Abstract
During the past 12 years, genome-wide association studies (GWASs) have uncovered thousands of genetic variants that influence risk for complex human traits and diseases. Yet functional studies aimed at delineating the causal genetic variants and biological mechanisms underlying the observed statistical associations with disease risk have lagged. In this review, we highlight key advances in the field of functional genomics that may facilitate the derivation of biological meaning post-GWAS. We highlight the evidence suggesting that causal variants underlying disease risk often function through regulatory effects on the expression of target genes and that these expression effects might be modest and cell-type specific. We moreover discuss specific studies as proof-of-principle examples for current statistical, bioinformatic, and empirical bench-based approaches to downstream elucidation of GWAS-identified disease risk loci.
Main Text
Introduction
For many decades after the discovery of the structure of DNA and the genetic code, the field of human genetics was largely focused on understanding the structure and function of protein-coding genes and how rare mutations in these genes cause disease. Indeed, the central dogma of molecular biology posits that genes are first transcribed into messenger RNA (mRNA), after which the mRNA is translated into protein.1 Because of the straightforward nature of the genetic code, it was easy to predict how alterations of the underlying DNA sequence would change the amino acid composition of the resulting protein.2 In addition, it was clear from Mendelian genetics that diseases that run in families in predictable patterns are caused by mutations in a single gene. Thus, beginning with the mapping of the genetic cause of the neurodegenerative disorder Huntington’s Disease in 1983,3 the causative mutations underlying many Mendelian diseases were elucidated by positional cloning,2 and an important hurdle was overcome in our understanding of the genetic bases of human disease.
Today, the genetic lesions responsible for many Mendelian diseases are known, and frequently researchers have determined how the mutation in question affects protein function, resulting in pathophysiology.2 However, many of the most common and burdensome diseases, such as cardiovascular disease, cancer, Alzheimer’s disease, Parkinson’s disease, and type 2 diabetes, are typically not (or never) caused by single mutations.4, 5 Such “complex traits” are instead influenced by a combination of multiple genetic and environmental risk factors, and thus do not follow Mendelian inheritance patterns.2 The departure from a “one-gene, one-mutation, one-outcome” model poses a formidable challenge to elucidating the biology of these diseases. Complex traits, by definition, are influenced by many genes (human height, for example, appears to be affected by genetic variation at several hundred loci across the genome),6 which may interact in additive or non-additive (i.e., epistatic) ways. Yet, while it may not always be necessary to understand the cause of a disease in order to successfully treat it, such a mechanistic understanding certainly increases the likelihood that a successful therapeutic intervention will be achieved.
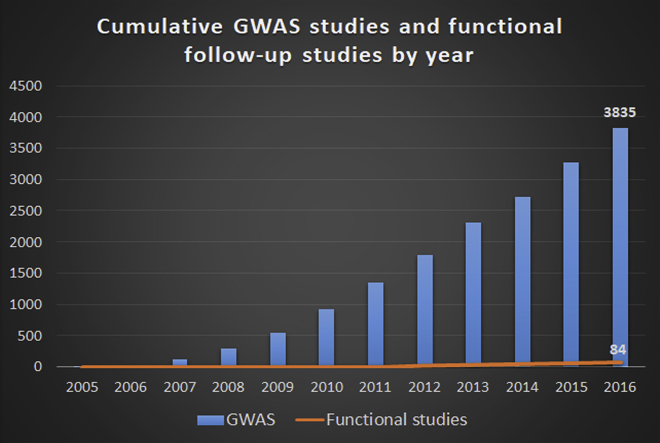
Starting with a set of 2005 studies linking genetic variation near the complement factor H gene to risk for age-related macular degeneration,7, 8, 9 researchers around the world have used the genome-wide association study (GWAS) to identify loci that harbor genetic variants (typically SNPs) that associate with risk for complex diseases and traits.10 The GWAS era has been successful in the sense that thousands of loci have been statistically associated with risk for diseases and traits, and a notable number of these loci are well-replicated, suggesting that they are true associations.11 However, several factors have made it difficult to bridge the gap between the statistical associations linking locus and trait and a functional understanding of the biology underlying disease risk. First, the association of a locus with disease does not specify which variant (or variants) at that locus is actually causing the association (the “causal variant”), nor which gene (or genes) is affected by the causal variant (the “target gene”). The former problem is due to the fact that there are often many co-inherited variants in strong linkage disequilibrium (LD) with the most significant (or “sentinel”) disease-associated variant, comprising a haplotype;12 within the haplotype, genetic variants in strong LD often have statistically indistinguishable associations with disease risk. As a consequence, empirical validation might be needed to determine which of the linked variants are functional.10, 13 The latter complication results from the fact that > 90% of disease-associated variants (daVs) are located in non-protein-coding regions of the genome, and many are far away from the nearest known gene.13, 14 What might these non-coding variants be doing? One clue arises from the observation that daVs, as well as variants in strong LD with them, are enriched in predicted transcriptional regulatory regions, called “cis-regulatory elements” (CREs).13, 14 This suggests that many loci implicated by GWASs to affect disease risk might do so by altering the genetic regulation of one or more target genes. However, the complex nature of eukaryotic transcriptional regulation15 can make it difficult to assign putative CREs (and any disease-associated variants within them) to their correct target genes,10 necessitating the use of genomic datasets and experimental approaches to help answer this question. Indeed, while several thousand GWASs have been performed, and many thousands of loci have been confirmed as bona fide disease risk factors,11 the number of studies that have investigated the mechanisms underlying particular associations is orders of magnitude fewer (Figure 1), and the number of studies that have functionally characterized candidate causal variants at a given locus in an objective (versus “cherry-picked”) manner is even fewer still (Table 1 lists the studies discussed in this review, but is by no means exhaustive). Thus, the purpose of this review is to present a general framework for the functional dissection of a disease-associated risk locus, and to highlight individual studies as proof-of-principle examples for the various approaches that have been used in mechanistic follow-up GWASs.
Figure 1.
Mechanistic Understanding of Disease Risk Loci Identified by GWASs Lags Far Behind the Discovery of New SNP-Trait Associations
The EBI GWAS catalog was used to determine the number of total GWASs reported from 2005 through the end of 2016, which are shown in blue. The number of post-GWAS functional studies reported each year were also identified (orange line) by (1) reviewing the titles, and in some cases, abstracts, of all research articles published in 23 relevant biomedical research journals∗, and (2) searching PubMed using the keywords “causal variant” or “functional variant.” Additional studies were identified through references from primary research or review articles found as described.
∗American Journal of Human Genetics, Cancer Cell, Cell, Cell Reports, Cell Stem Cell, eLife, Genome Biology, Genome Research, Human Molecular Genetics, Molecular Cell, Nature, Nature Biotechnology, Nature Communications, Nature Genetics, Nature Medicine, Nature Neuroscience, Nature Structural & Molecular Biology, Neuron, PLOS Genetics, PNAS, Science, Science Translational Medicine, Stem Cell Reports
Table 1.
Examples of Post-GWAS Functional Studies
| Report | Trait | Locus | Statistical Approaches | Bioinformatic Approaches | Bench-Based Testing Approaches | Causal Variant Mechanism | Target Gene Mechanism |
|---|---|---|---|---|---|---|---|
| Musunuru et al.35 | Low density lipoprotein cholesterol, myocardial infarction | 1p13 | QTL analyses | None | BAC sequencing, reporter assays, EMSAs, in vivo models | Altered TF binding & cis-regulation of SORT1 | Altered SORT1 levels affect plasma lipid and lipoprotein levels |
| Glubb et al.43 | Breast cancer | 5q11.2 | Imputation, conditional analyses, QTL analyses | Epigenomic prioritization | 3C, reporter assays, AS-ChIP-qPCR, cell-based assays | Altered TF binding & cis-regulatory activity | Unknown |
| Wu et al.44 | Adiponectin levels | WDR11-FGFR2 | Imputation, conditional analyses, QTL analyses | None | None | Unknown | Unknown |
| Guthridge et al.47 | Systemic lupus | 8p21 | Imputation, fine mapping | None | Reporter assays, EMSAs, cell-based assays | Altered promoter activity at BLK1 | Unknown |
| Vicente et al.54 | Allergy | 8q21 | Imputation, QTL analyses | Epigenomic prioritization | AS-3C, reporter assays, ChIP-qPCR | Altered TF binding and cis-regulation of PAG1 | Unknown |
| Wang et al.55 | Cardiac QT interval and QRS duration | 112 loci | QTL analyses | Epigenomic prioritization | Reporter assays, 4C, in vivo models | Altered enhancer function | Unknown |
| Bauer et al.62 | Fetal hemogloblin levels | BCL11A | N/A | Epigenomic prioritization | ChIP-seq, in vivo models, genome editing | Altered TF binding & cis-regulation of BCL11A | BCL11A represses fetal hemoglobin levelsa |
| Spisak et al.65 | Prostate cancer | 6q22.1 | N/A | Epigenomic prioritization | ChIP-qPCR, genome and epigenome editing, cell-based assays | Altered TF binding & cis-regulation of RFX6 | Altered RFX6 levels affects cell proliferation, migration and invasiona |
| Soldner et al.68 | Parkinson’s disease | SNCA | QTL analyses, conditional analyses | Epigenomic prioritization, in silico TF motif prediction | Genome editing, ChIP-qPCR, EMSAs | Altered TF binding & cis-regulation of SNCA | Increased SNCA may promote misfolding/ aggregationa |
| Smemo et al.73 | Obesity | FTO | QTL analyses | Epigenomic prioritization | 4C, 3C, in vivo models | Unknown, likely alters enhancer regulation of IRX3 | IRX3 levels affect body mass, composition, and metabolism |
| Claussnitzer et al.78 | Obesity | FTO | QTL analyses | Epigenomic prioritization | Reporter assays, genome editing, cell-based assays, in vivo models, EMSAs | Altered TF binding & cis-regulation of IRX3 and IRX5 | Altered IRX3 and IRX5 levels affect many obesity-related phenotypes |
| Stadhouders et al.81 | Fetal hemoglobin levels | MYB | N/A | Epigenomic prioritization | AS-ChIP-qPCR, AS-3C, reporter assays | Altered TF binding & cis-regulation of MYB | Unknown |
| Gallagher et al.85 | Fronto-temporal dementia | TMEM106B | QTL analyses, conditional analyses, fine mapping | Epigenomic prioritization | Reporter assays, EMSAs, Capture-C | Altered TF binding, chromatin architecture, & cis-regulation of TMEM106B | Altered TMEM106B levels affect lysosomal phenotypesa |
| Huang et al.64 | Prostate cancer | 6q22.1 | N/A | in silico TF motif prediction, epigenomic prioritization | EMSAs, AS-ChIP-qPCR, ChIP-seq, FAIRE-seq, reporter assays, cell-based assays | Altered TF binding & cis-regulation of RFX6 | Altered RFX6 levels affects cell proliferation, migration and invasion |
| Fogarty et al.90 | Type 2 diabetes | CDC123-CAMK1D | N/A | Epigenomic prioritization | Reporter assays, EMSA, DNA affinity capture, ChIP-qPCR | Altered TF binding & cis-regulatory activity | Unknown |
Studies discussed in the text as proof-of-principle examples for various statistical, bioinformatics, and experimental approaches are listed. QTL, quantitative trait loci; TF, transcription factor; BAC, bacterial artificial chromosome; EMSA, electromobility shift assay; 3C, chromosome conformation capture; 4C, circularized chromosome conformation capture; ChIP-qPCR, chromatin immunoprecipitation with quantitative PCR; ChIP-seq, chromatin immunoprecipitation with high-throughput sequencing; AS, allele-specific; FAIRE-seq, formaldehyde-assisted isolation of regulatory elements with high-throughput sequencing.
amechanism reported in prior work.
The Role of Gene Expression in Complex Traits
As mentioned above, the vast majority of daVs reside in noncoding regions of the genome, suggesting that these variants might affect gene expression through effects on transcription, splicing, or mRNA stability. Consistently, several studies have shown that daVs are enriched in predicted CREs, typically defined by chromatin accessibility (as determined by DNase-seq, FAIRE-seq, ATAC-seq, or MNase-seq), transcription factor (TF) binding, and/or histone marks known to be associated with transcriptional regulatory activity, such as H3K27ac, H3K4me1, and H3K4me3.13, 14 Intriguingly, daVs for a particular disease appear to be enriched in CREs active in disease-relevant cell types. For example, a study from Farh and colleagues (2015) examined the overlap of variants associated with 21 autoimmune diseases with six histone marks in multiple primary immune cell types and conditions.16 Importantly, the authors imputed the genotypes of variants not directly genotyped in their respective GWAS and determined which variants were most likely to be causal using an algorithm that incorporates the LD structure and association pattern at each locus. The authors found that candidate causal variants were enriched in predicted B and T cell enhancers (consistent with the expected cellular origin of autoimmune diseases) and that this enrichment increases with the likelihood that the variant is causal.16 When expanding this analysis to 18 additional traits and diseases and incorporating epigenetic data from additional cell and tissue types, the authors observed an enrichment of variants associated with neurological disease in predicted brain promoters and enhancers, whereas blood glucose risk variants were enriched in regulatory regions predicted to be active in pancreatic islets.16
Based on these results and other similar reports,14 many GWASs causal variants have been proposed to influence disease risk by altering the function of cell type-specific regulatory elements, with ensuing changes in target gene expression. This hypothesis is supported by the overlap of daVs with expression quantitative trait loci (eQTLs)—specifically, daVs are more likely to be associated with the expression (mRNA) levels of one or more genes than would be expected by chance (reviewed in17). Furthermore, the cell type in which the eQTL effect is observed often matches cell types already thought to be relevant to the disease in question or lends additional support to a role for a particular cell type in disease, consistent with the overlap of daVs with disease-relevant tissue-specific CREs. In a study by Raj and colleagues (2014), a large-scale eQTL analysis in primary T cells and monocytes, representing different “branches” of the immune response, was performed.18 The authors found a significant overlap between variants associated with gene expression in these cell types and variants associated with autoimmune diseases. Moreover, some additional daVs were only associated with gene-expression levels in one of the two immune cell types. For example, daVs for Alzheimer’s disease (AD) were associated with gene-expression levels only in monocytes. As AD genetic risk variants have also been reported to be enriched in predicted monocyte CREs,19 these studies might suggest an intriguing role for monocytes or cells resembling monocytes in AD.
While the summarized studies support a role of cell-type-specific cis-regulatory variation in complex disease pathogenesis, variants can also affect gene expression levels through post-transcriptional processes such as mRNA splicing and stability.20 Indeed, while outside of the scope of this review, we note that multiple studies have characterized functional daVs that may influence disease risk through these types of effects.21, 22, 23, 24 Other studies have associated genetic variants with altered levels of DNA methylation (mQTLs),25, 26 DNase hypersensitivity (dsQTLs),27 and TF binding (bQTLs),28, 29 and some of these reports show significant overlap of these variants with daVs as well.26, 29
Taken together, these observations suggest that the pathways by which many GWAS causal variants influence disease risk, whether by cis- or trans-acting mechanisms, converge on alteration of expression levels of a target gene, with ensuing effects on disease-relevant phenotypes. In this respect, eQTL studies have consistently shown that most eQTL effects are of relatively small magnitude (<2-fold change in expression),30, 31 agreeing with the results of large-scale experimental characterizations of putative regulatory variants.32, 33 However, while much of the work to date in functional genomics consists of identifying and characterizing functional cis-regulatory variants and their effects on gene expression, the mechanisms by which small changes in gene expression affect cellular or organismal phenotypes to influence disease risk are often not well understood.
Recent Advances in Genomic Annotation
As mentioned previously, the number of SNP-trait associations established by GWASs has increased astronomically in the last decade. The number of SNP-trait associations that have been functionally dissected in an unbiased and comprehensive manner, however, is still relatively low (Figure 1). The recent advent of large, publicly-available databases containing extensive genomic and epigenomic data might remove some of the hurdles that are frequently encountered in this important downstream functional work.
During the first five years after the first GWAS, information regarding the specific genetic variants that exist in human populations was limited, and the functions of the noncoding regions of the genome were largely unexplored. As a consequence, early post-GWAS functional studies first had to define the genetic variation spanning a disease-associated haplotype and then nominate causal variants based on biochemical assays or cell culture-based experiments.34, 35, 36 For example, Musunuru et al. (2010) used sequencing from bacterial artificial chromosomes (BACs) carrying different haplotypes to identify variants in a locus that had been associated with low density lipoprotein (LDL) levels. The authors then performed assays to demonstrate haplotype-specific effects on reporter gene expression, as well as expression of a handful of candidate target genes, including the gene whose expression they would link to both genetic variation at the GWAS-identified locus and the LDL phenotype of interest, SORT1.35 In the last 5–7 years, however, large-scale projects such as the International HapMap and 1000 Genomes projects37, 38 have extensively characterized genetic variation in numerous human populations, largely obviating the need for sequencing, permitting flexible definitions of haplotypes, and allowing for refinement and superior resolution of association signals.10
Many early post-GWAS functional studies also required labor-intensive work to identify potential CREs at disease-associated loci through characterization of epigenetic marks. However, data generated by the ENCODE Project,39 the NIH Roadmap EpiGenome Project,40 the FANTOM consortium41 and others, provide extensive epigenomic characterization, including annotations of putative CREs in hundreds of human cell types and tissues, through publicly available websites. In addition, a wealth of eQTL data is now available for dozens of cell and tissue types,17, 31 such that the association of a daV with gene-expression levels in multiple tissue types can be easily searched and potential CREs linking daVs and causative genes can be identified.
A General Framework for the Functional Dissection of a Genetic Risk Locus
The availability of data is not synonymous, however, with the presence of meaning. The challenge of our current times is to leverage the wealth of recently-available genomic and epigenomic data to derive true biological meaning from GWAS-implicated disease risk loci.
While each locus-trait association will certainly have unique features that require thoughtful “bespoke” delineations of appropriate post-GWAS functional studies, we outline here a general approach that might be applicable to many such studies. We moreover highlight reports exemplifying key steps in this approach.
Statistical Approaches
The resolution of microarray-based GWASs can be greatly increased by performing imputation of variants that were not directly genotyped, using population-based sequencing data, such as that from the 1000 Genomes Project.10 In this way, the significance of association of virtually all common (minor allele frequency ≥ 1%) variants with disease risk can be estimated.42 Conditional analyses can additionally be performed to determine if multiple weakly linked or unlinked causal variants are contributing to the association of the same locus with disease risk,10 as compared to a situation in which only one signal exists at the locus in question. In one example of the former situation, Glubb and colleagues (2015) performed a meta-analysis of breast cancer GWASs, finding a complex pattern of association involving at least three independent signals at and around the MAP3K1 locus.43 In an example of the latter situation, Wu and colleagues (2014) performed conditional analyses on seven loci associated with levels of adiponectin, an adipocyte-secreted protein associated with cardiovascular and metabolic traits.44 After conditioning on the sentinel GWAS SNP for each locus, six out of seven loci showed no residual association at any other variants, suggesting that these associations are driven by one or more strongly linked functional variants.44
To begin to prioritize daVs at a given association signal, Bayesian approaches can be used to determine the probability that each daV is causal for the association, resulting in a “credible set” of candidate causal variants, which might range in size from a single variant to hundreds of variants (reviewed in 45). Furthermore, because most GWASs are performed initially in genetically similar groups of cases and controls, leading to the association of traits with haplotypes as defined in these genetic groups, trans-ethnic fine-mapping can be used to refine the region of association. Specifically, the reduced LD and smaller haplotype blocks in certain populations, particularly Africans, may reduce the number of candidate causal variants.10, 46 For example, Guthridge et al. (2014) used such an approach, combined with re-sequencing of the candidate region, to reduce the number of candidate causal variants at a lupus-associated locus from 30 to 3.47 After employing these and other statistical methods, fine-mapped daVs can be investigated for association with gene expression levels in many cell and tissue types, using publicly available eQTL data.17 While studies integrating GWASs and eQTL data have reported that nearly half of all daVs are associated with gene mRNA levels in at least one cell type,48, 49 there are several other mechanisms by which a functional variant could influence disease risk. First, a variant could affect protein levels through effects on translation or protein stability without an effect on mRNA levels; indeed, up to 1/3 of variants that associate with protein levels (pQTLs) do not associate with the mRNA levels of the same gene,50 although only a few studies51, 52 have examined this overlap. In addition, a GWAS causal variant might alter the amino acid sequence of a protein, thereby affecting protein function rather than abundance.53 These possibilities can usually be excluded, however, if there are no daVs in exonic regions. In such a case, the association of a daV with mRNA expression levels of one or more potential target genes is important for downstream analyses. Namely, conditional and colocalization analyses can be performed using the sentinel GWAS and eQTL variants to determine if both effects are likely driven by the same underlying mechanism. If so, testable hypotheses regarding the function of the disease risk causal variant—that it either increases or decreases the expression of a specific gene or genes—follow naturally.
Incorporating Public Functional Genomics Data
To test such a genetic regulatory function hypothesis, one can capitalize on the wealth of previously-mentioned, publicly available epigenomic data to prioritize candidate causal variants. Specifically, overlap with accessible chromatin, TF binding, and/or histone marks associated with regulatory activity might all suggest a functional effect for a given candidate causal variant located within a predicted CRE. Moreover, the pattern of histone modifications observed at a putative CRE can help predict which type of regulatory element it may be (e.g., promoter, enhancer, insulator, etc.), guiding choice of functional assay. Such a “filtering” approach is exemplified in a study of the 8q21 locus associated with allergic diseases.54 The sentinel GWAS SNP was found to associate with the expression of PAG1 in B lymphoblasts, and ENCODE data was used to select 35 candidate causal SNPs (out of a total of 118 that are in moderate LD (r2 ≥ 0.6) with the sentinel SNP) overlapping four distinct regions of DNase I hypersensitivity and enhancer-associated histone marks in this cell type. These potential CREs were then investigated by multiple approaches, including chromosome conformation capture (3C) and reporter gene assays.54
In addition, epigenomic datasets have been used to investigate loci that are associated with disease risk by GWASs, but do not reach statistical significance after correction for multiple hypothesis testing.55 GWASs typically employ the Bonferroni correction method for multiple hypothesis testing, which might be overly conservative due to LD between nearby SNPs throughout the genome. Thus, some SNPs that do not reach the conventionally accepted genome-wide significance threshold (p < 5 × 10−8) might represent true disease risk loci. To identify such loci, a study by Wang et al. (2016) examined the overlap of SNPs associated with cardiac QT interval with epigenetic enhancer marks in cardiac and non-cardiac tissues. The authors found that both genome-wide significant SNPs (p < 5 × 10−8) and “sub-threshold” SNPs (5 × 10−8 ≤ p ≤ 1 × 10−4) were significantly enriched in predicted cardiac enhancers, and > 70% of enhancers harboring sub-threshold SNPs exhibit allele-specific regulatory activity in induced pluripotent stem cell (iPSC)-derived cardiomyocyte luciferase reporter assays.55 Furthermore, enhancer-associated sub-threshold SNPs were more strongly associated with QT interval than non-enhancer-associated sub-threshold SNPs, and the enhancer-associated SNPs were more likely to reach genome-wide significance in larger GWAS meta-analyses.55
Taken together, the prioritization of candidate causal variants based on epigenomic annotations may yield fruitful directions for downstream investigation. Moreover, the availability of “user-friendly” tools for this prioritization, recently reviewed elsewhere,56 make these types of analyses accessible to many types of scientists. We close this section, however, with a consideration of the limitations of these existing data. First, many gene-regulatory processes are known to be context-dependent. Because the vast majority of epigenomic and eQTL studies have been performed on resting (unstimulated) cells,17, 39, 40 these studies might be limited in their ability to identify context-dependent effects. Second, some cell and tissue types are more difficult to obtain and/or culture than others, which may preclude their incorporation into consortium-based, large-scale studies. Thus, for some diseases/loci involving these cell and tissue types in driver roles, the currently available datasets might be less useful. In such a scenario, approaches taking into account evolutionary conservation might be helpful in prioritizing candidate causal variants. In summary, limitations in causal variant identification might stem from the nature of existing epigenomic and eQTL datasets for some diseases and some loci. However, the bottleneck in our global understanding of risk loci found by GWAS is more likely to be due to a lack of disease-focused functional biological studies downstream of GWAS locus discovery than to a lack of epigenomic and eQTL datasets.
Testing the Function of a Regulatory Variant
Once a list of candidate CREs is identified, all containing one or more potential causal variants, various experimental approaches can be used to test the functions of these regions. A common approach involves in silico analysis to determine whether a particular variant is predicted to disrupt a TF binding motif, with the caveat that many causal variants that may in fact disrupt TF binding do not reside in known TF motifs. For example, only 10%–20% of predicted autoimmune GWAS causal cis-regulatory variants may reside in known TF motifs.16 An alternative approach is to functionally test all candidate CREs, using both the risk and protective alleles of the candidate causal variants. Cell culture-based reporter assays have been widely used for these purposes: the candidate CRE is cloned into a physiologically relevant position with respect to the reporter gene and transfected into a relevant cell type, and the activity of CREs containing alternate alleles (or haplotypes, if multiple daVs overlap the CRE) are compared. Because some CREs are not only cell type-specific, but signal-dependent,57 attention to the appropriate experimental conditions in which to test the variant is important.
Rather than testing reporter constructs one-by-one in cell culture contexts, several groups have developed massively parallel reporter assays (MPRAs), in which thousands of variants can be tested in a single experiment.58 For example, Tewhey and colleagues (2016) investigated ∼30,000 SNPs representing > 3,500 eQTL signals (eSNPs), testing each eSNP and all variants in perfect LD with it for enhancer activity in immortalized liver and B lymphoblast cell lines. ∼12% of the putative CREs displayed enhancer function in one or both of the cell types tested, and of these, ∼25% contained SNPs that caused significant changes in reporter gene expression.32 Importantly, ∼80% of the expression differences caused by these variants agreed with the direction of previously published eQTL effects in the same cell type.32 In addition, the majority of functional variants identified in this study altered reporter gene levels by less than 2-fold, consistent with eQTL effect sizes predicted by previous studies.30, 31, 33 These results underline the importance of investigating the cellular or organismal effects of modest changes in target gene expression.
While reporter assays are often useful in determining the function of a potential regulatory variant, they have several limitations. First, reporter assays can display a significant amount of transcriptional noise, and thus are not always reproducible.59 Second, small differences in reporter activity can result from small differences in the molar amounts of each plasmid that is transfected into cells, which is unavoidable even with the most accurate DNA concentration measurements. These issues can make small differences in expression difficult to distinguish statistically. Perhaps most importantly, reporter assays test the transcriptional function of a variant in the context of plasmid DNA, rather than the native genomic context in which the variant actually exists.58 This situation can produce false negative and false positive results, due to the intricate relationships between DNA, histones, transcription factors, noncoding RNAs, and long-range chromatin interactions.58
In light of these issues, a more physiologically-relevant method to confirm the function of a regulatory variant may be genome editing,60 pioneered through the use of zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), and more recently overtaken by the nucleic acid-based clustered regularly interspaced short palindromic repeat (CRISPR)-based systems.61 In one of the first applications of genome editing to a GWAS-nominated locus, Bauer and colleagues (2013) edited a region in the mouse ortholog of BCL11A. The orthologous region in humans harbors the top SNPs associated with fetal hemoglobin levels, for which BCL11A is a known repressor.62 Thus, the causal variant may function by regulating BCL11A, affecting downstream levels of fetal and embryonic β-globin—indeed, manipulating this pathway is an attractive prospect for treating β-hemoglobinopathies.63 The authors demonstrated that several top GWASs SNPs fall within three distinct regions of open chromatin and enhancer-associated histone marks that are specific to human erythroid cells, consistent with the erythroid-specific expression patterns of the globin genes, and the top candidate SNP was hypothesized to disrupt binding of the erythroid TFs GATA1 and TAL1. Using TALENs, this group deleted a 10kb intronic interval containing the putative causal variant in a murine erythroleukemia cell line, which resulted in dramatically reduced expression of Bc11a and concomitant increase of embryonic β-globin, thus establishing the region as a functional Bcl11a enhancer required for repression of embryonic β-globin.62
In the example above, a large genomic region was deleted to demonstrate the importance of that region to gene regulatory function. However, genome editing can also be used to make more precise changes, such as mutating an individual SNP from one allele to the other. In Spisák et al. (2015), the authors used TALEN-mediated homology directed repair (HDR) to confirm the functional role of a SNP previously reported to influence prostate cancer risk64 by modulating RFX6 expression.65 Specifically, they compared edited and unedited prostate cancer cell line clones, and demonstrated that the candidate causal variant altered RFX6 expression levels by ∼2-fold.65 Moreover, the authors characterized the regulatory potential of the region harboring the SNP by fusing a catalytically-inactive TALE array with either a VP64 transcriptional activation domain, or LSD1, a histone lysine-specific demethylase known to remove H3K4 methylation enhancer marks and decrease enhancer activity. As expected, site-specific recruitment of VP64 and LSD1 to the putative causal SNP increased and decreased RFX6 levels, respectively, establishing the region harboring the causal variant as a bona fide regulatory element.65 Thus, genome editing technologies can also be used to validate potential CREs by altering epigenetic state, rather than the underlying DNA sequence.66, 67
A more recent study used CRISPR/Cas9 gene editing to investigate candidate causal variants at the SNCA locus,68 which is associated with risk for Parkinson’s disease (PD), and which encodes α-synuclein, the protein that accumulates in the characteristic Lewy Body inclusions of PD. The authors demonstrate that the SNCA risk haplotype is associated with increased SNCA brain expression, which has previously been associated with PD pathogenesis, since families with duplications or triplications of the SNCA locus (resulting in increased SNCA levels) exhibit Mendelian forms of PD.69 After prioritizing candidate causal variants based on epigenetic signatures and in silico TF motif predictions, the authors deleted a 500bp putative enhancer at this locus containing two SNPs in human embryonic stem (ES) cells.68 They reinserted the 500bp region using HDR with either the risk or protective alleles of the two SNPs, and differentiated the ES cells into neural precursors and mixed neuronal cultures.68 Cell clones bearing the risk-associated alleles of the enhancer SNPs demonstrated significantly higher SNCA levels than clones bearing the protective alleles, and this effect was driven entirely by the variant predicted to be functional by in silico and experimental analyses.68
Analogous to the high-throughput reporter assays (MPRAs) mentioned above, several groups have also developed high-throughput CRISPR screens to identify essential genes, potential drug targets, or noncoding regulatory regions. As with MPRAs, high-throughput genome and epigenome editing screens may facilitate efficient testing of many candidate cis-regulatory regions and their associated variants. These approaches will likely be particularly useful for loci harboring multiple functional variants that all contribute to disease risk. While conditional analyses can oftentimes rule out the possibility of multiple unlinked (or weakly linked) causal variants, there is no a priori reason to assume that a given association signal is caused by just one functional variant. Indeed, computational studies have suggested that many disease risk loci are driven by multiple variants spanning multiple enhancers that target the same gene,70 further emphasizing the importance of experimentally testing all candidate causal variants at a given locus, rather than focusing on just one or two based on correlational data.
Determining the Target Gene(s) of a Regulatory Variant
While genome editing can confirm the allele-specific functions of a distal CRE, editing experiments alone cannot determine the mechanisms by which these elements affect transcription of their target genes. The last few years have seen an explosion in the number of studies investigating how the human genome is organized in the nucleus, both at small and large scales, and there is now abundant evidence that chromosomes can bend and form loops at kilobase and megabase scales, and that these loops play an important role in transcriptional regulation and disease. While the transcription of a gene occurs at the promoter, enhancers, and other distal regulatory elements appear to affect gene transcription by physically interacting with their target promoters, and oftentimes with each other, through chromatin looping interactions.71 Thus, physical contact between a distal regulatory element and a promoter may be considered evidence for a regulatory function of that element. The marriage of chromosome conformation capture (3C) techniques with high-throughput sequencing has allowed for the investigation of all long-range contacts in the genome (Hi-C, an “all-versus-all” approach), or, with superior depth and resolution, all long-range contacts involving a region of interest, such as a gene promoter (4C, Capture-C, or Capture Hi-C; “one-versus-all” approaches).72
The value that these approaches possess for post-GWAS functional studies (and for the study of eukaryotic transcriptional regulation in general) should be emphasized. Even in cases in which a likely causal variant is already known based on statistical association, allele-specific effects on reporter genes and TF binding, etc., it can be extremely difficult to know a priori what the target gene(s) of the CRE harboring the variant might be.10 “One-versus-all” approaches—in which a specific genomic region is captured or selectively amplified in conjunction with all interacting regions—are well-suited to identify the target promoters of enhancers and other distal elements, or conversely, all distal regulatory elements that interact with a given promoter, such that the regulatory effects of a variant can be linked to the correct gene(s).
A striking example of the importance of considering the three-dimensional organization of chromatin concerns the association of intronic genetic variants at the FTO locus with obesity.73 The FTO locus is the strongest known risk factor for obesity, with an odds ratio of > 1.4 for the sentinel SNP located on chromosome 16q12.2.74 Furthermore, Fto expression levels have been reported to affect body mass and composition in mice; thus, FTO was considered by many to be the gene responsible for conferring risk for obesity at this locus.75, 76, 77 However, while the top obesity-associated SNPs at this locus are all intronic, suggesting a regulatory effect for the causal variant, none of the SNPs have been associated with FTO expression levels.73 To resolve this conundrum, Smemo and colleagues (2014) performed 4C in mouse embryos and brain to determine if the obesity-associated interval interacts with any genes other than FTO. This analysis demonstrated strong interactions between the obesity-associated region and the Irx3 promoter, located several hundred kilobases away. As the obesity-associated region displays enhancer-associated histone marks, the authors then demonstrated enhancer activity for this region using transgenic mouse assays.73 Importantly, they also demonstrated an association between the obesity-associated SNPs and IRX3 expression in a large set of human brain samples, confirming IRX3 as a likely target gene of the FTO enhancer region. These results were then corroborated by mouse models demonstrating a role for lrx3 in body weight maintenance.73 An impressive follow-up investigation by Claussnitzer et al. (2015) identified the likely causal variant at this locus, using precise genome editing and several other approaches.78 This work identified an additional target gene of the obesity-associated region, IRX5, which also appears to affect obesity-related cellular phenotypes. Thus, 3C-based approaches were essential in identifying the genes responsible for conferring obesity risk through long-range regulatory effects of obesity-associated variants at the FTO locus.
It has been suggested that if the activity of a distal regulatory region is modified by a functional variant, and regulation of the target gene(s) by such a region involves long-range interactions, then regulatory variants might influence the long-range interactions themselves.79 Accordingly, 3C-based approaches have been used to identify allele-specific long-range interactions, typically using cell lines or tissues that are heterozygous for the disease-associated haplotype. Allele-specific long-range interactions can be detected by using SNP-specific primers or probes for PCR-based approaches (e.g. 3C or 4C),80 or, for approaches involving HT-seq (e.g., 4C or Hi-C), designing the experiment such that haplotype marker SNPs are present in the sequenced ligation products.79, 81, 82 In an example of the former approach, Stadhouders et al. (2014) used K562 cells to investigate long-range interactions involving a putative enhancer region that harbors variants associated with fetal hemoglobin levels.81 The authors performed 3C with a SNP-specific primer in order to demonstrate allele-specific chromatin looping between the putative enhancer and the promoter of MYB, which encodes a key hematopoietic and erythropoietic TF.81 With regards to the latter approach, several groups have attempted to identify allele-specific interactions by sequencing the 3C ligation products and determining whether SNPs contained in the ligated fragments deviate from either a 50/50 allelic ratio, or the allelic ratio present in a control sample or condition.54, 83, 84 Indeed, recent work from our group used such an approach to functionally dissect a 7p21 locus associated with the neurodegenerative disease frontotemporal dementia.85 Specifically, we linked a candidate causal variant to both disease risk and expression of the target TMEM106B and then demonstrated that this SNP affected recruitment of the mammalian chromatin organizing protein CTCF. To confirm our hypothesis that haplotype-dependent recruitment of CTCF would lead to haplotype-dependent participation in long-range chromatin interactions, we adapted a 3C-based technique to capture all interactions with our candidate SNP-containing CRE,86 demonstrating significant enrichment of the disease-associated haplotype in long-range chromatin contacts. Importantly, our causal SNP resides not in an enhancer or promoter, but a CTCF-bound architectural site,85 suggesting that daVs can affect not only genetic regulatory mechanisms dependent on long-range interaction (such as the enhancer-mediated FTO/IRX3 mechanism), but also the determinants of higher-order chromatin architecture themselves.
Determining the Molecular Function of a Regulatory Variant
If one or more of the approaches mentioned above succeed in identifying a functional cis-regulatory variant, the question remains as to how the variant affects the function of the CRE at the molecular level. Given the overwhelming evidence supporting the critical role of TFs and chromatin remodelers in transcriptional regulation,15 coupled with the significant overlap of daVs with mQTLs and bQTLs,26, 29 one may hypothesize that many causal cis-regulatory variants affect the ability of one or more trans-acting factors to bind the CRE. This effect may be direct (e.g., directly affecting binding of one or more TFs or chromatin-modifying proteins) or indirect (e.g., affecting DNA methylation).
The effect of a variant on TF binding can be confirmed by ChIP followed by qPCR (ChIP-qPCR) using allele-specific probes or primers, such that allelic differences in TF binding can be determined at the variant of interest in a heterozygous cell line or tissue, as exemplified by the demonstration of allele-specific binding of the transcription factor HOXB13 at a putative causal variant at the RFX6 locus.64 Alternatively, ChIP-seq experiments can be performed to investigate potential allele-specific TF binding. In samples heterozygous for the candidate functional variant, normalized sequencing reads covering the variant (or a linked proxy variant) are expected to be present in equal allelic ratios if the variant does not affect binding of that particular factor; conversely, deviations from a 50/50 allelic ratio have been used to infer function of candidate causal variants both at pre-determined loci62 and in genome-wide analyses.87
Electrophoretic mobility shift assays (EMSAs) are another method of determining whether a variant affects recruitment of a nuclear factor in vitro, although these assays can be difficult to interpret and lack biological context.88 Antibodies raised against candidate TFs can be added to the reaction to confirm TF binding to the variant; alternatively, purified recombinant protein can be used instead of nuclear extract to assess binding of specific TFs.88 In many cases it may be difficult to predict which TFs or other types of nuclear proteins are affected by the variant, in which case unbiased approaches such as EMSA followed by mass spectrometry can be helpful.89 For example, a study by Fogarty and colleagues (2014) combined these approaches to investigate a putative causal variant at the CDC123/CAMK1D Type 2 diabetes risk locus.90 After prioritizing candidate causal variants based on epigenomic annotations and identifying a variant that affects enhancer activity in cell-based reporter assays, the authors performed EMSAs to determine which trans-acting factors might be affected by the variant. Twenty-one base pair probes containing either the risk or protective allele of rs11257655, the candidate causal variant, were incubated with nuclear extract from HepG2 immortalized liver cells and two rodent insulinoma cell lines. In all three extract types, the authors observed one or more risk allele-specific probe/protein complexes that could be supershifted with antibodies raised against the enhancer-binding proteins FOXA1 and FOXA2.90 Consistent with this finding, rs11257655 is located within a predicted FOXA1/2 motif, and the protective allele alters a highly conserved “T” base pair within the motif. The authors further confirmed risk allele-specific binding of FOXA2 by performing a DNA affinity capture assay followed by mass spectroscopy.90
Linking Gene-Expression Changes to Complex Traits
While determining the molecular mechanism by which a disease-associated variant affects gene expression is important from a genetic regulatory standpoint, perhaps a more practical question is that of how small changes in a gene’s expression levels affect cellular and organismal phenotypes in a disease-relevant way. Indeed, while many studies have reported genetic variants that alter cis-regulatory function, the mechanisms by which the resulting alterations in gene expression influence disease risk are often not investigated, or are unknown. Some studies have functionally linked expression levels of the causative gene to disease-relevant phenotypes, but many of these studies relied upon imprecisely-controlled overexpression, strong knockdown, or knockout approaches.35, 64, 73, 91 Recapitulating the gene-expression differences relevant to a disease risk locus is difficult for at least two reasons: first, eQTL effect sizes are, in terms of fold expression change, typically unknown, not reported, or small; second, it is technically difficult to finely titrate the overexpression or knockdown of a gene. To overcome this issue, some studies have looked for correlations between the expression levels of the gene of interest and disease-relevant phenotypes, across samples or individuals. For example, Huang et al. (2014) characterized a functional variant at the RFX6 locus, which appears to increase prostate cancer risk by increasing enhancer-mediated RFX6 regulation. Knockdown of RFX6 impaired prostate cancer cell migration and invasion, and consistently, RFX6 expression levels were positively correlated with tumor aggressiveness and relapse across 128 prostate cancer samples.64 Some studies have also reported trait-relevant phenotypes that are distinguishable between cell lines of different genotypes, such as pigmentation in melanocytes,92 although this may not be a common phenomenon among complex traits.
The main limitation of the approaches discussed above is that they are correlational. Thus, to determine the phenotypic effects of allele-specific changes in gene expression, genome editing may again be the best approach. By mutating the causal variant from one allele to the other, the resulting changes in gene expression and cellular phenotypes are (1) more likely to be physiologically relevant than those seen in overexpression or knockdown experiments, and (2) can be causally linked to the variant in question. By using HDR to mutate the RFX6 causal variant, Spisák et al. (2015) demonstrate a 2-fold expression difference between risk and protective allele homozygote clones.65 Intriguingly, protective allele homozygote clones displayed notably different cellular morphology and impaired cellular adhesion compared with risk allele homozygotes. However, no effects on cell migration or invasion were seen, presumably because of the smaller changes in RFX6 expression compared to the previous Huang et al. study.65 In Claussnitzer et al. (2015), precise editing (and re-editing) of the FTO obesity causal variant in adipocytes not only resulted in the expected changes in target gene expression, but also affected metabolic rate, oxygen consumption, and thermogenesis, all pathways that are associated with obesity.78 While these initial results are promising, a more complete understanding of the mechanistic links between allele-specific changes in gene expression and risk for complex diseases and traits is needed. Important to this understanding will be the establishment of molecular, cellular, and organismal phenotypes tailored to the particular disease in question (i.e., LDL levels in cardiovascular disease) which, in turn, might benefit from disease-specialized knowledge.
Conclusions
GWASs have identified thousands of SNP-trait associations throughout the genome, linking common genetic variation to hundreds if not thousands of complex diseases and traits. However, only a small fraction of these statistical associations have been thoroughly investigated to determine (1) which variant or variants are causal, (2) what the molecular functions of the causal variants are, (3) which genes are affected by the causal variants, and (4) how changes in the function or regulation of the causal genes lead to altered disease risk. In our own disease area, that of neurodegeneration, GWASs have identified > 200 loci associated with the four major neurodegenerative diseases (Alzheimer’s Disease, Parkinson’s Disease, Amyotrophic Lateral Sclerosis, and FTD) and related phenotypes.11 However, only one of these loci, the SNCA locus, which was known to be involved in PD risk for years93, 94 prior to the advent of GWASs, was mechanistically investigated in detail prior to our own recent work on the TMEM106B locus.
We thus suggest that an increased emphasis on the downstream functional dissection of already-identified GWAS loci, rather than a search for ever more GWAS loci, might be most likely to benefit knowledge of pathophysiology. Indeed, as recently argued by Boyle, Li, and Pritchard,95 the advent of larger and larger GWASs yielding associations with smaller and smaller effects might result in the eventual finding of all genes expressed in disease-relevant cells as disease-associated loci, a case of clearly vanishing returns. To again use the example of FTD, in 2010, one GWAS, of modest numbers (∼500 cases), identified one risk locus of relatively large effect size (odds ratio > 1.6).96 In the years since, this locus has been conclusively linked to the target TMEM106B by multiple groups,97, 98 and TMEM106B has been shown to localize to and affect the function of lysosomes.99, 100, 101, 102 Moreover, genetic variation at the TMEM106B locus has been shown to modify phenotype in carriers of Mendelian mutations in GRN103 and C9orf72104 causal for FTD, as well as to modify risk for cognitive impairment and dementia in ALS,105 with, potentially, a more general role in brain “aging”106 and cognitive phenotypes.107 As a consequence of this active downstream investigation, multiple potential therapeutic avenues targeting TMEM106B—to reduce penetrance in GRN or C9orf72 mutation carriers, to decrease risk of dementia in ALS, to improve lysosomal activity in a way that might benefit cellular function—exist now. Should similar approaches be taken with the wealth of uncharacterized, but well-replicated, GWAS loci already implicated in neurodegeneration and, more broadly, in human disease, the benefits to human health promised for the last 12 years might begin to be realized.
Acknowledgments
M.D.G. was supported by the NIH (F31 NS090892), and A.S.C.-P. is supported by the NIH (R01 NS082265, UO1 NS097056, P30 AG010124) and the Alzheimer’s Association. We thank Christopher D. Brown for many helpful discussions.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S., Collins F.S. The geneticist’s approach to complex disease. Annu. Rev. Med. 1996;47:333–353. doi: 10.1146/annurev.med.47.1.333. [DOI] [PubMed] [Google Scholar]
- 3.Gusella J.F., Wexler N.S., Conneally P.M., Naylor S.L., Anderson M.A., Tanzi R.E., Watkins P.C., Ottina K., Wallace M.R., Sakaguchi A.Y. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 4.Hirschhorn J.N. Genetic approaches to studying common diseases and complex traits. Pediatr. Res. 2005;57:74R–77R. doi: 10.1203/01.PDR.0000159574.98964.87. [DOI] [PubMed] [Google Scholar]
- 5.Johnson G.C., Todd J.A. Strategies in complex disease mapping. Curr. Opin. Genet. Dev. 2000;10:330–334. doi: 10.1016/s0959-437x(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 6.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards A.O., Ritter R., 3rd, Abel K.J., Manning A., Panhuysen C., Farrer L.A. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 9.Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 10.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 13.Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lelli K.M., Slattery M., Mann R.S. Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albert F.W., Kruglyak L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 18.Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Q., Powles R.L., Abdallah S., Ou D., Wang Q., Hu Y., Lu Y., Liu W., Li B., Mukherjee S. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 2017;13:e1006933. doi: 10.1371/journal.pgen.1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pai A.A., Pritchard J.K., Gilad Y. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 2015;11:e1004857. doi: 10.1371/journal.pgen.1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paraboschi E.M., Rimoldi V., Soldà G., Tabaglio T., Dall’Osso C., Saba E., Vigliano M., Salviati A., Leone M., Benedetti M.D. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum. Mol. Genet. 2014;23:6746–6761. doi: 10.1093/hmg/ddu392. [DOI] [PubMed] [Google Scholar]
- 22.Richardson K., Nettleton J.A., Rotllan N., Tanaka T., Smith C.E., Lai C.Q., Parnell L.D., Lee Y.C., Lahti J., Lemaitre R.N. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am. J. Hum. Genet. 2013;92:5–14. doi: 10.1016/j.ajhg.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D., Poi M.J., Sun X., Gaedigk A., Leeder J.S., Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 2014;23:268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Liu Y., Song F., Zheng H., Hu L., Lu H., Liu P., Hao X., Zhang W., Chen K. Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA. 2011;108:13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplow I.M., MacIsaac J.L., Mah S.M., McEwen L.M., Kobor M.S., Fraser H.B. A pooling-based approach to mapping genetic variants associated with DNA methylation. Genome Res. 2015;25:907–917. doi: 10.1101/gr.183749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannon E., Spiers H., Viana J., Pidsley R., Burrage J., Murphy T.M., Troakes C., Turecki G., O’Donovan M.C., Schalkwyk L.C. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Degner J.F., Pai A.A., Pique-Regi R., Veyrieras J.B., Gaffney D.J., Pickrell J.K., De Leon S., Michelini K., Lewellen N., Crawford G.E. DNasecI sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Z., Ni Y., Timmer S.W., Lee B.K., Battenhouse A., Louzada S., Yang F., Dunham I., Crawford G.E., Lieb J.D. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 2014;10:e1004798. doi: 10.1371/journal.pgen.1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tehranchi A.K., Myrthil M., Martin T., Hie B.L., Golan D., Fraser H.B. Pooled ChIP-seq links variation in transcription factor binding to complex disease risk. Cell. 2016;165:730–741. doi: 10.1016/j.cell.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.I., Cooper G.M. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., Frazer K.A. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verlaan D.J., Berlivet S., Hunninghake G.M., Madore A.M., Larivière M., Moussette S., Grundberg E., Kwan T., Ouimet M., Ge B. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am. J. Hum. Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.1000 Genomes Project Consortium. Auton A., Brooks L.D. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International HapMap 3 Consortium. Altshuler D.M., Gibbs R.A. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.1000 Genomes Project Consortium. Abecasis G.R., Auton A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glubb D.M., Maranian M.J., Michailidou K., Pooley K.A., Meyer K.B., Kar S., Carlebur S., O’Reilly M., Betts J.A., Hillman K.M., GENICA Network. kConFab Investigators. Norwegian Breast Cancer Study Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am. J. Hum. Genet. 2015;96:5–20. doi: 10.1016/j.ajhg.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Gao H., Li H., Tabara Y., Nakatochi M., Chiu Y.F., Park E.J., Wen W., Adair L.S., Borja J.B. A meta-analysis of genome-wide association studies for adiponectin levels in East Asians identifies a novel locus near WDR11-FGFR2. Hum. Mol. Genet. 2014;23:1108–1119. doi: 10.1093/hmg/ddt488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spain S.L., Barrett J.C. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 2015;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell M.C., Tishkoff S.A. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guthridge J.M., Lu R., Sun H., Sun C., Wiley G.B., Dominguez N., Macwana S.R., Lessard C.J., Kim-Howard X., Cobb B.L. Two functional lupus-associated BLK promoter variants control cell-type- and developmental-stage-specific transcription. Am. J. Hum. Genet. 2014;94:586–598. doi: 10.1016/j.ajhg.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu J., Wolfs M.G., Deelen P., Westra H.J., Fehrmann R.S., Te Meerman G.J., Buurman W.A., Rensen S.S., Groen H.J., Weersma R.K. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hause R.J., Stark A.L., Antao N.N., Gorsic L.K., Chung S.H., Brown C.D., Wong S.S., Gill D.F., Myers J.L., To L.A. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. Am. J. Hum. Genet. 2014;95:194–208. doi: 10.1016/j.ajhg.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L., Candille S.I., Choi Y., Xie D., Jiang L., Li-Pook-Than J., Tang H., Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper G.M., Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 54.Vicente C.T., Edwards S.L., Hillman K.M., Kaufmann S., Mitchell H., Bain L., Glubb D.M., Lee J.S., French J.D., Ferreira M.A. Long-range modulation of PAG1 expression by 8q21 allergy risk variants. Am. J. Hum. Genet. 2015;97:329–336. doi: 10.1016/j.ajhg.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Tucker N.R., Rizki G., Mills R., Krijger P.H., de Wit E., Subramanian V., Bartell E., Nguyen X.X., Ye J. Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. eLife. 2016;5:e10557. doi: 10.7554/eLife.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tak Y.G., Farnham P.J. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8 doi: 10.1186/s13072-015-0050-4. 57-015-0050-4. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinz S., Romanoski C.E., Benner C., Glass C.K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue F., Ahituv N. Decoding enhancers using massively parallel reporter assays. Genomics. 2015;106:159–164. doi: 10.1016/j.ygeno.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown C.D., Mangravite L.M., Engelhardt B.E. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 2013;9:e1003649. doi: 10.1371/journal.pgen.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engel K.L., Mackiewicz M., Hardigan A.A., Myers R.M., Savic D. Decoding transcriptional enhancers: Evolving from annotation to functional interpretation. Semin. Cell Dev. Biol. 2016;57:40–50. doi: 10.1016/j.semcdb.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaj T., Sirk S.J., Shui S.L., Liu J. Genome-editing technologies: Principles and applications. Cold Spring Harb. Perspect. Biol. 2016;8:a023754. doi: 10.1101/cshperspect.a023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lettre G., Bauer D.E. Fetal haemoglobin in sickle-cell disease: from genetic epidemiology to new therapeutic strategies. Lancet. 2016;387:2554–2564. doi: 10.1016/S0140-6736(15)01341-0. [DOI] [PubMed] [Google Scholar]
- 64.Huang Q., Whitington T., Gao P., Lindberg J.F., Yang Y., Sun J., Väisänen M.R., Szulkin R., Annala M., Yan J. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet. 2014;46:126–135. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- 65.Spisák S., Lawrenson K., Fu Y., Csabai I., Cottman R.T., Seo J.H., Haiman C., Han Y., Lenci R., Li Q., GAME-ON/ELLIPSE Consortium CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat. Med. 2015;21:1357–1363. doi: 10.1038/nm.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mendenhall E.M., Williamson K.E., Reyon D., Zou J.Y., Ram O., Joung J.K., Bernstein B.E. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soldner F., Stelzer Y., Shivalila C.S., Abraham B.J., Latourelle J.C., Barrasa M.I., Goldmann J., Myers R.H., Young R.A., Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 70.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., Lupien M., Markowitz S., Scacheri P.C. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pombo A., Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 72.Denker A., de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–1382. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Church C., Moir L., McMurray F., Girard C., Banks G.T., Teboul L., Wells S., Brüning J.C., Nolan P.M., Ashcroft F.M., Cox R.D. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Brüning J.C., Rüther U. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 77.Gao X., Shin Y.H., Li M., Wang F., Tong Q., Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holwerda S.J., van de Werken H.J., Ribeiro de Almeida C., Bergen I.M., de Bruijn M.J., Verstegen M.J., Simonis M., Splinter E., Wijchers P.J., Hendriks R.W., de Laat W. Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res. 2013;41:6905–6916. doi: 10.1093/nar/gkt491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stadhouders R., Aktuna S., Thongjuea S., Aghajanirefah A., Pourfarzad F., van Ijcken W., Lenhard B., Rooks H., Best S., Menzel S. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunning A.M., Michailidou K., Kuchenbaecker K.B., Thompson D., French J.D., Beesley J., Healey C.S., Kar S., Pooley K.A., Lopez-Knowles E., EMBRACE. GEMO Study Collaborators. HEBON. kConFab Investigators Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ghoussaini M., Edwards S.L., Michailidou K., Nord S., Cowper-Sal Lari R., Desai K., Kar S., Hillman K.M., Kaufmann S., Glubb D.M., Australian Ovarian Cancer Management Group. Australian Ovarian Cancer Management Group Evidence that breast cancer risk at the 2q35 locus is mediated through IGFBP5 regulation. Nat. Commun. 2014;4:4999. doi: 10.1038/ncomms5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gallagher M.D., Posavi M., Huang P., Unger T.L., Berlyand Y., Gruenewald A.L., Chesi A., Manduchi E., Wells A.D., Grant S.F.A. A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am. J. Hum. Genet. 2017;101:643–663. doi: 10.1016/j.ajhg.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 87.Maurano M.T., Haugen E., Sandstrom R., Vierstra J., Shafer A., Kaul R., Stamatoyannopoulos J.A. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015;47:1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stead J.A., Keen J.N., McDowall K.J. The identification of nucleic acid-interacting proteins using a simple proteomics-based approach that directly incorporates the electrophoretic mobility shift assay. Mol. Cell. Proteomics. 2006;5:1697–1702. doi: 10.1074/mcp.T600027-MCP200. [DOI] [PubMed] [Google Scholar]
- 90.Fogarty M.P., Cannon M.E., Vadlamudi S., Gaulton K.J., Mohlke K.L. Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genet. 2014;10:e1004633. doi: 10.1371/journal.pgen.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kapoor A., Sekar R.B., Hansen N.F., Fox-Talbot K., Morley M., Pihur V., Chatterjee S., Brandimarto J., Moravec C.S., Pulit S.L., QT Interval-International GWAS Consortium An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am. J. Hum. Genet. 2014;94:854–869. doi: 10.1016/j.ajhg.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Visser M., Kayser M., Palstra R.J. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 94.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 95.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Van Deerlin V.M., Sleiman P.M., Martinez-Lage M., Chen-Plotkin A., Wang L.S., Graff-Radford N.R., Dickson D.W., Rademakers R., Boeve B.F., Grossman M. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finch N., Carrasquillo M.M., Baker M., Rutherford N.J., Coppola G., Dejesus-Hernandez M., Crook R., Hunter T., Ghidoni R., Benussi L. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van der Zee J., Van Langenhove T., Kleinberger G., Sleegers K., Engelborghs S., Vandenberghe R., Santens P., Van den Broeck M., Joris G., Brys J. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134:808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen-Plotkin A.S., Unger T.L., Gallagher M.D., Bill E., Kwong L.K., Volpicelli-Daley L., Busch J.I., Akle S., Grossman M., Van Deerlin V. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brady O.A., Zheng Y., Murphy K., Huang M., Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 2013;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Busch J.I., Unger T.L., Jain N., Tyler Skrinak R., Charan R.A., Chen-Plotkin A.S. Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum. Mol. Genet. 2016;25:2681–2697. doi: 10.1093/hmg/ddw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stagi M., Klein Z.A., Gould T.J., Bewersdorf J., Strittmatter S.M. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol. Cell. Neurosci. 2014;61:226–240. doi: 10.1016/j.mcn.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cruchaga C., Graff C., Chiang H.H., Wang J., Hinrichs A.L., Spiegel N., Bertelsen S., Mayo K., Norton J.B., Morris J.C., Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gallagher M.D., Suh E., Grossman M., Elman L., McCluskey L., Van Swieten J.C., Al-Sarraj S., Neumann M., Gelpi E., Ghetti B. TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 2014;127:407–418. doi: 10.1007/s00401-013-1239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vass R., Ashbridge E., Geser F., Hu W.T., Grossman M., Clay-Falcone D., Elman L., McCluskey L., Lee V.M., Van Deerlin V.M. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rhinn H., Abeliovich A. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst. 2017;4:404–415.e5. doi: 10.1016/j.cels.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 107.White C.C., Yang H.S., Yu L., Chibnik L.B., Dawe R.J., Yang J., Klein H.U., Felsky D., Ramos-Miguel A., Arfanakis K. Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med. 2017;14:e1002287. doi: 10.1371/journal.pmed.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]