Abstract
In vertebrates, mitochondria are tightly preserved energy producing organelles, which sustain nervous system development and function. The understanding of proteins that regulate their homoeostasis in complex animals is therefore critical and doing so via means of systemic analysis pivotal to inform pathophysiological conditions associated with mitochondrial deficiency. With the goal to decipher the role of the ATPase inhibitory factor 1 (IF1) in brain development, we employed the zebrafish as elected model reporting that the Atpif1a−/− zebrafish mutant, pinotage (pnttq209), which lacks one of the two IF1 paralogous, exhibits visual impairment alongside increased apoptotic bodies and neuroinflammation in both brain and retina. This associates with increased processing of the dynamin-like GTPase optic atrophy 1 (OPA1), whose ablation is a direct cause of inherited optic atrophy. Defects in vision associated with the processing of OPA1 are specular in Atpif1−/− mice thus confirming a regulatory axis, which interlinks IF1 and OPA1 in the definition of mitochondrial fitness and specialised brain functions. This study unveils a functional relay between IF1 and OPA1 in central nervous system besides representing an example of how the zebrafish model could be harnessed to infer the activity of mitochondrial proteins during development.
Introduction
The zebrafish (Danio rerio) represents a powerful model system for studying vertebrate development and the pathogenesis of human diseases (as reviewed in ref.1). Recently, it has also been employed as a tool for mitochondrial genetic and pharmacological studies2,3. Mitochondrial defects are commonly observed in a wide spectrum of human pathologies, such as cancer, diabetes and neurodegeneration. As in humans and mammals, the phenotypes observed in zebrafish include neuronal and synapse loss4, alterations in brain activity5, aberrant motor and sensory responses5, blood and vascular disorders6. Mitochondrial disorders can be caused by either defective mitochondrial bioenergetics or intracellular transport, which can result in abnormal subcellular localization of the organelle, increased production of reactive oxygen species (ROS) or impaired OXPHOS activity7. The zebrafish mitochondrial respiratory chain complexes have a high degree of identity with the human counterparts and their inhibition causes severe developmental defects, ranging from morphological and physiological abnormalities to embryonic arrest2. Brain development is tightly regulated by the metabolic state of mitochondria whose mass may change during neuronal differentiation. Indeed, while a decrease in mitochondrial activity has been observed as human embryonic stem cells (hESCs) differentiate into neural stem cells (NSC)8, mitochondrial biogenesis increases during the maturation of NSCs into motor neurons9. Trafficking of mitochondria is also crucially involved in the formation of axons, dendrites and synaptic connections10.
The F1Fo-ATPsynthase plays an important part in all this. Physiologically, its activity regulates cellular ATP provision11, mitochondrial ultrastructure12, Ca2+ handling and cell death13. Defects in the function and assembly of the F1Fo-ATPsynthase, are observed in postnatal and age-related neurometabolic disorders (i.e. Leigh syndrome, maternally inherited Leigh syndrome, Leber hereditary optic neuropathy and neuropathy, ataxia, and retinitis pigmentosa14), all of which characterized by early-onset and motor and sensory neurological symptoms, such movement disorders and visual impairments15. The activity of this enzyme is indeed involved in driving synapse formation and sustaining physiological brain activity, and its impairment associates with a decline in both neuronal performance and plasticity16.
Aging processes are not exception as loss of mitochondrial inner membrane organization following disassembly of F1Fo-ATPsynthase complexes is also described17.
The ATPase inhibitory factor 1 (IF1), which is the most characterized regulator of the F1Fo-ATPsynthase18,19, binds to the enzyme inhibiting its hydrolytic activity thereby protecting cells from ATP depletion during de-energized conditions18. A series of evidences suggests that, in cancer cells, the deregulated, enhanced binding of IF1 to the F1Fo-ATPsynthase activates oncogenic regulatory mechanisms20–22. IF1 though is mainly known for its protective role from hypoxic/ischaemic damage in tissues with high-energy demand, such as heart and brain23. However, its ubiquitous expression and high degree of sequence conservation underline a wider role, which has been recently associated with the regulation of stem cell differentiation24, hepatic cholesterol uptake25, haem synthesis6, cell proliferation and programmed demise26,27.
IF1 expression is sustained in neurons28, which highly rely on oxidative metabolism, which is per se indicative of a core role in energy provision, which is not reported in astrocytes29. The upregulation of IF1 expression is notably observed during brain preconditioning, involving adaptation of both mitochondrial metabolism30 and quality control23,31–33. Equally, deregulation of IF1 activity could harm these processes, leading to the onset of pathological conditions. We therefore set to investigate this using the Atpif1a−/− zebrafish mutant pinotage (pnttq209), which is indeed lacking of the a gene type of the Atpif1 paralogues.
The zebrafish genome contains two copies of the Atpif1 gene, Atpif1a and Aatpif1b6, a duplication that is likely to originate in teleosts34 and so encode for two proteins with partially redundant functions6. We have recently used the zebrafish mutant pnttq209 to study the involvement of IF1 in haem synthesis reporting reduced rate of ferrochelatase-dependent iron incorporation into protoporphyrin6 leading to decreased haem content and erythrocyte volume6. Whether IF1 expression affects other systemic functions and organ physiology during development is nonetheless unknown. Here we report that alterations in IF1 expression lead to visual impairments in both zebrafish larvae and mice providing evidences in complex model systems of an underlying interplay between IF1 and OPA1 in the definition of mitochondrial homoestasis exploited during development and reflected systemically.
Results
Reduced IF1 levels lead to increased apoptosis and neuroinflammation in the brain and retina
The analysis of the effect of reduced IF1 levels on zebrafish neurodevelopment started with an examination of nervous system cell survival and proliferation during embryogenesis in the pnttq209 mutant. Indeed, an imbalance between proliferation and apoptosis leads to defective clonal expansion of progeny cells, abnormal organ growth and functional impairments. For the purpose, we first determined the number of apoptotic bodies in both brain and retina of pnttq209 mutants and normal siblings (Sbs) at 72 h post fertilisation (hpf).
Larvae were subjected to TUNEL staining prior to HuC/D immunostaining to simultaneously visualize apoptotic bodies and neuronal cells, respectively (Fig. 1a). The analysis, conducted with a confocal microscope, revealed an increase in the number of apoptotic bodies in both brain and retina of pnttq209 mutants (Fig. 1b), even though no obvious differences in the pattern of the neuronal marker HuC/D were observed between mutants and Sb (Fig. 1a).
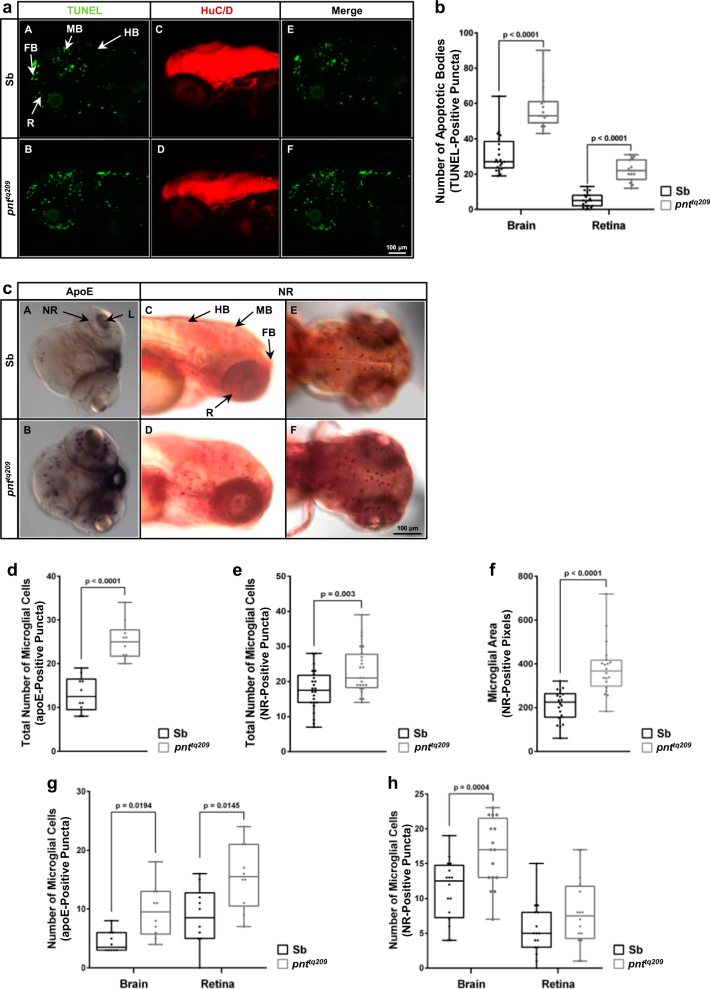
Fig. 1. Apoptosis and microglial activation are increased in the CNS of pnttq209 larvae.
a TUNEL assay (A, B) and HuC/D (C, D) immunostaining of PTU-treated normal Sb and pnttq209 mutant zebrafish at 72 hpf to detect apoptotic cells and differentiated neurons, respectively. The merge between the two fluorescent signals is shown in E and F. b Quantification of apoptotic bodies, detected with the TUNEL assay, in Sb and pnttq209 zebrafish, showing a significant increase in the total number of apoptotic bodies in the brain and retina of mutant larvae (number of apoptotic bodies (TUNEL-positive puncta) in the brain, Sb: 31.10 ± 2.35, pnttq209: 57.53 ± 3.12; in the retina, Sb: 5.33 ± 0.85, pnttq209: 21.80 ± 1.52; results are reported as mean ± S.E.M. (n = 15–20)). c apoE in situ hybridization (A, B) and NR staining (C–F) were carried out in PTU-treated, 72 hpf zebrafish to visualise microglia. d, e Quantification of apoE-positive (d) and NR-positive (e) cells in the whole CNS. A significant increase in the number of microglial cells characterizes pnttq209 zebrafish ((number of apoE-positive puncta, Sb: 13.10 ± 1.28, pnttq209: 25.20 ± 1.38; number of NR-positive puncta, Sb: 17.50 ± 1.18, pnttq209: 23.13 ± 1.35; results are reported as mean ± S.E.M. (apoE: n = 10; NR: n = 24)). f Analysis of the size of NR puncta. pnttq209 zebrafish showed a significant increase in the size of microglia when compared to Sb (microglial area (NR-positive pixels), Sb: 210.45 ± 15.65, pnttq209: 379.35 ± 26.73; results are reported as mean ± S.E.M. (n = 15)). g, h Evaluation of the number of cells stained with apoE (g) and NR (h) in the brain and retina, separately. The results demonstrate that, in pnttq209 larvae, the increase in microglial cell population is also extended to the retina ((number of apoE-positive puncta in the brain, Sb: 4.40 ± 0.56, pnttq209: 9.60 ± 1.38; in the retina, Sb: 8.70 ± 1.58, pnttq209: 15.60 ± 1.73; number of NR-positive puncta in the brain, Sb: 11.19 ± 1.10, pnttq209: 16.50 ± 1.21; in the retina, Sb: 5.44 ± 0.93, pnttq209: 7.94 ± 1.23; results are reported as mean ± S.E.M. (apoE: n = 10; NR: n = 15)). FB forebrain, HB hindbrain, MB midbrain, R retina
Considering the increased rate of programmed cell death observed in the pnttq209 mutants, we hypothesized that this could lead to a neuroinflammatory response. Therefore, we monitored the level of microglial activation in both mutants and Sb. For the purpose, we employed in situ hybridisation (ISH) of apolipoprotein E (apoE) mRNA and neutral red (NR) vital dye staining35,36 to visualize microglia in the pnttq209 zebrafish brain (Fig. 1c). Interestingly, we found an increase in both apoE-positive (Fig. 1d, g) and NR-positive (Fig. 1e, h) cells in the brain and retina of pnttq209 larvae. Microglial cells did also appear significantly larger (Fig. 1f), thus implying an activated state37. Taken together, these data suggest the induction of an inflammatory response in pnttq209 zebrafish as ablation of Atpif1a expression may lead to cell loss and a neuroinflammatory phenotype in zebrafish.
IF1 loss causes visual impairment in pnttq209 zebrafish and Atpif1−/− mice
Excessive, uncontrolled microglial activation is a major cause of inflammation-mediated neurodegeneration38. As higher levels of apoptotic bodies were detected in both brain and retina of pnttq209 mutants, and chronic neuroinflammation is frequently associated with retinal neurodegeneration39, we decided to further explore the phenotypic outcome of Atpif1a loss by assessing the visual function in pnttq209 larvae. For this purpose, the optokinetic response (OKR) of pnttq209 and normal Sb zebrafish larvae was measured. The OKR is the ocular movement induced by changes in the visual surround, and is commonly used to obtain an accurate quantitative readout of visual ability in zebrafish larvae40. At 72 hpf, the larval OKR was assessed by counting the number of eye movements under normal conditions or during constant visual stimulation. Each record lasted 3 min, and visual stimulation was achieved by moving black and white stripes around the test chamber (Fig. 2a). Remarkably, the analysis revealed a different response to the moving striped pattern between pnttq209 mutants and normal Sb. Even though pnttq209 larvae did not show any noticeable alteration under normal conditions, they were significantly less sensitive to constant light changes in the surrounding environment (Fig. 2b, c). This indicates that Atpif1a deficiency is associated with mild vision impairment, which affects the responsiveness of pnttq209 larvae to visual stimuli.
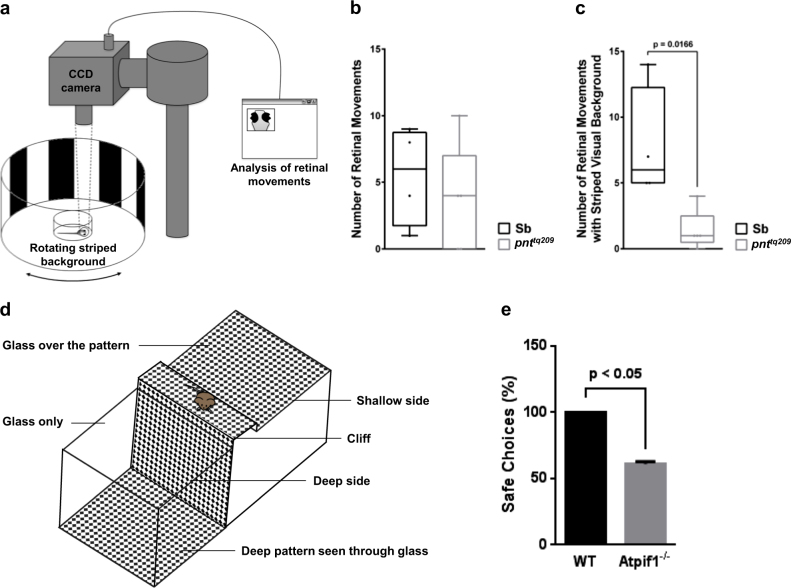
Fig. 2. Vision is impaired in zebrafish and mice lacking Atpif1.
a Schematic representation of the device used to measure the OKR of zebrafish larvae. b, c OKR in Sb and pnttq209 zebrafish at 72 hpf. While normal and mutant larvae have a comparable number of retinal movements in normal conditions (b), the latter exhibit a significantly reduced response to visual stimulation, which was achieved by a rotating striped disc (c) (number of retinal movements in normal conditions, Sb: 5.5 ± 1.85, pnttq209: 3.5 ± 1.58; with striped visual background, Sb: 7.75 ± 2.14, pnttq209: 1.40 ± 0.40; results are reported as mean ± S.E.M. (n = 5)). d Model of the visual cliff apparatus. e Visual cliff test in WT and Atpif1−/− mice. The quantification of safe choices (avoidance of the drop) indicates that visual acuity is mildly compromised in mutant mice (percentage safe choices, WT: 100.00 ± 0.00, Atpif1−/−: 61.00 ± 1.73); results are presented as mean ± S.E.M. (n = 5))
To corroborate this finding, we monitored the visual acuity in 4-month-old Atpif1−/− mice. This was investigated by using the visual cliff apparatus, monitoring the propensity by mice to step towards the platform side (safe choice) and avoid the drop (Fig. 2d). Notably, the loss of Atpif1 correlated with alterations in the spatial perception and a substantial reduction in the number of positive choices was observed in Atpif1−/− mice (Fig. 2e), a response that also characterizes strains affected by retinal degeneration31.
The data collected so far confirm that Atpif1 contributes to correct visual function in vertebrates. The protein can therefore be ascribed among the mitochondrial factors that play a role in the development and maintenance of the neural retina, among which the dynamin-like protein optic atrophy 1 (OPA1) is the most known37. Mutations in the OPA1 gene that impair the expression or activity of the protein are associated with hereditary optic neuropathies related with mitochondrial dysfunction, such as autosomal-dominant optic atrophy (ADOA)41,42. Interestingly, although ubiquitously expressed throughout the body, during embryonic development IF1 is significantly upregulated in the retina of both zebrafish6 and mouse43,44. Furthermore, IF1 shares a similar chronological expression profile during development with OPA145 and recently we described a functional relay between IF1 and OPA1, which shields the latter from the processing mediated by the Metalloendopeptidase OMA146.
In order to investigate whether the vision defects detected in zebrafish and mice lacking IF1 expression were related to OPA1 inactivation, we first monitored the levels of OPA1 in the brain and retina of pnttq209 and normal Sb zebrafish at 72 hpf. The analysis was carried out by whole-mount immunofluorescence and revealed an extensive decrease in the expression profile of the protein in both brain (midbrain-hindbrain, in particular) and retina of pnttq209 mutants when compared to Sb (Fig. 3a, b). Despite this, no prominent differences in the eye morphology (size and shape) were detected between pnttq209 and wild-type (WT) larvae (Fig. 3c, d), implying that Atpif1a loss does not cause any major anatomical eye defects, and that mainly affects the neural part of the organ. To confirm this observation, the levels of both OPA1 isoforms were quantified via western blotting in the whole embryos (Fig. 3e, f). This analysis reports a decrease in the levels of the profusion protein, which prevalently affects the short isoforms. However, we should consider that the samples were obtained from the whole pnttq209 larvae, entailing technical limitations in protein extraction by homogenization.
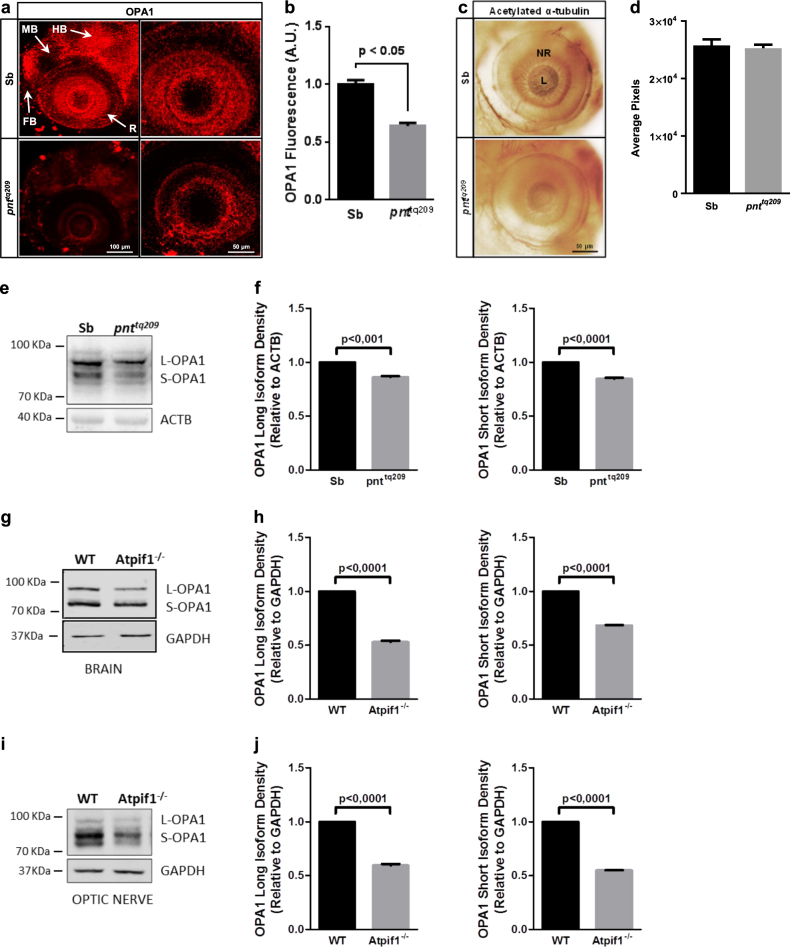
Fig. 3. Atpif1 deficiency causes a decline in the retinal expression of OPA1.
a, b Fluorescent IHC of whole-mount 72 hpf zebrafish, stained with anti-OPA1 antibody. Representative images (a) and quantification of OPA1 fluorescence intensity (b) are reported, showing a reduction in the OPA1 expression levels in the brain and retina of pnttq209 larvae (OPA1 fluorescence (A.U.), Sb: 1.00 ± 0.01, pnttq209: 0.70 ± 0.09; results are presented as mean ± S.E.M. (n = 3)). c, d Analysis of eye morphology in Sb and pnttq209 zebrafish obtained through anti-acetylated α-tubulin IHC of whole-mount 72 hpf zebrafish. No major morphological differences were observed between normal and mutant larvae, as shown in the prototypical images (c) and relative quantification of eye size (d) (eye size (pixels), Sb: 2.58 ± 0.10, pnttq209: 2.53 ± 0.06; results are presented as mean ± S.E.M. (n = 10)). e, f Quantification of OPA1 levels via western blotting analysis in 72 hpf wild type and pnttq209 larvae. The representative membrane blotted for both OPA1 isoform (e) and the bar chart (f) show a significant decrease in the levels of the short isoform of the protein in pnttq209 larvae. g–j (OPA1 band density relative to ACTB, OPA1 long isoform Sb: 1.00 ± 0.01, pnttq209:0.97 ± 0.01; OPA1 short isoform Sb: 1.00 ± 0.01, pnttq209:0.84 ± 0.01; results are presented as mean ± S.E.M. (n = 3), Quantitative western blot analysis of OPA1 levels in the brain (frontal cortex/hippocampus) (j, k) and isolated optic nerve (l, m) of WT and Atpif1−/− mice. Representative blots (j, l) and quantitated band densities relative to GAPDH (k, m) are reported. Significantly lower levels of short and long OPA1 isoforms expression were found in the brain and, specifically, in the optic nerve of mutant mice (OPA1 band density relative to GAPDH, OPA1 long isoform WT: 1.00 ± 0.01, Atpif1−/−: 0.52 ± 0.01 in the frontal cortex/hippocampus; in the optic nerve, WT: 1.00 ± 0.05, Atpif1−/−: 0.59 ± 0.09; in the frontal cortex/hippocampus OPA1 short isoform WT: 1.00 ± 0.01, Atpif1−/−: 0.68 ± 0.01; in the optic nerve, WT: 1.00 ± 0.05, Atpif1−/−: 0.548 ± 0.01; results are presented as mean ± S.E.M. (n = 3). FB forebrain, HB hindbrain, MB midbrain, NR neural retina, L lens
A tissue-specific analysis was instead obtained using extracts from WT and Atpif1−/−mouse. In these, OPA1 isoforms have been monitored in the brain (frontal cortex/hippocampus), optic nerve and liver homogenates (Fig. 3g-j, SFigure 1a,b). Even in this case, the expression of short and long OPA1 isoforms was downregulated in Atpif1−/− mice (almost halved when compared to WT mice). This data suggest that, even though Atpif1 deficiency causes a tissue-specific phenotypic alteration leading to visual impairment6,47, the physiological OPA1 processing seems to be retained in each tissue investiagated. Considering the close link between OPA1 loss-of-function and defects in optic nerve morphogenesis, which are a direct consequence of impaired mitochondrial function48, we further investigated the effect of IF1 loss on mitochondrial respiratory capacity. For the purpose, we evaluated the levels and assembly of mitochondrial respiratory chain complexes by two-dimensional blue native/SDS gel electrophoresis. Mitochondrial bioenergetic efficiency and adaptive capacity rely on the dynamic supramolecular organization of respiratory chain complexes in supercomplexes (SCs) or respirasomes. The analysis of the stoichiometric composition of mitochondrial respirasomes gives significant information on the organelle bioenergetic homoeostasis.
We attempted to measure the relative levels of SCs in extracts from WT and Atpif1−/− murine liver and brain, as well as WT and IF1 knockdown human neuroblastoma SHSY-5Y cells. The analysis revealed that IF1 expression levels alters the respirasome assembly in both murine and human derived lines (S Figs. 1 and 2). Most notably, differences appear prominent in the layout of the complexes in which association between complexes I and III is far less in extracts in which the Atpif1 gene is ablated or the protein product downregulated. We also note an overall reduction of the complex IV levels, which could per se reduce the respiratory efficiency, as well as favouring the accumulation of ROS and therefore trigger apoptosis.
All this is nonetheless indicative of an underlying mitochondrial phenotype associated with Atpif1 loss and aberrant processing of OPA1, which could lead to the defects in visual capacity observed in both zebrafish and mice.
Analysis of locomotor ability in pnttq209 zebrafish
Based on the results described above, we examined whether locomotion could also be affected by IF1 deficiency due to axonal alterations in the caudal region of the larvae. The number and morphology of spinal cord motor axons were monitored via immunostaining of acetylated α-tubulin (Fig. 4a). The absence of noticeable differences between pnttq209 and normal Sb zebrafish larvae suggests that IF1 loss does not significantly affect the neurogenesis of spinal cord motor neurons. Nonetheless, we carried out a quantitative assay of larvae locomotion to assess whether motor neuron function was compromised. The total distance moved by mutant and WT larvae, together with their mean and maximum velocity, were analysed (Fig. 4b-d). The assay demonstrated no significant motility alteration of pnttq209 mutants, indicating that zebrafish lacking Atpif1a does not exhibit typical signs of motor neuron degeneration.
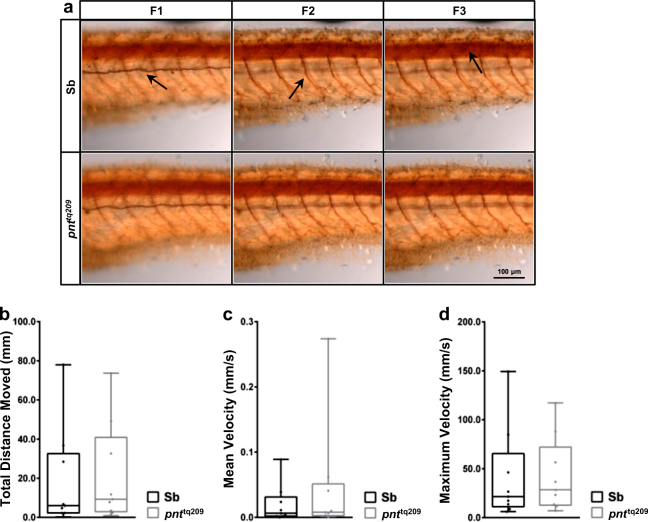
Fig. 4. pnttq209 zebrafish show normal development of the spinal cord and locomotor ability.
a Anti-acetylated α-tubulin IHC of whole-mount 72-hpf zebrafish. No marked differences in axonal growth and morphology were observed between Sb and pnttq209 larvae. Images were acquired at three different focal planes to visualize the lateral line (F1) and the ventral and dorsal motor axons (F2 and F3, respectively). b–d Locomotor activity assay in 72-hpf zebrafish. Total distance moved (b), mean velocity (c) and maximum velocity (d) were measured (average total distance moved (mm), Sb: 18.39 ± 8.60, pnttq209: 21.22 ± 8.46; average mean velocity (mm/s), Sb: 0.02 ± 0.01, pnttq209: 0.05 ± 0.03; average maximum velocity (mm/s), Sb: 41.57 ± 15.74, pnttq209: 42.47 ± 12.62; results are reported as mean ± S.E.M. (n = 9)). No variances were observed between Sb and pnttq209 larvae
Discussion
This study examined the role of IF1 in the development of the zebrafish nervous system during embryogenesis. Our interest stemmed from the notion that IF1 expression, which is maintained at high levels in adult neurons30,49, undergoes timed regulation during differentiation and maturation of stem cells24,50, suggesting a possible role for the protein in vertebrate development. This hypothesis appears to be confirmed by the defects in erythroblast differentiation associated with Atpif1a loss in zebrafish, which cause a severe form of sideroblastic anaemia14. Nonetheless, little is still known about the effect of pathological alterations in IF1 expression during embryogenesis. The zebrafish is an extremely valuable model system for studying vertebrate biology, also enabling developmental genetic and functional studies51. Moreover, considering the high level of conservation of genetic sequences, molecular processes and organ systems between zebrafish and other vertebrates, including humans, zebrafish is emerging as preferred model for studying developmental pathology52. In this work, the use of the pnttq209 zebrafish mutant allowed the identification of alterations in the neural system, attributable to loss of Atpif1a expression. Specifically, we detected cell loss (i), induction of a pro-inflammatory environment (ii) and vision impairment (iii).
During brain maturation, apoptosis plays a key role and any defect in the process, such as irregular, premature and/or excessive apoptosis, can lead to neurodegeneration53. We recently reported that the level of IF1 expression defines the cellular response to apoptotic stress preventing cellular demise22,26. Here we found that the pnttq209 mutant zebrafish is characterized by a significant increase in the number of apoptotic bodies in both brain and retina, even though no significant alterations were observed in the pattern of differentiated neurons between mutant and WT larvae (Fig. 1).
The abnormal levels of apoptosis that characterize pnttq209 zebrafish seem to have an impact on the activity of microglial cells, which represent the resident macrophages of the CNS and are involved in the maintenance of cell homeostasis by clearing dead and dysfunctional neurons. pnttq209 zebrafish are characterized by higher microglial activity (Fig. 1), which can be a direct consequence of the increased programmed demise both in the brain and retina of pnttq209 mutants. Though microglia play a neuroprotective role36, their excessive activity is frequently observed in damaged brain tissues and neurodegenerative disorders38,54.
By using specific microglial markers, we identified an increase in both number and size of microglial cells in pnttq209 mutant zebrafish confirming that phagocytic activity of microglia is enhanced in the absence of Atpif1a and causes neuroinflammatory damage. Notably, activated microglial cells are localized at high levels in the retina of the larvae (Fig. 1).
The concomitant rise in both cell death and microglia-induced neuroinflammation detected in the retina of pnttq209 larvae suggests that organ function may also be compromised. By testing the OKR of pnttq209 larvae (Fig. 2), we gained evidence that IF1 deficiency associates with visual impairment noting a significant reduction in retinal movements triggered by changes in the surrounding environment (Fig. 2). A decline in visual acuity was also observed in Atipif1−/− mice, corroborating that IF1 may have a conserved role in the development of vertebrate vision (Fig. 2).
The retinal photoreceptor cells, which comprise two different types of neurons, cones and rods, both specialized in phototransduction, are metabolically active cells characterized by high-mitochondrial content55,56. Tissues with high-energy demand, such as the neural retina, are frequently affected by the presence of mutations in genes that encode for mitochondrial proteins57. The impairments in visual capacity caused by IF1 deficiency in zebrafish and mouse may be therefore related to retinal mitochondrial alterations. Indeed, IF1 controls different aspects of mitochondrial homeostasis6,20,26,58 and mitochondrial defects are commonly associated with eye disorders59.
A mitochondrial protein that is highly involved in vertebrate embryonic development is the dynamin-like GTPase OPA160,61. OPA1, which exists as long, inner membrane-bound, and short, soluble, isoforms, forms high-molecular weight complexes that localize at the cristae junctions and control both the fusion of the inner membranes of two merging mitochondria and the shaping of mitochondrial cristae62. OPA1 also plays a prominent part in cristae remodelling and cytochrome c release during apoptosis63. Recently, we discovered that IF1 has similar activity that relies on stabilization of the mitochondrial ultrastructure22, a function that exerted in coordination with OPA147. Alterations in mitochondrial dynamics and metabolism have been previously proposed as a possible mechanism of the OPA1-type ADOA pathogenesis64. Moreover, OPA1 deficiency causes developmental defects in zebrafish, including abnormal cardiac function and blood circulation60. With our study, we provide further proof of shared pathological mechanisms between IF1 and OPA1. Both are highly expressed in the developing vertebrate brain and retina43–45,65 and we learnt that IF1 interplays with OPA1 in the control of mitochondrial adaptation to programmed cell death46. Here we show that impairment of this relay, which is core to mitochondrial structure and functions affects the visual capacity in both models of analysis leaving unaffected other parametres such as the locomotion at least at the state of analysis here adoboted66,67.
Beyond the pathophysiological outcome, which we linked to OPA1, the modifications in the mitochondrial complexes and SCs assembly recorded via the in-gel analysis corroborate how IF1 is intimately linked with mitochondrial OXPHOS homeostasis (S Figs. 1 and 2) beyond the preservation of ATP pools during reversion of the ATPsynthase28.
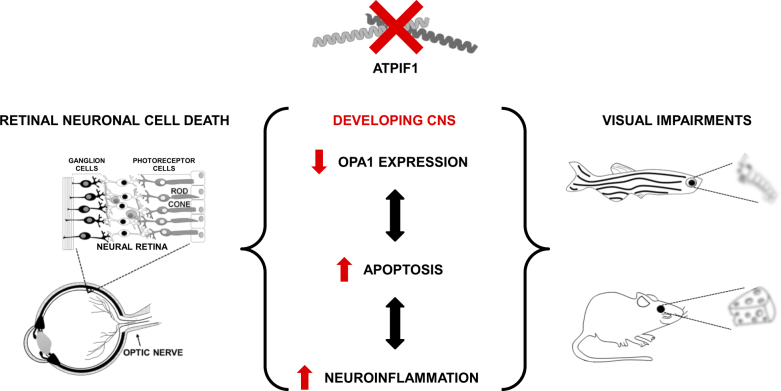
Though in zebrafish the loss of IF1 induces detectable alterations solely in their visual capacity, we cannot exclude endogenous compensatory mechanisms mediated by the Atpif1a-related paralogue, Atpif1b, which might reduce or mask the effects of the IF1 loss in other tissues. However, the lethal phenotype of the double Atpif1a/b KO zebrafish mutant6 prevented further in-depth analysis. The merit of this work does lie in its capacity to reveal how IF1 takes part the developmental of central nervous system widening the relevance of this mitochondrial protein (see working model depicted in Fig. 5) beyond the pathological processes to which it is prevalently associated26,32,68.
Fig. 5. Involvement of IF1 in vertebrate eye development.
The loss of IF1 activity causes abnormally high apoptosis and microglial activation in the CNS and retina during zebrafish embryogenesis, an event which suggests increased levels of neuroinflammation. IF1 deficiency is also associated with a marked decline in the brain and retinal levels of OPA1, the expression of which is fundamental for guaranteeing the correct development of the vertebrate eye. As a consequence, IF1 KO zebrafish and mouse models are both characterized by mild visual impairment that does not interfere with the normal animal functions, and which is possibly caused by apoptosis- and neuroinflammation-mediated neuro-retinal damage
Materials and methods
Animal models
Zebrafish were housed in a multi-rack aquarium system at the Royal Veterinary College and kept on a constant 14/10 h light/dark cycle at 27–29 °C69. The pnttq209 mutant zebrafish line6 was obtained from the Tübingen Stock Center. Zebrafish embryos, obtained by natural spawning, were examined and manually dechorionated under a Nikon SMZ1500 microscope (Nikon, Kingston upon Thames, UK). To prevent pigmentation, embryos were raised in water containing 0.003% 1-phenyl-2-thiourea (PTU), starting at 24 hpf. At 72 hpf, larvae were categorized as either normal Sb or pnttq209 mutants depending on their phenotype6. All zebrafish experiments were locally ethically approved by the Royal Veterinary College, and nationally approved by the UK Home Office under the Animal (Scientific Procedures) Act 1986.
C57 BL/6 (WT) and C57/BL6J (Atpif1−/−) mice47 were used in accordance with national and European (86/609/EEC) guidelines.
TUNEL and anti-HuC/D immunofluorescence double-labelling assay
DeadEnd™ Fluorometric TUNEL System (Promega, G3250) was used for the specific detection and quantitation of apoptotic cells in PTU-treated and dechorionated 72 hpf zebrafish. The catalytic incorporation of fluorescein-12-dUTP at 3′-OH DNA ends was developed following the principle of the TUNEL (TdT-mediated dUTP Nick-End Labelling) assay and the manufacturer recommendations. Briefly, 72 hpf zebrafish were fixed in 4% paraformaldehyde (PFA) in PBS (overnight incubation at 4 °C). After fixation, larvae were washed three times in PBS + 0.1% Triton™ X-100 (PBT) and then subjected to proteinase K (Sigma-Aldrich, P2308) digestion (15 ng/ml for 1 h in PBT). Subsequently, larvae were fixed for 20 min in 4% PFA in PBS, blocked in equilibration buffer for 30 min, and finally incubated for 1 h at 37 °C in TdT reaction mix (equilibration buffer + nucleotide mix and rTdT enzyme in a 5:1 ratio). Unincorporated fluorescein-12-dUTP was removed through three washes in PBT. After TUNEL assay was performed, larvae were incubated for 1 h at room temperature in blocking solution (10% normal goat serum plus 1% dimethyl sulphoxide in PBT), and then overnight at 4 °C with a mouse anti-HuC/D antibody (1:1000; Molecular Probes, A21271). Following several washes in PBT, the larvae were incubated overnight at 4 °C with a goat anti-mouse Alexa Fluor®568-conjugated antibody (1:400; Invitrogen, A11004). Fluorescently-labelled larvae were imaged using a laser-scanning confocal microscope (LeicaTM SP5). All images were acquired without changing the microscope settings and processed in the same way. The number of apoptotic bodies in the brain and retina was quantified using ImageJ software (NIH).
Immunohistochemistry (IHC) of whole-mount zebrafish larvae
IHC was performed in whole-mount PTU-treated and dechorionated 72 hpf zebrafish as previously described70. Larvae immunostained with anti-acetylated α-tubulin (1:1000; Sigma-Aldrich, T7451)70 were imaged with a Zeiss Axiovert inverted microscope.
For anti-OPA1 fluorescent IHC, PTU-treated and dechorionated 72 hpf zebrafish were fixed in 4% PFA in PBS and treated with proteinase K (15 ng/ml for 1 h in PBS + 0.1% Triton™ X-100). After leaving the larvae in blocking solution for 1 h, they were incubated overnight at 4 °C with a mouse anti-OPA1 antibody (1:500; BD Biosciences, 612607). Following a wash in PBT for several hours, larvae were then incubated overnight at 4 °C with a goat anti-mouse Alexa Fluor®568-conjugated antibody (1:400; Invitrogen, A11004). Larvae were cleared in 70% glycerol in PBS and imaged with a laser-scanning confocal microscope (LeicaTM SP5). OPA1 fluorescent intensity was quantified using ImageJ software.
NR assay
Microglial cells were detected in living dechorionated zebrafish larvae by staining with NR, a vital dye that is accumulated in the lysosomes through endocytosis35. PTU-treated and dechorionated 56 hpf zebrafish were incubated in the dark overnight (to reach the larval stage) at 28–30 °C in 2.5 μg/mL NR solution. After two rinses with fish water, larvae were fixed in 2% low melting-point agarose in fish water. Live imaging was carried out using a Zeiss Axiovert inverted microscope. The number and size of NR-positive cells in the brain and retina were quantified using ImageJ software.
Whole-mount ISH of apoE mRNA
ISH was carried out on PTU-treated and dechorionated 72 hpf zebrafish using an apoE riboprobe (a generous gift from Dr. Francesca Peri, European Molecular Biology Laboratory, Heidelberg, Germany), as previously described36. The number of apoE-positive cells in the brain and retina was quantified using ImageJ software (NIH).
Western blot analysis of mouse brain extracts
Four-month-old mice were killed by cervical dislocation and, prior to dissection, sterilized with ethanol. The skull was opened to release the brain, which was immediately stored on ice. The frontal cortex, hippocampus and optic nerve were then isolated and lysed in radioimmunoprecipitation assay buffer (50 Mm Tris, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1% Triton™ X-100, 0.25% sodium deoxycholate, 0.1% SDS, pH 7.4) supplemented with protease/phosphatase inhibitors (Roche Diagnostics, 04693132001). The lysates obtained from the frontal cortex and hippocampus were then combined. The lysates were sonicated and centrifuged at 17,000×g at 4 °C for 20 min, after which the supernatants were collected and stored at −80 °C.
The protein concentration was estimated using a BCA protein assay reagent (Thermo Scientific). Equal amounts of protein (20 μg) were resolved in 8% polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was blocked in 3% non-fat dry milk in TBST (50 mM Tris, 150 mM NaCl, 0.05% Tween 20 (Sigma-Aldrich), pH 7.5) for 1 h and then incubated with the appropriate diluted primary antibody at 4 °C overnight: mouse anti-OPA1 (1:1000; BD Biosciences, 612607), mouse anti-GAPDH (1:10000; Abcam, ab 8245). After three washes in TBST, the membrane was incubated with goat anti-mouse HRP-conjugated antibody for 1 h at room temperature. Amersham ECL Prime Western Blotting Detection Reagent kit (GE Healthcare Life Sciences, RPN2232) was used to develop the membrane. Immunoreactive bands were analysed with ImageJ software (NIH).
In-gel analysis of mitochondrial respirasomes
The method was adapted from Nijtmans et al.71, Wittig et al.72 and Calvaruso et al.73. Mice were killed by cervical dislocation. Then, brain and liver tissues were collected on ice and homogenized using a glass-Teflon potter homogenizer in mitochondrial isolation buffer (440 mM sucrose, 20 mM Mops, 1 mM EDTA) with 0.2 mM phenylmethylsulfonyl fluoride. Protein extracts were separated by Blue Native Bis-Tris PAGE 4–16% (NativePage Thermo Fisher scientific).
Homogenates were centrifuged at 20,000×g for 15 min at 4 °C. After that the pellet was homogenized in a solution composed of 1 M aminocaproic acid and 50 mM Bis-Tris-HCL pH 7.0. Digitonin 4 mg/mL was added to the homogenate and incubated for 20 min on ice. After centrifugation at 100,000×g for 15 min, the supernatant was collected and combined with Serva blue G 5% in 1 M aminocaproic acid.
The Coomassie Blue Staining was obtained by immersing the gels in staining solution 0.1% Serva G in 40% methanol for 30 min at room temperature. Then the staining solution was removed and the gels were incubated with destaining solution 40% methanol and 10% acetic acid for 30 min twice. At this point the gels were scanned.
Locomotor activity assay
Dechorionated 72 hpf zebrafish were individually placed into the wells of a 96-well plate. A DMx21AF04 digital camera was used with Noldus 2010 Media Recorder software to record 20 min videos with a frame rate of 30 frames/s and 640 × 480 resolution. The videos were processed using EthoVision XT8 software (Noldus) to track the movements of each zebrafish larva. Quantitive data was produced using three movement parameters: total distance moved (mm), mean velocity (mm/s) and maximum velocity (mm/s).
OKR measurements
Three minute, 30 fps digital recordings of 72 hpf zebrafish were taken using a DMx21AF04 digital camera with Noldus 2010 Media Recorder software. Each animal was recorded with and without a moving striped visual background made from black and white cards, where each stripe was ~2 cm thick. In recordings using the striped visual background, card was rotated around the 96-well plate in small back and forth movements in quick succession. One retinal movement includes one or both retinae.
Visual cliff test
Visual acuity was tested in C57 BL/6 (WT) and C57/BL6J Atpif1−/− mice using the visual cliff test according to Carlezon and colleagues74. The apparatus comprised a 1 m height checkered pattern platform covered with a clear piece of acrylic glass, extended by 50 cm from the edge of the platform, and a checkered pattern sheet positioned below the extending glass. A raised dais was placed between the platform and the extending glass, creating the illusion of a cliff. Each mouse was placed on the raised dais and allowed to step off to either the shallow side (safe choice) or the deep side (unsafe choice). Each mouse was tested in ten trials, and choices were manually recorded as safe if the mouse stepped towards the platform and unsafe if the mouse stepped towards the extending acrylic glass.
Statistics
Statistical analyses were performed using GraphPad Prism 6 software. Two groups were compared using the unpaired t-test. Comparison of three or more groups was performed by one-way ANOVA. A p-value <0.05 was considered significant.
Electronic supplementary material
Acknowledgements
Thank you to all members of the Claire Russell and Michelangelo Campanella research groups for the ongoing support.
The research led by C.R. was funded as follows: E.A. and H.B.R. were undergraduate project students supported by bench fees from UCL and RVC; L.A. was an undergraduate summer student visiting from the Universidad Autónoma de Madrid; R.M.-J. is a postdoctoral researcher funded by a research grant to C.R. and M.C. from SPARKS children’s charity.
The research activities led by M.C. are supported by the following funders, which are gratefully acknowledged: Biotechnology and Biological Sciences Research Council (grant numbers BB/M010384/1 and BB/N007042/1); the Medical Research Council [grant number G1100809/2], Bloomsbury Colleges Consortium PhD Studentship Scheme; The Petplan Charitable Trust; Umberto Veronesi Foundation Young Investigator Research Programme; Marie Curie Actions [TSPO & Brain (304165)], LAM-Bighi Grant Initiative. FIRB-Research Grant Consolidator Grant 2 [grant number: RBFR13P392], Italian Ministry of Health [IFO14/01/R/52].
The greatest acknowledgement of all should go to the late Prof. Barry Paw who called for our attention on the zebrafish mutant, pinotage (pnttq209). This work is therefore dedicated to his vision, collaborative spirit and now living memory.
Author contributions
M.C. and C.R. designed and coordinated the experiments. M.C. supervised the work on murine models run by D.F., C.F. and D.S., whilst C.R. supervised those on zebrafish experiments performed by R.M.-J., E.A., H.B.R. and L.A. R.M.-J. carried out zebrafish TUNEL, immunofluorescence and western blot experiments, with aid from L.A. for the OPA1 IHC. apoE ISH and NR stain were performed by H.B.R. with analysis by D.F. The zebrafish functional assays were executed by E.A. Figures were finalised by D.F. D.F., R.M.-J., C.R. and M.C. contributed to manuscript preparation. Funding was secured by C.R. and M.C.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Rebeca Martín-Jiménez, Danilo Faccenda.
Co-senior Authors: Claire Russell, Michelangelo Campanella.
Edited by D. Bano
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41419-018-0578-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Martin-Jimenez R, Campanella M, Russell C. New zebrafish models of neurodegeneration. Curr. Neurol. Neurosci. Rep. 2015;15:33. doi: 10.1007/s11910-015-0555-z. [DOI] [PubMed] [Google Scholar]
- 2.Pinho BR, et al. How mitochondrial dysfunction affects zebrafish development and cardiovascular function: an in vivo model for testing mitochondria-targeted drugs. Br. J. Pharmacol. 2013;169:1072–1090. doi: 10.1111/bph.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanes F, et al. The compound BTB06584 is an IF1 -dependent selective inhibitor of the mitochondrial F1 Fo-ATPase. Br. J. Pharmacol. 2014;171:4193–4206. doi: 10.1111/bph.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo C, Flinn LJ, Bandmann O. Heterozygous mutations in the FGF8, SHH and nodal/transforming growth factor beta pathways do not confer increased dopaminergic neuron vulnerability--a zebrafish study. Neurosci. Lett. 2013;532:55–58. doi: 10.1016/j.neulet.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 5.Zdebik AA, et al. Epilepsy in kcnj10 morphant zebrafish assessed with a novel method for long-term EEG recordings. PLoS ONE. 2013;8:e79765. doi: 10.1371/journal.pone.0079765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah DI, et al. Mitochondrial Atpif1 regulates haem synthesis in developing erythroblasts. Nature. 2012;491:608–612. doi: 10.1038/nature11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMauro S. Mitochondrial diseases. Biochim. Biophys. Acta. 2004;1658:80–88. doi: 10.1016/j.bbabio.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Birket MJ, et al. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell Sci. 2011;124:348–358. doi: 10.1242/jcs.072272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien LC, Keeney PM, Bennett JP., Jr. Differentiation of human neural stem cells into motor neurons stimulates mitochondrial biogenesis and decreases glycolytic flux. Stem Cells Dev. 2015;24:1984–1994. doi: 10.1089/scd.2015.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson TJ, Slupe AM, Strack S. Cell signaling and mitochondrial dynamics: Implications for neuronal function and neurodegenerative disease. Neurobiol. Dis. 2013;51:13–26. doi: 10.1016/j.nbd.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida M, Muneyuki E, Hisabori T. ATP synthase--a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 12.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl Acad. Sci. USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonora M, et al. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene. 2015;34:1475–1486. doi: 10.1038/onc.2014.96. [DOI] [PubMed] [Google Scholar]
- 14.Hejzlarova K, et al. Nuclear genetic defects of mitochondrial ATP synthase. Physiol. Res. 2014;63:S57–S71. doi: 10.33549/physiolres.932643. [DOI] [PubMed] [Google Scholar]
- 15.Chinnery, P. F. Mitochondrial Disorders Overview GeneReviews® [Internet]. (Seattle, WA: University of Washington, Seattle; 1993-2018). [PubMed]
- 16.Veas-Perez de Tudela M, et al. Regulation of Bcl-xL-ATP synthase interaction by mitochondrial cyclin B1-cyclin-dependent kinase-1 determines neuronal survival. J. Neurosci. 2015;35:9287–9301. doi: 10.1523/JNEUROSCI.4712-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daum B, Walter A, Horst A, Osiewacz HD, Kuhlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl Acad. Sci. USA. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faccenda D, Campanella M. Molecular regulation of the mitochondrial F(1)F(o)-ATPsynthase: physiological and pathological significance of the inhibitory factor 1 (IF(1)) Int J. Cell Biol. 2012;2012:367934. doi: 10.1155/2012/367934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Bermudez J, Cuezva JM. The ATPase inhibitory factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochim. Biophys. Acta. 2016;1857:1167–1182. doi: 10.1016/j.bbabio.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Domenis R, Bisetto E, Rossi D, Comelli M, Mavelli I. Glucose-modulated mitochondria adaptation in tumor cells: a focus on ATP synthase and inhibitor Factor 1. Int. J. Mol. Sci. 2012;13:1933–1950. doi: 10.3390/ijms13021933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Arago M, Formentini L, Garcia-Bermudez J, Cuezva JM. IF1 reprograms energy metabolism and signals the oncogenic phenotype in cancer. Cell Cycle. 2012;11:2963–2964. doi: 10.4161/cc.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faccenda D, Tan CH, Duchen MR, Campanella M. Mitochondrial IF(1) preserves cristae structure to limit apoptotic cell death signaling. Cell Cycle. 2013;12:2530–2532. doi: 10.4161/cc.25840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matic I, Strobbe D, Frison M, Campanella M. Controlled and impaired mitochondrial quality in neurons: molecular physiology and prospective pharmacology. Pharmacol. Res. 2015;99:410–424. doi: 10.1016/j.phrs.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Arago M, Garcia-Bermudez J, Martinez-Reyes I, Santacatterina F, Cuezva JM. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013;14:638–644. doi: 10.1038/embor.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genoux A, et al. Mitochondrial inhibitory factor 1 (IF1) is present in human serum and is positively correlated with HDL-cholesterol. PLoS ONE. 2011;6:e23949. doi: 10.1371/journal.pone.0023949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faccenda D, Tan CH, Seraphim A, Duchen MR, Campanella M. IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodelling. Cell Death Differ. 2013;20:686–697. doi: 10.1038/cdd.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol. Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Campanella M, et al. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Volkenhoff A, et al. Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 2015;22:437–447. doi: 10.1016/j.cmet.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Formentini L, et al. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014;33:762–778. doi: 10.1002/embj.201386392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox MW. The visual cliff test for the study of visual depth perception in the mouse. Anim. Behav. 1965;13:232–233. doi: 10.1016/0003-3472(65)90040-0. [DOI] [PubMed] [Google Scholar]
- 32.Matic I, et al. Neuroprotective coordination of cell mitophagy by the ATPase Inhibitory Factor 1. Pharmacol. Res. 2016;103:56–68. doi: 10.1016/j.phrs.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Arago M, et al. Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis. 2013;2:e46. doi: 10.1038/oncsis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postlethwait JH, et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 35.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 36.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 37.Kozlowski C, Weimer RM. An automated method to quantify microglia morphology and application to monitor activation state longitudinally in vivo. PLoS ONE. 2012;7:e31814. doi: 10.1371/journal.pone.0031814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry VH, Holmes C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 2014;10:217–224. doi: 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- 39.Madeira MH, Boia R, Santos PF, Ambrosio AF, Santiago AR. Contribution of microglia-mediated neuroinflammation to retinal degenerative diseases. Mediat. Inflamm. 2015;2015:673090. doi: 10.1155/2015/673090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brockerhoff SE, et al. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc. Natl Acad. Sci. USA. 1995;92:10545–10549. doi: 10.1073/pnas.92.23.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander C, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 42.Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 43.Diez-Roux G, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magdaleno S, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aijaz S, Erskine L, Jeffery G, Bhattacharya SS, Votruba M. Developmental expression profile of the optic atrophy gene product: OPA1 is not localized exclusively in the mammalian retinal ganglion cell layer. Invest. Ophthalmol. Vis. Sci. 2004;45:1667–1673. doi: 10.1167/iovs.03-1093. [DOI] [PubMed] [Google Scholar]
- 46.Faccenda D, et al. Control of mitochondrial remodeling by the ATPase inhibitory factor 1 unveils a pro-survival relay via OPA1. Cell Rep. 2017;18:1869–1883. doi: 10.1016/j.celrep.2017.01.070. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura J, Fujikawa M, Yoshida M. IF1, a natural inhibitor of mitochondrial ATP synthase, is not essential for the normal growth and breeding of mice. Biosci. Rep. 2013;33:e00067. doi: 10.1042/BSR20130078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delettre C, Lenaers G, Pelloquin L, Belenguer P, Hamel CP. OPA1 (Kjer type) dominant optic atrophy: a novel mitochondrial disease. Mol. Genet. Metab. 2002;75:97–107. doi: 10.1006/mgme.2001.3278. [DOI] [PubMed] [Google Scholar]
- 49.Campanella M, Parker N, Tan CH, Hall AM, Duchen MR. IF(1): setting the pace of the F(1)F(o)-ATP synthase. Trends Biochem. Sci. 2009;34:343–350. doi: 10.1016/j.tibs.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez-Martin A, et al. The mitochondrial H(+)-ATP synthase and the lipogenic switch: new core components of metabolic reprogramming in induced pluripotent stem (iPS) cells. Cell Cycle. 2013;12:207–218. doi: 10.4161/cc.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingham PW. Zebrafish genetics and its implications for understanding vertebrate development. Hum. Mol. Genet. 1997;6:1755–1760. doi: 10.1093/hmg/6.10.1755. [DOI] [PubMed] [Google Scholar]
- 52.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 53.Okouchi M, Ekshyyan O, Maracine M, Aw TY. Neuronal apoptosis in neurodegeneration. Antioxid. Redox Signal. 2007;9:1059–1096. doi: 10.1089/ars.2007.1511. [DOI] [PubMed] [Google Scholar]
- 54.Luo XG, Ding JQ, Chen SD. Microglia in the aging brain: relevance to neurodegeneration. Mol. Neurodegener. 2010;5:12. doi: 10.1186/1750-1326-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ames A, 3rd, Li YY, Heher EC, Kimble CR. Energy metabolism of rabbit retina as related to function: high cost of Na+transport. J. Neurosci. 1992;12:840–853. doi: 10.1523/JNEUROSCI.12-03-00840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okawa H, Sampath AP, Laughlin SB, Fain GL. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr. Biol. 2008;18:1917–1921. doi: 10.1016/j.cub.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 58.Barbato S, Sgarbi G, Gorini G, Baracca A, Solaini G. The inhibitor protein (IF1) of the F1F0-ATPase modulates human osteosarcoma cell bioenergetics. J. Biol. Chem. 2015;290:6338–6348. doi: 10.1074/jbc.M114.631788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schrier SA, Falk MJ. Mitochondrial disorders and the eye. Curr. Opin. Ophthalmol. 2011;22:325–331. doi: 10.1097/ICU.0b013e328349419d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahn JJ, Stackley KD, Chan SS. Opa1 is required for proper mitochondrial metabolism in early development. PLoS ONE. 2013;8:e59218. doi: 10.1371/journal.pone.0059218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olichon A, et al. Mitochondrial dynamics and disease, OPA1. Biochim. Biophys. Acta. 2006;1763:500–509. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim. Biophys. Acta. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Frezza C, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 64.Olichon A, et al. Effects of OPA1 mutations on mitochondrial morphology and apoptosis: relevance to ADOA pathogenesis. J. Cell Physiol. 2007;211:423–430. doi: 10.1002/jcp.20950. [DOI] [PubMed] [Google Scholar]
- 65.Thisse B, et al. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/S0091-679X(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 66.Saint-Amant L, Drapeau P. Time course of the development of motor behaviors in the zebrafish embryo. J. Neurobiol. 1998;37:622–632. doi: 10.1002/(SICI)1097-4695(199812)37:4<622::AID-NEU10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 67.Brustein E, et al. Steps during the development of the zebrafish locomotor network. J. Physiol. Paris. 2003;97:77–86. doi: 10.1016/j.jphysparis.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Rouslin W, Pullman ME. Protonic inhibition of the mitochondrial adenosine 5’-triphosphatase in ischemic cardiac muscle. Reversible binding of the ATPase inhibitor protein to the mitochondrial ATPase during ischemia. J. Mol. Cell. Cardiol. 1987;19:661–668. doi: 10.1016/S0022-2828(87)80374-7. [DOI] [PubMed] [Google Scholar]
- 69.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) Oregon: Univ of Oregon Press; 2007. [Google Scholar]
- 70.Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/S0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- 71.Nijtmans LGJ, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 72.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat. Prot. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 73.Calvaruso MA, Smeitink J, Nijtmans L. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods. 2008;46:281–287. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 74.Van’t Veer A, et al. Ablation of kappa-opioid receptors from brain dopamine neurons has anxiolytic-like effects and enhances cocaine-induced plasticity. Neuropsychopharmacology. 2013;38:1585–1597. doi: 10.1038/npp.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lefebvre V, et al. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013;9:1770–1779. doi: 10.4161/auto.25413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.