Abstract
Genome-wide association studies (GWASs) have reproducibly associated variants within intergenic regions of 1p36.12 locus with osteoporosis, but the functional roles underlying these noncoding variants are unknown. Through an integrative functional genomic and epigenomic analyses, we prioritized rs6426749 as a potential causal SNP for osteoporosis at 1p36.12. Dual-luciferase assay and CRISPR/Cas9 experiments demonstrate that rs6426749 acts as a distal allele-specific enhancer regulating expression of a lncRNA (LINC00339) (∼360 kb) via long-range chromatin loop formation and that this loop is mediated by CTCF occupied near rs6426749 and LINC00339 promoter region. Specifically, rs6426749-G allele can bind transcription factor TFAP2A, which efficiently elevates the enhancer activity and increases LINC00339 expression. Downregulation of LINC00339 significantly increases the expression of CDC42 in osteoblast cells, which is a pivotal regulator involved in bone metabolism. Our study provides mechanistic insight into how a noncoding SNP affects osteoporosis by long-range interaction, a finding that could indicate promising therapeutic targets for osteoporosis.
Keywords: osteoporosis, 1p36.12, eQTL, long-range, enhancer, chromatin interaction, rs6426749, LINC00339, loop, TFAP2A
Introduction
Genome-wide association studies (GWASs) have successfully identified numerous genetic variants for human complex diseases or traits. However, many of the identified variants are located in the noncoding regions of human genome.1 It is particularly challenging to identify the precise gene targets for these noncoding variants and elucidate their functional mechanisms involved in disease pathophysiology. The traditional annotation of GWAS hits usually focuses on the nearest or most biologically plausible gene candidate, which may not be the true target gene and therefore might result in expensive and time-consuming efforts to explore the function of non-causal genes. Strikingly, recent studies have found that some of the noncoding GWAS SNPs are within potential regulatory or functional elements to regulate expressions of distal genes by long-range genome interactions.2, 3 For example, Gupta et al.3 prioritized a functional variant (rs9349379) at 6p24 associated with five vascular diseases. They further validated that rs9349379 specifically regulates expression of EDN1 (MIM: 131240), a long-range target gene (>600 kb) with known function on the vasculature.3 These studies provide us promising insights into deciphering the relationship between noncoding SNPs and diseases. Addressing these knowledge gaps is critical to help translate GWAS findings into clinically useful information.
Osteoporosis (MIM: 166710) is one of the most common metabolic skeletal diseases characterized by low bone mass, poor bone quality, and an increased predisposition to fracture.4 The clinical diagnosis and assessment of osteoporosis is mainly based on bone mineral density (BMD),5 which has a high heritability of 0.6–0.8.6 Previous GWASs have successfully identified more than 60 genetic loci for BMD and osteoporosis.7, 8 Some of these loci are mapped to genes with important function on bone, such as RANK-RANKL-OPG (RANK [MIM: 603499], RANKL [MIM: 602642], OPG [MIM: 602643]),9, 10 ESR1 (MIM: 133430),10 and LRP5 (MIM: 603506).11 However, some loci are localized to genes not known to have a role in bone biology. For example, 1p36.12 was identified by an initial large-scale BMD GWAS10 and further replicated by multiple GWAS meta-analyses.9, 12, 13, 14, 15, 16 The reported SNPs within 1p36.12 are located in the noncoding region, indicating that they may reside within putative regulatory elements. The closest gene is ZBTB40 (MIM: 612106), which is more than 60 kb away and has unknown function or connection with bone biology. Interestingly, the genes upstream of ZBTB40—WNT4 (392 kb [MIM: 603490]) and CDC42 (399 kb [MIM: 116952])—both have potential connection with bone or osteoporosis. WNT4 could attenuate bone loss in osteoporosis and skeletal aging mouse models by inhibiting nuclear factor-κB (NF-κB) via noncanonical Wnt signaling.17 CDC42 is an effector molecule involved in bone metabolism and skeletal development.18, 19 Therefore, it is extremely interesting to find out the true target gene and investigate how the susceptibility SNPs at 1p36.12 affect disease risk.
In this study, we hypothesized that SNPs at 1p36.12 might act as distal regulatory element to influence the expression of target genes and modulate bone metabolism via long-range interaction. To achieve this aim, we implemented a series of computational analyses using data from sources including Hi-C (high-throughput 3C), expression quantitative trait locus (eQTL), and epigenomic annotation, then followed by various functional validation experiments, with the flowchart shown in Figure 1. We demonstrate that an intergenic SNP (rs6426749) at 1p36.12 acts as strong long-range enhancer to regulate expression of a long noncoding RNA (lncRNA, LINC00339) through chromatin loop formation. LINC00339 could interact with CDC42, which is an important regulator involved in bone metabolism. Our findings provide a mechanistic basis for how a noncoding SNP affects osteoporosis by long-range interaction, which would be a potential and promising therapeutic target for osteoporosis.
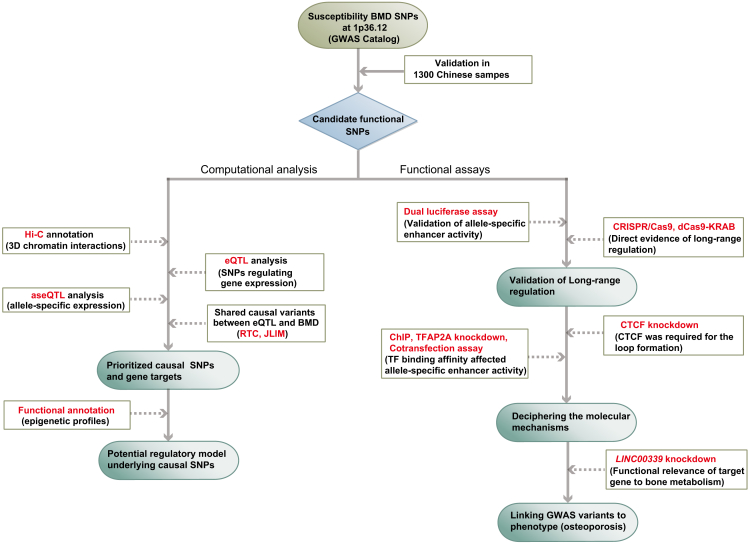
Figure 1.
Flowchart for the Integrative Analyses Approach
Flowchart for the identification of functional BMD SNPs at 1p36.12 followed by experimental validation.
Material and Methods
Study Subjects
The study was approved by the Institutional Review Board of School of Life Science and Technology at Xi’an Jiaotong University. Signed informed consent documents were obtained from all study participants before recruitment. We enrolled 1,300 unrelated Midwestern Chinese subjects of Han ethnicity from the city of Xi’an and its neighboring areas. The inclusion and exclusion criteria have been detailed in our previous publication.20 BMD (g/cm2) values at lumbar spine (LS) and femoral neck (FN) were measured with dual energy X-ray absorptiometry (DXA) using Hologic 4500 W machine (Hologic) that was calibrated daily.
Genotyping and Association Analysis
Genomic DNA was extracted from peripheral blood leukocytes using a commercial DNA isolation kit (Gentra systems) according to the protocol of the kit. For eight BMD-associated SNPs at 1p36.12 collected from the National Human Genome Research Institute (NHGRI) GWAS Catalog,21 we conducted SNP genotyping using MALDI-TOF mass spectrometry on a MassARRAY system (Sequenom) with iPLEX assay. Genotype calling was performed in real time with MassARRAY RT software v.3.0.0.4 and analyzed using the MassARRAY Typer software v.3.4 (Sequenom). All eight SNPs for the Chinese cohort were successfully genotyped.
Before association analyses, we adjusted raw BMD values using significant covariates, including age, sex, height, and weight. Association analyses with BMD were conducted using linear regression model implemented with PLINK22 assuming an additive inheritance model. For significant SNPs, we performed conditional association analysis by fitting each SNP genotype as a covariate and testing for secondary association on the remaining ones. We also checked the association signals in the GEFOS (Genetic Factors for Osteoporosis Consortium) dataset. GEFOS is the largest GWAS meta-analysis on BMD so far, including 17 GWASs and 32,965 individuals of 9 European cohorts.16
LD and Haplotype Analysis
BMD-associated SNPs at 1p36.12 were collected from the NHGRI GWAS Catalog21 (October 2016). Linkage disequilibrium (LD) and haplotype analysis were conducted using Haploview v.4.223 in different populations (European, East Asian, and African) from the 1000 Genomes V3 genotype data.24
Hi-C and TAD Analysis
Hi-C data on IMR90 cells were collected from 4DGenome database.25, 26 Hi-C or capture Hi-C data on GM12878 and CD34 cells were obtained from several studies.27, 28 We also collected capture Hi-C data on 17 human primary blood cell types from a recently published large-scale genome-wide chromatin study.29 DNase Hi-C data on human embryonic stem cells (H1-hESC) were retrieved from GEO database (GSE56869).30 TAD data on IMR90 cells were acquired from GEO database (GSE35156).31 The original ChIA-PET data and newly improved ChIA-PET data on six cell lines (K562, NB4, HCT-116, HeLa-S3, GM12878, and MCF-7)32 were retrieved from the UCSC ENCODE download portal and GEO database (GSE72816), separately. All data used were summarized in Table S2. Bedtools33 was used to extract our prioritized SNPs and/or genes within the same pair of Hi-C interaction regions. We only reported SNP-gene pairs within the same TAD region.
cis-eQTL Analysis
We obtained matched SNP genotyping and RNA-seq data for 462 unrelated human lymphoblastoid cell lines (LCLs) samples from ArrayExpress at EBML-EBI (ArrayExpress: E-GEUV-1). Since there is no direct osteoporosis-related human tissues with large sample size, we chose LCLs to explore the eQTL analysis. Previous studies have shown that there is a large overlap in the transcriptomic effects of genetic variation between human osteoblasts and LCLs,34, 35 and LCLs has been widely used as surrogate for eQTL analysis in genetic studies on osteoporosis.36, 37, 38, 39 cis-eQTL analysis was conducted between each of BMD SNPs and expression of nearby transcripts in a 1 Mb region using ANOVA test implemented in R software. Eta-squared (η2) was calculated to measure the effect size [η2 = SSfactor/(SSfactor + SSerror)].
AseQTL Analysis
Matched SNP genotyping and allele-specific expression (ASE) data were collected from GTEx40 in dbGap (phs000424.v6.p1). We conducted aseQTL analysis by testing for correlations between heterozygosity of rs6426749 and allelic imbalance at LINC00339 or CDC42 expression using Wilcoxon rank sum test. As described by Oldridge et al.,41 we defined allelic fractions as min (A, B) / (A + B), where A or B is different alleles of a synonymous exonic SNP in the target gene.
Analysis of Shared Causal Genetic Variants
Two complementary methods were used to explore whether eQTL signal and GWAS BMD association were driven by the same causal variants. We downloaded recombination hotspot intervals as defined by McVean et al.42 from HapMap web site and converted it to hg19 genome assembly using liftOver software. We used the regulatory trait concordance (RTC) test from Nica et al.43 to distinguish between shared causal effects and coincidental overlaps, which is a rank-based score system testing for association between cis-eQTL and GWAS effect. GWAS SNP with the lowest genome-wide association (GWA) meta p value in GEFOS data with LS or FN BMD within corresponding recombination hotspot intervals was extracted to calculate RTC scores for the tested cis-eQTL SNP. For the joint likelihood mapping (JLIM) analysis,44 we ran JLIM (v.1.0.2) with default parameters.
Functional Annotation
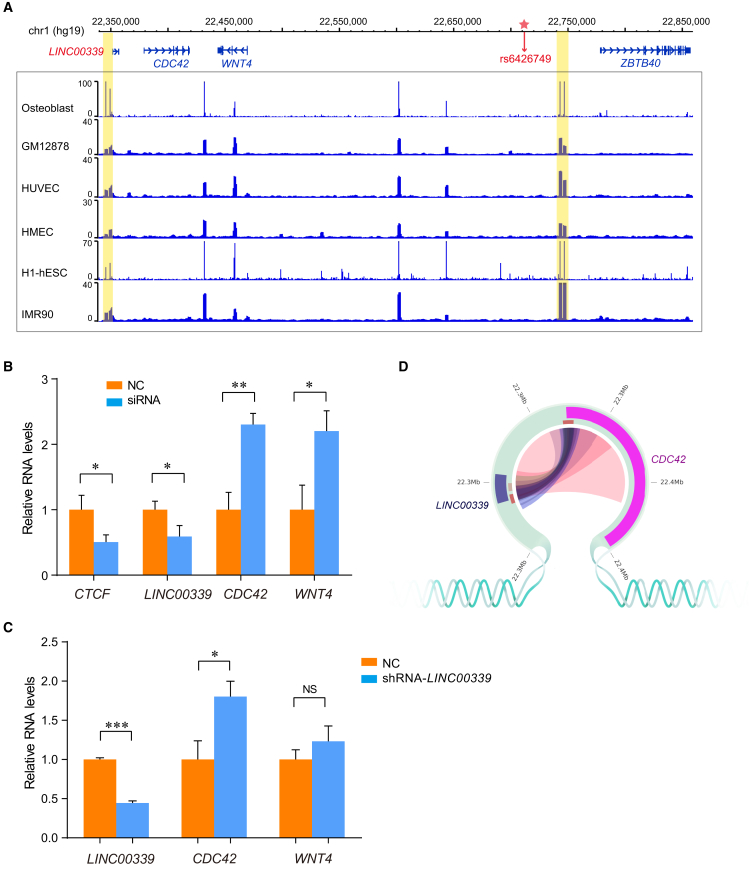
We annotated epigenetic regulatory features for SNPs and genomic regions of interest using ChIP-seq data from ENCODE, including CTCF insulator marks on six healthy cells (Osteoblast, GM12878, HUVEC, HMEC, H1-hESC, IMR90) and enhancer markers (H3k4me1, H3K4me3, H3K27ac, p300) on osteoblast cells. All data were displayed using WashU EpiGenome Browser.
Motif Analysis
The effect of rs6426749 on transcription factor binding motifs was analyzed using HaploReg v4.123 and MEME Suite toolkit45 with TF motifs available from three public motif databases: JASPAR, HOCOMOCO, and SwissRegulon.46, 47, 48 Motifs with at least three hits by different databases were reported. ChIP-seq data were retrieved from ENCODE and GEO database (GSE44257)49, 50 to validate the motif prediction.
Culture of Cell Lines
The hFOB 1.19 cells were obtained from the Institute of Biochemistry and Cell Biology of Shanghai (Shanghai, China) and cultured in DMEM/F12 supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 0.1 mg/mL streptomycin in 5% CO2 at 37°C. The human embryonic kidney 293T cells (HEK293T) were purchased from the American Type Culture Collection (ATCC) and cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 0.1 mg/mL streptomycin in 5% CO2 at 37°C. All cell lines were free of mycoplasma.
Dual-Luciferase Reporter Assays and Site-Directed Mutagenesis
For rs6426749, rs34963268, or rs6684375, we chose a 991-bp, 944-bp, or 995-bp fragment surrounding each SNP as the putative enhancer element, separately. A 1,077-bp fragment upstream of LINC00339 TSS was selected as the promoter for LINC00339. Both enhancer and promoter fragments were PCR amplified from human genomic DNA using the primers listed in Table S7. In order to obtain either the major or minor allele at three SNPs, site-directed mutagenesis was performed with the Quick Change II Site-Directed Mutagenesis Kit (Agilent Technology) according to the manufacturer’s instruction, with primers listed in Table S7. All constructs were validated by sequencing and did not contain any other sequence variations. Constructs were co-transfected into hFOB 1.19 or HEK293T cells along with pRL-TK vector containing Renilla luciferase (Promega) using X-treme GENE HP DNA transfection reagent (Roche). After 48 hr of transfection, the cells were harvested and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega), with Renilla luciferase (Rluc) reporter gene as the internal reference. Results were obtained from three independent experiments and each experiment was done in triplicate.
Enhancer Deletion and Repression
To delete enhancer fragment containing rs6426749 (749 bp) in hFOB 1.19 cells, the pCas9-dual sgRNA vector containing two sgRNAs was transfected into target cells by using Lipofectamine 2000 transfection reagent (Invitrogen). After selection with puromycin (1 mg/mL) for 1 week, the remaining cells were isolated as clones and verified by PCR sequencing. To repress enhancer activity surrounding rs6426749 in hFOB 1.19 cells, the pdCas9-KRAB vector and hU6 sgRNA vector containing distal sgRNA (sgRNA-1: 315 bp upstream rs6426749) or proximal sgRNA (sgRNA-2: 46 bp upstream rs6426749; sgRNA-3: 67 bp downstream rs6426749) were cotransfected into target cells by using Lipofectamine 2000 transfection reagent (Invitrogen). All sgRNA primers are listed in Table S7.
Genotyping of rs6426749
PCR-RFLP method was used to obtain genotype of rs6426749 in hFOB 1.19 cells and HEK293T cells. A 991-bp sequence centered on rs6426749 was first PCR amplified from human genomic DNA using primers the same with Luciferase Reporter assays (Table S7). The amplified DNAs were digested using the restriction enzyme (Sac I), which were subsequently subjected to the electrophoresis assay.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed in HEK293T cells with the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to the manufacturer’s instruction. In brief, approximately 3 × 107 cells were cross-linked with 1% formaldehyde for 10 min. After quenching with glycine solution, cells were rinsed, pelleted in cold PBS, and then resuspended and pelleted twice with buffer A and B, respectively. Micrococcal Nuclease (2,000 gel units/μL) was then added for nucleus digesting. After stopping digesting by EDTA, the nuclei fractions were pelleted by centrifugation, with sediment resuspended in ChIP buffer using protease inhibitor cocktail. The lysate was sonicated with the VirTis Virsonic 100 Ultrasonic Homogenizer/Sonicator for 3 pulses, after clarifying lysates by centrifugation, and the supernatant was collected. The supernatant containing sheared chromatin was immunoprecipitated with TFAP2a antibody (ab52222, Abcam) or normal immunoglobulin G (IgG) as a negative control and precleared with agarose beads. DNA protein complex was then precipitated with agarose beads, eluted from the beads, and reversely cross-linked by 5M NaCl and Proteinase K. The DNA fragments enriched in ChIP assays were purified for downstream RT-qPCR analysis, with primers listed in Table S7.
siRNA and shRNA Knockdown
siRNA knockdown experiments for CTCF (MIM: 604167) and TFAP2A (MIM: 107580) were conducted in hFOB 1.19 cells, separately. The siRNAs targeting CTCF or TFAP2A with related negative controls were synthesized by GenePharma. All siRNA sequences are listed in Table S7. Transfection of siRNAs was carried out in triplicate using the X-tremeGENE siRNA Transfection Reagent (Roche) according to the manufacturer’s instructions. For LINC00339 knockdown, we constructed the miR30-based short hairpin RNA (shRNA) expression vectors by using two oligonucleotides targeting LINC00339. These two oligonucleotides were connected with miR30 backbone and inserted into XhoI and EcoRI site of pcDNA3.1- plasmid (Invitrogen), with shRNA and negative control (NC) sequences shown as follows: shRNA-1: 5′-TGAGATCACTACCCAATGA-3′, shRNA-2: 5′-GACCTGATATCCACACAAA-3′, NC: 5′-GTTCTCCGAACGTGTCACGT-3′. Transfection was performed in triplicate according to the manufacturer’s instructions. Briefly, hFOB 1.19 cells were seeded into 6-well plates and cultured to reach the confluence of 80%. Each well was transfected with 3 μg DNA using ViaFect Transfection Reagent (Promega). Cells were collected after 72 hr for further experiments.
Total RNA Isolation and Quantitative Real-Time PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen), and complementary DNA (cDNA) was synthesized using the Super Scripts II First-Strand cDNA synthesis kit (Invitrogen). RT-qPCR was performed by BIO-RAD CFX Connect Real-Time System, with primers listed in Table S7. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control.
Mendelian Randomization Analysis
Two complementary Mendelian randomization (MR) methods were used to explore the causal relationship between LINC00339 expression and BMD. The GWAS data on BMD were downloaded from a recent published large-scale GWAS.51 The eQTL association data on whole blood were extracted from the GTEx database.40 We first performed a summary-data-based Mendelian Randomization (SMR) analysis52 (v.0.66) with default parameters which assigned the top cis-eQTL as instrumental variable. To exclude potential biased causal effect estimates deriving from invalid instrument variables, we also performed a multi-instrument based MR analysis using R package MendelianRandomization,53 including inverse-variance weighted method and median-based method for causal test. The cis-eQTLs (p < 0.01) were first pruned for independence (r2 < 0.2) by PLINK,22 and the remaining cis-eQTLs were used as instrumental variables.
Results
Validation of GWAS SNPs in BMD Locus 1p36.12 in Chinese Population
1p36.12 was identified by an initial large-scale BMD GWAS10 and replicated by multiple GWAS meta-analyses.9, 10, 12, 13, 14, 15, 16 According to the GWAS Catalog,21 there are eight SNPs at 1p36.12 reported by seven different GWASs.9, 10, 12, 13, 14, 15, 16 Since most of GWAS samples are of European descent, we further examined associations of these eight SNPs with BMD in a Chinese cohort of 1,300 subjects. The basic characteristics of our sample are summarized in Table S1. The detailed association results are summarized in Table 1. These eight SNPs are all located in noncoding regions, which can be classified into three spatial clusters, including intron region of WNT4 (rs3765350, rs2235529), intergenic region near WNT4 (rs7521902 and rs3920498, more than 46 kb), and intergenic region near ZBTB40 (rs7524102, rs34920465, rs6696981, and rs6426749, more than 67 kb) (Figure 2A). Four SNPs near ZBTB40 were successfully validated for association with both lumbar spine (LS) and femoral neck (FN) BMD (p < 0.05, β > 0, Table 1). However, no significant signals were detected for the other four SNPs at 1p36.12. We also compared association signals with GEFOS meta-analysis dataset.16 Consistently, only the four SNPs near ZBTB40 achieved genome-wide significance level (p < 5 × 10−8) (Table 1). We performed LD and haplotype analyses. As shown in Figure 2A, two blocks in high LD were identified. The four SNPs near ZBTB40 were in strong LD with each other (r2 > 0.7) and belonged to one block. This block was highly conserved among diverse populations (European, East Asian, and African samples). Conditional analysis using any of these four SNPs as covariate obliterated association signals for the remaining three SNPs (data not shown), suggesting strong correlations of them.
Table 1.
Association Results of Eight BMD SNPs at 1p36.12
| Closest Gene/Candidate | SNP | Positiona | A1/A2b |
Chinese Cohort |
GEFOS |
Referencesc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | P-LS | β-LS | P-FN | β-FN | MAF | P-LS | β-LS | P-FN | β-FN | |||||
| WNT4 intron | rs3765350 | 22447316 | G/A | 0.280 | 0.546 | −0.003 | 0.651 | 0.003 | 0.273 | 0.098 | −0.018 | 0.004 | −0.027 | Kemp et al.15 |
| rs2235529 | 22450487 | T/C | 0.488 | 0.375 | 0.001 | 0.464 | 0.004 | 0.179 | 0.024 | −0.028 | 0.003 | −0.032 | Kemp et al.15 | |
| WNT4 proximal | rs7521902 | 22490724 | A/C | 0.486 | 0.142 | 0.004 | 0.734 | 0.006 | 0.253 | 0.002 | −0.034 | 6.60E−05 | −0.037 | Estrada et al.13 |
| rs3920498 | 22492887 | C/G | 0.450 | 0.475 | −0.002 | 0.740 | −0.003 | 0.217 | 2.91E−06 | −0.054 | 1.07E−05 | −0.043 | Kemp et al.15 | |
| ZBTB40 proximal | rs7524102∗ | 22698447 | G/A | 0.214 | 0.004∗ | 0.015 | 0.013∗ | 0.015 | 0.198 | 2.41E−14∗ | 0.090 | 7.36E−17∗ | 0.084 | Styrkarsdottir et al.;9, 10 Rivadeneira et al.;12 Estrada et al.;13 Zhang et al.;14 Zheng et al.16 |
| rs34920465∗ | 22700351 | G/A | 0.213 | 0.003∗ | 0.015 | 0.014∗ | 0.015 | 0.216 | 1.57E−09∗ | 0.084 | 1.81E−12∗ | 0.080 | Zhang et al.;14 Zheng et al.16 | |
| rs6696981∗ | 22702858 | T/G | 0.210 | 0.005∗ | 0.015 | 0.019∗ | 0.014 | 0.143 | 9.02E−09∗ | 0.080 | 4.78E−08∗ | 0.065 | Styrkarsdottir et al.;9, 10 Rivadeneira et al.;12 Estrada et al.;13 Zheng et al.16 | |
| rs6426749∗ | 22711473 | C/G | 0.211 | 0.006∗ | 0.015 | 0.021∗ | 0.014 | 0.192 | 1.15E−13∗ | 0.088 | 5.90E−16∗ | 0.082 | Rivadeneira et al.;12 Estrada et al.;13 Zhang et al.;14 Zheng et al.16 | |
Significant SNPs and p values are indicated with an asterisk (∗). Abbreviations: MAF, minor allele frequency; LS, lumbar spine BMD; FN, femoral neck BMD; Chinese cohort, 1,300 in-house Chinese cohort; GEFOS, Genetic Factors for Osteoporosis Consortium.
Position is relative to the hg19 version of the human genome
A1 is the minor allele according to 1000 Genomes
Refs included the large-scale GWASs and meta-analysis for BMD and osteoporosis
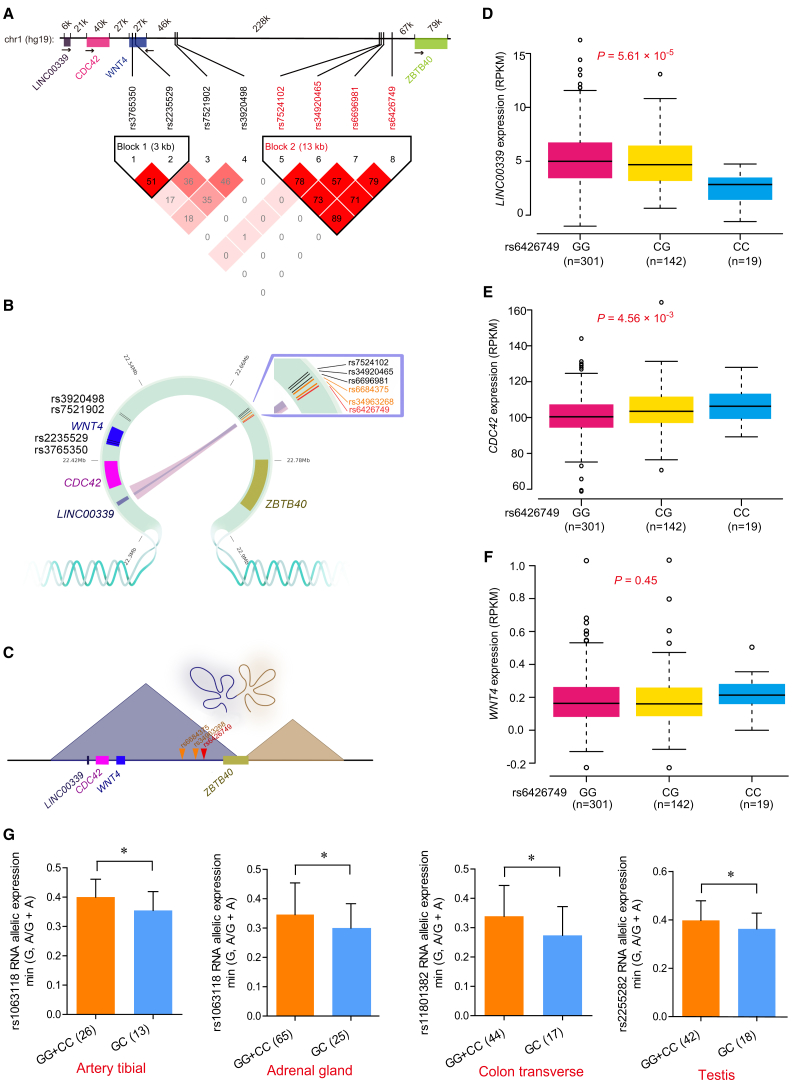
Figure 2.
Integrating Analyses Indicate the Long-Range Interaction between rs6426749 and LINC00339
(A) LD blocks for eight BMD SNPs. The upper bar shows genomic positions for eight BMD SNPs in 1p36.12 and nearby genes, with distance between genes and (or) SNPs displayed above (kb). The bottom inverted triangle shows the LD blocks for eight BMD SNPs at 1p36.12, with each diamond representing the r2 measure of LD using standard color scheme, where the darker shades of red represent greater values.
(B) Hi-C interactions between eQTLs and promoters of target genes, and different color of lines indicated different Hi-C dataset (pink: Hi-C data on IMR90 cells from 4DGenome;25 blue: DNase Hi-C data on H1-hESC cells30). SNP rs6426749 overlapped with Hi-C regions is labeled in red. Another two SNPs in strong LD with rs6426749 within the same Hi-C interaction regions are labeled in orange.
(C) The loop between rs6426749 and LINC00339 is located within a 600 kb topologically associated domain (TAD) in IMR90 cells.
(D–F) Boxplot for LINC00339 (D) or CDC42 (E) or WNT4 (F) expression in samples with different genotypes of rs6426749 (GG, CG, CC) taken from 462 LCLs samples.54 Sample counts are shown.
(G) Allele-specific expression (ASE) analysis between rs6426749 and LINC00339, using monoallelic gene expression data from GTEx.40 Four significant tissues (p < 0.05) are shown. The horizontal axis represents individuals with homozygous or homozygous genotypes for rs6426749. The vertical axis represents the exonic SNP chosen as a measurement of allelic expression of LINC00339. Error bars, SD; ∗p < 0.05 as determined by Wilcoxon rank sum test.
Integrating Hi-C and eQTL Analyses to Identify Regulatory SNPs at 1p36.12 and Their Target Genes
To identify the potential gene targets and evaluate functional role of above noncoding BMD SNPs at 1p36.12, we investigated the long-range chromatin interactions surrounding them using various available Hi-C and ChIA-PET datasets (Table S2).25, 26, 27, 28, 29, 30, 31, 32 We identified seven candidate target genes for six SNPs (Table S3). To validate the predicted gene targets, we conducted cis-eQTL analyses using data from 462 unrelated human LCLs samples.54 Through combining Hi-C and cis-eQTL results, we found that only rs6426749 fulfilled both criteria (Table S3), which had long-range chromatin interactions with LINC00339 promoter in IMR90 cells26 and H1-hESC cells30 (Figure 2B), and this loop was located inside a conserved TAD with a size of 600 kb in IMR90 cells (Figure 2C). Moreover, rs6426749-G allele was significantly associated with increased LINC00339 expression (p = 5.61 × 10−5, η2 = 0.042) (Figure 2D and Table S4). Analysis of eQTL from GTEx project40 further validated the association between rs6426749-G allele and increased LINC00339 expression in LCLs (n = 118, p = 0.02, Figure S1A). However, there was no chromatin interaction between the other three SNPs in block 2 and LINC00339, implying that rs6426749 might be an independent regulatory SNP for LINC00339 in this block. We noticed that rs6426749-G allele was also associated with decreased expression of CDC42 in 462 LCL samples (p = 4.56 × 10−3, η2 = 0.023, Figure 2E and Table S4),54 and no significant association between rs6426749 and WNT4 expression was found in either 462 LCL samples (p = 0.45, Figure 2E and Table S4)54 or GTEx LCL tissue (p = 0.76, Figure S1C).40 We also found some significant associations between rs6426749 and CDC42 or WNT4 expression in several other tissues from GTEx, which are shown in Figure S1.
A different 3D chromatin interaction loop might indicate independent regulatory circuitry affecting expression of target genes.55 We found that compared with all other SNPs within the same Hi-C interaction region, rs6426749 showed the strongest eQTL association with LINC00339 and that another two SNPs (rs6684375, rs34963268) in high LD with rs6426749 (r2 > 0.8) had relative weaker eQTL association with LINC00339 (p = 4.25 × 10−4, η2 = 0.033) (Table S4). However, no secondary eQTL signals remained after adjusting residual effect of rs6426749 (Figure S2B), indicating that rs6426749 was the primary eQTL SNP within the Hi-C interaction region. We further performed conditional eQTL analysis for rs6426749 by adjusting the residual effect of each SNP within 1M region surrounding LINC00339 (Figure S2A) and found that significant eQTL signal for rs6426749 on LINC00339 was retained (p < 0.05, Figure S2C). Together, these data indicated the potential independent long-range regulation on LINC00339 for rs6426749.
Validation of cis-eQTL Regulation on LINC00339
To further validate the cis-eQTL effect on LINC00339, we conducted allele-specific expression (ASE) analysis56 for rs6426749 on LINC00339 or CDC42 expression using matched ASE data and genotype data from GTEx.40 Individuals with heterozygous genotype for aseQTL should have more imbalanced ASE than those homozygous ones. As expected, we observed significantly higher imbalanced LINC00339 expression in individuals heterozygous for rs6426749 (GC) than individuals homozygous for rs6426749 (GG + CC) in four different tissues (p < 0.05, Figure 2G), which provides strong independent validation of the cis-eQTL regulation of rs6426749 on LINC00339. However, no aseQTL effects on CDC42 were detected for rs6426749, indicating that CDC42 might not be the direct target. We also detected significant aseQTL effect on LINC00339 instead of CDC42 for rs6684375 and rs34963268 in six different tissues (p < 0.05, Figures S3A and S3B), further supporting the long-range cis-regulation on LINC00339.
Evidence of Shared Causal Variants for BMD Association and LINC00339 Expression
It is important to distinguish whether the overlap between GWAS signal and cis-eQTLs are coincidental or true shared causal variants. Therefore, we applied two different methods to identify whether associations with BMD and gene expression were driven by the same causal variants. The first is the regulatory trait concordance (RTC) method.43 We detected high RTC scores for three eQTL SNPs (rs6426749: RTC = 0.97; rs6684375: RTC = 0.98; rs34963268, RTC = 0.98) with LINC00339 expression, indicating strong evidence of shared causal effects between the eQTLs for LINC00339 and the BMD GWAS SNPs at 1p36.12. Another method is the joint likelihood mapping (JLIM) analysis,44 which could assess whether association signals between cis-eQTLs and BMD at 1p36.12 were due to the same underlying effect. We detected significant associations between eQTLs for LINC00339 and BMD at LS (p = 0.04) or FN (p = 0.04), further supporting the functional relevance of eQTL regulation for LINC00339 with BMD.
Evaluation of Allele-Specific Enhancer Activity for rs6426749
Noncoding regions of DNA may influence expression of distant genes by acting as enhancers to physically interact with target gene. Enhancers are identifiable by the presence of active epigenetic histone modifications, such as H3K4me1, H3K4me3, and H3K27ac, as well as co-activator and acetyltransferase (CBP/p300).57 Therefore, we used publicly available ChIP-seq datasets from the ENCODE Project58 to evaluate the potential regulatory function of regions around rs6426749, rs6684375, and rs34963268. We observed that the regions surrounding these three SNPs overlapped with many enhancer marks, including H3K4me1, H3K4me3, H3K27ac, and p300 in human osteoblast cells (Figure 3A).
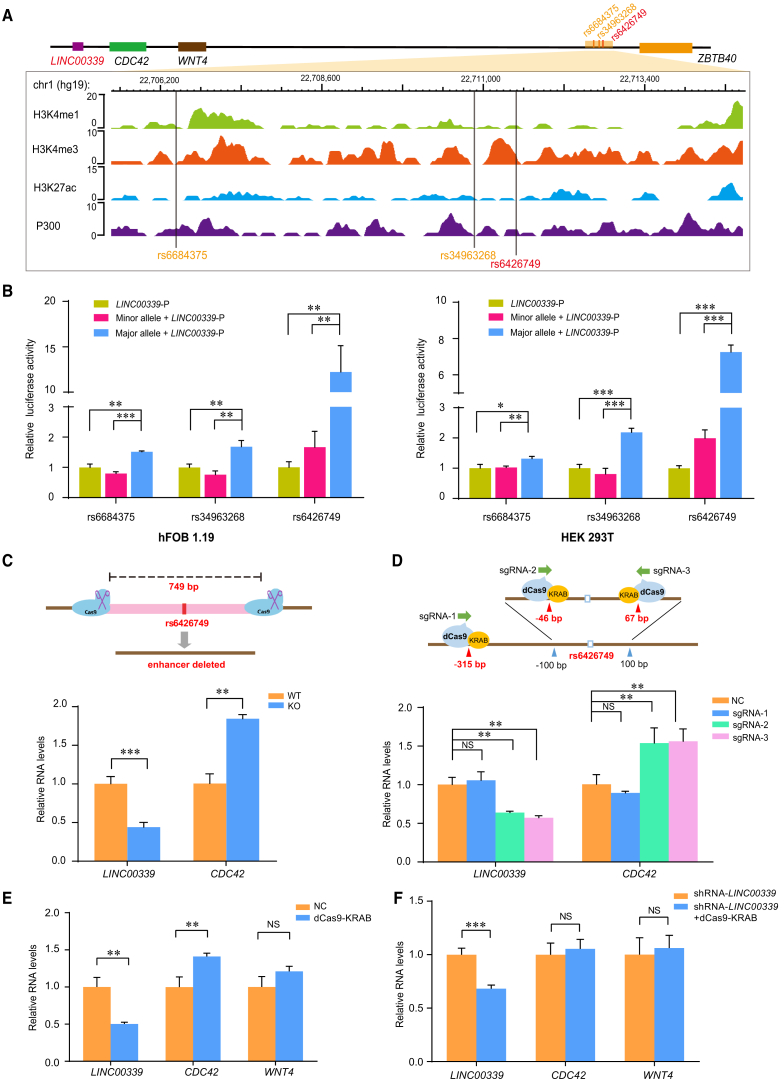
Figure 3.
The Region Containing rs6426749 Acts as Strong Allele-Specific Enhancer on the LINC00339 Promoter
(A) Epigenetic annotation for region surrounding rs6684375, rs34963268, and rs6426749 in osteoblast cells. The data are obtained from ENCODE Project taken from WashU EpiGenome Browser, including active histone modification (H3k4me1, H3K4me3, H3k27ac) as well as acetyltransferase (P300).
(B) The dual-luciferase assay for LINC00339 promoter (LINC00339-P) containing rs6684375, rs34963268, or rs6426749 locus with either the major or minor allele, or individual LINC00339-P was measured in hFOB 1.19 cells or HEK293T cells. The individual LINC00339-P was used as baseline control. Luciferase signal was normalized to Renilla signal. Error bars, SD. n ≥ 3. ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.
(C) Comparison of LINC00339 and CDC42 expression between rs6426749 region deleted hFOB 1.19 cells (KO) mediated by CRISPR/Cas9 and normal cells (WT).
(D) Comparison of LINC00339 and CDC42 expression between rs6426749-locus repressed hFOB 1.19 cells using dCas9-KRAB and normal cells (NC, negative control). One distal sgRNA (sgRNA-1) and two proximal sgRNAs (sgRNA-2, sgRNA-3) were designed.
(E) Effect of rs6426749-locus repression in hFOB 1.19 cells using dCas9-KRAB (sgRNA-3) on LINC00339, CDC42, and WNT4 expression.
(F) RT-qPCR for LINC00339, CDC42, and WNT4 expressions in hFOB 1.19 cells after silencing of both LINC00339 using shRNA and rs6426749-locus using dCas9-KRAB (blue) as compared with LINC00339 silenced cells (orange), respectively. Error bars, SD. n ≥ 3. NS: not significant, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.
To further validate the allele-specific enhancer activity for these three SNPs on target gene LINC00339, we cloned rs6684375, rs34963268, and rs6426749 locus with the major or minor allele of corresponding SNP, and inserted into a luciferase reporter vector, upstream of the LINC00339 promoter, respectively. Upon transfection of these constructs into hFOB 1.19 cells, the major or minor allele of rs6426749 exhibited the greatest different effect on LINC00339 promoter activity (Figure 3B). The rs6426749-G allele had significant increase in luciferase expression as compared with the rs6426749-C allele (p < 0.005, Fold = 7.32) or the LINC00339 promoter-only construct (p < 0.005, Fold = 12.23) (Figure 3B). However, there was no significant difference between the minor allele of rs6426749 and the LINC00339 promoter constructs. The consistent results were obtained in HEK293T cells (Figure 3B). In contrast, only modest increase in luciferase expression was detected between the major and minor allele of another two LD SNPs in hFOB 1.19 cells (p < 0.001, Fold = 1.91 for rs6684375; p < 0.01, Fold = 2.22 for rs34963268) or HEK293T cells (p < 0.01, Fold = 1.29 for rs6684375; p < 0.001, Fold = 2.70 for rs34963268) (Figure 3B). Together, our data demonstrated that rs6426749 could act as strong allele-specific functional enhancer for LINC00339.
Validation of Enhancer Activity for rs6426749 via CRISPR/Cas9 and dCas9-KRAB
Genotyping of rs6426749 revealed that HEK293T cells are heterozygous (G/C) and hFOB 1.19 cells are homozygous (G/G). To directly validate the long-range regulation between rs6426749 and LINC00339, we deleted a 749-bp enhancer region containing rs6426749 using CRISPR/Cas9 in hFOB 1.19 cells. As shown in Figure 3C, significantly decreased LINC00339 expression (p < 0.005) while increased CDC42 expression (p < 0.01) were detected in enhancer-deleted cells (KO) compared with the normal cells (WT), indicating that LINC00339 was the direct target gene underlying distal enhancer-promoter regulation. To further validate the central role of rs6426749 in controlling enhancer activity, we designed two proximal sgRNAs (sgRNA-2: 46 bp upstream or sgRNA-3: 67 bp downstream) and one distal sgRNA (sgRNA-1: 315 bp upstream) targeting the rs6426749 locus using dCas9-KRAB in hFOB 1.19 cells, respectively. As shown in Figure 3D, we detected significantly reduced LINC00339 expression (p < 0.01) while elevated CDC42 expression (p < 0.01) on the proximal sgRNAs. However, the expression of WNT4 was not changed by using the proximal sgRNA-3 in hFOB 1.19 cells (p > 0.05, Figure 3E), indicating that WNT4 might not be the direct target of rs6426749. Moreover, we inhibited LINC00339 expression using shRNA in hFOB 1.19 cells and then inhibited the enhancer region containing rs6426749 using dCas9-KRAB. As compared with the LINC00339 knockdown cells, we detected significantly decreased expression of LINC00339 (p < 0.01) while no perturbation on either CDC42 or WNT4 expression (p > 0.05, Figure 3F), suggesting that LINC00339 was the direct target gene of rs6426749, instead of CDC42 or WNT4.
Analysis of Transcription Factor Binding at rs6426749 Region
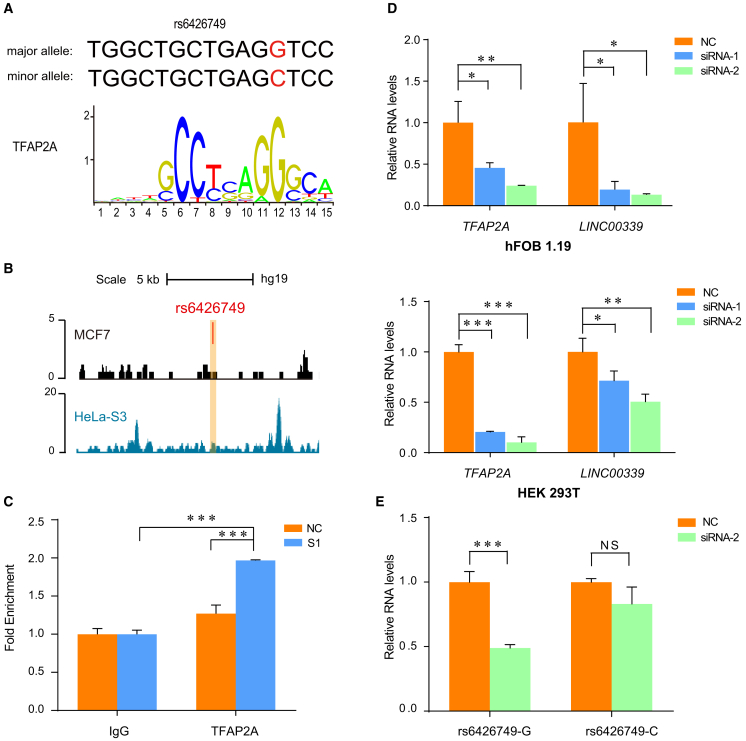
We investigated the functional mechanism for rs6426749 underlying the strongest enhancer activity. The allele-specific activity of enhancer might be due to the different binding affinity of transcription factor (TF). We conducted motif analysis using multiple databases and identified two motifs, TFAP2A and TFAP2C of TFAP2 family, as candidate factors specifically binding to rs6426749-G (Figures 4A and S4A). RNA expression analysis revealed that TFAP2A is expressed much higher than TFAP2C in hFOB 1.19 cells (Figure S4B). ChIP-seq data from ENCODE49 and GEO50 identified that TFAP2A could bind to region surrounding rs6426749 in HeLa-S3 cells and MCF7 cells (Figure 4B), and the binding signal was much higher than the background in both HeLa-S3 cells (Fold = 2.9) and MCF7 cells (Fold = 3.1) (Figure S5). To experimentally verify the motif prediction, we performed ChIP-qPCR. Significant enrichment of TFAP2A binding was observed on the rs6426749 region compared with the negative control in HEK293T cells (p < 0.001, Figure 4C). We suppressed TFAP2A expression by siRNA in both HEK293T cells and hFOB 1.19 cells, which resulted in significant reduction of LINC00339 expression (p < 0.05, Figure 4D). We further provided evidence of allele-specific binding affinity of TFAP2A using cotransfection assays: the TFAP2A knockdown diminished LINC00339 expression in rs6426749-G allele, while it had no effect on rs6426749-C allele in hFOB 1.19 cells (p < 0.001, Figure 4E). Taken together, these data suggest that rs6426749 modulates TFAP2A binding to regulate LINC00339 transcription.
Figure 4.
Identification of Transcription Factors Required for the Activity of Enhancer Containing rs6426749
(A) Motif analysis indicated that TFAP2A motif exclusively binds to G allele of rs6426749.
(B) TFAP2A binding surrounding rs6426749 was observed in MCF7 cells (GEO: GSE44257)50 and HeLa-S3 cells (ENCODE Project, taken from UCSC Genome browser).
(C) ChIP-qPCR of TFAP2A binding at rs6426749 region and negative control region in HEK293T cells. Primers targeting rs6426749 region (S1) or RPL30 exon (NC) are used. The binding of TFAP2A is shown as fold enrichment over IgG.
(D) The siRNA-mediated depletion of TFAP2A diminished LINC00339 expression. RT-qPCR for TFAP2A and LINC00339 expression in hFOB 1.19 cells or HEK293T cells after knockdown of TFAP2A (siRNA-1 and siRNA-2: two different siRNAs, blue and green) compared to NC siRNA-treated cells (NC: negative control, orange), respectively.
(E) The siRNA-mediated depletion of TFAP2A specifically diminished activity of enhancer containing rs6426749 on LINC003339 expression. The pGL3 basic vector containing rs6426749-G (C) allele locus and LINC00339 promoter (see also Figure 2B), as well as the TFAP2A silencer (siRNA-2) or negative control was cotransfected into the hFOB 1.19 cells. Error bars, SD. n ≥ 3. NS: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.
CTCF Is Involved in Mediating Long-Range Chromatin Interaction between rs6426749 and LINC00339
CCCTC-binding factor (CTCF) is the best characterized insulator-binding protein, which is abundant in loop anchors and essential for loop formation and maintenance.27, 32, 59, 60 Using the annotation data from ENCODE, we found specific enrichment of CTCF binding at LINC00339 promoter and rs6426749 nearby region (Figure 5A), which suggested that CTCF might play a role in mediating long-range loop interaction between rs6426749 and LINC00339. In this case, downregulation of CTCF could result in destruction of loop structure and decrease in the expression of target gene. Therefore, to validate the role of CTCF involved in the loop formation, we suppressed the expression of CTCF by siRNA in hFOB1.19 cells. As shown in Figure 5B, knockdown of CTCF significantly decreased the expression of LINC00339 (p < 0.05) while it increased the expression of both CDC42 (p < 0.01) and WNT4 (p < 0.05), which means that CTCF is required for the loop formation to facilitate the regulatory element approaching and activating the expression of LINC00339.
Figure 5.
CTCF Modulated Long-Range Loop Formation between cis-eQTLs and LINC00339 and LINC00339 Negatively Regulated CDC42
(A) CTCF binding sites surrounding rs6426749 or LINC00339 from six different healthy cells taken from WashU EpiGenome Browser are displayed, with focal peak regions highlighted by yellow colors.
(B) The siRNA-mediated depletion of CTCF diminished LINC00339 expression while it elevated both CDC42 and WNT4 expression. RT-qPCR for CTCF, LINC00339, and CDC42 expression in hFOB 1.19 cells after knockdown of CTCF (siRNA, blue) compared to NC siRNA-treated cells (NC: negative control, orange).
(C) RT-qPCR for LINC00339, CDC42, and WNT4 expressions in hFOB 1.19 cells after silencing of LINC00339 using shRNA (blue) compared to NC-treated cells.
(D) Hi-C annotation revealed interaction between LINC00339 and CDC42. Different shade of colors represents the strength of long-range interactions, and different colors indicated different Hi-C dataset (pink: Hi-C data on IMR90 cells from 4DGenome;25 blue: DNase Hi-C data on H1-hESC cells;30 purple: ChIA-PET data taken from ENCODE on three cell lines [HeLa-S3, K562, and MCF-7]49 or CEO database on GM12878 cells32). Error bars, SD. n ≥ 3. NS: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 as determined by an unpaired, two-tailed Student’s t test.
LINC00339 Influences Bone Metabolism by Modulating Expression of CDC42
Next, we investigated the potential function for LINC00339 expression involved in bone metabolism. We estimated the coding probability of LINC00339 using the Coding Potential Assessment Tool.61 The score was 0.0079 (with a score >1 indicating a potential coding gene), supporting the non-protein-coding nature of LINC00339. Using RNA expression data from GTEx Project40 and FANTOM5 Project,62 we found that LINC00339 was ubiquitously expressed across all 54 various tissues and all 69 primary cells in human comparable levels (Figures S6A and S6B). The FANTOM5 Project defined significant trait-associated genes by systematically annotating susceptibility variants surrounding 59,110 genes.62 We found strong association for LINC00339 with bone resorption disease (p = 3.0 × 10−12) and abnormality of bone mineral density diseases (p = 3.0 × 10−12), suggesting the functional relevance of LINC00339 involved in bone metabolism.
We have demonstrated the direct effect of rs6426749 on LINC00339. The above functional assays (including CRISPR/Cas9, dCas9-KRAB, and CTCF knockdown) all imply a negative correlation between LINC00339 and CDC42 expressions (Figures 3C–3E and 5B). Given that CDC42 has been reported to play an important role in bone metabolism18 and knockout of Cdc42 in mouse results in severe skeletal abnormalities,63, 64 LINC003339 might have potential regulatory correlation with CDC42. To verify this hypothesis, we conducted co-expression analysis using expression data from GTEx Project40 and found that LINC00339 was negatively correlated with CDC42 in 12 tissues and positively correlated with CDC42 in another 17 tissues (p < 0.05, Table S5). It was notable that more than 70% of positive tissues (12/17) were brain-related tissues, and LINC00339 was expressed relatively much weaker (mean RPKM < 5) among more than 80% (14/17) of them. We inhibited the expression of LINC00339 in hFOB 1.19 cells. Similarly, knockdown of LINC00339 significantly increased the expression of CDC42 (p < 0.005) while it had no effect on WNT4 expression (Figure 5C), revealing that CDC42 instead of WNT4 was negatively regulated by LINC00339 (Figure 5C). We further analyzed the chromatin interaction between LINC00339 and CDC42 using multiple Hi-C and ChIA-PET data (Table S2). Strong long-range interaction was observed between LINC00339 and CDC42 in six cells (K562, GM12878, H1-hESC, IMR90, MCF7, and HeLa-S3; Figure 5D and Table S6). Together, our results suggest that LINC00339 might influence bone metabolism by modulating expression of CDC42.
Causal Relationship between LINC00339 Expression and BMD
We applied Mendelian randomization (MR) analysis to characterize the causal association between LINC00339 expression and BMD. The SMR analysis using the top cis-eQTL on LINC00339 as instrumental variable (rs2255282) detected significant association between LINC00339 expression and BMD (PSMR = 0.02). To exclude potential biased causal effect estimates deriving from invalid instrument variables, we further performed a multi-instrument-based MR analysis.53 A total of 44 purified cis-eQTL SNPs were selected as instrumental variables with the detailed information summarized in Table S8. We detected robust causal association between LINC00339 expression and BMD based on either inverse variance-weighted method (p = 0.009) or median-based method (p = 0.0001). A scatterplot of genetic association with LINC003339 against association with BMD are shown in Figure S7. Collectively, these data consistently suggested the causal relationship between LINC00339 expression and BMD.
Discussion
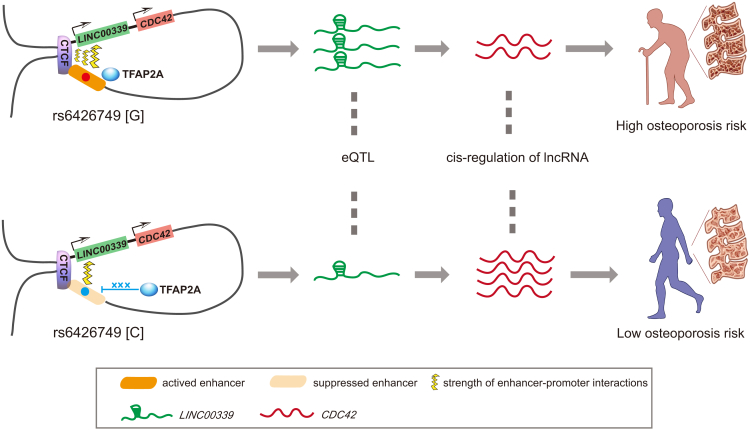
Most of the BMD-associated SNPs identified by GWASs are located in the non-coding regions of genome. The molecular mechanisms underlying the causal actions and biological effects of BMD-associated SNPs are largely unknown. Our study provides extensive evidence that an intergenic SNP (rs6426749) at 1p36.12 acts as a strong long-range allele-specific enhancer to regulate the expression of LINC00339. In particular, we demonstrate that the distal enhancer interacts with LINC00339 via long-range chromatin loop, and CTCF plays a critical role in this loop formation and maintenance. Moreover, the activity of the enhancer containing rs6426749 is mediated by the transcription factor TFAP2A. The rs6426749-G allele robustly recruits TFAP2A, which efficiently elevates the enhancer activity and increases the LINC00339 expression. The target gene LINC00339 could negatively modulate the expression of CDC42, which is an important gene involved in bone metabolism (Figure 6). Taken together, we elucidate a potential mechanistic basis for the genetic association between rs6426749 and osteoporosis, which highlights the regulatory effect of noncoding SNPs underlying the pathogenesis of diseases.
Figure 6.
Potential Regulatory Model between rs6426749, LINC00339, and CDC42
A schematic representation elucidating how genetic variant (rs6426749) affects disease predisposition (osteoporosis). In the top panel, rs6426749-G allele robustly binds to TFAP2A, which elevates the activity of enhancer containing rs6426749 and increases LINC00339 expression. Overexpressed LINC00339 acts as a cis-regulatory element to suppress CDC42 expression, whose relative low expression level is a risk factor to decrease BMD and increase osteoporosis incidence. In the bottom panel, in contrast, rs6426749-C allele is absent from TFAP2A binding, which represses the enhancer activity, resulting in relatively lower LINC00339 expression, which further increases the CDC42 expression. The relatively high expression level of CDC42 decreases the risk to osteoporosis incidence.
Our analysis reveals that a distal enhancer could regulate the expression of LINC00339 via long-range chromatin loop formation. A looped genomic architecture is mediated by some DNA-binding proteins, which facilitate the folding of the 3D genome and bring the distal regulatory elements and promoters into proximity. CTCF is one of the most widely characterized proteins in mediating long-range loop formation.59, 60, 65, 66 CTCF has been shown to bind to distal enhancer and promoter regions to activate enhancer-promoter interactions.27, 32 Our study also found robust CTCF binding near the boundaries of Hi-C regions involving rs6426749 and LINC00339, supporting the potential role of CTCF involved in loop formation. It has been reported that depletion of CTCF could cause global reduction of intradomain interactions.67 Consistent with this finding, knockdown of CTCF efficiently repressed LINC00339 expression in our functional assays, indicating that CTCF is required for the loop formation. Our finding is comparable to the report by Xiang et al.,68 in which they found that CTCF was specifically enriched near MYC locus, and knockdown of CTCF reduced chromatin interaction frequencies between the MYC promoter and its enhancers as well as MYC expression. Therefore, we speculate that loss of CTCF could disrupt the loop structure and restrict the enhancer from approaching the promoter, and therefore inhibit the expression of target gene LINC00339.
Here we provide the key mechanistic insight that rs6426749 acts as an allele-dependent enhancer to functionally contribute to differences in allelic gene expression, which demonstrates that genetic variation in regulatory elements can have a strong influence on common human phenotypic traits. It is generally believed that the enhancer regulates target gene transcription via altering TFs occupancy.69 Our results indicated that TFAP2A could particularly bind to rs6426749-G allele to increase the expression of LINC00339. TFAP2A is a transcriptional activator that can bind to enhancer regions to elevate the enhancer activities.70, 71 Our knockdown experiment found that downregulation of TFAP2A efficiently repressed LINC00339 expression in osteoblast cells, which provides functional evidence to support the role of transcriptional activation for TFAP2A.
Our study implicates LINC00339 as the target for a noncoding susceptibility SNP rs6426749 located at the well-described BMD locus 1p36.12. The nearest gene of this susceptibility SNP is ZBTB40, which has unknown function or connection with bone metabolism. Our results reveal that the nearest gene may not be the true target gene for the susceptibility SNPs identified by GWASs, especially for SNPs located in the intergenic region. The number of lncRNAs neighboring those noncoding SNPs far exceeds that of protein-coding genes. The FANTOM5 Project62 recently elucidated nearly 20,000 potential functional lncRNAs overlapping trait-associated variants or eQTL SNPs, implying the importance of lncRNA in disease development. Another recent study has identified a set of lncRNAs regulated by noncoding SNPs in prostate cancer (MIM: 176807),72 highlighting the importance of investigating the functional link between the noncoding SNPs and lncRNAs. LINC00339 is ubiquitously expressed in various tissues and cells with hardly any coding potential. Some variants in LINC00339 have been identified for association with endometriosis (MIM: 131200) and ovarian cancers (MIM: 167000).73, 74 A recent study found that the leading endometriosis risk SNPs within noncoding region at 1p36.12 could act through inverse regulation of CDC42 and LINC00339,75 supporting the functionality of LINC00339 and potential negative regulation between LINC00339 and CDC42, which is consistent with our study. The function of LINC00339 in bone metabolism is unknown. However, considering that lncRNA could regulate expression of target genes in cis,76 we found that downregulation of LINC00339 can significantly increase the expression of CDC42 in osteoblast cells. CDC42 (cell division cycle 42) is a small Rho GTPase and key regulator of cytoskeletal components. Moreover, CDC42 is a crucial component of the MAPK (mitogen-activated protein kinase) pathway, which is a pivotal mediator of bone metabolism and plays essential roles in osteoblast differentiation and skeletal development.18 Previous studies have revealed the important roles of CDC42 in bone modeling and remodeling.77 We induced the human umbilical cord mesenchymal stem cells (hUCMSCs) into osteoblast and adipocyte cells, respectively. We found that the expression level of CDC42 was significantly increased during osteoblast differentiation, but the expression level of WNT4 was negligible compared with CDC42 or LINC00339, supporting the important role of CDC42 in bone metabolism (Figure S8). However, no BMD variants in CDC42 have been reported, indicating that this gene might be regulated by remote BMD susceptibility SNPs. Our study implicates the functional connection between rs6426749 and CDC42. Previous GWASs have identified rs6426749-G as the risk allele for BMD. Our data posit that rs6426749-G can enhance the expression of LINC00339 and therefore suppress the expression of CDC42. Deletion of Cdc42 in mice could lead to increased adipocyte differentiation and decreased bone formation,19 as well as severe skeletal abnormalities,63 which gives us a strong support that the target gene CDC42 of rs6426749-G could affect the bone formation and increase the risk of osteoporosis. Future investigations are encouraged to elucidate the precise molecular mechanisms.
Our study also has limitations. First, we leveraged eQTL to prioritize functional GWAS variants. However, due to the smaller sample size and disease or cell type relevance of current eQTL data, there might exist unbalanced signals between eQTLs and GWAS SNPs. We therefore reinforce the need of functional assays to validate the findings indicated by eQTL analysis. Second, it is worth noting that our regulatory model could not exclude the contribution of other genetic variants, but instead highlights the results of the study at hand, and might be useful in developing hypotheses for future experimentation. Finally, our study highlights the regulatory effect of noncoding SNPs on osteoporosis through LINC00339. Future functional experiments are encouraged to investigate the detailed molecular mechanism between LINC00339 and osteoporosis.
In summary, through an integrative analysis combining various computational analyses and functional assays, we elucidate a potential mechanistic basis for a functional susceptibility SNP (rs6426749) with long-range target genes (LINC00339, CDC42) at 1p36.12. We anticipate that many other BMD-associated variants in noncoding regions may have similar mechanisms. The integrative approach described in this study can be further used to assign function to more noncoding SNPs in future studies, which is the primary task in our post-GWAS period.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31771399, 81573241, 31471188, 31701095), China Postdoctoral Science Foundation (2016M602797), Natural Science Basic Research Program Shaanxi Province (2016JQ3026), and the Fundamental Research Funds for the Central Universities. We thank the GTEx Consortium. We obtained GTEx data through dbGaP authorized access at https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login with the accession number of phs000424.v6.p1. We would also like to thank the School of Life Science and Technology at Xi’an Jiaotong University for the sharing platform of laboratory apparatus.
Published: April 26, 2018
Footnotes
Supplemental Data include eight figures and eight tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.001.
Contributor Information
Yan Guo, Email: guoyan253@mail.xjtu.edu.cn.
Tie-Lin Yang, Email: yangtielin@mail.xjtu.edu.cn.
Web Resources
1000 Genomes V3 genotype data, ftp://ftp.trace.ncbi.nih.gov/1000genomes/ftp/release/20130502/
4DGenome, https://4dgenome.research.chop.edu/
ArrayExpress, https://www.ebi.ac.uk/arrayexpress/
FANTOM5, http://fantom.gsc.riken.jp/5/data
GEFOS, http://www.gefos.org/
GTEx Portal, https://www.gtexportal.org/home/
GWAS Catalog, http://www.ebi.ac.uk/gwas/
HaploReg, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
International HapMap Project, ftp://ftp.ncbi.nlm.nih.gov/hapmap/
MEME Suite, http://meme-suite.org/
Mouse Genome Informatics, http://www.informatics.jax.org/
OMIM, http://www.omim.org/
R statistical software, https://www.r-project.org/
UCSC ENCODE download portal, https://genome.ucsc.edu/encode/downloads.html
WashU EpiGenome Browser, http://epigenomegateway.wustl.edu/browser/
Supplemental Data
References
- 1.Frazer K.A., Murray S.S., Schork N.J., Topol E.J. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 2.Liu N.Q., Ter Huurne M., Nguyen L.N., Peng T., Wang S.Y., Studd J.B., Joshi O., Ongen H., Bramsen J.B., Yan J. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat. Commun. 2017;8:14418. doi: 10.1038/ncomms14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta R.M., Hadaya J., Trehan A., Zekavat S.M., Roselli C., Klarin D., Emdin C.A., Hilvering C.R.E., Bianchi V., Mueller C. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170:522–533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanis J.A., Delmas P., Burckhardt P., Cooper C., Torgerson D., The European Foundation for Osteoporosis and Bone Disease Guidelines for diagnosis and management of osteoporosis. Osteoporos. Int. 1997;7:390–406. doi: 10.1007/BF01623782. [DOI] [PubMed] [Google Scholar]
- 5.Johnell O., Kanis J.A., Oden A., Johansson H., De Laet C., Delmas P., Eisman J.A., Fujiwara S., Kroger H., Mellstrom D. Predictive value of BMD for hip and other fractures. J. Bone Miner. Res. 2005;20:1185–1194. doi: 10.1359/JBMR.050304. [DOI] [PubMed] [Google Scholar]
- 6.Peacock M., Turner C.H., Econs M.J., Foroud T. Genetics of osteoporosis. Endocr. Rev. 2002;23:303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y.J., Zhang L., Papasian C.J., Deng H.W. Genome-wide association studies for osteoporosis: A 2013 update. J. Bone Metab. 2014;21:99–116. doi: 10.11005/jbm.2014.21.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards J.B., Zheng H.-F., Spector T.D. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat. Rev. Genet. 2012;13:576–588. doi: 10.1038/nrg3228. [DOI] [PubMed] [Google Scholar]
- 9.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Snorradóttir S., Center J.R. New sequence variants associated with bone mineral density. Nat. Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 10.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Center J.R., Nguyen T.V. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 11.Richards J.B., Rivadeneira F., Inouye M., Pastinen T.M., Soranzo N., Wilson S.G., Andrew T., Falchi M., Gwilliam R., Ahmadi K.R. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivadeneira F., Styrkársdottir U., Estrada K., Halldórsson B.V., Hsu Y.H., Richards J.B., Zillikens M.C., Kavvoura F.K., Amin N., Aulchenko Y.S., Genetic Factors for Osteoporosis (GEFOS) Consortium Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M., Amin N., Kemp J.P. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., Choi H.J., Estrada K., Leo P.J., Li J., Pei Y.-F., Zhang Y., Lin Y., Shen H., Liu Y.-Z. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum. Mol. Genet. 2014;23:1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp J.P., Medina-Gomez C., Estrada K., St Pourcain B., Heppe D.H.M., Warrington N.M., Oei L., Ring S.M., Kruithof C.J., Timpson N.J. Phenotypic dissection of bone mineral density reveals skeletal site specificity and facilitates the identification of novel loci in the genetic regulation of bone mass attainment. PLoS Genet. 2014;10:e1004423. doi: 10.1371/journal.pgen.1004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H.F., Forgetta V., Hsu Y.H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., AOGC Consortium. UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu B., Chang J., Liu Y., Li J., Kevork K., Al-Hezaimi K., Graves D.T., Park N.H., Wang C.Y. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-κB. Nat. Med. 2014;20:1009–1017. doi: 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt M.B., Shim J.-H., Glimcher L.H. Mitogen-activated protein kinase pathways in osteoblasts. Annu. Rev. Cell Dev. Biol. 2013;29:63–79. doi: 10.1146/annurev-cellbio-101512-122347. [DOI] [PubMed] [Google Scholar]
- 19.Wuerfel C., Hoffmann C., Kawelke N., Aszodi A. Deletion of cdc42 in osteoblast progenitors leads to increased adipocyte differentiation and decreased bone formation. Bone. 2012;50 [Google Scholar]
- 20.Yang T.L., Guo Y., Liu Y.J., Shen H., Liu Y.Z., Lei S.F., Li J., Tian Q., Deng H.W. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos. Int. 2012;23:781–787. doi: 10.1007/s00198-011-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng L., He B., Wang J., Tan K. 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics. 2015;31:2560–2564. doi: 10.1093/bioinformatics/btv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mifsud B., Tavares-Cadete F., Young A.N., Sugar R., Schoenfelder S., Ferreira L., Wingett S.W., Andrews S., Grey W., Ewels P.A. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 29.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., BLUEPRINT Consortium Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma W., Ay F., Lee C., Gulsoy G., Deng X., Cook S., Hesson J., Cavanaugh C., Ware C.B., Krumm A. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods. 2015;12:71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwan T., Grundberg E., Koka V., Ge B., Lam K.C.L., Dias C., Kindmark A., Mallmin H., Ljunggren O., Rivadeneira F. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu Y.-H., Zillikens M.C., Wilson S.G., Farber C.R., Demissie S., Soranzo N., Bianchi E.N., Grundberg E., Liang L., Richards J.B. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang J.Y., Lee S.H., Go M.J., Kim B.J., Kou I., Ikegawa S., Guo Y., Deng H.W., Raychaudhuri S., Kim Y.J. Meta-analysis identifies a MECOM gene as a novel predisposing factor of osteoporotic fracture. J. Med. Genet. 2013;50:212–219. doi: 10.1136/jmedgenet-2012-101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang T.L., Guo Y., Zhang J.G., Xu C., Tian Q., Deng H.W. Genome-wide survey of runs of homozygosity identifies recessive loci for bone mineral density in Caucasian and Chinese populations. J. Bone Miner. Res. 2015;30:2119–2126. doi: 10.1002/jbmr.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang R., Wei Y., Li Z., Chen H., Miao Q., Bian Z., Zhang H., Wang Q., Wang Z., Lian M. A common variant in CLDN14 is associated with primary biliary cirrhosis and bone mineral density. Sci. Rep. 2016;6:19877. doi: 10.1038/srep19877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu D.L., Guo Y., Zhang Y., Dong S.S., Xu W., Hao R.H., Chen X.F., Yan H., Yang S.Y., Yang T.L. A functional SNP regulated by miR-196a-3p in the 3'UTR of FGF2 is associated with bone mineral density in the Chinese population. Hum. Mutat. 2017;38:725–735. doi: 10.1002/humu.23216. [DOI] [PubMed] [Google Scholar]
- 40.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldridge D.A., Wood A.C., Weichert-Leahey N., Crimmins I., Sussman R., Winter C., McDaniel L.D., Diamond M., Hart L.S., Zhu S. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McVean G.A., Myers S.R., Hunt S., Deloukas P., Bentley D.R., Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 43.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chun S., Casparino A., Patsopoulos N.A., Croteau-Chonka D.C., Raby B.A., De Jager P.L., Sunyaev S.R., Cotsapas C. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat. Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp335. W202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Soboleva A.V., Kasianov A.S., Ashoor H., Ba-Alawi W., Bajic V.B., Medvedeva Y.A., Kolpakov F.A., Makeev V.J. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2016;44(D1):D116–D125. doi: 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pachkov M., Balwierz P.J., Arnold P., Ozonov E., van Nimwegen E. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2013;41:D214–D220. doi: 10.1093/nar/gks1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogachek M.V., Chen Y., Kulak M.V., Woodfield G.W., Cyr A.R., Park J.M., Spanheimer P.M., Li Y., Li T., Weigel R.J. Sumoylation pathway is required to maintain the basal breast cancer subtype. Cancer Cell. 2014;25:748–761. doi: 10.1016/j.ccr.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kemp J.P., Morris J.A., Medina-Gomez C., Forgetta V., Warrington N.M., Youlten S.E., Zheng J., Gregson C.L., Grundberg E., Trajanoska K. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017;49:1468–1475. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 53.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corradin O., Cohen A.J., Luppino J.M., Bayles I.M., Schumacher F.R., Scacheri P.C. Modeling disease risk through analysis of physical interactions between genetic variants within chromatin regulatory circuitry. Nat. Genet. 2016;48:1313–1320. doi: 10.1038/ng.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calo E., Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol. Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Splinter E., Heath H., Kooren J., Palstra R.-J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W.H., Ye C., Ping J.L.H., Mulawadi F. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Park H.J., Dasari S., Wang S., Kocher J.-P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt006. e74–e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aizawa R., Yamada A., Suzuki D., Iimura T., Kassai H., Harada T., Tsukasaki M., Yamamoto G., Tachikawa T., Nakao K. Cdc42 is required for chondrogenesis and interdigital programmed cell death during limb development. Mech. Dev. 2012;129:38–50. doi: 10.1016/j.mod.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Blake J.A., Eppig J.T., Kadin J.A., Richardson J.E., Smith C.L., Bult C.J., the Mouse Genome Database Group Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45(D1):D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Majumder P., Gomez J.A., Chadwick B.P., Boss J.M. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majumder P., Boss J.M. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell. Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuin J., Dixon J.R., van der Reijden M.I.J.A., Ye Z., Kolovos P., Brouwer R.W.W., van de Corput M.P.C., van de Werken H.J.G., Knoch T.A., van IJcken W.F.J. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiang J.-F., Yin Q.-F., Chen T., Zhang Y., Zhang X.-O., Wu Z., Zhang S., Wang H.-B., Ge J., Lu X. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 70.Mitchell P.J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 71.Williams T., Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 72.Guo H., Ahmed M., Zhang F., Yao C.Q., Li S., Liang Y., Hua J., Soares F., Sun Y., Langstein J. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016;48:1142–1150. doi: 10.1038/ng.3637. [DOI] [PubMed] [Google Scholar]
- 73.Albertsen H.M., Chettier R., Farrington P., Ward K. Genome-wide association study link novel loci to endometriosis. PLoS ONE. 2013;8:e58257. doi: 10.1371/journal.pone.0058257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuchenbaecker K.B., Ramus S.J., Tyrer J., Lee A., Shen H.C., Beesley J., Lawrenson K., McGuffog L., Healey S., Lee J.M., EMBRACE. GEMO Study Collaborators. Breast Cancer Family Registry. HEBON. KConFab Investigators. Australian Cancer Study (Ovarian Cancer Investigators) Australian Ovarian Cancer Study Group. Consortium of Investigators of Modifiers of BRCA1 and BRCA2 Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015;47:164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powell J.E., Fung J.N., Shakhbazov K., Sapkota Y., Cloonan N., Hemani G., Hillman K.M., Kaufmann S., Luong H.T., Bowdler L. Endometriosis risk alleles at 1p36.12 act through inverse regulation of CDC42 and LINC00339. Hum. Mol. Genet. 2016;25:5046–5058. doi: 10.1093/hmg/ddw320. [DOI] [PubMed] [Google Scholar]
- 76.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 77.Ito Y., Teitelbaum S.L., Zou W., Zheng Y., Johnson J.F., Chappel J., Ross F.P., Zhao H. Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J. Clin. Invest. 2010;120:1981–1993. doi: 10.1172/JCI39650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.