Abstract
We examined the expression of a family of Arabidopsis response regulators (ARR) and found that the steady-state levels of RNA for most are elevated very rapidly by cytokinin. Using nuclear run-on assays we demonstrated that this increase in ARR transcript levels in response to cytokinin is due, at least in part, to increased transcription. The start site of transcription for the ARR5 gene was identified using primer extension analysis. A DNA fragment comprised of 1.6 kb upstream of the ARR5 transcript start site conferred cytokinin-inducible gene expression when fused to a β-glucuronidase reporter, confirming that the transcription rate of ARR5 is elevated by cytokinin. This reporter construct was also used to examine the spatial pattern of ARR5 expression. The highest levels of expression were observed in the root and shoot apical meristems, at the junction of the pedicle and the silique, and in the central portion of mature roots. The expression of ARR5 in the apical meristems was confirmed by whole mount in situ analysis of seedlings and is consistent with a role for cytokinin in regulating cell division in vivo.
Cytokinins are a class of plant hormones originally identified by their ability to stimulate cell division in concert with auxin, and to act antagonistically to auxin in the control of shoot and root initiation in culture (Miller et al., 1955, 1956). Cytokinins have subsequently been implicated in many other plant growth and development processes including shoot organogenesis, leaf senescence, sink/source relationships, vascular development, lateral bud release, and photomorphogenic development (Mok and Mok, 1994).
Cytokinins affect the expression of many different genes in a variety of plant species. However, most of these genes are also regulated by other stimuli and/or are induced after a relatively long lag period (>1 h) (Crowell and Amasino, 1994; Hare and Staden, 1997; Schmülling et al., 1997). Two exceptions are the Arabidopsis response regulator (ARR)4 and ARR5 genes (previously called IBC7 and IBC6; Brandstatter and Kieber, 1998), which were identified in a differential display screen for cytokinin-regulated transcripts. ARR4 and ARR5 display properties of cytokinin primary-response genes: The elevation of the steady-state level of transcript occurs within 10 min of exogenous cytokinin application, the rapid induction is specific for cytokinins, and it is resistant to inhibition of protein synthesis (Brandstatter and Kieber, 1998).
The sequences of ARR4 and ARR5 are similar to bacterial two-component response regulators (for review, see Stock et al., 1990; Parkinson, 1993; Hoch and Silhavy, 1995). Response regulators act downstream of sensor His kinases in bacterial two-component signaling systems. The signal is detected by the input domain of the sensor kinase, which in turn regulates the dimerization and His autophosphorylation of the transmitter domain. The signal is transmitted by transfer of a phosphate from the phospho-His of the transmitter domain to an Asp residue contained within the receiver domain of a cognate response regulator, which in turn commonly regulates the function of a fused output domain that acts as a transcription factor.
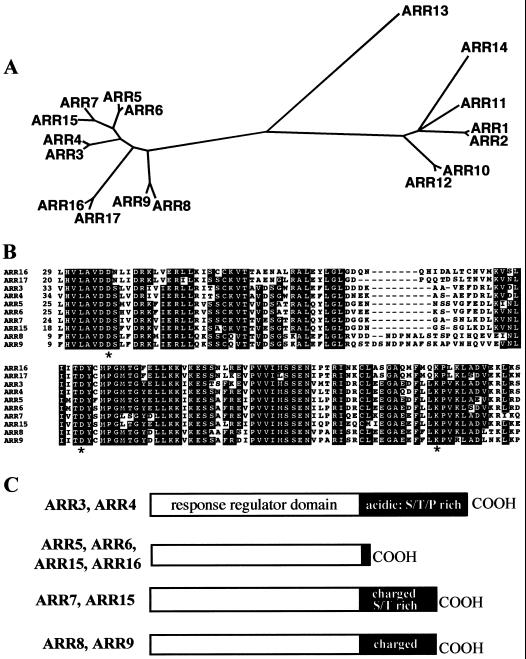
In addition to ARR4 and ARR5, the Arabidopsis genome encodes several other response regulator homologs (Fig. 1; Brandstatter and Kieber, 1998; Taniguchi et al., 1998; Imamura et al., 1999; Sakakibara et al., 2000). This ARR gene family has been divided into two groups, called type A and type B, which differ in their sequence and domain structure (Brandstatter and Kieber, 1998; Imamura et al., 1999). Furthermore, several of the type A genes have been found to be responsive to exogenous cytokinin treatment, but the type B genes do not appear to be regulated by cytokinin (Brandstatter and Kieber, 1998; Taniguchi et al., 1998; Imamura et al., 1999; Kiba et al., 1999). Type A-like ARR genes have also been found in maize, rice, and cotton (Brandstatter and Kieber, 1998; Sakakibara et al., 1998).
Figure 1.
The ARR gene family. A, Phylogenetic tree of ARR receiver domains. The tree was generated using the AllAll program at Molecular Biology Computational Resource at Baylor college of Medicine (http://cbrg.ing.ethz.ch/subsection3_1_1.html). The amino acid sequences of only the receiver domains of each gene was used (see B). This program uses a least-squared, heuristic method to generate trees. The accession numbers for all but ARR15, ARRR16, and ARR17 are presented in D'Agostino and Kieber (1999). The accession numbers for the novel ARR-predicted amino acid sequences are as follows: ARR15: AF305720; ARR16: AF305721; ARR17: AF305722. B, Alignment of type A ARR receiver domain amino acid sequences. Residues identical in >6 ARRs are blocked in black. The three residues that are invariant among all response regulators are marked with an asterisk, including the predicted Asp phosphorylation site, embedded in a conserved TDY sequence. C, A cartoon representation of the domain structures of the various type A ARR proteins. The ARRs corresponding to each of the four different structures is indicated to the left of each cartoon. The response regulator domains are represented by white rectangles and the C-terminal extensions by black rectangles. The different domains are depicted to scale and the various features of the C-terminal extensions are noted within the black rectangles.
Receiver domain homologs are also found fused to sensor kinases in Arabidopsis (Chang and Stewart, 1998). These are referred to as hybrid sensor kinases and have been found in most eukaryotic and some prokaryotic two-component systems (Loomis et al., 1997; Perraud et al., 1999). In Arabidopsis the hybrid sensor kinase family includes several of the ethylene receptors, as well as CKI1, which has been implicated in cytokinin action (Kakimoto, 1996).
The type A ARRs are similar in structure to CheY, a bacterial response regulator involved in chemotaxis, in that they lack a typical output domain (D'Agostino and Kieber, 1999). The type B ARRs (ARR1, 2, and 10–14) have a large C-terminal extension that has characteristics of an output domain. The type B, but not the type A, ARRs contain potential nuclear localization signals in the carboxy-terminal domain, and ARR10 and ARR11 have been confirmed to be nuclear-localized proteins using transient transformation of green fluorescent protein-fusions into parsley protoplasts (Lohrmann et al., 1999). Furthermore, the C-terminal domain of ARR11, but not the C terminus of ARR4, can activate transcription in yeast when fused to the GAL4 DNA-binding domain (Lohrmann et al., 1999). Thus, it is likely that the type B ARRs are transcription factors and that the carboxy-terminal portions of these proteins act as output domains.
In this study we examined the kinetics of the response of the type A ARR genes to cytokinin. The increase in the steady-state level of ARR mRNA in response to cytokinin could be due to increased transcription, stabilization of the existing message, or a combination of both mechanisms. There are several examples of transcriptional and post-transcriptional mechanisms of cytokinin-induced mRNA accumulation (Flores and Tobin, 1988; Langridge et al., 1989; Dominov et al., 1992; Lu et al., 1992; Silver et al., 1996; Dowens and Crowell, 1998). Here we demonstrate, using nuclear run-on analyses and fusions to reporter constructs, that the induction of several of the type A ARR genes by cytokinin is at least partially due to increased transcription levels. We also examined the kinetics of induction of several new type A ARR genes and determined the pattern of expression of ARR5. These studies begin to shed light on the role of these genes in Arabidopsis.
RESULTS
The ARR Gene Family in Arabidopsis
Fourteen response regulator homologs (ARR1–ARR14) were previously identified in Arabidopsis (D'Agostino and Kieber, 1999; Imamura et al., 1999). A search of the Arabidopsis genomic sequence with the ARR3-ARR9 predicted amino acid sequences revealed three additional type A ARR family members (Fig. 1). We have called these new type A genes ARR15, ARR16, and ARR17 (genomic sequences are from accession nos. [AC008263/AC013258], AC007660, and AL163972, located on chromosome 1, 2, and 3, respectively). Comparison of the amino acid sequences predicted from the annotation of these genomic sequences to the other type A ARR predicted peptides suggested that ARR15 and ARR16 were annotated correctly, but that some of the 3′-coding sequences were missing from the predicted coding region of ARR17. Sequence analysis of reverse transcriptase-PCR products from each of these three genes is consistent with the genome database splicing predictions of the ARR15 and ARR16 genes, and the larger ARR17 product (accession nos. AF305720, AF305721, and AF305722, respectively; data not shown). Furthermore, the predicted sizes of ARR15 and ARR16 coding regions are consistent with the apparent mRNA sizes for these genes as determined by northern analysis (data not shown).
Ten type A ARR genes have now been identified in Arabidopsis. All contain the highly conserved Lys and two Asp residues found in other receiver domains, including the predicted Asp phosphorylation site (Fig. 1B). Phylogenetic analysis reveals that the type A genes are very distinct from the type B genes, and that the 10 type A genes fall into five pairs of related sequences (Fig. 1). This pairing of the type A genes is consistent with a recent evolutionary duplication postulated to have occurred in the Arabidopsis genome (Pickett and Meeks-Wagner, 1995) and suggests that there may be genetic redundancy within this gene family. The sequences among the type A genes display from 60% to 93% amino acid identity, but the type A and type B receiver domains display less than 30% amino acid identity. The amino acid sequences of the receiver domains fused to the Arabidopsis sensor kinases ETR1 (Chang et al., 1993) and CKI1 (Kakimoto, 1996) are only distantly related (< 25% amino acid identity) to the ARRs.
The type A ARR proteins are most similar to each other in their receiver domains. The C-terminal extensions are significantly shorter (<100 amino acids) than those of the type B proteins (some as large as 500 amino acids) and they lack properties consistent with output domains. Although the predicted amino acid sequences of the C-terminal extensions of the type A genes are more variable than those of the receiver domains, within each ARR pair these C-terminal sequences tend to be similar (Fig. 1C). ARR3 and ARR4 have the longest predicted C-terminal domains, which are highly acidic and Ser, Thr, and Pro rich. The C-terminal extensions of ARR8, ARR9, ARR7, and ARR15 contain many charged residues, and those of ARR7 and ARR15 are also rich in Ser and Thr. ARR5, ARR6, ARR16, and ARR17 have very short (<30 amino acids) C-terminal domains.
Characterization of Cytokinin Induction of ARRs
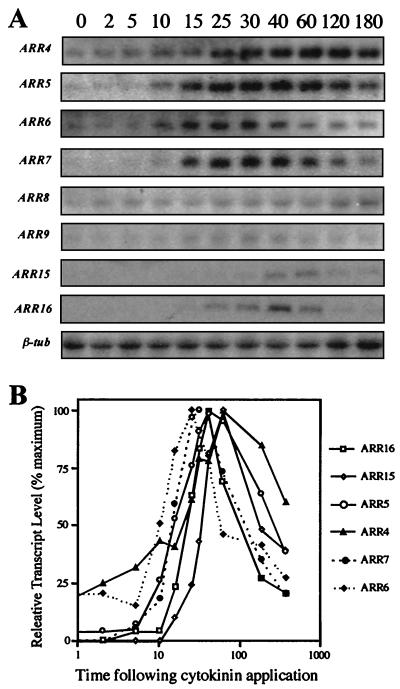
The expression of the 10 Arabidopsis type A ARR genes in response to exogenous cytokinin in etiolated Arabidopsis seedlings was analyzed using northern blotting (Fig. 2). Consistent with previous results (Brandstatter and Kieber, 1998), ARR4 and ARR5 were induced rapidly by cytokinin. The steady-state levels of ARR5 and ARR4 transcripts were elevated within 10 min of application of exogenous cytokinin, reached a maximal induction at 30 to 40 min (ARR4) or 60 min (ARR5), and then slowly declined. The steady-state levels of ARR6, ARR7, ARR15, and ARR16 mRNA were also elevated by exogenous cytokinin, with kinetics generally similar to that of ARR5. Control seedlings treated with dimethyl sulfoxide (DMSO) for 15, 45, 120, and 360 min demonstrated no significant increase in the steady-state level of ARR transcripts (data not shown). We detected little or no increase in steady-state level of ARR8 or ARR9 mRNA following cytokinin treatment (Fig. 2), and were unable to detect ARR3 or ARR17 transcripts on the blots. It should be emphasized that the relative hybridization signals among the different blots may not reflect the relative transcript levels of these genes as the hybridization probes may have distinct specific activities and the exposure times for each blot varied.
Figure 2.
Kinetics of ARR gene induction. A, Northern-blot analysis of 15 μg of total RNA from 3-d-old etiolated seedlings treated with 5 μm BA for various times (indicated in minutes above each lane) and hybridized with the indicated ARR probe (at left). The β-tubulin shown is a representative image and is not the loading control for all of the blots (see methods). B, Quantification of transcript levels from blot depicted in A. The signals from the northern blots were quantified with a PhosphorImager and normalized to the β-tubulin loading control. The highest level of expression for each probe was assigned a value of 100% and all other points were normalized to it (percentage of maximum).
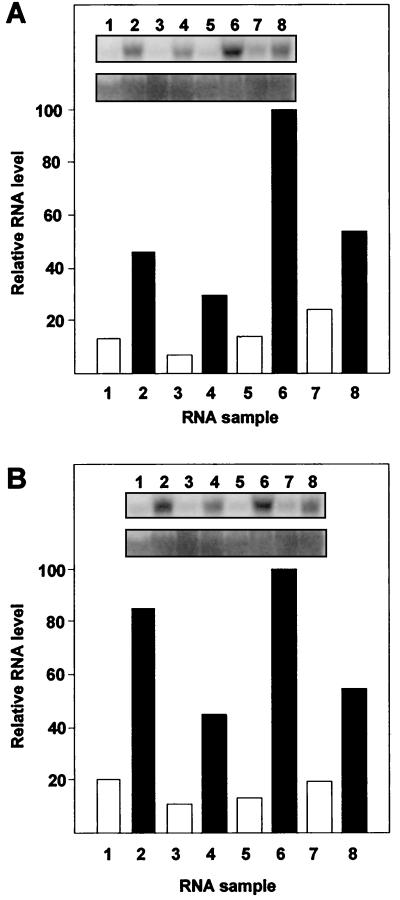
We also examined ARR6 and ARR7 expression in various adult tissues in the presence and absence of cytokinin (Fig. 3). Flowers, inflorescence stems, leaves, and soil-grown roots from 2-week-old plants were treated with 5 μm Benzyladenine (BA) or DMSO for 50 min. Similar to the ARR4 and ARR5 genes (Brandstatter and Kieber, 1998), the steady-state levels of ARR6 and ARR7 mRNA were elevated after cytokinin treatment in all tissues tested. ARR6 and ARR7 displayed similar patterns of expression, the major difference being that ARR7 showed a greater induction in flowers.
Figure 3.
Tissue-specific expression of ARR6 and ARR7. A and B, RNA was extracted from tissues treated with 5 μm BA (lanes 2, 4, 6, and 8) or buffer control (lanes 1, 3, 5, and 7) for 50 min. Fifteen micrograms of total RNA was separated by agarose gel electrophoresis, blotted to a nylon membrane, and hybridized with an ARR6 (A), ARR7 (B), or 18S rDNA (bottom of each inset) probe. Tissues analyzed were buds and young flowers (lanes 1 and 2), stems of young inflorescences (lanes 3 and 4), leaves from 2-week-old adult plants (lanes 5 and 6), and soil-grown roots (lanes 7 and 8). The signals were quantified using a PhosphorImager and the expression level in each tissue normalized to the 18S loading control. The highest signal was set to 100% and the other samples were normalized to it.
Effect of Cycloheximide on ARR Gene Expression
To determine if the elevation of the steady-state level of ARR mRNA in response to cytokinin was dependent on de novo protein synthesis we examined the effect of the protein synthesis inhibitor cycloheximide. As shown in Table I, cycloheximide treatment itself results in a rise in the steady-state level of most of the ARR genes. This is characteristic of many primary-response genes, including many of the auxin-induced indole-3-acetic acid (IAA) genes (Franco et al., 1990; Abel et al., 1995). Cycloheximide failed to block the induction of these genes by cytokinin, indicating that the proteins required to transduce the cytokinin signal to elevate ARR transcript levels are present before application of the hormone.
Table I.
Effect of cycloheximide treatment on ARR gene expression in response to cytokinin
| Gene | Control | +BA | +Chxa | +BA and +Chx |
|---|---|---|---|---|
| ARR4 | 25 | 53 | 74 | 100 |
| ARR5 | ndb | 48 | 74 | 100 |
| ARR6 | 15 | 58 | 46 | 100 |
| ARR7 | nd | 41 | 64 | 100 |
| ARR8 | 20 | 27 | 59 | 100 |
| ARR9 | 45 | 39 | 51 | 100 |
| ARR15 | nd | 32 | 59 | 100 |
| ARR16 | nd | nd | 51 | 100 |
Seedlings were treated with nothing (control) or a combination of 50 μm cycloheximide and subsequently BA as described in “Materials and Methods” and the total RNA was analyzed by northern blotting. The blots were hybridized to the indicated probes. The intensity of each band was quantified using a phosphor imager and the relative intensity was normalized to a tubulin loading control. The highest signal for each probing from each treatment was assigned a value of 100% and the others are presented as a percentage of this maximum.
Chx, Cycloheximide.
nd, No signal detected above the background.
Cytokinin Increases the Rate of Transcription of the ARR Genes
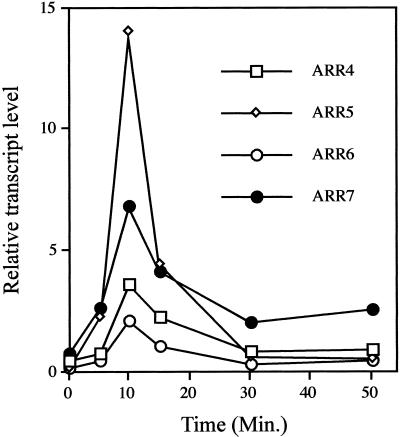
We used nuclear run-on assays to examine the rate of transcription of ARR4, ARR5, ARR6, and ARR7 in response to exogenous cytokinin in fully expanded leaves (Fig. 4). The ARR7 and ARR5 genes displayed a slight increase in the rate of transcription within 5 min of cytokinin treatment and all four genes showed maximal induction 10 min following cytokinin treatment. ARR4 demonstrated the highest rate of transcription in response to cytokinin at the peak of induction and ARR6 showed the lowest level of transcription. After 30 min, the rates of transcription for ARR4, ARR5, and ARR6 returned to basal levels. The rate of ARR7 transcription also declined, but still remained elevated above the basal level after 50 min. There was no increase in the rate of ARR transcription in control leaves treated with DMSO for 50 min (data not shown). Because all nascent RNA transcripts are labeled equally in the in vitro transcription reactions, the relative amplitudes shown in Figure 4 accurately depict differences in the rates of transcription among the ARR genes.
Figure 4.
Nuclear run-on analysis of ARR genes in cytokinin-treated tissue. Leaves from 2-week-old adult plants were incubated with 5 μm BA for the indicated times. Intact nuclei were purified and used for in vitro transcription. Nuclear transcripts were hybridized to duplicate slot blots containing 200 ng of ARR5, ARR4, ARR6, ARR7, or β-tubulin cDNAs. The hybridizations were quantified using a PhosphorImager and the ARR signals normalized to the tubulin control. The resultant relative transcript levels were plotted as a function of time.
Identification of the Transcriptional Start Site of ARR5
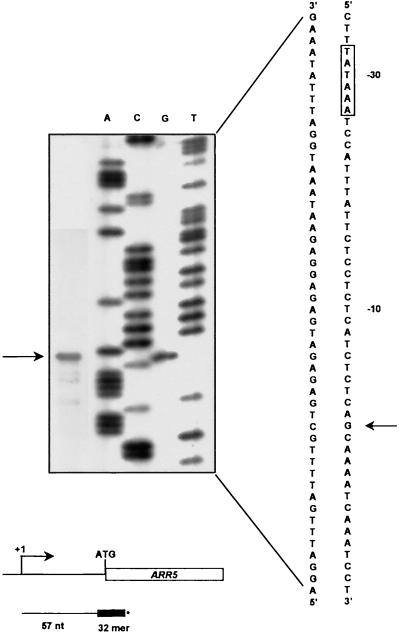
The transcription initiation site for ARR5 was determined using a primer extension assay (Fig. 5). Using a 32-bp primer that encompassed the presumed start site of translation, a single primer extension product was identified. The start site of transcription was found to be 68 bp upstream of the translation start codon. There are no other potential ATG initiation codons within this 68-bp 5′-untranslated region. A sequence consistent with the consensus for a TATA box is located 27 bp upstream of the transcription start site.
Figure 5.
Identification of the transcription initiation site of the ARR5 gene. Primer extension analysis. A [32P]-labeled ARR5 primer (see “Materials and Methods”) was hybridized with 50 μg of total RNA and extended using reverse transcriptase. The product was resolved on a sequencing gel (far left lane), and the mobility compared with an ARR5 genomic DNA sequencing reaction using the same ARR5 primer (lanes A, C, G, and T). The nucleotide sequence in the vicinity of the start site of transcription is shown to the right of the gel insert. The arrow marks the deduced transcription start site and the boxed residues mark the predicted TATA box. Below the gel inset is a schematic diagram of the ARR5 primer relative to the transcription and translation start sites and the primer extended product.
Cytokinin Activates Transcription of a β-Glucuronidase (GUS) Reporter Fused to the ARR5 Promoter
To confirm that induction of these genes is due to transcriptional activation, a GUS reporter gene was cloned downstream of the ARR5 promoter. This construct contained 1.6 kb of ARR5 upstream sequences, starting at position −1 (where the deduced transcription start site is +1). The resulting pIB-1.6TC plasmids were transformed into Arabidopsis. GUS activity was strongly elevated by growth in the presence of cytokinin for multiple independent pIB-1.6TC transformants (Fig. 6). Because this construct contained no ARR5 transcribed sequences, this confirms that cytokinin elevates the rate of transcription from the ARR5 promoter. However, this does not preclude the possibility that post-transcriptional mechanisms may also regulate ARR5 in response to cytokinin.
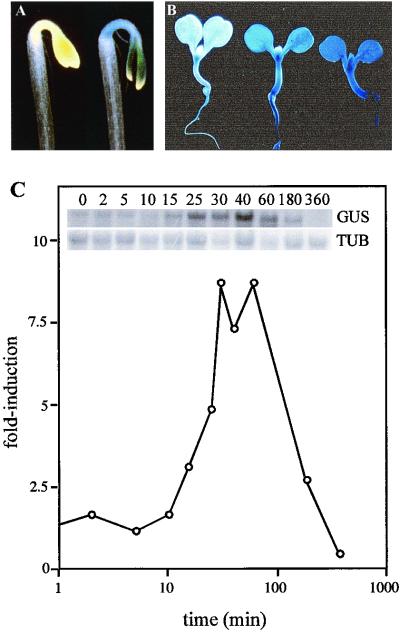
Figure 6.
Transcriptional induction of ARR5 by cytokinin as demonstrated by promoter GUS fusions in transgenic plants. A, GUS staining of etiolated transgenic seedlings harboring an ARR5 promoter, but lacking the 5′-untranslated region fused to GUS (pIB-1TC). Seedlings were grown for 3 d in the dark on MS with no added hormone (left seedling) in the presence of 2.5 μm BA (right seedling). B, GUS staining of pIB-1TC transgenic seedlings were grown for 6 d in the light on MS containing no (left seedling), 0.5 μm BA (center seedling), or 5 μm BA (right seedling). C, Northern analysis of GUS expression in pIB-1TC transgenic seedlings following cytokinin treatment. Leaves from 2-week-old pIB-1TC plants were treated with 5 μm BA for various times and total RNA isolated. The RNA was analyzed by northern blotting and hybridized with a GUS (top inset, GUS), or β-tubulin (bottom inset, TUB). The signals were quantified using a PhosphorImager and the GUS signal was normalized to the β-tubulin loading control. The relative signal at time 0 was assigned a value of 1, and the subsequent time points plotted relative to it (fold-induction).
Northern analysis was used to analyze the kinetics of GUS induction by cytokinin (Fig. 6). The steady-state level of GUS mRNA begins to rise in the pIB-1.6TC transformants within 15 min following application of exogenous cytokinin and peaks at 30 to 60 min. These induction kinetics are similar to those for the endogenous ARR5 gene, which suggests that this 1.6-kb fragment is sufficient to confer cytokinin regulation and that the elevation of ARR5 transcripts is mediated, at least in part, via elevated transcription.
Pattern of ARR5 Expression
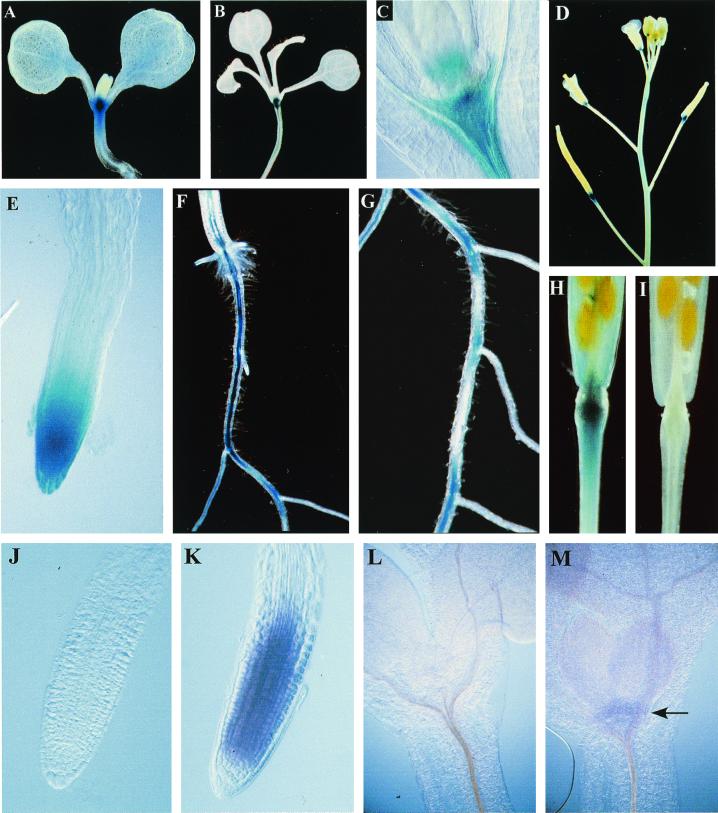
We examined transgenic plants harboring a T-DNA containing a fusion of 1.6 kb of the ARR5 promoter region to a GUS reporter (pIB-1.6TC) to examine the pattern of ARR5 expression. Plants at various stages of development were stained with 5-bromo-4-choloro-3-indolyl-glucuronidide to reveal reporter gene expression (Fig. 7, A–I). Qualitatively similar patterns of expression were observed in multiple, independent lines and the common features of this staining will be described here. The earliest and most prominent GUS expression was seen in the root and shoot meristem regions. In 15-d-old plants grown on Murashige and Skoog (MS) media staining is also very strong in the vasculature of the older portions of the primary root and moderate staining is present in the vasculature of the hypocotyl. Increased staining is seen as the primary root ages (Fig. 7F). Patchy, intermittent vascular staining is observed in mid portions of the root, especially at the junction of lateral roots (Fig. 7G). There is a high level of GUS expression in the primary root tip (Fig. 7E) and also in the tips of the lateral root. No GUS expression is detectable in very young leaves, but weak, diffuse staining appears in older leaves and cotyledons. Strong staining is evident in the abscission zone of flowers and immature siliques (Fig. 7, D and H), but as siliques turn yellow expression is no longer detectable (Fig. 7I).
Figure 7.
Pattern of ARR5 expression. A through I, Gus staining of stable Arabidopsis transformants harboring a pIB1.6-TC-GUS T-DNA. Apical portion of 7-d-old (A) and 15-d-old (B) seedlings grown in the light on MS. C, Close-up of the region corresponding to the apical meristem in a 10-d-old light-grown plant. D, Inflorescence of 4-week-old plants. E, Close-up of primary root tip from 7-d-old seedlings grown in the light on MS. F, Mature root/hypocotyl junction from 15-d-old seedlings grown in the light on MS. G, Central portion of root from same seedling as in F. H, Silique from 4-week-old plant, mature, but fully green. I, Silique that had just turned yellow from a 4-week-old plant. J through M, Whole mount in situ hybridization with ARR5. Five-day-old light-grown seedlings grown on MS were fixed and hybridized with a sense (J and L) or an antisense (K and M) digoxygenin-labeled ARR5 RNA probe (see “Materials and Methods”). The sense probe is a negative control and should not hybridize to the endogenous ARR5 transcript. Purple color indicates a positive reaction. The arrow in M indicates the shoot apical meristem staining evident with the antisense probe.
To confirm the endogenous pattern of ARR5 expression seen with the GUS staining, whole mount in situ hybridization was performed. Arabidopsis plants were grown in culture for 5 d, fixed, and hybridized with sense and antisense ARR5 probes (Fig. 7, J–M). ARR5 expression as revealed by this technique was generally consistent with the pattern of GUS staining in transgenic seedlings harboring pIB-1.6TC (see above). Hybridization with an ARR5 antisense probe resulted in strong staining in the root apical meristem, and weak, but consistent staining in the shoot apical meristem (Fig. 7). However, staining in the root tip cells did not extend into the columella and root cap cells as did the GUS stain. Hybridization with an ARR5 sense probe resulted in little or no staining under these conditions.
DISCUSSION
ARR4 and ARR5 (previously called IBC7 and IBC6) were the first cytokinin primary response genes identified (Brandstatter and Kieber, 1998). They are members of the Arabidopsis type A family of two-component response regulator homologs, several others of which had been previously reported as cytokinin-inducible (Taniguchi et al., 1998; Kiba et al., 1999). Here we provide data that indicates that several additional type A ARRs are also cytokinin primary response genes and present a more detailed analysis of their response to cytokinin. We have also determined the pattern of ARR5 gene expression.
Using northern analysis, a rise in the steady-state level of ARR gene expression is first detected 10 min after application of exogenous cytokinin. Similar to ARR4 and ARR5, the steady-state levels of ARR6, ARR7, ARR15, and ARR16 transcripts increase very rapidly (within 10 min) in response to cytokinin. Analysis of extension of nascent transcripts using a nuclear run-on assay indicates that increased transcription of these genes may first occur as rapidly as 5 min following cytokinin application. This resembles the kinetics of the auxin response of the PS-IAA, IAA, and SAUR auxin primary-response genes (Theologis et al., 1985; Koshiba et al., 1995). The discrepancy between the rise in the steady-state level and the elevation of transcription rates can be attributed to the time it takes for RNA processing and/or the time it takes to accumulate sufficient transcript to a level that is detectably higher than the baseline.
These rapid induction kinetics of the type A ARRs raises the question as to why expression of these genes is elevated following hormone perception. In many bacterial systems, response regulators are the first genes up-regulated upon perception of stimulus, resulting in a self-activating feedback loop that amplifies and sustains the response (Stock et al., 1990). If the ARR genes function as positive regulators of cytokinin action, then in a similar manner their induction may serve to amplify the signal flux initiated by cytokinin perception. In an alternate manner, the ARR proteins could act as negative regulators of cytokinin action, in which case their induction by exogenous cytokinin could act to attenuate the response to elevated cytokinin levels.
The induction of the ARR genes in response to cytokinin is resistant to inhibition of protein synthesis by cycloheximide. This, coupled with the induction kinetics, is a defining characteristic of primary response genes and indicates that the response to cytokinin is achieved via pre-existing components. It is interesting that these genes are induced by cycloheximide itself, which also occurs with other primary response genes (Theologis et al., 1985; Herschman, 1991; Abel et al., 1995). Elevation of steady-state RNA levels by cycloheximide has been postulated to be the result of the elimination of a short-lived transcription repressor protein or stabilization of existing message due to a degradation of a short-lived, transcript-specific RNase (Koshiba et al., 1995).
ARR8 and ARR9 are not significantly induced by cytokinin, although there does appear to be some increase in the steady-state level of their mRNA in the presence of cycloheximide and a slight further increase in response to BA and cycloheximide. An increase in the steady-state level of ARR8 and ARR9 mRNA in response to cytokinin in the absence of cycloheximide has previously been reported (Kiba et al., 1999). This discrepancy could be due to differences in experimental conditions such as the tissues treated and the amount and/or type of cytokinin used. In addition, the induction in the previous report was not quantified, making definitive conclusions as to the level of induction difficult.
The induction of the ARR genes by cytokinin can be explained by an increase in transcription or by a post-transcriptional stabilization of the existing message. The steady-state level of mRNA represents a balance between ongoing transcription and message degradation. Run-on transcription in isolated nuclei represents the elongation of previously initiated transcripts by bound RNA polymerases. The nuclear run-on analyses show that the induction of ARR4, ARR5, ARR6, and ARR7 is regulated, at least in part, at the level of transcription. This does not exclude the possibility that changes in the half-life of the message also contribute to increases in the steady-state level of these mRNAs in response to cytokinin.
The transcriptional activation of the ARR5 gene in response to cytokinin was confirmed using transcriptional fusions of the ARR5 promoter to a GUS reporter gene. In addition, these studies indicate that an ARR5 1.6-kb upstream fragment contains cis-acting elements sufficient to confer cytokinin responsiveness, and a detailed analysis of this region should define a cytokinin responsive element, which will begin to bridge the gap between cytokinin perception and gene activation.
We used the ARR5 promoter-GUS fusions and whole mount in situ analysis to examine the expression pattern of the ARR5. In response to exogenous cytokinin, ARR5 expression is evident throughout the entire seedlings (Fig. 6A), indicating that most cells are developmentally capable of expressing ARR5. Expression of ARR5 in untreated seedlings was highest in the apical meristems, which is consistent with ARR5, and by inference cytokinin, having a role in the regulation of cell division (Hare and Staden, 1997). However, ARR5 expression was also present in non-dividing cells and not detectable in some dividing cell types, most notably the floral meristems. It is interesting that the pattern of ARR5 expression in the root tip that is observed coincides closely with the pattern of incorporation of [3H]thymidine into Arabidopsis roots (Dolan et al., 1993). Incorporation of thymidine reflects endogenous cell division rates. Cytokinins accumulate in mitotically active areas such as the root and shoot apical meristem (Mok and Mok, 1994; Dewitte et al., 1999) and expression of ARR5 in these regions may reflect these high levels of endogenous cytokinin.
Comparison of GUS and ARR5 RNA levels using northern analysis indicates similar temporal levels of expression, suggesting that GUS expression is a reliable reflection of the expression of ARR5. The slight differences between the GUS and in situ signals in the root tip could be due to tissue-specific differences in the stability of GUS and ARR5 mRNA, resistance of certain cells to permeabilization required for whole mount in situ hybridization, the long half-life of GUS protein, and/or a missing ARR5 regulatory sequence.
Three additional type A ARR genes were identified using in silico analysis of the Arabidopsis genome sequence. Only one other type A-like ARR gene is present in the nearly completed Arabidopsis genomic sequence database. However, this additional gene (accession no. AC006217.3) is likely to be a pseudogene it encodes only a carboxy-terminal fragment of an ARR. Thus the final size of the A ARR gene family is likely to be 10. An obvious question is why so many genes? The high conservation of the receiver domains among the type A genes and their generally similar induction kinetics suggest that there may be overlapping function within the family. It is possible that the various type A genes have distinct temporal or spatial patterns of expression. It is also possible that the large number of genes may reflect the requirement for the ARRs to signal to numerous, distinct downstream effectors, which would be consistent with the multitude of roles that cytokinins are hypothesized to play in plants. If this is indeed the case, the variable C-terminal domain may mediate signaling to these distinct effectors.
There are additional response regulator homologs present in Arabidopsis, including a homolog of the oilseed rape SAC13 gene and additional type B-like ARRs (from in silico analysis of the Arabidopsis genomic sequence; data not shown), as well as several receiver domains that are missing the putative Asp phosphorylation site, which have been termed pseudo-response regulators (Makino et al., 2000). The predicted amino acid sequences of these genes are only distantly related to those of the type A genes (data not shown), and only the SAC13 homolog has a type A domain structure. The type B ARRs are not affected by exogenous cytokinin treatment, and it is not known whether the oilseed rape homolog or the pseudo-response regulator genes are responsive to cytokinin. This suggests that these genes may not function in a cytokinin response pathway, but perhaps in the response to other stimuli such as ethylene (Chang and Stewart, 1998) or osmotic stress (Urao et al., 1998).
A definitive in vivo role for these genes awaits identification of loss-of-function mutations, possibly via targeted gene disruption that is now feasible as the result of recent developments in insertional mutagenesis in Arabidopsis. In any case these genes should provide valuable tools for the dissection of the molecular basis for cytokinin action and may help elucidate the role of this hormone in plant growth and development.
MATERIALS AND METHODS
Plant Material
The Arabidopsis ecotype used in this study was Wassilewskija. Plants were grown in Metro-Mix 200 (Grace, Boca Raton, FL) at 23°C under continuous illumination with fluorescent lights. BA was obtained from Gibco-BRL (Gaithersburg, MD). For plants grown under sterile conditions, seeds were surface sterilized and plated on MS media (Gibco-BRL) containing 0.8% (w/v) agarose as described (Brandstatter and Kieber, 1998). The seeds were incubated at 4°C for 4 d and then transferred to a 23°C chamber and grown in constant light.
Northern-Blot Analysis
Surface sterilized seeds were suspended in MS media containing 0.8% (w/v) agarose and plated on sterile filter paper placed on top of MS agar. After 3 d in the dark the seedlings were removed with the filter paper and immersed in liquid MS containing 5 μm BA or liquid MS containing an equal volume of DMSO (control) and harvested at the indicated times. For adult tissue, tissue was excised from 2-week-old plants and submerged in liquid MS containing 5 μm BA or an equal volume of DMSO for 50 min. Total RNA was extracted with phenol-chloroform, precipitated using LiCl, and fractionated on a glyoxyl-agarose gel (Ausubel et al., 1994). The RNA was capillary blotted to a Hybond-N membrane (Amersham-Pharmacia, Piscataway, NJ) and hybridized with DNA probes made by random-hexamer-labeling (Ausubel et al., 1994). Full-length cDNAs used for the probes were obtained by PCR using gene-specific primers. The GUS gene was isolated from the plasmid pBI101.1 (Jefferson et al., 1987). Signals were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Blots were stripped by incubating with boiling 0.2% (w/v) SDS and the absence of a signal was confirmed using a PhosphorImager. Each blot was not probed more than three times, twice using ARRs and once with β-tubulin (or 18S rDNA). The β-tubulin (or 18S) signal from each blot was used to quantify the induction of the corresponding ARRs used to probe that blot. To determine whether the ARRs cross hybridize, 50 ng of ARR4, ARR5, ARR6, and ARR7 were dot blotted onto a Hybond N membrane (Amersham-Pharmacia) and hybridized with each cDNA probe using conditions identical to those used for the northern blots. This analysis revealed that they was no detectable cross-hybridization among these probes (data not shown).
For cycloheximide treatment, 3-d-old etiolated seedlings were immersed in liquid MS at 22°C for 30 min with rocking in the presence or absence of 50 μm cycloheximide. Five micromolar BA or an equal volume of DMSO was then added and the seedlings incubated an additional 30 min. RNA was extracted and analyzed by northern RNA blots as above.
Isolation of Nuclei and in Vitro Transcription
Two-week-old adult leaves were treated with 5 μm BA for the indicated times and the nuclei were harvested essentially as described (Folta and Kaufman, 2000). In brief, 2 g of tissue were immersed in approximately 4 volumes of ice-cold anhydrous ethyl ether, chopped with scissors while in the ether, and incubated on ice for 5 min. After the ether was removed, the tissue was washed with 3 volumes of extraction buffer [1 m hexylene glycol (2-methyl-2, 4-pentadiol)/0.5 m PIPES (1,4-piperazinediethanesulfonic acid)-KOH, pH 7.0/10 mm MgCl2/5 mm β-mercaptoethanol] and resuspended in 2 volumes of extraction buffer. The tissue was homogenized on ice for 5 to 10 min using a Polytron tissue homogenizer (PT 10–35, Brinkman, Westbury, NY) set at the lowest speed. The homogenate was passed through two layers of cheesecloth soaked in extraction buffer, and extraction buffer was added up to a total volume of 40 mL. To lyse organellar membranes, Triton X-100 was added, while stirring, to a final concentration of 1% (v/v). The lysates were then centrifuged at 3,000 rpm and 4°C for 30 min. The nuclei, which are present as a thin powdery layer over the pellet, were gently resuspended in 5 mL of ice-cold gradient buffer (0.5 m hexylene glycol/0.5 m PIPES-KOH, pH 7.0/10 mm MgCl2/5 mm β-mercaptoethanol/1% [v/v] Triton X-100) using a camel hair paint brush. The nuclei-containing fraction was transferred to a new tube, diluted with 35 mL of ice-cold gradient buffer, and centrifuged again. The nuclei were again resuspended in 5 mL of gradient buffer and centrifuged. The final pellet was resuspended in 500 μL of nuclear storage buffer and 50 μL was used for the nuclear run-on assay.
For in vitro transcription assays, 50 μL of isolated nuclei were pre-incubated with 20 units of RNase inhibitor (Roche Molecular Biochemicals, Indianapolis) at 30°C for 10 min. A transcription assay cocktail [20 μL of 5× transcription buffer (250 mm Tris [tris(hydroxymethyl)-aminomethane]-HCl, pH 7.8/375 mm NH4Cl/50 mm MgCl2/50% [w/v] glycerol)], 5 μL each of UTP, GTP, and ATP, and 10 μL of water, and 10 μL (100 μCi) of [32P]CTP was pre-warmed and added to each sample. The in vitro transcription was allowed to progress for 1 h at 30°C. The reaction was terminated by incubation with 10 units of RNase free DNase I (30°C for 10 min), addition of termination mix (7.5 m urea/0.5% [w/v] SDS/20 mm EDTA, pH 7.5/100 mm LiCl), and extraction with 300 μL of phenol:chloroform. The aqueous phase was added to 100 μL of 4 m ammonium acetate containing 200 μg/mL yeast tRNA and placed in an ethanol/dry ice bath for 10 min. After centrifugation, the pellet was resuspended in 500 μL of hybridization buffer (50% [w/v] formamide/5× sodium chloride/sodium phosphate/EDTA/5× Denhardt's/0.1% [w/v] SDS/0.1 mg/mL salmon sperm DNA). Labeled transcripts were hybridized to 200 ng of cDNA, which was generated by PCR using gene-specific primers and slot-blotted to GeneScreen membranes (NEN-DuPont, Wilmington, DE). The membranes were pre-incubated with hybridization buffer at 42°C for 6 h before addition of the probes and hybridization was done overnight at 42°C. After hybridization, the blots were washed twice at room temperature with 1× SSC/0.1% (w/v) SDS and three times at 65°C with 0.1× SSC/0.1% (w/v) SDS for 15 min. Signals were visualized and quantified using a PhosphorImager (Molecular Dynamics).
Primer Extension
The start site of transcription for ARR5 was determined using primer extension analysis. An ARR5 specific primer (5′-GGGACGCAAAACCTCAGCCATATCAAGAAGAG-3′; the ATG start codon is underlined), complementary to nucleotides +57 to +88, was end-labeled using [32P]ATP (Chaconas and van de Sande, 1980). The unincorporated phosphate was removed using a Nick Column (Sephadex G-50, Amersham-Pharmacia). The labeled oligonucleotide was incubated with 50 μg of total RNA isolated from 3-d-old etiolated seedling treated with 5 μm BA in 15 μL of hybridization buffer (0.15 m KCl/10 mm Tris-HCl, pH 8.3/1 mm EDTA) at 65°C for 90 min, then cooled slowly to room temperature. For the primer extension, 30 μL of reaction solution (30 mm Tris-Cl, pH 8.3/15 mm MgCl2/8 mm dithiothreitol/225 μg/mL actinomycin D/200 μm dNTPs/66U Superscript reverse transcriptase) was added to the hybridization mixture and incubated at 42°C for 1 h. RNase reaction mixture (10 mm Tris-Cl, pH 7.5/1 mm EDTA/300 mm NaCl/100 μg/mL salmon sperm DNA/20 μg/mL RNase A) was added to a total volume of 150 μL and incubated at 37°C for 15 min. The sample was extracted with phenol:chloroform:isoamyl alcohol (24:24:1) and precipitated with 10% (v/v) 3 m sodium acetate and 2 volumes of absolute ethanol. The sample was analyzed on a 6% (w/v) acrylamide/7 m urea sequencing gel together with an ARR5 35S dideoxy sequencing reaction, primed with the ARR5 primer described above, and catalyzed by Sequenase DNA polymerase, as described by the manufacturer (U.S. Biochemicals, Cleveland).
Plasmid Construction
An approximately 1.6-kb fragment of the presumed promoter region of the ARR5 gene were cloned into the Agrobacterium tumefaciens binary plasmid vector, pBI101.1 (Jefferson et al., 1987). The ARR5 promoter fragment was generated by PCR using primers designed such that fragment would begin just before the start site of transcription (TC), but contain the predicted TATA box. The primer 5′-aaggatccTGAGAGATGAGAGGAGAATAA-3′ (lowercase letters are non-ARR5 sequences designed to incorporate a BamHI site in the amplified fragment) is located at the start site of transcription in the antisense direction and was used with the sense primer, 5′-aaggatccGGAAACC- AATAAAGCATATTTG-3′ (approximately 1.6 kb upstream of the start site of translation) to amplify the 1.6-kb fragment. This ARR5 fragment was cloned into the BamHI site of pBI101.1 to create the pIB-1.6TC plasmid.
Plant Transformation and Culture Conditions
Each plasmid was transformed into A. tumefaciens strain C58 and grown overnight to saturation. Two-week-old Wassilewskija plants were transformed using the floral dip method (Clough and Bent, 1998). The Agrobacteria were centrifuged and resuspended in 1 to 2 volumes (starting culture) of 5% (w/v) Suc/0.05% (v/v) Stilwet L-77. The plants were dipped into the solution for 3 min at room temperature. The process was repeated 7 d later. T1 seeds were plated on MS medium containing kanamycin to screen for the presence of the pIB plasmids. Activation of the GUS reporter gene was detected by submerging plant material in GUS staining solution (100 mm KPO4, 10 mm EDTA, 0.1% [w/v] Tween, 0.5 mm K·Ferricyanide, 0.5 mm K·Ferrocyanide, 2 mm 5-bromo-4-choloro-3-indolyl-glucuronidide) for 2 to 12 h at 37°C.
Whole-Mount in Situ Hybridization
An ARR5 gene-specific probe was generated using PCR amplification of the 295 bp at the 3′ end of ARR5 transcript (nucleotides 581–876 of ARR5 cDNA) using the oligonucleotide primers 5′-CTACTCGCAGCTAAAACGC-3′ and 5′-TAATTAACTTCCAAAAATAACAACACC-3′. The resultant PCR fragment was cloned in the pTOPO TA cloning vector (Invitrogen, Carlsbad, CA) and then subcloned into the pBluescript-KS vector (Stratagene, La Jolla, CA) as an EcoRI fragment. Riboprobes were prepared in vitro using a digoxigenin RNA labeling mix (Roche Molecular Biochemicals, Basel) and then digested to an average length of 150 nucleotides by controlled alkaline hydrolysis (Cox et al., 1984). In situ hybridization was performed as described (de Almeida Engler et al., 1994), except that digestion with 2% (w/v) driselase (Sigma, St. Louis) was added to the prehybridization treatment. Seedlings were mounted on microscope slides, visualized under a microscope (Optiphot, Nikon, Tokyo) equipped with Nomarski optics and photographed.
ACKNOWLEDGMENTS
We thank Kevin Folta for assistance with the nuclear run-on assays and members of the Kieber laboratory for critically reading the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB-9996354 to J.J.K.).
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Brandstatter I, Kieber JJ. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–1020. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, van de Sande JH. 5′ 32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65:75–88. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker A, Meyerowitz E. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Stewart RC. The two-component system: regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–713. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cox K, Deleon DV, Angerer LM, Angerer RC. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Amasino RM. Cytokinins and plant gene regulation. In: Mok DWS, Mok MC, editors. Cytokinins: Chemistry, Activity and Function. Boca Raton, FL: CRC Press; 1994. pp. 233–242. [Google Scholar]
- D'Agostino IB, Kieber JJ. The emerging family of plant response regulators. Trends Biochem. 1999;24:452–456. doi: 10.1016/s0968-0004(99)01465-6. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J, van Montagu M, Engler G. Hybridization in situ of whole-mount messenger RNA in plants. Plant Mol Biol. 1994;31:419–427. [Google Scholar]
- Dewitte W, Chiapetta A, Azmi A, Witters E, Strnad M, Rembur J, Noin M, Chriqui D, Van Onckelen H. Dynamics of cytokinins in apical shoot meristems of a day-neutral tobacco during floral transition and flower formation. Plant Physiol. 1999;119:111–121. doi: 10.1104/pp.119.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead PJ, Poethig S, Roberts K, Scheres B. Cellular organization of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Dominov J, Stenzler L, Lee S, Schwarz J, Leisner S, Howell S. Cytokinins and auxins control the expression of a gene in Nicotiana plumbaginifolia cells by feedback regulation. Plant Cell. 1992;4:451–461. doi: 10.1105/tpc.4.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowens BP, Crowell DN. Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;37:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Flores S, Tobin EM. Cytokinin modulation of LHCP mRNA levels: the involvement of post-transcriptional regulation. Plant Mol Biol. 1988;11:409–415. doi: 10.1007/BF00039021. [DOI] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS. Preparation of transcriptionally active nuclei from etiolated Arabidopsis thaliana. Plant Cell Rep. 2000;19:504–510. doi: 10.1007/s002990050764. [DOI] [PubMed] [Google Scholar]
- Franco A, Gee M, Guilfoyle T. Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem. 1990;265:15845–15849. [PubMed] [Google Scholar]
- Hare PD, Staden Jv. The molecular basis of cytokinin action. Plant Growth Regul. 1997;23:41–78. [Google Scholar]
- Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:199–211. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- Hoch JA, Silhavy TJ. Two-Component Signal Transduction. Washington, DC: ASM Press; 1995. [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999;40:733–742. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase is a sensitive and versatile fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- Kiba T, Taniguchi M, Imamura A, Ueguchi C, Mizuno T, Sugiyama T. Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:767–771. doi: 10.1093/oxfordjournals.pcp.a029604. [DOI] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong L-M, Theologis A. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum) J Mol Biol. 1995;253:396–413. doi: 10.1006/jmbi.1995.0562. [DOI] [PubMed] [Google Scholar]
- Langridge WHR, Fitzgerald KJ, Koncz C, Schnell J, Szalay AA. Dual promoter of Agrobacterium tumefaciens mannopine synthase is regulated by plant hormones. Proc Natl Acad Sci USA. 1989;86:3219–3223. doi: 10.1073/pnas.86.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Buchholz G, Keitel C, Sweere C, Kircher S, Bäurle I, Kudla J, Harter K. Differentially-expressed and nuclear-localized response regulator-like proteins from Arabidopsis thaliana with transcription factor properties. J Plant Biol. 1999;1:495–506. [Google Scholar]
- Loomis WF, Shaulsky G, Wang N. Histidine kinases in signal transduction pathways of eukaryotes. J Cell Sci. 1997;110:1141–1145. doi: 10.1242/jcs.110.10.1141. [DOI] [PubMed] [Google Scholar]
- Lu J-L, Ertl JR, Chen C-M. Transcriptional regulation of nitrate reductase mRNA levels by cytokinin-abscisic acid interactions in etiolated barley leaves. Plant Physiol. 1992;98:1255–1260. doi: 10.1104/pp.98.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- Medford J, Horgan R, El-Sawi Z, Klee H. Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentyl transferase gene. Plant Cell. 1989;1:403–413. doi: 10.1105/tpc.1.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Okomura FS, von Saltza MH, Strong FM. Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc. 1956;78:1345–1350. [Google Scholar]
- Miller CO, Skoog F, Von Saltza MH, Strong F. Kinetin, a cell division factor from deoxyribonucleic acid. J Am Chem Soc. 1955;77:1392–1293. [Google Scholar]
- Mok DWS, Mok MC. Cytokinins: Chemistry, Activity and Function. Boca Raton, FL: CRC Press; 1994. [Google Scholar]
- Parkinson J. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Perraud A-L, Weiss V, Gross R. Signaling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- Pickett FB, Meeks-Wagner DR. Seeing double: appreciating genetic redundancy. Plant Cell. 1995;7:1347–1356. doi: 10.1105/tpc.7.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Suzuki M, Takei K, Deji A, Taniguchi M, Sugiyama T. A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998;14:337–344. doi: 10.1046/j.1365-313x.1998.00134.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Taniguchi M, Sugiyama T. His-Asp phosphorelay signaling: a communication avenue between plants and their environment. Plant Mol Biol. 2000;42:273–278. doi: 10.1023/a:1006334926388. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;72:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T, Schäfer S, Romanov G. Cytokinins as regulators of gene expression. Physiol Plant. 1997;100:505–519. [Google Scholar]
- Silver DL, Pinaev A, Chen RJ, de Bruijn FJ. Post-transcriptional regulation of the Sesbania rostrata early nodulin gene SrEnod2 by cytokinin. Plant Physiol. 1996;112:559–567. doi: 10.1104/pp.112.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JF, Stock AN, Mottonen JM. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett. 1998;429:259–262. doi: 10.1016/s0014-5793(98)00611-5. [DOI] [PubMed] [Google Scholar]
- Theologis A, Huynh TV, Davis RW. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol. 1985;183:53–68. doi: 10.1016/0022-2836(85)90280-3. [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427:175–178. doi: 10.1016/s0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]