SUMMARY
Tardigrades are microscopic animals that survive a remarkable array of stresses, including desiccation. How tardigrades survive desiccation has remained a mystery for more than 250 years. Trehalose, a disaccharide essential for several organisms to survive drying, is detected at low levels or not at all in some tardigrade species, indicating that tardigrades possess potentially novel mechanisms for surviving desiccation. Here we show that tardigrade-specific intrinsically disordered proteins (TDPs) are essential for desiccation tolerance. TDP genes are constitutively expressed at high levels or induced during desiccation in multiple tardigrade species. TDPs are required for tardigrade desiccation tolerance, and these genes are sufficient to increase desiccation tolerance when expressed in heterologous systems. TDPs form non-crystalline amorphous solids (vitrify) upon desiccation, and this vitrified state mirrors their protective capabilities. Our study identifies TDPs as functional mediators of tardigrade desiccation tolerance, expanding our knowledge of the roles and diversity of disordered proteins involved in stress tolerance.
In Brief
Tardigrades (water bears) survive a number of extreme stresses, including desiccation. Boothby et al. show that tardigrade disordered proteins are required for desiccation tolerance and exogenously protect cells and purified enzymes from drying. When dry, these proteins form glasses, the integrity of which correlates with their protective capabilities.
INTRODUCTION
Tardigrades (water bears) comprise a phylum of microscopic animals renowned for their ability to survive a vast array of environmental extremes, including essentially complete desiccation for up to a decade (Goldstein and Blaxter, 2002). Although they have fascinated scientists for more than 250 years, little is known about how tardigrades survive such extreme environmental stresses, and no molecular mediators of tardigrade desiccation tolerance have been experimentally confirmed. The disaccharide trehalose has been proposed and often assumed to play a role in mediating desiccation tolerance in tardigrades (Hengherr et al., 2008; Jönsson and Persson, 2010; Westh and Ramløv, 1991). Trehalose is essential for some organisms to survive desiccation and is thought to protect organisms by vitrifying their cellular contents (Erkut et al., 2011; Sakurai et al., 2008; Tapia and Koshland, 2014). However, some desiccation-tolerant animals do not require or even appear to make this sugar (Lapinski and Tunnacliffe, 2003). Currently, the use and presence of trehalose in tardigrades are unclear; some studies report low levels of this sugar, while others failed to identify trehalose at all in the same species (Guidetti et al., 2011; Hengherr et al., 2008; Jönsson and Persson, 2010; Westh and Ramløv, 1991).
In addition to trehalose and other sugars, a number of protein families and classes have been implicated in mediating desiccation tolerance in other systems, including heat-shock proteins, antioxidant enzymes, and some intrinsically disordered protein (IDP) families (Hincha and Thalhammer, 2012; Hoekstra et al., 2001). This latter class of proteins is enigmatic, in that unlike typical globular proteins, they lack persistent tertiary structure. In the past two decades, myriad cellular roles for IDPs have emerged, including roles in transcription, post-translational modification, development, cellular organization, and abiotic stress tolerance (Chakrabortee et al., 2012; Garay-Arroyo et al., 2000; Hincha and Thalhammer, 2012; Iakoucheva et al., 2004; Nott et al., 2015; Xie et al., 2007; Zhang et al., 2015). However, the role of IDPs in tardigrade desiccation tolerance remains untested.
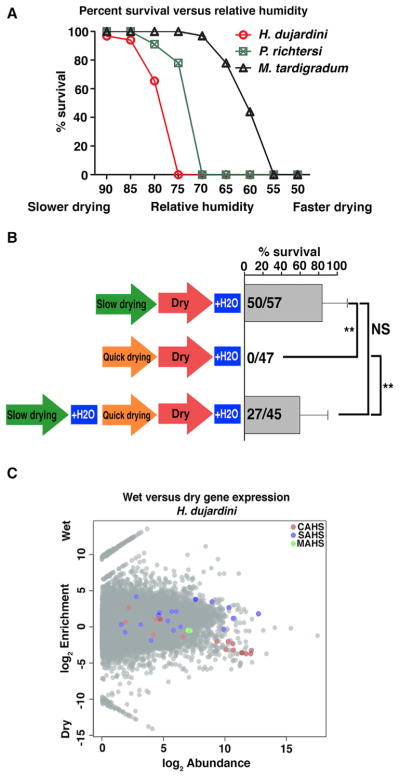
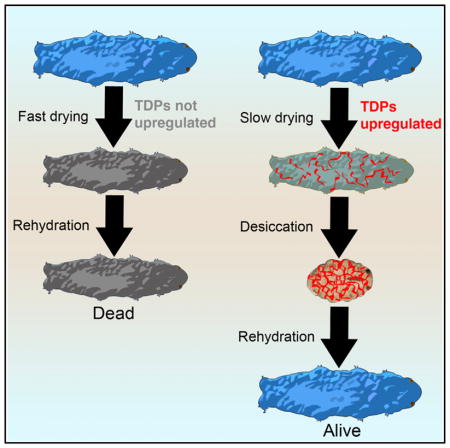
Although no molecular mediators of desiccation tolerance have been identified in tardigrades, one clue as to how these animals survive desiccation comes from the observation (Figure 1A) that different tardigrade species survive drying at different rates, but all species tested die if dried too quickly (Wright, 1989). This trend suggests that tardigrades need time to produce protectants, a theory supported by the recent evidence that de novo transcription and translation are required for the tardigrade Hypsibius dujardini to robustly survive desiccation (Kondo et al., 2015).
Figure 1. Tardigrades Upregulate Genes Encoding Tardigrade-Specific IDPs as They Dry.
(A) Published data on the survival versus relative humidity for Hypsibius dujardini (red), Paramacrobiotus richtersi (green), and Milnesium tardigradum (black). Data from Table 1 in Wright (1989). Animals desiccated at lower relative humidity experience increased rates of drying compared with those desiccated at higher relative humidity.
(B) Survival of H. dujardini after slow drying (95% relative humidity), quick drying (70% RH), and slow followed by quick drying. t test: NS, not significant; **p < 0.001.
(C) MA plot showing enrichment (log2 fold change) versus abundance (log2 CPM [count per million reads]) of expressed H. dujardini genes under hydrated and dry conditions. Colored circles indicate CAHS (red), SAHS (blue), and MAHS (green) genes encoding tardigrade-specific IDPs.
Here we present evidence that tardigrades upregulate the expression of genes encoding tardigrade-specific IDPs in response to drying. Disruption of gene function for several TDPs, using RNAi, results in severely diminished desiccation tolerance. Furthermore, the heterologous expression of TDPs in both prokaryotic and eukaryotic cells is sufficient to increase desiccation tolerance in these systems, and purified TDPs protect desiccation-sensitive proteins in vitro. Like trehalose, purified and heterologously expressed TDPs vitrify upon desiccation, and this vitreous state mirrors the protective capabilities of these proteins. These findings identify TDPs as functional mediators of tardigrade desiccation tolerance and expand our understanding of the diversity and roles of IDPs. We anticipate that these findings will build a foundation for pursuing long-term goals of the desiccation tolerance field, including the engineering of desiccation-tolerant crops and the development of technologies for the dry preservation of pharmaceuticals, cells, and tissues.
RESULTS AND DISCUSSION
Identification of Likely Mediators of Tardigrade Desiccation Tolerance
To test whether tardigrades produce protectants that are sufficient to protect against desiccation, we assayed whether slowly dried (preconditioned) tardigrades can survive subsequent drying at higher, typically non-survivable, rates. Specimens of the tardigrade H. dujardini that had been dried slowly could subsequently survive more rapid desiccation (Figure 1B), suggesting that a sufficient protectant(s) was made during slow drying. This finding, in addition to the fact that H. dujardini requires de novo transcription and translation to robustly survive desiccation (Kondo et al., 2015), makes H. dujardini attractive for differential gene expression studies.
To identify potential mediators of desiccation tolerance, genes induced by drying, in an unbiased fashion we sequenced and performed differential gene expression analysis on transcriptomes of hydrated and slowly drying H. dujardini specimens in triplicate (Data S1). Consistent with a previous report (Mali et al., 2010), our transcriptome as well as multiple independent H. dujardini genome and transcriptome assemblies (Bemm et al., 2016; Boothby et al., 2015; Koutsovoulos et al., 2016; Levin et al., 2016) did not contain a homolog for trehalose phosphatase, the enzyme required to produce trehalose, despite the apparent completeness of these assemblies (Figure S1). The discordant reports of trehalose in tardigrades, coupled with lack of evidence for a trehalose biosynthetic pathway from sequencing resources, suggest that H. dujardini uses potentially novel molecules for protection against the harmful effects of drying (Hengherr et al., 2008; Jönsson and Persson, 2010; Westh and Ramløv, 1991).
Our differential gene expression analysis revealed that 11 of 17 cytosolic abundant heat soluble (CAHS) protein transcripts expressed by H.dujardini (Figure 1C) are enriched 4- to 22-fold during desiccation relative to hydrated conditions (cutoff: p ≤ 0.05 and false discovery rate ≤ 0.05). H. dujardini expresses 19 secreted abundant heat soluble (SAHS) protein transcripts, and although only two are enriched 2- to 5-fold during drying (Figure 1C), several SAHS transcripts are expressed constitutively at extremely high levels. For example, one SAHS transcript was the sixth most abundant transcript detected (Figure 1C). H. dujardini expresses two mitochondrial abundant heat soluble (MAHS) protein transcripts, neither of which is particularly abundant or differentially expressed between hydrated and dry conditions (Figure 1C).
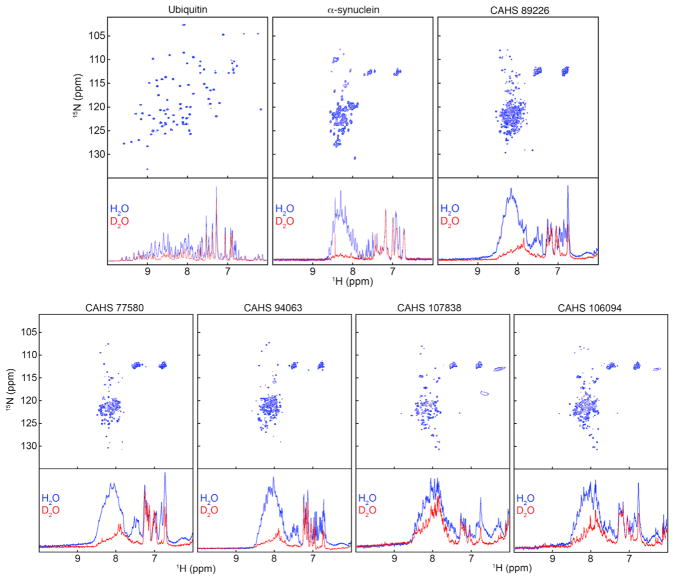
These gene families, CAHS, SAHS, and MAHS, were previously identified in a proteomic analysis of tardigrades (Tanaka et al., 2015; Yamaguchi et al., 2012). On the basis of heat solubility experiments and circular dichroism spectropolarimetry, all three families are thought to encode IDPs (Tanaka et al., 2015; Yamaguchi et al., 2012). Previous bioinformatic studies suggest that not all members of disordered protein families are natively unfolded (Hundertmark and Hincha, 2008). Thus, we took a bioinformatic approach to characterize the widespread disorder of members of tardigrade IDP families. These predictions strongly support previous evidence that CAHS and MAHS proteins are largely disordered, while by this approach SAHS proteins appear less disordered (Supplemental Information). IDPs lack persistent secondary structure (Theillet et al., 2014; Yamaguchi et al., 2012), which we confirmed for CAHS proteins using nuclear magnetic resonance spectroscopy (NMR). To do this we mapped the chemical environment of the covalent bond between each backbone amide nitrogen and its attached proton based on the heteronuclear single-quantum coherence (HQSC) spectra of the protein. In this experiment, each bond gives rise to a feature called a crosspeak at the chemical shift coordinates of the two nuclei for each non-proline residue. For structured proteins such as ubiquitin, the crosspeaks occur over a range of ~7.5 to ~10 ppm in the proton dimension (Figure 2, top). For α-synuclein, a known disordered protein, and for CAHS proteins, the crosspeaks occur over a narrower window (Figure 2, top), from ~8.0 to ~8.6 ppm, which coincides with the range for amide protons in the central residue of unstructured tripeptides (Schwarzinger et al., 2000). To further test our conclusion that these proteins are disordered, we assessed backbone proton-deuterium exchange. Amide protons in tripeptides exchange with deuterons from D2O in seconds (Bai et al., 1993) but are protected in the interior of stable globular proteins for days to weeks (Englander and Kallenbach, 1983). After acquiring the HSQC spectra (Figure 2), we removed two aliquots from each sample. One aliquot was diluted 10-fold with H2O, and the other was diluted 10-fold with D2O. For the disordered proteins tested (α-synuclein and CAHS proteins), nearly all the amide protons were exchanged for deuterons within 20 min, as shown by the decrease in intensity of the one-dimensional proton spectrum (Figure 2). In contrast, little exchange was observed for the structured protein ubiquitin in 20 min (Figure 2). These data, combined with heat solubility, circular dichroism, and bioinformatics approaches, show that many, if not all, tardigrade CAHS, and likely MAHS and SAHS proteins, are disordered. We refer to these families (CAHS, SAHS, MAHS) of tardigrade-specific IDPs as TDPs to distinguish them from other IDPs, because, at the sequence level, no homologs of TDPs are found outside the phylum tardigrade (Tanaka et al., 2015; Yamaguchi et al., 2012).
Figure 2. Tardigrade Cytosolic Abundant Heat Soluble Proteins Are Intrinsically Disordered.
Top: two-dimensional 15N-1H HSQC spectra of ubiquitin (a globular protein), α-synuclein (a known disordered protein), and tardigrade CAHS proteins in 90:10 (vol/vol) H2O:D2O 50 mM sodium phosphate (pH 7.0). Bottom: after the spectra were acquired, two aliquots were diluted 10-fold with either buffered 90:10 (vol/vol) H2O:D2O or buffered D2O and one-dimensional proton spectra acquired 20 min later.
Several families of IDPs, such as late embryogenesis abundant (LEA) proteins and hydrophilins, have known or suspected roles in stress tolerance in organisms spanning all kingdoms of life (Chakrabortee et al., 2012; Dang et al., 2014; Garay-Arroyo et al., 2000), and a recent study demonstrated slightly improved hyperosmotic tolerance in cells heterologously expressing an MAHS protein (Tanaka et al., 2015). These observations, coupled with the fact that TDPs are induced by drying in H. dujardini, suggests that they play a role in tardigrade stress tolerance (Yamaguchi et al., 2012; Tanaka et al., 2015). However, until now no studies have been conducted to directly examine the effect of environmental conditions on the expression of genes encoding TDPs or their involvement in tardigrade desiccation tolerance.
Constitutive Expression or Enrichment of TDPs during Desiccation Is Conserved among Eutardigrades
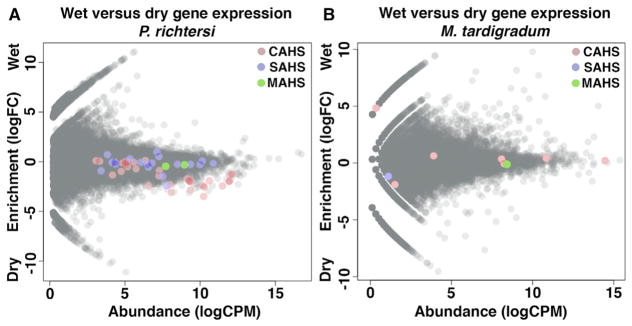
We hypothesized that high levels of TDP transcripts in drying H. dujardini are a characteristic of desiccation-tolerant tardigrades more generally. To test this hypothesis, we sequenced hydrated and dry transcriptomes from a second desiccation-tolerant tardigrade species, Paramacrobiotus richtersi, which also cannot tolerate rapid drying (Wright, 1989). The results recapitulated our H. dujardini observations, with 20 of 31 CAHS transcripts, 2 of 19 SAHS transcripts, and 0 of 2 MAHS transcripts enriched in dry P. richtersi (Figure 3A).
Figure 3. Constitutive Expression and Enrichment of TDPs during Desiccation Is Conserved among Eutardigrade Species.
(A and B) MA plots for P. richtersi (A) and M. tardigradum (B) showing enrichment (log2 fold change) versus abundance (log2 CPM) of expressed genes under hydrated and dry conditions. Colored circles indicate CAHS (red), SAHS (blue), and MAHS (green) genes encoding tardigrade-specific IDPs.
To test if the extent to which a tardigrade species requires preconditioning mirrors the induction of TDPs upon desiccation, we assembled and analyzed the transcriptome (from publically available short reads) of a third tardigrade species, Milnesium tardigradum (Figure 1A), which requires much less preconditioning (Wright, 1989). M. tardigradum did not significantly enrich expression of any TDPs during desiccation (Figure 3B). However, several CAHS transcripts were expressed at constitutively high levels (Figure 3B). For example, one CAHS transcript was the third most abundant transcript identified (Figure 3B).
Taken together, these data demonstrate that the expression level of TDPs in different tardigrade species mirrors the degree to which that species requires preconditioning. In species requiring extensive preconditioning (H. dujardini and P. richtersi), many TDPs are upregulated upon desiccation, while in a tardigrade requiring relatively little preconditioning (M. tardigradum), these genes do not respond to drying but are constitutively expressed at high levels.
Tardigrade-Specific IDPs Are Required for Desiccation Tolerance
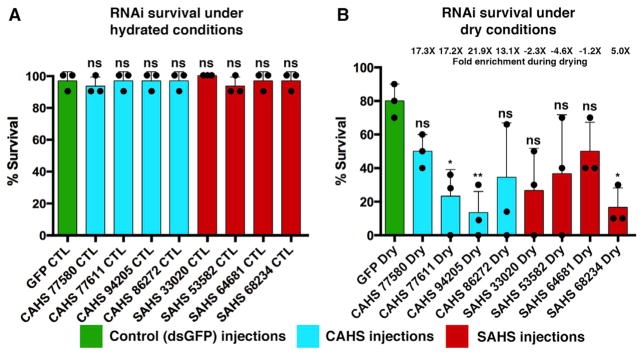
To test if TDPs are required for tardigrades to survive desiccation, we performed RNAi (Tenlen et al., 2013) to disrupt the function of specific genes. We targeted both highly induced (CAHSs and SAHSs) and constitutively active (SAHSs) TDPs and tested the ability of H. dujardini to survive under control (hydrated) and dry conditions. For all treatments, under hydrated conditions there were no significant decreases in survival (Figure 4A). However, targeting two of four highly induced (13- to 22-fold) CAHS genes significantly (p < 0.01) reduced survival after desiccation compared with a control treatment, GFP RNAi (Figure 4B). Additionally, RNAi targeting of an induced (5-fold) SAHS gene resulted in a significant (p < 0.01) decrease in survival after desiccation compared with the GFP RNAi controls (Figure 4B). These results demonstrate that some TDPs expressed at high levels in drying tardigrades are also essential for tardigrades to survive desiccation.
Figure 4. TDPs Are Essential for Efficient Survival of Desiccation.
(A and B) Survival after RNAi injection targeting GFP (control), CAHS, or SAHS transcripts in (A) hydrated and (B) dry Hypsibius dujardini specimens. Dots represent individual trials. N = 10 for each individual trial (30 total). t test: ns, not significant; *p < 0.01; **p < 0.001. RNA abundance fold change values given above each bar (e.g., 17×) indicate the increase in abundance in dry relative to hydrated conditions. Error bars, SD.
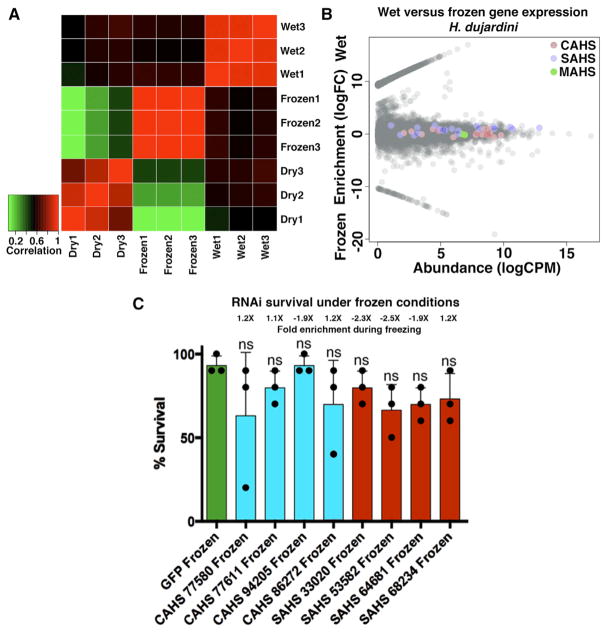
It has been suggested that tardigrades may have first evolved the ability to survive drying and acquired resistances to other stresses (cross-tolerance) as a byproduct of desiccation tolerance (Jönsson, 2003). Cross-tolerance in other systems has been studied, but there is no consensus despite multiple studies in a variety organisms (Levis et al., 2012; Tammariello et al., 1999). This makes extending conclusions from other organisms to tardigrades difficult. If tardigrades do possess cross-tolerance, one would anticipate that different forms of stress would induce similar changes in gene expression (Sinclair et al., 2013). To test this idea, we sequenced transcriptomes of gradually frozen H. dujardini specimens and compared changes in gene expression induced by freezing with those induced by drying. Changes in expression under these stress conditions were divergent, with gene expression in either stress condition (frozen or dry) being more similar to control conditions (hydrated) than to the other stress condition (Figure 5A). Additionally, only 2 of 17 CAHS transcripts were enriched during freezing (as opposed to 11 of 17 under drying conditions), and these genes were expressed at relatively low levels and underwent small changes in expression (Figure 5B). No SAHS or MAHS transcripts were enriched during freezing in H. dujardini (Figure 5B). Interestingly, none of our CAHS or SAHS RNAi treatments significantly decreased survival of frozen tardigrades relative to double-stranded GFP RNAi controls (Figure 5C). Our RNAi results, coupled with the observed divergence between frozen and drying transcriptomes, suggest that different stresses are less mechanistically linked than previously suspected.
Figure 5. Divergence in H. dujardini’s Response to Drying and Freezing.
(A) Heatmap showing correlation between expression profiles of transcriptomes derived from dry, frozen, and hydrated H. dujardini specimens.
(B) MA plots showing the enrichment (log2 fold change) versus abundance (log2 CPM) of transcripts under control (hydrated) and frozen conditions in H. dujardini. Colored circles represent CAHS (red), SAHS (blue), and MAHS (green) TDPs.
(C) Survival under frozen conditions of H. dujardini specimens injected with RNAi constructs targeting control (green), CAHS (blue), and SAHS (red) genes. Dots represent individual trials with n = 10 for each individual trial (30 total). t test: ns = not significant. RNA abundance fold change values given above each bar (e.g., 1.2×) indicate the increase in abundance of that transcript in frozen relative to hydrated conditions.
Error bars, SD.
Tardigrade-Specific IDPs Increase Desiccation Tolerance in Heterologous Systems and In Vitro
A long-term goal of many projects studying cryptobiotic organisms is the identification of proteins and molecules that can protect or stabilize sensitive biological material. Previous studies have identified desiccation-related IDPs, such as LEA proteins, as sufficient protectants in both in vivo and in vitro systems (Dang et al., 2014; Goyal et al., 2005). To test if TDPs might be good protectants, we assessed their ability to increase the desiccation tolerance of other systems by quantifying the desiccation tolerance (percent survival) of yeast and bacteria engineered to exogenously express CAHS proteins (Figure 6). In cases in which heterologously expressed genes were found to slow growth, these genes were excluded from our analysis, as slowed growth correlates with increased desiccation tolerance. Several CAHS proteins were sufficient to increase the desiccation tolerance of yeast nearly 100-fold (Figure 6A). Similar results were obtained in bacteria, with exogenous expression of some CAHS proteins resulting in increases of more than 2 orders of magnitude in desiccation tolerance (Figure 6B). Importantly, α-synuclein, a protein that exists as a disordered monomer in cells (Fauvet et al., 2012; Theillet et al., 2016) and has no known connection to stress tolerance (Drescher et al., 2012; Theillet et al., 2014), did not increase survival under drying conditions (Figure 6B), demonstrating that something beyond intrinsic disorder of TDPs is essential for their protective capabilities.
Figure 6. CAHS Proteins Are Sufficient to Increase Desiccation Tolerance in Cells and Protect Proteins In Vitro.
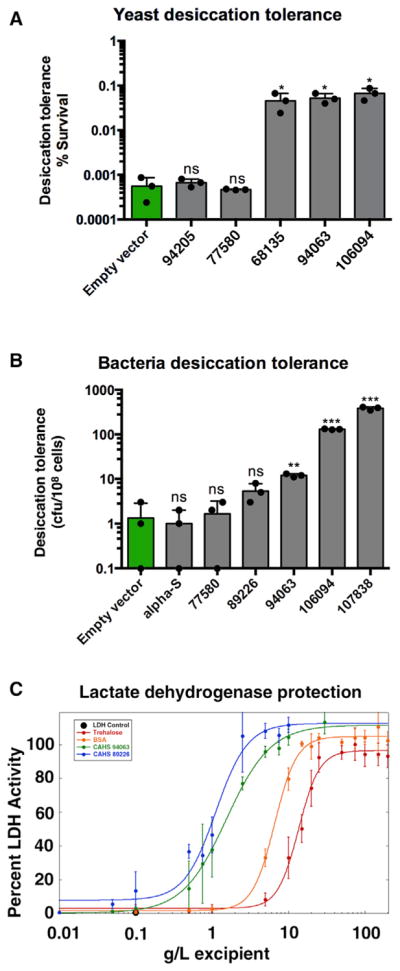
(A) Desiccation tolerance (percent survival) of yeast expressing CAHS genes.
(B) Desiccation tolerance (number of colony forming units/108 cells) of E. coli BL21 bacteria expressing CAHS or control (α-synuclein) IDPs. Dots represent individual trials. t test: ns, not significant; *p < 0.01; **p < 0.001; ***p < 0.0001.
(C) Lactate dehydrogenase enzyme (LDH) was dehydrated and rehydrated in the presence of tardigrade CAHS proteins and known excipients trehalose and BSA and its activity assessed. Experiments were performed in triplicate.
Error bars, SD.
Similarly, we tested the ability of purified CAHS proteins to preserve the activity of the enzyme lactate dehydrogenase (LDH) subjected to desiccation and rehydration. When desiccated and rehydrated by itself, the activity of LDH is reduced to <1% (Figure 6C). However, when mixed with CAHS protein prior to desiccation, LDH activity was preserved as a function of increasing CAHS protein concentration (Figure 6C). Importantly, our experiments show that at a sufficient concentration CAHS proteins (Figure 6C) preserve 100% of LDH activity and do so up to an order of magnitude better than trehalose and BSA (the bovine homolog of U.S. Food and Drug Administration-approved excipient human serum albumin).
Tardigrade CAHS Proteins Vitrify upon Desiccation
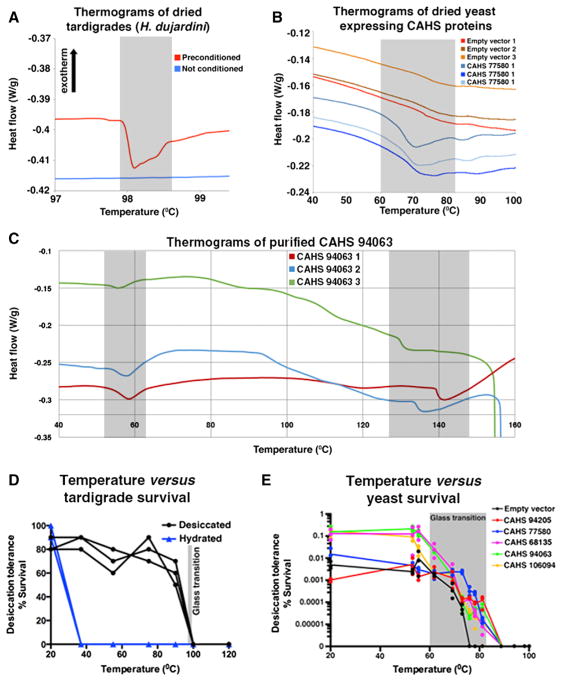
In organisms that use trehalose to survive desiccation, there are two competing, but not mutually exclusive, hypotheses about the mechanism of protection (Crowe, 2002; Crowe et al., 1987, 1988; Green and Angell, 1989; Levine and Slade, 1992; Sakurai et al., 2008). The vitrification hypothesis suggests that trehalose forms a glass-like matrix within cells, physically preventing protein denaturation, protein aggregation, and membrane fusion. The water replacement hypothesis posits that hydrogen bonds between water and cellular components are replaced by trehalose as cells dry, which would also prevent protein denaturation, aggregation, and membrane fusion. It has been suggested that like trehalose, highly hydrophilic molecules (such as IDPs), may play a role in either or both mechanisms (Sakurai et al., 2008). Tardigrades are known to vitrify upon desiccation (Hengherr et al., 2009) (Figure 7A), and the absence, or at best low levels, of trehalose in tardigrades suggests that other molecule(s) are likely responsible for this vitrified state.
Figure 7. Vitrification of Tardigrade CAHS Proteins.
(A) Differential scanning calorimetry (DSC) thermograms comparing preconditioned (slowly dried) and non-conditioned (quickly dried) tardigrades.
(B) DSC thermograms showing a novel glass transition in yeast induced by the expression of a CAHS protein.
(C) DSC thermograms of a purified dried CAHS protein showing glass transitions.
(D and E) The survival of dry tardigrades (D) and yeast expressing CAHS proteins (E) was assessed below, at, and above their glass transition temperatures.
Gray boxes denote range of glass transitions.
TDPs are prime candidates for mediators of vitrification and water replacement in tardigrades, because they are highly hydrophilic, abundantly expressed, and essential as well as sufficient for desiccation tolerance. Therefore, we tested the ability of CAHS proteins to vitrify using differential scanning calorimetry and found that when dried, CAHS proteins vitrify in vitro and when heterologously expressed in yeast (Figures 7B, 7C, and S2). It should be noted that the glass transition temperatures of dried H. dujardini specimens, yeast expressing tardigrade IDPs, and purified IDPs differ significantly. This is to be expected, as the glass transition temperature of a material is affected by cosolutes and humidity (Kasapis et al., 2003; Sakurai et al., 2008), and it is reasonable to assume that the contents of a tardigrade cell differ from those in yeast and thus would affect the biophysical properties of a vitrified material differently. Our data show that CAHS proteins, like trehalose, vitrify when dried and suggest that tardigrades may rely on the vitrification of these highly abundant proteins much in the same way that other desiccation-tolerant organisms rely on trehalose.
The vitreous state of a material can be disrupted by heating it up past its glass transition temperature (similar to heating a crystalline solid past its melting temperature), the temperature at which a vitrified solid transitions to a rubbery state. The vitrified state of dried tardigrades correlates with their viability (Hengherr et al., 2009), which we confirmed for H. dujardini (Figure 7D). To test specifically if the vitrified state of CAHS proteins mirrors their protective capabilities, we heated desiccated yeast expressing CAHS proteins, below, within, and above their glass transition temperatures. We observed that dry yeast with CAHS proteins tolerated temperatures ~10°C higher than wild-type yeast (Figure 7E). Furthermore, survival of CAHS expressing yeast was largely unaffected at temperatures below their CAHS-induced glass transition temperature, while survival declined sharply at temperatures within the glass transition range, and no yeast survived temperatures beyond the glass transition range (Figure 7E). Thus, as with trehalose, the protective capabilities of tardigrade CAHS proteins mirror their glass transition temperatures, with specimens displaying robust survival up to, but not beyond the point at which vitrification is disrupted.
In summary, we have demonstrated that tardigrades express many TDPs in response to drying and/or constitutively express TDPs at high levels. The level of TDP enrichment during drying mirrors different tardigrade species’ requirement for preconditioning (slow drying) to survive desiccation, with species that require less preconditioning expressing TDPs constitutively at high levels. We find that several TDPs contributed functionally to H. dujardini’s ability to robustly survive desiccation. Additionally, this study shows that in tardigrades, changes in gene expression induced by different stress conditions are more divergent than suspected. Our study demonstrates that exogenous expression of CAHS proteins in both prokaryotic and eukaryotic cells is sufficient to increase desiccation tolerance in these systems. TDPs represent, to our knowledge, the first functional mediators of tardigrade desiccation tolerance to be identified. Our findings provide evidence that proteins, and IDPs in particular, may protect biological material through vitrification. The fact that trehalose and CAHS proteins appear to function by the same mechanism demonstrates the elegance of convergent evolution and the ability to arrive at similar biological mechanisms via vastly different means (a sugar versus a protein mediator). We speculate that vitrification protects desiccation-sensitive macromolecules by trapping them within the pores of an amorphous matrix that prevents their denaturation, aggregation, fusion, and fragmentation. Our mechanistic and functional data, coupled with the previous observations and speculation that denatured globular proteins and IDPs vitrify upon desiccation (Hoseney et al., 1986; Sochava and Smirnova, 1993), hints that the phenomenon of desiccation tolerance via vitrification of proteins may be more widespread than appreciated. More broadly, our study highlights the diversity and functional role of disordered proteins in mediating stress tolerance. We anticipate that this work will serve as a foundation for pursuing long-term goals of the desiccation tolerance field, including the engineering of drought-resistant crops and the stabilization of sensitive pharmaceuticals and cells in a dry state.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli BL21 (DE3) – CAHS 77580 | This study | NA |
| E. coli BL21 (DE3) – CAHS 89226 | This study | NA |
| E. coli BL21 (DE3) – CAHS 94063 | This study | NA |
| E. coli BL21 (DE3) – CAHS 106094 | This study | NA |
| E. coli BL21 (DE3) – CAHS 107838 | This study | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol | ThermoFisher | 15596026 |
| Roche cOmplete EDTA-free protease inhibitor tablet | Sigma-Aldrich | 4693159001 |
| Lactate dehydrogenase | Sigma-Aldrich | 10127230001 |
| BSA | Sigma-Aldrich | B4287-25G |
| D-Trehalose | Sigma-Aldrich | 6138-23-4 |
| NADH | Sigma-Aldrich | 10128023001 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | 74104 |
| TruSeq kit | Illumina | RS-122-2001 |
| MasterPure RNA purification kit | Epicenter | MCR85102 |
| Deposited Data | ||
| Assemblies generated in this study | NCBI BioProject database | PRJNA369152 |
| Raw RNaseq reads generated in this study | NCBI Search Read Archive | SRR5217912, SRR5217911, SRR5217848, SRR5217652, SRR5217467 |
| Experimental Models: Organisms/Strains | ||
| Hypsibius dujardini | Sciento | Z151 |
| Paramacrobiotus richtersi | Collected for study | N/A |
| S. cerevisiae CAHS 94205 | This study | N/A |
| S. cerevisiae CAHS 77580 | This study | N/A |
| S. cerevisiae CAHS 68135 | This study | N/A |
| S. cerevisiae CAHS 94063 | This study | N/A |
| S. cerevisiae CAHS 106094 | This study | N/A |
| Software and Algorithms | ||
| Trinity | Haas et al. (2013) | https://github.com/trinityrnaseq/trinityrnaseq/wiki |
| RSEM | Li and Dewey (2011) | http://deweylab.github.io/RSEM/ |
| edgeR | Robinson et al. (2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| CEGMA | Parra et al. (2007) | https://github.com/KorfLab/CEGMA_v2/ |
| Excel 2008 for Mac V12.3 | Microsoft | NA |
| Other | ||
| Tzero aluminum pans | Fisher | NC9553081 |
METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagent may be directed to the Lead Contact Thomas Boothby (tboothby@live.unc.edu; tboothby@gmail.com).
Experimental Model and Subject Details
Tardigrade species used in this study include H. dujardini and P. richtersi, and were obtained and cultured as described below.
Bacterial strains were all made from BL21 star (DE3) E. coli. The construction of bacterial strains is detailed below.
Yeast strains were derived from a MAT α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nth1::G418R can1::PTDH3-AGT1 strain. Details on strain construction are given below.
METHOD DETAILS
Tardigrade Culture and Collection
H. dujardini specimens were obtained originally from Sciento and were cultured in glass petri-dishes filled with spring water (Deer Park) and fed unicellular Chlorococcum sp. algae similar to (Gabriel et al., 2007). P. richtersi was extracted from hazel leaf litter collected at Formigine (Northern Italy; N 44°34.253′, E 10°50.892′, 80 m a.s.l.). Dry leaf litter was stored at −80°C until specimen collection. To isolate P. richtersi, leaf litter was sprinkled with tap water for 15 min, and then submerged in water for 30 min. Active P. richtersi specimens were then extracted by sieves (250 μm and 37 μm mesh) under running water, and animals were isolated via direct microscopic observation. M. tardigradum short reads were downloaded from NCBI (Accessions SRA: SRX426237-SRX426240).
H. dujardini RNA Extraction and Library Preparation
For RNaseq experiments three biological replicates were used for each condition: wet, drying, or frozen. To isolate RNA from desiccating specimens, 400 μL of Trizol was used to wash specimens from dishes into a 1.5 mL Eppendorf tube. For frozen and wet specimens, excess liquid was removed from pelleted animals and 400 μL of Trizol was added directly to the tubes. Plastic pestles were placed in tubes and the tubes dipped into liquid nitrogen. The frozen samples were ground with pestles and allowed to thaw. Five rounds of freeze-thaw homogenization were conducted. An additional 100 μL of Trizol was used to wash the pestles. Chloroform (100 μl) was mixed with each sample. Tubes were capped, shaken for 20 s, and allowed to sit at room temperature for 3 min. Samples were then centrifuged at 10,000g for 18 min at 4°C. The clear top layer was removed to a fresh tube and an equal volume of 100% ethanol was added. Samples were then processed using QIAGEN’s RNeasy Mini Kit (QIAGEN, Cat# 74104) according to manufacturer’s instructions. RNA samples were used for library construction using the Illumina mRNA TruSeq v2 kit.
P. richtersi RNA Extraction and Library Preparation
We isolated RNA from biological replicates of P. richtersi specimens (three wet replicates and two dry replicates) by methods to those used for H. dujardini. RNA was extracted using the Epicenter MasterPure RNA Purification kit (Cat# MCR85102). RNA samples were used for library construction using the Illumina mRNA TruSeq v2 kit.
Transcriptome Sequencing, Assembly and Differential Expression Analysis
RNaseq libraries were multiplexed and sequenced on the Illumina HighSeq 2000 platform. Raw transcriptome reads for M. tardigradum were obtained from NCBI’s SRA database (Accessions SRA: SRX426237-SRX426240). Pooled reads (H. dujardini – wet + drying + frozen; P. richtersi – wet + dry; M. tardigradum - Accessions SRA: SRX426237-SRX426240) were used for de novo assembly of transcripts using the program Trinity (Haas et al., 2013). Read mapping was performed for each RNaseq library using RSEM (Li and Dewey, 2011) against the appropriate reference transcriptome, followed by normalization. For M. tardigradum, differential expression analysis was performed comparing active (SRA: SRX426237) and inactive (SRA: SRX426238) read counts. For H. dujardini and P. richtersi a transcript/gene was considered ‘expressed’ if it had a sum across all sequencing libraries of mapped read counts of 100 or more. Mapped read counts were used to perform differential expression for expressed genes using the program edgeR (Robinson et al., 2010). A transcript was deemed differentially expressed (enriched) if it had both a p value and a false discovery rate of < 0.05.
Protein Expression and Purification
E. coli codon optimized gBlocks encoding tardigrade CAHS proteins were synthesized (Integrated DNA Technologies) and cloned into the pET28b expression vector. BL21 star (DE3) E. coli were transformed with pET28b + CAHS plasmids.
A single bacterial colony was used to inoculate 10 mL of Lennox broth (LB, 10 g/L, tryptone, 5 g/L yeast extract, 5 g/L NaCl) supplemented with 60 μg/mL of kanamycin. The culture was shaken at 37°C overnight (New Brunswick Scientific Innova I26, 225 rpm). Three of these cultures were used to inoculate 1 L of supplemented M9 media (50 mM Na2HPO4, 20 mM KH2PO4, 9 mM NaCl, 4 g/L glucose, 1 g/L 15NH4Cl, 0.1 mM CaCl2 2 mM MgSO4, 10 mg/L thiamine, 10 mg/L biotin, and 60 μg/mL of kanamycin).
The 1 L cultures were shaken at 37°C until the optical density at 600 nm reached 0.5. IPTG (1 mM final concentration) was then added to induce expression. After 4 hr, the cells were pelleted at 1,000g at 10°C for 30 min. The cell pellets were stored at −20°C. Pellets were resuspended in 12.5 mL of 50 mM HEPES, 50 mM NaCl (pH 8.0) supplemented with half a Roche cOmplete EDTA-free protease inhibitor tablet (Sigma-Aldrich Cat. #4693159001). Cells were then lysed by heat shock at 95°C for 15 min. Lysates were cooled at room temperature for 30 min. Insoluble components were removed by centrifugation at 20,000g and 10°C for 30 min.
MgCl2 (final concentration 2 mM) was added to the heat soluble fraction before digestion with 1250 units of Benzonase (Sigma-Aldrich) at 37°C for 1 hr. Benzonase was then inactivated by heating to 95°C. After cooling to room temperature, the lysate was sterile filtered using a 0.45 μm filter and transferred to 10,000 MWCO dialysis tubing. Samples were dialyzed against 50 mM sodium phosphate (pH 7.0) overnight followed by dialysis against three changes of 17 MΩ cm−1 H2O for at least 3 hr each. The dialysate was again filtered before being flash frozen in CO2(s)/ethanol and lyophilized for 48 hr (Labconco FreeZone). Purity was determined by SDS-PAGE, DNA electrophoresis, and an ethidium bromide fluorescence assay. Alpha-synuclein and ubiquitin were purified according to Smith et al. (2015) and Wang et al. (2012), respectively.
NMR
Purified CAHS proteins were dissolved at 10 g/L in 50 mM sodium phosphate (pH 7.0), 90:10 (vol/vol) H2O:D2O by boiling and then centrifuged at 14,000g for 10 min to remove undissolved material. 15N-1H HSQC spectra were acquired at 298 K on an 850 MHz Bruker Avance III spectrometer equipped with a TCI cryoprobe. Sweep widths were 11,000 Hz and 3,500 Hz in the 1H and 15N dimensions, respectively. Each spectrum comprised 256 increments of 24 scans per increment. One-dimensional spectra were taken 20 min after sample preparation using a 1H sweep width of 13,500 Hz and comprised 128 scans. Each pair of H2O/D2O spectra was normalized using the methyl resonances at 0.8 ppm.
Purified ubiquitin (2 mM) was resuspended in 50 mM sodium phosphate (pH 7.0), 95:5 (vol/vol) H2O:D2O and centrifuged at 20,000g for 5 min to remove undissolved material. 15N-1H HSQC spectra were acquired at 298 K on the 850 MHz spectrometer. Sweep widths were 14,000 Hzand3,500 Hzin the 1H and 15N dimensions, respectively. Each spectrum comprised 256 increments of 4 scans per increment. One-dimensional spectra were taken 20 min after sample preparation using a 1H sweep width of 14,000 Hz and comprised 128 scans. Each one-dimensional spectrum was normalized using the methyl resonance at −0.15 ppm, and all spectra are referenced to DSS.
Purified α-synuclein (0.1 mM) was resuspended in 50 mM sodium phosphate (pH 7.0), 95:5 (vol/vol) H2O:D2O and centrifuged at 20,000g for 5 min to remove undissolved material. 15N-1H HSQC spectra were acquired at 298 K on the 850 MHz spectrometer. Sweep widths were 14,000 Hz and 3,500 Hz in the 1H and 15N dimensions, respectively. Each spectrum comprised 256 increments of 4 scans per increment. One-dimensional spectra were taken 20 min after sample preparation using a 1H sweep width of 14,000 Hz and comprised 128 scans. Each one dimensional spectrum was normalized using the methyl resonance at 1 ppm, and all spectra are referenced to DSS.
Trehalose Phosphatase Identification
In animals the final step in trehalose production is mediated by the enzyme trehalose phosphatase (EC 3.1.3.12). C. elegans tps-1&2 (trehalose-6-phosphates) as well as gob-1 (trehalose phosphatase) and the D. melanogaster Tps1 (a trehalose-6-phosphate/trehalose phosphatase fusion) protein sequences were obtained from http://www.wormbase.org/#012-34-5 and http://www.flybase.org, respectively, and used as query sequences for BLAST analysis against a database derived from our transcriptome assembly with an E-value cutoff of 1E-10. No transcript (from our or the Yanai lab’s transcriptome (Levin et al., 2016)) or predicted sequences from genome assemblies (Bemm et al., 2016; Boothby et al., 2015; Koutsovoulos et al., 2016) aligned to gob-1 with an E-value less than or equal to 1E-10. Several H. dujardini predicted protein sequences aligned to tps-1, tps-2, Tps1 – but these sequences were not expressed in either hydrated or desiccated specimens and lacked a conserved trehalose phosphatase and HAD domain, which are found in both C. elegans and D. melanogaster trehalose phosphatases. Additionally, these sequences reciprocally BLAST to C. elegans tps genes not gob-1. Together, the detection of low levels of trehalose (at best) in some tardigrade species, coupled with the lack of expression of genes involved in trehalose anabolism and the absence of trehalose phosphatases suggests that H. dujardini does not rely on trehalose to survive desiccation.
CEGMA Analysis
The percent of core eukaryotic genes contained within our assembly was determined as a means of assessing the completeness of the transcriptome using CEGMA (Parra et al., 2007) with default settings.
Identification of TDP-encoding Transcripts
Transcript sequences were used as BLASTx queries and searched against NCBI’s non-redundant protein database. Reciprocal best BLAST was performed with an E-value cutoff of 1E-10.
RNA Interference
Double stranded RNA (dsRNA) was made and microinjections performed with slight modification of a published protocol (Tenlen et al., 2013). dsRNAs were diluted to a concentration of 1 μg/μl in nuclease-free water. Specimens were not sedated with levamisole as previously described (Tenlen et al., 2013) to reduce the number of factors potentially influencing survival. Injected specimens were transferred to 30 mm plastic dishes filled with fresh spring water and left overnight. The next day, specimens were either left in spring water with fresh food added (control), desiccated, or frozen. For each RNAi treatment and stress condition three individual trials were performed, with ten tardigrades injected per trial.
H. dujardini Desiccation
After injection (RNAi studies) or directly from larger cultures used for RNaseq, H. dujardini specimens were transferred to 35 mm plastic petri dishes filled with fresh spring water without algal food. Specimens were starved for 24 hr. Melted 2% agar (300 μl) was used to evenly coat the lid of 35 mm dishes and excess agar removed. After solidification, tardigrades were transferred to the center of coated lids. Using a mouth pipette, excess water was removed and lids were placed in humidified chambers. The relative humidity (95% for slow drying and 70% for quick drying) of each chamber was established using a mixture of glycerol and water (Forney and Brandl, 1992) and monitored using a hygrometer. Tardigrades were dried overnight, enough time for tun formation to occur, and then removed and exposed to laboratory conditions (~35% relative humidity) for 24 hr to allow for further desiccation. Rehydration was achieved by pipetting 1.5 mL of spring water into dishes. Rehydrated samples were left for 2 hr before observation and quantification of survival. Coordinated movement was used to score survival.
P. richtersi Desiccation
P. richtersi specimens were desiccated by placing each group of animals on a Whatman filter paper (25 mm2 or 1 cm2) with mineral water (9 μl or 30 μl, respectively) and exposing them initially to 80% relative humidity (RH) and 18°C for 4 hr, then to 50% RH at 18°C for 4 hr in a climatically controlled chamber, and finally to 0%–3% RH at room temperature for 12 hr (Altiero et al., 2011). At the end of this treatment animals exhibit the typical tun shape.
H. dujardini Freezing
After injection (RNAi studies) or directly from larger cultures (RNaseq), H. dujardini specimens were transferred to 35 mm plastic petri dishes filled with fresh spring water without algal food. Specimens were starved for 24 hr. Specimens were then transferred to 1.5 mL microcentrifuge tubes, and the volume of spring water adjusted to 1 mL. The tubes were centrifuged briefly to move specimens to the bottom and then placed in a styrofoam box at −80°C for 24 hr. For RNAi studies, thawing was achieved by moving tubes to ambient laboratory conditions (~20°C) for 2 hr. Following thawing the contents of each tube were transferred to a new 35 mm dish for observation and quantification of survival. Coordinated movement was used to score for survival. For RNaseq, thawing was accelerated by warming the specimens by hand and then rapidly moving on to RNA extraction.
Bacterial Heterologous Expression and Desiccation Survival Assay
Cloning and transformation of bacteria was performed as described above. For expression, 10 mL cultures were grown overnight. The following day an aliquot of overnight culture was added to fresh culture media at a ratio of 1:200. Cultures were grown to log phase (OD600 0.4–0.8). Expression of CAHS genes was then induced with 1 mM IPTG and the cultures grown for an additional 4 hr. Optical densities were measured again and approximately 108 cells were transferred to 1.5 mL microcentrifuge tubes and spun at 4,000g for 20 min. Excess culture media was removed, and cells were washed with water and re-pelleted. Water was quickly removed with a pipette and pellets were dried overnight in a SpeedVac (Savant SpeedVac SC100). The tubes, caps open, were transferred to a sealed desiccator filled with Drierite (Sigma-Aldrich, Cat. #238937) for 1 week.
Rehydration and pellet dispersal was achieved by adding 1 mL of culture media to dry pellets and vortexing for 10 min. Cells were then transferred to kanamycin plates and grown overnight at 37°C. The following day colonies were counted and survival reported as colony forming units/108 cells plated.
Yeast Heterologous Expression and Desiccation Survival Assay
The strain MAT α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nth1::G418R can1::PTDH3-AGT1 was used. This strain is a haploid alpha strain, with the nth1 trehalase gene deleted and replaced with G418 and with the AGT1 trehalose transporter under a constitutive highly expressed TDH3 promoter.
Tardigrade CAHS coding sequences were cloned into the p413-GPD plasmid. Tardigrade genes were under the same TDH3 promoter on CEN plasmids, with histidine selection.
Standard yeast propagation and transformation procedures were used. Strains were grown in selective, synthetic complete, media (2% glucose without histidine). Cultures were grown to logarithmic phase from an overnight culture by incubation overnight at 30°C. Cultures were re-diluted to an OD600 of ~0.05 and allowed to reach mid-log phase (OD600 0.4 – 0.6).
Desiccation tolerance assays were performed as follows. Approximately 107 cells were withdrawn from liquid cultures, washed twice in water and brought to a final volume of 1 ml. Undesiccated controls were plated for colony counting. Aliquots (200 μl) were then transferred to a 96-well tissue culture plate (Becton Dickinson, 353075), centrifuged and most of the water removed without disturbing the cell pellet. Cells were desiccated in a 23°C incubator with a constant 60% RH, with the lid raised, for 48 hr. Samples were resuspended in water and plated for colony counting.
Lactate Dehydrogenase Enzyme Activity Assay
Assays for lactate dehydrogenase (LDH) activity were largely based on the procedure outlined by Goyal et al. (2005). LDH from rabbit muscle (Roche) was diluted to 0.1 g/L in 100 μL of 25 mM Tris/HCl (pH 7) containing various concentrations of CAHS D, CAHS G, BSA (Sigma), or D-trehalose (Aldrich Chemical Company). Half of each sample was stored at 4°C while the other half was dehydrated in an EZ-2 Personal Evaporator (Genevac) for 16 hr on the aqueous setting with the heat lamp off. Dehydrated samples were re-dissolved in 250 μL of water, control samples were diluted with 200 μL of water, and all samples were kept on ice until their activity was measured. To determine the enzyme activity of LDH, 10 μL of LDH/excipient solution was added to 990 μL of 100mM sodium phosphate (pH 6), 100 μM NADH and 2mM pyruvate. The absorbance at 340 nm was measured every 0.1 s for 1 min in a Cary Series UV-Vis Spectrophotometer (Agilent Technologies). The percent activity was determined by comparing the initial, linear reaction rate of each dehydrated and rehydrated sample compared to its corresponding unstressed control. Experiments were performed in triplicate. For LDH assays, protein expression was performed as described above, except that 10 mL of overnight cultures were used to inoculate 1 L of LB.
Differential Scanning Calorimetry
For dried tardigrades, CAHS proteins, and modified yeast strains, samples (6–9 mg) were hermetically sealed in Tzero aluminum pans (TA Instruments) and accessed using a Discovery Series differential scanning calorimeter (TA Instruments). Samples were equilibrated at −20°C, and then ramped to 250°C at a rate of 5°C/min. The temperature profile was cycled once.
Yeast Glass Transition Survival Assay
Samples were prepared and analyzed as described for desiccation tolerance assays (above). 200 μL aliquots were transferred to 48-well PCR plates and pelleted at 4,000 rpm for 4 min in a table-top centrifuge. Supernatant was aspirated and the PCR plates were placed into a vacuum desiccator (Centrivap Concentrator; Labconco, Kansas City, MO) connected to a rotary vane vacuum pump (Labconco) producing 135 kPa of vacuum O/N. After desiccation the samples were placed in a thermal cycler (Biorad, C1000) and heat shocked at temperatures ranging from 52°–100°C for 30 min.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differential Expression Analysis
Differential expression analysis was performed using edgeR (Robinson et al., 2010) and the script run_DE_analysis.pl provided with Trinity (Haas et al., 2013). Expressed transcripts (≥100 raw read counts) were considered differentially expressed if they had a p value ≤ 0.05 and false discovery rate ≤ 0.05.
Tardigrade, Yeast, and Bacterial Survival
For tardigrade, yeast, and bacterial survival assays 3 trials performed, unless otherwise indicated. For tardigrades each trial used 10 animals (unless indicated). For bacteria, approximately 108 cells were used per trial and colony forming units scored. For yeast, approximately 107 cells were used per trial and colony forming units scored. Statistical analysis for survival was performed using Prism 6 software. t test were performed and bars show SD. Significance was defined as a p value ≤ 0.01.
LDH Activity Assay
LDH activity assays were performed in triplicate. Error bars show SD.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNaseq datasets reported in this study is NCBI BioProject: PRJNA369152. The accession numbers for the raw RNaseq reads reported in this paper are NCBI (Sequence Read Archive) SRA: SRR5217912, SRR5217911, SRR5217848, SRR5217652, and SRR5217467.
Supplementary Material
Highlights.
Tardigrade intrinsically disordered proteins (TDPs) are enriched during desiccation
TDPs are required for tardigrades to survive desiccation
Expression of TDPs increases desiccation tolerance in heterologous systems
TDPs vitrify, and this vitrified state mirrors their protective capabilities
Acknowledgments
This work was supported by NASA (NNX15AB44G to T.C.B.) and the National Science Foundation (MCB 1410854 and CHE 1607359 to G.J.P., IOS 1557432 and 1257320 to B.G.). L.R. and I.G. were supported by Young Researchers International Mobility of the University of Modena and Reggio Emilia and Fondo di Ateneo per la Ricerca (2015). We acknowledge the Harold and Leila Y. Mathers Charitable Foundation for supporting H.T. and the Simons Foundation of the Life Sciences Research Foundation for supporting T.C.B.
Footnotes
Supplemental Information includes two figures and two data files and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.02.018.
AUTHOR CONTRIBUTIONS
Conceptualization, (Lead) T.C.B. (supporting) B.G., G.J.P.; Investigation, T.C.B., H.T., A.H.B., S.P., A.E.S., I.G.; Resources, I.G., L.R., H.T., D.K.; Writing – Original Draft, T.C.B., S.P., G.J.P.; Writing – Review & Editing, T.C.B., H.T., A.H.B, S.P., A.E.S., I.G., L.R., G.J.P., D.K., B.G., Supervision, T.C.B., L.R., D.K., B.G., G.J.P.
References
- Altiero T, Guidetti R, Caselli V, Cesari M, Rebecchi L. Ultraviolet radiation tolerance in hydrated and desiccated eutardigrades. J Zoolog Syst Evol Res. 2011;49(Suppl 1):104–110. [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemm F, Weiß CL, Schultz J, Förster F. Genome of a tardigrade: Horizontal gene transfer or bacterial contamination? Proc Natl Acad Sci U S A. 2016;113:E3054–E3056. doi: 10.1073/pnas.1525116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby TC, Tenlen JR, Smith FW, Wang JR, Patanella KA, Nishimura EO, Tintori SC, Li Q, Jones CD, Yandell M, et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A. 2015;112:15976–15981. doi: 10.1073/pnas.1510461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Tripathi R, Watson M, Schierle GS, Kurniawan DP, Kaminski CF, Wise MJ, Tunnacliffe A. Intrinsically disordered proteins as molecular shields. Mol Biosyst. 2012;8:210–219. doi: 10.1039/c1mb05263b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe LM. Lessons from nature: the role of sugars in anhydrobiosis. Comp Biochem Physiol A Mol Integr Physiol. 2002;131:505–513. doi: 10.1016/s1095-6433(01)00503-7. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Stabilization of dry phospholipid bilayers and proteins by sugars. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Carpenter JF, Rudolph AS, Wistrom CA, Spargo BJ, Anchordoguy TJ. Interactions of sugars with membranes. Biochim Biophys Acta. 1988;947:367–384. doi: 10.1016/0304-4157(88)90015-9. [DOI] [PubMed] [Google Scholar]
- Dang NX, Popova AV, Hundertmark M, Hincha DK. Functional characterization of selected LEA proteins from Arabidopsis thaliana in yeast and in vitro. Planta. 2014;240:325–336. doi: 10.1007/s00425-014-2089-z. [DOI] [PubMed] [Google Scholar]
- Drescher M, Huber M, Subramaniam V. Hunting the chameleon: structural conformations of the intrinsically disordered protein alpha-synuclein. ChemBioChem. 2012;13:761–768. doi: 10.1002/cbic.201200059. [DOI] [PubMed] [Google Scholar]
- Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Erkut C, Penkov S, Khesbak H, Vorkel D, Verbavatz JM, Fahmy K, Kurzchalia TV. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol. 2011;21:1331–1336. doi: 10.1016/j.cub.2011.06.064. [DOI] [PubMed] [Google Scholar]
- Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, Tsika E, Coune P, Prudent M, Lion N, et al. α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem. 2012;287:15345–15364. doi: 10.1074/jbc.M111.318949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forney CF, Brandl DG. Control of Humidity in Small Controlled environment Chambers using Glycerol-Water Solutions. Horttechnology. 1992;2:52–54. [Google Scholar]
- Gabriel WN, McNuff R, Patel SK, Gregory TR, Jeck WR, Jones CD, Goldstein B. The tardigrade Hypsibius dujardini, a new model for studying the evolution of development. Dev Biol. 2007;312:545–559. doi: 10.1016/j.ydbio.2007.09.055. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275:5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Blaxter M. Tardigrades. Curr Biol. 2002;12:R475. doi: 10.1016/s0960-9822(02)00959-4. [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Angell CA. Phase relations and vitrification in saccharide-water solutions and the trehalose anomaly. J Phys Chem. 1989;93:2880–2882. [Google Scholar]
- Guidetti R, Altiero T, Rebecchi L. On dormancy strategies in tardigrades. J Insect Physiol. 2011;57:567–576. doi: 10.1016/j.jinsphys.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengherr S, Heyer AG, Köhler HR, Schill RO. Trehalose and anhydrobiosis in tardigrades—evidence for divergence in responses to dehydration. FEBS J. 2008;275:281–288. doi: 10.1111/j.1742-4658.2007.06198.x. [DOI] [PubMed] [Google Scholar]
- Hengherr S, Worland MR, Reuner A, Brümmer F, Schill RO. High-temperature tolerance in anhydrobiotic tardigrades is limited by glass transition. Physiol Biochem Zool. 2009;82:749–755. doi: 10.1086/605954. [DOI] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A. LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans. 2012;40:1000–1003. doi: 10.1042/BST20120109. [DOI] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6:431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- Hoseney RC, Zeleznak K, Lai CS. Wheat gluten-a glassy polymer. Cereal Chem. 1986;63:285–286. [Google Scholar]
- Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson KI. Causes and consequences of excess resistance in cryptobiotic metazoans. Physiol Biochem Zool. 2003;76:429–435. doi: 10.1086/377743. [DOI] [PubMed] [Google Scholar]
- Jönsson KI, Persson O. Trehalose in three species of desiccation tolerant tardigrades. Open Zool J. 2010;3:1–5. [Google Scholar]
- Kasapis S, Al-Marhoobi IM, Mitchell JR. Molecular weight effects on the glass transition of gelatin/cosolute mixtures. Biopolymers. 2003;70:169–185. doi: 10.1002/bip.10427. [DOI] [PubMed] [Google Scholar]
- Kondo K, Kubo T, Kunieda T. Suggested involvement of PP1/PP2A activity and de novo gene expression in anhydrobiotic survival in a tardigrade, Hypsibius dujardini, by chemical genetic approach. PLoS ONE. 2015;10:e0144803. doi: 10.1371/journal.pone.0144803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci U S A. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinski J, Tunnacliffe A. Anhydrobiosis without trehalose in bdelloid rotifers. FEBS Lett. 2003;553:387–390. doi: 10.1016/s0014-5793(03)01062-7. [DOI] [PubMed] [Google Scholar]
- Levin M, Anavy L, Cole AG, Winter E, Mostov N, Khair S, Senderovich N, Kovalev E, Silver DH, Feder M, et al. The mid-developmental transition and the evolution of animal body plans. Nature. 2016;531:637–641. doi: 10.1038/nature16994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine H, Slade L. Another view of trehalose for drying and stabilizing biological materials. BioPharm. 1992;5:36–40. [Google Scholar]
- Levis NA, Yi SX, Lee RE., Jr Mild desiccation rapidly increases freeze tolerance of the goldenrod gall fly, Eurosta solidaginis: evidence for drought-induced rapid cold-hardening. J Exp Biol. 2012;215:3768–3773. doi: 10.1242/jeb.076885. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali B, Grohme MA, Förster F, Dandekar T, Schnölzer M, Reuter D, Wełnicz W, Schill RO, Frohme M. Transcriptome survey of the anhydrobiotic tardigrade Milnesium tardigradum in comparison with Hypsibius dujardini and Richtersius coronifer. BMC Genomics. 2010;11:168. doi: 10.1186/1471-2164-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Furuki T, Akao K, Tanaka D, Nakahara Y, Kikawada T, Watanabe M, Okuda T. Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilum vanderplanki. Proc Natl Acad Sci U S A. 2008;105:5093–5098. doi: 10.1073/pnas.0706197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J Biomol NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- Sinclair BJ, Ferguson LV, Salehipour-shirazi G, MacMillan HA. Cross-tolerance and cross-talk in the cold: relating low temperatures to desiccation and immune stress in insects. Integr Comp Biol. 2013;53:545–556. doi: 10.1093/icb/ict004. [DOI] [PubMed] [Google Scholar]
- Smith AE, Zhou LZ, Pielak GJ. Hydrogen exchange of disordered proteins in Escherichia coli. Protein Sci. 2015;24:706–713. doi: 10.1002/pro.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochava IV, Smirnova OI. Heat capacity of hydrated and dehydrated globular proteins. Denaturation increment of heat capacity. Food Hydrocoll. 1993;6:513–524. [PubMed] [Google Scholar]
- Tammariello SP, Rinehart JP, Denlinger DL. Desiccation elicits heat shock protein transcription in the flesh fly, Sarcophaga crassipalpis, but does not enhance tolerance to high or low temperatures. J Insect Physiol. 1999;45:933–938. doi: 10.1016/s0022-1910(99)00073-6. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tanaka J, Miwa Y, Horikawa DD, Katayama T, Arakawa K, Toyoda A, Kubo T, Kunieda T. Novel mitochondria-targeted heat-soluble proteins identified in the anhydrobiotic Tardigrade improve osmotic tolerance of human cells. PLoS ONE. 2015;10:e0118272. doi: 10.1371/journal.pone.0118272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia H, Koshland DE. Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol. 2014;24:2758–2766. doi: 10.1016/j.cub.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Tenlen JR, McCaskill S, Goldstein B. RNA interference can be used to disrupt gene function in tardigrades. Dev Genes Evol. 2013;223:171–181. doi: 10.1007/s00427-012-0432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet FX, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, et al. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs) Chem Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet FX, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, Selenko P. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ. Macromolecular Crowding and Protein Stability. J Am Chem Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- Westh P, Ramløv H. Trehalose accumulation in the tardigrade Adorybiotus coronifer during anhydrobiosis. J Exp Zool. 1991;258:303–311. [Google Scholar]
- Wright JC. Desiccation tolerance and water-retentive mechanisms in tardigrades. J Exp Biol. 1989;142:267–292. [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1 Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Tanaka S, Yamaguchi S, Kuwahara H, Takamura C, Imajoh-Ohmi S, Horikawa DD, Toyoda A, Katayama T, Arakawa K, et al. Two novel heat-soluble protein families abundantly expressed in an anhydrobiotic tardigrade. PLoS ONE. 2012;7:e44209. doi: 10.1371/journal.pone.0044209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.