Abstract
Exposing the adolescent brain to drugs of abuse is associated with increased risk for adult onset psychopathologies. Cannabis use peaks during adolescence, with largely unknown effects on the developing brain. Cannabis’ major psychoactive component, Δ9-tetrahydrocannabinol (THC) alters neuronal, astrocytic, and microglial signaling. Therefore, multiple cellular and signaling pathways are affected with a single dose of THC. The endogenous cannabinoids (eCBs), N-arachidonoyl ethanolamine (AEA) and 2-arachidonoyl glycerol (2-AG) are members of an interconnected lipidome that includes an emerging class of AEA structural analogs, the lipoamines, additional 2-acyl glycerols, free fatty acids, and prostaglandins (PGs). Lipids in this lipidome share many biosynthetic and metabolic pathways, yet have diverse signaling properties. Here, we show that acute THC drives age-dependent changes in this lipidome across 8 regions of the female mouse brain. Interestingly, most changes are observed in the adult, with eCBs and related lipids predominately decreasing. Analysis of THC and metabolites reveals an unequal distribution across these brain areas; however, the highest levels of THC were measured in the hippocampus (HIPP) in all age groups. Transcriptomic analysis of the HIPP after acute THC showed that like the lipidome, the adult transcriptome demonstrated significantly more changes than the adolescent. Importantly, the regulation of 31 genes overlapped between the adolescent and the adult, suggesting a conserved transcriptomic response in the HIPP to THC exposure independent of age. Taken together these data illustrate that the first exposure to a single dose of THC has profound effects on signaling in the CNS.
Keywords: Lipidomics, THC, transcriptomic, adolescent brain, lipoamine, endocannabinoid
1. Introduction
Cannabis (aka Marijuana) is the most widely used illicit drug [1, 2]. Δ9-tetrahydrocannabinol (THC) is an active compound produced in cannabis that is primarily responsible for its psychoactive effects [3]. The use of cannabis peaks in adolescence [1], and heavy adolescent cannabis use has been correlated with an increased risk of psychopathology in adulthood [4–10]. Illustrating the urgency to understand how THC affects both the adolescent and adult brain, the average concentration of THC in street cannabis increased from 3 to 12% in the past 20 years [11, 12]. A central mechanism of action of THC is through the cannabinoid receptor, CB1, which is widely expressed in the brain in both neurons and glia [13, 14]. CB1, likewise, has ubiquitous endogenous ligands, called endogenous cannabinoids (eCBs). Two of the most studied eCBs are N-arachidonoyl ethanolamine (AEA) [15], also known as anandamide, and 2-arachidonoyl glycerol (2-AG) [16, 17].
AEA and 2-AG belong to families of N-acyl ethanolamines (NAEs) and 2-acyl glycerols, respectively. Whereas AEA and 2-AG are formed with arachidonic acid (AA), other NAEs and 2-acyl glycerols are formed with the conjugation of other fatty acids and ethanolamine or glycerol, respectively [18, 19]. Additional variations of the NAEs occur with a substitution of the ethanolamine group for other amines such as amino acids, which can conjugate with different fatty acids (Supplemental Figure 1). The resultant molecules form the larger family of secondary amines called lipoamines [20–23]. We routinely measure AEA, its lipoamine structural analogs, 2-AG, its 2-acyl glycerol structural analogs, AA-derived prostaglandins (PGs), and free fatty acids in a variety of tissues and show that they are members of wider, interconnected lipidome in the brain and throughout the body [21, 23–28]. We have shown that this interconnected lipidome is regulated by eCB system-related enzymes and receptors such as N-acyl phosphatidylethanolamine-specific phospholipase D (NAPE-PLD) [23], fatty acid amide hydrolase (FAAH), monoacylglycerol lipase (MAGL), and CB1 [21], as well as in a variety of disease models [27, 29–33]. These data show that regulation of this lipidome has broader signaling implications than just the eCB system; yet eCB signaling appears to play a primary role in how this integration occurs. THC use during adolescence may drive changes in this brain lipidome that have lasting effects on signaling in the adult.
Adolescence is a critical period for brain development [4, 12, 34]. The use of THC during adolescence disrupts the eCB system in the brain, and these disruptions may persist into adulthood [8]. Given that levels of eCBs are dynamic throughout development [8, 35, 36], the wider lipidome may also be dynamic during adolescence; therefore, the effect of THC on lipid signaling may, likewise, change with age. Testing the hypothesis that THC affects lipid signaling molecules beyond AEA and 2-AG in a developmentally-dependent manner, we measured the effects of an acute dose of THC on female mice in adolescence (post-natal day (PND) 35 and PND 50) and adulthood (PND 130-134) in a targeted brain lipidome in 8 specific brain regions. We also tested the hypotheses that THC distribution and THC metabolism are not equally distributed throughout the brain. Finally, we tested the hypothesis that acute THC would have differential effects on the hippocampal transcriptome in the adolescent versus adult. Our findings support these hypotheses showing that developmental age significantly changes how the first exposure to THC drives changes in the brain lipidome, levels of THC compartmentalization in the brain, and the hippocampal transcriptome. These data highlight the fact that THC has wide-ranging effects on CNS signaling that evolve with age.
2. Methods
2.1 Mice and Drug Injections
The Bloomington Institutional Animal Care and Use Committee of Indiana University reviewed and approved the procedures used here and all comply with ARRIVE guidelines [37]. All mice used were female from the CD1 strain. The CD1 strain is the genetic background for the line of CB1 knockout mice used in our published baseline lipidomics screen [21]. Females were chosen because, although the effects of THC during adolescence are sex-dependent [7, 38], they appear to be more severe in females [39]. For each lipidomics experiment, 8 drug-treated mice were compared to 8 vehicle-treated mice. In total 48 mice were used for the THC lipidomics study: 16 PND 35 mice, 16 PND 50 mice, and 16 adult mice. The adult mice were ~ 4 months old. Mice were given a single i.p. injection of either 3 mg/kg THC or vehicle (1:1:18 cremophor: ethanol: saline). It is estimated that a 2.5 mg/kg THC injection in rodents is equivalent to smoking 1 joint for humans [40], meaning 3 mg/kg is a reasonable level to model human use. The 3 mg/kg dose is non-aversive to mice, but is still sufficiently able to alter eCB-dependent synaptic plasticity [41]. For the transcriptomics experiment, the transcriptome from female CD1 mice given a single injection of 3 mg/kg THC was compared to vehicle-treated mice within each age group. In total, 30 mice were used for the transcriptomics study: 14 PND 35 mice, 6 given vehicle and 8 given THC, and 16 adult mice (PND 105-106), 7 given vehicle and 9 given THC.
2.2 Tissue Collection and Extraction
2.2.1 Tissue Collection
2 hours after the injection, mice were sacrificed via rapid decapitation. The 2 hour time point was chosen because THC would still be present in the brain at this time [42], allowing us to test the hypothesis that the distribution of THC differs by brain region. Brains were immediately removed and placed in liquid nitrogen, and then stored in a −80°C freezer until dissections were performed. Brains were dissected while frozen on an ice-cold dissection plate into the following regions: striatum (STR), hippocampus (HIPP), cerebellum (CER), thalamus (THAL), cortex (CTX), hypothalamus (HYP), midbrain (MID), and brainstem (STEM) as previously described [29]. These abbreviations for these brain areas will be used exclusively when discussing the results that are generated by these specific types of dissections. Each dissected area was immediately placed in liquid nitrogen and then stored at −80°C until used for lipid or RNA extraction. The entire STR, HIPP, CER, THAL, HYP, MID and STEM from both hemispheres were used for lipid extraction, whereas a single hemisphere of the CTX was randomly selected for lipid extraction. The whole, bilateral HIPP was processed for RNA extraction.
2.2.2 Lipid Extraction
Tissue lipid extracts were performed on each of the 8 brain areas as previously described [21, 23, 43–45]. In brief, to begin the lipid extraction, samples were shock frozen in liquid nitrogen, and weighed before being transferred to a centrifuge tube. The mass of the largest sample was multiplied by 50 to determine how many mL of high-pressure liquid chromatography (HPLC)-grade methanol (Avantor Performance Materials, Inc., Center Valley, PA, USA) was added to the tube. Then, samples were spiked with 500 picomoles of deuterium-labeled N-arachidonoyl glycine (d8NAGly; Cayman Chemical, Ann Arbor, MI, USA) as an internal standard to determine extraction efficiency. Samples were placed on ice in darkness for 2 hours then individually homogenized. Homogenates were centrifuged at 19,000g for 20 minutes at 20°C. Supernatants were decanted and diluted with HPLC water (purified in house) to make a 75% water, 25% supernatant solution. Partial purification was achieved using C-18 solid phase extraction columns (Agilent, Palo Alto, CA, USA). A series of 4 elutions with 1.5 mL of 60%, 75%, 85%, and 100% methanol were collected [23, 45]. Vials of eluants were stored at −80°C until they were ready for analysis.
2.2.3 RNA Extraction
To isolate a purified sample of total RNA from the HIPP, flash-frozen hippocampi were weighed and processed using the Absolutely RNA Miniprepkit (Agilent) according to the manufacturer’s instructions and under RNase free conditions. This extraction involves a DNase treatment as one of the steps, decreasing the likelihood of DNA contamination [46]. The quality of the RNA was checked using a NanoDrop spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA), which measures the ratio of absorbance at 260 and 280 nm to identify contamination with protein or with RNA extraction reagents [47]. Only samples with a 260/280 ratio close to 2 were used for analysis, as this indicates a pure RNA sample [48]. The RNA samples, dissolved in 60 μL of elution buffer (10mM Tris-HCl, pH 7.5, 0.1 mM EDTA), were then stored at −80°C.
These samples were further analyzed using a Tape Station (Agilent), which determines the concentration of RNA, checks that strands of mRNA are intact, and provides an RNA integrity number (RIN) as a measure of degradation [49]. Only intact RNA of sufficient concentration was used for transcriptomics analysis. Supplemental Table 1 shows the 260/280 ratios, RNA yield, and the RINs from the PND 35 and adult HIPP RNA extractions. At least 4 vehicle-treated and at least 4 THC-treated hippocampi from each age group were submitted to create the cDNA library necessary for RNA-Seq.
2.3 High-Pressure Liquid Chromatography Coupled with Tandem Mass Spectrometry (HPLC/MS/MS)
Lipid extracts were analyzed using an Applied Biosystems API 3000 triple quadrupole mass spectrometer with electrospray ionization (Foster City, CA, USA). 20μL from each elution were chromatographed using XDB-C18 reversed phase HPLC analytical column (Agilent) and optimized mobile phase gradients. Mobile phase A: 20% methanol, 80% water (v/v) and 1 mM ammonium acetate (Sigma, St. Louis, MO, USA). Mobile phase B: 100% methanol, 1 mM ammonium acetate. Two Shimadzu 10ADvp pumps (Columbia, MD, USA) provided the pressure for gradient elution. Every method run began with 0% mobile phase B, reached a state of 100% mobile phase B flowing at 0.2 mL per minute, and gradually returned to 0% mobile phase B.
2.3.1 HPLC/MS/MS Data Analysis and Statistical Procedures
Levels of each compound were determined by running each sample using a multiple reactions monitoring (MRM) method tailored for each group of structurally similar compounds (Supplemental Figure 2). Analysis of the HPLC/MS/MS data was performed using Analyst software (Applied Biosystems) as previously described [21, 23, 43–45]. Chromatograms were generated by determining the retention time of analytes with a [M−1] or [M+1] parent peak and a fragmentation peak corresponding to the programmed values. The retention time was then compared to the retention time of a standard for the suspected compound. If the retention times matched, then the concentration of the compound was determined by calculating the area under the curve for the unknown and comparing it to the calibration curve obtained from the standards. Therefore, unknown lipids are matched to known standards according to retention time from the analytical column and their mass fingerprint.
Extraction efficiency was calculated with the d8NAGly spiked recovery vial as a standard as previously described [21, 23, 29, 43–45]. For each individual lipid in each of the areas, concentrations in moles per gram adjusted for percent recovery from the drug-treated animals were compared to vehicle concentrations using a one-way ANOVA. All statistical tests were carried out using SPSS Statistics (IBM, Armonk, NY, USA). Statistical significance was defined as p < 0.05 and p < 0.10. For analysis of the distribution of THC and its metabolites, the main effect of brain area on levels of THC, 11-OH-THC and (±)-11-nor-9-carboxy THC was tested using a one-way ANOVA with SPSS Statistics software (IBM). Statistical significance was defined as p < 0.05.
Analyzed lipidomics data are represented in tabular format illustrating both the direction and magnitude of change (Supplemental Figure 3). To determine the magnitude change and therefore the number of arrows to assign each significant difference, the mean level of a particular lipid in a specific region of the THC mice was divided by that same lipid’s mean level in the same brain region of the corresponding vehicle mice. For example, the average level of PGE2 in the HIPP of PND 35 THC mice was 6.68×10−10 moles per gram and the average level of PGE2 in the HIPP of the vehicle was 5.9×10−10 moles per gram; 6.68×10−10 divided by 5.9×10−10 equals 1.13, assigning it 1 up arrow in the figures because the magnitude of change was between 1 and 1.5 times higher than vehicle. For decreases the process was very similar: the mean level in the THC group was divided by the mean level in the vehicle; however, the reciprocal of the decimal was taken to express a fold decrease (if the level in the THC mouse is ½ of the vehicle level then that is a 2 fold decrease). As an example the mean level of AEA was 4.76×10−11 moles per gram in the HIPP of the adult THC mice and 5.48×10−11 moles per gram in the corresponding vehicle HIPP. 4.76×10−11 divided by 5.48×10−11 is 0.87 and the reciprocal of 0.87 is 1.15, meaning that the decrease is between 1 and 1.5 times vehicle levels and giving it 1 down arrow on our scale.
2.4 RNA Sequencing
The extracted RNA was purified into poly-A RNA using oligo-dT binding [50] and fragmented into 30-400 base pair segments [51]. Then, the RNA fragments were reverse transcribed into cDNA using random primers [51, 52]. Adaptors were placed on the ends of the cDNA fragments that were read by the sequencer [52]. The Illumina IG sequencer (San Diego, CA, USA) has been well validated for RNA-Seq purposes [51–53] and Illumina kits for library preparation were used as previously described [54]. The Illumina applies ‘sequencing by synthesis’ [53]. The cDNA library was placed in lanes in a flow-cell, where individual cDNA fragments were amplified. A linearization enzyme spliced the cDNA into single strands. One base at a time, fluorescently labeled nucleotides complemented the template. The sequencer captured the signal from the change in fluorescence [53, 54], which can be translated into a nucleotide base.
Raw reads from the sequencer were checked for quality using FastQC software (Babraham Bioinformatics, Cambridge, UK) [55]. Adapter sequences were trimmed from the reads using Trimmomatic software (Usadel Lab, Aachen University, Germany) [56]. Up to 19,809,240 raw reads were made from each sample. After trimming, around 99% of the reads remained for further analysis (Supplemental Table 2). Trimmed reads from the sequencer were mapped onto the Mus musculus GRCm38.75 reference transcriptome using tophat2 software (Johns Hopkins University Center for Computational Biology, Baltimore, MD, USA) to generate a base expression profile for each gene [52]. Typically, over 90% of the reads were mapped. 2 samples, Adult V1 and PND 35 THC6, had poor mapping onto the mouse reference transcriptome (Supplemental Table 2) and were excluded from the differential expression analysis. This yielded an n of 4 for PND 35 vehicle, an n of 5 for PND 35 THC, an n of 5 for Adult vehicle, and an n of 4 for Adult THC. Bioinformaticians mostly agree that a sample size of 3 per group is acceptable if the experiment was well controlled [57], meaning that we have sufficient sample size for differential expression analysis.
Normalization of sequence read count is an important issue for RNA-Seq studies using mice, because a few genes are very abundantly expressed and can bias the analysis [58]. The DESeq software package (Simon Anders, European Molecular Biology Laboratory, Heidelberg, Germany) for normalization of count data [59] has been applied to several data sets before [54, 59] and adequately controls for differences in library composition. A counts table containing read counts for each gene, to be used as input for DESeq2, was generated using htseq-count 0.5.4p1 (Simon Anders) [60]. The DE-Seq2 software package [57] was used for normalization of count data and to determine genes that were differentially expressed within each age group in THC-treated HIPP versus vehicle-treated HIPP at a 5% false discovery rate (FDR) [59]. The ability of RNA-Seq to detect differences between groups in levels of weakly expressed transcripts and its thoroughness of coverage makes it a very attractive choice for studies that want to identify differentially expressed genes between groups [58]. It isn’t possible to quantify every differentially expressed gene between groups, but RNA-Seq gives an excellent overview of genes that are broadly affected by experimental condition. An RNA-Seq approach may identify novel pathways contributing to differences in lipid levels that may be missed when taking a targeted approach.
2.4.1 Pathway Analysis
In order to better understand the biological consequences of the set of genes affected by acute THC in the HIPP, pathway analysis was performed with Ingenuity Pathway Analysis (IPA) software (Qiagen, Hilden, Germany) [61]. Genes with differential expression between THC and vehicle with significance defined at 1% FDR in Adult and PND 35 hippocampal RNA samples were analyzed using IPA, to find a list of diseases, developmental processes, and cellular functions significantly associated with the set of upregulated or downregulated transcripts.
3. Results
3.1 Overall Effects of Acute THC on the CD1 WT Female Mouse Brain Lipidome
All of the NAE, PG, free fatty acid, and 2-acyl glycerol species analyzed here were detected in all brain regions. Of the 67 lipoamines in our screening library (Supplemental Figure 2), over 50 were detected in most brain regions in both the vehicle and THC treated mice at each developmental time point. Together this lipidomics data analysis totals approximately 62,000 data points analyzed. The most lipids in the library were detected in larger brain regions such as the cortex (CTX), cerebellum (CER), and brainstem (STEM). The fewest were detected in smaller brain regions such as the hypothalamus (HYP), which likely represents a technical detection limit and not specific signaling properties of the region. The detailed list of levels of lipids detected in each of the eight brain regions assayed from each experiment and the statistical analyses of each are available in the Supplemental Tables 3-50.
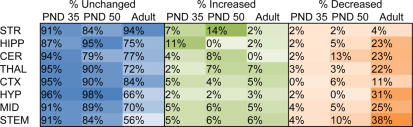
As a first level of analysis, we have calculated the percentage of significant changes in lipid levels as a result of THC exposure at each developmental time point (Figure 1). In PND 35 mice, out of the 656 possible changes (82 lipids in each brain area) in lipid concentrations, 33 significant changes were detected across all 8 brain areas (Figure 1; Supplemental Figure 4). Most of the changes in lipid levels were increases in a lipid’s concentration (23 increases total, compared to 10 decreases total). The brain area with the most robust effect of THC on the lipidome in PND 35 mice was the hippocampus (HIPP), which had increases in levels of 6 lipids and a decrease in 1 lipid. The brain region least impacted by acute THC in the PND 35 mice was the HYP, with an increase in N-oleoyl glycine and a decrease in 2-AG representing the only changes in the lipidome detected in that brain region.
Figure 1. Percentage of significant changes in the CD1 female mouse brain lipidome in post-natal day (PND) 35, PND 50, and adult mice 2 hours after a 3 mg/kg systemic Δ9-tetrahydrocannabinol (THC) injection.
The left, blue part of the figure represents the percentage of the lipids detected within each of brain region (row) within each age group (column) with concentrations that were unaffected by acute THC. The middle, green part represents the percentage of lipids detected that increased with THC treatment and the right, orange proportion represents the percentage of lipids detected that decreased with THC treatment relative to vehicle. STR=striatum; HIPP=hippocampus; CER=cerebellum; THAL=thalamus; CTX=cortex; HYP=hypothalamus; MID=midbrain; STEM=brainstem
In PND 50 mice, levels of 58 lipids were significantly modified. Unlike in the PND 35 mice, the numbers of increases and decreases were nearly equivalent (30 increases and 28 decreases; Figure 1; Supplemental Figure 5). However, there were distinctly different patterns of lipid changes in different brain areas. For example, in the HYP all the changes with acute THC were increases in a lipid’s concentration and in the HIPP all the changes were decreases in a lipid’s concentration. The brain area with the most robust effect of THC on the lipidome was the CER, which had increases in levels of 5 lipids and decreases in levels of 8 lipids. The brain region least impacted by acute THC in the PND 50 mice was the HYP, with an increase in PGF2α representing the only change in the lipidome.
Adult brains showed the most changes in lipid levels across the 8 brain regions assayed, with levels of 116 lipids changing in concentration in the THC group compared to the vehicle group. Unlike in the adolescent brain, most of the changes in lipid levels were decreases in a lipid’s concentration relative to vehicle (99 decreases, 17 increases; Figure 1; Supplemental Figure 6). The CER stands out in the adult experiment wherein all 14 changes in lipid levels were decreases. The region most affected by acute THC in the adult brain was the STEM, where levels of 28 different lipids were altered by THC (24 decreases, 4 increases). In contrast, the least affected brain region was the striatum (STR) with only 3 lipids affected (2 decreases, 1 increase).
3.2 Specific Lipid Modifications across the Brain with Acute THC are Age-Dependent
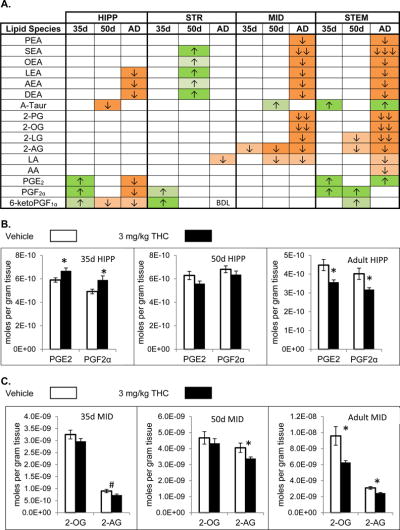
Maps of all significant changes, shown in Supplemental Figures 4-6, contain many orphan lipids of unknown function. Examples shown here are lipids that have the most known signaling pathways and the most understood biosynthetic and metabolic pathways. The transition from predominately increases to predominately decreases in specific lipid levels from early adolescence to adult is highlighted in the HIPP regarding the levels of PGE2 and PGF2α. Acute THC drives an increase in these PGs in the PND 35 mouse, no change in the PND 50 mouse and a decrease in the adult (Figure 2A; Figure 2B). PGs are emerging as important molecules for hippocampal development [62] and eCB signaling appears to play a critical role in these pathways [63–65].
Figure 2. Effects of systemic 3 mg/kg Δ9-tetrahydrocannabinol (THC) on levels of targeted lipids 2 hours after injection in the post-natal day (PND) 35, PND 50, and adult CD1 female mouse hippocampus (HIPP), striatum (STR), midbrain (MID), and brainstem (STEM).
Panel A: Cells with shaded arrows indicate a change for that lipid in the THC-exposed brain area relative to the same vehicle-exposed area within each age group (35d = PND 35, 50d = PND 50, and AD = Adult). The arrow color indicates the direction of a significant result relative to control. Green colors represent increases, with darker green representing a significant (p < 0.05) increase and lighter green representing a trending (p < 0.1) increase. Orange colors represent decreases in a lipid’s concentration, with darker orange indicating a significant (p < .05) decrease and light orange representing a trending (p < .01) decrease. The number of arrows indicates the magnitude of the difference between THC and vehicle. One arrow indicates a magnitude difference of less than 1.5 fold, 2 arrows indicates a 1.5-1.99 fold change, and 3 arrows indicate a 2-2.99 fold change. BDL stands for “Below Detection Limit,” whereas a blank cell indicates that there was no change in the lipid’s level due to THC. See Methods and Supplemental Figure 3 for more detailed description of analysis. Lipids listed in this figure are N-palmitoyl ethanolamine (PEA), N-stearoyl ethanolamine (SEA), N-oleoyl ethanolamine (OEA), N-linoleoyl ethanolamine (LEA), N-arachidonoyl ethanolamine (AEA), N-docosahexaenoyl ethanolamine (DEA), N-arachidonoyl taurine (A-Taur), 2-palmitoyl glycerol (2-PG), 2-oleoyl glycerol (2-OG), 2-linoleoyl glycerol (2-LG), 2-arachidonoyl glycerol (2-AG), linoleic acid (LA), arachidonic acid (AA), prostaglandin E2 (PGE2), prostaglandin F2α (PGF2α), and 6-keto prostaglandin F1α (PGF1α).
Panel B: Bar graphs showing mean levels of prostaglandin E2 (PGE2) and prostaglandin F2α (PGF2α) in the post-natal day 35 hippocampus (35d HIPP), post-natal day 50 hippocampus (50d HIPP), and adult hippocampus (adult HIPP) 2 hours after a vehicle injection (open bars) or a 3 mg/kg THC injection (black bars). The units on the y-axis are moles of lipid per gram of tissue. Error bars are ± standard error. Asterisk (*) represents a difference of p < 0.05 between THC and vehicle groups. In the post-natal day 35 hippocampus levels of these prostaglandins are higher in the THC-treated group (corresponding to an green cell with 1 up arrow in panel A), whereas there are no differences in the post-natal day 50 hippocampus, and levels of these prostaglandins are lower in the THC-treated adult hippocampus (corresponding to an orange cell with 1 down arrow in panel A).
Panel C: Bar graphs showing mean levels of 2-oleoyl glycerol (2-OG) and 2-arachidonoyl glycerol (2-AG) in the post-natal day 35 midbrain (35d MID), post-natal day 50 midbrain (50d MID), and adult midbrain (adult MID) 2 hours after a vehicle injection (open bars) or a 3 mg/kg THC injection (black bars). The units on the y-axis are moles of lipid per gram of tissue. Error bars are ± standard error. Asterisk (*) represents a difference of p < 0.05 between THC and vehicle groups and the pound sign (#) represents a difference of p < 0.1. In the post-natal day 35 and post-natal day 50 midbrain, there were no significant differences between groups in levels of 2-OG, whereas 2-OG levels are lower in the THC-treated adult midbrain (corresponding to an orange cell with 2 down arrows in panel A). In all 3 age groups, levels of 2-AG were lower in the THC-exposed midbrain (corresponding to orange cells with 1 down arrow in panel A).
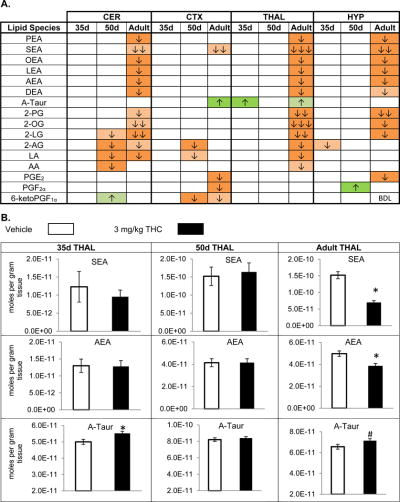
THC has a particular impact on the eCBs AEA and 2-AG and their structural analogs, but only in the PND 50 and adult mice. Only 2 changes, decreases in 2-AG in the midbrain (MID) and HYP, were observed in eCBs in the PND 35 mice to acute THC and the only scenario in which a change in a lipid’s concentration was the same in all three developmental stages was a decrease in 2-AG in the MID (Figure 2C). Importantly, the eCB, AEA, and its structural analogs the NAEs were only significantly increased in the PND 50 STR; however, these NAEs were significantly decreased in the adult in all brain areas except the STR and CTX (Figure 2A; Figure 3A; Figure 3B). Finally, the ubiquitous transient receptor potential vanilloid (TRPV) 4 agonist, N-arachidonoyl taurine (A-Taur) [66] was modulated in 5 of the 8 brain regions, where it was primarily increased after THC administration (Figure 2A; Figure 3A; Figure 3C).
Figure 3. Effects of systemic 3 mg/kg Δ9-tetrahydrocannabinol (THC) on levels of targeted lipids 2 hours after injection in the post-natal day (PND) 35, PND 50, and adult CD1 female mouse cerebellum (CER), cortex (CTX), thalamus (THAL), and hypothalamus (HYP).
Panel A: Cells with shaded arrows indicate a change for that lipid in the THC-exposed brain area relative to the same vehicle-exposed area within each age group (35d = PND 35, 50d = PND 50, and AD = Adult). The arrow color indicates the direction of a significant result relative to control. Green colors represent increases, with darker green representing a significant (p < 0.05) increase and lighter green representing a trending (p < 0.1) increase. Orange colors represent decreases in a lipid’s concentration, with darker orange indicating a significant (p < .05) decrease and light orange representing a trending (p < .01) decrease. The number of arrows indicates the magnitude of the difference between THC and vehicle. One arrow indicates a magnitude difference of less than 1.5 fold, 2 arrows indicates a 1.5-1.99 fold change, and 3 arrows indicate a 2-2.99 fold change. BDL stands for “Below Detection Limit,” whereas a blank cell indicates that there was no change in the lipid’s level due to THC. See Methods and Supplemental Figure 3 for more detailed description of analysis. Lipids listed in this figure are N-palmitoyl ethanolamine (PEA), N-stearoyl ethanolamine (SEA), N-oleoyl ethanolamine (OEA), N-linoleoyl ethanolamine (LEA), N-arachidonoyl ethanolamine (AEA), N-docosahexaenoyl ethanolamine (DEA), N-arachidonoyl taurine (A-Taur), 2-palmitoyl glycerol (2-PG), 2-oleoyl glycerol (2-OG), 2-linoleoyl glycerol (2-LG), 2-arachidonoyl glycerol (2-AG), linoleic acid (LA), arachidonic acid (AA), prostaglandin E2 (PGE2), prostaglandin F2α (PGF2α), and 6-keto prostaglandin F1α (PGF1α).
Panel B: Bar graphs showing mean levels of N-stearoyl ethanolamine (SEA), N-arachidonoyl ethanolamine (AEA), and N-arachidonoyl taurine (A-Taur) in the post-natal day 35 thalamus (35d THAL), post-natal day 50 thalamus (50d THAL), and adult thalamus (adult THAL) 2 hours after a vehicle injection (open bars) or a 3 mg/kg THC injection (black bars). The units on the y-axis are moles of lipid per gram of tissue. Error bars are ± standard error. Asterisk (*) represents a difference of p < 0.05 between THC and vehicle groups and the pound sign (#) represents a difference of p < 0.1. In the post-natal day 35 and the post-natal day 50 thalamus, there are no significant differences in SEA or AEA levels between groups, but there is a significant decrease in both SEA (corresponding to an orange cell with 3 down arrows in panel A) and AEA (corresponding to an orange cell with 1 down arrow in panel A) in the adult thalamus. Levels of A-Taur were significantly higher in the THC-exposed post-natal day 35 thalamus (corresponding to a green cell with 1 up arrow in panel A) and were trending higher in the adult thalamus (corresponding to a lighter green cell with 1 up arrow in panel A).
3.3 Differential THC Distribution by Brain Region
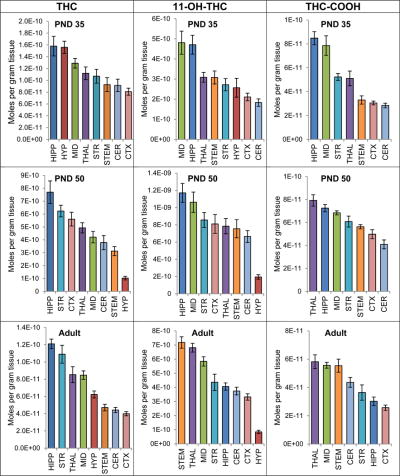
Levels of THC and its 2 main metabolites were measured in each of the 8 brain regions 2 hours after acute THC injection. In all developmental ages, levels of THC varied by brain region (Figure 4; Supplemental Figure 7A; Supplemental Tables 51-59). As experiments using different age groups were conducted several months apart, analysis of the main effect of age on THC levels across brain areas was not possible. However, within each age group, a one-way ANOVA revealed a significant main effect of brain area on levels of THC (p < 0.001) indicating that the distribution of THC throughout the brain is not uniform. The only exception is that the levels of THC were highest in the HIPP in all 3 age groups (Figure 4A). On the other end of the spectrum, levels of THC were lowest in the CTX in both PND 35 and the adult mice; however, levels of THC were lowest in the HYP in the PND 50 mice.
Figure 4. Levels of Δ9-tetrahydrocannabinol (THC) and the THC metabolites 11-OH-THC and (±)-11-nor-9-carboxy-THC (THC-COOH) in 8 brain regions of female mice 2 hours after an acute 3 mg/kg THC injection.
The top row shows levels of THC and metabolites for the post-natal day 35 mice (PND 35), the middle row shows levels for the post-natal day 50 mice (PND 50) and the bottom row shows levels for the adult mice. Units on the y-axis are moles of lipid per gram of tissue. Error bars are ± standard error. Brain areas are shown on the x-axis. STR=striatum; HIPP=hippocampus; CER=cerebellum; THAL=thalamus; CTX=cortex; HYP=hypothalamus; MID=midbrain; STEM=brainstem. In each graph, brain areas are ordered to correspond with their levels of lipid, with the area having the highest concentration shown on the left and the area with the lowest on the furthest right.
3.4 Differential Distribution of THC Metabolites by Brain Region and Development
Levels of THC metabolites also varied by brain region: a one-way ANOVA revealed a main effect of brain area on levels of both THC metabolites in the screening library (p < 0.001). Levels of the psychoactive metabolite [67] 11-OH-THC were highest in the MID of the PND 35 group, the HIPP of the PND 50 mice, and the STEM of the adult mice, whereas levels of 11-OH-THC were lowest in the CER of the PND 35 mice and lowest in the HYP in the PND 50 and adult mice (Figure 4B; Supplemental Figure 7B; Supplemental Tables 51-59). Levels of the non-psychoactive metabolite (±)-11-nor-9-carboxyTHC [68], likewise, varied by brain region and by development (Figure 4; Supplemental Figure 7C; Supplemental Tables 51-59). Despite this compound’s non-psychoactive nature, it is long lasting [69] and is the compound that is typically used by forensic investigators to determine recent cannabis use [68]. Levels of this compound were highest in the HIPP of the PND 35 mice and in the thalamus (THAL) of both PND 50 and adult mice; however, were lowest in the CER of the PND 35 and PND 50 mice and the CTX of the adult mice. (±)-11-nor-9-carboxyTHC was not detected in every sample from the HYP, so this brain area was not analyzed.
3.5 Transcriptome of Hippocampus Following Acute Systemic THC Administration
3.5.1 Analysis of PND 35 Hippocampal Transcriptome
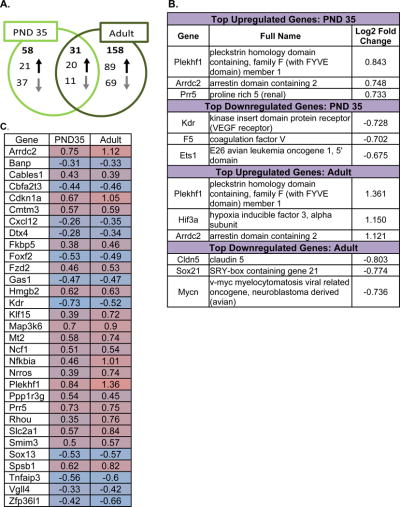
Acute THC treatment in PND 35 female mice caused 89 transcripts to be differentially expressed in the HIPP, wherein 48 were downregulated and 41 were upregulated (Supplemental Figure 8). 58 of these changes were unique to the PND 35 HIPP (Figure 5A; Supplemental Figure 8). When considering only the 58 uniquely changed transcripts, 21 of these were upregulated and 37 were downregulated, which is a higher proportion of downregulations than when changes shared with the adult are considered (Figure 5A). The gene with the largest upregulation following acute THC in the PND 35 HIPP was pleckstrin homology domain containing, family F (with FYVE domain) member 1 (Plekhf1), with a log2 fold change of 0.84; whereas the gene with the most robust downregulation was kinase insert domain protein receptor (kdr) with a log2 fold change of −0.73 (Figure 5B). For full statistical analysis of the significant differences found in the PND 35 THC HIPP versus the PND 35 vehicle HIPP, see Supplemental Table 60.
Figure 5. Effects of systemic 3 mg/kg Δ9-tetrahydrocannabinol (THC) on the transcriptome in the hippocampus of in the post-natal day (PND) 35 and adult female mice 2 hours post-injection.
Panel A: Venn diagram showing the number of significant changes in expression of transcripts due to acute THC relative to vehicle that were unique to each age group (PND 35 and adult) and the number of significant changes in transcript levels shared by each age group. The directionality of expression differences in the THC-treated group relative to the vehicle group is also shown. Full lists of the transcripts affected by acute THC in each age group can be found in Supplemental Figures 8 and 9.
Panel B: The top portion of this panel shows the transcripts with the largest magnitude significant differences in expression in the PND 35 THC-treated hippocampus versus the PND 35 vehicle-treated hippocampus. The full names for the 3 genes with the largest magnitude increase in expression and the 3 genes with the largest magnitude decrease in expression are written out and the log2 fold change is also shown here. The lower portion of this panel shows the transcripts with the largest magnitude significant differences in expression in the adult THC-treated hippocampus versus the adult vehicle-treated hippocampus. The full names for the 3 genes with the largest magnitude increase in expression and the 3 genes with the largest magnitude decrease in expression are written out and the log2 fold change is also shown here.
Panel C: Heat map of transcripts significantly affected by acute THC in the hippocampus of both PND 35 and adult mice. As shown in panel A of Figure 5, there were 31 transcripts with altered expression in both the PND 35 and the adult hippocampus. This panel shows the direction and magnitude of change for each of these 31 transcripts in the PND 35 and in the adult hippocampus. The shading color represents the direction of change: pink colors represent increases in expression in the THC group relative to vehicle, whereas blue colors indicate decreases in expression. The magnitude of change is expressed by the fold change in expression, which is written in each cell. For more information on the 31 transcripts affected by acute THC in both age groups, please refer to Supplemental Table 62, as well as Supplemental Figures 8-9.
3.5.2 Analysis of Adult Hippocampal Transcriptome
Compared to the PND 35 mice, acute THC caused a higher degree of transcript regulation in the adult HIPP. 189 transcripts were differentially expressed in the HIPP of adult THC-treated mice compared to adult vehicle-treated mice (Supplemental Figure 9). 109 of these changes were increases in expression and 80 of these changes were decreases in a transcript’s expression (Supplemental Figure 9). 158 of these changes were unique to the adult HIPP wherein 89 changes were increases and 69 were decreases in a transcript’s expression (Figure 5A). Just like in the PND 35 HIPP, the gene with the largest upregulation following acute THC in the adult HIPP was Plekhf1, with a log2 fold change of 1.36 (Figure 5B). Notably, the magnitude of the change was larger in the adult HIPP. The gene with the most robust downregulation in expression after acute THC in the adult HIPP was claudin 5 (cldn5) with a log2 fold change of −0.80 (Figure 5B). For full statistical analysis of the significant differences found in the adult THC HIPP versus the adult vehicle HIPP, see Supplemental Table 61.
3.5.3 Shared Differentially Expressed Genes between PND 35 and Adult
Suggesting a unique transcriptomic signature following acute THC in the HIPP, 31 genes were affected by acute THC in the same direction, regardless of age (Figure 5C). Most of these transcripts are found in multiple cell types in the HIPP: neurons, astrocytes, and microglia (Supplemental Table 62). An Ensembl [70] and DAVID [71, 72] search revealed a wide variety of functions of these transcripts (Supplemental Table 62). In the HIPP of both PND 35 and adult mice, expression of 20 of these transcripts was significantly upregulated and of 11 of these transcripts was significantly downregulated (Figure 5A; Figure 5C). Although the changes in expression were in the same direction in both age groups, the magnitude of the change in expression was consistently larger in the adult HIPP than the PND 35 HIPP (Figure 5C). IPA indicated that a subset of diseases, developmental processes, and cellular functions were significantly associated with the set of transcripts affected by acute treatment with THC. Typically, more functions were associated with the set of THC-affected transcripts in the adult HIPP (Supplemental Figure 10).
4. Discussion
Exogenous and endogenous cannabinoids share receptor targets in the mammalian brain; however, little is known about how exogenous cannabinoids interact with eCBs and even less is known about how THC affects the wider eCB lipidome. Here we show that the first administration of a single-dose of THC regulates the ligands of the eCB system and members of the broader lipidome, that THC and its metabolites are differentially compartmentalized in the brain, that a single dose of THC drives changes in the hippocampal transcriptome, and that each of these effects are developmentally and region-dependent. Given the emerging data on how adolescent exposure to THC has longer-lasting outcomes on behavior than the same level of exposure in the adult [4–10], we hypothesized that there would be more effects on the brain lipidome and transcriptome in the adolescent compared to the adult. Contrary to our expectations, the adult brain experienced more THC-induced changes in lipid and RNA transcript levels than the adolescent brain. While on the surface this may appear to suggest that adolescent brains are protected from THC exposure, it may also represent a lack of a rapid response to THC during the first experience with THC that may underlie the adult brain’s ability to show fewer long-term effects with THC exposure.
4.1 Developmental Trajectory of eCB Signaling
One explanation for the age-dependent differences in the lipidome could be baseline differences in lipid production and signaling that occur with age. Not much is known about developmental changes in eCBs in the CNS and even less about changes in female subjects. In one study Ellgren and colleagues examined the developmental trajectory of eCB levels in male rats. Levels of AEA were increased by age in the prefrontal cortex (PFC), but did not change in the nucleus accumbens (NAc) or caudate putamen. In contrast, 2-AG concentrations are highest in the early adolescent in the NAc, whereas levels in the PFC were highest in the early adolescent, reached a minimum during mid adolescence and then rose again in the late adolescent, not quite reaching the early adolescent levels [73]. One other study compared levels of NAEs (AEA, N-palmitoyl ethanolamine (PEA), and N-oleoyl ethanolamine (OEA)), as well as activity of the NAE-degrading enzyme FAAH [74], at PND 25, 35, 45 and 70 in the hippocampus, hypothalamus, PFC, and amygdala of male rats. In all 4 brain regions, there were effects of developmental age on levels of AEA, whereby levels of AEA were elevated in PND 35 and PND 70 time-points compared to PND 45 and PND 25. Levels of PEA and OEA followed a similar pattern. FAAH activity somewhat correlated with the lipidomics results, meaning that an increase in FAAH-mediated hydrolysis of NAEs can partially explain why levels of NAEs are lower at some time points [36].
CB1 expression is also dynamic, generally being highest in the rat brain around PND 25 and decreasing by adulthood. However, this reduction in CB1 expression follows a slightly different trajectory in different cortical areas. For example, this decrease is delayed until PND 40 in sensorimotor areas, and the decrease is more pronounced in the medial PFC [75]. Therefore, both the ligands and receptors of the eCB system are dynamic throughout development, with a certain degree of regional specialization. Unfortunately, we could not directly compare eCB levels between age groups because experiments were conducted at very different times and our HPLC/MS/MS methodology does not allow for absolute comparisons from samples run several months apart. Given that levels of eCBs and their metabolic enzymes appear to be dynamic throughout adolescence, a follow-up study could examine how the wider eCB lipidome changes throughout development.
4.2 Changes in eCB signaling with THC: Potential Effects on Synaptic Plasticity
A hallmark of eCB signaling is its modulatory effects on activity-dependent neural plasticity [76]. Here, we show that THC in effect downregulates the eCB system, but in a region and development-dependent manner. 2-AG levels were reduced in 4 brain regions of adult and PND 50 and 2 areas of the PND 35 THC-treated mice. AEA levels were reduced in 6 brain regions of adult mice, but were only reduced in 2 regions of the PND 50 mice and were unaffected in the PND 35 mice. It has previously been shown that a single dose of 3 mg/kg THC dysregulates eCB-mediated plasticity in the hippocampus and midbrain [41]. 2-AG exerts its effects on plasticity by multiple processes. Some of these processes involve 2-AG as a retrograde messenger, which typically serves to decrease neurotransmitter release [77]. These processes include depolarization-induced suppression of inhibition or excitation, metabotropic-induced suppression of inhibition or excitation, and the longer-lasting eCB-long term depression [76]. Highlighting the diversity of eCB signaling, eCBs can also signal in a non-retrograde manner and interact with other signaling systems in the CNS [77].
The complexity of eCB signaling means that the effects of THC on synaptic transmission are not always as predicted, especially because CB1 is located on both excitatory and inhibitory terminals in the hippocampus [78]. On glutamatergic axon terminals in the Schaffer collateral of the hippocampus, THC typically serves to decrease glutamate release via its actions at CB1, which is hypothesized to underlie some of the deleterious effects of THC on memory. However, there are also adenosine receptors on the same axon terminals, and blocking adenosine receptors augmented THC’s ability to decrease glutamate release [79]. THC was found to disrupt both glutamate release and long-term potentiation (LTP) in the hippocampus, possibly contributing to the memory impairment seen after a single dose of THC. However, THC’s ability to disrupt glutamate release and LTP was not as strong as synthetic cannabinoids with higher efficacy at CB1 [78]. Given that 2-AG is usually considered a full agonist at CB1, whereas THC is usually considered a partial agonist, it is possible that 2-AG would have a stronger ability to disrupt hippocampal function than THC. For example, in cultured hippocampal neurons, Kelley and Thayer found that THC antagonizes the effects of 2-AG on synaptic transmission. In that study, 1 μM 2-AG could inhibit spiking in hippocampal neurons at an EC50 of 63 nM. When 100nM THC was added to the culture, the EC50 shifted to 1430 nM, meaning that THC made 2-AG less effective at modulating synaptic plasticity. The authors hypothesized that the ability of THC to antagonize 2-AG signaling is due to the lower intrinsic efficacy of THC at CB1 compared to 2-AG [80]. However, our data suggest that a potential mechanism for the ability of THC to block 2-AG signaling is by lowering levels of 2-AG. Although it seems like the younger brains should be more protected from the negative effects of THC on the eCB system because there were fewer changes in eCB levels, it might be that this lack of rapid 2-AG downregulation augments THC’s ability to affect synaptic regulation at a younger age.
4.3 THC and Lipidome Signaling at Non-Cannabinoid Receptors and Proteins
THC does not exclusively activate cannabinoid receptors CB1 and CB2. For example, THC activates the G-protein coupled receptors (GPCRs) GPR55 [81] and GPR18 [82], and the ion channel TRPV2 [83], and inhibits CaV3.1, CaV3.2, and CaV3.3 type calcium channels [84]. Therefore, we cannot determine from this study whether acute THC’s effects on the lipidome are due to its actions at receptors of the canonical eCB system or due to its actions at another protein target. GPR55 and GPR18 have activity in vasculature [81] and microglia [82] respectively. More recent data illustrates that GPR55 is also important for synaptic plasticity in the hippocampus [85, 86]. CaV3 (also known as T-type) calcium channels are implicated in several clinical conditions, such as insomnia, epilepsy, neuropathic pain, and hypertension [84]. Notably, these receptor targets are also targets for some of the lipids in the screening library. For example, N-arachidonoyl glycine (NAGly) activates GPR18 [87, 88] and GPR55 [89], and N-palmitoyl tyrosine is a TRPV2 agonist [29]. Cazade and colleagues showed that AEA, N-docosahexaenoyl glycine, NAGly, N-arachidonoyl GABA, N-arachidonoyl serine, N-arachidonoyl alanine, A-Taur, and N-linoleoyl glycine all inhibited CaV3.3 currents in transfected HEK cell lines, albeit by varying degrees [90]. Several of these endogenous GPCR and T-type calcium channel modulators were affected here by acute THC. In the adult brain, the reduction in AEA levels may counteract the inhibition of T-type calcium channels caused by THC. The PND 35 brain lacks the reductions in CaV3.3 inhibitors observed here in the adult, and therefore the activity of the CaV3.3 channel may be more inhibited in the PND 35 brain than in the adult brain.
4.4 Age and Regional Differences in THC and THC Metabolites
Another unique feature of this data set is the analysis of the regional distribution of THC and THC metabolites in the CNS. We show that the only commonality among the age groups was that the levels of THC were highest in HIPP; however, the levels of metabolites might be an important indicator of continued effects on hippocampal function in adolescents. The PND 35 HIPP had the 2nd highest levels of 11-OH-THC and the highest levels of (±)-11-nor-9-carboxy-THC relative to other brain regions. Similarly, the PND 50 HIPP had the highest levels of 11-OH-THC and the 2nd highest levels of (±)-11-nor-9-carboxy-THC. Relative levels of these cannabinoids in the adult showed that HIPP levels ranked 5th for 11-OH-THC and 6th for (±)-11-nor-9-carboxy-THC. This suggests that the high levels of THC in the adolescent HIPP are not the product of a lack of metabolism. Yet a lack of metabolism converting THC to 11-OH-THC and (±)-11-nor-9-carboxy-THC is a potential explanation for high THC levels in the HIPP that is specific to the adult brain. The conversion of THC to 11-OH-THC and (±)-11-nor-9-carboxyTHC is carried out by the cytochrome P450 family of enzymes, mainly CYP2C9 [91] and CYP3A4 [92]. Most of the metabolism of THC appears to take place in the liver [93], where expression of P450 enzymes is very high; however, these enzymes are also expressed in the brain suggesting that local metabolism is also likely [94]. The contribution of these more localized enzymes to THC metabolism needs to be investigated, and may vary by developmental stage.
4.5 The eCB system in the Hippocampus
The hippocampus is home to specialized machinery for eCB signaling [9] and studies performed in the hippocampus have been crucial to our understanding of the eCB system [95]. Indeed, CB1 expression is enriched in the hippocampus relative to other brain areas. However, suggesting specialization of the eCB system even within a brain area, levels of CB1 mRNA vary widely by hippocampal cell type, being highest in cholecystokinin-containing GABAergic interneurons that are distributed throughout the hippocampal formation [96, 97]. Compared to expression on glutamate axon terminals, the expression of CB1 on GABAergic axon terminals is much higher. The difference in expression levels means that THC can act as a full agonist at CB1 receptors on GABA neurons in the hippocampus, whereas THC acts as a partial agonist at CB1 located on glutamatergic neurons [98]. Increased expression of CB1 and elevated levels of THC found in the HIPP after acute THC exposure may make this region especially vulnerable to acute THC.
4.6 Hippocampal Signature of THC: A Signature of Cognitive Impairment?
Our data on the relative levels of THC being the highest in HIPP in all age groups is corroborated by the fact that THC can have profound effects on short-term memory [99, 100]. Acute THC impairs memory in both animals and in humans[101]. One of the memory processes supported by the hippocampus is spatial memory, assessed in animals using the Morris water maze. Varvel and colleagues examined the effects of acute THC on spatial working memory by giving male C57 mice varying doses of THC and testing performance on the Morris water maze 30 minutes later. A dose of just 3 mg/kg significantly disrupted spatial working memory without producing cannabimimetic effects in the tetrad. This suggests that working memory is particularly sensitive to THC [102]. A dose of 3 mg/kg THC impaired novel object recognition, another task reliant on the hippocampus, 24 hours after dosing [103]. This behavioral data combined with our lipidomics data validates the choice of the hippocampus for our analysis for transcriptomics and furthermore validates our choice of dose.
4.7 THC Exposure on the Hippocampus Suggests a Role for Cell Death
Transcripts impacted by acute THC in the PND 35 and adult HIPP were not traditional members of the eCB system (Figure 5; Supplemental Figures 8 and 9); yet the changes measured likely have important consequences for signaling. A deeper look at the types of transcripts suggests a specific role for cell survival (Supplemental Figure 10) that may be a key component for understanding long-term consequences of THC exposure. Although the effects of acute THC on the transcriptome were very broad in terms of pathways, the number of transcripts significantly affected by acute THC was relatively low considering the very wide coverage of the RNA-Seq data; however, this may point to a directed set of changes in signaling parameters. In a microarray study, acute THC altered expression of only 28 out of 24,500 possible genes in the library [104]. That we measured 189 differentially expressed transcripts in the adult HIPP might be explained by the differences in techniques. Older, hybridization-based technology, like microarrays, required knowledge of the sequences for specific transcripts hypothesized to change in the experiment. In contrast, RNA-Seq does not rely on hybridization to a targeted sequence, but instead requires knowledge of the transcriptome for analysis [52]. Thus, microarrays are targeted, whereas RNA-Seq is less biased. As there is no upper-limit for quantification, the dynamic range of RNA-Seq is very large [52], especially compared to microarrays. This facilitates detection of differences between groups in transcripts expressed at low levels [54], which likely explain the discovery of these novel transcript changes here.
4.8 THC and Neurogenesis
Another important aspect of the hippocampus is that it is a site of adult neurogenesis [105]. Specifically, neural progenitor cells originate from the dentate gyrus of the hippocampus [106]. New neurons are constantly being produced and many of them are integrated into existing brain networks with an estimated 700 new neurons being added in the adult human hippocampus each day [108]. Neurogenesis is impaired in psychopathology, one example being major depression, and anti-depressant therapies are known to stimulate neurogenesis [109, 110].
Although THC may not directly affect neurogenesis [111], challenging the eCB system with THC may produce changes in gene expression that support a decrease in neurogenesis, such as a downregulation of CB1, contributing to the increased risk of psychopathology in people who heavily used cannabis during their youth [112, 113]. Indeed, chronic exposure of male rats to the synthetic cannabinoid WIN55,212-2 during adolescence caused deficits in spatial learning and memory that were more severe than those seen when the drug was administered during adulthood and were correlated with a stronger reduction in dorsal hippocampal neurogenesis [114]. Given that AEA and 2-AG can promote neurogenesis [115], the fact that levels of eCBs were reduced in the adult HIPP after acute THC may indicate a lipid environment less supportive of neurogenesis. However, it must be noted that we did not measure levels of eCBs in the dentate gyrus specifically, and so it is not known whether the changes in lipid levels are occurring where the new granule neurons are being produced [116]. Prior studies have shown that acute THC did not increase the number of neural progenitor cells in the dentate gyrus, but these studies did not measure neurogenesis, leaving open the possibility that THC is affecting neurogenesis via cell proliferation or cell survival [117].
One of the factors that induces cell proliferation and promotes the survival of immature neurons is vascular endothelial growth factor (VEGF) [118]. Here, we show that acute THC significantly lowered mRNA encoding the receptor for VEGF, Kdr, in both the adult and PND 35 HIPP. A consequence of this decreased Kdr expression may be that the survival of new neurons is compromised. Some of the genes altered by acute THC had restricted expression to sites of neurogenesis. For example, expression of cyclin dependent kinase inhibitor 1A (Cdkn1a) is restricted to immature hippocampal neurons in the subgranular zone of the dentate gyrus, a site of adult neurogenesis. Cdkn1a can keep immature neurons in a quiescent state [119]. Interestingly, Cdkn1a was one of the transcripts upregulated by acute THC in the HIPP of mice from both age groups. Therefore, the changes seen with acute THC may support a decrease in neurogenesis. Follow-up studies could directly test the hypothesis that acute THC impairs neurogenesis in the hippocampus. These decreases in neurogenesis may be sustained with repeated use, which could contribute to psychopathologies. For example, Realini and colleagues uncovered a depressive-like phenotype in female rats chronically exposed to THC in adolescence that was correlated with a reduction in the number of proliferating neurons in the hippocampus [120]. Furthermore, there could be unique consequences to decreasing neurogenesis for the adolescent brain.
4.9 Acute THC induced changes in the CNS may be an indicator of the relative resilience of older brains
Although more lipids and transcripts changed in expression with acute exposure to THC in the adult brain, these alterations may be an indication of the relative resilience of older brains to the effects of THC. For example, the effect of acute THC on PG levels in the HIPP completely switched direction with developmental age; levels of PGs increased in the PND 35 HIPP, whereas they decreased in the adult HIPP. Increased levels of PGE2 in the HIPP have been linked with THC-induced memory impairment [62], and may be indicative of a more pro-inflammatory effect of acute THC in the HIPP of younger mice because an increase in PG production is often considered a marker of inflammation [62, 121].
Changes in the transcriptome may also be indicative of the relative resilience of older brains. A regulator of neurogenesis, glycogen synthase kinase 3β (GSK3β) has been implicated as a target for medications treating psychopathology, such as depression. In mice, inhibiting GSK3β causes a reduction in depressive-like behaviors and increasing GSK3β activity causes increased depressive-like behaviors. FK506 binding protein 5 (FKBP5) can inhibit GSK3β activity to produce anti-depressant effects and deleting FKBP5 makes anti-depressants less effective [122]. The acute exposure to 3 mg/kg THC upregulated FKBP5 message in the HIPP of both age groups, but the increase was of a larger magnitude in the adult brain. FKBP5 can be considered a stress-induced gene because acute stress upregulates FKBP5 in the CA1 region and the dentate gyrus of the hippocampus [123]. However, exposures to some of the molecular alterations driven by acute stress may be beneficial in conferring resilience to stress-related psychopathology. Gassen and colleagues hypothesized that the acute stress-induced effects of increasing FKBP5 could support coping with stress by inhibiting GSK3β [122]. Stress also likely has different effects in different developmental contexts, with developing brains appearing more vulnerable to stressors [124].
5. Conclusions
These experiments emphasize how the effects of the first exposure to a single dose of THC on the brain are widespread, region-dependent, and, most importantly, developmental age-dependent. That both the broader lipidome and the transcriptome are significantly affected just 2 hours after THC treatment highlights the finding that AEA and 2-AG and CB1 and CB2 are not the only signaling systems altered by acute THC. While drug addiction is a disease that develops with persistent, chronic drug use, it is the acute effects produced by drugs of abuse like cannabis that are the first step towards the development of substance use disorders. Current treatments for drug abuse do not take into consideration the biochemical changes that accompany the transition from the first exposure to a drug to the development of addiction. Delineating the signaling changes that occur during the transition to addiction represents a novel avenue to identify mechanisms for pharmacological interventions. The adolescent hippocampus appears particularly vulnerable to the impairing effects of THC exposure [125, 126]; the adult hippocampus may be relatively more resistant to developing these psychopathologies. Perhaps some of the acute THC induced changes in the CNS observed here may be a clue as to why these outcomes are so divergent.
Supplementary Material
Acknowledgments
6.1 Funding
RNA-Seq data was generated with the help of the Center for Genomics and Bioinformatics, Indiana University and the transcriptomics portion of the study was supported by Clinical and Translational Sciences Institute (CTSI) grant #UL1 TR001108. The work was supported in part by National Institutes of Health grant numbers DA021696, DA039463, and DA041208.
Abbreviations
- 2-AG
2-arachidonoyl glycerol
- AA
arachidonic acid
- AEA
N-arachidonoyl ethanolamine
- A-Taur
N-arachidonoyl taurine
- Cdkn1a
cyclin dependent kinase inhibitor 1A
- CER
cerebellum specific for this study
- cldn5
claudin 5
- CTX
cortex specific for this study
- d8NAGly
deuterium-labeled N-arachidonoyl glycine
- eCB
endogenous cannabinoid
- FAAH
fatty acid amide hydrolase
- FDR
false discovery rate
- FKBP5
FK506 binding protein 5
- GPCR
G-protein coupled receptor
- GSK3β
glycogen synthase kinase 3β
- HIPP
hippocampus specific for this study
- HPLC
high pressure liquid chromatography
- HPLC/MS/MS
high pressure liquid chromatography coupled with tandem mass spectrometry
- HYP
hypothalamus specific for this study
- IPA
Ingenuity Pathway Analysis
- kdr
kinase insert domain protein receptor
- LTP
long term potentiation
- MAGL
mono acyl glycerol lipase
- MID
midbrain specific for this study
- MRM
multiple reactions monitoring
- NAc
nucleus accumbens
- NAE
N-acyl ethanolamine
- NAGly
N-arachidonoyl glycine
- NAPE-PLD
N-acyl phosphatidyl ethanolamine specific phospholipase D
- OEA
N-oleoyl ethanolamine
- PEA
N-palmitoyl ethanolamine
- PFC
pre-frontal cortex
- PG
prostaglandin
- Plekhf1
pleckstrin homology domain containing, family F (with FYVE domain) member 1
- PND
post-natal day
- RIN
RNA integrity number
- STEM
brainstem specific for this study
- STR
striatum specific for this study
- THAL
thalamus specific for this study
- THC
Δ9-tetrahydrocannabinol, TRPV, transient receptor potential vanilloid
- VEGF
vascular endothelial growth factor
Footnotes
6.2 Conflict of Interest
The author, Heather B Bradshaw, of this manuscript is on the Advisory Board for Phytecs and consults on how endogenous cannabinoids function in the central nervous system. Phytecs had no financial contribution to the current work.
6.3 Ethical Approval
All procedures using animals were approved by the Bloomington Institutional Animal Care and Use Committee of Indiana University and complied with ARRIVE guidelines. This article does not contain any studies with human participants performed by any of the authors.
References
- 1.Zamberletti E, Gabaglio M, Prini P, Rubino T, Parolaro D. Cortical neuroinflammation contributes to long-term cognitive dysfunctions following adolescent delta-9-tetrahydrocannabinol treatment in female rats. Eur Neuropsychopharmacol. 2015 doi: 10.1016/j.euroneuro.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 2.S. Abuse, Mental Health Services Administration (SAMHSA) Results from the 2013 National Survey on Drug Use and Health: Mental health findings. Vol. 2014. Rockville, MD: SAMHSA; 2015. [Google Scholar]
- 3.Console-Bram L, Marcu J, Abood ME. Cannabinoid receptors: nomenclature and pharmacological principles. Progress in neuro-psychopharmacology & biological psychiatry. 2012;38:4–15. doi: 10.1016/j.pnpbp.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubino T, Parolaro D. Long lasting consequences of cannabis exposure in adolescence. Molecular and cellular endocrinology. 2008;286:S108–113. doi: 10.1016/j.mce.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Rubino T, Parolaro D. Cannabis abuse in adolescence and the risk of psychosis: a brief review of the preclinical evidence. Progress in neuro-psychopharmacology & biological psychiatry. 2014;52:41–44. doi: 10.1016/j.pnpbp.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Rubino T, Parolaro D. The Impact of Exposure to Cannabinoids in Adolescence: Insights from Animal Models. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Rubino T, Parolaro D. Sex-dependent vulnerability to cannabis abuse in adolescence. Front Psychiatry. 2015;6:56. doi: 10.3389/fpsyt.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubino T, Prini P, Piscitelli F, Zamberletti E, Trusel M, Melis M, Sagheddu C, Ligresti A, Tonini R, Di Marzo V, Parolaro D. Adolescent exposure to THC in female rats disrupts developmental changes in the prefrontal cortex. Neurobiol Dis. 2015;73:60–69. doi: 10.1016/j.nbd.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Rubino T, Vigano D, Realini N, Guidali C, Braida D, Capurro V, Castiglioni C, Cherubino F, Romualdi P, Candeletti S, Sala M, Parolaro D. Chronic delta 9-tetrahydrocannabinol during adolescence provokes sex-dependent changes in the emotional profile in adult rats: behavioral and biochemical correlates. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2008;33:2760–2771. doi: 10.1038/sj.npp.1301664. [DOI] [PubMed] [Google Scholar]
- 10.Murphy M, Mills S, Winstone J, Leishman E, Wager-Miller J, Bradshaw HB, Mackie K. Chronic adolescent Δ9-tetrahydrocannabinol treatment of mice leads to long-term cognitive and behavioral dysfunction, which are prevented by concurrent cannabidiol treatment. Cannabis and cannabinoid research. 2017;2:235–246. doi: 10.1089/can.2017.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volkow ND, Compton WM, Weiss SR. Adverse health effects of marijuana use. The New England journal of medicine. 2014;371:879. doi: 10.1056/NEJMc1407928. [DOI] [PubMed] [Google Scholar]
- 12.Wu CS, Jew CP, Lu HC. Lasting impacts of prenatal cannabis exposure and the role of endogenous cannabinoids in the developing brain. Future Neurol. 2011;6:459–480. doi: 10.2217/fnl.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devane WA, Dysarz Fr, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Molecular pharmacology. 1988;34:605–613. [PubMed] [Google Scholar]
- 14.Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 15.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 16.Sugiura T, Kondo S, Sukagawa A, Nakane S, Shinoda A, Itoh K, Yamashita A, Waku K. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochemical and biophysical research communications. 1995;215:89–97. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- 17.Mechoulam R, Ben-Shabat S, Hanus L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochemical pharmacology. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- 18.Kuehl FA, Jacob TA, Ganley OH, Ormond RE, Meisinger MAP. The Identification of N-(2-Hydroxyethyl) -Palmitamide as a Naturally Occurring Anti-Inflammatory Agent. J Am Chem Soc. 1957;79:5577–5578. [Google Scholar]
- 19.Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. A key role for diacylglycerol lipase-alpha in metabotropic glutamate receptor-dependent endocannabinoid mobilization. Mol Pharmacol. 2007;72:612–621. doi: 10.1124/mol.107.037796. [DOI] [PubMed] [Google Scholar]
- 20.Huang SM, Bisogno T, Petros TJ, Chang SY, Zavitsanos PA, Zipkin RE, Sivakumar R, Coop A, Maeda DY, De Petrocellis L, Burstein S, Di Marzo V, Walker JM. Identification of a new class of molecules, the arachidonyl amino acids, and characterization of one member that inhibits pain. The Journal of biological chemistry. 2001;276:42639–42644. doi: 10.1074/jbc.M107351200. [DOI] [PubMed] [Google Scholar]
- 21.Leishman E, Cornett B, Spork K, Straiker A, Mackie K, Bradshaw HB. Broad impact of deleting endogenous cannabinoid hydrolyzing enzymes and the CB1 cannabinoid receptor on the endogenous cannabinoid-related lipidome in eight regions of the mouse brain. Pharmacol Res. 2016 doi: 10.1016/j.phrs.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leishman E, Kunkler PE, Manchanda M, Sangani K, Stuart JM, Oxford GS, Hurley JH, Bradshaw HB. Environmental toxin acrolein alters levels of endogenous lipids, including TRP agonists: A potential mechanism for headache driven by TRPA1 activation. Neurobiology of Pain. 2017 doi: 10.1016/j.ynpai.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leishman E, Mackie K, Luquet S, Bradshaw HB. Lipidomics profile of a NAPE-PLD KO mouse provides evidence of a broader role of this enzyme in lipid metabolism in the brain. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbalip.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raboune S, Stuart JM, Takacs S, Rhodes BP, Jameyfield E, Leishman E, McHugh D, Bradshaw H. Novel endogenous N-acyl amides activate TRPV1-4 receptors, BV-2 microglia, and are regulated in brain an acute model of inflammation. Frontiers in Cellular Neuroscience. 0 doi: 10.3389/fncel.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rimmerman N, Bradshaw HB, Hughes HV, Chen JS, Hu SS, McHugh D, Vefring E, Jahnsen JA, Thompson EL, Masuda K, Cravatt BF, Burstein S, Vasko MR, Prieto AL, O’Dell DK, Walker JM. N-palmitoyl glycine, a novel endogenous lipid that acts as a modulator of calcium influx and nitric oxide production in sensory neurons. Molecular pharmacology. 2008;74:213–224. doi: 10.1124/mol.108.045997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradshaw HB, Rimmerman N, Hu SS, Burstein S, Walker JM. Novel endogenous N-acyl glycines identification and characterization. Vitamins and hormones. 2009;81:191–205. doi: 10.1016/S0083-6729(09)81008-X. [DOI] [PubMed] [Google Scholar]
- 27.Smoum R, Bar A, Tan B, Milman G, Attar-Namdar M, Ofek O, Stuart JM, Bajayo A, Tam J, Kram V, O’Dell D, Walker MJ, Bradshaw HB, Bab I, Mechoulam R. Oleoyl serine, an endogenous N-acyl amide, modulates bone remodeling and mass. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17710–17715. doi: 10.1073/pnas.0912479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradshaw H, Allard CM. Endogenous cannabinoid production in the rat female reproductive tract is regulated by changes in the hormonal milieu. Pharmaceuticals. 2011;4:17. [Google Scholar]
- 29.Raboune S, Stuart JM, Leishman E, Takacs SM, Rhodes B, Basnet A, Jameyfield E, McHugh D, Widlanski T, Bradshaw HB. Novel endogenous N-acyl amides activate TRPV1-4 receptors, BV-2 microglia, and are regulated in brain in an acute model of inflammation. Frontiers in cellular neuroscience. 2014;8:195. doi: 10.3389/fncel.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balakrishna S, Song W, Achanta S, Doran SF, Liu B, Kaelberer MM, Yu Z, Sui A, Cheung M, Leishman E, Eidam HS, Ye G, Willette RN, Thorneloe KS, Bradshaw HB, Matalon S, Jordt SE. TRPV4 inhibition counteracts edema and inflammation and improves pulmonary function and oxygen saturation in chemically induced acute lung injury, American journal of physiology. Lung cellular and molecular physiology. 2014;307:L158–172. doi: 10.1152/ajplung.00065.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowe MS, Leishman E, Banks ML, Gujjar R, Mahadevan A, Bradshaw HB, Kinsey SG. Combined inhibition of monoacylglycerol lipase and cyclooxygenases synergistically reduces neuropathic pain in mice. British journal of pharmacology. 2015;172:1700–1712. doi: 10.1111/bph.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koppel J, Bradshaw H, Goldberg TE, Khalili H, Marambaud P, Walker MJ, Pazos M, Gordon ML, Christen E, Davies P. Endocannabinoids in Alzheimer's disease and their impact on normative cognitive performance: a case-control and cohort study. Lipids in health and disease. 2009;8:2. doi: 10.1186/1476-511X-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. The Journal of clinical investigation. 2010;120:803–815. doi: 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leishman E, Kokesh KJ, Bradshaw HB. Lipids and addiction: how sex steroids, prostaglandins, and cannabinoids interact with drugs of abuse. Annals of the New York Academy of Sciences. 2013;1282:25–38. doi: 10.1111/nyas.12081. [DOI] [PubMed] [Google Scholar]
- 35.Berrendero F, Sepe N, Ramos JA, Di Marzo V, Fernandez-Ruiz JJ. Analysis of cannabinoid receptor binding and mRNA expression and endogenous cannabinoid contents in the developing rat brain during late gestation and early postnatal period. Synapse. 1999;33:181–191. doi: 10.1002/(SICI)1098-2396(19990901)33:3<181::AID-SYN3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Lee TT, Hill MN, Hillard CJ, Gorzalka BB. Temporal changes in N-acylethanolamine content and metabolism throughout the peri-adolescent period. Synapse. 2013;67:4–10. doi: 10.1002/syn.21609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. Animal research: reporting in vivo experiments: the ARRIVE guidelines. British journal of pharmacology. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Rodriguez AB, Llorente-Berzal A, Garcia-Segura LM, Viveros MP. Sex-dependent long‐term effects of adolescent exposure to THC and/or MDMA on neuroinflammation and serotoninergic and cannabinoid systems in rats. British journal of pharmacology. 2014;171:1435–1447. doi: 10.1111/bph.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burston JJ, Wiley JL, Craig AA, Selley DE, Sim-Selley LJ. Regional enhancement of cannabinoid CB₁ receptor desensitization in female adolescent rats following repeated Delta-tetrahydrocannabinol exposure. British journal of pharmacology. 2010;161:103–112. doi: 10.1111/j.1476-5381.2010.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamberletti E, Prini P, Speziali S, Gabaglio M, Solinas M, Parolaro D, Rubino T. Gender-dependent behavioral and biochemical effects of adolescent delta-9-tetrahydrocannabinol in adult maternally deprived rats. Neuroscience. 2012;204:245–257. doi: 10.1016/j.neuroscience.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 41.Mato S, Chevaleyre V, Robbe D, Pazos A, Castillo PE, Manzoni OJ. A single in-vivo exposure to delta 9THC blocks endocannabinoid-mediated synaptic plasticity. Nature neuroscience. 2004;7:585–586. doi: 10.1038/nn1251. [DOI] [PubMed] [Google Scholar]
- 42.Wiley JL, Burston JJ. Sex differences in Delta(9) -tetrahydrocannabinol metabolism and in vivo pharmacology following acute and repeated dosing in adolescent rats. Neurosci Lett. 2014;576:51–55. doi: 10.1016/j.neulet.2014.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan B, Bradshaw HB, Rimmerman N, Srinivasan H, Yu YW, Krey JF, Monn MF, Chen JS, Hu SS, Pickens SR, Walker JM. Targeted lipidomics: discovery of new fatty acyl amides. The AAPS journal. 2006;8:E461–465. doi: 10.1208/aapsj080354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tortoriello G, Rhodes BP, Takacs SM, Stuart JM, Basnet A, Raboune S, Widlanski TS, Doherty P, Harkany T, Bradshaw HB. Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One. 2013;8:e67865. doi: 10.1371/journal.pone.0067865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuart JM, Paris JJ, Frye C, Bradshaw HB. Brain levels of prostaglandins, endocannabinoids, and related lipids are affected by mating strategies. International journal of endocrinology. 2013;2013:436252. doi: 10.1155/2013/436252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Ghaffari N, Johnson CD, Braga-Neto UM, Wang H, Chen R, Zhou H. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC bioinformatics. 2011;12(Suppl 10):S5. doi: 10.1186/1471-2105-12-S10-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nature protocols. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raz T, Kapranov P, Lipson D, Letovsky S, Milos PM, Thompson JF. Protocol dependence of sequencing-based gene expression measurements. PLoS One. 2011;6:e19287. doi: 10.1371/journal.pone.0019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. Journal of biomedicine & biotechnology. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:btu 170. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloe D, Le Gall C, Schaeffer B, Le Crom S, Guedj M, Jaffrezic F. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Briefings in Bioinformatics. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 59.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014:btu638. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis (ipa) Bioinformatics. 2013:btt703. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen R, Zhang J, Fan N, Teng ZQ, Wu Y, Yang H, Tang YP, Sun H, Song Y, Chen C. Delta9-THC-caused synaptic and memory impairments are mediated through COX-2 signaling. Cell. 2013;155:1154–1165. doi: 10.1016/j.cell.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lenz KM, McCarthy MM. A starring role for microglia in brain sex differences. Neuroscientist. 2015;21:306–321. doi: 10.1177/1073858414536468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCarthy MM. How it’s made: Organisational effects of hormones on the developing brain. Journal of Neuroendocinology. 2010;22 doi: 10.1111/j.1365-2826.2010.02021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amateau SK, McCarthy MM. A novel mechanism of dendritic spine plasticity involving estradiol induction of prostaglandin-E2. Journal of Neuroscience. 2002;22:8586–8596. doi: 10.1523/JNEUROSCI.22-19-08586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry. 2006;45:9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- 67.Lemberger L, Crabtree RE, Rowe HM. 11-hydroxy-9-tetrahydrocannabinol: pharmacology, disposition, and metabolism of a major metabolite of marihuana in man. Science. 1972;177:62–64. doi: 10.1126/science.177.4043.62. [DOI] [PubMed] [Google Scholar]
- 68.Schwilke EW, Schwope DM, Karschner EL, Lowe RH, Darwin WD, Kelly DL, Goodwin RS, Gorelick DA, Huestis MA. Delta9-tetrahydrocannabinol (THC), 11-hydroxy-THC, and 11-nor-9-carboxy-THC plasma pharmacokinetics during and after continuous high-dose oral THC. Clin Chem. 2009;55:2180–2189. doi: 10.1373/clinchem.2008.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson JR, Jennison TA, Peat MA, Foltz RL. Stability of delta 9-tetrahydrocannabinol (THC), 11-hydroxy-THC, and 11-nor-9-carboxy-THC in blood and plasma. J Anal Toxicol. 1984;8:202–204. doi: 10.1093/jat/8.5.202. [DOI] [PubMed] [Google Scholar]
- 70.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T. The Ensembl genome database project. Nucleic acids research. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 72.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellgren M, Artmann A, Tkalych O, Gupta A, Hansen HS, Hansen SH, Devi LA, Hurd YL. Dynamic changes of the endogenous cannabinoid and opioid mesocorticolimbic systems during adolescence: THC effects. Eur Neuropsychopharmacol. 2008;18:826–834. doi: 10.1016/j.euroneuro.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 75.Heng L, Beverley JA, Steiner H, Tseng KY. Differential developmental trajectories for CB1 cannabinoid receptor expression in limbic/associative and sensorimotor cortical areas. Synapse. 2011;65:278–286. doi: 10.1002/syn.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu HC, Mackie K. An Introduction to the Endogenous Cannabinoid System. Biol Psychiatry. 2016;79:516–525. doi: 10.1016/j.biopsych.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castillo PE, Younts TJ, Chávez AE, Hashimotodani Y. Endocannabinoid signaling and synaptic function. Neuron. 2012;76:70–81. doi: 10.1016/j.neuron.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoffman AF, Lycas MD, Kaczmarzyk JR, Spivak CE, Baumann MH, Lupica CR. Disruption of hippocampal synaptic transmission and long-term potentiation by psychoactive synthetic cannabinoid ‘Spice’compounds: comparison with Δ9-tetrahydrocannabinol, Addiction biology. 2017;22:390–399. doi: 10.1111/adb.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoffman AF, Laaris N, Kawamura M, Masino SA, Lupica CR. Control of cannabinoid CB1 receptor function on glutamate axon terminals by endogenous adenosine acting at A1 receptors. Journal of Neuroscience. 2010;30:545–555. doi: 10.1523/JNEUROSCI.4920-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kelley BG, Thayer SA. Delta 9-tetrahydrocannabinol antagonizes endocannabinoid modulation of synaptic transmission between hippocampal neurons in culture. Neuropharmacology. 2004;46:709–715. doi: 10.1016/j.neuropharm.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 81.Ryberg E, Larsson N, Sjogren S, Hjorth S, Hermansson NO, Leonova J, Elebring T, Nilsson K, Drmota T, Greasley PJ. The orphan receptor GPR55 is a novel cannabinoid receptor. British journal of pharmacology. 2007;152:1092–1101. doi: 10.1038/sj.bjp.0707460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McHugh D, Page J, Dunn E, Bradshaw HB. Delta(9) -Tetrahydrocannabinol and N-arachidonyl glycine are full agonists at GPR18 receptors and induce migration in human endometrial HEC-1B cells. British journal of pharmacology. 2012;165:2414–2424. doi: 10.1111/j.1476-5381.2011.01497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Petrocellis L, Ligresti A, Moriello AS, Allara M, Bisogno T, Petrosino S, Stott CG, Di Marzo V. Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. British journal of pharmacology. 2011;163:1479–1494. doi: 10.1111/j.1476-5381.2010.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ross HR, Napier I, Connor M. Inhibition of recombinant human T-type calcium channels by Delta9-tetrahydrocannabinol and cannabidiol. The Journal of biological chemistry. 2008;283:16124–16134. doi: 10.1074/jbc.M707104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hurst K, Badgley C, Ellsworth T, Bell S, Friend L, Prince B, Welch J, Cowan Z, Williamson R, Lyon C, Anderson B, Poole B, Christensen M, McNeil M, Call J, Edwards JG. A putative lysophosphatidylinositol receptor GPR55 modulates hippocampal synaptic plasticity. Hippocampus. 2017;27:985–998. doi: 10.1002/hipo.22747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marichal-Cancino BA, Fajardo-Valdez A, Ruiz-Contreras AE, Mendez-Díaz M, Prospero-García O. Advances in the physiology of GPR55 in the central nervous system. Current Neuropharmacology. 2017;15:771–778. doi: 10.2174/1570159X14666160729155441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McHugh D, Hu SS, Rimmerman N, Juknat A, Vogel Z, Walker JM, Bradshaw HB. N-arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC neuroscience. 2010;11:44. doi: 10.1186/1471-2202-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McHugh D, Roskowski D, Xie S, Bradshaw HB. Δ9-THC and N-arachidonoyl glycine regulate BV-2 microglial morphology and cytokine release plasticity: implications for signaling at GPR18. Frontiers in pharmacology. 2014;4:162. doi: 10.3389/fphar.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Console-Bram L, Ciuciu SM, Zhao P, Zipkin RE, Brailoiu E, Abood ME. N-arachidonoyl glycine, another endogenous agonist of GPR55. Biochemical and biophysical research communications. 2017;490:1389–1393. doi: 10.1016/j.bbrc.2017.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cazade M, Nuss CE, Bidaud I, Renger JJ, Uebele VN, Lory P, Chemin J. Cross-modulation and molecular interaction at the Cav3.3 protein between the endogenous lipids and the T-type calcium channel antagonist TTA-A2. Mol Pharmacol. 2014;85:218–225. doi: 10.1124/mol.113.089581. [DOI] [PubMed] [Google Scholar]
- 91.Watanabe K, Matsunaga T, Yamamoto I, Funae Y, Yoshimura H. Involvement of CYP2C in the metabolism of cannabinoids by human hepatic microsomes from an old woman. Biol Pharm Bull. 1995;18:1138–1141. doi: 10.1248/bpb.18.1138. [DOI] [PubMed] [Google Scholar]
- 92.Watanabe K, Yamaori S, Funahashi T, Kimura T, Yamamoto I. Cytochrome P450 enzymes involved in the metabolism of tetrahydrocannabinols and cannabinol by human hepatic microsomes. Life Sci. 2007;80:1415–1419. doi: 10.1016/j.lfs.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 93.Varvel SA, Wiley JL, Yang R, Bridgen DT, Long K, Lichtman AH, Martin BR. Interactions between THC and cannabidiol in mouse models of cannabinoid activity. Psychopharmacology (Berl) 2006;186:226–234. doi: 10.1007/s00213-006-0356-9. [DOI] [PubMed] [Google Scholar]
- 94.Meyer RP, Gehlhaus M, Knoth R, Volk B. Expression and function of cytochrome p450 in brain drug metabolism. Curr Drug Metab. 2007;8:297–306. doi: 10.2174/138920007780655478. [DOI] [PubMed] [Google Scholar]
- 95.Lupica CR, Hu Y, Devinsky O, Hoffman AF. Cannabinoids as hippocampal network administrators. Neuropharmacology. 2017 doi: 10.1016/j.neuropharm.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 96.Mackie K. Understanding cannabinoid psychoactivity with mouse genetic models. PLoS Biol. 2007;5:e280. doi: 10.1371/journal.pbio.0050280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsou K, Mackie K, Sanudo-Pena MC, Walker JM. Cannabinoid CB1 receptors are localized primarily on cholecystokinin-containing GABAergic interneurons in the rat hippocampal formation. Neuroscience. 1999;93:969–975. doi: 10.1016/s0306-4522(99)00086-x. [DOI] [PubMed] [Google Scholar]
- 98.Laaris N, Good CH, Lupica CR. Delta9-tetrahydrocannabinol is a full agonist at CB1 receptors on GABA neuron axon terminals in the Hippocampus. Neuropharmacology. 2010;59:121–127. doi: 10.1016/j.neuropharm.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han J, Kesner P, Metna-Laurent M, Duan T, Xu L, Georges F, Koehl M, Abrous DN, Mendizabal-Zubiaga J, Grandes P, Liu Q, Bai G, Wang W, Xiong L, Ren W, Marsicano G, Zhang X. Acute cannabinoids impair working memory through astroglial CB1 receptor modulation of hippocampal LTD. Cell. 2012;148:1039–1050. doi: 10.1016/j.cell.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 100.Abel EL. Marijuana and memory. Nature. 1970;227:1151–1152. doi: 10.1038/2271151b0. [DOI] [PubMed] [Google Scholar]
- 101.Lichtman AH, Dimen KR, Martin BR. Systemic or intrahippocampal cannabinoid administration impairs spatial memory in rats. Psychopharmacology (Berl) 1995;119:282–290. doi: 10.1007/BF02246292. [DOI] [PubMed] [Google Scholar]
- 102.Varvel S, Hamm R, Martin B, Lichtman A. Differential effects of Δ [sup 9]-THC on spatial reference and working memory in mice. Psychopharmacology. 2001;157 doi: 10.1007/s002130100780. [DOI] [PubMed] [Google Scholar]
- 103.Puighermanal E, Marsicano G, Busquets-Garcia A, Lutz B, Maldonado R, Ozaita A. Cannabinoid modulation of hippocampal long-term memory is mediated by mTOR signaling. Nature neuroscience. 2009;12:1152–1158. doi: 10.1038/nn.2369. [DOI] [PubMed] [Google Scholar]
- 104.Kittler JT, Grigorenko EV, Clayton C, Zhuang SY, Bundey SC, Trower MM, Wallace D, Hampson R, Deadwyler S. Large-scale analysis of gene expression changes during acute and chronic exposure to [Delta]9-THC in rats. Physiol Genomics. 2000;3:175–185. doi: 10.1152/physiolgenomics.2000.3.3.175. [DOI] [PubMed] [Google Scholar]
- 105.Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- 106.Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proceedings of the National Academy of Sciences. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human Hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 108.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Bostrom E, Westerlund I, Vial C, Buchholz BA, Possnert G, Mash DC, Druid H, Frisen J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanson ND, Owens MJ, Nemeroff CB. Depression, antidepressants, and neurogenesis: a critical reappraisal. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2011;36:2589–2602. doi: 10.1038/npp.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Surget A, Tanti A, Leonardo ED, Laugeray A, Rainer Q, Touma C, Palme R, Griebel G, Ibarguen-Vargas Y, Hen R, Belzung C. Antidepressants recruit new neurons to improve stress response regulation. Molecular psychiatry. 2011;16:1177–1188. doi: 10.1038/mp.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wolf SA, Bick-Sander A, Fabel K, Leal-Galicia P, Tauber S, Ramirez-Rodriguez G, Muller A, Melnik A, Waltinger TP, Ullrich O, Kempermann G. Cannabinoid receptor CB1 mediates baseline and activity-induced survival of new neurons in adult hippocampal neurogenesis. Cell Commun Signal. 2010;8:12. doi: 10.1186/1478-811X-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Giron L, Befort K. Cannabinoids in Health and Disease. InTech, Place Published; 2016. Cannabinoids: Drug or Medication? [Google Scholar]
- 113.Volkow ND, Baler RD, Compton WM, Weiss SR. Adverse health effects of marijuana use. The New England journal of medicine. 2014;370:2219–2227. doi: 10.1056/NEJMra1402309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Abboussi O, Tazi A, Paizanis E, El Ganouni S. Chronic exposure to WIN55,212-2 affects more potently spatial learning and memory in adolescents than in adult rats via a negative action on dorsal hippocampal neurogenesis. Pharmacol Biochem Behav. 2014;120:95–102. doi: 10.1016/j.pbb.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 115.Prenderville JA, Kelly ÁM, Downer EJ. The role of cannabinoids in adult neurogenesis. British journal of pharmacology. 2015;172:3950–3963. doi: 10.1111/bph.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cameron HA, Mckay RD. Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. Journal of Comparative Neurology. 2001;435:406–417. doi: 10.1002/cne.1040. [DOI] [PubMed] [Google Scholar]
- 117.Kochman LJ, dos Santos AA, Fornal CA, Jacobs BL. Despite strong behavioral disruption, Δ 9-tetrahydrocannabinol does not affect cell proliferation in the adult mouse dentate gyrus. Brain research. 2006;1113:86–93. doi: 10.1016/j.brainres.2006.07.080. [DOI] [PubMed] [Google Scholar]
- 118.Cao L, Jiao X, Zuzga DS, Liu Y, Fong DM, Young D, During MJ. VEGF links hippocampal activity with neurogenesis, learning and memory. Nature genetics. 2004;36:827–835. doi: 10.1038/ng1395. [DOI] [PubMed] [Google Scholar]
- 119.Aizawa S, Yamamuro Y. Valproate administration to mice increases hippocampal p21 expression by altering genomic DNA methylation. Neuroreport. 2015;26:915–920. doi: 10.1097/WNR.0000000000000448. [DOI] [PubMed] [Google Scholar]
- 120.Realini N, Vigano D, Guidali C, Zamberletti E, Rubino T, Parolaro D. Chronic URB597 treatment at adulthood reverted most depressive-like symptoms induced by adolescent exposure to THC in female rats. Neuropharmacology. 2011;60:235–243. doi: 10.1016/j.neuropharm.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 121.Bhattacharya S. Delta-9-tetrahydrocannabinol (THC) increases brain prostaglandins in the rat. Psychopharmacology. 1986;90:499–502. doi: 10.1007/BF00174068. [DOI] [PubMed] [Google Scholar]
- 122.Gassen NC, Hartmann J, Zannas AS, Kretzschmar A, Zschocke J, Maccarrone G, Hafner K, Zellner A, Kollmannsberger LK, Wagner KV, Mehta D, Kloiber S, Turck CW, Lucae S, Chrousos GP, Holsboer F, Binder EB, Ising M, Schmidt MV, Rein T. FKBP51 inhibits GSK3beta and augments the effects of distinct psychotropic medications. Molecular psychiatry. 2016;21:277–289. doi: 10.1038/mp.2015.38. [DOI] [PubMed] [Google Scholar]
- 123.Scharf SH, Liebl C, Binder EB, Schmidt MV, Müller MB. Expression and regulation of the Fkbp5 gene in the adult mouse brain. PLoS One. 2011;6:e16883. doi: 10.1371/journal.pone.0016883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Romeo RD. Adolescence: a central event in shaping stress reactivity. Developmental psychobiology. 2010;52:244–253. doi: 10.1002/dev.20437. [DOI] [PubMed] [Google Scholar]
- 125.Quinn HR, Matsumoto I, Callaghan PD, Long LE, Arnold JC, Gunasekaran N, Thompson MR, Dawson B, Mallet PE, Kashem MA, Matsuda-Matsumoto H, Iwazaki T, McGregor IS. Adolescent rats find repeated Delta(9) -THC less aversive than adult rats but display greater residual cognitive deficits and changes in hippocampal protein expression following exposure. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2008;33:1113–1126. doi: 10.1038/sj.npp.1301475. [DOI] [PubMed] [Google Scholar]
- 126.Rubino T, Realini N, Braida D, Guidi S, Capurro V, Vigano D, Guidali C, Pinter M, Sala M, Bartesaghi R, Parolaro D. Changes in hippocampal morphology and neuroplasticity induced by adolescent THC treatment are associated with cognitive impairment in adulthood. Hippocampus. 2009;19:763–772. doi: 10.1002/hipo.20554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.