Abstract
Objective
To evaluate the microbial load and the inflammatory response in the distal and proximal parts of the cervical mucus plug.
Design
Experimental research.
Population
Twenty women with a normal, singleton pregnancy.
Sample
Vaginal swabs and specimens from the distal and proximal parts of the cervical mucus plug. Methods. Immunohistochemistry, enzyme-linked immunosorbent assay, quantitative polymerase chain reaction and histology.
Results
The total bacterial load (16S rDNA) was significantly lower in the cervical mucus plug compared with the vagina (p = 0.001). Among women harboring Ureaplasma parvum, the median genome equivalents/g were 1574 (interquartile range 2526) in the proximal part, 657 (interquartile range 1620) in the distal part and 60 240 (interquartile range 96 386) in the vagina. Histological examinations and quantitative polymerase chain reaction revealed considerable amounts of lactobacilli and inflammatory cells in both parts of the cervical mucus plug. The matrix metalloproteinase-8 concentration was decreased in the proximal part of the plug compared with the distal part (p = 0.08).
Conclusion
The cervical mucus plug inhibits, but does not block, the passage of Ureaplasma parvum during its ascending route from the vagina through the cervical canal.
Keywords: Cervical mucus plug, 16S rDNA quantitative polymerase chain reaction, Lactobacillus species, antimicrobial properties, intra-amniotic infection
Introduction
The cervical mucus plug (CMP) is a viscoelastic, gel-like structure that fills the cervical canal during pregnancy1. It exhibits intense antimicrobial activity against Staphylococcus saprophyticus, S. aureus, Escherichia coli, Pseudomonas aeruginosa, Enterococcus faecium, Streptococcus pyogenes and S. agalactiae, and is widely accepted as a key component of the innate immune response protecting against infections ascending from the vagina to the uterus2. Such ascending infections have been causally linked to spontaneous preterm birth and long-term complications in the newborn3.
Ureaplasma spp. are the bacteria most commonly identified in the amniotic fluid using culture or molecular microbiological techniques3–8. This is the case in patients with spontaneous preterm labor with intact membranes9, with preterm premature rupture of membranes10, and with a short cervix11. Furthermore, studies in monkeys (rhesus macaques) and sheep have shown that intrauterine inoculation of U. parvum as a single microorganism can induce preterm labor as well as a fetal inflammatory response syndrome12,13. However, the antimicrobial activity of the CMP against this microorganism remains to be studied.
If the CMP has intense antimicrobial activity, one would expect a bacterial gradient to be present through the plug, i.e. the bacterial load in the distal part (toward the vagina) of the CMP should exceed that of the proximal part (near the uterus and chorioamniotic membranes). This hypothesis is supported by previous studies of spontaneously shed, intact CMPs in which such a gradient has been described, but uncertainty about the true orientation of CMPs collected under these circumstances represents a challenge to the interpretation of these findings2. Furthermore, the gradient has not been studied with regard to Ureaplasma spp.
Lactobacillus spp. are the predominant bacteria in the healthy vaginal microbiome14, and protect the vagina against pathogenic microorganisms15. For this reason, it is interesting to explore whether Lactobacillus spp. are present in the CMP and are possibly contributing to an antimicrobial environment through a similar mechanism. Matrix metalloproteinase-8 (MMP-8), also known as neutrophil collagenase, is produced by neutrophils, and its presence in high concentrations may indicate an acute inflammatory process. High concentrations of MMP-8 in the cervical mucus have been considered as a sign of an intense, localized inflammation16–18. A possible difference in the MMP-8 concentrations between the proximal and distal parts of the CMP has not been studied.
The objective of this study was to assess the load of bacteria, in general, as well as those of Ureaplasma spp. and Lactobacillus spp., and the inflammatory response in the distal and proximal parts of the CMP.
Materials and methods
Participants for the study were recruited at the Department of Obstetrics and Gynecology at Aarhus University Hospital in Aarhus, Denmark. Seventeen women were recruited in late pregnancy before induction of labor (gestational age 38+0 to 42+0 weeks+days), and three women were recruited in the first trimester before termination of pregnancy (gestational age 7+6 to 9+0 weeks+days). The women recruited in late pregnancy were between 24 and 40 years of age, their parity ranged from 0 to 3, the length of the vaginal part of the cervix at specimen collection was from 1 to 3 cm (measured by the midwife during vaginal examination) and cervical external os dilatation was from closed to 2 cm. The first-trimester participants were between 27 and 39 years old and had a parity range from 0 to 2. Exclusion criteria were vaginal examination within the last 3 days, isolation of group B streptococcus from the vagina/urine, any use of antibiotics during the current pregnancy, diabetes, treatment with prostaglandins and cervical dilatation beyond 2 cm. The project was approved by the Central Denmark Region Committee on Biomedical Research Ethics (Project ID: 2005-0053) and informed consent was obtained from each participant.
One vaginal swab and two CMP specimens, one from the distal part and one from the proximal part of the plug, were obtained from each participant. Before collection of specimens the area around the external os was cleaned for any visible mucus or vaginal fluid. A vaginal swab was obtained from the posterior vaginal fornix. The distal specimen of the CMP was then obtained with a sterile 3.1-mm-thick catheter (Aspirette® Endocervical Aspirator; Cooper Surgical, Trumbull, CT, USA) inserted into the cervical canal (approximately 3 mm). The distal specimen of the CMP was aspirated into the catheter by pulling back the piston. The thick viscoelastic consistency of the CMP required placement of a needle holder on the catheter before withdrawal (Figure 1). The proximal specimen was aspirated 3–4 cm up the cervical canal by means of another catheter. The piston in that catheter ensured only minimal contamination during the passage through the distal part of the CMP. The specimens were stored in the catheters at −80 °C until analyzed. Because of the small size of the specimens, it was not possible to perform all analyses on every specimen. Twelve specimens were analyzed only by polymerase chain reaction (PCR); three specimens were analyzed by histology, immunohistochemistry and enzyme-linked immunosorbent assay (ELISA) for MMP, and five specimens underwent analyses for ELISA, PCR and histology and immunohistochemistry.
Figure 1.

The catheter used for specimen collection containing a cervical mucus plug specimen. The needle holder is placed on the catheter tip.
Specimens were fixed in 10% neutral formalin overnight and then paraffin-embedded. Each block was cut to generate 5-μm sections, which were dried at 56–60 °C for at least 30 min before staining. Each tissue was stained with hematoxylin & eosin and by the Gram method. Immunohistochemistry was conducted using antibodies against cytokeratin-7 (1:2000 dilution, mouse monoclonal; DAKO, Glostrup, Denmark), CK 5/6, CD 68, CD3 and CD20. Immunohistochemistry was performed using an automatic immunostainer (Ventana Discovery; Ventana Medical Systems, Tucson, AZ, USA). Microscopic examination included cell counts in 10 high-power fields (HPF) (×400) for each slide. If the specimens were too small to allow the examination of 10 HPF, as many HPF as possible were examined, and the total count was calculated equivalent to 10 HPF. Bacteria detected by Gram stain were analyzed with oil immersion at ×1000 to identify morphology.
Protein extraction and MMP-8 ELISA
This extraction procedure was specifically developed and validated for MMPs from the CMP16. Two mechanical extractions and one heat extraction were performed because the validation process demonstrated detectable amounts of MMP when analyzing the third extraction separately. The two mechanical extractions overnight were followed by a heat extraction because heating has been shown to result in extraction of considerably more collagenase from tissue homogenates of wounds compared with other extraction procedures19.
Specimens were weighed, and the amount of buffer needed to create a 1:30 dilution was calculated. Each specimen was manually homogenized with the calculated volume of extraction buffer (50 mM Tris–HCl, 10 mM CaCl2, 0.05% Brij 35 and 1 mM phenylmethyl-sulfonyl fluoride, pH 7.4). An aliquot from this 1:30 dilution was extracted by rotation at 4° C. This was followed by centrifugation (20 min at 16 000 g). The supernatant was pipetted and stored until the next day (first extraction). Buffer was added to the pellet, which was re-homogenized and re-extracted overnight, centrifuged and pipetted (second extraction). During the final extraction step, buffer was again added to the pellet, which was then heated for 4 min at 60° C, centrifuged and pipetted (third extraction). Supernatants from the three separate extraction procedures were pooled, and buffer was added to ensure a final dilution of 1:100 (total volume 2 mL). The specimens were stored at −80° C until analyzed. We used a commercially available ELISA kit to measure the concentrations of MMP-8 (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK, product number RPN2619). The specimens were analyzed in duplicate and measured in a 0.25–4 ng/mL range.
DNA extraction and quantitative PCR
Bacterial DNA was extracted using a bead-beating proto-col20 and subsequently with a Qiagen DNA Mini Kit, essentially as described by the manufacturer (QIAGEN GmbH, Hilden, Germany) from both vaginal swabs and CMP specimens. The CMP specimens were dissolved in 200 μL lysis buffer (30 mM Tris–HCl pH 8.0; 1 mM EDTA, 15 mg/mL lysozyme, 20 μg/ml proteinase K) and 200 μL AL buffer (supplied in the Qiagen DNA mini kit) to a final volume of 400 μL in an incubator at 56° C until the mucus was completely dissolved. The specimens were then transferred to a 2-mL tube containing 200 μL bead suspension (Zirkonia/Silica Beads 0.1 mm; Roth, Karlsruhe, Germany) in TE buffer (30 mM Tris–HCl pH 8.0; 1 mM EDTA). The specimens were homogenized at a speed setting 7000 for 70 s in a MagNALyzer (Roche, Hvidovre, Denmark), centrifuged at 30 000 g for 5 min to eliminate the foam, inverted carefully to re-suspend the beads, then spun for 15 s. Subsequent steps were carried out as described by the manufacturer.
The detection of microorganisms was performed using quantitative PCR (qPCR) designed to detect the 16S rDNA microbial gene21. Table 1 describes the primers and probes used for each organism. Standard curves were generated by analyzing 10-fold dilutions of genomic target DNA; for the 16S bacterial load assay, DNA from Legionella pneumophila was used for the standard curve. The qPCR for L. iners was a TaqMan™ probe-based assay, whereas the remaining Lactobacillus species were detected in SYBR-Green assays. Species of Ureaplasma were detected using a multiplex PCR assay able to identify both U. urealyticum and U. parvum in the same assay. An internal processing control was included in the Ureaplasma and Mycoplasma hominis assays to control for sample inhibition.
Table 1.
Primers and probes used for quantitative polymerase chain reaction.
| Target | Forward primer | Reverse primer | Probe |
|---|---|---|---|
| 16S rDNA | TCCTRCGGGAGGCWGCAGT | GGACTACCAGGGTATCTAATCCTGTT | FAM-CGTATTACCGCGGCTGCTGGCAC-BHQ1 |
| Ureaplasma urealyticum | GCAAGAAGACGTTTAGCTAGAGGTTT | CACGAGCAGATTCGATTAAGCAG | FAM-TAATTACTGACCACGTAGTGGA-MGB |
| Ureaplasma parvum | GCAAGAAGACGTTTAGCTAGAGGTTT | CGAGCAGATTGCATTAGGTCAG | VIC-TTTAATTACTGATCATGTAATGGA-MGB |
| Mycoplasma hominis | CATGCATGTCGAGCGAGGTT | CCATGCGGTTCCATGCGT | FAM-CATTGTTTCCAATGGGT-MGB |
| Lactobacillus iners | CGAGTCTGCCTTGAAGATCGG | GTTATCCCGATCTCTTGGGCA | FAM-CTTGCACTCTGTGAAACAAGATACAGGCTAGC-BHQ |
| Lactobacillus jenseniijesenii | CCTTAAGTCTGGGATACCATT | ACGCCGCCTTTTAAACTTCTT | SYBR-Green assay |
| Lactobacillus gasseri | GCTTAGCTCAGATGGGAGAGCG | TTCAACAGCCTTAACAGCTCTTGA | SYBR-Green assay |
| Lactobacillus crispatus | AGCGAGCGGAACTAACAGATTTAC | AGCTGATCATGCGATCTGCTT | SYBR-Green assay |
| Lambda IPC Taqman r-probe: | Specific primer for target | Specific primer for target | TAMRA-TCCTTCGTGATATCGGACGTTGGCTG-BHQ2 |
IPC, internal processing control.
Statistical analysis
All statistical analyses were performed using SIGMAPLOT 12.0 (Systat Software, San Jose, CA, USA). The data did not meet the criteria for normal distribution, and therefore a nonparametric, paired test (Wilcoxon Signed Rank Test) was used when comparing two groups. Results are presented as median values and interquartile range (IQR) unless otherwise stated. Values of p that were <0.05 were considered significant.
Results
For CMP microscopic examination eight plug sets comprising eight proximal and eight distal biopsies were analyzed. Hematoxylin & eosin-stained slides of the CMP specimens showed a cell-free eosinophilic, proteinous-like material in which epithelial and inflammatory cells were co-localized in clusters (Figure 2). The number of cells and composition in the distal and proximal parts of the CMP did not differ.
Figure 2.
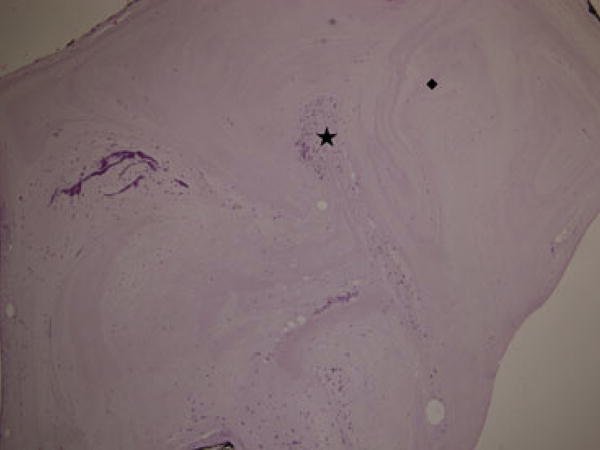
Hematoxylin & eosin-stained slide of the proximal part of a cervical mucus plug with cell clusters (*) and cell-free areas (◆).
Immunohistochemistry revealed that neutrophil leukocytes and macrophages (CD68) were the most abundant cell types. In the distal specimens, the median number of neutrophil leukocytes was 520 per 10 HPF (IQR 1097), and the median number of macrophages was 228 per 10 HPF (IQR 222), whereas it was 584 (IQR 1137) for neutrophils and 220 (IQR 107) for macrophages in the proximal specimens. T cells (CD3) were present in 7/8 distal slides and in 5/8 proximal slides. B cells (CD20) were identified in 1/8 distal slides and in 1/8 proximal slides. The number of epithelial cells from the exocervix (squamous, superficial epithelial cells) exceeded the number of cells from the endocervix (columnar cells). The Gram-stain was positive for bacteria in 5/8 distal slides and in 4/8 proximal slides. The morphology of the bacteria was consistent with long and short Gram-positive rods compatible with Lactobacillus spp. No differences could be observed between CMPs from first-trimester and from women at term.
Concentrations of MMP-8 were lower in the proximal than in the distal parts of the CMP, but the difference was not significant (p = 0.08) (Table 2).
Table 2.
Bacterial load and maxtrix metalloproteinase-8 concentrations.
| 16SrDNA geq/g CMP | Lactobacillus crispatus geq/g CMP | Lactobacillus jensenii geq/g CMP | Lactobacillus gasseri geq/g CMP | Lactobacillus iners geq/g CMP | Ureaplasma parvum geq/g CMP | MM P-8 μg/g CMP | |
|---|---|---|---|---|---|---|---|
| Number of women with positive analysisa | 13 of 13 | 13 of 13 | 13 of 13 | 13 of 13 | 13 of 13 | 7 of 13 | 8 of 8 |
| Vaginal swab; median (IQR) | 842 (1110) × 106 | 879 (171 000) × 103 | 139 (32 900) × 103 | 0.0 (106) × 103 | 49 (85 142) × 103 | 60 (96) × 103 | Not measured |
| p-value (vaginal vs. distal) | 0.001 | 0.001 | 0.001 | 0.08 | 0.001 | 0.02 | – |
| Distal CMP; median (IQR) | 100 (299) × 106 | 1.5 (20 000) × 103 | 0.44 (9.6) × 103 | 0.0 (2.0) × 103 | 0.20 (2.0) × 103 | 0.66 (1.6) × 103 | 18 (20) |
| p-value (distal vs. proximal) | 0.001 | 0.02 | 0.01 | 0.13 | 0.47 | 0.19 | 0.08 |
| Proximal CMP; median (IQR) | 5.0 (299) × 106 | 2.8 (4850) × 103 | 0.0 (1.0) × 103 | 0.0 (0.05) × 103 | 0.13 (2.7) × 103 | 1.5 (2.5) × 103 | 11 (7.8) |
| p-value (vaginal vs. proximal) | 0.001 | 0.001 | 0.001 | 0.06 | 0.001 | 0.02 |
CMP, cervical mucus plug; geq, genome equivalents; IQR, interquartile range.
Only women with a positive analysis (defined as identification of the analyte in at least one of the three compartments) were included in the median values given In the table. Because of the small size of the specimens, it was not possible to perform all analyses on every specimen, i.e. not all eight women in the matrix metalloproteinase group had their specimens analyzed by quantitative polymerase chain reaction.
The qPCR analyses revealed 16Sr DNA in all vaginal specimens as well as in distal and proximal specimens of the CMP (Table 2). The median bacterial load as measured by 16SrDNA genome equivalents/g (geq/g) in both the distal CMP specimens and in the proximal specimens was significantly lower than that in the vagina (p = 0.001). The median bacterial load was 842 × 106 (IQR 1110 × 106) in the vagina, 100 × 106 (IQR 299 × 106) in the distal part of the CMP and 5 × 106 (IQR 299 × 106) in the proximal part. The bacterial load was significantly lower in the proximal CMP than in the distal CMP (p = 0.01).
The bacterial load of L. iners, L. crispatus and L. jensenii decreased significantly from the vagina to the distal as well as the proximal part of the CMP. Both L. crispatus and L. jensenii decreased significantly from the distal to the proximal CMP (Table 2). U. parvum was detected in 7/13 vaginal specimens, in 6/13 distal specimens and in 5/13 proximal specimens (Table 2). Among those women harboring U. parvum, the median concentration of this species in both the distal and in the proximal CMP specimens was significantly lower compared with that in the vagina (p = 0.02 for both). The median load of U. parvum was 60 241 (IQR 96 386) geq/g in the vagina, 657 (IQR 1620) geq/g in the distal part of the CMP and 1574 (IQR 2526) geq/g in the proximal specimens. The distal CMP from one woman was positive for both U. parvum and U. urealyticum. Two were positive for M. hominis in the vaginal swabs and in their distal specimens, but not in the proximal specimens.
Discussion
This study of CMPs from pregnant women shows that bacteria, predominantly Lactobacillus species, and inflammatory cells in the CMP are co-localized in clusters within otherwise cell-free areas of mucus. The median bacterial load (16S rDNA) in the specimens obtained 3–4 cm up the cervical canal (proximal specimens) was significantly lower than that in the distal specimens. Among women harboring U. parvum, the load of this species in the distal specimens was less than 1% of that in the vaginal specimens. The strength of this study lies in the specimen collection. A thin catheter with a piston was used, ensuring minimal contamination of the proximal CMP with material from the distal CMP. This method provided an opportunity to examine and compare the two parts of the CMP. On the other hand, the number of specimens did not allow us to determine the distribution of the less common pathogens U. urealyticum and M. hominis.
Specific assays for genital mycoplasmas showed that U. parvum is the most abundant mycoplasma species found in the CMP. This bacterium has been found as the predominant species detected in the amniotic cavity in cases of intra-amniotic infection22. In a prospective study, Kataoka et al.23 examined 877 asymptomatic pregnant women at approximately 11 weeks of gestation with molecular techniques. Vaginal fluid was positive for U. parvum in 52% (456/877) of women, and 96% (440/456) led to a term delivery. This underscores our results of ureaplasmas as a normal finding in the vagina of healthy women. However, it seems that if ureaplasmas ascend from the vagina to the amniotic cavity they are pathogenic and may cause preterm labor. Several studies show a correlation between Ureaplasma spp. and M. hominis in the amniotic fluid and preterm labor9,10,12,13. Our study shows that the amount of U. parvum decreases from the vagina and through the CMP, as an indication of the CMP’s protective barrier. However, the CMP is not an absolute barrier against U. parvum because the bacteria were also present in the proximal part of the CMP. Surprisingly, the median load of U. parvum in geq/g was not different in the proximal part compared with the distal part of the CMP (p = 0.19). Most likely, this finding is a result of the low number of U. parvum-positive specimens. On the other hand, it can be speculated whether this organism, in particular, is capable of escaping the immune barriers of the CMP. The women in this study who were positive for U. parvum in their proximal CMP did not deliver preterm. This may be explained by the fact that our proximal CMP specimens may not represent the last centimeter of the CMP, keeping in mind that the catheter was inserted 3–4 cm up the cervix when obtaining this specimen, or that the fetal membranes constitute an additional barrier24.
Lactobacillus species, L. crispatus and L. jensenii in particular, are known to dominate a healthy vaginal microbial environment, hence protecting the vagina against pathogens13. It is tempting to speculate that Lactobacillus spp. could play a similar role in the CMP, thereby contributing to protection against ascending infections. According to the qPCR analyses in this study, the concentration of Lactobacillus species in the CMP specimens was approximately 2900 times that of the Ureaplasma concentration. In addition to the well-known protective properties of Lactobacillus spp. in the vagina, it is possible that they also contribute to the physicochemical properties of the CMP by creating an acidic environment. It has been proposed that a pH change in the cervical mucus of non-pregnant women is responsible for a modification in the mucin structure that occurs during ovulation, and that a low pH makes the mucus less susceptible to invasion by sperm and microorganisms25. Under these circumstances, lactobacillus-induced pH changes may help the CMP structure to remain thick and resistant to microbial migration. It is, therefore, likely that the bacterial load in the mucus plug described is physiological rather than indicative of ascending infection. Although the number of women studied was small, it was evident that different Lactobacillus spp. dominated in different women. This is in good agreement with studies of the vaginal microbiome, where various community states have been described among asymptomatic women26. The community states were associated with race and also with vaginal pH; in particular, women with L. crispatus-dominated flora had lower vaginal pH than those women in whom other species dominated.
The histological and immunohistochemical analyses of the CMP, showing massive inflammatory cell invasion by neutrophil leukocytes and macrophages, few T cells and Gram-positive rods as the only bacteria present, are consistent with previous descriptions by Hein et al2. However, our study, based on both histology and qPCR, could not confirm the absence of bacteria and inflammatory cells in the proximal part of the CMP. The concentration of neutrophil leukocytes was also evenly distributed in the two compartments of the plug. This difference between results can be attributed to the different technique used by Hein et al. to define the proximal and distal parts of the CMP ex situ. Alternatively we did not obtain the proximal CMP specimens from the most proximal part of the cervical canal as we inserted the catheter only 3–4 cm.
In conclusion, the CMP inhibits bacteria in general, but it does not block the passage of U. parvum from the vagina to the lower pole of the uterus. Future studies are required to examine the concentration of different bacteria rather than the presence or absence of microorganisms. It is tempting to assume that Lactobacillus species through pH adjustment contribute to the environment in the CMP that protects against ascending infection by other microorganisms.
Key Message.
The cervical mucus plug contains bacteria and inflammatory cells colocalized in clusters within cell-free areas. The median concentrations of 16S rDNA and Ureaplasma parvum genome equivalents/g were much lower in the cervical mucus plug compared with the vagina.
Acknowledgments
We wish to thank staff at the Department of Obstetrics and Gynecology, Aarhus University Hospital, Denmark for help during the collection of the CMP specimens. We acknowledge the technical assistance of the staff at the STI Research and Development group, Statens Serum Institute, Denmark.
Funding
This work was supported, in part, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, DHHS. Furthermore, grants from the Danish Council for Independent Research – Medical Sciences, Augustinus Fonden, Den Bøhmske Fond, Margot and John Fribergs Fond and Torben and Alice Frimodts Fond supported this work.
Abbreviations
- CMP
cervical mucus plug
- geq
genome equivalents
- HPF
highpower fields
- IQR
interquartile range
- MMP
matrix metalloproteinase
- qPCR
quantitative polymerase chain reaction
References
- 1.Becher N, Adams Waldorf K, Hein M, Uldbjerg N. The cervical mucus plug: structured review of the literature. Acta Obstet Gynecol Scand. 2009;88:502–13. doi: 10.1080/00016340902852898. [DOI] [PubMed] [Google Scholar]
- 2.Hein M, Petersen AC, Helmig RB, Uldbjerg N, Reinholdt J. Immunoglobulin levels and phagocytes in the cervical mucus plug at term of pregnancy. Acta Obstet Gynecol Scand. 2005;84:734–42. doi: 10.1111/j.0001-6349.2005.00525.x. [DOI] [PubMed] [Google Scholar]
- 3.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 2010;362:529–35. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 4.Viscardi RM. Ureaplasma species: role in diseases of prematurity. Clin Perinatol. 2010;37:393–409. doi: 10.1016/j.clp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–7. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 6.Gerber S, Vial Y, Hohlfeld P, Witkin SS. Detection of Ureaplasma urealyticum in second-trimester amniotic fluid by polymerase chain reaction correlates with subsequent preterm labor and delivery. J Infect Dis. 2003;187:518–21. doi: 10.1086/368205. [DOI] [PubMed] [Google Scholar]
- 7.Waites KB, Schelonka RL, Xiao L, Grigsby PL, Novy MJ. Congenital and opportunistic infections: Ureaplasma species and Mycoplasma hominis. Semin Fetal Neonatal Med. 2009;14:190–9. doi: 10.1016/j.siny.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Kundsin RB, Leviton A, Allred EN, Poulin SA. Ureaplasma urealyticum infection of the placenta in pregnancies that ended prematurely. Obstet Gynecol. 1996;87:122–7. doi: 10.1016/0029-7844(95)00376-2. [DOI] [PubMed] [Google Scholar]
- 9.Yoon BH, Chang JW, Romero R. Isolation of Ureaplasma urealyticum from the amniotic cavity and adverse outcome in preterm labor. Obstet Gynecol. 1998;92:77–82. doi: 10.1016/s0029-7844(98)00122-7. [DOI] [PubMed] [Google Scholar]
- 10.Romero R, Yoon BH, Mazor M, Gomez R, Gonzalez R, Diamond MP, et al. A comparative study of the diagnostic performance of amniotic fluid glucose, white blood cell count, interleukin-6, and gram stain in the detection of microbial invasion in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 1993;169:839–51. doi: 10.1016/0002-9378(93)90014-a. [DOI] [PubMed] [Google Scholar]
- 11.Hassan S, Romero R, Hendler I, Gomez R, Khalek N, Espinoza J, et al. A sonographic short cervix as the only clinical manifestation of intra-amniotic infection. J Perinat Med. 2006;34:13–19. doi: 10.1515/JPM.2006.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novy MJ, Duffy L, Axthelm MK, Sadowsky DW, Witkin SS, Gravett MG, et al. Ureaplasma parvum or Mycoplasma hominis as sole pathogens cause chorioamnionitis, preterm delivery, and fetal pneumonia in rhesus macaques. Reprod Sci. 2009;16:56–70. doi: 10.1177/1933719108325508. [DOI] [PubMed] [Google Scholar]
- 13.Moss TJM, Nitsos I, Ikegami M, Jobe AH, Newnham JP. Experimental intrauterine Ureaplasma infection in sheep. Am J Obstet Gynecol. 2005;192:1179–86. doi: 10.1016/j.ajog.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 14.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, et al. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG. 2011;118:533–49. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skarin A, Sylwan J. Vaginal lactobacilli inhibiting growth of Gardnerella vaginalis, Mobiluncus and other bacterial species cultured from vaginal content of women with bacterial vaginosis. Acta Pathol Microbiol Immunol Scand B. 1986;94:399–403. doi: 10.1111/j.1699-0463.1986.tb03074.x. [DOI] [PubMed] [Google Scholar]
- 16.Becher N, Hein M, Danielsen CC, Uldbjerg N. Matrix metalloproteinases and their inhibitors in the cervical mucus plug at term of pregnancy. Am J Obstet Gynecol. 2004;191:1232–9. doi: 10.1016/j.ajog.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 17.Becher N, Hein M, Uldbjerg N, Danielsen CC. Balance between matrix metalloproteinases (MMP) and tissue inhibitors of metalloproteinases (TIMP) in the cervical mucus plug estimated by determination of free non-complexed TIMP. Reprod Biol Endocrinol. 2008;6:45. doi: 10.1186/1477-7827-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becher N, Hein M, Danielsen CC, Uldbjerg N. Matrix metalloproteinases in the cervical mucus plug in relation to gestational age, plug compartment, and preterm labor. Reprod Biol Endocrinol. 2010;8:113. doi: 10.1186/1477-7827-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agren MS, Taplin CJ, Woessner JF, Jr, Eaglstein WH, Mertz PM. Collagenase in wound healing: effect of wound age and type. J Invest Dermatol. 1992;99:709–14. doi: 10.1111/1523-1747.ep12614202. [DOI] [PubMed] [Google Scholar]
- 20.De Boer R, Peters R, Gierveld S, Schuurman T, Kooistra-Smid M, Savelkoul P. Improved detection of microbial DNA after bead-beating before DNA isolation. J Microbiol Methods. 2010;80:209–11. doi: 10.1016/j.mimet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–66. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 22.Kasper DC, Mechtler TP, Reischer GH, Witt A, Langgartner M, Pollak A, et al. The bacterial load of Ureaplasma parvum in amniotic fluid is correlated with an increased intrauterine inflammatory response. Diagn Microbiol Infect Dis. 2010;67:117–21. doi: 10.1016/j.diagmicrobio.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Kataoka S, Yamada T, Chou K, Nishida R, Morikawa M, Minami M, et al. Association between preterm birth and vaginal colonization by mycoplasmas in early pregnancy. J Clin Microbiol. 2006;44:51–5. doi: 10.1128/JCM.44.1.51-55.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjaergaard N, Hein M, Hyttel L, Helmig RB, Schønheyder HC, Uldbjerg N, et al. Antibacterial properties of human amnion and chorion in vitro. Eur J Obstet Gynecol Reprod Biol. 2001;94:224–9. doi: 10.1016/s0301-2115(00)00345-6. [DOI] [PubMed] [Google Scholar]
- 25.Brunelli R, Papi M, Arcovito G, Bompiani A, Castagnola M, Parasassi T, et al. Globular structure of human ovulatory cervical mucus. FASEB J. 2007;21:3872–6. doi: 10.1096/fj.07-8189com. [DOI] [PubMed] [Google Scholar]
- 26.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]


