Abstract
Small extracellular vesicles (sEVs) show promise as natural nano-devices for delivery of therapeutic RNA, but efficient loading of therapeutic RNA remains a challenge. We have recently shown that the attachment of cholesterol to small interfering RNAs (siRNAs) enables efficient and productive loading into sEVs. Here, we systematically explore the ability of lipid conjugates—fatty acids, sterols, and vitamins—to load siRNAs into sEVs and support gene silencing in primary neurons. Hydrophobicity of the conjugated siRNAs defined loading efficiency and the silencing activity of siRNA-sEVs complexes. Vitamin-E-conjugated siRNA supported the best loading into sEVs and productive RNA delivery to neurons.
Keywords: lipid-conjugated siRNA, hydrophobicity, small extracellular vesicles
Graphical Abstract
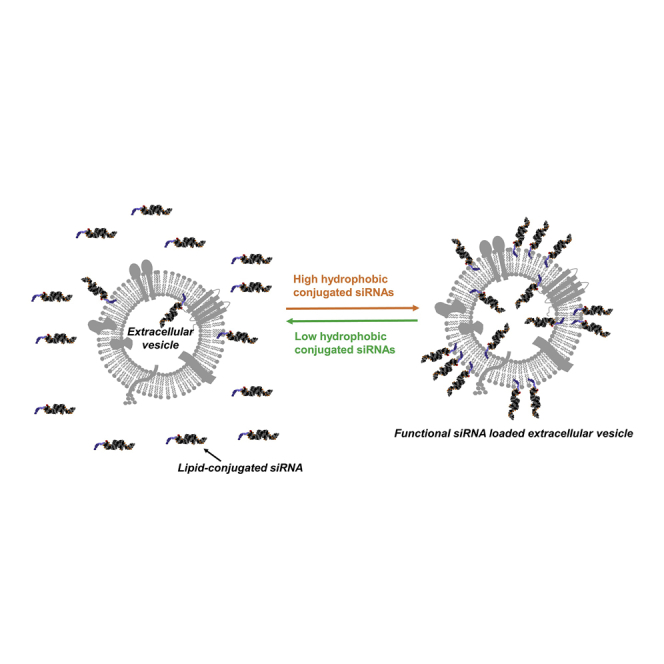
Biscans et al. developed a library of 14 lipid-conjugated siRNAs and evaluated their impact on small extracellular vesicle (sEV) loading and productive neuronal transfer. Four chemically diverse conjugates that support efficient RNA cargo loading into sEVs and target gene silencing in primary neurons were identified.
Introduction
Small extracellular vesicles (sEVs) are produced by most cell types and present in most body fluids (e.g., blood, saliva, urine, cerebrospinal fluid, and milk).1, 2, 3 They possess the ability to transport RNA, including mRNA and microRNA, over short and long intercellular distances, and thus empower sequence-specific, phenotype-modulating RNA types to act as a messenger.4, 5, 6, 7 The intercellular RNA trafficking mechanism via sEVs would make a powerful tool to fight disease when used to deliver therapeutic RNA.
Delivery of small interfering RNAs (siRNAs) to target cells remains an important challenge to their development as therapeutics.8, 9 Nanoparticle carriers have been explored as siRNA delivery vehicles.10, 11 Despite some clinical success, the characteristic toxicity, immunogenicity, and poor trafficking of nanoparticles has hampered further development as therapeutic RNA delivery vehicles.12, 13 By contrast, the natural RNA trafficking properties, low toxicity and immunogenicity, high stability in circulation, and target-cell specificity,14 of sEVs offer a promising alternative for efficient and selective delivery of siRNA to target cells.15, 16, 17, 18, 19
Loading RNA into sEVs remains a bottleneck for clinical application of sEVs as delivery vesicles for therapeutic RNA. The two most common loading strategies have been direct electroporation into the vesicles19, 20, 21 and transfection into EV source cells.22, 23 However, electroporation may induce vesicle damage and siRNA aggregation,24 transfection may disrupt EV integrity,19 and both methods lack robust batch-to-batch reproducibility and scalability. Recent studies have shown that the covalent conjugation of siRNA to a hydrophobic cholesterol moiety can drive efficient and controllable loading of siRNAs to sEVs, yielding thousands of copies of RNAs per vesicle.17, 18, 25 Conjugation of other hydrophobic moieties (e.g., α-tocopherol or docosahexaenoic acid) to siRNAs has been performed and tested for in vivo delivery to liver and brain.26, 27, 28, 29 However, hydrophobic moieties other than cholesterol have never been used in EV-mediated delivery of siRNAs.
We have synthesized a panel of lipid-conjugated hydrophobically modified siRNAs (hsiRNAs) to be loaded into sEVs to evaluate how the lipids affect the hsiRNA sEV loading efficiency. We found that hydrophobicity drives loading of lipid-conjugated hsiRNAs into sEVs. Moreover, the ability of sEV-loaded hsiRNAs to silence Huntingtin mRNA in primary murine cortical neurons correlates with the amount of lipid-conjugated hsiRNAs loaded into sEVs.
Results
Generation of a Library of Diverse Lipid-Conjugated hsiRNAs
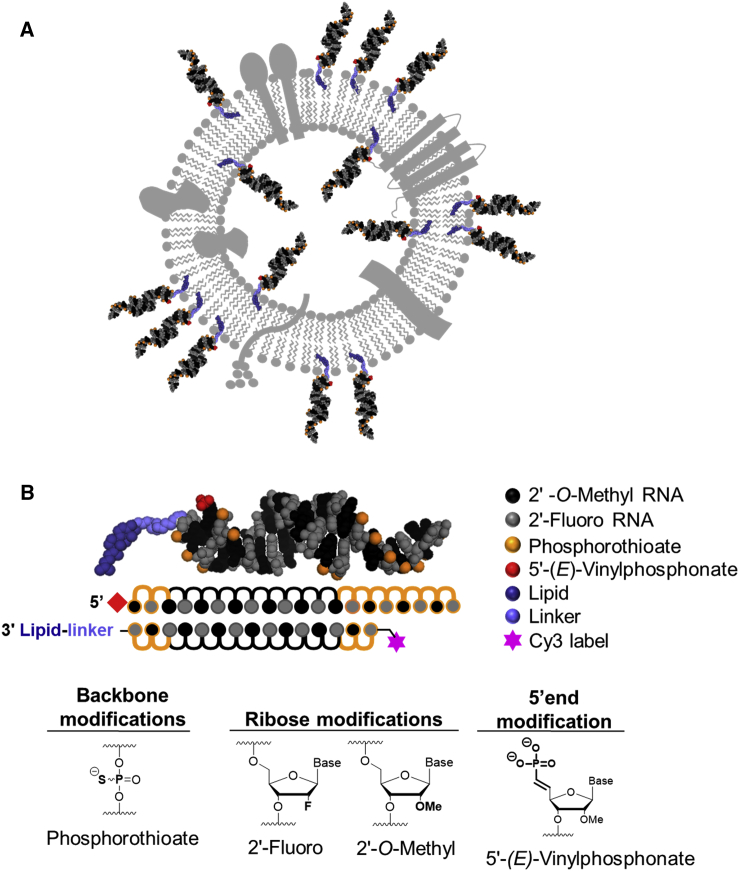
We have recently shown that cholesterol-conjugated chemically stabilized siRNAs efficiently associate with sEVs.18 We hypothesized that the hydrophobicity of cholesterol is the driving force behind loading of cholesterol-conjugated siRNAs into membranes (Figure 1A). To define the structure-function relationship between lipid conjugate and its ability to drive siRNA loading capacity into sEVs, we synthesized a library of lipid-conjugated siRNAs with a broad range of hydrophobicity. In each case, we used the same fully chemically stabilized asymmetric siRNA scaffold (i.e., hydrophobically modified siRNA or hsiRNA).30 hsiRNAs have a short duplex region (15 bp) and single-stranded fully phosphorothioate-modified tail that assists membrane association.31, 32 All riboses are fully chemically modified using an alternating 2′-O-methyl and 2′-fluoro modification pattern, which confers stability and minimizes innate immune activation.33, 34, 35 Moreover, the antisense strand is modified with a 5′-(E)-vinylphosphonate (E-VP) group that mimics the 5′-phosphate of the antisense strand to promote recognition by RNA-induced silencing complex (RISC)36, 37 and provides stability against phosphatases and exonucleases.38, 39, 40 Full chemical stabilization of hsiRNAs improves EV-mediated delivery of hsiRNAs (R.A.H., R.M., M.-C.D., A.B., J.F. Alterman, M.R. Hassler, L.R., D.E., E. Sapp, M. DiFiglia, N.A., and A.K., unpublished data). Compounds were labeled with Cy3 at the 5′ end of the sense strand, which allows visualization and quantification of hsiRNAs loaded into sEVs. The lipid conjugates were attached at the 3′ end of the sense strand (Figure 1B).
Figure 1.
Cholesterol-Conjugated hsiRNAs Load into sEVs
(A) Representation of sEV membrane loaded with cholesterol-conjugated hsiRNAs. Cholesterol is the driving force for efficient loading of siRNA into sEVs. (B) Schematic of hydrophobically modified siRNAs (hsiRNAs) is shown.
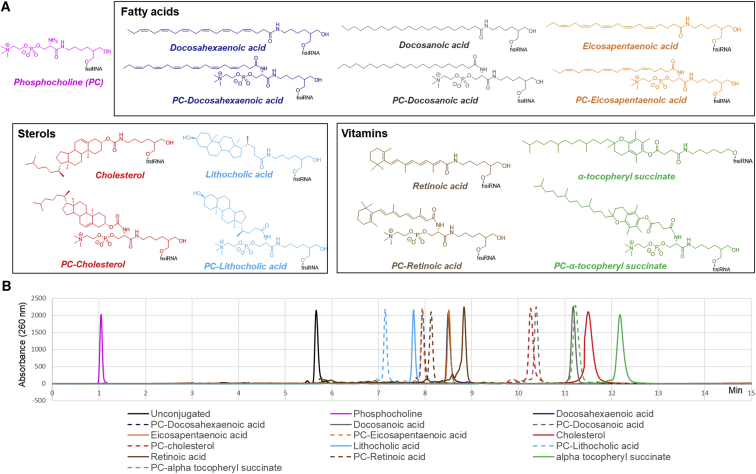
A wide range of natural lipids, such as fatty acids, sterols, and vitamins, were conjugated to the sense strand of hsiRNAHtt, which targets the Huntington’s disease gene (Table S1).41 In nature, many lipids are esterified (mostly phosphatidyl choline esters), which contributes to the specificity of cellular membrane interactions.42 To explore how esterification of the lipid conjugate affects hsiRNAs loading into sEVs, all lipid-conjugated hsiRNAs were synthesized with or without a phosphocholine (PC) head group. Because an ester bond is labile and incompatible with solid-phase oligonucleotide synthesis, we have recently developed a synthetic approach that allows a phosphocholine group to be attached to lipid moieties using an amide bond.29 The chemical compositions of synthesized lipid-conjugated hsiRNAs are depicted in Figure 2A. All lipid-conjugated hsiRNAs (with the exception of α-tocopheryl succinate hsiRNAs and PC-α-tocopheryl succinate hsiRNAs) were synthesized by using a functionalized solid support (Schemes S1A and S1B).28, 29 For phosphocholine-modified variants, the Fmoc (fluorenylmethyloxycarbonyl)-protected PC group (Scheme S1B, compound 7) was first attached to the C7-amino-functionalized solid support via a peptide bond followed by conjugation of respective lipids (Scheme S1B, solid supports 9).28, 29 Both α-tocopheryl succinate variants were synthesized using a post-synthetic conjugation between an amino group present at the 3′ end of the sense strand and the NHS (N-hydroxysuccinimide)-α-tocopheryl-succinate compound (Materials and Methods; Scheme S1C). For the synthesis of α-tocopheryl succinate sense strands, a commercial C6 amino solid support was used to synthesize the strands. For PC-α-tocopheryl succinate sense strands, the PC amino solid support 8 (Scheme S1B) was used to synthesize the strands.
Figure 2.
The Chemical Compositions of Lipid Conjugates Significantly Affect hsiRNA Hydrophobicity
(A) Library of lipophilic moieties attached to hsiRNAsHtt. (B) HPLC retention time of lipid-conjugated Cy3-hsiRNAHtt sense strands is shown. C18, buffer A, 0.1 M triethylammonium acetate in water; buffer B, acetonitrile; gradient, 0%–100% in B in 15 min; temperature, 60°C; flow, 1 mL/min.
The relative hydrophobicity of a molecule can be determined by its retention time in reverse-phase high-performance liquid chromatography (HPLC). Hydrophobicity increases with retention time.43 Figure 2B shows HPLC traces for synthesized sense strands, with retention times varying between 1 and 13 min, indicating a broad range of hydrophobicity.44 The structure of the conjugate principally contributed to hsiRNA retention time and hydrophobicity. Compounds with a saturated carbon chain (cholesterol, docosanoic acid, and α-tocopheryl succinate) were more hydrophobic than those with an unsaturated carbon chain (docosahexaenoic acid and eicosapentaenoic acid) or cyclic structure (lithocholic acid and retinoic acid). The incorporation of a polar head (phosphocholine group) decreased the hydrophobicity of all lipid-conjugated hsiRNAs. This synthetic library of 15 different lipid-conjugated hsiRNAs, covering a broad range of lipid structures and hydrophobicity, allowed us to examine how lipid structure affects efficiency of hsiRNA loading into sEVs.
Loading Efficiency of Conjugated hsiRNAs into sEVs Correlates with hsiRNA Hydrophobicity
sEVs were isolated by differential ultracentrifugation from Wharton’s-jelly-derived (umbilical cord) mesenchymal stem cells.45 They displayed uniform size distribution (mean, 140 nm; Figure S2A). Small EVs appeared as lipid-bilayer-surrounded vesicles on transmission electron microscopy (Figure S2B). Western blot (Figure S2C) and liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Figure S2D) showed enrichment in positive EV marker proteins (CD63, CD81, CD9, Alix, tetraspanin-14, and Tsg101) and depletion in negative EV marker proteins (calnexin, calreticulin, cytochrome C, and HNRPK). Thus, small EVs used in this study are bona fide extracellular vesicles according to the guidelines established by International Society of Extracellular Vesicles.46
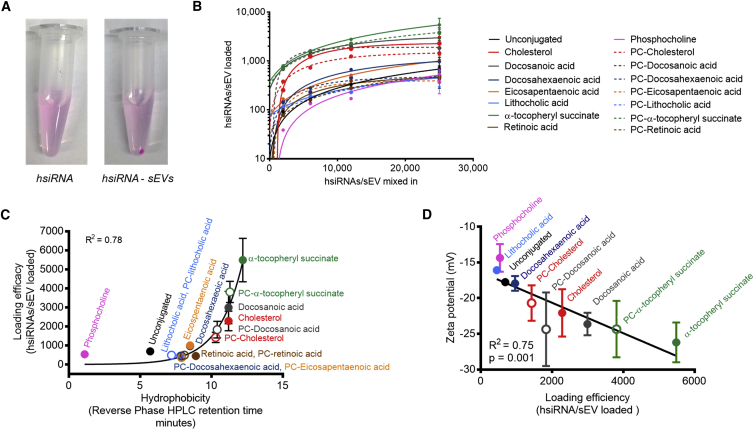
Cholesterol-conjugated hsiRNAs were more efficient at inducing Huntingtin mRNA silencing in neurons when they were delivered via small EVs (e.g., 100,000 g fraction of differential ultracentrifugation protocol) compared to cholesterol-hsiRNA alone (Figure S3). Cholesterol-hsiRNAs did not silence target mRNA when delivered via large EVs (e.g., 10,000 g fraction; Figure S3). Therefore, we used small EVs (e.g., 100,000 g fraction) to test delivery of all conjugated hsiRNA to neurons. sEVs were co-incubated with increasing concentrations of Cy3-hsiRNAHtt conjugated to the above described lipids (1:2,000, 1:6,000, 1:12,000, and 1:25,000 sEV-to-hsiRNA ratios). Ultracentrifugation of Cy3-hsiRNA-sEV mixture resulted in a fluorescent pink pellet, revealing the association of labeled hsiRNA with sEVs (Figure S4). Ultracentrifugation of hsiRNAsHtt without sEV did not generate a pellet, indicating the absence of hsiRNA aggregation (Figure 3A). The integrity of the sEV membrane after hsiRNA loading was confirmed by using transmission electron microscopy (Figure S2B).
Figure 3.
The Number of hsiRNAs Loaded into sEVs Depends on the Hydrophobicity of the Conjugate
(A) Ultracentrifugation of conjugated Cy3-hsiRNAs incubated without sEV (left) showing absence of pellet and after co-incubation of Cy3-hsiRNAs and sEVs (right) showing formation of pellet. Representative pictures are shown. (B) Cy3-hsiRNA accumulation in pellet following co-incubation of hsiRNAs and sEVs with varying hsiRNA:sEV ratio is shown (n = 3; mean ± SEM for the last point). (C) Exponential relationship between loading efficiency and hydrophobicity of conjugated hsiRNA is shown (n = 3; mean ± SEM). (D) Linear correlation between the surface charge of hsiRNA-loaded sEVs and hsiRNA loading efficiency (n = 2; mean ± SEM).
The efficiency of Cy3-hsiRNA loading into sEVs was quantified by spectrophotometry (Figure 3B). Increasing hsiRNA-to-sEV ratios yielded higher loading efficiencies with saturation kinetics: at a 1:25,000 sEV-to-hsiRNA ratio, loading was nearly saturated for each lipid-conjugated hsiRNA. The loading efficiency depended on the structure of the lipid conjugate attached to the hsiRNA. A strong exponential correlation was observed between the hydrophobicity of the lipid-conjugated hsiRNA and the sEV loading efficiency (Figure 3C). Thus, hydrophobicity of a hsiRNA directly predicts the number of molecules that can be loaded into sEVs. Efficient loading (at least 1,700 hsiRNAs per vesicle for a 1:25,000 ratio) required the presence of a highly hydrophobic conjugate (cholesterol, PC-cholesterol, docosanoic acid, PC-docosanoic acid, α-tocopheryl succinate, and PC-α-tocopheryl succinate) attached to the hsiRNA. Unsaturated fatty acid chains (docosahexaenoic acid and eicosapentaenoic acid) conferred less hydrophobicity and therefore less vesicle loading efficiency to hsiRNAs. Conjugation of α-tocopheryl succinate (vitamin E) to hsiRNA yielded the best sEV loading efficiency, outperforming cholesterol. Docosanoic acid was as effective as cholesterol at loading hsiRNAs into sEVs. Despite being structurally similar to cholesterol, the reverse sterol conjugate formed by lithocholic acid is not as efficient as the cholesterol conjugate, indicating that the saturated carbon tail of cholesterol is important for sEV loading. These data suggest the model that anchoring of hsiRNA to the surface of EV is mediated by the insertion of the saturated carbon chain into the vesicular membrane.
Consistent with our previous studies,18 the loading of lipid-conjugated hsiRNAs reduced the zeta potential of sEVs (Figure 3D), indicating the presence of negatively charged hsiRNAs on the surfaces of vesicles. We observed a linear correlation between zeta potential and hsiRNA loading efficiency: the lower the surface charge, the higher the loading efficiency. α-tocopheryl succinate (vitamin E)-conjugated hsiRNA-loaded sEVs had the highest hsiRNA-to-sEV ratio and displayed a zeta potential of −35 mV, a significant change relative to unloaded sEVs (−13 mV). We have shown previously that the majority of hsiRNAs are bound to the outside of the sEVs.18 However, because the hsiRNAs are stable against RNases in vitro and in vivo,33, 38 the presence of hsiRNAs on the surface of sEVs should not reduce siRNA activity.
Of 15 lipid-conjugated hsiRNA variants evaluated, we identified five additional lipid-conjugated hsiRNAs that associate with sEVs as well as or better than cholesterol-conjugated hsiRNA: PC-cholesterol; docosanoic acid; PC-docosanoic acid; α-tocopheryl succinate; and PC-α-tocopheryl succinate hsiRNAs. The degree of hydrophobicity of the lipid-conjugated hsiRNA seems to define the efficiency of loading into sEVs. Loading efficiency of lipid-conjugated hsiRNAs correlates with a decrease in sEV surface charge, which indicates the amount of hsiRNAs bound to the surface of the sEV.
Lipid-Conjugated hsiRNA-Loaded sEVs Induce Gene Silencing in Primary Mouse Neurons
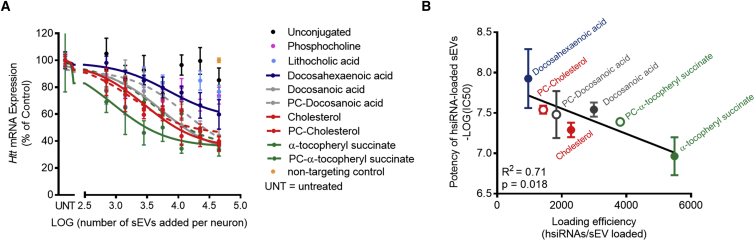
We next asked whether the change in the charge and perhaps other properties of sEV membranes loaded with hsiRNA affect the trafficking of sEVs to target cells. We incubated primary mouse neurons with increasing concentrations of sEVs loaded with lipid-conjugated hsiRNAHtt for one week and measured Htt mRNA levels (Figure 4A). We assigned lipid-conjugated hsiRNAs to groups based on their hydrophobicity and chose to test only a subset of representative hsiRNAs: (1) low hydrophobicity: phosphocholine, lithocholic acid, and unconjugated hsiRNAs; (2) medium hydrophobicity: docosahexaenoic acid; and (3) high hydrophobicity: docosanoic acid, PC-docosanoic acid, cholesterol, PC-cholesterol, α-tocopheryl succinate, and PC-tocopheryl succinate. Only lipid-conjugated hsiRNAs with high hydrophobicity resulted in a visible pink pellet upon loading (Figure S4). Dose-dependent silencing of Htt mRNA was observed for medium- and high-hydrophobicity lipid-conjugated hsiRNAs loaded into sEVs (Figure 4A). Non-conjugated hsiRNAs or lipid-conjugated hsiRNAs with low hydrophobicity did not induce silencing when loaded into sEVs. These results are consistent with hsiRNA levels detected in neurons (Figure S5). Medium and high hydrophobicity enabled the accumulation of 2- to 6-fold more hsiRNAs in neurons than low-hydrophobic compounds when delivered via sEVs.
Figure 4.
Silencing Activity of hsiRNA-Loaded sEVs Correlates with the Loading Efficiency of hsiRNAs
(A) Htt mRNA levels in primary mouse neurons incubated with increasing concentrations of hsiRNAHtt-loaded sEVs (sEV:hsiRNA ratio, 1:25,000) for one week. Htt mRNA levels were normalized to Hprt (hypoxanthine-guanine phosphoribosyl transferase) and presented as percent of untreated control (n = 3; mean ± SEM). UNT, untreated. (B) Correlation between IC50 of hsiRNA-loaded sEVs and loading efficiency of hsiRNAs is shown (n = 2).
In general, only hydrophobic conjugates that support loading of more than 1,000 hsiRNAs per vesicle enabled productive silencing (Figure 3B). sEVs loaded with lipid-conjugated hsiRNAs induced sequence-specific silencing, because sEVs loaded with non-targeting control hsiRNAs of similar chemical composition were ineffective (Figure 4A). We observed a linear correlation between the half-maximal inhibitory concentration (IC50) of hsiRNA-loaded sEVs and the amount of loaded hsiRNAs, which defines the activity of hsiRNA-loaded sEVs (Figure 4B). Thus, hydrophobic conjugates that efficiently load hsiRNAs into sEVs induce productive silencing of Htt mRNA in primary neurons. The direct correlation between efficiency of loading and silencing indicates that, independent of the structure of the conjugate, loaded hsiRNAs can induce functional silencing.
Discussion
The simple and scalable loading of cholesterol-conjugated siRNAs provides an attractive strategy for loading RNA cargo into sEVs.17, 18, 25 We used the principle to expand the range of lipid conjugates that can drive efficient loading of therapeutic RNA into sEVs. Notably, docosanoic acid and α-tocopheryl succinate and their PC derivatives supported siRNA loading into sEVs as well as or better than cholesterol.
Because loading into sEVs is proportional to hydrophobicity, a range of available lipid conjugates enables dynamic modulation of RNA cargo levels in sEVs for a range of applications. Particle charge may influence pharmacokinetic behavior in vivo or interfere with the natural trafficking pathways of sEVs. Titrating the amount of RNA cargo in sEVs will therefore be essential to accurately set the charge and function of the vesicle.
We expect that sEVs purified from various sources will be loaded by lipid-conjugated siRNAs. Indeed, in addition to Wharton’s-jelly-derived mesenchymal stem cells (umbilical cord) used in this study, we have successfully used cholesterol-conjugated siRNAs to load sEVs from U87 glioblastoma cells,18 bEND3 endothelial polyoma cells, and bone-marrow-derived and adipose-derived mesenchymal stem cells. Nevertheless, because hsiRNA loading is driven by lipid-conjugate hydrophobicity, the exact level of loading may depend on the specific membrane composition and therefore cell source47, 48 of the sEVs. Future work is needed to identify the optimal hsiRNA for a given sEV source.
The structure-activity relationship between lipid conjugates and sEV loading of siRNA creates a framework for the rational design of RNA cargo for sEV delivery. The covalent lipid-conjugation strategy could be used to load sEVs with other types of therapeutic oligonucleotides, including CRISPR guide RNAs or microRNAs (miRNAs). Using lipid-conjugated small RNA tethers, the loading of larger oligonucleotide species might also be possible.
Finally, we observed a correlation between the sEV loading efficiency of hsiRNAs and target gene silencing in neurons. We believe that sEVs loaded with lipid-conjugated siRNA could be used to deliver therapeutic nucleic acids to recipient cells other than neurons. Indeed, sEVs administered by injection efficiently distribute in the brain18, 49, 50, 51 and other tissues.14, 52, 53, 54 Thus, a simple and scalable method of efficiently loading therapeutic oligonucleotides into sEVs is a significant advance toward the treatment of neurodegenerative disorders,55 inflammatory diseases,56, 57 and cancer.52
Materials and Methods
Additional detailed material and methods can be found in Supplemental Information.
Preparation of Oligonucleotides
Oligonucleotides were synthesized on an Expedite ABI DNA/RNA Synthesizer following standard protocols. Each synthesis was done at a 1-μmol scale using the different synthesized lipophilic conjugated CPG28, 29 (Schemes S1A and S1B, compound 4, 8, or 9) or a commercial C6 amino CPG (ChemGenes, Wilmington, MA; for the post-synthetic conjugation of α-tocopheryl succinate) for the sense strand or at a 10-μmol scale using a Unylinker terminus (ChemGenes, Wilmington, MA) for the antisense strand. 2′-O-methyl phosphoramidites (ChemGenes, Wilmington, MA), 2′-fluoro phosphoramidites (BioAutomation, Irving, Texas), Cy3-labeled phosphoramidites (Gene Pharma, Shanghai, China), and synthesized E-Vinyl Phosphonate phosphoramidites38 were prepared as 0.15 M solutions in acetonitrile. Phosphoramidite coupling time was 250 s for all amidites using 5-(benzylthio)-1H-tetrazole (BTT) 0.25 M in acetonitrile as coupling activator. Detritylations were performed using 3% dichloroacetic acid (DCA) in dichloromethane for 80 s, and capping was done with a 16% N-methylimidazole in tetrahydrofuran (THF) (CAP A) and THF:acetic anhydride:2,6-lutidine, (80:10:10, v/v/v; CAP B) for 15 s. Sulfurizations were carried out with 0.1 M solution of 1,2,4-dithiazole-5-hione (DDTT) in acetonitrile for 3 min. Oxidation was performed using 0.02 M iodine in THF:pyridine:water (70:20:10; v/v/v) for 80 s.
Deprotection and Purification of Oligonucleotides
Sense strands were cleaved and deprotected using 1 mL of 40% aqueous (aq.) methylamine at 45°C for 1 hr. Antisense strands were first deprotected with a solution of bromotrimethylsilane/pyridine (3:2; v/v) in dichloromethane (5 mL) for the E-Vinyl Phosphonate deprotection and then cleaved and deprotected with 10 mL of 40% aq. methylamine at 45°C for 1 hr. For both sense and antisense strands, the oligonucleotide solutions were frozen in liquid nitrogen for a few minutes and dried under vacuum in a Speedvac overnight. The resulting pellets were suspended in water and purified using an Agilent Prostar System (Agilent Technologies, Santa Clara, CA). For the sense strand, a Hamilton HxSil C18 column (150 × 21.2) was used (conditions: buffer A: 50 mM sodium acetate in water with 5% acetonitrile; buffer B: acetonitrile; gradient = 90% A, 10% B to 10% A, 90% B in 18 min; temperature: 70°C; flow rate: 5 mL/min), and for the antisense strand, a Dionex NucleoPac PA-100 (9 × 250) was used (conditions = buffer A: 30% acetonitrile in water; buffer B: 1 M perchlorate de sodium in water with 30% acetonitrile; gradient: 100% A to 20% A, 80% B in 30 min; temperature: 65°C; flow: 10 mL/min). The pure oligonucleotides were collected, desalted by size-exclusion chromatography using a Sephadex G25 column (GE Healthcare Life Sciences, Marlborough, MA) and lyophilized. For the attachment of α-tocopheryl succinate variants, the 3′ end C6 amino or the 3′ PC amino sense strands were dissolved in water and a solution of 1 M sodium bicarbonate (pH 8.5) was added to obtain a 0.1 M sodium bicarbonate final concentration. Then, a solution of N-hydroxysuccinimide-α-tocopheryl succinate (10–100 equivalents; Scheme S1) in dimethylformamide (DMF) was added to the solutions containing the sense strand. The mixtures were incubated overnight at room temperature. A solution of 3 M sodium acetate (pH 5.2) was added to obtain a 0.3 M sodium acetate final concentration. Then, 3× of ethanol (95%) of the whole volume were added. The mixtures were vortexed, placed at 80°C for 1 hr, and centrifuged 30 min at 5,200 g. The supernatants were removed, and the lipid-conjugated sense strands were purified and desalted as described previously.
Analysis of Oligonucleotides
The identity of oligonucleotides was established by LC-MS analysis on an Agilent 6530 accurate-mass Q-TOF LC/MS (Agilent, Santa Clara, CA) using the following conditions: buffer A (9 mM triethylamine/100 mM hexafluoroisopropanol in water); buffer B (9 mM triethylamine/100 mM hexafluoroisopropanol in MeOH); column: Agilent AdvanceBio oligonucleotides 2.1 × 50 mm (Agilent, Santa Clara, CA); gradient for sense strand: 0–2 min (1% B–40% B) and 2–10.5 min (40% B–100% B); and gradient for the antisense strand: 0–2 min (1% B–12% B), 2–10.5 min (12% B–30% B), and 10.5–11 min (30% B–100% B). Sample LC-MS chromatograms of both sense strand and antisense strand are shown in Figure S1.
Cell Culture
Umbilical cord, Wharton’s-jelly-derived mesenchymal stem cells (ATCC PCS-500-010) were cultured in appropriate stem cell medium (ATCC PCS-500-030) in the presence of 2% fetal bovine serum (FBS) and growth factors (ATCC PCS-500-040) at 37°C, 5% CO2. Medium was changed every three days and cells expanded until passage 9 to reach a total of 3,000 cm2 surface in T500 triple flasks.
Isolation of sEVs
Medium on umbilical-cord-derived mesenchymal stem cells was changed to RPMI (Gibco RPMI 1640; Thermo Fisher Scientific) with no FBS or other supplements added for 24 hr. sEVs were then purified from this conditioned RPMI supernatant via differential ultracentrifugation as described previously.18 Briefly, cell debris was pelleted at 300 g (10 min). Then microvesicles were pelleted at 10,000 g (30 min), supernatant filtered through a 0.2-μm membrane (Nalgene aPES; Thermo Fisher Scientific), and sEVs pelleted at 100,000 g (90 min). sEVs were then washed with PBS once and finally frozen for later use in 0.1 M sucrose and 1× protease inhibitor (cOmplete Mini; Sigma-Aldrich) in PBS.
Characterization of sEVs
Concentration and size distribution of sEVs were measured by Nanoparticle Tracking Analysis (NanoSight NS300; Malvern). Briefly, samples were diluted in PBS 1:100–1:1,000, manually injected into the instrument and videos acquired at ambient temperature at camera level 9 for 1 min per sample, n = 3. sEVs were then frozen at −80°C in 0.1 M sucrose and protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO; No. P8340) until further use. Surface charge of unloaded or loaded sEVs was measured using a Zetasizer Nano ZS (Malvern) following dilution of samples in 1 mL water in a universal glass cuvette Dip Cell kit (Malvern; No. ZEN1002). Transmission electron microscopy of sEVs was conducted at Mass General Hospital using a JEOL 1100 transmission electron microscope (JEOL, Peabody, MA) at 60 kV as described previously.47 LC-MS/MS proteomics of sEVs and source cells was conducted at the Mass Spectrometry and Proteomics Facility of University of Massachusetts Medical School as described previously (Figure S2).47 For western blot analyses, sEV or cell pellets were suspended in radioimmunoprecipitation assay (RIPA) buffer (Pierce 899000; Thermo Fisher Scientific, Waltham, MA) containing PMSF (36978; Thermo Fisher Scientific) and protease inhibitor cocktail (cOmplete Mini; 11836153001; Roche, Indianapolis, IN), and samples were sonicated for 15 min. Insoluble material was pelleted by centrifugation for 15 min at 10,000 g, 4°C. Proteins (10–50 μg) were loaded onto NuPAGE 4%–12% Bis-Tris gels (Thermo Fisher Scientific, Waltham, MA). After transfer to polyvinylidene fluoride (PVDF) (Bio-Rad, Hercules, CA), membranes were incubated with antibodies and washed, and images were captured using an Odyssey system (LI-COR Biosciences, Bad Homburg, Germany) according to manufacturer’s instructions. Primary antibodies used were calnexin (C5C9; Cell Signaling Technology, Danvers, MA), CD63 (H5C6; BD BioSciences, San Jose, CA), and CD81 (B11; Santa Cruz Biotechnology, Dallas, TX). In order to facilitate replication, the sEV methods can be found in EV-TRACK.58
sEV Loading with hsiRNAs
sEVs were thawed at room temperature and co-incubated with known amount of hsiRNA at 37°C for one hour in 500 μL PBS. Then, the sEV-hsiRNA mixture was centrifuged at 100,000 g for 90 min and supernatant containing unloaded hsiRNA removed (supernatant). Pellet was taken up in 500 μL PBS for fluorescence measurement or in 300 μL Neural Q medium for treatment of primary neurons. To quantify loading, a 200-μL aliquot taken from resuspended loaded sEV pellet or from the supernatant fluorescence was measured at 550 nm excitation, 570 nm emission on TECAN instrument. Percent of loaded hsiRNA was calculated as follows: pellet/(pellet + supernatant). To estimate hsiRNA copy number per sEV, the following formula was used: (percent of loaded hsiRNA) × (amount of hsiRNA initially mixed in with sEVs [mol]) × (Avogadro number)/(number of sEVs initially mixed in).
Preparation of Primary Cortical Neurons
Primary cortical neurons were isolated from embryonic day 15.5 (E15.5) mouse embryos of wild-type FVBNj mice. Pregnant females were anesthetized by intraperitoneal injection of ketamine (100 mg/kg; KETASET; Zoetis, Kalamazoo, MI)-xylazine (10 mg/kg; AnaSed; AKORN, Laker Forest, IL; No. NDC59399-111-50) followed by cervical dislocation. Embryos were removed and transferred to ice-cold DMEM/F12 medium (Invitrogen, Carlsbad, CA; No. 11320). Brains were removed, and meninges were carefully detached. Cortices were isolated and transferred into pre-warmed papain solution for 25 min at 37°C, 5% CO2 to dissolve the tissue. Papain (Worthington, Lakewood, NJ; No. 54N15251) was dissolved in 2 mL Hibernate E (Brainbits, Springfield, IL; No. HE) and supplemented with 0.25 mL of 10 mg/mL DNase1 (Worthington, Lakewood, NJ; No. 54M15168) in Hibernate E. After 30 min incubation, the papain solution was removed and 1 mL NeuralQ (Sigma-Aldrich, St. Louis, MO; No. N3100) supplemented with 2.5% FBS was added to the tissue. Tissues were then dissociated by trituration through a fire-polished, glass Pasteur pipet. Neurons were counted and diluted at 106 cells/mL. 105 neurons per well were plated on 96-well plates pre-coated with poly-L-lysine (BD BIOCOAT, Corning, NY; No. 356515). After overnight incubation at 37°C, 5% CO2, an equal volume of NeuralQ supplemented with anti-mitotics, 0.484 μL/mL of 5′UtP (Sigma, St Louis, MO; No. U6625) and 0.2402 μL/mL of 5′FdU (Sigma, St Louis, MO; No. F3503) was added to prevent the growth of non-neuronal cells. Half of the volume of media was replaced with fresh NeuralQ containing anti-mitotic every 48 hr until the experiments were performed.
Measurement of hsiRNA Silencing Activity in Neurons
Neurons were treated with hsiRNA-loaded sEVs and incubated for 7 days at 37°C, 5% CO2 post-treatment. Neurons were then lysed and mRNA quantification was performed using the QuantiGene 2.0 assay kit (Affymetrix; No. QS0011) as described previously.59 Catalog numbers for probes used in QuantiGene 2.0 assay kit are as follows: mouse Htt (Affymetrix; No. SB-14150) and mouse Hprt (Affymetrix; No. SB-15463). Datasets were normalized to housekeeping gene Hprt. Each measurement was run in triplicates.
Measurement of hsiRNA Uptake to Neurons
hsiRNA guide strands in neuron cell lysates were quantified using a peptide-nucleic acid (PNA) hybridization assay.28, 29, 38, 60 PNAs are oligonucleotides, where the sugar-phosphate backbone is replaced with a polyamide backbone. PNAs have no charge and have a high hybridization energy to RNA. SDS from leftover neuron lysates after mRNA quantification was precipitated with 3 M KCl and pelleted at 4,000 × g for 15 min. hsiRNA guide strands in cleared supernatant were hybridized to fully complementary Cy3-labeled PNA strands (PNABio, Thousand Oaks, CA). hsiRNA-PNA duplexes were injected into HPLC DNAPac PA100 anion exchange column (Thermo Scientific, Carlsbad, CA), and Cy3 fluorescence was monitored and peaks integrated. The mobile phase for HPLC was 50% water, 50% acetonitrile, 25 mM Tris-HCl (pH 8.5), and 1 mM EDTA, and the salt gradient was 0–800 mM NaClO4. For the calibration curve, a known amount of hsiRNA duplex was spiked into cell lysis solution.
Author Contributions
A.B. designed and A.B. and D.E. synthesized all compounds. R.A.H., R.M., and M.-C.D. conducted all sEVs experiments. M.N. takes part in the development of DHA compounds and L.R. in the development of E-VP antisense strands. A.K., N.A., A.B., and R.A.H. came up with the concept. A.B. wrote the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by an NIH UH3 grant (TR 000888 05), NIH grants (RO1GM10880304, RO1NS10402201, and S10 OD020012), and the CHDI Foundation (Research Agreement A-6119, JSC A6367). We thank Darryl Conte for help with the manuscript writing and editing.
Footnotes
Supplemental Information includes one scheme, five figures, and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.03.019.
Contributor Information
Neil Aronin, Email: neil.aronin@umassmed.edu.
Anastasia Khvorova, Email: anastasia.khvorova@umassmed.edu.
Supplemental Information
References
- 1.Simpson R.J., Jensen S.S., Lim J.W. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 2.van Niel G., Porto-Carreiro I., Simoes S., Raposo G. Exosomes: a common pathway for a specialized function. J. Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 3.Henderson M.C., Azorsa D.O. The genomic and proteomic content of cancer cell-derived exosomes. Front. Oncol. 2012;2:38. doi: 10.3389/fonc.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 5.Muralidharan-Chari V., Clancy J.W., Sedgwick A., D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitt J.M., Kroemer G., Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J. Clin. Invest. 2016;126:1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simons M., Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Nair J.K., Willoughby J.L.S., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 9.Burnett J.C., Rossi J.J., Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antimisiaris S., Mourtas S., Papadia K. Targeted si-RNA with liposomes and exosomes (extracellular vesicles): how to unlock the potential. Int. J. Pharm. 2017;525:293–312. doi: 10.1016/j.ijpharm.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 11.Parlea L., Puri A., Kasprzak W., Bindewald E., Zakrevsky P., Satterwhite E., Joseph K., Afonin K.A., Shapiro B.A. Cellular delivery of RNA nanoparticles. ACS Comb. Sci. 2016;18:527–547. doi: 10.1021/acscombsci.6b00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue H.Y., Liu S., Wong H.L. Nanotoxicity: a key obstacle to clinical translation of siRNA-based nanomedicine. Nanomedicine (Lond.) 2014;9:295–312. doi: 10.2217/nnm.13.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akhtar S. Cationic nanosystems for the delivery of small interfering ribonucleic acid therapeutics: a focus on toxicogenomics. Expert Opin. Drug Metab. Toxicol. 2010;6:1347–1362. doi: 10.1517/17425255.2010.518611. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vader P., Mol E.A., Pasterkamp G., Schiffelers R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;106(Pt A):148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Greco K.A., Franzen C.A., Foreman K.E., Flanigan R.C., Kuo P.C., Gupta G.N. PLK-1 silencing in bladder cancer by siRNA delivered with exosomes. Urology. 2016;91:241.e1–241.e7. doi: 10.1016/j.urology.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 17.O’Loughlin A.J., Mäger I., de Jong O.G., Varela M.A., Schiffelers R.M., El Andaloussi S., Wood M.J.A., Vader P. Functional delivery of lipid-conjugated siRNA by extracellular vesicles. Mol. Ther. 2017;25:1580–1587. doi: 10.1016/j.ymthe.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Didiot M.C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R. Exosome-mediated delivery of hydrophobically modified siRNA for Huntingtin mRNA silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahlgren J., De L Karlson T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 21.Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akao Y., Iio A., Itoh T., Noguchi S., Itoh Y., Ohtsuki Y., Naoe T. Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 2011;19:395–399. doi: 10.1038/mt.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shtam T.A., Kovalev R.A., Varfolomeeva E.Y., Makarov E.M., Kil Y.V., Filatov M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kooijmans S.A., Stremersch S., Braeckmans K., de Smedt S.C., Hendrix A., Wood M.J., Schiffelers R.M., Raemdonck K., Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release. 2013;172:229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Stremersch S., Vandenbroucke R.E., Van Wonterghem E., Hendrix A., De Smedt S.C., Raemdonck K. Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release. 2016;232:51–61. doi: 10.1016/j.jconrel.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 27.Murakami M., Nishina K., Watanabe C., Yoshida-Tanaka K., Piao W., Kuwahara H., Horikiri Y., Miyata K., Nishiyama N., Kataoka K. Enteral siRNA delivery technique for therapeutic gene silencing in the liver via the lymphatic route. Sci. Rep. 2015;5:17035. doi: 10.1038/srep17035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikan M., Osborn M.F., Coles A.H., Godinho B.M., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids. 2016;5:e344. doi: 10.1038/mtna.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikan M., Osborn M.F., Coles A.H., Biscans A., Godinho B.M.D.C., Haraszti R.A., Sapp E., Echeverria D., DiFiglia M., Aronin N., Khvorova A. Synthesis and evaluation of parenchymal retention and efficacy of a metabolically stable O-Phosphocholine-N-docosahexaenoyl-l-serine siRNA conjugate in mouse brain. Bioconjug. Chem. 2017;28:1758–1766. doi: 10.1021/acs.bioconjchem.7b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassler M.R., Turanov A.A., Alterman J.F., Haraszti R.A., Coles A.H., Osborn M.F., Echeverria D., Nikan M., Salomon W.E., Roux L. Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018;46:2185–2196. doi: 10.1093/nar/gky037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Ly S., Navaroli D.M., Didiot M.C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017;45:15–25. doi: 10.1093/nar/gkw1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 34.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 38.Haraszti R.A., Roux L., Coles A.H., Turanov A.A., Alterman J.F., Echeverria D., Godinho B.M.D.C., Aronin N., Khvorova A. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45:7581–7592. doi: 10.1093/nar/gkx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parmar R., Willoughby J.L.S., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA–GalNAc conjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 40.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically modified siRNAs silence Huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith M., Jungalwala F.B. Reversed-phase high performance liquid chromatography of phosphatidylcholine: a simple method for determining relative hydrophobic interaction of various molecular species. J. Lipid Res. 1981;22:697–704. [PubMed] [Google Scholar]
- 44.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 45.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 46.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haraszti R.A., Didiot M.-C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao F., Narain N.R., DiFiglia M., Kiebish M.A. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffiths S.G., Cormier M.T., Clayton A., Doucette A.A. Differential proteome analysis of extracellular vesicles from breast cancer cell lines by chaperone affinity enrichment. Proteomes. 2017;5:E25. doi: 10.3390/proteomes5040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frühbeis C., Fröhlich D., Kuo W.P., Amphornrat J., Thilemann S., Saab A.S., Kirchhoff F., Möbius W., Goebbels S., Nave K.A. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Abels E.R., Redzic J.S., Margulis J., Finkbeiner S., Breakefield X.O. Potential transfer of polyglutamine and CAG-repeat RNA in extracellular vesicles in Huntington’s disease: background and evaluation in cell culture. Cell. Mol. Neurobiol. 2016;36:459–470. doi: 10.1007/s10571-016-0350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krämer-Albers E.M. Ticket to ride: targeting proteins to exosomes for brain delivery. Mol. Ther. 2017;25:1264–1266. doi: 10.1016/j.ymthe.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen S., Dooner M., Cheng Y., Papa E., Del Tatto M., Pereira M., Deng Y., Goldberg L., Aliotta J., Chatterjee D. Mesenchymal stromal cell-derived extracellular vesicles rescue radiation damage to murine marrow hematopoietic cells. Leukemia. 2016;30:2221–2231. doi: 10.1038/leu.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beltrami C., Besnier M., Shantikumar S., Shearn A.I., Rajakaruna C., Laftah A., Sessa F., Spinetti G., Petretto E., Angelini G.D., Emanueli C. Human pericardial fluid contains exosomes enriched with cardiovascular-expressed microRNAs and promotes therapeutic angiogenesis. Mol. Ther. 2017;25:679–693. doi: 10.1016/j.ymthe.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuyama K., Sun H., Mitsutake S., Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012;287:10977–10989. doi: 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotzerke K., Mempel M., Aung T., Wulf G.G., Urlaub H., Wenzel D., Schön M.P., Braun A. Immunostimulatory activity of murine keratinocyte-derived exosomes. Exp. Dermatol. 2013;22:650–655. doi: 10.1111/exd.12230. [DOI] [PubMed] [Google Scholar]
- 57.Sheng H., Hassanali S., Nugent C., Wen L., Hamilton-Williams E., Dias P., Dai Y.D. Insulinoma-released exosomes or microparticles are immunostimulatory and can activate autoreactive T cells spontaneously developed in nonobese diabetic mice. J. Immunol. 2011;187:1591–1600. doi: 10.4049/jimmunol.1100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Deun J., Mestdagh P., Agostinis P., Akay Ö., Anand S., Anckaert J., Martinez Z.A., Baetens T., Beghein E., Bertier L., EV-TRACK Consortium EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods. 2017;14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 59.Coles A.H., Osborn M.F., Alterman J.F., Turanov A.A., Godinho B.M., Kennington L., Chase K., Aronin N., Khvorova A. A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 2016;26:86–92. doi: 10.1089/nat.2015.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roehl, I., Schuster, M., and Seiffert, S. August 2011. Oligonucleotide detection method. U.S. patent application US20110201006A1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.