oil immersion lens on an upright DM60 Leica microscope the same as for the nasal epithelia cells, at 500 frames per second at 37°C. The CiliaFA plugin in ImageJ was used for assessment of cilia beat frequency in Paramecium.58
Paramecium TEM and Immunofluorescence Analysis
The sample fixation and processing protocol for TEM in Paramecium was as previously described.59 Images of cilia cross-sections were acquired with a digital camera (AMT 16X, Deben UK). The numbers of the missing IDA and ODA arms were recorded per cross-section by an observer blinded to the condition. For immunofluorescence studies, paramecia were fixed in 2% paraformaldehyde in PHEM buffer (Pipes 60 mM, HEPES 25 mM, EGTA 10 mM, MgCl2 2 mM, adjusted to pH 6.9 with NaOH) for 15 min. After fixation, cells were permeabilized for 15 min in 1% Triton X-100 in PHEM. Cells were washed 3 times in PBS/BSA 3%. Immunostaining used polyglutamylated tubulin antibodies (1/500) as previously described.60
Phylogenetic Comparisons
The presence or absence of C11orf70 orthologs was determined by searches of selected organism genomes (at the genus level) using BLASTp at NCBI with the Chlamydomonas ortholog protein FBB5 sequence (GenBank: XP_001694050) as query. Apparent hits were confirmed only if they also gave FBB5 as the top hit in a reciprocal BLAST search. This procedure was also used to establish the presence or absence of two ODA subunits (HCgamma and IC2), two IDA subunits (I1HCalpha and IC140), two IFTA subunits (IFT122 and IFT140), four IFTB subunits (IFT46, IFT52, IFT88, and IFT172), ODA-DC2, and ODA16. Retention of additional IFT subunits by selected organisms was taken from van Dam et al.61
Functional Characterizations of C11orf70 in Paramecium
The Paramecium C11orf70 ortholog (GSPATT00011350001) was cloned upstream of GFP into the SpeI-XhoI sites of pPXV-GFP plasmid which contains the constitutive regulators of the Paramecium calmodulin gene.62 Three glycine codons were added between C11orf70 and the GFP sequence. The IFT46-tagged GFP gene was expressed under its own regulators. The IFT46 (GSPATG00024708001) putative promotor was cloned between the BamHI and SphI sites of pPZZ-GFP02, a modified pPXV-GFP vector (gift of J. Cohen), upstream of the GFP sequence. The full-length IFT46 was cloned between KpnI and SmaI sites of pZZ-GFP02 after the GFP sequence. Individual ND7-1 mutant cells unable to discharge their trichocysts were then transformed with the expression vectors, by micro-injection into their macronucleus, with filtered and concentrated DNA containing a mixture of the linearized plasmids of interest (5 μg/μL) and of plasmid DNA directing the expression of wild-type ND7. 3/4 divisions after the injections, the cell lines issued from the injected cells were screened for their ability to discharge their trichocysts and to express the fluorescent protein. Microinjection was made under an inverted Nikon phase-contrast microscope, using a Narishige micromanipulation device and an Eppendorf air pressure microinjector. Transformants with the same growth rate as untransformed cells and various plasmid DNA copy numbers were selected for experiments. Knockdown of IFT139 in transformants expressing C11orf70-GFP was performed by the feeding method, using bacteria expressing double-strand RNA homologous to 536 bp of IFT139 sequence (PTET.51.1.G0310064) from base 40 to 576. After 2 or 3 divisions (24 hr) on feeding medium, the cells were fixed in 2% paraformaldehyde in PHEM buffer for 15 min. To deciliate the paramecia, they were placed in 10 mM Tris (pH 7.4), 1 mM CaCl2, 5% ethanol for 30 s with vortexing. To allow ciliary regrowth, cells were incubated in growth medium for various times from 15 to 30 min. Ciliary growth was stopped by fixation and the cells processed for IF. For biochemistry, paramecia were deciliated according to Adoutte et al.63 Cilia were recovered by centrifugation at 28,000 × g for 30 min and either dissolved in SDS-Laemmli buffer or extracted in 20 mM Tris (pH 7.5), 1 mM EDTA, and 0.2% Triton X-100. Soluble proteins were precipitated using 9 volumes of methanol before resuspension in SDS-Laemmli buffer. Insoluble proteins were directly solubilized in the same volume of SDS-Laemmli buffer.
Functional Characterization of C11orf70 in Chlamydomonas
Genomic sequences for Chlamydomonas C11orf70 ortholog FBB5 (Cre12.g556300; GenBank: XM_001693998.1) and intron/exon boundaries were based on genome assembly annotation version 5.5 and obtained from the Phytozome server. Alignments with FBB5 protein (GenBank: XP_001694050) orthologs were generated with MEGA version 6.64 The expression vector pDIC2linkerHA was created by replacing the Chlamydomonas PsaD promoter and 5′ UTR in pGenDlinkerHA19 with the promoter and 5′ UTR of the Chlamydomonas ODA-IC2 gene.65 The resulting expression vector retains a unique EcoRI site between the IC2 ATG start codon and sequences that encode three copies of an HA epitope tag. Two overlapping gBlocks Gene Fragments (Integrated DNA Technologies) of 464 and 458 bp were used to insert FBB5 coding sequences at this EcoRI site. The resulting FBB5::HA minigene retains the first intron of the endogenous FBB5 gene. In the resulting protein, the first three amino acids of native FBB5 are replaced with vector-encoded amino acids (MTA -> MVG) and the C-terminal residue (W) is followed immediately by a 9 amino acid linker (AFPRGGISR) and the three HA tag sequences. This expression construct, pFBB5::HA, was co-transformed with ARG plasmid pJD67 into an arg2 mutant Chlamydomonas strain. Of 72 randomly selected ARG colonies, 8 expressed an HA tagged protein of the expected size (35 kDa), as seen by blots of whole cell samples probed with rat anti-HA McAb 3F10 (Roche), and one strain was selected for all subsequent work. Mutant strains expressing FBB5::HA were generated by standard genetic crosses, using blots of whole cell samples to select tetrad products that expressed the FBB5::HA transgene.
Chlamydomonas flagellar isolation by dibucaine deflagellation and flagellar fractionation by detergent followed methods previously described by Mitchell et al.66 Chlamydomonas strains ida1, oda3, oda6, oda8, oda10, oda16, and fla10-1ts67 are available from the Chlamydomonas Resource Centre, University of Minnesota. An ift46 mutant strain expressing an N-terminally truncated IFT46 transgene and a Guinea pig antibody that recognizes the IFT46 C terminus were generously provided by George Witman, UMASS Medical School, Worcester, MA.68 To test the effects of loss of IFT proteins from flagella, wild-type and temperature-sensitive IFT kinesin mutant fla10ts strains expressing FBB5::HA were incubated at 31°C for 1.5 hr prior to flagellar isolation. For flagellar growth experiments, a wild-type strain expressing FBB5::HA was deflagellated by pH shock, allowed to regenerate for 60 min, and deflagellated a second time. Flagellar samples were adjusted to identical protein concentrations before loading onto gels. Anti-IC2 mouse McAb C11.469 and anti-ODA16 rabbit sera40 have been previously characterized.
Results
Targeted Next Generation Sequencing Identifies C11orf70 Mutations in Individuals with PCD
A targeted next-generation sequencing gene panel was used for mutational analysis in a cohort of affected individuals with PCD. As previously described, the panel contains all the currently known genes which, when mutated, cause PCD in addition to a set of other candidate genes predicted to play a role in cilia motility, based upon collaborative information derived from past PCD genetic mutation screening projects and a number of ciliate organism genome and proteome projects.31 Screening of 161 unrelated individuals with a confirmed or presumed diagnosis of PCD revealed variants in a previously uncharacterized candidate gene, C11orf70 (GenBank: NM_032930.2, NP_116319), in two unrelated case subjects where PCD diagnosis had been confirmed by a number of diagnostic clinical testing modalities.42
First, we identified a homozygous C11orf70 missense variant (GenBank: NM_032930.2; c.776A>G [p.His259Arg]) in one of two affected siblings in a Pakistani consanguineous family where the parents are first cousins. Sanger sequencing confirmed the homozygous status of this mutation in the affected individual and in their affected sibling as well as the carrier status of both parents and an unaffected sibling (Figure 1A). This variant is found only once in the ExAC control exome database in a heterozygous carrier from the South Asian population, showing an overall allele frequency in unaffected controls of 8.242e−06. It is absent from the EVS, 1000G, and dbSNP control databases. Phylogenetic analysis revealed that the His259 amino acid is a highly conserved residue, located within the DUF4498 domain of C11orf70 (Figures 1B and S1). Using PolyPhen-2 software, this mutation is predicted to be damaging, with a score of 0.997 (sensitivity: 0.27; specificity: 0.98). It is also predicted to have a damaging effect on the protein using the SIFT tool. MutationTaster predicts it to be a disease-causing mutation and it has a CADD score of 23.9 (CADD score of ≥20 indicates a variant is among the top 1% of deleterious variants in the genome).
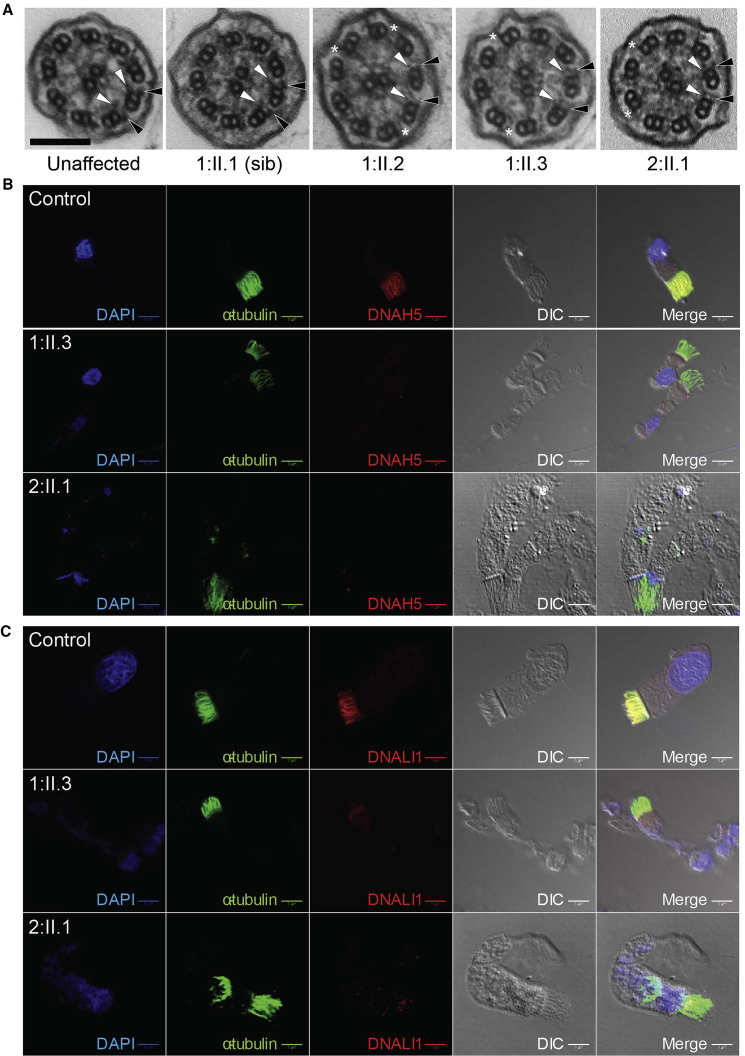
Figure 1.
Bi-allelic C11orf70 Mutations Identified in Individuals with Primary Ciliary Dyskinesia
(A) Pedigree structures of affected family 1 and 2. Electropherograms show variant regions in family 1 individual I-1 and I-2 (carrier father and mother), II-1 (carrier sibling), II-2 and I-3 (affected siblings), family 2 individual I-1 and I-2 (carrier father and mother), II-1 (affected). Familial segregation shows recessive inheritance in the affected siblings in family 1 (II-2 and II-3) both of whom carry a homozygous missense mutation c.776A>G (p.His259Arg) and in the affected child in family 2 (II-1) who carries compound heterozygous nonsense mutations c.154C>T (p.Gln52∗) and c.361C>T (p.Arg121∗).
(B) The genomic structure of C11orf70 and the C11orf70 protein shows the locations of the identified disease variants. The only known conserved domain is a “domain of unknown function” (DUF4498). The phylogenetic conservation of the mutated amino acid histidine 259 across different mammalian and other ciliate species is highlighted in blue below the protein. Uniprot accession numbers are indicated; XENTR, Xenopus tropicalis; DANRE, Danio rerio; CHLRE, Chlamydomonas reinhardtii.
(C) Clinical characteristics of individuals carrying biallelic C11orf70 mutations. NRD, neonatal respiratory distress; SS, situs solitus; SI, situs inversus totalis; nNO, nasal nitric oxide level (normal cutoff is >77 nL/min); HSVM, high-speed video microscopy; n.t., not tested as subject too young; IDA, inner dynein arm; ODA, outer dynein arm.
Second, we identified two compound heterozygous C11orf70 stop-gain (nonsense) variants in an affected child from an Indian non-consanguineous family (GenBank: NM_032930.2; c.154C>T [p.Gln52∗] inherited from the father and GenBank: NM_032930.2; c.361C>T [p.Arg121∗] inherited from the mother) (Figure 1A). Sanger sequencing confirmed the mutations in the affected individual’s DNA and the carrier status of the parents. In the ExAC database, the p.Gln52∗ mutation (rs767760877) is found in two heterozygote carriers from the South Asian population with a total allele frequency of 1.653e−05 and the p.Arg121∗ mutation (rs561237622) is found only once in a heterozygote carrier from the South Asian population with a total allele frequency of 8.254e−06. Both stop-gain variants are also located within the C11orf70 DUF4498 domain (Figure 1B) and are expected to cause a major deleterious effect on the protein. Their respective CADD scores are 36 (c.154C>T [p.Gln52∗]) and 39 (c.361C>T [p.Arg121∗]).
C11orf70 Mutations Cause PCD with Combined Loss of Outer and Inner Dynein Arms
The affected individuals from both families presented with typical PCD symptoms from early life, including a positive history of neonatal respiratory distress, and displayed other disease features of chronic wet cough and rhinitis (Figure 1C). Only the eldest affected child in family 1 was of an age appropriate for measurement of nasal nitric oxide levels, which were highly reduced (below the diagnostic cut off of 77 nL/min).70 Dextrocardia or situs inversus were observed in both the affected families. High-speed video microscopy (HSVM) examination of nasal brushing biopsies of the affected individuals showed that the respiratory epithelium cells had completely immotile cilia (Movies S1, S2, and S3) compared to the effective ciliary beating observed in both the unaffected sibling II-3 from family 1 (Movie S4) and control subjects (Movie S5).
Ultrastructural studies using transmission electron microscopy (TEM) analysis of nasal brushing biopsies, with quantification of outer and inner dynein arms, was performed within a formal clinical diagnostic setting, typically evaluating 50–70 cilia per individual. The 9+2 microtubule pattern of the cilia appeared to be undisturbed overall but the survey revealed a significant loss of both the inner and outer dynein arm structures from respiratory epithelial cell cilia that affected both families equivalently. Outer dynein arms were absent in 67% and 69% of cross-sections in family 1 individuals II-2 and II-3 and in 94% of cross-section in the family 2 case II-1. The inner dynein arms were lost in 75% and 95% of cilia cross-sections in family 1 II-2 and II-3 and in 89% of cilia cross-sections in family 2 II-1. These data are shown in Figure S2, with representative images shown in Figure 2A.
Figure 2.
Affected Individuals with C11orf70 Mutations Display Loss of the Cilia Outer and Inner Dynein Arms in Respiratory Cells
(A) Representative images of transmission electron micrographs of respiratory cilia in cross-section reveals a normal 9+2 pattern of the axoneme but with a loss of both the outer (black arrowheads) and inner (white arrowheads) dynein arms from the affected individuals in family 1 and 2 (right three panels), compared to a healthy control (far left) and the unaffected carrier sibling in family 1 (second left). Asterisks indicate remnants of short outer dynein arms that were observed to be retained in some cross sections. Scale bar, 100 nm.
(B and C) Immunofluorescence staining of respiratory epithelial cells for an antibody marker of the ciliary axoneme (acetylated alpha-tubulin, green) and the outer dynein arm antibody marker DNAH5 (red) shows highly reduced staining in affected individuals from both families indicating a loss of the outer dynein arms in agreement with the TEM data (B). Double antibody staining for acetylated alpha-tubulin (green) and the inner dynein arm antibody marker DNALI1 (red) shows a highly reduced staining in affected individuals from both families for DNALI1, indicating also a loss of the inner dynein arms (C). Differential interference contrast (DIC) imaging shows the outline of the cell and cilia in each case and the nuclei are in blue (4’,6-diamidino-2-phenylindole stain, DAPI). Scale bars, 5 μM.
The composition of the cilia in the respiratory epithelial cell samples in these affected individuals was also examined by immunofluorescence (IF) analysis compared to control subjects. Respiratory epithelial cells were double-labeled with antibodies directed against the axoneme (acetylated alpha-tubulin) and markers of the outer dynein and inner dynein arms. This showed a significantly reduced level (absence) of staining for the established diagnostic outer dynein arm marker DNAH5 (Figure 2B) and inner dynein arm marker DNALI1 (MIM: 602135) (Figure 2C) in the affected individual’s respiratory cilia compared to control samples from an unaffected healthy individual. In agreement with the TEM ultrastructure results, immunofluorescent staining with markers of other cilia structures including the radial spokes (RSPH4A [MIM: 612647] antibody71) and nexin-dynein regulatory complexes (GAS8 [MIM; 605178] antibody72) did not display altered distribution in the cilia (data not shown).
C11orf70 Has a Characteristic Transcriptional Profile during Mucociliary Differentiation
Previous gene expression profiling studies on human tissues showed higher expression of C11orf70 in multiciliated tissues compared to tissues with just primary, non-motile cilia.73, 74 We analyzed the transcriptional profile of C11orf70 during human motile ciliogenesis, by quantitative PCR of mRNA isolated from air-liquid interface cultures of human ciliated epithelial cells using qPCR primers listed in Table S1. We found that the C11orf70 has a similar expression profile to DNAH5 (Figure 3A) encoding the DNAH5 ODA heavy chain dynein, which is the gene most commonly mutated in PCD.9, 75 The expression of both genes increased over the time of culture in parallel, with a sharp rise around day 19 at the point when the cells start to ciliate, then falling to a plateau phase around day 24.
Figure 3.
C11orf70 Is Widely Conserved and Distributed in Both the Cytoplasm and Cilia
(A) Quantitative RT-PCR analysis of C11orf70 expression during multi-ciliogenesis in ALI culture over 30 days (D) in culture. The qPCR analysis compared expression of C11orf70 to that of DNAH5. A similar transcriptional profile was seen over time in ALI culture, from around day 10 with a peak at day 19 followed by a plateau from day 24. Error bars indicate SEM.
(B) Phylogenomic distribution of C11orf70 orthologs compared with axonemal dyneins and dynein trafficking proteins. A single C11orf70 copy is retained (filled circles) in organisms with motile cilia (last column, curved organelle) that use outer dynein arms (ODA column) unless IFT-B complexes are also completely missing (open circles) or compromised by missing subunits (half-filled circles). Each circle represents the results of reciprocal BLAST searches of two or more proteins in each complex, starting with the Chlamydomonas sequences (see Material and Methods for details).
(C) Stained gels (top) and blot probed for HA (bottom) of cell body and flagellar fractions of Chlamydomonas cells expressing tagged C11orf70 ortholog FBB5-HA. More than 90% of FBB5-HA is cytoplasmic, but a small fraction is present in flagella.
(D) Gel (top) and blot (bottom) showing fractionation of isolated Chlamydomonas flagella by detergent. FBB5-HA is quantitatively released into the detergent-soluble membrane-matrix fraction.
(E) Western blot of deciliated wild-type paramecia (Cell body) expressing C11orf70-GFP as well as cilia fraction of the same cells loaded with a 2- or 10-fold excess of cilia, probed with anti-GFP and anti-PolyE tubulin antibodies. C11orf70 is enriched in the cytoplasm, despite 2% is found in the cilia fraction.
(F) Cilia fraction in Paramecium solubilized using Triton X-100 buffer. The Triton X-100 soluble (Sol) and insoluble (Insol) fractions were loaded at equal stoichiometry. C11orf70 is found in the soluble fraction corresponding to the matrix and membrane fraction, while PolyE tubulin is highly enriched in the axonemal fraction.
C11orf70 Is Highly Conserved and Expressed in Species that Require Dynein Arms for Cilia Motility
C11orf70 is a highly conserved protein across species (Figure S1). Cross species phylogenetic analysis showed that C11orf70 orthologs are retained in most species that have motile cilia with a few notable exceptions (Figure 3B). C11orf70 is not found in the genomes of nematode and lower plants that lack outer arms (e.g., Caenorhabditis, Selaginella, Physcomitrella) and is also lost from organisms that have outer dynein arms but that do not use IFT for flagellar assembly (e.g., Plasmodium, Toxoplasma, Thalassiosira). ODA16, an IFT-interacting protein involved in dynein trafficking, shows a similar distribution. Most of the genomes contained a single C11orf70 copy, with the exception of genomes such as Xenopus tropicalis that have undergone duplications. Although Paramecium tetraurelia has undergone at least three whole-genome duplications, its C11orf70 gene is unique (duplications could be followed by deletion).
C11orf70 Localizes to Both the Cilia and Cell Body in Both Chlamydomonas and Paramecium
Since C11orf70 is a previously uncharacterized human protein, we next proceeded to investigate its wider biological role in cilia motility. Two unicellular model organisms were employed for this analysis: Chlamydomonas, a biflagellate alga commonly used to model PCD disease processes,19, 22, 23 and the ciliate Paramecium tetraurelia. Paramecium is a unicellular multiciliated organism that is easy to cultivate and, like Chlamydomonas, allows for ciliary molecular and biochemical analyses. Moreover, RNAi by feeding in Paramecium is a very robust and easy method for functional characterization of potential candidate PCD proteins.76
Both the Chlamydomonas and Paramecium genomes encode a single, well-conserved C11orf70 ortholog. In Chlamydomonas, it was previously designated as FBB5 based on its phylogenomic distribution with other genes encoding flagellar and basal body proteins.77, 78 FBB5 is upregulated following deflagellation79, 80 but is not present in any published ciliary proteomes. To explore the function of C11orf70 in Chlamydomonas and Paramecium, we expressed a C-terminal hemagglutinin (HA)-tagged version in wild-type Chlamydomonas and a C-terminal GFP-tagged version of the Paramecium C11orf70 ortholog in wild-type Paramecium. Western blots of deflagellated cells and isolated flagella from Chlamydomonas revealed that FBB5::HA is predominantly cytoplasmic, with only about 2% appearing in the flagellar fraction (Figure 3C). Consistent with these results, GFP-tagged Paramecium C11orf70 is present mostly in the cytoplasm with about 2% in cilia (Figure 3E). Chlamydomonas flagellar FBB5::HA was quantitatively released by detergent treatment, which placed it in the soluble membrane and matrix fraction (Figure 3D). Similarly, cilia purification followed by Triton X-100 extraction in Paramecium released the ciliary C11orf70 into the soluble fraction (Figure 3F).
C11orf70 Has a Conserved Role in Dynein Arm Assembly in Paramecium
We further assessed the involvement of C11orf70 as a gene that may be associated with PCD etiology and its role in cilia motility by gene silencing in Paramecium. RNAi knockdown of the Paramecium C11orf70 reduced the expression of C11orf70 by >80% (Figures 4A and S3). As a control, the ND7 gene,53 which plays no role in cilia motility, was subjected to RNAi in parallel. C11orf70 knockdown led to an evident phenotypic defect in the Paramecium swimming pattern with severely reduced velocity after 72 hr (Figure 4B). The average swimming velocity of the C11orf70-RNAi cells (0.58 ± 0.22 mm/s) was reduced to 68% of the control ND7-RNAi cells (0.85 ± 0.20 mm/s) (Figure 4C). This decrease in the Paramecium swimming speed after C11orf70 RNAi was confirmed not to be a consequence of an alteration in cell size, cilia length, or number of cilia per cell (Figure S4). When HSVM analysis of the cilia motility of C11orf70 RNAi-silenced Paramecium was performed, a significant decrease in cilia beat frequency (CBF) was observed in the C11orf70 knockdown cells compared to the control ND7 knockdown cells (Figure 4D). The mean CBF of the control ND7-RNAi Paramecium was 24.3 ± 2 Hz while the cilia of the C11orf70-RNAi Paramecia were beating at a frequency averaging 20.3 ± 4 Hz. Despite the modest reduction of cilia beat frequency, a close examination of the movies showed that some cilia were not beating. The cilia waveform was also affected by C11orf70 knockdown with a variable phenotype ranging from a slight reduction in amplitude to a half-beat (Movies S6 and S7).
Figure 4.
Paramecium Gene Knockdown Shows a Conserved Function for C11orf70 in Dynein Arm Assembly
(A) Quantitative RT-PCR analysis of C11orf70 expression in both the C11orf70 knockdown cells and the ND7 knockdown control. The expression level of Paramecium C11orf70 is reduced by about 80% in the C11orf70 knockdown cells compared to its expression in the control ND7 knockdown cells.
(B) Paramecium swimming motility captured by dark-field microscopy, shown by the Z projection of track recordings, captured using a 10× objective after 72 hr of RNAi treatment. Wider spaced dots mark quicker moving Paramecium, dots tighter together mark slower moving or immobile Paramecium. C11orf70 silenced cells (bottom) show a severe motility phenotype compared to the control ND7 silenced cells (top).
(C) Analysis of the swimming velocity of C11orf70 and ND7 knockdown cells. The swimming velocity of C11orf70 knockdown cells is reduced by 32% of that of the control ND7 knockdown cells. At least 90 organisms evaluated per condition; p < 0.001, independent samples t test.
(D) At 72 hr, of RNAi the ciliary beat frequency of C11orf70 knockdown cells is reduced by about 20% compared to ND7 control knockdown cells. 7–10 paramecia evaluated per condition; p < 0.05, Mann-Whitney U test.
(E) Transmission electron micrographs of Paramecium cilia in cross-section showing normal 9+2 arrangement for the control ND7 knockdown (left) and absence of the outer (white arrows) and inner (gray arrows) dynein arms in the C11orf70 knockdown cells (right). Scale bar, 100 nm.
(F) Quantification of TEM dynein arms counts across >40 cross-sections per strain showed significant alteration of numbers of ODA and IDA in C11orf70 knockdown cells compared to ND7 control knockdown cells. p > 0.001, independent samples t test.
All error bars indicate SEM.
These results led us to examine the presence of cilia ODA and IDA by TEM after C11orf70 knockdown, since ODA are supposed to be involved in the beat frequency and IDA in the waveform.68, 69 TEM examination of cilia cross-sections showed a similar phenotype to that seen in individuals carrying C11orf70 mutations (Figure 4E), with a statistically significant reduction of both IDA and ODA numbers per cilia cross-section observed in the C11orf70 knockdown cells compared to the ND7 knockdown strain. The mean numbers of IDA and ODA were 7.8 ± 1.1 and 7.9 ± 1.3 in the control Paramecium cilia cross-sections compared to 4.9 ± 1.8 and 7.2 ± 2.2 in the C11orf70-silenced Paramecium cilia cross-sections, respectively (Figure 4F).
Overall, these phenotypic findings in Paramecium after C11orf70 ortholog silencing indicate the high level of evolutionary conservation of its gene function across ciliated species, with cilia defects that are consistent with the diagnostic findings in individuals affected with PCD that carry mutations in C11orf70.
C11orf70 Has a Conserved Intracellular Distribution Pattern Similar to IFT-Associated Proteins
To gain insight into the mechanism by which C11orf70 might behave like an IFT-associated protein and potentially interact with the ciliary IFT system to affect inner and outer dynein arm assembly, we next studied its behavior in Paramecium after ablation of IFT and also during cilia re-growth. We expressed GFP-tagged C11orf70 in cells, generating several transformant clones showing a wild-type growth phenotype that expressed various levels of C11orf70-GFP. All of them showed the same results, with C11orf70-GFP protein found mostly in the cytoplasm with a small amount found in cilia (Figure 5A, left), in agreement with the Chlamydomonas and Paramecium cell fractionation experiments in Figures 3C and 3E. We then applied RNAi to the C11orf70-GFP transformed cells, to knock down the function of IFT139 (TTC21B), one member of the IFT-A complex (involved in IFT-associated retrograde transport out of cilia). Depletion of IFT139 led to a strikingly aberrant accumulation of C11orf70 protein at the tips of the cilia, suggesting that it behaves like an IFT protein with its ciliary transport governed by IFT-A trafficking (Figure 5A, right).
Figure 5.
C11orf70-GFP Accumulates at the Ciliary Tip during Reciliation and Its Flagellar Abundance Is IFT Dependent
(A) Left: paramecia expressing the Paramecium C11orf70-GFP show a very high level of cytoplasmic GFP staining with protein also present in the cilia. The cell body was overexposed in order to allow the detection of cilia. Right: RNAi knockdown of IFT139/TTC21B disrupting intraflagellar transport, causes the overexpressed C11orf70 protein to accumulate aberrantly in the cilia toward the tip regions, as shown also in higher magnification in the white boxes. Scale bars, 20 μm.
(B) Paramecium expressing either GFP alone (left), C11orf70-GFP (middle), or IFT46-GFP (right) were subject to deciliation after which the cilia were allowed to regrow for 15 min, in order to study the localization of the tagged proteins during reciliation. Direct GFP fluorescence (green, top) and monoclonal antibody staining against polyglutamylated tubulin (red, middle) are shown along with the merge (bottom). Paramecia expressing GFP are used as a negative control since the GFP protein enters the cilia passively (<27 kDA). The GFP staining is found distributed homogeneously along the entire length of the cilia. In contrast, IFT46-GFP accumulates at the ciliary tips during ciliary regrowth, as expected for an IFT-B family member. C11orf70-GFP is found at much higher levels in the cytoplasm than IFT46-GFP and is also present in the cilia. Upon ciliary regrowth, C11orf70- GFP behaves in a similar fashion to IFT46 GFP in cilia with an accumulation of both proteins observed at the ciliary tips. Scale bars, 10 μm. White boxes show higher magnification.
(C) Coomassie-stained gels (above) and western blots (below) show that the flagellar fraction abundance of Chlamydomonas FBB5 is similar in strains carrying mutations in components of the outer or inner dynein arms (oda6, ida1), the ODA-DC (oda3) and late-stage ODA assembly factors (oda8, oda10). However, its abundance is reduced by 75% in the absence of ODA16, an IFT-associated dynein transport factor (left). Blots of IFT46 used as a marker of flagellar IFT, shows that IFT is reduced by 50% in oda16. Dotted line indicates removal of an intervening lane on the blot.
(D) Gels (top) and blots (bottom) of whole cells and flagella from wild-type and fla10ts strains of Chlamydomonas. Similar levels of FBB5-HA are expressed in both strains (left panels, whole cells). Flagella from fla10ts cells incubated at the restrictive temperature lack IFT46 and have also lost FBB5-HA, whereas elevated temperature has no effect on wild-type flagella (right panels).
(E) Gels (top) and blots (bottom) of whole cells and flagella from FBB5-HA-expressing wild-type Chlamydomonas and an ift46 mutant strain that also express an N-terminally truncated IFT46 transgene. Similar levels of IFT46, IFT46ΔN, ODA-IC2, ODA16, and FBB5-HA are expressed in whole cells of both strains (left). The truncated IFT46 supports IFT-dependent flagellar assembly, but the flagella show reduced levels of ODAs (IC2) and ODA16. FBB5-HA levels, however, are not affected (right).
(F) Gel (top) and blots (bottom) of non-regenerating flagella, and regenerating flagella isolated 60 min after deflagellation, show no change in abundance of IFT proteins, but increased abundance of FBB5::HA in the growing flagella.
We next compared C11orf70-GFP behavior to GFP alone, which enters cilia by diffusion, and IFT46-GFP, an IFT-B anterograde transport protein involved in IFT-associated anterograde transport of ODA into cilia, using transformant clones expressing these proteins individually. We assessed their localization compared to C11orf70, after deciliation followed by cilia re-growth of the transformed Paramecium cells. After 15 min of cilia re-growth, when IFT transport is particularly active, we observed that IFT-46 GFP accumulates at the ciliary tips (Figure 5B, right panels) compared to a uniform distribution of fluorescence in the cilia of cells expressing GFP alone (Figure 5B, left panels). Similarly to IFT46-GFP, the C11orf70-GFP protein showed a comparable cilia tip accumulation during ciliogenesis (Figure 5B, middle panels). These findings suggest that C11orf70 is actively transported in cilia and recycled back to the cell, in a fashion consistent with involvement in IFT-based transport.
Flagellar C11orf70 Abundance Is IFT Dependent
We next took advantage of Chlamydomonas genetics to further investigate the functional role of C11orf70 in IFT-related cilia motility by expressing the HA-tagged FBB5 ortholog in various axonemal assembly mutant strains. We found that flagellar FBB5::HA levels were not affected by mutations in outer or inner dynein arm subunits (oda6, ida1 mutant strains),81, 82 the ODA-DC (oda3),83 or late-stage cytoplasmic assembly factors (oda8, oda10).38, 83 However, FBB5::HA levels were found to be decreased by a loss of IFT-associated dynein transport factor ODA16 (oda16)40, 41, 84 (Figure 5C), with confirmation in the oda16 strain that FBB5::HA was expressed at equivalent levels to the other tested strains (Figure S5). Since this suggests a role for FBB5 in IFT-dependent dynein transport, we further tested the role of IFT in transport of FBB5 to the flagellar matrix by expressing FBB5::HA in the temperature-sensitive IFT kinesin mutant deficient in anterograde IFT, fla10ts.67 After shifting to the restrictive temperature (32°C) for 1.5 hr, the fla10ts flagella were found to be depleted of IFT complexes (monitored by the abundance of IFT46) and of FBB5::HA (Figure 5D). Since ODA transport factor ODA16 (human ortholog DAW1) interacts with IFT4640 and specifically depends on an interaction with the N-terminal domain of IFT46 for ODA transport,68, 84 we tested the effects of deleting the IFT46 N terminus on flagellar abundance of FBB5. Flagella of an ift46 mutant strain that also expressed an N-terminally truncated IFT46 protein68 had reduced levels of ODA16 and greatly reduced assembly of ODAs (as monitored by the abundance of ODA intermediate chain IC2) but showed no reduction in FBB5::HA (Figure 5E). As an additional test of the role of FBB5 in assembly-related processes, we tested the relative abundance of FBB5::HA in non-regenerating full-length flagella and in regenerating half-length flagella. Although the abundance of IFT139 (IFT-A subunit) and IFT81 (IFT-B subunit) did not change, the abundance of FBB5::HA increased in the assembling flagella, as expected of a factor associated with ciliary precursors during transport (Figure 5F).
Discussion
Here we report mutations in a previously uncharacterized gene, C11orf70, in individuals with PCD and situs inversus, demonstrating the potential power of using a targeted NGS approach for discovery of PCD-causing mutations in previously unrecognized disease genes. C11orf70 mutations give rise to immotile respiratory cilia that have a normal 9+2 ultrastructure with a combined lack of both inner and outer dynein arms.
Cilia axonemal structure is highly conserved across species and various model organisms have been used in the past to characterize candidate genes for PCD. Phylogenetic analysis showed that C11orf70 is a highly conserved protein across anciently diverging species, being present today only in organisms with motile cilia/flagella. It is lost in organisms that do not use IFT assembly of their cilia/flagella. It has a specific distribution across different species, following a similar pattern to that of IFT-associated dynein assembly factors like ODA16 (WDR69).40, 41, 68 Together these findings support the importance of C11orf70 in the cytoplasmic assembly, maturation, and IFT-B-based transport into cilia of dynein arm motors that are required for cilia motility in multiciliated cells.
Transcriptional profiling of C11orf70 shows that it is enriched in tissues bearing motile cilia,73, 74 although it was not identified in proteomic profiling of human respiratory cilia.85 Microarray-based gene expression profiling has shown that C11orf70 expression is upregulated during mucociliary differentiation.86 This is consistent with our finding that the C11orf70 expression profile follows a similar pattern during ciliogenesis to that of other genes that cause PCD when mutated, with a peak around the time of cilia emergence that reaches a plateau afterward. Interestingly, the Chlamydomonas C11orf70 ortholog, FBB5, has been identified as among the flagellar-basal body group of genes,77, 78 upregulated following deflagellation,79, 80 but it is not present in any ciliary proteomes. The Paramecium C11orf70 ortholog, GSPATG00011350001, was also not detected within the cilium proteome87 or during cilia regrowth after deciliation of Paramecium.88
To study the role of C11orf70 in cilia motility, we knocked down its ortholog in Paramecium. We first confirmed the efficiency of silencing at the RNA level and then studied the phenotypic consequences of gene silencing on cilia motility and ultrastructure. We found that C11orf70 RNAi knockdown led to a significant reduction in swimming velocity as a secondary effect of a decrease in cilia beating frequency. Analysis of the ciliary ultrastructural phenotype demonstrated the highly conserved, ancient function of C11orf70 in dynein arm assembly since C11orf70-silenced Paramecium cells mimic the combined outer and inner dynein arm loss of affected individuals with PCD carrying C11orf70 mutations, consistent with their immotile respiratory cilia. This provides evidence that C11orf70 plays a role in the dynein arm assembly/transport process. This work highlights the utility of Paramecium for studies of cilia motility and its emergence as a suitable organism to model the cell biology of disease mechanisms underlying PCD.
In Chlamydomonas we found that the distribution of the C11orf70 ortholog FBB5 between the cytoplasm and cilia/flagella was similar to other IFT-associated dynein assembly proteins such as ODA1640 and ODA8.38 A similar distribution was seen for Paramecium C11orf70. Our expression studies in both Chlamydomonas and Paramecium showed that C11orf70 is a soluble, mainly cytoplasmic protein, with a small amount also present in the matrix fraction of cilia/flagella. The protein was around 40× more abundant in the cytoplasm than the cilia/flagella in both organisms. This is similar to ODA16, which is concentrated around the basal bodies with ∼2% found in flagella.40, 41 The anterograde IFT partner of ODA16, IFT-B family member IFT46, shows a similar pattern, also concentrated around basal bodies with ∼5% present in flagella. ODA16 and IFT46 are known to interact together to mediate the transport of outer dynein arms into the cilia.68 Among the other PCD-associated dynein assembly factors, the distribution of C11orf70 is reminiscent of two, ZMYND10 and C21orf59, that are considered predominantly cytoplasmic with a small component potentially also detectable within cilia.33, 35
The flagellar localization of Chlamydomonas FBB5-HA was unaffected by most mutations that disrupt ODA assembly but was reduced by loss of the IFT46-associated transport factor ODA16, although we found that its abundance in the axoneme was not dependent on the N terminus of IFT46 (we note that this effect could result from a general reduced abundance of IFT in oda16 flagella rather than being a direct consequence of ODA16 mutation). Flagellar localization was dependent on IFT, and FBB5::HA abundance increased during flagellar growth, a time when high levels of precursor transport are needed.89 Paramecium work supported these findings, since after deciliation followed by cilia regrowth C11orf70 accumulates in the cilia tips in Paramecium in the same way as IFT46. IFT46 is an IFT-B family component that governs anterograde (cilia base to tip) transport in the cilium and is very active during ciliation. IFT46 is released at the tip of the cilium where it accumulates during active IFT and C11orf70 may behave similarly. We further found in Paramecium that depletion of an IFT-A retrograde (cilia tip to base) transport component, IFT139/TTC21B, affected the ciliary localization of C11orf70, causing it to accumulate abnormally at the ciliary tips. This is to be expected if the protein, like IFT components, is recycled out of cilia by the IFT-A transport system.
Overall, our data implicate C11orf70 as playing a role within the chaperone-mediated sequential protein folding and trafficking pathways that are required for the cytoplasmic preassembly of dynein arm motors and their subsequent import into the motile ciliary axoneme.18, 23, 31, 82, 90 As is the case for other putative dynein assembly factors acting to correctly assemble the dynein motor complexes responsible for cilia motility, it is not yet clear how a protein can act as an adaptor for both ODA and IDA cilia assembly and whether it would need to recognize a component of both inner and outer dynein arms. Since we find that C11orf70 is transported into cilia, interacts with IFT-A, and behaves similarly to an IFT component, we propose that it could provide chaperone-related functions within the cytoplasm, continuing until the dynein arm complex is anchored in cilia, then being recycled back to the cytoplasm via retrograde IFT (IFT-A).
Together, these data identify mutations in C11orf70 as a cause of PCD with combined outer and inner dynein loss, cilia immotility, and situs inversus. This has important clinical implications for improved understanding of the genetic basis of the disease and counselling of affected families. The combined evidence from human and distantly related unicellular organisms support a role for C11rof70 as an additional dynein assembly/transport factor, its abundance in the cilium dependent upon IFT.
Acknowledgments
We are very grateful to the families who participated in this study and thank the UK PCD Family Support Group for their continued support. We thank Ms. Dani Lee and Prof. Chris O’Callaghan (UCL Great Ormond Street Institute of Child Health) for help with assessment of Paramecium motility and Dr. Dale Moulding (GOS-ICH Confocal Microscopy Core Facility) for experimental assistance. Funding for this study was provided by Action Medical Research (GN2101; H.M.M.) and Great Ormond Street Children’s Charity grant (V4515; H.M.M.) and Leadership awards (V1299, V2217; H.M.M.). M.R.F. is supported by the British Council Newton-Mosharafa Fund and the Ministry of Higher Education in Egypt. We acknowledge support from the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London (Doctoral Trainee Support Award; M.R.F.). Work by A.S. is independent research funded by a postdoctoral research fellowship from the National Institute of Health Research and Health Education England. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute of Health Research, or the Department of Health. We would like to thank C. Mathon for generating plasmid constructs, L. Shi for the IFT46 construct, J. Bonaventure for his help in RNA isolation, and C. Janke for his generous gift of PolyE antibodies. The authors acknowledge the contribution of the BEAT-PCD COST Action. The authors participate in the COST Action BEAT-PCD: Better Evidence to Advance Therapeutic options for PCD network (BM1407) and this work was supported by two BM1407 COST Action STSM Grants awarded to M.R.F. and P.L.B. The authors alone are responsible for the content and writing of the paper.
Published: May 3, 2018
Footnotes
Supplemental Data include five figures, three tables, and seven movies and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.024.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
BLAST/BLOSUM62, https://blast.ncbi.nlm.nih.gov/Blast.cgi
Chlamydomonas Resource Center, http://chlamycollection.org/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
Human Splicing Finder, http://www.umd.be/HSF3/
MAPP, http://mendel.stanford.edu/sidowlab/downloads/MAPP/index.html
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
ParameciumDB, http://paramecium-archive.i2bc.paris-saclay.fr/
PhastCons, http://compgen.cshl.edu/phast/
Phytozome, https://phytozome.jgi.doe.gov/pz/portal.html
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Primer-BLAST, https://www.ncbi.nlm.nih.gov/tools/primer-blast/
Supplemental Data
References
- 1.Mitchison H.M., Valente E.M. Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 2.Satir P., Christensen S.T. Structure and function of mammalian cilia. Histochem. Cell Biol. 2008;129:687–693. doi: 10.1007/s00418-008-0416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milla C.E. The evolving spectrum of ciliopathies and respiratory disease. Curr. Opin. Pediatr. 2016;28:339–347. doi: 10.1097/MOP.0000000000000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennekamp P., Menchen T., Dworniczak B., Hamada H. Situs inversus and ciliary abnormalities: 20 years later, what is the connection? Cilia. 2015;4:1. doi: 10.1186/s13630-014-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fliegauf M., Olbrich H., Horvath J., Wildhaber J.H., Zariwala M.A., Kennedy M., Knowles M.R., Omran H. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas J.S., Chetcuti P., Copeland F., Hogg C., Kenny T., Moya E., O’Callaghan C., Walker W.T. Overcoming challenges in the management of primary ciliary dyskinesia: the UK model. Paediatr. Respir. Rev. 2014;15:142–145. doi: 10.1016/j.prrv.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Hosie P.H., Fitzgerald D.A., Jaffe A., Birman C.S., Rutland J., Morgan L.C. Presentation of primary ciliary dyskinesia in children: 30 years’ experience. J. Paediatr. Child Health. 2015;51:722–726. doi: 10.1111/jpc.12791. [DOI] [PubMed] [Google Scholar]
- 8.Noone P.G., Leigh M.W., Sannuti A., Minnix S.L., Carson J.L., Hazucha M., Zariwala M.A., Knowles M.R. Primary ciliary dyskinesia: diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med. 2004;169:459–467. doi: 10.1164/rccm.200303-365OC. [DOI] [PubMed] [Google Scholar]
- 9.Hornef N., Olbrich H., Horvath J., Zariwala M.A., Fliegauf M., Loges N.T., Wildhaber J., Noone P.G., Kennedy M., Antonarakis S.E. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am. J. Respir. Crit. Care Med. 2006;174:120–126. doi: 10.1164/rccm.200601-084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onoufriadis A., Shoemark A., Munye M.M., James C.T., Schmidts M., Patel M., Rosser E.M., Bacchelli C., Beales P.L., Scambler P.J., UK10K Combined exome and whole-genome sequencing identifies mutations in ARMC4 as a cause of primary ciliary dyskinesia with defects in the outer dynein arm. J. Med. Genet. 2014;51:61–67. doi: 10.1136/jmedgenet-2013-101938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panizzi J.R., Becker-Heck A., Castleman V.H., Al-Mutairi D.A., Liu Y., Loges N.T., Pathak N., Austin-Tse C., Sheridan E., Schmidts M. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat. Genet. 2012;44:714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King S.M., Patel-King R.S. The oligomeric outer dynein arm assembly factor CCDC103 is tightly integrated within the ciliary axoneme and exhibits periodic binding to microtubules. J. Biol. Chem. 2015;290:7388–7401. doi: 10.1074/jbc.M114.616425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjeij R., Onoufriadis A., Watson C.M., Slagle C.E., Klena N.T., Dougherty G.W., Kurkowiak M., Loges N.T., Diggle C.P., Morante N.F., UK10K Consortium CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet. 2014;95:257–274. doi: 10.1016/j.ajhg.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onoufriadis A., Paff T., Antony D., Shoemark A., Micha D., Kuyt B., Schmidts M., Petridi S., Dankert-Roelse J.E., Haarman E.G., UK10K Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles M.R., Leigh M.W., Ostrowski L.E., Huang L., Carson J.L., Hazucha M.J., Yin W., Berg J.S., Davis S.D., Dell S.D., Genetic Disorders of Mucociliary Clearance Consortium Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;92:99–106. doi: 10.1016/j.ajhg.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horani A., Ferkol T.W., Dutcher S.K., Brody S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai P.B., Dean A.B., Mitchell D.R. Cytoplasmic preassembly and trafficking of axonemal dyneins. In: King S., editor. Dyneins. Second Edition. Academic Press; 2018. pp. 140–161. [Google Scholar]
- 19.Duquesnoy P., Escudier E., Vincensini L., Freshour J., Bridoux A.M., Coste A., Deschildre A., de Blic J., Legendre M., Montantin G. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan-Brown J., Schottenfeld J., Okabe N., Hostetter C.L., Serluca F.C., Thiberge S.Y., Burdine R.D. Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev. Biol. 2008;314:261–275. doi: 10.1016/j.ydbio.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loges N.T., Olbrich H., Becker-Heck A., Häffner K., Heer A., Reinhard C., Schmidts M., Kispert A., Zariwala M.A., Leigh M.W. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am. J. Hum. Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N.T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchison H.M., Schmidts M., Loges N.T., Freshour J., Dritsoula A., Hirst R.A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 2012;44:381–389, S1–S2. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrasekar G., Vesterlund L., Hultenby K., Tapia-Páez I., Kere J. The zebrafish orthologue of the dyslexia candidate gene DYX1C1 is essential for cilia growth and function. PLoS ONE. 2013;8:e63123. doi: 10.1371/journal.pone.0063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarkar A., Loges N.T., Slagle C.E., Francis R., Dougherty G.W., Tamayo J.V., Shook B., Cantino M., Schwartz D., Jahnke C., UK10K DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat. Genet. 2013;45:995–1003. doi: 10.1038/ng.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diggle C.P., Moore D.J., Mali G., zur Lage P., Ait-Lounis A., Schmidts M., Shoemark A., Garcia Munoz A., Halachev M.R., Gautier P. HEATR2 plays a conserved role in assembly of the ciliary motile apparatus. PLoS Genet. 2014;10:e1004577. doi: 10.1371/journal.pgen.1004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horani A., Druley T.E., Zariwala M.A., Patel A.C., Levinson B.T., Van Arendonk L.G., Thornton K.C., Giacalone J.C., Albee A.J., Wilson K.S. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horani A., Ferkol T.W., Shoseyov D., Wasserman M.G., Oren Y.S., Kerem B., Amirav I., Cohen-Cymberknoh M., Dutcher S.K., Brody S.L. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS ONE. 2013;8:e59436. doi: 10.1371/journal.pone.0059436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inaba Y., Shinohara K., Botilde Y., Nabeshima R., Takaoka K., Ajima R., Lamri L., Takeda H., Saga Y., Nakamura T., Hamada H. Transport of the outer dynein arm complex to cilia requires a cytoplasmic protein Lrrc6. Genes Cells. 2016;21:728–739. doi: 10.1111/gtc.12380. [DOI] [PubMed] [Google Scholar]
- 30.Kott E., Duquesnoy P., Copin B., Legendre M., Dastot-Le Moal F., Montantin G., Jeanson L., Tamalet A., Papon J.F., Siffroi J.P. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2012;91:958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olcese C., Patel M.P., Shoemark A., Kiviluoto S., Legendre M., Williams H.J., Vaughan C.K., Hayward J., Goldenberg A., Emes R.D., UK10K Rare Group X-linked primary ciliary dyskinesia due to mutations in the cytoplasmic axonemal dynein assembly factor PIH1D3. Nat. Commun. 2017;8:14279. doi: 10.1038/ncomms14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paff T., Loges N.T., Aprea I., Wu K., Bakey Z., Haarman E.G., Daniels J.M.A., Sistermans E.A., Bogunovic N., Dougherty G.W. Mutations in PIH1D3 cause X-linked primary ciliary dyskinesia with outer and inner dynein arm defects. Am. J. Hum. Genet. 2017;100:160–168. doi: 10.1016/j.ajhg.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore D.J., Onoufriadis A., Shoemark A., Simpson M.A., zur Lage P.I., de Castro S.C., Bartoloni L., Gallone G., Petridi S., Woollard W.J. Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:346–356. doi: 10.1016/j.ajhg.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zariwala M.A., Gee H.Y., Kurkowiak M., Al-Mutairi D.A., Leigh M.W., Hurd T.W., Hjeij R., Dell S.D., Chaki M., Dougherty G.W. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am. J. Hum. Genet. 2013;93:336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Austin-Tse C., Halbritter J., Zariwala M.A., Gilberti R.M., Gee H.Y., Hellman N., Pathak N., Liu Y., Panizzi J.R., Patel-King R.S. Zebrafish ciliopathy screen plus human mutational analysis identifies C21orf59 and CCDC65 defects as causing primary ciliary dyskinesia. Am. J. Hum. Genet. 2013;93:672–686. doi: 10.1016/j.ajhg.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaffe K.M., Grimes D.T., Schottenfeld-Roames J., Werner M.E., Ku T.S., Kim S.K., Pelliccia J.L., Morante N.F., Mitchell B.J., Burdine R.D. c21orf59/kurly controls both cilia motility and polarization. Cell Rep. 2016;14:1841–1849. doi: 10.1016/j.celrep.2016.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean A.B., Mitchell D.R. Late steps in cytoplasmic maturation of assembly-competent axonemal outer arm dynein in Chlamydomonas require interaction of ODA5 and ODA10 in a complex. Mol. Biol. Cell. 2015;26:3596–3605. doi: 10.1091/mbc.E15-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai P.B., Freshour J.R., Mitchell D.R. Chlamydomonas axonemal dynein assembly locus ODA8 encodes a conserved flagellar protein needed for cytoplasmic maturation of outer dynein arm complexes. Cytoskeleton. 2015;72:16–28. doi: 10.1002/cm.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Klena N.T., Gabriel G.C., Liu X., Kim A.J., Lemke K., Chen Y., Chatterjee B., Devine W., Damerla R.R. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520–524. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed N.T., Gao C., Lucker B.F., Cole D.G., Mitchell D.R. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J. Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao C., Wang G., Amack J.D., Mitchell D.R. Oda16/Wdr69 is essential for axonemal dynein assembly and ciliary motility during zebrafish embryogenesis. Dev. Dyn. 2010;239:2190–2197. doi: 10.1002/dvdy.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucas J.S., Barbato A., Collins S.A., Goutaki M., Behan L., Caudri D., Dell S., Eber E., Escudier E., Hirst R.A. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017;49:49. doi: 10.1183/13993003.01090-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trump N., McTague A., Brittain H., Papandreou A., Meyer E., Ngoh A., Palmer R., Morrogh D., Boustred C., Hurst J.A. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J. Med. Genet. 2016;53:310–317. doi: 10.1136/jmedgenet-2015-103263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrison, E., and Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907.
- 46.Munye M.M., Shoemark A., Hirst R.A., Delhove J.M., Sharp T.V., McKay T.R., O’Callaghan C., Baines D.L., Howe S.J., Hart S.L. BMI-1 extends proliferative potential of human bronchial epithelial cells while retaining their mucociliary differentiation capacity. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017;312:L258–L267. doi: 10.1152/ajplung.00471.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirst R.A., Rutman A., Williams G., O’Callaghan C. Ciliated air-liquid cultures as an aid to diagnostic testing of primary ciliary dyskinesia. Chest. 2010;138:1441–1447. doi: 10.1378/chest.10-0175. [DOI] [PubMed] [Google Scholar]
- 48.Shoemark A., Dixon M., Corrin B., Dewar A. Twenty-year review of quantitative transmission electron microscopy for the diagnosis of primary ciliary dyskinesia. J. Clin. Pathol. 2012;65:267–271. doi: 10.1136/jclinpath-2011-200415. [DOI] [PubMed] [Google Scholar]
- 49.Shoemark A., Frost E., Dixon M., Ollosson S., Kilpin K., Patel M., Scully J., Rogers A.V., Mitchison H.M., Bush A., Hogg C. Accuracy of immunofluorescence in the diagnosis of primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2017;196:94–101. doi: 10.1164/rccm.201607-1351OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chilvers M.A., Rutman A., O’Callaghan C. Functional analysis of cilia and ciliated epithelial ultrastructure in healthy children and young adults. Thorax. 2003;58:333–338. doi: 10.1136/thorax.58.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 52.Beisson J., Bétermier M., Bré M.H., Cohen J., Duharcourt S., Duret L., Kung C., Malinsky S., Meyer E., Preer J.R., Jr., Sperling L. Silencing specific Paramecium tetraurelia genes by feeding double-stranded RNA. Cold Spring Harb. Protoc. 2010;2010:t5363. doi: 10.1101/pdb.prot5363. [DOI] [PubMed] [Google Scholar]
- 53.Skouri F., Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz F., Vayssié L., Klotz C., Sperling L., Madeddu L. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell. 1998;9:931–943. doi: 10.1091/mbc.9.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beisson J., Bétermier M., Bré M.H., Cohen J., Duharcourt S., Duret L., Kung C., Malinsky S., Meyer E., Preer J.R., Jr., Sperling L. Mass culture of Paramecium tetraurelia. Cold Spring Harb. Protoc. 2010;2010:t5362. doi: 10.1101/pdb.prot5362. [DOI] [PubMed] [Google Scholar]
- 56.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bell W.E., Hallworth R., Wyatt T.A., Sisson J.H. Use of a novel cell adhesion method and digital measurement to show stimulus-dependent variation in somatic and oral ciliary beat frequency in Paramecium. J. Eukaryot. Microbiol. 2015;62:144–148. doi: 10.1111/jeu.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith C.M., Djakow J., Free R.C., Djakow P., Lonnen R., Williams G., Pohunek P., Hirst R.A., Easton A.J., Andrew P.W., O’Callaghan C. ciliaFA: a research tool for automated, high-throughput measurement of ciliary beat frequency using freely available software. Cilia. 2012;1:14. doi: 10.1186/2046-2530-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bengueddach H., Lemullois M., Aubusson-Fleury A., Koll F. Basal body positioning and anchoring in the multiciliated cell Paramecium tetraurelia: roles of OFD1 and VFL3. Cilia. 2017;6:6. doi: 10.1186/s13630-017-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aubusson-Fleury A., Cohen J., Lemullois M. Ciliary heterogeneity within a single cell: the Paramecium model. Methods Cell Biol. 2015;127:457–485. doi: 10.1016/bs.mcb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 61.van Dam T.J., Townsend M.J., Turk M., Schlessinger A., Sali A., Field M.C., Huynen M.A. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. USA. 2013;110:6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hauser K., Haynes W.J., Kung C., Plattner H., Kissmehl R. Expression of the green fluorescent protein in Paramecium tetraurelia. Eur. J. Cell Biol. 2000;79:144–149. doi: 10.1078/S0171-9335(04)70016-3. [DOI] [PubMed] [Google Scholar]
- 63.Adoutte A., Ramanathan R., Lewis R.M., Dute R.R., Ling K.Y., Kung C., Nelson D.L. Biochemical studies of the excitable membrane of Paramecium tetraurelia. III. Proteins of cilia and ciliary membranes. J. Cell Biol. 1980;84:717–738. doi: 10.1083/jcb.84.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang Y., Mitchell D.R. An intronic enhancer is required for deflagellation-induced transcriptional regulation of a Chlamydomonas reinhardtii dynein gene. Mol. Biol. Cell. 1998;9:3085–3094. doi: 10.1091/mbc.9.11.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitchell B.F., Pedersen L.B., Feely M., Rosenbaum J.L., Mitchell D.R. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell. 2005;16:4509–4518. doi: 10.1091/mbc.E05-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kozminski K.G., Beech P.L., Rosenbaum J.L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou Y., Witman G.B. The N-terminus of IFT46 mediates intraflagellar transport of outer arm dynein and its cargo-adaptor ODA16. Mol. Biol. Cell. 2017;28:2420–2433. doi: 10.1091/mbc.E17-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitchell D.R., Rosenbaum J.L. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J. Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leigh M.W., Hazucha M.J., Chawla K.K., Baker B.R., Shapiro A.J., Brown D.E., Lavange L.M., Horton B.J., Qaqish B., Carson J.L. Standardizing nasal nitric oxide measurement as a test for primary ciliary dyskinesia. Ann. Am. Thorac. Soc. 2013;10:574–581. doi: 10.1513/AnnalsATS.201305-110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Castleman V.H., Romio L., Chodhari R., Hirst R.A., de Castro S.C., Parker K.A., Ybot-Gonzalez P., Emes R.D., Wilson S.W., Wallis C. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olbrich H., Cremers C., Loges N.T., Werner C., Nielsen K.G., Marthin J.K., Philipsen M., Wallmeier J., Pennekamp P., Menchen T. Loss-of-function GAS8 mutations cause primary ciliary dyskinesia and disrupt the nexin-dynein regulatory complex. Am. J. Hum. Genet. 2015;97:546–554. doi: 10.1016/j.ajhg.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Geremek M., Bruinenberg M., Ziętkiewicz E., Pogorzelski A., Witt M., Wijmenga C. Gene expression studies in cells from primary ciliary dyskinesia patients identify 208 potential ciliary genes. Hum. Genet. 2011;129:283–293. doi: 10.1007/s00439-010-0922-4. [DOI] [PubMed] [Google Scholar]
- 75.Lucas J.S., Burgess A., Mitchison H.M., Moya E., Williamson M., Hogg C., National PCD Service, UK Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 2014;99:850–856. doi: 10.1136/archdischild-2013-304831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galvani A., Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- 77.Li J.B., Gerdes J.M., Haycraft C.J., Fan Y., Teslovich T.M., May-Simera H., Li H., Blacque O.E., Li L., Leitch C.C. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 78.Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stolc V., Samanta M.P., Tongprasit W., Marshall W.F. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albee A.J., Kwan A.L., Lin H., Granas D., Stormo G.D., Dutcher S.K. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 (Bethesda) 2013;3:979–991. doi: 10.1534/g3.113.006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kamiya R., Kurimoto E., Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fowkes M.E., Mitchell D.R. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 1998;9:2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koutoulis A., Pazour G.J., Wilkerson C.G., Inaba K., Sheng H., Takada S., Witman G.B. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taschner M., Mourão A., Awasthi M., Basquin J., Lorentzen E. Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46. J. Biol. Chem. 2017;292:7462–7473. doi: 10.1074/jbc.M117.780155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blackburn K., Bustamante-Marin X., Yin W., Goshe M.B., Ostrowski L.E. Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J. Proteome Res. 2017;16:1579–1592. doi: 10.1021/acs.jproteome.6b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ross A.J., Dailey L.A., Brighton L.E., Devlin R.B. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007;37:169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- 87.Arnaiz O., Malinowska A., Klotz C., Sperling L., Dadlez M., Koll F., Cohen J. Cildb: a knowledgebase for centrosomes and cilia. Database (Oxford) 2009;2009:bap022. doi: 10.1093/database/bap022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arnaiz O., Goût J.F., Bétermier M., Bouhouche K., Cohen J., Duret L., Kapusta A., Meyer E., Sperling L. Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC Genomics. 2010;11:547. doi: 10.1186/1471-2164-11-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wren K.N., Craft J.M., Tritschler D., Schauer A., Patel D.K., Smith E.F., Porter M.E., Kner P., Lechtreck K.F. A differential cargo-loading model of ciliary length regulation by IFT. Curr. Biol. 2013;23:2463–2471. doi: 10.1016/j.cub.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.King S.M. Axonemal dynein arms. Cold Spring Harb. Perspect. Biol. 2016;8:8. doi: 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.