Abstract
Neurogenesis is the process through which neural stem and progenitor cells generate neurons. During the development of the mouse neocortex, stem and progenitor cells sequentially give rise to neurons destined to different cortical layers and then switch to gliogenesis resulting in the generation of astrocytes and oligodendrocytes. Precise spatial and temporal regulation of neural progenitor differentiation is key for the proper formation of the complex structure of the neocortex. Dynamic changes in gene expression underlie the coordinated differentiation program, which enables the cells to generate the RNAs and proteins required at different stages of neurogenesis and across different cell types. Here, we review the contribution of epigenetic mechanisms, with a focus on Polycomb proteins, to the regulation of gene expression programs during mouse neocortical development. Moreover, we discuss the recent emerging concept of epigenetic and transcriptional pre-patterning in neocortical progenitor cells as well as post-transcriptional mechanisms for the fine-tuning of mRNA abundance.
Keywords: gene regulation, histone methylation, neocortical development, neural progenitor cell, Polycomb, epigenetics, chromatin, neurogenesis
Introduction
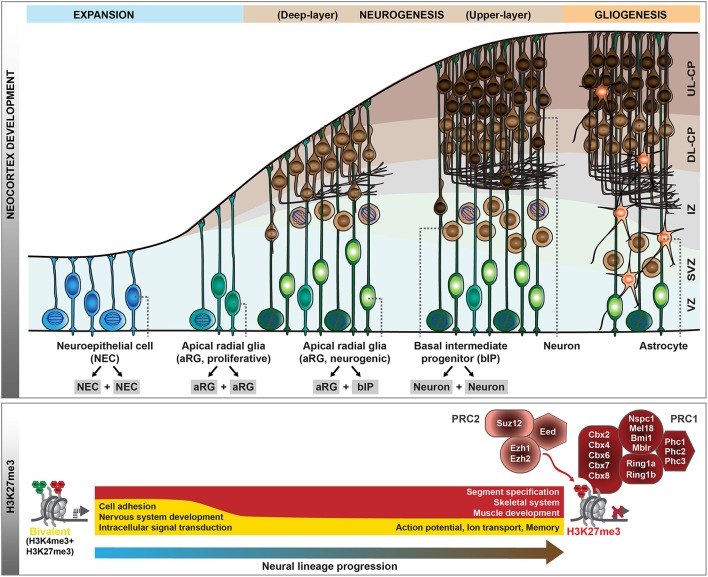
The generation of neocortical neurons during mouse development is the result of balanced proliferative and differentiative divisions of neural stem and progenitor cells (Götz and Huttner, 2005; Lui et al., 2011; Florio and Huttner, 2014). In the early developing central nervous system, neuroepithelial cells (NECs) function as the primary neural stem cells which show apico-basal polarity and undergo symmetric proliferative divisions to expand the stem cell pool (Figure 1). With the onset of neurogenesis at around mouse embryonic day (E) 10, NECs transform into apical radial glia (aRG), which retain apico-basal polarity and become more elongated. Their cell bodies reside in the ventricular zone, whereas their long basal processes extend to the basal lamina and provide a scaffold for neuronal migration to the cortical plate. aRG are characterized by their ability to self-renew and to simultaneously give rise to neurons, mainly indirectly through basal intermediate progenitors (bIPs). bIPs delaminate from the ventricular surface and reside in the subventricular zone. They lack apico-basal polarity and in mouse typically divide symmetrically to produce two neurons. Neocortical neurons are organized into six horizontal layers, with the deep-layer neurons born first during neurogenesis followed by the generation of upper-layer neurons. At around E17, neurogenesis is terminated and the remaining neural stem and progenitor cells switch to gliogenesis. Thus, throughout mouse neocortical development, the potential of neural progenitor cells (NPCs) for proliferation and differentiation changes as NPCs pass through phases of expansion, deep- and upper-layer neurogenesis, and gliogenesis. In this review, we will discuss the dynamic changes in transcriptional programs and epigenetic information that accompany and guide these transitions. We will mainly focus on post-translational modifications of histones, as the role of other epigenetic pathways, including DNA modifications and chromatin remodeling, in neocortex development are reviewed elsewhere (see Sokpor et al., 2018; Stricker and Gotz, 2018, in this Research Topic).
Figure 1.
Polycomb-mediated histone methylation during mouse neocortex development. During the development of the mouse neocortex, neural progenitor cells pass through consecutive stages of expansion, neurogenesis, and gliogenesis (top scheme). aRG undergoing neurogenic divisions give rise to bIPs, which are the main source of neurons in the mouse. As neural progenitor cells transition from proliferation to neurogenic divisions, their histone methylation profiles change dynamically. Whereas, many genes are in a bivalent configuration in early proliferative progenitor cells, many of these poised domains are resolved with progressive lineage commitment. The gene ontology categories characteristic of the genes marked by H3K27me3 (red) or bivalent modifications (yellow) during early neurogenesis and in neurons are indicated (bottom scheme). In addition, the core components of Polycomb repressive complex 1 (PRC1) and 2 (PRC2) are shown. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; DL, deep-layer; UL, upper-layer.
Trithorax and polycomb complexes
Epigenetic information, in concert with transcription factors, coordinates the instruction of specific cellular identities from the genomic DNA template, and as such plays an essential role in the transition of cell fates during development. Post-translational histone modifications represent one major epigenetic system, among others. In particular, chromatin modifiers of the Trithorax (TrxG) and Polycomb (PcG) groups were identified as part of an evolutionary conserved epigenetic memory system that acts antagonistically to maintain active and repressed gene expression states, important during stem cell differentiation and embryonic development (reviewed in Piunti and Shilatifard, 2016; Schuettengruber et al., 2017). PcG proteins assemble into two major complexes, PRC1 and PRC2 (Figure 1), which catalyze mono-ubiquitination of histone 2A lysine 119 (H2AK119ub1) and tri-methylation of histone 3 lysine 27 (H3K27me3), respectively. These complexes have also been shown to regulate gene expression during neocortical development, and importantly, are one of the major determinants of the ability of NPCs to either self-renew or to give rise to neurons or glial cells (Tyssowski et al., 2014; Mitrousis et al., 2015; Yao et al., 2016).
The transition from expansion to neurogenesis
During early development, the neural tube is formed by NECs that divide symmetrically to expand the neural stem cell pool. Following this initial expansion phase, NECs turn into neurogenic aRG, characterized by the appearance of glial hallmarks, a change in the mitotic behavior and a more restricted progenitor fate (Götz and Huttner, 2005; Taverna et al., 2014; Subramanian et al., 2017). This transition is accompanied by a major redistribution of the PcG-mediated H3K27me3 mark (Albert et al., 2017), which is associated with transcriptional gene silencing (Comet et al., 2016). Several tight junction-associated genes convert to a more repressive chromatin configuration, whereas the genes encoding the glial-specific glutamate transporter (Slc1a3/Glast) and the brain lipid-binding protein (Fabp7/Blbp) acquire H3K4me3 (Albert et al., 2017), a hallmark of TrxG-associated gene activation (Schuettengruber et al., 2017). Notably, in line with NECs representing the earliest and least committed neural stem cells of the developing neocortex, the majority of the genes marked by H3K27me3 in NECs carry H3K4me3 in addition (Albert et al., 2017), a configuration which has been termed “bivalent” (Bernstein et al., 2006). Such bivalent domains are abundant in embryonic and adult stem cells (Shema et al., 2016), where they decorate genes implicated in cell-fate determination and development (Schuettengruber et al., 2017). This has been hypothesized to keep future lineage choices open (Bernstein et al., 2006). With the transition of NECs to aRG, a large fraction of bivalent domains is resolved, either to H3K27me3 at promoters of genes involved in the development of other organs (Figure 1), or to H3K4me3 at genes involved in nervous system development, cell adhesion and cell surface signaling (Albert et al., 2017). Thus, the switch of NPCs from the initial expansion phase to the neurogenic phase is accompanied by major epigenetic changes.
The neurogenic phase
During the neurogenic phase, aRG have the potential to either proliferate or to self-renew and generate basal progenitors or, rarely, neurons. PcG complexes have been shown to contribute to the regulation of this balance between proliferation and differentiation. The PRC2 histone methyltransferase Ezh2, which generates H3K27me3, is highly expressed in NPCs of the mouse developing neocortex, particularly during early neurogenesis (Pereira et al., 2010; Piper et al., 2014). Specific deletion of Ezh2 in the developing neocortex from E9.5 results in a loss of H3K27me3 and up-regulation of gene expression, consequently shifting aRG fate from self-renewal toward differentiation (Pereira et al., 2010). This shift results in an overproduction of bIPs and neurons at the expense of aRG, ultimately reducing the neuronal output and leading to a substantially smaller neocortex (Pereira et al., 2010). In light of this, it is interesting to note that the promoters of many transcription factors involved in bIP generation and neuronal differentiation (like Insm1, Eomes, Neurog1/2, and Neurod1/2) are H3K27me3-positive during the expansion phase of NPCs (Albert et al., 2017), and a loss of this repressive state might contribute to the precocious activation of these genes. In addition, the PRC1 component Bmi1 has been shown to regulate the self-renewal and differentiation of NPCs (Fasano et al., 2007, 2009; Yadirgi et al., 2011).
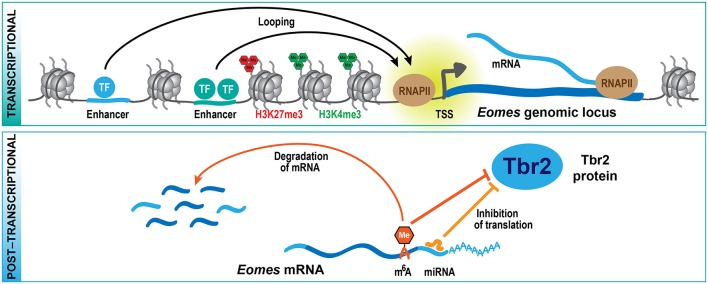
From these genetic studies, it is clear that PcG proteins contribute to the regulation of the balance between self-renewal vs. differentiation during neocortex development, but what are the underlying molecular mechanisms? Epigenome profiling in specific cell populations isolated at mid-neurogenesis (E14.5) has shown that H3K4me3 and H3K27me3 marks are highly dynamic during neocortical lineage progression (Albert et al., 2017). In particular, several transcription factors involved in cell fate commitment during neurogenesis display transient changes in histone methylation at their promoters, potentially involved in cell type-specific induction of gene expression. Notably, the promoter of the Eomes gene, which encodes the key transcription factor Tbr2 implicated in the generation of bIPs (Arnold et al., 2008; Sessa et al., 2008), changes from a repressive configuration marked by H3K27me3 in proliferative aRG to an active configuration marked by H3K4me3 in aRG undergoing neurogenic divisions (Albert et al., 2017). As these changes likely occur within one cell-cycle, it is conceivable that the H3K27me3 mark is actively removed, most likely by the histone demethylase Jmjd3, which is expressed in the developing neocortex (Sessa et al., 2017) and has been shown to act on Eomes gene regulatory regions (Kartikasari et al., 2013). The active configuration of the Eomes promoter is largely maintained in bIPs, whereas H4K4me3 levels decline and H3K27me3 is re-established in neurons (Albert et al., 2017), in which Eomes is no longer expressed (Florio et al., 2015). Thus, Eomes is one example of a gene that undergoes dynamic changes in histone methylation during neocortical differentiation (Figure 2), and these changes correlate well with the gene expression dynamics. In addition, the regulation of other transcription factors that control progenitor proliferation or differentiation has been linked to various histone methylation states, including H3K4me3 and H3K79me3 (Büttner et al., 2010; Yang et al., 2012).
Figure 2.
Multi-layered regulation of gene expression. At the transcriptional level (top scheme), cell type-specific expression of genes is regulated by transcription factors that bind to regulatory sequences, including distal enhancers that contact their respective target genes by looping. Histone methylation, mediated by TrxG (H3K4me3) and PcG (H3K27me3) proteins, is part of an epigenetic system that also contributes to the regulation of specific gene expression. At the post-transcriptional level (bottom scheme), the translation and stability of mRNAs is regulated by miRNAs and epitranscriptomic mechanisms including m6A, providing a multi-layered system for the control of protein expression during development. These mechanisms are exemplified here for the key transcription factor Tbr2 (Eomes), which regulates bIP generation during neocortex development.
But are the histone methylation patterns instrumental for the correct expression of the related genes in the developing neocortex? Previous studies, which applied CRISPR/Cas9-based genome editing in vivo to disrupt Eomes expression in NPCs during neocortical development, showed that this acute targeting results in a reduction in bIPs and an increase in neuronal differentiation (Kalebic et al., 2016). Importantly, CRISPR/Cas9-based epigenome editing at the Eomes locus in the developing neocortex has shown that the decrease in H3K27me3 in neurogenic NPCs is required for normal Tbr2 expression and bIP regulation (Albert et al., 2017). These results underscore the importance of epigenetic information in the regulation of specific gene expression and as facilitator of cell fate transitions during development.
The H3K27me3 mark is recognized by different “reader” proteins, one of which is the chromatin remodeler Chd5 expressed in neurons of the developing neocortex (Egan et al., 2013). Depletion of Chd5 during neurogenesis results in a block of neuronal differentiation, which can be rescued by Chd5 only if the latter contains an intact double chromodomain mediating H3K27me3 binding. In addition, components of the PRC1 complex can bind to H3K27me3, and at the majority of genomic target sites, H3K27me3 and PRC1 are found to co-localize, even though this traditional model of sequential binding of PRC2 followed by PRC1 complexes has been challenged by several studies (Puschendorf et al., 2008; Blackledge et al., 2015; Kloet et al., 2016). Deletion of Ring1b, an integral component of PRC1 (Leeb and Wutz, 2007), specifically in the mouse developing neocortex during the neurogenic phase results in altered neuronal subtype specification (Morimoto-Suzki et al., 2014). By mediating the timed termination of Fezf2 expression, Ring1b regulates the number of subcerebral projection neurons produced. These data suggest that PcG complexes and associated proteins control several aspects of cortical neurogenesis, including the balance between self-renewal and differentiation of aRG as well as the switch from deep- to upper-layer neurogenesis in NPCs.
The transition to the gliogenic phase
In mouse, the neurogenic phase is followed by a period of gliogenesis, during which astrocytes and oligodendrocytes are generated. The timing of the switch from neurogenic to gliogenic fate of NPCs is critical for brain development, as it is one of the determinants of the final number of cortical neurons produced. In addition to extracellular cues, cell-intrinsic programs regulate NPC fate, to which epigenetic mechanisms are thought to contribute. The PcG proteins have been demonstrated to play an important role in the timing of the neurogenic to gliogenic transition. Depletion of PcG proteins during the neurogenic period leads to a prolonged neurogenic phase of NPCs and a delayed onset of astrogenesis (Hirabayashi et al., 2009; Sparmann et al., 2013). Toward the time when neurogenesis is normally terminated, several genes associated with the neurogenic lineage are selectively derepressed in PcG-mutant NPCs, including neurogenin 1 (Neurog1), a key proneural transcription factor that can suppress astrocytic differentiation (Hirabayashi et al., 2009).
Interestingly, deletion of Ezh2 before, or at, the onset of neurogenesis has the opposite effect, leading to a shorter neurogenic period and precocious astrocyte generation (Pereira et al., 2010; Sparmann et al., 2013). In NPCs in vitro, PcG proteins mediate the suppression of the key astrogenic marker Gfap (Mohn et al., 2008; Sparmann et al., 2013), which has been proposed to prevent the premature onset of gliogenesis (Sparmann et al., 2013). In the developing neocortex, however, the promoters of Gfap as well as of other genes characteristic of astrocytes are not marked by H3K27me3 at mid-neurogenesis (ENCODE Project Consortium, 2012; Albert et al., 2017), which is in agreement with other reports suggesting a role for alternative repressive pathways, including DNA and H3K9 methylation, in the regulation of astrocyte-specific genes (Takizawa et al., 2001; Song and Ghosh, 2004; Fan et al., 2005; Hatada et al., 2008). Future research should be aimed at identifying PcG target genes underlying the context- and stage-dependent role of PcG proteins in NPCs during different phases of neocortex development, and should provide a more general view beyond the limited number of well-characterized known regulators.
Cell type- and stage-specific roles of polycomb proteins
Previous studies in mouse and human embryonic stem cells (Mikkelsen et al., 2007; Mohn et al., 2008; Burney et al., 2013; Ziller et al., 2015) and the developing mouse neocortex (Albert et al., 2017) have shown that H3K27me3 levels are highly dynamic at different NPC stages and during neuronal differentiation, raising the question of how PcG target gene specificity is achieved. One way to dynamically control PcG function is by altering the composition of PcG complexes, which in mammals, as opposed to flies, is highly diverse, enabling the assembly of various sub-complexes with different chromatin binding affinities and interaction partners (Piunti and Shilatifard, 2016; Schuettengruber et al., 2017).
In embryonic stem cells, the interchange of Chromobox (Cbx) family proteins, part of PRC1, has been reported to modulate the balance between self-renewal and lineage commitment (Morey et al., 2012; O'Loghlen et al., 2012; Santanach et al., 2017), and different Cbx paralogs are required for different cell lineages (Luis et al., 2011; Klauke et al., 2013). Of note, the Cbx paralogs are differentially expressed in neural sub-populations of the developing neocortex (Florio et al., 2015). Moreover, chromatin remodelers of the chromodomain helicase DNA-binding (Chd) family, which have been reported to interact with PcG complexes, also show differential expression during neocortex development. Whereas Chd5 is expressed in neurons and controls neuronal differentiation (Egan et al., 2013), Chd4 is expressed in NPCs during early neurogenesis where it has been proposed to function in PcG-mediated inhibition of astroglial differentiation (Sparmann et al., 2013). This switch in subunit composition may contribute to the re-targeting of PcG complexes during neocortex development.
PcG complexes themselves bind relatively unspecifically to CG-rich regions lacking DNA methylation (Schuettengruber et al., 2017). In addition, the chromatin targeting of PRC2 is stabilized by interactions with transcription factors, non-coding RNAs and other chromatin factors resulting in increased binding and H3K27me3 deposition at specific regions. The highly restricted expression pattern of many of these factors and RNAs during neocortex development (Aprea et al., 2013; Molyneaux et al., 2015; Liu et al., 2016) provides a potential mechanistic explanation for cell type-specific PcG targeting. Moreover, the H3K27me3-specific histone demethylase Jmjd3 has been implicated in the activation of neuronal gene expression (Jepsen et al., 2007; Park et al., 2014), and associates with the transcription factor Tbr2 in the developing neocortex (Sessa et al., 2017), further contributing to the dynamic regulation of H3K27me3.
Transcriptional pre-patterning
During recent years, there have been massive efforts to characterize the transcriptomic signatures of the various NPC subtypes in the mouse developing neocortex, but also in other mammalian species including the ferret, macaque and human (Ayoub et al., 2011; Fietz et al., 2012; Aprea et al., 2013; Arcila et al., 2014; Miller et al., 2014; Pollen et al., 2014; Camp et al., 2015; De Juan Romero et al., 2015; Florio et al., 2015; Johnson et al., 2015; Liu et al., 2016; Telley et al., 2016; Nowakowski et al., 2017; Zhong et al., 2018). From these studies, a variety of gene expression differences have been uncovered that underlie specific cell biological features, proliferative capacities and differentiation potential of the distinct NPC types (reviewed in Silver, 2016; Florio et al., 2017). Interestingly, several of these studies described the expression of genes in aRG whose protein products are well-known to function only downstream in the lineage, in bIPs or neurons (Florio et al., 2015; Telley et al., 2016; Nowakowski et al., 2017), raising the possibility that there is a delay in translation for certain mRNAs.
One example of such a gene that is expressed already in aRGs, specifically those undergoing neurogenic divisions, is Eomes (Florio et al., 2015), which gives rise to the bIP transcription factor Tbr2 (Arnold et al., 2008; Sessa et al., 2008). What is it that keeps the Eomes mRNA from being translated in aRG? The Tbr2 protein has been shown to be repressed by the microRNAs (miRNAs) miR-92 and miR-92b, and both miRNAs regulate bIP specification in the developing neocortex (Bian et al., 2013; Nowakowski et al., 2013). Interestingly, miR-92 and miR-92b are specifically expressed in aRG undergoing neurogenic divisions, where the Eomes mRNA is highly expressed (Florio et al., 2015). In contrast, bIPs, which express Tbr2 protein, have low levels of both miRNAs (Nowakowski et al., 2013; Florio et al., 2015). Of note, many other miRNAs display unique profiles of expression in the developing neocortex (Barca-Mayo and De Pietri Tonelli, 2014; Rajman and Schratt, 2017), and among their validated target genes are several cell cycle and neurogenesis regulators (Arcila et al., 2014; Fei et al., 2014), indicating that miRNA-mediated control of RNA translation (Figure 2) may play a widespread role during neocortex development and also evolution. Moreover, two components of the miRNA microprocessor complex, Drosha and DGCR8, were shown to regulate gene expression in the developing neocortex in a miRNA-independent fashion (Knuckles et al., 2012; Marinaro et al., 2017), further adding to the complexity of post-transcriptional gene regulation.
In addition, recently a new epitranscriptomic mechanism has been identified that regulates the metabolism and translation of mRNAs, which involves the post-transcriptional modification of mRNAs by N6-methyladenosine (m6A) (reviewed in Zhao et al., 2017). Depletion of m6A during neocortex development leads to a prolonged cell cycle of aRGs and extends neuron production to postnatal stages, suggesting that m6A regulates cortical neurogenesis (Yoon et al., 2017). Among the transcripts that are tagged by m6A, several encode transcription factors regulating NPC fate, such as Pax6, Sox2, Neurog2, and Tbr2. The presence of m6A on these transcripts promotes their rapid turnover, and in absence of the m6A methyltransferase complex component Mettl14, several neuronal lineage proteins, like Neurod1 and Tbr2, are precociously expressed in aRG. This observation led to the proposal of the novel concept of transcriptional pre-patterning during cortical neurogenesis, whereby a subset of neuronal lineage genes is already expressed in aRG but their levels actively suppressed post-transcriptionally by m6A-dependent decay (Yoon et al., 2017). A second study that analyzed the role of m6A during neurogenesis found that Mettl14 deletion results in decreased radial glia proliferation and premature differentiation (Wang et al., 2018). The authors of this study ascribed the observed phenotypes to genome-wide changes in histone methylation patterns which may result from the destabilization of transcripts that encode histone-modifying enzymes. While further mechanistic studies are required to dissect the role of m6A in specific neural subpopulations, the two studies (Yoon et al., 2017; Wang et al., 2018) describe a novel post-transcriptional mechanism regulating protein expression during neurogenesis (Figure 2).
Epigenetic pre-patterning
Whereas, transcriptome analysis provides a snapshot of a cell's gene expression pattern at a specific point in time, the corresponding epigenetic information captures gene regulatory mechanisms, developmental origins, and potential future responses to developmental stimuli (Mo et al., 2015). Transcription factors, which are thought to be instrumental for the specification of cell type-specific gene expression programs, bind to DNA in the context of chromatin, which carries multiple post-translational modifications, and these affect transcription factor binding (Shlyueva et al., 2014; Yin et al., 2017). As such, the epigenetic landscape can permit the transcription of certain genes, while rendering others inaccessible to most transcription factors.
That said, the transition from “closed” to “open” chromatin, and vice versa, is determined by regulatory proteins, most prominently a special class of transcription factors, called pioneer factors (Shlyueva et al., 2014). These factors can bind to repressed chromatin and recruit chromatin remodelers to evict nucleosomes to open up the region, thereby making the DNA accessible to other transcription factors. During neural differentiation, such pioneer factors have been proposed to remodel the binding site repertoire for proneural factors at the NPC stage by changing the epigenetic landscape at their respective target sites (Ziller et al., 2015). This is also thought to ensure proper further lineage specification by restricting the binding capacity of proneural and other transcription factors toward appropriate sites.
Differential gene expression in specific cell types is mainly controlled by distal cis-regulatory elements, among which enhancers are the most abundant (Spitz and Furlong, 2012; de Laat and Duboule, 2013). Enhancer sequences contain short DNA motifs that serve as binding sites for sequence-specific transcription factors. In a given tissue, active enhancers are brought into spatial proximity of their respective target gene by looping (Shlyueva et al., 2014). Our understanding of how chromatin is organized and folded within the nucleus, and how this affects gene regulation and cell fate decisions, has greatly expanded during recent years, mainly due to technological advances in detecting chromatin contacts in 3D (Bonev and Cavalli, 2016; Franke et al., 2016).
During neural differentiation, both in vitro and in the mouse developing neocortex, chromatin interactions change dynamically, frequently related to neural transcription factors that contribute to chromosome reorganization (Bonev et al., 2017). In addition, PcG proteins have been proposed to facilitate neural induction by establishing physical interactions between poised enhancers and their target genes in embryonic stem cells (Cruz-Molina et al., 2017). These preformed contacts are thought to provide a permissive topology that facilitates the timely and robust induction of major neural genes upon differentiation. The importance of understanding chromosome conformation has been underscored by recent studies in the human developing neocortex, which have revealed regulatory relationships relevant to the evolution of human cognition but also to diseases (Won et al., 2016; de la Torre-Ubieta et al., 2018).
Conclusions
It is well-established that epigenetic mechanisms contribute to the regulation of gene expression during stem cell differentiation and development. In this review, we have summarized recent advances in our understanding of the role of Polycomb proteins during mouse neocortex development. In particular, recent epigenome profiling has shed further light on the context-dependent functions of Polycomb proteins during the proliferative and neurogenic phase of neocortex development. It remains to be shown on a genome-wide scale how PcG targets change with the transition to the gliogenic phase. Moreover, in the future, it will be interesting to apply the emerging CRISPR/Cas9-based epigenome editing tools (Pulecio et al., 2017) to dissect the role of epigenetic changes at gene regulatory regions of important regulators of neocortex development. In a proof of principle study, the role of H3K27me3 has been analyzed in vivo during neocortex development at the Eomes gene promoter (Albert et al., 2017). From such epigenome editing experiments, further functional insights into chromatin-mediated gene regulation can be expected. Importantly, such studies will allow to move the field forward beyond correlations of epigenetic information and gene expression to interrogating the functional relevance of histone modifications at regulatory regions in specific neural cell types and at various periods of neocortex development. Recent technological advances have revealed important insights into the 3D genome organization during neocortex development and have led to the identification of distal regulatory elements. With CRISPR/Cas9-based genome and epigenome editing techniques at hand, the functional interplay of histone modifications, genome organization, and gene expression can now be unraveled.
The epigenetic landscape provides a framework within which many transcription factors operate, but which, in turn, is modulated by the action of transcription factors and gene expression itself. During development, epigenetic patterning is important for the correct spatio-temporal regulation of gene expression. In addition, the translation of expressed mRNAs is regulated by miRNAs and novel epitranscriptomic modifications, providing a multi-layered mechanism to precisely control the dynamic expression of genes, both at the mRNA and protein level. The challenge for the future will be to integrate the different layers of transcriptional and post-transcriptional gene regulation into a comprehensive framework that allows to link the different mechanisms and to understand the cross-talk between these systems.
Author contributions
MA designed the figures. MA and WH wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Huttner lab for discussions and critical reading of the manuscript. MA acknowledges support from the Christiane Nüsslein-Volhard Foundation. WH was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (SFB 655, A2), the European Research Council (250197), and ERA-NET NEURON (MicroKin).
References
- Albert M., Kalebic N., Florio M., Lakshmanaperumal N., Haffner C., Brandl H., et al. (2017). Epigenome profiling and editing of neocortical progenitor cells during development. EMBO J. 36, 2642–2658. 10.15252/embj.201796764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J., Prenninger S., Dori M., Ghosh T., Monasor L. S., Wessendorf E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. 10.1038/emboj.2013.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcila M. L., Betizeau M., Cambronne X. A., Guzman E., Doerflinger N., Bouhallier F., et al. (2014). Novel primate miRNAs coevolved with ancient target genes in germinal zone-specific expression patterns. Neuron 81, 1255–1262. 10.1016/j.neuron.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. J., Huang G. J., Cheung A. F., Era T., Nishikawa S., Bikoff E. K., et al. (2008). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479–2484. 10.1101/gad.475408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub A. E., Oh S., Xie Y., Leng J., Cotney J., Dominguez M. H., et al. (2011). Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 14950–14955. 10.1073/pnas.1112213108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O., De Pietri Tonelli D. (2014). Convergent microRNA actions coordinate neocortical development. Cell Mol. Life Sci. 71, 2975–2995. 10.1007/s00018-014-1576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Bian S., Hong J., Li Q., Schebelle L., Pollock A., Knauss J. L., et al. (2013). MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406. 10.1016/j.celrep.2013.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge N. P., Rose N. R., Klose R. J. (2015). Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16, 643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B., Cavalli G. (2016). Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678. 10.1038/nrg.2016.112 [DOI] [PubMed] [Google Scholar]
- Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G. L., Lubling Y., et al. (2017). Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557.e24–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burney M. J., Johnston C., Wong K. Y., Teng S. W., Beglopoulos V., Stanton L. W., et al. (2013). An epigenetic signature of developmental potential in neural stem cells and early neurons. Stem Cells 31, 1868–1880. 10.1002/stem.1431 [DOI] [PubMed] [Google Scholar]
- Büttner N., Johnsen S. A., Kügler S., Vogel T. (2010). Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 7042–7047. 10.1073/pnas.0912041107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J. G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U.S.A. 112, 15672–15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comet I., Riising E. M., Leblanc B., Helin K. (2016). Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803–810. 10.1038/nrc.2016.83 [DOI] [PubMed] [Google Scholar]
- Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolic M., Fueyo R., et al. (2017). PRC2 facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell 20, 689.e9–705.e9. 10.1016/j.stem.2017.02.004 [DOI] [PubMed] [Google Scholar]
- De Juan Romero C., Bruder C., Tomasello U., Sanz-Anquela J. M., Borrell V. (2015). Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 34, 1859–1874. 10.15252/embj.201591176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W., Duboule D. (2013). Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502, 499–506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- de la Torre-Ubieta L., Stein J. L., Won H., Opland C. K., Liang D., Lu D., et al. (2018). The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172, 289.e18–304.e18. 10.1016/j.cell.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan C. M., Nyman U., Skotte J., Streubel G., Turner S., O'Connell D. J., et al. (2013). CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell 26, 223–236. 10.1016/j.devcel.2013.07.008 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Martinowich K., Chin M. H., He F., Fouse S. D., Hutnick L., et al. (2005). DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132, 3345–3356. 10.1242/dev.01912 [DOI] [PubMed] [Google Scholar]
- Fasano C. A., Dimos J. T., Ivanova N. B., Lowry N., Lemischka I. R., Temple S. (2007). shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell 1, 87–99. 10.1016/j.stem.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Fasano C. A., Phoenix T. N., Kokovay E., Lowry N., Elkabetz Y., Dimos J. T., et al. (2009). Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 23, 561–574. 10.1101/gad.1743709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J. F., Haffner C., Huttner W. B. (2014). 3' UTR-dependent, miR-92-mediated restriction of Tis21 expression maintains asymmetric neural stem cell division to ensure proper neocortex size. Cell Rep. 7, 398–411. 10.1016/j.celrep.2014.03.033 [DOI] [PubMed] [Google Scholar]
- Fietz S. A., Lachmann R., Brandl H., Kircher M., Samusik N., Schröder R., et al. (2012). Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc. Natl. Acad. Sci. U.S.A. 109, 11836–11841. 10.1073/pnas.1209647109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Florio M., Borrell V., Huttner W. B. (2017). Human-specific genomic signatures of neocortical expansion. Curr. Opin. Neurobiol. 42, 33–44. 10.1016/j.conb.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Florio M., Huttner W. B. (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., et al. (2016). Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Götz M., Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788. 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- Hatada I., Namihira M., Morita S., Kimura M., Horii T., Nakashima K. (2008). Astrocyte-specific genes are generally demethylated in neural precursor cells prior to astrocytic differentiation. PLoS ONE 3:e3189. 10.1371/journal.pone.0003189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Suzki N., Tsuboi M., Endo T. A., Toyoda T., Shinga J., et al. (2009). Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63, 600–613. 10.1016/j.neuron.2009.08.021 [DOI] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R. J., Kim H. J., Glass C. K., et al. (2007). SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450, 415–419. 10.1038/nature06270 [DOI] [PubMed] [Google Scholar]
- Johnson M. B., Wang P. P., Atabay K. D., Murphy E. A., Doan R. N., Hecht J. L., et al. (2015). Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646. 10.1038/nn.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N., Taverna E., Tavano S., Wong F. K., Suchold D., Winkler S., et al. (2016). CRISPR/Cas9-induced disruption of gene expression in mouse embryonic brain and single neural stem cells in vivo. EMBO Rep. 17, 338–348. 10.15252/embr.201541715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari A. E., Zhou J. X., Kanji M. S., Chan D. N., Sinha A., Grapin-Botton A., et al. (2013). The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 32, 1393–1408. 10.1038/emboj.2013.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke K., Radulović V., Broekhuis M., Weersing E., Zwart E., Olthof S., et al. (2013). Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat. Cell Biol. 15, 353–362. 10.1038/ncb2701 [DOI] [PubMed] [Google Scholar]
- Kloet S. L., Makowski M. M., Baymaz H. I., van Voorthuijsen L., Karemaker I. D., Santanach A., et al. (2016). The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 23, 682–690. 10.1038/nsmb.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Vogt M. A., Lugert S., Milo M., Chong M. M., Hautbergue G. M., et al. (2012). Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 15, 962–969. 10.1038/nn.3139 [DOI] [PubMed] [Google Scholar]
- Leeb M., Wutz A. (2007). Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 178, 219–229. 10.1083/jcb.200612127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. J., Nowakowski T. J., Pollen A. A., Lui J. H., Horlbeck M. A., Attenello F. J., et al. (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17:67. 10.1186/s13059-016-0932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J. H., Hansen D. V., Kriegstein A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18–36. 10.1016/j.cell.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N. M., Morey L., Mejetta S., Pascual G., Janich P., Kuebler B., et al. (2011). Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233–246. 10.1016/j.stem.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Marinaro F., Marzi M. J., Hoffmann N., Amin H., Pelizzoli R., Niola F., et al. (2017). MicroRNA-independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. 18, 603–618. 10.15252/embr.201642800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. A., Ding S. L., Sunkin S. M., Smith K. A., Ng L., Szafer A., et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrousis N., Tropepe V., Hermanson O. (2015). Post-translational modifications of histones in vertebrate neurogenesis. Front. Neurosci. 9:483. 10.3389/fnins.2015.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A., Mukamel E. A., Davis F. P., Luo C., Henry G. L., Picard S., et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F., Weber M., Rebhan M., Roloff T. C., Richter J., Stadler M. B., et al. (2008). Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 30, 755–766. 10.1016/j.molcel.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Molyneaux B. J., Goff L. A., Brettler A. C., Chen H. H., Brown J. R., Hrvatin S., et al. (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. 10.1016/j.neuron.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L., Pascual G., Cozzuto L., Roma G., Wutz A., Benitah S. A., et al. (2012). Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Morimoto-Suzki N., Hirabayashi Y., Tyssowski K., Shinga J., Vidal M., Koseki H., et al. (2014). The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development 141, 4343–4353. 10.1242/dev.112276 [DOI] [PubMed] [Google Scholar]
- Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski T. J., Fotaki V., Pollock A., Sun T., Pratt T., Price D. J. (2013). MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc. Natl. Acad. Sci. U.S.A. 110, 7056–7061. 10.1073/pnas.1219385110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loghlen A., Muñoz-Cabello A. M., Gaspar-Maia A., Wu H. A., Banito A., Kunowska N., et al. (2012). MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10, 33–46. 10.1016/j.stem.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. H., Hong S. J., Salinas R. D., Liu S. J., Sun S. W., Sgualdino J., et al. (2014). Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 8, 1290–1299. 10.1016/j.celrep.2014.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J. D., Sansom S. N., Smith J., Dobenecker M. W., Tarakhovsky A., Livesey F. J. (2010). Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 107, 15957–15962. 10.1073/pnas.1002530107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M., Barry G., Harvey T. J., McLeay R., Smith A. G., Harris L., et al. (2014). NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J. Neurosci. 34, 2921–2930. 10.1523/JNEUROSCI.2319-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A. (2016). Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058. 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulecio J., Verma N., Mejía-Ramírez E., Huangfu D., Raya A. (2017). CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell 21, 431–447. 10.1016/j.stem.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschendorf M., Terranova R., Boutsma E., Mao X., Isono K., Brykczynska U., et al. (2008). PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat. Genet. 40, 411–420. 10.1038/ng.99 [DOI] [PubMed] [Google Scholar]
- Rajman M., Schratt G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144, 2310–2322. 10.1242/dev.144337 [DOI] [PubMed] [Google Scholar]
- Santanach A., Blanco E., Jiang H., Molloy K. R., Sansó M., LaCava J., et al. (2017). The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat. Commun. 8:1235. 10.1038/s41467-017-01464-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H. M., Di Croce L., Cavalli G. (2017). Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Sessa A., Ciabatti E., Drechsel D., Massimino L., Colasante G., Giannelli S., et al. (2017). The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cereb. Cortex 27, 3378–3396. 10.1093/cercor/bhx209 [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C. A., Hadjantonakis A. K., Klein W. H., Broccoli V. (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56–69. 10.1016/j.neuron.2008.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E., Jones D., Shoresh N., Donohue L., Ram O., Bernstein B. E. (2016). Single-molecule decoding of combinatorially modified nucleosomes. Science 352, 717–721. 10.1126/science.aad7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D., Stampfel G., Stark A. (2014). Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286. 10.1038/nrg3682 [DOI] [PubMed] [Google Scholar]
- Silver D. L. (2016). Genomic divergence and brain evolution: how regulatory DNA influences development of the cerebral cortex. Bioessays 38, 162–171. 10.1002/bies.201500108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G., Abbas E., Rosenbusch J., Staiger J. F., Tuoc T. (2018). Transcriptional and epigenetic control of mammalian olfactory epithelium development. Mol. Neurobiol. [Epub ahead of print]. 10.1007/s12035-018-0987-y [DOI] [PubMed] [Google Scholar]
- Song M. R., Ghosh A. (2004). FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7, 229–235. 10.1038/nn1192 [DOI] [PubMed] [Google Scholar]
- Sparmann A., Xie Y., Verhoeven E., Vermeulen M., Lancini C., Gargiulo G., et al. (2013). The chromodomain helicase Chd4 is required for Polycomb-mediated inhibition of astroglial differentiation. EMBO J. 32, 1598–1612. 10.1038/emboj.2013.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E. E. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Stricker S. H., Götz M. (2018). DNA-Methylation: master or slave of neural fate decisions? Front. Neurosci. 12:5. 10.3389/fnins.2018.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L., Bershteyn M., Paredes M. F., Kriegstein A. R. (2017). Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat. Commun. 8:14167. 10.1038/ncomms14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., et al. (2001). DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1, 749–758. 10.1016/S1534-5807(01)00101-0 [DOI] [PubMed] [Google Scholar]
- Taverna E., Götz M., Huttner W. B. (2014). The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502. 10.1146/annurev-cellbio-101011-155801 [DOI] [PubMed] [Google Scholar]
- Telley L., Govindan S., Prados J., Stevant I., Nef S., Dermitzakis E., et al. (2016). Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446. 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- Tyssowski K., Kishi Y., Gotoh Y. (2014). Chromatin regulators of neural development. Neuroscience 264, 4–16. 10.1016/j.neuroscience.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R. J., et al. (2018). N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206. 10.1038/s41593-017-0057-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H., de la Torre-Ubieta L., Stein J. L., Parikshak N. N., Huang J., Opland C. K., et al. (2016). Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527. 10.1038/nature19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadirgi G., Leinster V., Acquati S., Bhagat H., Shakhova O., Marino S. (2011). Conditional activation of Bmi1 expression regulates self-renewal, apoptosis, and differentiation of neural stem/progenitor cells in vitro and in vivo. Stem Cells 29, 700–712. 10.1002/stem.614 [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Baltus A. E., Mathew R. S., Murphy E. A., Evrony G. D., Gonzalez D. M., et al. (2012). Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 151, 1097–1112. 10.1016/j.cell.2012.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Christian K. M., He C., Jin P., Ming G. L., Song H. (2016). Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549. 10.1038/nrn.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356:eaaj2239. 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K. J., Ringeling F. R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m6a methylation. Cell 171, 877.e17–889.e17. 10.1016/j.cell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S., Roundtree I. A., He C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Ziller M. J., Edri R., Yaffe Y., Donaghey J., Pop R., Mallard W., et al. (2015). Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature 518, 355–359. 10.1038/nature13990 [DOI] [PMC free article] [PubMed] [Google Scholar]