Abstract
Adoptive cell therapy (ACT) is becoming a prominent alternative therapeutic treatment for cancer patients relapsing on traditional therapies. In parallel, antibodies targeting immune checkpoint molecules, such as cytotoxic-T-lymphocyte-associated antigen 4 (CTLA-4) and cell death protein 1 pathway (PD-1), are rapidly being approved for multiple cancer types, including as first line therapy for PD-L1-expressing non-small-cell lung cancer. The combination of ACT and checkpoint blockade could substantially boost the efficacy of ACT. In this study, we generated a novel self-delivering small interfering RNA (siRNA) (sdRNA) that knocked down PD-1 expression on healthy donor T cells as well as patient-derived tumor-infiltrating lymphocytes (TIL). We have developed an alternative chemical modification of RNA backbone for improved stability and increased efficacy. Our results show that T cells treated with sdRNA specific for PD-1 had increased interferon γ (IFN-γ) secreting capacity and that this modality of gene expression interference could be utilized in our rapid expansion protocol for production of TIL for therapy. TIL expanded in the presence of PD-1-specific sdRNA performed with increased functionality against autologous tumor as compared to control TIL. This method of introducing RNAi into T cells to modify the expression of proteins could easily be adopted into any ACT protocol and will lead to the exploration of new combination therapies.
Keywords: PD-1, checkpoint blockade, adoptive cell therapy, rapid expansion protocol, RNA interference, sdRNA, sd-rxRNA
Ligtenberg et al. show that chemically modified self-deliverable siRNA conjugates (sdRNAs) allow for efficient T cell transfection. Using sdRNA to silence PD-1 expression on tumor-infiltrating lymphocytes increases their anti-tumor functionality. This methodology can be adopted into any expansion protocol, helping release the full potential of adoptively transferred T cells.
Introduction
Immunotherapy of malignant diseases is rapidly expanding the therapeutic options for cancer patients. Adoptive cell therapy (ACT) is a potent method of harnessing autologous immune cells, allowing for ex vivo manipulation of T cells or natural killer (NK) cells prior to their re-infusion into the patient. ACT includes therapy based on peripheral blood mononuclear cells (PBMCs) engineered to become tumor specific or on expansion of tumor-infiltrating lymphocytes (TILs) cultured from a surgical resection of the tumor. Clinical trials have shown promising results with TIL therapy of malignant melanoma, yielding an overall response (OR) rate around 30%–50%.1, 2 T cells engineered to express T cell receptors (TCRs) specific for tumor antigens in solid tumors have demonstrated a clinical response with an OR rate of 45%–70%.3, 4 The first ACT with chimeric antigen receptor (CAR) T cells engineered to express CD19 for treatment of relapsing B cell acute lymphoblastic leukemia (ALL) was recently approved by the US Food and Drug Administration (FDA) (ClincalTrials.gov ID: NCT02435849). CAR-based ACTs have seen complete responses (CRs) ranging from 68% to 100% for adult and pediatric B cell malignancies in multiple independent clinical trials.5 The experience from CAR therapy of solid tumors is, however, much more limited, with several major challenges remaining. The safety profiles for different types of ACTs are significantly different, with TILs having a relatively benign safety profile and most adverse events being due to the high-dose interleukin-2 (IL-2) administered. With TCR- or CAR-engineered T cell therapies, a number of more severe adverse events, ranging from tumor lysis syndrome, cytokine storm, and even fatal neurotoxicities, have been reported.3, 6, 7
The other major arm of immunotherapy recently being harnessed by oncologists is that of checkpoint-inhibiting antibodies (CIA). Antibody blockade of the checkpoints cytotoxic-T-lymphocyte-associated antigen 4 (CTLA-4) and the programmed cell death protein 1 pathway (PD-1/PD-L1) have demonstrated efficacy in a number of malignancies.8 The first FDA-approved CIA (ipilimumab) is responsible for blocking the inhibitory T cell signal mediated by CTLA-4 during the priming of naive T cells in lymph nodes. This allows the expansion of the T cell repertoire, including also the tumor-reactive T cell clones. Although ipilimumab was shown to produce a durable response in 20% of the patients, adverse events are frequent but manageable.9, 10 The clinical use of ipilimumab has now been largely replaced by antibodies targeting either the PD-1 receptor, expressed mainly by T cells, or the ligand PD-L1, expressed by antigen-presenting cells (APCs) or the tumor itself. It is important to note that PD-1/PD-L1 is a checkpoint involved in controlling peripheral tissue damage after an inflammatory response but hijacked by the tumor to effectively suppress anti-tumoral responses. Monotherapy with PD-1 blockade has resulted in better response rates (35%) and overall survival in advanced melanoma patients, with combination checkpoint blockade further increasing the overall survival.11 PD-1 blockade is currently standard of care for melanoma and has been FDA approved for use in non-small-cell lung carcinoma, renal cell carcinoma, and urothelial carcinoma.
Combining adoptive cell therapy with CIA is an attractive possibility already pursued in clinical trials (ClincalTrials.gov IDs: NCT02621021, NCT02926833, and NCT02757391), because blocking inhibitory checkpoint receptors concomitantly with adoptive T cell transfer has been shown to lead to a better tumor control in pre-clinical studies as well as in one recent clinical observation.12, 13 PD-1 binding can force a T cell into a state of senescence and even directly into apoptosis, whereas interference of the PD-1/PD-L1 axis by antibody therapy may allow the adoptively transferred T cells to continue their anti-tumor activity. The combination of ACT with CIA may, however, result in systemic serious adverse events caused by CIA acting on autoreactive T cell clones derived from ex vivo activated and expanded T cells in the TILs or genetically engineered T cell preparations. Furthermore, the injected CIA may not adequately penetrate into the immunosuppressive tumor microenvironment (TME), where the transferred T cells are supposed to perform their effector functions.
We therefore reasoned that an attractive alternative to the combination of ACT with antibody-mediated checkpoint blockade will be to silence PD-1 expression in the T cells prior to their transfer to the patient. This would allow ex vivo quantification of the PD-1 expression on the T cells prior to transfer and ensure that they are functionally enhanced in their tumor-recognizing capacity. To be able to produce TILs in which PD-1 expression is selectively silenced, we utilized self-delivering RNAi (self-delivering [sd] small interfering RNA [siRNA]) platform,14, 15 together with our good manufacturing practice (GMP)-compatible ACT TIL production pipeline.16 The sdRNA platform is based on extensive chemical modifications of siRNAs, conferring the resulting hydrophobically modified siRNA molecules (sdRNA and hsiRNA) the ability to penetrate all cell types ex vivo and in vivo and achieve long-lasting specific target gene knockdown without any additional delivery formulations or techniques (this class of compounds is referred to as sd-rxRNA by RXi Pharmaceuticals that develops them for human therapeutic and diagnostic applications).14, 15, 17, 18, 19 Our results demonstrate that this approach led to markedly reduced extracellular and intracellular PD-1 protein levels in the majority of the human primary T cells and also resulted in increased capacity of T cells to secrete interferon γ (IFN-γ) upon polyclonal stimulation. Furthermore, when this sdRNA technology was applied to a GMP TIL production pipeline, the PD-1 expression levels on the produced TILs were found to be reduced and their capacity to proliferate and produce cytokines to be enhanced.
Results
Identification of Sequence for the Potent Silencing of PDCD1 Gene Expression in Lymphocytes
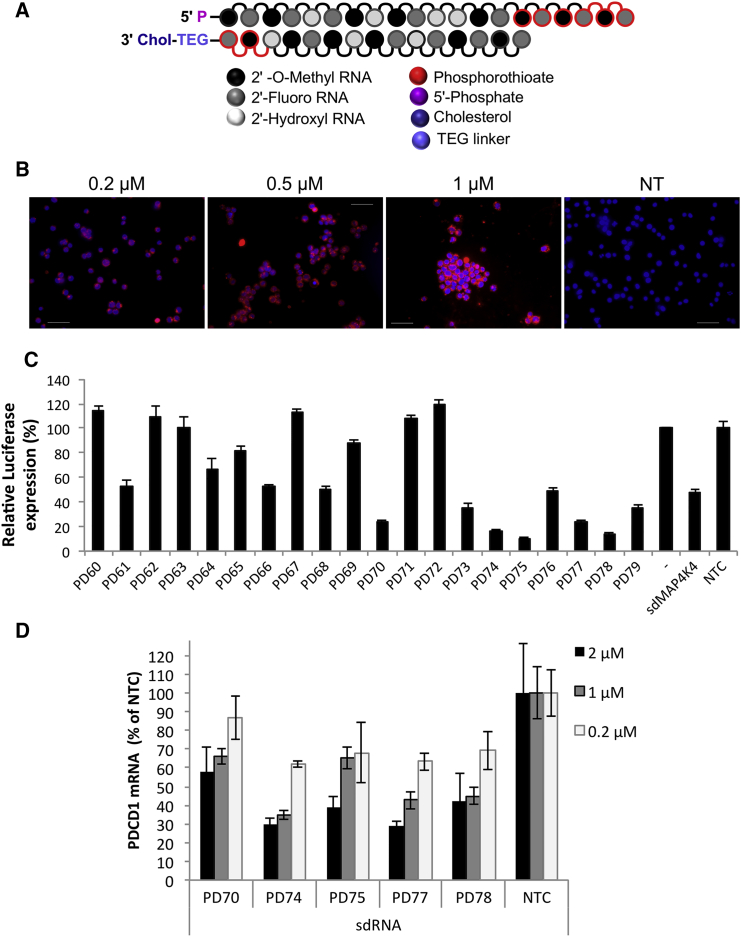
The self-deliverable RNAi molecule is a chemically synthesized asymmetric siRNA duplex consisting of the 20-nt antisense (guide) strand and 13–15 base sense (passenger) strand conjugated to cholesterol at its 3′ end using tetraethylenglycol (TEG) linker (Figure 1A). Most or all 2′OH positions of ribose residues are substituted with 2′OMe or 2′F modifications, conferring sdRNA molecule resistance to nuclease degradation in both extra- and intracellular environment. Additional nuclease protection is provided by phosphorothioate modifications at 3′ ends of guide and passenger strands. The combination of these modifications within an asymmetric siRNA scaffold is essential for its self-delivering properties and long-term knockdown activity.
Figure 1.
Development of PD1 Targeting Compounds
(A) Schematic structure of sdRNA. sdRNA is an asymmetric chemically modified siRNA duplex, containing 2′OMe, 2′F, and phosphorothioate backbone modifications and cholesterol conjugated to 3′ end of the passenger strand. (B) Fluorescent sdRNA uptake by Jurkat T cell lymphoma is shown. Cells were treated with Cy3-conjugated MAP4K4 sdRNA (red) for 48 hr in RPMI medium supplemented with 3% FBS. Cells were then incubated with Hoechst dye (blue), washed with PBS, and imaged live under fluorescent microscope Olympus BX-60. NT, non-treated cells. Representative images are shown. The scale bars represent 50 μm. (C) Lead compounds selection in a reporter luciferase assay is shown. Silencing is measured in cells expressing Renilla-PD1 fusion transcript and treated with PD1 sdRNAs at 1 μM for 48 hr. Data were normalized for Firefly luciferase and expressed as percent of non-treated “vector-only” cells (n = 3; mean ± SD). sdRNA targeting sequences are listed in Table S1. (D) PD1 mRNA knockdown by selected compounds in primary T cells is shown. Cells were activated with anti-CD3/CD28 beads for 4 days prior to treatment with sdRNA for another 72 hr. Cells were transfected at 200,000 cells/well in 96-well plate in RPMI medium with 1,000 U/mL IL-2. PD1 mRNA knockdown was quantified by qPCR using Taqman probes and normalized to GAPDH mRNA levels. Data are expressed as percent of non-targeting control (n = 4; mean ± SD).
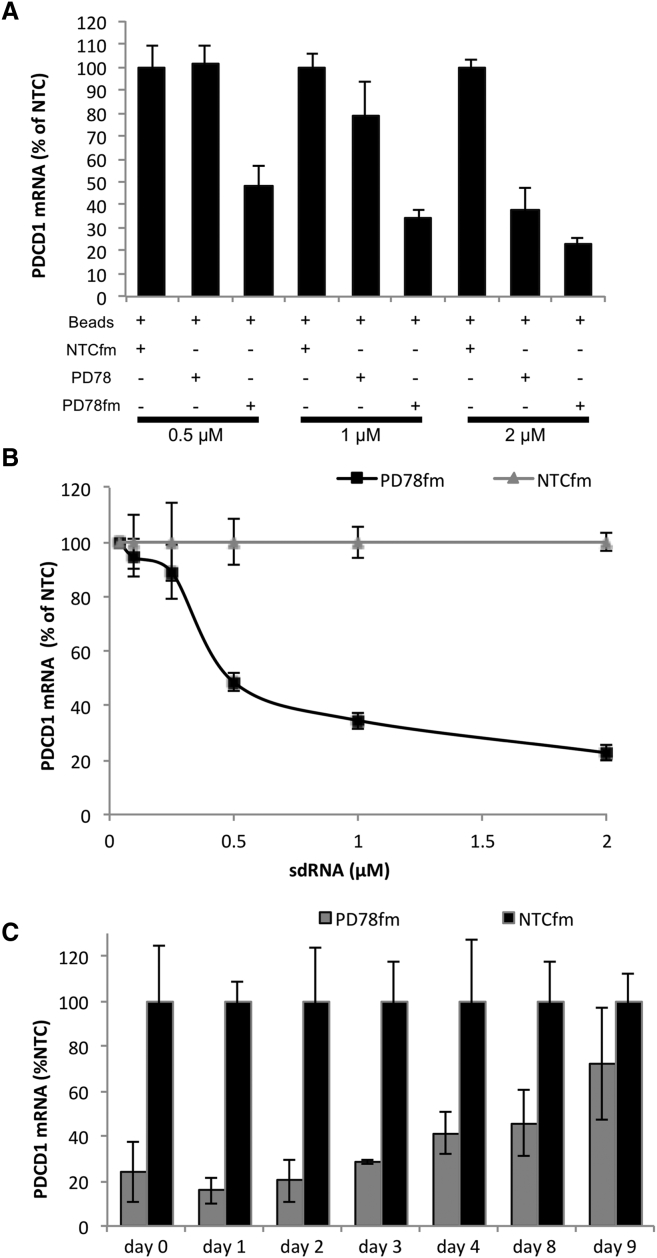
To evaluate sdRNA uptake by lymphocytes, fluorescent Cy3-conjugated sdRNA was first transfected into Jurkat lymphoma cells. Significant staining was observed at as low as 0.2 μM sdRNA concentration, and bright labeling was observed at 1 μM (Figure 1B). The uptake of fluorescent sdRNA was then measured in primary T cells showing greater than 95% transfection efficiency (Figure S1). To screen for sdRNA sequences that would inhibit PD-1 expression, we designed a luciferase reporter system based on dual-luciferase vector that included the sequence of PDCD1 gene and the target region of previously validated sdRNA against MAP4K4 downstream of Renilla luciferase expression cassette (Figure S2). We predicted and generated 20 sdRNA compounds specific for PDCD1 and compared their efficacy by determining relative luminescence in HeLa cells transiently expressing constructed plasmid. Multiple lead compounds that reduced PDCD1 reporter expression to around 20% (Figure 1C) were selected for evaluation of endogenous gene silencing. The addition of these sdRNAs (PD70, PD74, PD75, PD77, and PD78) to cultures of primary human T cells reduced PDCD1 gene expression by five-fold at 2 μM (Figure 1D). To further enhance the silencing efficiency of the sdRNA, we converted the identified sequences into a fully modified form,14 replacing all remaining ribose residues with either 2′F or 2′OMe modifications. These fully modified sdRNAs (sdRNAfm) proved to be more potent and led to larger inhibition of target mRNA expression than the initial sdRNA (Figure 2A) in the activated T cells (Figure S3). To determine the concentration of sdRNA to use in further experiments, the most potent sdRNA PD78fm and non-targeting control were titrated into primary T cell cultures and PDCD1 gene expression was validated (Figure 2B), with 2 μM sdRNAfm concentration showing the most potent effect after 72 hr post-introduction of sdRNA into the culture medium. The presence of neither PD78fm nor non-targeting control (NTC) had any significant effect on viability of T cells (Figure S4). Whereas short-term silencing of PDCD1 mRNA validated the knockdown effect of PD78fm sdRNA, the longevity of the effect in rapidly dividing cells would be an essential parameter in ACT optimization. To establish this, primary T cells were cultured in medium containing PD78fm or NTCfm sdRNA for 72 hr followed by removing the sdRNA-containing media, washing cells, and re-suspension in culture medium containing no sdRNAs. PDCD1 expression was monitored by qPCR over a period of 9 days, demonstrating that PDCD1 mRNA was still significantly suppressed as late as day 8 but started to return to base line levels at day 9 (Figure 2C).
Figure 2.
Improved Efficacy of PD1 sdRNA Knockdown by Additional Backbone Stabilization
(A) PD1 mRNA from activated T cells treated with selected (PD78) or chemically improved (PD78fm) sdRNA for 72 hr in the presence of anti-CD3/CD28 magnetic beads. Gene expression was quantified in a multiplex qPCR using GAPDH as a housekeeping gene (n = 3; mean ± SD). (B) Dose response analysis of PD1 mRNA in cells treated with improved PD78fm is shown. Effective dose was IC50 = 556 nmol/L. Transfection conditions and gene expression analysis were as in (A). (C) Longevity of PD1 knockdown in dividing T cells is shown. T cells activated with anti-CD3/CD28 beads for 5 days were then treated with 2 μM PD78fm for 72 hr in the presence of the activation beads. Cells were washed (point “0”) and incubated in complete AIM-V/IL-2 medium without sdRNA. Cell aliquots were collected at specified times for gene expression analysis. Shown is quantified by qPCR as in 1D (n = 3; mean ± SD).
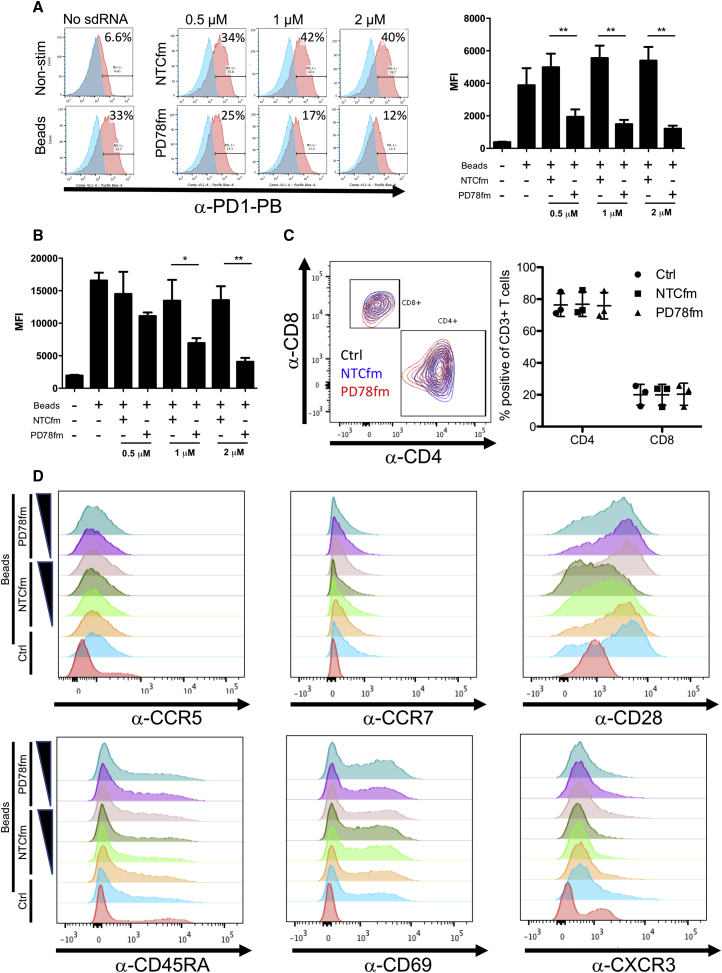
PD-1 Protein Levels Are Decreased in Primary Human T Cells by sdRNA Specific for PDCD1 while Minimally Impacting Other Cell Surface Markers
Next, we investigated the ability of PF78fm sdRNA to specifically reduce the protein levels of PDCD1. To this end, freshly isolated T cells were cultured with fully modified PD78 (PD78fm) or NTC (NTCfm) at varying concentrations followed by stimulation with anti-CD3/CD28 beads. Four days post-stimulation, extra- and intracellular levels of PD-1 were evaluated by flow cytometry. PD78fm sdRNA significantly decreased PD-1 cell surface expression over all three concentrations of sdRNA (Figure 3A) as well as intracellular PD-1 expression at 1 μM and 2 μM (Figures 3B and S5). Stimulation with CD3/CD28 beads did not alter the CD4/CD8 ratio in any of the conditions tested (Figure 3C). We also considered that the introduction of exogenous RNA may trigger natural antiviral mechanisms that activate T cells. To investigate this possibility, differentiation and activation markers on the sdRNA and control-treated T cells were stained with fluorescent antibodies and were acquired by fluorescence-activated cell sorting (FACS). Indeed, increasing concentrations of both NTCfm and PD78fm sdRNA resulted in slightly elevated CD69 expression on bead-stimulated T cells. Increasing levels of NTCfm sdRNA led to decreased expression of CD28 whereas this was maintained on PD78fm sdRNA (Figure 3D).
Figure 3.
Characterization of Healthy Donors’ T Cells after Treatment with Increasing Concentrations of PD78fm or the Control sdRNA NTCfm
T cells were cultured for four days in the presence of PD78fm at varying concentrations or non-targeting control (NTCfm) followed by stimulation with CD3/CD28 beads. (A and B) Extracellular (n = 3; A) and intracellular (n = 3; B) PD-1 expression levels are shown. (C) Staining of CD4 and CD8 on T cells (left panel) and quantification (n = 3) is shown. (D) Expression of differentiation and activation markers is shown. Error bars represent mean ± SD.
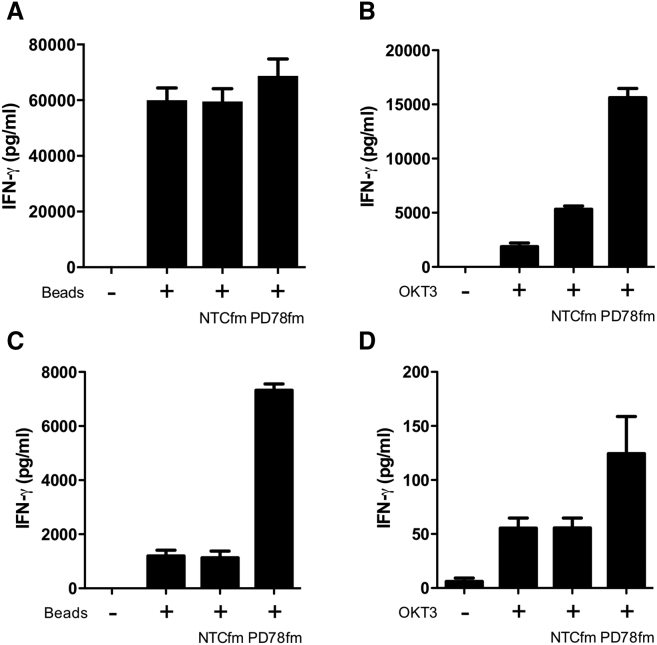
PD78fm sdRNA-Treated Primary T Cells Secrete More IFN-γ upon Activation
We then interrogated the consequences of sdRNA silencing of PD-1 for T cell functionality. Because T cells overexpress PD-L1 upon activation, we expected that PD-1 knockdown would result in stronger activation. Human T cells were cultured in the presence of 2 μM of sdRNA (PD78fm and NTCfm) and activated with CD3/CD28 beads or OKT3 antibody. Supernatants from PD78fm-treated T cells activated with beads were found to contain moderately more IFN-γ (Figure 4A). This effect was amplified when milder activation with OKT3 antibodies was used (Figure 4B). Most likely the stronger activation with CD3/CD28 beads overcame the inhibitive effect of PD-1/PD-L1 interaction between T cells. Even though CD3/CD28-activated T cells expressed some PD-L1 (Figure S6A), a more tumor-relevant model to study induction of T cell suppression by the PD-1/PD-L1 axis was then established. The cell line (KADA) previously established in our lab from a patient with advanced melanoma was found to express high levels of PD-L1 (Figure S6B). KADA tumor cells were seeded into plates and co-cultured with PD78fm or NTCfm pretreated or untreated OKT3 activated T cells. T cells pretreated with PD78fm secreted significantly more IFN-γ than non-sdRNA-treated and NTCfm sdRNA-treated T cells (Figure 4C). We therefore concluded that, in the presence of PD-L1-expressing tumor cells, the sdRNA-mediated blockade of PD-1 expression clearly enhanced the ability of activated T cells to secrete IFN-γ.
Figure 4.
IFN-γ Production by sdRNA-Treated Healthy Donor’s T Cells after In Vitro Activation
(A and B) T cells were seeded in RPMI+2% FBS in 96 U well plate—0.2M/100 μL/well, activated with CD3/CD28 beads (A) or OKT3 (30 ng/mL; B) and incubated with 2 μM of particular sdRNA for 4 days in total. The second day, FBS was added up to 10%. Supernatants were harvested on day 4, and IFN-γ level was measured by ELISA. (C and D) T cells were seeded in RPMI+2% FBS in 96 U well plate—0.2M/100 μL/well, activated by 0.1 μL/well beads (C) or 30 ng/mL OKT-3 (D) and incubated with 2 μM of particular sdRNA for 4 days. The second day, FBS was added up to 10%. On day 4, T cells were harvested and seeded in 96 F well plates with KADA cells at 1:1 ratio and incubated for 24 hr. Error bars represent mean ± SD.
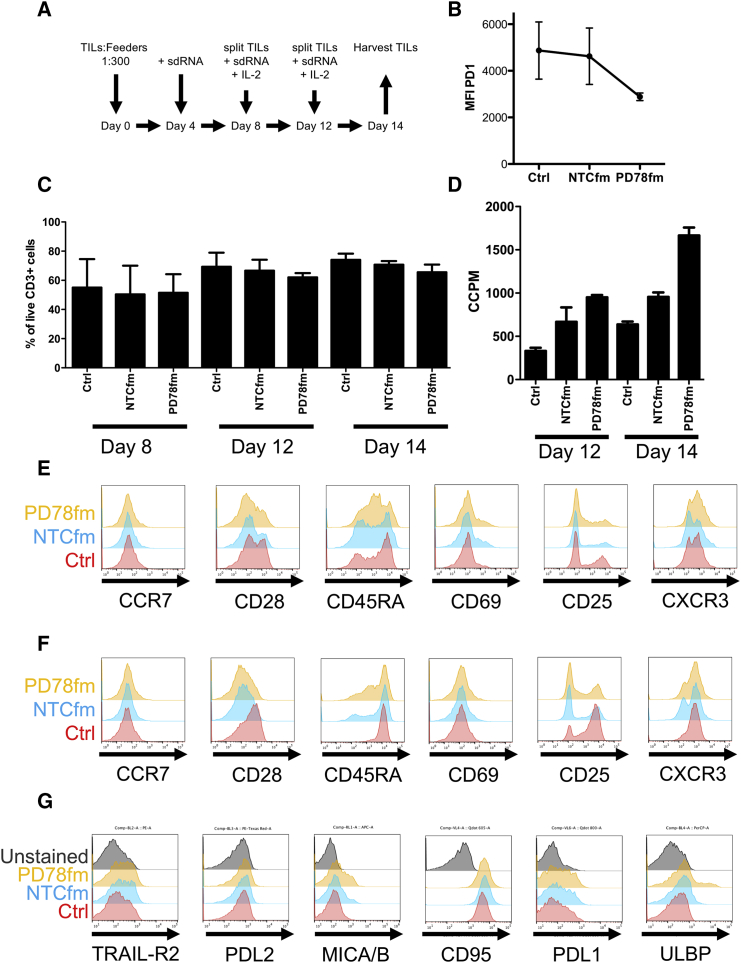
Silencing PD-1 by sdRNA Results in Enhanced Proliferation and IFN-γ Production in TILs Undergoing Rapid Expansion Protocol
For the treatment of melanoma patients with TIL therapy, large quantities of TILs are commonly produced by a standard rapid expansion protocol (REP).16 We therefore investigated whether sdRNA could be applied to a REP and lead to an improved TIL product in this clinically relevant protocol. A standard REP protocol used routinely also in our GMP facility (ClinicalTrials.gov; NCT01946373) was applied but with addition of sdRNA on days 4, 8, and 12, as schematically represented in Figures 5A and 5B. The addition of sdRNA every fourth day was motivated by the findings above demonstrating that PD-1 gene expression levels start to increase after this time. We monitored the viability of TILs on days 8, 12, and 14 post-initiation of REP (Figure 5C) and found that TILs undergoing REP in the presence of NTCfm and PD78fm tended to have slightly lower percent CD3+ live cells compared to control cultured TILs, ±70% and ±60% viability, respectively, compared to ±80% viability for control TILs. On days 12 and 14, the proliferative capacity of TILs was evaluated by thymidine incorporation. PD78fm-treated T cells had significantly increased thymidine incorporation on day 14 compared to control and NTCfm-treated TILs (Figure 5D). On day 14, TILs were harvested from the rapid expansion protocol and would typically be quality controlled prior to patient therapy. Harvested TILs were stained for differentiation and activation markers, and whereas most markers in both CD4+ TILs (Figure 5E) and CD8+ TILs (Figures 5E and 5F) were similar in TILs treated with each of the three products, there was a strong decrease in CD25+, CD4+, and CD8+ T cells in TILs treated with PD78fm or NTCfm (Figures 5E and 5F, fifth panel). Motivated by the difference in proliferative potential and a trend toward decreased viability, we stained TILs with a panel of antibodies for cell surface stress and activation markers. The control-, NTCfm-, and PD78fm-treated CD3+ TILs all expressed very little PD-L2 (Figure 5G, second panel). Furthermore, comparable levels of Trail-R2 (Figure 5G, first panel), PD-L1 (Figure 5G, fifth panel), and CD95 (Figure 5G, fourth panel) were detected. In contrast, PD78fm-treated TILs had increased levels of major histocompatibility complex (MHC) class I chain-related protein A and protein B (MICA/B) as well as a bright population of UL16 binding protein (ULBP)-positive cells that was not detected in the NTCfm-treated or control TILs (Figure 5G, third and sixth panels). Additionally, CD4/CD8 ratios were calculated and no significant differences were observed between control-treated and PD78fm-treated TILs (Figure S7).
Figure 5.
Characterization of TIL Expanded in the Presence of sdRNA
T cells were expanded for two weeks in CellGro medium supplemented with 2% human AB serum, 30 ng/mL OKT-3, and 300 U/mL IL-2. sdRNA was added at a 2 μM concentration on days 4, 8, and 12. Cell samples were collected for characterization on days 8, 12, and 14, at the end of the expansion protocol. Data are the result of three independent expansions from one single donor (A–D); (E–G) representative plots from one expansion are shown. (A) Schematic representation of the rapid expansion protocol (REP) is shown. (B) Intracellular expression of PD1 on day 14 (n = 3) is shown. (C) Viability of T cells during the expansion protocol as determined by Trypan Blue staining is shown. (D) T cell proliferation capacity during REP is shown. (E) Expression of differentiation and activation markers in CD4 T cells is shown. (F) Expression of differentiation and activation markers in CD8 T cells is shown. (G) Expression of cell surface stress and activation markers is shown. Error bars represent mean ± SD.
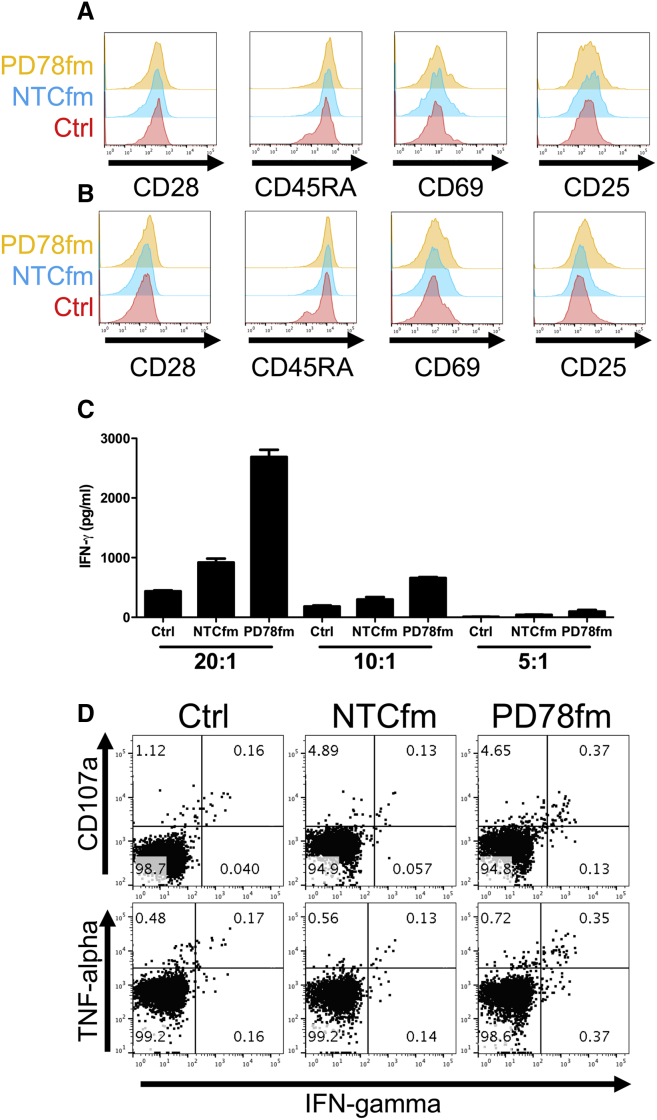
PD78fm-Cultured TILs Have Enhanced Ability to Recognize Autologous Tumors
Modulation of PD-1 protein expression in TILs can potentially lead to a better tumor recognition capacity. TILs expanded using the rapid expansion protocol and treated with either PD78fm, NTCfm sdRNA, or untreated control TILs were co-cultured with autologous tumor cell lines and evaluated for the cell surface differentiation and activation markers CD28, CD45RA, CD69, and CD25. All three products had similar expression of these markers on CD8+ T cells after 6 hr and 24 hr of co-culture (Figures 6A and 6B). Supernatants were collected from co-cultures and found to contain significantly more IFN-γ when TILs were expanded in the presence of sdPD78fm, but not NTCfm, sdRNA compared to control TILs (Figure 6C). This was further validated by intracellular staining for IFN-γ and tumor necrosis factor alpha (TNF-α) as well as staining for degranulation marker CD107a, where PD78fm-treated TILs degranulated and produced more cytokines than NTCfm or control TILs (Figure 6D).
Figure 6.
Effects of sdRNA on the Recognition of Autologous Tumor Cell Lines by Expanded TIL
T cells were co-cultured for 24 hr with autologous cell lines and harvested for characterization. (A and B) Expression of surface differentiation and activation markers after 6 hr (A) and 24 hr (B) is shown. (C) IFN-γ measured in the supernatant of co-cultured T cells at different tumor cell:T cell ratios after 24 hr is shown. (D) Cytokine production as measured by intracellular cytokine staining (ICS) after 24 hr (effector-to-target [E:T] ratio, 2:1) is shown. Error bars represent mean ± SD.
Discussion
Targeting the PD-1/PD-L1 axis can enhance the ex vivo expansion rate and in vivo longevity and functionality of T cells to be applied as ACT in cancer patients. Here, we investigate the potential of improving TIL therapy by applying a novel technology of self-delivering RNAi molecules specific to PD-1.
The use of RNAi to protect/enhance action of immune cells in ACT has been proposed previously,20 with the major hurdle being the difficulty of utilizing RNAi to transfect T cells. The commonly used transfection methods, such as lipid-mediated delivery or electroporation, result in significant loss of T cell viability and non-specific activation.
Self-deliverable RNAi technology based on the chemical modification of siRNAs successfully resolves the cellular delivery issue. The combination of backbone modifications with asymmetric siRNA structure and a hydrophobic ligand (Figure 1 and Khvorova and Watts14) allows sdRNAs to penetrate cultured mammalian cells without additional formulations and methods by simple addition to the culture media, capitalizing on the nuclease stability of sdRNAs. This stability allows the support of constant levels of RNAi-mediated knockdown of target gene activity simply by maintaining the active concentration of sdRNA in the media. The backbone stabilization of sdRNA allows it to exert long-lived gene knockdown effect, which can last for months in non-dividing cells.14, 17, 18
This technology was applied both to freshly isolated donor T cells and to a clinically relevant protocol, which is now regarded as the gold standard for expansion of large numbers of TIL.21 We demonstrated over 95% transfection efficiency of T cells and knockdown of the target PDCD1 gene by various specific sdRNAs. Originally identified sdRNAs containing several unmodified ribose residues were replaced with fully modified sequences to increase their potency and the longevity of RNAi effect. We have demonstrated the persistence for 8 days after transfection of more than 70% knockdown of PD-1 expression in rapidly dividing T cells. This knockdown would provide infused T cells with a head start, avoiding immediate suppression by the PD1/PD-L1 pathway. Additionally, and due to the benign safety profile involved, multiple infusions of a TIL product (as suggested by Rosenberg et al.22) would ensure that a potent in vivo effect could be established by the use of sdRNA. Altogether, PD-1 knockdown efficacy, persistence, and safety profile present sdRNA silencing of PD-1 as an attractive method to use for production of TILs for clinical trials.
Our findings are of direct clinical relevance for ACT of cancer patients. They are similar to the results obtained with antibodies blocking PD-1 when T cells are co-cultured in the presence of antigen-expressing tumor cells,23, 24, 25 where the secretion of cytokines (IFN-γ in particular) is augmented.26, 27, 28 We found that this was true for PD78fm-treated T cells both for low dose non-specific OKT3 stimulation as well as stimulation in the presence of a PD-L1 high tumor cell line. In line with these results, we found a potent increase in IFN-γ secretion when TILs were treated with PD78fm during activation with OKT3 prior to co-culture with autologous tumor cells.
We also observed an increased proliferation of sdRNA PD78fm-treated TILs during REP. This is not surprising, as PD-1 has been implicated directly in suppressing T cell proliferation by upregulation of p27 and p15 and resulting in induction of cell cycle arrest through suppression of Cdc25a.29 Growing TILs in the presence of anti-PD-1 was shown to increase yields, potentially by accelerated TIL proliferation during REP.25, 30 We consistently found an increased proliferative state in the sdRNA PD78fm compared to control sdRNA NTCfm-treated TILs. This led us to investigate whether PD78fm-treated TILs expressed increased activation-induced cell death (AICD) ligands. Whereas CD95 expression was similar among cells subjected to all three conditions, MICA/B and ULBPs were found to be slightly upregulated on PD78fm (and NTC)-treated TILs, which may in part be attributed to the non-specific activation of toll-like receptors by sdRNAs. It has indeed been found that activation of T cells leads to the upregulation of NKG2D ligands, potentially facilitating AICD.31, 32 During the REP, AICD may be responsible for the slightly decreased CD3+ T cell viability observed in sdRNA-treated TILs, because we added sdRNA at regular intervals to maintain low expression of PD-1. Complete genetic absence of PD-1 has been shown to lead to terminally differentiated phenotype and increased turnover of T cells.33 In support of this, we found a slight upregulation of the activation markers CD69 and CD25 when treating T cells with sdRNA. Because a non-specific activation was noted also with non-targeting sdRNA, we speculate that this could be due to cytosolic nucleic acid sensors recognizing the sdRNA, resulting in the activation of T cells.34
The exact mechanisms by which PD-1 inhibits the function of T cells is still a matter of debate. In contrast to earlier findings of the TCR signaling as the target of PD-1, Hui et al.35 recently elucidated that the de-phosphorylation of CD28 is the main target of PD-1 recruited and activated Shp2. In T cells activated with CD3/CD28 beads, we found a decrease in CD28 expression on cells cultured with NTCfm sdRNA, but not in T cells cultured with PD78fm sdRNA. High CD28 expression in the PD78fm sdRNA-treated T cells could be essential to facilitate their survival and retain their functional capacity.36
Currently, there is great interest in modification of T cells with gene editing tools, such as CRISPR-CAS and TALENs, and successful efforts have been made to generate T cells knockout for PD-1.37, 38, 39, 40 Furthermore, there are multiple clinical trials starting soon utilizing these technologies (NCT03044743 and NCT02793856) to target PD-1 and adoptively transfer T cells back into patients. There is intrinsically high risk that goes paired with this type of permanent genetic modification, namely, if an off-target event were to hit a tumor suppressor gene this may lead to the adoptive transfer of malignant cells. Off-target cleavage of the CRISPR-CAS system has been well documented,41, 42, 43 which makes this concern particularly valid. In addition to the off-target effects of genome editing, unintended long-term on-target effects may also lead to toxicities in the patients that will be receiving PD-1 knockout (KO) T-cell-based therapies. Indeed, it has been well described in mouse models that PD-1 KO T cells have a tendency to induce autoimmunity when confronted with viral challenge.44, 45 Similar results were found in PD-L1 KO mice, which succumb to immunopathology of over-activated T cells.46 RNAi could also have an off-target effect but one that would be limited by the transient nature of the therapy. Furthermore, the transient nature of sdRNA could in certain contexts be considered beneficial compared to permanently knocking out genes in T cells. In the case of PD-1, this will allow, after 10 days when we find the effect of PD-1-specific sdRNA to wane (in rapidly dividing cells), the T cells to enter their transition from effector to memory cells. PD-1 has been shown to be, in part, responsible for controlling this differentiation process, which is essential for the natural functioning of adoptively transferred cells.47, 48 This may also be the case for other regulators of T cell functionality, where it would be preferential to have a transient effect and not permanently disturb the natural T cell cycle. Finally, introduction of CAS9 into T cells has been achieved with electroporation of either nucleic acids encoding CAS9 or CAS9 protein guide RNA (gRNA) complexes.37, 38, 40, 49 Electroporation is known to have a highly detrimental effect on the viability of the cells, and many efforts have been made to optimize efficiency of gene transfer as well as minimization of cell death.50, 51, 52 Considering that TIL products have already been through considerably more stress than healthy donor-derived T cells, electroporation with the goal of gene editing may yield a much inferior T cell product for the patients than desired. In contrast, sdRNA technologies could be plugged into any ongoing clinical protocol with only minor modifications.
With the treatment of patients with checkpoint inhibitors, it is becoming clear that resistance is becoming an issue, just as was earlier found with small-molecule inhibitors. Resistance to PD-1 therapy may be prevented by targeting of additional T cell inhibitory molecules. Tumor-specific T cells can be identified by PD-1 but also by other exhaustion makers, such as TIM-3 and LAG-3.53 Koyama et al.54 have shown that patients relapsing while on PD-1 therapy had increased T cell exhaustion TIM-3, LAG-3, and CTLA-4. By sequentially treating with anti-TIM-3, they were able to extend the survival of mice in their mouse model. TIGIT similarly is co-expressed with PD-1, and dual blockade of TIGIT with PD-1 facilitated increased tumor-antigen-specific T cell proliferation, cytokine release, and degranulation.55 The rationale for combining multiple immune checkpoint blockade inhibitors has been well established,56 and utilizing the flexibility and ease of targeting several molecules simultaneously with specific sdRNA will facilitate this strategy. For example, patients receiving combination of CTLA4 and PD1 blockade have experienced radical tumor regressions in very short periods of time57 but at a greater risk of immune-related adverse events. By targeting multiple inhibitory molecules, we could potentially alleviate the short-lived effect of sdRNA, benefitting from faster response times. We believe that sdRNA technology will greatly facilitate the development of functional RNAi14 that targets inhibitors of T cell functions, helping to release the full potential of adoptively transferred T cells.
Materials and Methods
sdRNA Selection
20 sdRNA sequences targeting PDCD1 gene (NM_005018) were selected based on Advirna’s proprietary selection algorithm, designed on the basis of a functional screen of over 500 sdRNA sequences. Regression analysis was used to establish a correlation between the frequency of occurrence of specific nucleotide and modification at any specific position in sdRNA duplex and its functionality in gene suppression assay. Selected sequences were synthesized (TriLink Biotechnologies) in a 0.2-μmol scale and dissolved in sterile RNase-, DNase-free water for injection (CalBiochem; 4.86505). Duplexes were annealed by heating up at 95°C for 5 min and gradually cooling down to room temperature.
Cells and Patient Material
PBMCs were isolated from healthy donor buffy coats (Karolinska University Hospital; ethical permit No. 20010305,01-50) by gradient centrifugation (Ficoll-Paque Plus; GE Healthcare). T cells were purified using the Pan T cell isolation kit (Miltenyi Biotec) following manufacturer’s instructions. Peripheral blood CD3+ Pan T cells from AllCells (PB009-1F) were used for sdRNA validation and chemical structure optimization studies. HeLa cells (ATCC) were routinely maintained at 50%–80% confluence for at most 15 passages.
Tumor biopsies and PBMCs were obtained from advanced melanoma patients undergoing treatment at the Department of Oncology, Karolinska Hospital. The early-passage KADA melanoma cell line was established from a stage III melanoma patient undergoing treatment in the oncology clinic at Karolinska University Hospital as previously described.58 The Karolinska Institutet review board approved the protocol (2011/143-32/1), and all patients provided written informed consent in accordance with the Declaration of Helsinki.
sdRNA Direct Delivery (Passive Uptake)
Oligonucleotides were diluted in serum-free medium and dispensed into 96-well culture plate in triplicates. Cells were seeded in appropriate culture medium containing reduced FBS in the plate with pre-diluted compounds for indicated time. HeLa cells were transfected in Eagle’s minimal essential medium (EMEM) medium with 3% FBS at 10,000 cells/well. Primary human T cells (AllCells, CA) were cultured in complete AIM-V (Gibco) medium containing 500 IU/mL IL-2 (ProSpec). Cells were activated with anti-CD3/CD28 Dynabeads (Gibco; 11131) according to the manufacturer’s instructions for at least 4 days prior to transfection. T cells were transfected in 5% FBS at 100,000 cells/well without removing the Dynabeads, unless otherwise specified.
Fluorescent images were obtained from live cells transfected with Cy3-conjugated sdRNA using Olympus BX-60 microscope. Nuclear staining was obtained with Hoechst 33342 (Molecular Probes; H1398) added to transfected cells for 30 min. Images were processed with ImageJ (1.47v) software.
Lead sdRNA Compound Identification
Luciferase reporter plasmid was constructed by inserting PDCD1 targeting regions into psiCheck2 plasmid (Promega; C8021) downstream Renilla luciferase sequence. Previously validated MAP4K4 sdRNA sequence was also inserted as a positive control.
For the screening, HeLa cells were transfected with the cloned plasmid using Fugene HD (Promega; E2311) according to the manufacturer’s instructions. Briefly, cells were seeded at 2.5 × 106 cells/10 cm2 dish in EMEM (ATCC; 30-2003) medium without antibiotics and transfected 6 hr later with the plasmid at 2.5:1 FuGENE:DNA ratio. Cells were incubated for 16–18 hr, washed 3 times with PBS, trypsinized, and seeded into 96-well plate with pre-diluted sdRNA compounds at final concentration 1 μM sdRNA/10,000 cells/100 μL EMEM with 3% FBS. Cells were treated with sdRNA for 48 hr to facilitate passive cellular uptake of compounds, lysed with Glo lysis buffer (Promega; E266A), and assayed for Renilla and Firefly luciferase expression. For that, 20-μL aliquots of each lysate were added into duplicate opaque 96-well plates and mixed with either Matthews (Renilla) assay buffer59 or Firefly luciferase assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4 [pH 7.8], and 1 mM D-Luciferin). The substrates D-Luciferin (Promega; E1605) and h-Coelenterazine (NanoLight; 301) were added immediately prior to use. Luminescence was measured on SpectraMax i3 (Molecular Devices), normalized (Renilla/Firefly), and expressed as a percent untreated control.
mRNA Quantification by qPCR
Total RNA was isolated from transfected cells using the PureLink Pro96 Purification Kit (Invitrogen; 12173-011A) according to the manufacturer’s recommendations. Dilutions of non-transfected (NT) cells of 1:5 and 1:25 were routinely prepared for a standard curve generation. Gene expression was analyzed in a one-step multiplex qPCR by mixing 20–40 ng purified RNA with Quanta qScript qRT-PCR ToughMix (VWR; 89236672) and Taqman probes—PDCD1-FAM (Taqman; Hs01550088_m1) and GAPDH-VIC (Applied Biosystems; 4326317E) in the same reaction. Samples were amplified using Quanta’s recommended settings in a StepOnePlus qPCR machine (Applied Biosystems). PDCD1 expression was normalized to GAPDH, adjusted to the standard curve, and expressed as a percent of NTC-transfected cells.
Antibodies and Flow Cytometry
Antibody details are provided in Table S2. PBMCs were stained at 4°C according to the manufacturer’s recommendations, after proper titration in order to obtain an optimal signal to noise ratio. Dead cells were excluded with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen). Cells were analyzed using an LSRII flow cytometer (BD Biosciences) and FlowJo software (Treestar, Ashland, OR), using a non-stained control for each sample. Quality control of the flow cytometer’s performance and coefficient of variation (CV) values were monitored on a day-to-day basis using CS&T beads (BD Biosciences).
Cell Viability Assay
Primary T cells activated with anti-CD3/CD28 beads and seeded in triplicates in a 96-well plate were transfected with sdRNA oligonucleotides at various doses for 72 hr. Cells were washed and incubated with 1:10 diluted CellTiter-Blue reagent (Promega; G808A) for 1 hr at 37°C. Plates were brought to room temperature and fluorescence recorded at 530 nm ex/590 nm em. Linear range was confirmed by plating 4 series of 2-fold cells dilutions in the same conditions and plotting fluorescence readings.
TIL Isolation
TILs were isolated as previously described (PMID: 24993563). Briefly, 1 mm3 pieces of tumor were incubated in 24-well plates with CellGro medium supplemented with 6,000 U/mL IL-2 (PeproTech) and 2% human AB serum for approximately 2 weeks, when expanding cells were pooled and counted.
Rapid Expansion Protocol
Purified TILs from advanced melanoma patients were thawed, washed, and seeded in flasks at 6 × 103 cells/mL along with 1.75 × 106 irradiated feeder PBMCs and OKT3 (Miltenyi) at 30 ng/mL. Rapid expansion was carried out in CellGro medium (CellGenix) supplemented with 2% human AB serum and 300 U/mL IL-2 (PeproTech) and 2 μM of either PD78fm sdRNA or the control sdRNA NTCfm. Medium was replaced every four days, and cell samples were collected on days 4, 8, and 12. The final product was harvested on day 14 and used immediately.
Thymidine Incorporation Assay
TILs harvested on days 12 and 14 during the rapid expansion protocol were seeded in triplicate on a 96-well plate (104 cells/well) in CellGro medium supplemented with 2% human AB serum. After 1 hr, 1 μCi/well of [methyl-3H] thymidine (PerkinElmer, Waltham, MA) was added to each well and incubated for four hours. Cells were then harvested and 3H-thymidine incorporation was measured in a Trilux 1450 microBeta liquid scintillation counter (Wallac).
IFN-γ Secretion of sdRNA-Treated Cells
IFN-γ production by stimulated T cells was measured in the supernatant using the Human IFN-γ ELISA development kit (Mabtech) as per manufacturer’s instructions. Purified T cells from healthy donors were stimulated with OKT3 (30 ng/mL) and treated with 2 μM sdRNA for four days. After this period, the supernatant was collected for ELISA analysis and T cells were harvested. Harvested T cells were further co-cultured with early passage melanoma cell line KADA for 24 hr, when the supernatant was again collected for IFN-γ determination.
Intracellular Cytokine Staining
Purified expanded TILs were co-cultured with autologous KADA cell line (2:1 effector:target ratio; 105 TILs/well) in the presence of fluorescein isothiocyanate (FITC)-labeled anti-CD107a antibody (BioLegend). After two hours, cytokine secretion was blocked with GolgiPlug/GolgiStop (BD Biosciences) and further incubated for an additional four hours. Intracellular levels of IFN-γ and TNF-α (Table S2) were measured after cellular permeabilization with cytofix/cytoperm (BD Biosciences) in an LSRII flow cytometer (BD Biosciences).
Author Contributions
Design and/or Interpretation of the Reported Experiments, M.A.L., Y.P.d.C., Y. Yoshimoto, A.D.W., T.S., A.V.E., and R.K.; Acquisition of Data, T.S., M.B.-B., Y. Yoshimoto, Y. Yang, I.T., M.A.L., and Y.P.d.C.; Analysis and Interpretation of Data, T.S., A.D.W., Y. Yoshimoto, I.T., M.A.L., Y.P.d.C., and R.K.; Drafting and Revising the Manuscript, A.D.W., T.S., A.V.E., M.A.L., Y.P.d.C., Y. Yoshimoto, Y. Yang, I.T., and R.K.; Obtained of Regulatory Approvals, R.K.
Conflicts of Interest
A.D.W., T.S., and M.B.-B. are employees of Advirna, a company developing sdRNAs for research applications. R.K. is a scientific advisor to RXi Pharmaceuticals and owns stock options in this company. A.V.E. and A.D.W. are shareholders of RXi Pharmaceuticals, a company that develops sdRNAs for human therapeutic and diagnostic applications.
Acknowledgments
This work was partially supported by grants from the NIH (5R44HG006788-03), The Swedish Cancer Society (2013/379), The Cancer Society in Stockholm and The King Gustaf V’s Jubilee Foundation (144102), The Swedish Medical Research Council (521-2013-4100), Stockholm City Council project grant 2016-1376 (ALF Medicin 2016-1376), and the “Knut and Alice Wallenberg Foundations.” The authors would like to thank Disha Rao for collaborating in several aspects of this work. This work was performed on sites in Stockholm, Sweden and Cambridge, MA, USA.
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.04.015.
Supplemental Information
References
- 1.Besser M.J., Shapira-Frommer R., Itzhaki O., Treves A.J., Zippel D.B., Levy D., Kubi A., Shoshani N., Zikich D., Ohayon Y. Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin. Cancer Res. 2013;19:4792–4800. doi: 10.1158/1078-0432.CCR-13-0380. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg S.A., Yang J.C., Sherry R.M., Kammula U.S., Hughes M.S., Phan G.Q., Citrin D.E., Restifo N.P., Robbins P.F., Wunderlich J.R. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg S.A., Restifo N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson L.A., Morgan R.A., Dudley M.E., Cassard L., Yang J.C., Hughes M.S., Kammula U.S., Royal R.E., Sherry R.M., Wunderlich J.R. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivière I., Sadelain M. Chimeric antigen receptors: a cell and gene therapy perspective. Mol. Ther. 2017;25:1117–1124. doi: 10.1016/j.ymthe.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan R.A., Yang J.C., Kitano M., Dudley M.E., Laurencot C.M., Rosenberg S.A. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 2010;18:843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Berg J.H., Gomez-Eerland R., van de Wiel B., Hulshoff L., van den Broek D., Bins A., Tan H.L., Harper J.V., Hassan N.J., Jakobsen B.K. Case report of a fatal serious adverse event upon administration of T cells transduced with a MART-1-specific T-cell receptor. Mol. Ther. 2015;23:1541–1550. doi: 10.1038/mt.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pico de Coaña Y., Choudhury A., Kiessling R. Checkpoint blockade for cancer therapy: revitalizing a suppressed immune system. Trends Mol. Med. 2015;21:482–491. doi: 10.1016/j.molmed.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Hodi F.S., O’Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert C., Thomas L., Bondarenko I., O’Day S., Weber J., Garbe C., Lebbe C., Baurain J.F., Testori A., Grob J.J. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 11.Larkin J., Hodi F.S., Wolchok J.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015;373:1270–1271. doi: 10.1056/NEJMc1509660. [DOI] [PubMed] [Google Scholar]
- 12.John L.B., Devaud C., Duong C.P.M., Yong C.S., Beavis P.A., Haynes N.M., Chow M.T., Smyth M.J., Kershaw M.H., Darcy P.K. Anti-PD-1 antibody therapy potently enhances the eradication of established tumors by gene-modified T cells. Clin. Cancer Res. 2013;19:5636–5646. doi: 10.1158/1078-0432.CCR-13-0458. [DOI] [PubMed] [Google Scholar]
- 13.Chong E.A., Melenhorst J.J., Lacey S.F., Ambrose D.E., Gonzalez V., Levine B.L., June C.H., Schuster S.J. PD-1 blockade modulates chimeric antigen receptor (CAR)-modified T cells: refueling the CAR. Blood. 2017;129:1039–1041. doi: 10.1182/blood-2016-09-738245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrne M., Tzekov R., Wang Y., Rodgers A., Cardia J., Ford G., Holton K., Pandarinathan L., Lapierre J., Stanney W. Novel hydrophobically modified asymmetric RNAi compounds (sd-rxRNA) demonstrate robust efficacy in the eye. J. Ocul. Pharmacol. Ther. 2013;29:855–864. doi: 10.1089/jop.2013.0148. [DOI] [PubMed] [Google Scholar]
- 16.Poschke I., Lövgren T., Adamson L., Nyström M., Andersson E., Hansson J., Tell R., Masucci G.V., Kiessling R. A phase I clinical trial combining dendritic cell vaccination with adoptive T cell transfer in patients with stage IV melanoma. Cancer Immunol. Immunother. 2014;63:1061–1071. doi: 10.1007/s00262-014-1575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.-C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikan M., Osborn M.F., Coles A.H., Godinho B.M., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids. 2016;5:e344. doi: 10.1038/mtna.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ly S., Navaroli D.M., Didiot M.-C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017;45:15–25. doi: 10.1093/nar/gkw1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris K., Castanotto D., Al-Kadhimi Z., Jensen M., Rossi J., Cooper L.J.N. Enhancing siRNA effects in T cells for adoptive immunotherapy. Hematology. 2005;10:461–467. doi: 10.1080/10245330500233569. [DOI] [PubMed] [Google Scholar]
- 21.Dudley M.E., Yang J.C., Sherry R., Hughes M.S., Royal R., Kammula U., Robbins P.F., Huang J., Citrin D.E., Leitman S.F. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J. Clin. Oncol. 2008;26:5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg S.A., Yang J.C., Robbins P.F., Wunderlich J.R., Hwu P., Sherry R.M., Schwartzentruber D.J., Topalian S.L., Restifo N.P., Filie A. Cell transfer therapy for cancer: lessons from sequential treatments of a patient with metastatic melanoma. J. Immunother. 2003;26:385–393. doi: 10.1097/00002371-200309000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank C., Kuball J., Voelkl S., Wiendl H., Becker B., Walter B., Majdic O., Gajewski T.F., Theobald M., Andreesen R., Mackensen A. Blockade of PD-L1 (B7-H1) augments human tumor-specific T cell responses in vitro. Int. J. Cancer. 2006;119:317–327. doi: 10.1002/ijc.21775. [DOI] [PubMed] [Google Scholar]
- 24.Wong R.M., Scotland R.R., Lau R.L., Wang C., Korman A.J., Kast W.M., Weber J.S. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. Int. Immunol. 2007;19:1223–1234. doi: 10.1093/intimm/dxm091. [DOI] [PubMed] [Google Scholar]
- 25.Saunders P.A., Hendrycks V.R., Lidinsky W.A., Woods M.L. PD-L2:PD-1 involvement in T cell proliferation, cytokine production, and integrin-mediated adhesion. Eur. J. Immunol. 2005;35:3561–3569. doi: 10.1002/eji.200526347. [DOI] [PubMed] [Google Scholar]
- 26.Blank C., Brown I., Peterson A.C., Spiotto M., Iwai Y., Honjo T., Gajewski T.F. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells. Cancer Res. 2004;64:1140–1145. doi: 10.1158/0008-5472.can-03-3259. [DOI] [PubMed] [Google Scholar]
- 27.Dulos J., Carven G.J., van Boxtel S.J., Evers S., Driessen-Engels L.J.A., Hobo W., Gorecka M.A., de Haan A.F., Mulders P., Punt C.J. PD-1 blockade augments Th1 and Th17 and suppresses Th2 responses in peripheral blood from patients with prostate and advanced melanoma cancer. J. Immunother. 2012;35:169–178. doi: 10.1097/CJI.0b013e318247a4e7. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Jie H.B., Lei Y., Gildener-Leapman N., Trivedi S., Green T., Kane L.P., Ferris R.L. PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer Res. 2015;75:508–518. doi: 10.1158/0008-5472.CAN-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patsoukis N., Sari D., Boussiotis V.A. PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle. 2012;11:4305–4309. doi: 10.4161/cc.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall M., Liu H., Malafa M., Centeno B., Hodul P.J., Pimiento J., Pilon-Thomas S., Sarnaik A.A. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. J. Immunother. Cancer. 2016;4:61. doi: 10.1186/s40425-016-0164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerboni C., Zingoni A., Cippitelli M., Piccoli M., Frati L., Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK- cell lysis. Blood. 2007;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 32.Molinero L.L., Fuertes M.B., Rabinovich G.A., Fainboim L., Zwirner N.W. Activation-induced expression of MICA on T lymphocytes involves engagement of CD3 and CD28. J. Leukoc. Biol. 2002;71:791–797. [PubMed] [Google Scholar]
- 33.Odorizzi P.M., Pauken K.E., Paley M.A., Sharpe A., Wherry E.J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan Y.K., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hui E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boomer J.S., Green J.M. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2010;2:a002436. doi: 10.1101/cshperspect.a002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren J., Zhang X., Liu X., Fang C., Jiang S., June C.H., Zhao Y. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget. 2017;8:17002–17011. doi: 10.18632/oncotarget.15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumann K., Lin S., Boyer E., Simeonov D.R., Subramaniam M., Gate R.E., Haliburton G.E., Ye C.J., Bluestone J.A., Doudna J.A., Marson A. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 2015;112:10437–10442. doi: 10.1073/pnas.1512503112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menger L., Sledzinska A., Bergerhoff K., Vargas F.A., Smith J., Poirot L., Pule M., Hererro J., Peggs K.S., Quezada S.A. TALEN-mediated inactivation of PD-1 in tumor-reactive lymphocytes promotes intratumoral T-cell persistence and rejection of established tumors. Cancer Res. 2016;76:2087–2093. doi: 10.1158/0008-5472.CAN-15-3352. [DOI] [PubMed] [Google Scholar]
- 40.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura H., Nose M., Hiai H., Minato N., Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 45.Frebel H., Nindl V., Schuepbach R.A., Braunschweiler T., Richter K., Vogel J., Wagner C.A., Loffing-Cueni D., Kurrer M., Ludewig B., Oxenius A. Programmed death 1 protects from fatal circulatory failure during systemic virus infection of mice. J. Exp. Med. 2012;209:2485–2499. doi: 10.1084/jem.20121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barber D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 47.Charlton J.J., Tsoukatou D., Mamalaki C., Chatzidakis I. Programmed death 1 regulates memory phenotype CD4 T cell accumulation, inhibits expansion of the effector memory phenotype subset and modulates production of effector cytokines. PLoS ONE. 2015;10:e0119200. doi: 10.1371/journal.pone.0119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charlton J.J., Chatzidakis I., Tsoukatou D., Boumpas D.T., Garinis G.A., Mamalaki C. Programmed death-1 shapes memory phenotype CD8 T cell subsets in a cell-intrinsic manner. J. Immunol. 2013;190:6104–6114. doi: 10.4049/jimmunol.1201617. [DOI] [PubMed] [Google Scholar]
- 49.Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chicaybam L., Sodre A.L., Curzio B.A., Bonamino M.H. An efficient low cost method for gene transfer to T lymphocytes. PLoS ONE. 2013;8:e60298. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Johnson C., Fujimura S., Teque F., Levy J.A. Transfection optimization for primary human CD8+ cells. J. Immunol. Methods. 2011;372:22–29. doi: 10.1016/j.jim.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y., Zheng Z., Cohen C.J., Gattinoni L., Palmer D.C., Restifo N.P., Rosenberg S.A., Morgan R.A. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol. Ther. 2006;13:151–159. doi: 10.1016/j.ymthe.2005.07.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gros A., Robbins P.F., Yao X., Li Y.F., Turcotte S., Tran E., Wunderlich J.R., Mixon A., Farid S., Dudley M.E. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koyama S., Akbay E.A., Li Y.Y., Herter-Sprie G.S., Buczkowski K.A., Richards W.G., Gandhi L., Redig A.J., Rodig S.J., Asahina H. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 2016;7:10501. doi: 10.1038/ncomms10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chauvin J.-M., Pagliano O., Fourcade J., Sun Z., Wang H., Sander C., Kirkwood J.M., Chen T.H., Maurer M., Korman A.J., Zarour H.M. TIGIT and PD-1 impair tumor antigen-specific CD8+ T cells in melanoma patients. J. Clin. Invest. 2015;125:2046–2058. doi: 10.1172/JCI80445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smyth M.J., Ngiow S.F., Ribas A., Teng M.W.L. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 2016;13:143–158. doi: 10.1038/nrclinonc.2015.209. [DOI] [PubMed] [Google Scholar]
- 57.Chapman P.B., D’Angelo S.P., Wolchok J.D. Rapid eradication of a bulky melanoma mass with one dose of immunotherapy. N. Engl. J. Med. 2015;372:2073–2074. doi: 10.1056/NEJMc1501894. [DOI] [PubMed] [Google Scholar]
- 58.Selvan S.R., Carbonell D.J., Fowler A.W., Beatty A.R., Ravindranath M.H., Dillman R.O. Establishment of stable cell lines for personalized melanoma cell vaccine. Melanoma Res. 2010;20:280–292. doi: 10.1097/CMR.0b013e3283390696. [DOI] [PubMed] [Google Scholar]
- 59.Matthews J.C., Hori K., Cormier M.J. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977;16:85–91. doi: 10.1021/bi00620a014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.