Abstract
We have characterized developmental, environmental, and genetic regulation of abscisic acid-insensitive (ABI)4 gene expression in Arabidopsis. Although expressed most strongly in seeds, ABI4 transcripts are also present at low levels in vegetative tissue; vegetative expression is not induced by abscisic acid (ABA) or stress treatments. Comparison of transcript levels in mature seeds of ABA-insensitive, ABA-hypersensitive, ABA-deficient, or heterochronic mutants indicates that ABI4 expression is altered in only two of the backgrounds, the ABA-insensitive mutants abi1-1 and abi3-1. To determine whether ABI4 is necessary and/or sufficient for ABA response, we assayed the effects of loss of ABI4 function and ectopic ABI4 expression on growth and gene expression. We examined genetic interactions among three ABA response loci, ABI3, ABI4, and ABI5, by comparing phenotypes of mutants, ectopic expression lines, mutants carrying an ectopically expressed transgene, and the corresponding wild-type lines. Our results indicate some cross-regulation of expression among ABI3, ABI4, and ABI5 and suggest that they function in a combinatorial network, rather than a regulatory hierarchy, controlling seed development and ABA response.
Abscisic acid (ABA) regulates many aspects of plant growth and development, including embryogenesis, water relations, and tolerance of a variety of environmental stresses (for review, see Leung and Giraudat, 1998). Many lines of evidence indicate that there are multiple ABA perception and signaling mechanisms. Multiple receptor types are implicated by the variation in stereospecificity among ABA responses, studies of response to impermeant ABA-bovine serum albumin conjugates (Jeannette et al., 1999), and microinjection studies showing that ABA has both intra- and extra-cellular sites of action (for review, see Leung and Giraudat, 1998). Many likely signaling intermediates correlated with ABA response (e.g. ABA-activated or -induced kinases and DNA-binding proteins that specifically bind ABA responsive elements) have been identified by molecular and biochemical studies (for review, see Leung and Giraudat, 1998), but the relationships among these proteins are unclear. Genetic studies, especially in Arabidopsis, have identified many loci involved in ABA response and analyzed their functional roles (for review, see Bonetta and McCourt, 1998; Leung and Giraudat, 1998).
The maize viviparous 1 (vp1) and Arabidopsis ABA-insensitive (abi) and enhanced response to ABA (era) mutants are the most extensively characterized ABA response mutants. The vp1 mutations of maize have pleiotropic effects on seed development, not all of which can be ascribed to defects in ABA response (Robertson, 1955), suggesting that VP1 mediates responses to a variety of signals including ABA. Four Arabidopsis loci affecting ABA sensitivity have also been extensively characterized: ABI1, ABI2, ABI3, and ERA1 (for review, see Finkelstein and Zeevaart, 1994; Bonetta and McCourt, 1998). Mutations at these three ABI loci result in reduced seed dormancy and a reduction in sensitivity to exogenous ABA for inhibition of germination. In contrast, the era1 mutants have increased dormancy and sensitivity to exogenous ABA for inhibition of seedling root growth and induction of stomatal closing (Cutler et al., 1996; Pei et al., 1998). Mutants at two additional Arabidopsis loci, ABI4 and ABI5, were also selected for their ability to germinate on ABA concentrations inhibitory to wild-type germination (Finkelstein, 1994). Initial physiological and genetic analyses suggested that the ABI3, ABI4, and ABI5 loci were likely to be acting in the same seed-specific signaling pathway; mutants at all three loci exhibited defects in seed ABA sensitivity and accumulation of at least one late embryogenesis abundant transcript, but displayed normal vegetative growth (Finkelstein and Somerville, 1990; Finkelstein, 1994). Furthermore, in studies of digenic mutants, mutations at all three loci greatly enhanced the ABA-resistance of abi1 mutants and mutations in the ABI3 and ABI5 loci also significantly enhanced the ABA-resistance of abi2 mutants. In contrast, combination of a weak abi3 allele (abi3-1) with either abi4-1 or abi5-1 resulted in only slightly greater resistance. Prior to a molecular description of the abi4-1 and abi5-1 mutations it was not possible to determine the likely order of gene action in this proposed pathway because the abi4 and abi5 mutations might have been either null or leaky mutations. In fact, the observation that abi4-1 and abi5-1 mutant seeds were desiccation tolerant and only slightly resistant to ABA (Finkelstein, 1994), whereas severe abi3 alleles resulted in production of desiccation intolerant highly ABA-insensitive “green seeds” (Nambara et al., 1992), lent support to the view that the abi4-1 and abi5-1 mutations were leaky. However, we have now cloned ABI4 and ABI5 and found that the abi4-1 and abi5-1 alleles have very severe mutations (Finkelstein et al., 1998; Finkelstein and Lynch, 2000).
The ABI3, ABI4, and ABI5 genes have been cloned and found to encode putative transcription factors of the B3 domain, AP2 domain, and bZIP factor classes, respectively (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000). Although mutations in ABI3, ABI4, and ABI5 have their greatest impact on gene expression during seed maturation, all three genes are expressed to a limited degree in vegetative tissues (Finkelstein et al., 1998; Rohde et al., 1999; Finkelstein and Lynch, 2000), suggesting they may play a role in vegetative ABA response. Consistent with this, additional abi4 mutants have recently been isolated on the basis of exhibiting salt-resistant germination (Quesada et al., 2000) or sugar-insensitive seedling growth (Gibson et al., 1999; Huijser et al., 1999; Arenas-Huertero et al., 2000).
Ectopic expression of ABI3 has been shown to confer ABA-inducible vegetative expression of several “seed-specific” genes, leading to the suggestion that ABI3 expression is sufficient for the seed developmental program and ABA sensitivity. However, changes in seed ABA sensitivity during embryogenesis cannot be explained by fluctuations in ABI3 content, which remains relatively constant (on a per cell basis) through embryogenesis (Parcy et al., 1994). This indicates that although required for ABA response, ABI3 content is not the only factor determining the degree of ABA sensitivity for any specific response.
In addition to the genetically defined transcription factors involved in seed and ABA response described above, many factors presumed to regulate ABA-inducible and embryonic gene expression have been identified biochemically (for review, see Leung and Giraudat, 1998). Although it is likely that many of these transcription factors regulate some of the same genes, the majority of specific target genes for most regulatory factors are unknown. Furthermore, for most factors it is not known whether regulation of common target genes is accomplished by independent binding to distinct cis-acting sites, activation of a regulatory cascade, combinatorial action of factors, or a combination of these mechanisms. Many of these questions can be addressed by a molecular analysis of lines with loss or gain of specific regulatory factors, resulting from mutations or ectopic expression.
In the present study we have characterized the developmental, environmental, and genetic regulation of ABI4 expression. In addition, we have compared different abi4 mutant alleles, ABI4 ectopic expression lines, and wild-type plants to assay the role of ABI4 function in regulating gene expression in embryos and seedlings, cold, saline, and osmotic stress sensitivity, and sensitivity to ABA inhibition of root growth. We have also tested whether the previously described “hypersensitivity” to ABA due to ectopic ABI3 expression is dependent on ABI4 or ABI5 function. Our results indicate some cross-regulation among these ABI loci, but the observed alterations in gene expression in the mutants are more consistent with combinatorial control by these three loci than a simple linear pathway.
RESULTS
Function of the ABI4 Protein and Its Mutant Alleles
Molecular cloning of the ABI4 gene identified it as a member of the APETALA2 domain family and thus presumed to encode a transcriptional regulator. Many transcription factors have been shown to have transcription activation function in hybrid proteins expressed in heterologous systems such as yeast. To determine whether ABI4 is capable of transcription activation in yeast we constructed a fusion with the GAL4 DNA-binding domain (GAL4-BD) and tested its ability to activate a GAL4-responsive promoter in yeast. A fusion protein containing all but the first two and last amino acids of ABI4 (residues 3–327) directed high level expression of a β-galactosidase reporter gene (Table I), consistent with function as a transcriptional activator.
Table I.
Transcriptional activation by GAL4BD-ABI4 fusions in yeast
| Construct | ABI4 Amino Acids Present | Putative Activation Domains Present | β-Galactosidase Activity (Miller units)a |
|---|---|---|---|
| pGBD | None | None | 5.2 ± 2.9 |
| pGBD-ABI4 | 3–327 | Q-rich, P-rich, acidic | 224.5 ± 12.4 |
| pGBD-ABI4 Bam | 3–287 | Q-rich, P-rich | 66.7 ± 0.1 |
| pGBD-ABI4 Sac | 3–224 | Q-rich | 42.5 ± 4.8 |
| pGBD-ABI4 Nla | 3–156 | None | 6.8 ± 4.4 |
| pGBD-ABI4 C-term | 157–327 | Q-rich, P-rich, acidic | 2.6 ± 0.4 |
β-Galactosidase activity produced by activation of a GAL7-lacZ reporter gene by GAL4BD fusions containing various domains of ABI4. Data presented are averages ± sd of assays with at least duplicate extracts.
The original abi4-1 mutant allele contains a frameshift at codon 157 (of 328), resulting in a protein containing the presumed DNA binding and dimerization domains, but lacking the presumed activation domain(s). Additional mutant abi4 alleles have recently been isolated in screens for sugar-insensitive (Gibson et al., 1999; Huijser et al., 1999; Arenas-Huertero et al., 2000) or salt-resistant (Quesada et al., 2000) seedling growth, leading Quesada et al. to suggest that the abi4-1 mutant still had residual ABI4 function. We have compared transcription activation function of full-length and truncated ABI4 products in a one-hybrid assay in yeast. A fusion containing only amino acids 3 through 156 was no more active than the product of the vector alone (Table I). This indicates that the abi4-1 encoded protein is inactive as a transcriptional activator in yeast, although it may still be capable of forming a DNA-binding complex.
The C-terminal half of the ABI4 protein contains several regions that are candidates for transcription activation domains: a Gln-rich (amino acids 188–208), a Pro-rich (amino acids 275–289), and an acidic domain (amino acids 295–326). To determine whether any of these are required for transcription activation we have also tested fusions containing larger segments of ABI4. The longest of these contained all but the C-terminal acidic domain and its transcription activation function was reduced 5-fold relative to the full-length protein fusion. A fusion containing only the Q-rich domain had only approximately 18% the activity of the full-length fusion. These results suggest that all three putative activation domains contribute to activation function. Although necessary, the C-terminal portion is not sufficient; a fusion containing residues 157 through 327 does not activate reporter expression. However, we cannot rule out the possibility that the inactive fusions are misfolded or unstable.
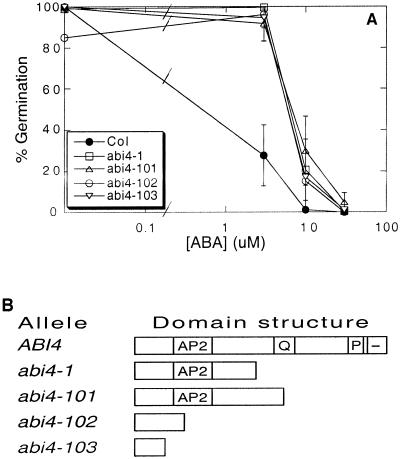
As an independent test of abi4-1 protein function we have compared an allelic series of abi4 mutants with respect to their sensitivities to ABA inhibition of germination. In addition to abi4-1 we tested alleles containing nonsense mutations such that the proteins are truncated at amino acids 193, 80, and 39 (in the abi4-101[sis5-1], abi4-102 [sis5-2], and abi4-103 [sis5-3] alleles, respectively). Thus the abi4-103 mutant (Laby et al., 2000) is truncated five amino acids earlier than the frame-shift in abi4-2 (san5) (Quesada et al., 2000), lacks the entire AP2 domain, as well as the presumed activation domain, and is therefore the most likely allele to produce a biochemical null. Comparison of ABA sensitivities for germination inhibition shows no statistically significant difference among these alleles (Fig. 1). Similar germination percentages were obtained with ABI4 anti-sense transgenes in wild-type and abi4-1 mutant backgrounds, although the lines containing an antisense transgene and the mutation germinated slightly faster (data not shown). However, the fact that the abi4-1 and abi4-103 mutants show equivalent sensitivities to ABA suggests that neither product has significant residual activity.
Figure 1.
Comparison of sensitivities to ABA inhibition of germination for seeds of wild type and an abi4 allelic series. Seeds were plated on the indicated concentrations of ABA and germination was scored after 7 d (A). Domain structures of these abi4 alleles are shown in B. Data presented are averages ± sd of at least duplicate assays.
Regulation of ABI4 Expression
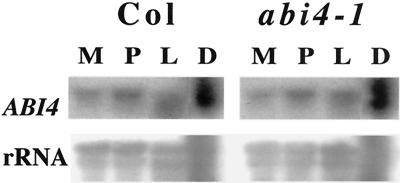
Our initial characterization of ABI4 expression indicated that transcript levels were highest in pooled stages of developing siliques, but reduced in the abi4-1 mutant. In addition, very low ABI4 transcript levels were detected in 11-d-old plants, indicating that expression was not seed specific (Finkelstein et al., 1998). We have now expanded our analysis to include developmental, environmental, and genetic regulation of ABI4 expression. We have used a combination of RNA gel-blot analyses and assays of β-glucuronidase (GUS) activity derived from a variety of ABI4 promoter-GUS fusions. These studies confirmed that expression is highest in seeds, with transcript levels fairly stable through the latter half of embryogenesis, then increasing slightly at seed maturity (Fig. 2). We were surprised to find that ABI4 transcript levels were quite similar in wild-type and abi4-1 seeds. However, we had previously compared RNA from pooled stages of siliques and a slight bias toward dry seeds in the wild-type pool would have made the ABI4 transcript appear more abundant in this sample.
Figure 2.
ABI4 transcript accumulation during the latter half of embryogeny. RNA was isolated from Col or abi4-1 siliques at 8 to 11 days post anthesis (dpa) (M, maturation), 12 to 16 dpa (P, post-abscission), 17 to 21 dpa (L, late embryogenesis), and >21 dpa (D, dry seeds) of each genotype. Each lane contains approximately 8 μg of total RNA.
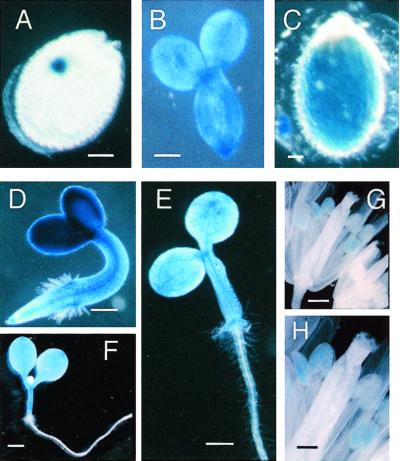
Studies of GUS fusion lines indicate that the ABI4 promoter is active from globular stage onward and its activity is limited to the embryo, where it is uniformly expressed (Fig. 3, A–C). Additional histochemical staining showed limited patches of ABI4 promoter activity in vegetative tissue, especially in vascular tissue, consistent with the low levels of ABI4 transcript observed after germination (Fig. 3, D–F). No significant change in GUS activity was detected in 5-d-, 11-d-, or 4-week-old plants subjected to a variety of stress (drought, saline, high Glc, or cold) or ABA treatments (data not shown). Although GUS activity was observed in mature anthers of transgenic lines (Fig. 3, G and H), but not in those of untransformed controls (data not shown), activity of the endogenous ABI4 promoter was not confirmed by reverse transcriptase-PCR analyses of RNA isolated from floral buds (data not shown).
Figure 3.
ABI4-driven GUS expression. Histochemical staining of globular embryo in seed (A), excised 5-d embryo (B), and mature seed (C); 2-d (D), 3-d (E), and 5-d (F) seedlings, and transgenic (G and H) flowers. Bar = 100 μm in A and C; bar = 150 μm in B; bar = 300 μm in D; bar = 500 μm in E and G; bar = 1 mm in F; bar = 250 μm in H.
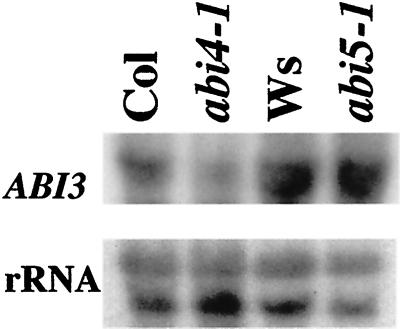
ABI4 is one of many genes with pleiotropic effects on seed development. To determine whether ABI4 is likely to be part of a regulatory hierarchy with other known seed regulatory loci, we measured ABI4 transcript levels in dry seeds of a variety of heterochronic (fus3 and lec1), ABA response (abi and era1), or ABA biosynthetic (aba1) mutants. ABI4 transcript accumulation was nearly normal in most of the genotypes tested, but was significantly reduced in abi1-1 and abi3-1 (Fig. 4). Although the fus3 and lec1 dry seeds are dead, even a slight reduction in ABI4 transcript levels would be consistent with normal expression until viability is lost (at late embryogenesis stage).
Figure 4.
ABI4 transcript accumulation in mature dry seeds of ABA response (abi), ABA biosynthetic (aba), or seed regulatory (fus3, lec1, and era1) mutants. Mutant lines are grouped according to their ecotype backgrounds along with the appropriate wild-type line. Each lane contains approximately 5 μg of total RNA. Top row is ABI4 transcript; bottom row is rRNA.
Role of ABI4 in Vegetative Growth Regulation
The similar, but relatively weak, ABA resistance of the abi4 mutants suggests that ABI4 plays a minor role in ABA response or acts redundantly with other ABA response loci. To determine whether ABI4 is necessary and/or sufficient for ABA response, we have assayed the effects of loss of ABI4 function and ectopic ABI4 expression on growth and gene expression.
The abi4-1 mutant was isolated on the basis of ABA-resistant germination and our initial characterization suggested its effects were seed specific (Finkelstein, 1994). Upon cloning ABI4 we found that it was expressed at low levels in vegetative tissue (Finkelstein et al., 1998). Furthermore, new mutant abi4 alleles have been identified in seedling screens (Gibson et al., 1999; Huijser et al., 1999; Arenas-Huertero et al., 2000; Quesada et al., 2000), indicating that ABI4 function is required during seedling growth under some conditions. To analyze the role of ABI4 in regulation of vegetative growth, we compared the effects of loss of ABI4 function and ectopic ABI4 expression on two additional physiological responses: ABA sensitivity of root growth and growth under saline or osmotic stress. The ectopic expression lines analyzed had a broad range of ABI4 expression levels and associated phenotypes.
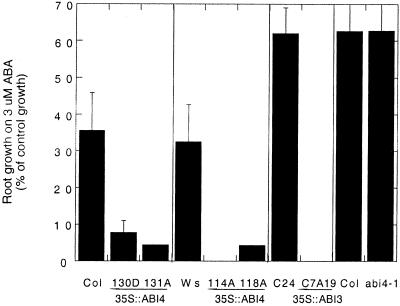
Although loss of ABI4 function had little effect on root growth, ectopic ABI4 expression resulted in hypersensitivity to root growth inhibition by ABA (Fig. 5). Growth of the transgenic line with strongest ABI4 expression (114A) was stunted on hormone-free medium (approximately 40% of wild type) and completely blocked by 3 μm ABA, a concentration permitting approximately 35% of control growth in wild-type lines. The more weakly expressing transgenic lines also showed significantly less root growth than wild-type plants when exposed to ABA, but grew normally on hormone-free medium. For comparison, root growth in an ABA-hypersensitive line produced by ectopic constitutive expression of ABI3 (ABI3c) was completely blocked by 3 μm ABA, but was normal on hormone-free medium. In contrast to the consistent increase in ABA-sensitivity of root growth produced by ectopic ABI4 expression, the ABI4 transgenes had very limited effects on saline or osmotic stress sensitivity of plant growth (data not shown). These results show that increased ABI4 function is sufficient for increased ABA sensitivity of vegetative tissues, but has negligible effects on stress tolerance.
Figure 5.
Effects of ABI4 or ABI3 function on ABA sensitivity of root growth. After 2 d of growth on hormone-free medium, seedlings were transferred to fresh media containing 0 or 3 μm ABA. New growth was measured after 4 d and expressed as a percentage of growth on the control medium. Each set represents the average of at least 10 replicates within an individual experiment; standard deviations are expressed as percentages of the control growth.
ABI4-Regulated Gene Expression
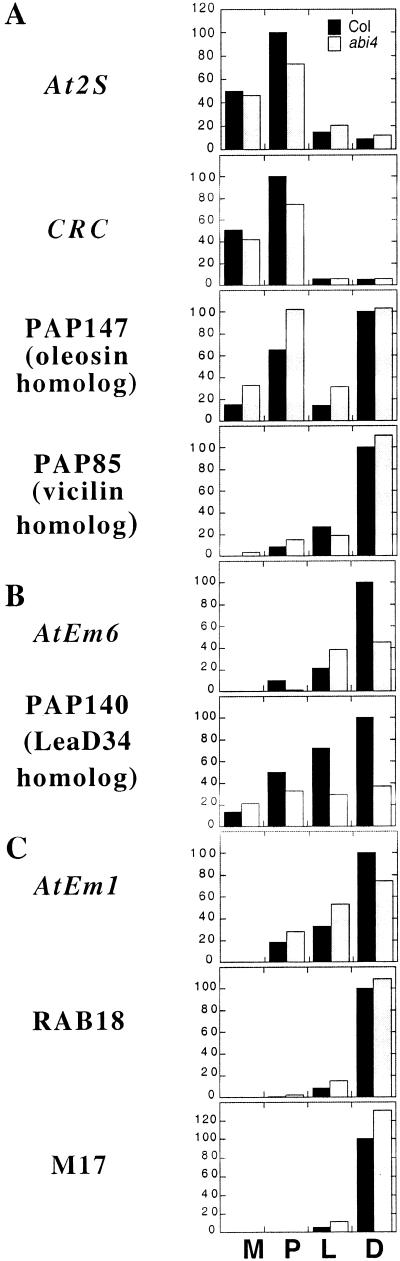
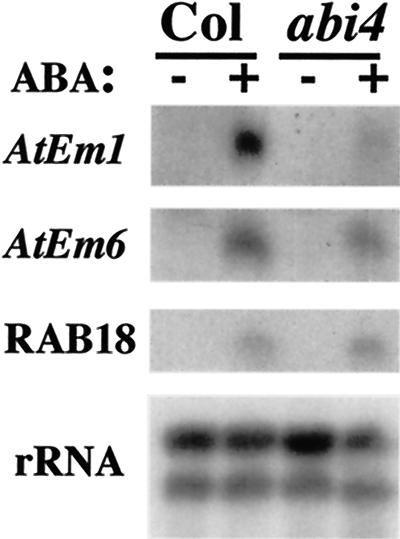
Our initial characterization of the abi4-1 mutant showed that AtEm6 transcript accumulation was reduced in dry seeds (Finkelstein, 1994). We have extended this analysis to include eight additional genes whose expression was assayed through the latter half of embryogenesis. These results show that the abi4-1 mutant has only minor effects on expression of most of these genes (Fig. 6). Although there is a substantial reduction in dry seed transcript levels of AtEm6 and the LeaD34 homolog, AtEm1 expression is only slightly reduced and some of these transcripts (e.g. AtEm1 and AtEm6) appear to accumulate earlier in the mutant than in wild-type seeds. The storage protein genes, At2S3 and CRC, also show only minimal changes in expression (less than 25% reduction) in the abi4-1 mutant, whereas accumulation of other transcripts (e.g. RAB18 and the vicilin homolog) is essentially unaffected in this mutant (within 10% of the wild type). In contrast, expression of some genes (M17 and mid-embryogenesis stage expression of the oleosin homolog) is increased in the abi4-1 mutant, as observed for the abi5-1 mutant (Finkelstein and Lynch, 2000), rather than decreased as in the abi3 mutants (Parcy et al., 1994). Several of the LEA genes are ABA-inducible in vegetative tissues, but ABI4 is required for only a subset of these (AtEm1 and AtEm6) to be fully expressed (Fig. 7). The relatively minor effects of the abi4-1 mutation on transcript accumulation indicate that ABI4 is not the major regulator of embryonic expression for most of these genes, but that it does play a role in seedling and vegetative gene regulation.
Figure 6.
Comparison of transcript accumulation in developing siliques of wild-type versus abi4-1 mutants. Relative abundance of MAT (A), LEA (B), and LEA-A (C) class transcripts from four developmental stages (M, P, L, and D; stages as defined in Fig. 2) is shown for Col and abi4, with peak level in wild type arbitrarily set at 100. Hybridization to the indicated probes was quantified from RNA gel blots by either phosphorimaging or densitometry of autoradiograms, then normalized relative to total RNA quantified by phosphorimaging of rRNA hybridization.
Figure 7.
RNA gel-blot comparison of ABA-inducible LEA and LEA-A transcript accumulation in 13-d plantlets of wild-type versus abi4-1 mutants. RNA was extracted from 13-d seedlings incubated for 2 d on plates ±50 μm ABA, then analyzed by RNA gel blots hybridized to cloned probes for the indicated transcripts.
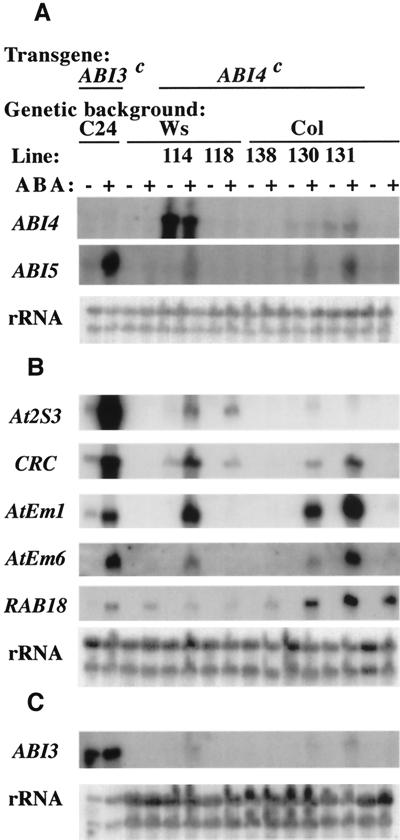
To test whether ABI4 is sufficient for expression of any of these genes we measured their transcript levels in five independent transgenic lines ectopically overexpressing ABI4, designated ABI4c (Fig. 8). Ectopic ABI3 expression had previously been shown to confer ABA-inducible expression of several “seed-specific” storage protein and lea genes in vegetative tissues (Parcy et al., 1994); therefore, we included a line with a strongly expressed ABI3 transgene, designated ABI3c, for comparison. The ABI4c transgenic lines displayed a broad range of ABI4 expression and many of them showed ABA-inducible vegetative expression of the same “seed-specific” genes as in the ABI3 ectopic expression lines. In addition, constitutive expression of ABI4 resulted in ABA-inducible ABI3 expression and hyperinduction of ABI5. As previously shown for ABI3 transgenic lines (Parcy et al., 1994), there was not a simple correlation between the level of expression for any of the ABI genes and the magnitude of target gene induction. Furthermore, the importance of any given regulator varied depending on the target gene. For example, AtEm1 and AtEm6 induction correlated fairly well with ABI5 expression, consistent with their strong dependence on ABI5 function for expression in seeds (Finkelstein and Lynch, 2000). In contrast, the storage protein genes At2S3 and CRC were far more strongly induced by the ABI3c transgene than by the ABI4c transgenes, consistent with their strong dependence on ABI3 for embryonic expression. However, even the highest ABI3 transcript levels in the ABI4c lines were approximately 100-fold lower than in the ABI3c line and were therefore unlikely to be sufficient for promoting the observed level of storage protein gene expression. It is possible that the ABI4c expression potentiates the effects of ABI3 and ABI5 in promoting At2S3 and CRC expression. The differential response of the storage protein and lea genes to ABI4c expression alternatively might reflect either ecotype or transgene differences since lines 114 and 118 are in a different genetic background and have a much shorter 5′-untranslated region (UTR) than lines 130 and 131. However, regardless of the magnitude of response in individual transgenic lines, these results indicate that ectopic expression of ABI3 or ABI4 is sufficient for ABA-inducible vegetative expression of some LEA and storage protein genes and suggest that the transgenes' effects may be partially mediated by ABI5.
Figure 8.
Effects of ectopic ABI4 expression on ABA-inducible vegetative accumulation of transcripts normally expressed primarily in seeds. RNA was extracted from 13-d seedlings incubated for 2 d on plates ±50 μm ABA, then analyzed by RNA gel blots hybridized to cloned probes for the indicated transcripts. A, All lanes contain 10 μg of total RNA. B, All lanes contain 5 μg of RNA. C, Lanes with RNA from ABI3 ectopic expression line contain 1 μg of RNA; all others contain 10 μg of RNA.
Cross-Regulation of ABI3, ABI4, and ABI5 Function and Expression
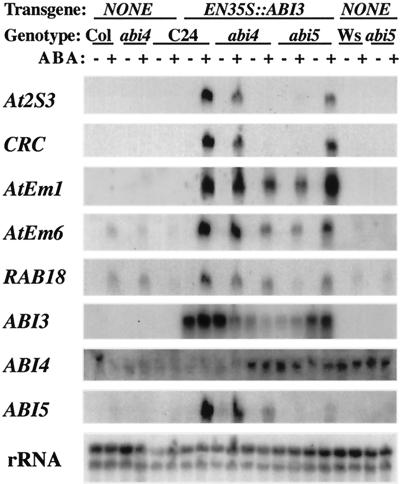
To determine whether the effects of ectopic ABI3 expression were dependent on ABI4 or ABI5 function, a strongly expressed ABI3c transgene was backcrossed into abi4-1 and abi5-1 mutant backgrounds. Because the transgene and the abi mutations were all in different genetic backgrounds, multiple segregants were analyzed to ensure that we were not monitoring random segregation of unknown modifying genes in the various backgrounds. Although ABI3 expression was controlled by a constitutive doubly enhanced cauliflower mosaic virus (CaMV)35S promoter, ABI3 transcript accumulation varied among segregants and marker gene expression correlated fairly well with the level of ABI3 transcript in the ABA-treated plants (Fig. 9). However, in the abi5 line with ABI3 expression comparable with that in the wild-type (C24) background, At2S3, CRC, AtEm6, and RAB18 expression appear slightly reduced. These results suggest that although all three ABI genes monitored may promote ABA-inducible vegetative expression of the storage protein and lea genes, strong expression of one (e.g. ABI3) can reduce reliance on the others. We do not yet know whether ectopic ABI5 expression has similar effects on ABA-inducible responses, or if ABI3 function is required for ABI4c expression to be effective.
Figure 9.
Effects of the abi4-1 and abi5-1 mutations on ABA-induced, EN35S::ABI3-dependent vegetative accumulation of primarily seed-expressed transcripts. RNA was extracted from 13-d seedlings incubated for 2 d on plates ±50 μm ABA, then analyzed by northern blots hybridized to cloned probes for the indicated transcripts. Ten genotypes were compared: Col, abi4-1, Ws, abi5-1, and C24 with no transgenes, a strongly expressed EN35S::ABI3 transgene in the C24 background, and two independent EN35S::ABI3 transgenic segregants in each of the abi4-1 and abi5-1 backgrounds, illustrating the range of transgene and marker gene expression observed among the segregants.
RNA gel-blot analyses have provided evidence of cross-regulation among the ABI loci. ABI5 expression is reduced to varying extents in aba1-1, abi1-1, abi2–1, abi3-1, abi4-1, and abi5-1 mutant seeds (Finkelstein and Lynch, 2000) and hyperinduced by ABA in transgenic plants ectopically expressing either ABI3 or ABI4 (Fig. 8). ABI4 expression is reduced in abi1-1 and abi3-1 seeds (Fig. 4). ABI3 expression is induced by ABA in ABI4c lines (Fig. 8). To determine whether ABI3 expression in seeds is regulated by ABI4 or ABI5 we assayed activity of an ABI3::GUS transgene, as well as transcript accumulation. ABI3 promoter activity was reduced approximately 3-fold in abi5-1 seeds and 4-fold in abi4-1 seeds (Table II). ABI3 transcript levels are slightly reduced in abi4-1 mutant seeds, but appear similar in abi5-1 and Wassilewskija (Ws) (wild-type) seeds (Fig. 10). These results indicate that, whereas ABI4 and ABI5 may regulate ABI3 promoter activity, ABI3 transcript accumulation is controlled by additional factors, possibly including mRNA stability.
Table II.
ABI3 promoter activity in abi4 and abi5 mutants
| Genotype | GUS Activity (units/seed)a |
|---|---|
| Wild type (C24) | 8,565 ± 297 |
| abi4-1 | 1,892 ± 227 |
| abi5-1 | 3,109 ± 907 |
GUS activity in mature seeds carrying an ABI3∷GUS transgene in wild-type (C24), abi4, and abi5-1 mutant backgrounds. Data presented are averages ± sd of assays with at least two independent extracts (C24) or segregants per genotype in the mutant backgrounds.
Figure 10.
ABI3 expression in abi4-1 and abi5-1 seeds. RNA gel-blot analysis of ABI3 transcript accumulation in abi4-1 and abi5-1 seeds relative to their wild-type progenitors. Each lane contains 5 μg of total RNA.
DISCUSSION
ABI4 Is a Transcriptional Activator
ABI4 is a member of the APETALA2 (AP2) domain family of transcriptional regulators. Sequence comparisons with other family members show extremely limited homology outside the conserved AP2 domain, which has been shown to be involved in DNA binding (Ohme-Takagi and Shinshi, 1995) and has been suggested to participate in dimerization of other AP2 domain proteins (Okamuro et al., 1997). Although transcription activation functions have been demonstrated for other family members (Buttner and Singh, 1997; Stockinger et al., 1997), it was not clear whether ABI4 alone could activate transcription. Assays of reporter gene expression have demonstrated that ABI4 does supply a strong activation function to a fusion with the GAL4-BD. In contrast, fusions with a series of truncated portions of ABI4 have little or no activity in this assay. These results indicate that loss of the C-terminal acidic domain and the adjacent Pro-rich domain severely disrupt the transcription activation function of ABI4. Both of these domains are missing in all of the available mutant alleles.
The fact that severe mutations in the ABI4 gene produce relatively weak phenotypic effects suggests that defects in ABI4 function may be masked by genetic redundancy. Although no close ABI4 homologs have been reported, we have cloned a gene corresponding to the most strongly hybridizing fragment detected by reduced stringency genomic DNA gel blots (data not shown). The predicted products of this gene and the closest match to ABI4 currently available in the database are highly similar to ABI4 within the AP2 domain (54% and 60% identical in predicted amino acid sequence, respectively). However, they are very divergent from ABI4 outside this domain and cluster more closely with the DREB2 subfamily (Liu et al., 1998) in multiple sequence alignments. Neither has been associated with a mutant phenotype and it is not known whether either plays a role in ABA signaling.
Seed Sensitivity to ABA Shows Little Variation among abi4 Alleles
Mutant abi4 alleles have been identified in a variety of screens including ABA-resistant germination (Finkelstein, 1994), salt-resistant germination (Quesada et al., 2000), and sugar-resistant seedling growth (Gibson et al., 1999; Huijser et al., 1999; Arenas-Huertero et al., 2000).
Comparison of sensitivities to ABA inhibition of germination among these mutants has produced a surprising range of results. Although the abi4-2 mutant was described as germinating almost completely on 30 μm ABA (Quesada et al., 2000), we found that all of the other alleles were blocked from germinating by this ABA concentration. All were approximately 3-fold less resistant to ABA than was reported for abi4-2, including a mutant producing a more severely truncated ABI4 protein than that in the abi4-2 mutant. It is not clear why the abi4-2 mutant appeared so much more resistant to ABA than any of the other alleles, but this result cannot be simply ascribed to the relative lengths of the mutant gene products.
ABI4 Function in Seeds
A combination of RNA gel-blot analyses and measurements of GUS activity driven by an ABI4 promoter fragment indicate that ABI4 is expressed throughout the embryo, from globular stage onward, but transcript levels are highest at seed maturity. Consistent with this expression pattern, the most severe defects of the mutants are observed in mature seeds: decreased sensitivity to ABA, salt and osmotic inhibition of germination, and significantly reduced expression of some lea genes. Although there are some alterations in transcript accumulation for genes expressed at earlier stages, most of these are relatively subtle changes, suggesting that ABI4 plays only a minor role in regulating their expression. It is interesting that some of the transcripts (e.g. M17) show increased accumulation, indicating that ABI4 may function as an activator or a repressor of different subsets of genes. Similar results have been described for regulation by ABI5 (Finkelstein and Lynch, 2000). Although M17 is expressed at high levels in late embryogenesis, its expression is repressed in young seedlings by ABA, salt, or drought treatment (Raynal et al., 1999). It is possible that ABI3 is essential for its induction late in embryogenesis, whereas ABI4 and ABI5 mediate the repression in response to ABA and stresses.
ABI4 expression is near normal in seeds of a variety of ABA-insensitive, ABA-hypersensitive, ABA-deficient, and heterochronic mutants, but slightly reduced in the abi1-1 and abi3-1 mutants. This contrasts with control of ABI5 expression, which is regulated by ABA and all of the ABI loci (Finkelstein and Lynch, 2000). Although ABI4 transcript accumulation is not regulated by many of these ABA- or seed response loci, it is still possible that ABI4 protein accumulation or activity is dependent on some of these loci.
Role of ABI4 in Post-Germinative Growth
ABI4 is expressed constitutively, but at very low levels, in vegetative tissues. Histochemical staining of ABI4-driven GUS activity shows low expression throughout the hypocotyl, roots, and cotyledons of young seedlings, which may be residual from the comparatively high expression levels in embryogenesis. Additional expression is observed in newly formed vegetative organs, but the localization patterns are variable and do not correlate with any specific tissues or structures.
Although previously described as ABA-inducible in 4-week-old plants (Wang et al., 1999), we saw no difference in ABI4-driven GUS expression in 5-d-, 11-d-, or 4-week-old plants treated with or without 100 μm ABA. Consistent with this, we saw no effect of ABA treatment on ABI4 transcript accumulation in 11-d-old plants. We also saw no difference in ABI4-driven GUS expression between control-treated plants and those exposed to drought, saline, or cold stresses. Although ABI4 expression was recently reported to be “Glc-induced,” based on reverse transcriptase-PCR comparison of transcript levels in plants grown for 15 d with or without 7% (w/v) Glc (Arenas-Huertero et al., 2000), ABI4-driven GUS activity shows no effect of 24 to 48 h exposure to 7% (w/v) Glc in 1- or 2-week-old plants. It is quite likely that the reported “Glc induction” reflects residual ABI4 transcript in wild-type plants whose development is arrested by exposure to high exogenous Glc.
Although ABI4 expression is not regulated by assorted stresses, ABI4 function appears to be important for controlling a number of stress responses in young plants, including suppression of growth and induction of anthocyanin synthesis. Although abi4 mutants appear “tolerant” of salt, osmotic, or sugar stresses in germination and early seedling growth, this does not result in improved growth (i.e. weight gain) relative to wild type over long term exposure to saline or osmotic stresses.
Ectopic ABI4 expression also resulted in hypersensitivity to ABA-inhibition of root growth. In fact, the lines with greatest ABI4 expression had stunted root growth even in the absence of applied ABA, possibly due to excessive response to their endogenous ABA levels. These plants were stunted overall, presumably due to the limited surface area of their roots and consequent limitations of nutrient and water uptake.
Interactions among ABI3, ABI4, and ABI5 in a Regulatory Network
Comparisons of embryonic gene expression in severe mutants of ABI3, ABI4, and ABI5 have shown that these transcription factors regulate many of the same genes, but in different ways. In general, loss of ABI3 function results in extreme reductions of embryonic gene expression (Parcy et al., 1994). In contrast, loss of ABI4 or ABI5 function produces relatively minor changes in expression of many ABI3-regulated embryonic genes (Fig. 6; Finkelstein and Lynch, 2000). The genes showing significantly altered expression in abi4 or abi5 mutants may be positively or negatively regulated by ABI4 and ABI5. Such differences in regulatory patterns are not consistent with action of these ABI genes in a simple regulatory hierarchy.
We have also examined interactions of these three ABI loci in controlling LEA and storage protein gene expression in vegetative tissues ectopically expressing either ABI3 or ABI4. Ectopic expression of either confers ABA-inducible expression of a subset of these “seed-specific” genes in 11-d-old plants. In addition, both sets of transgenic lines show hyperinduction of ABI5 expression in response to ABA treatment. Although the ABI4 ectopic expression lines also induce ABI3 expression when exposed to ABA, this regulation is not reciprocated: ABI4 transcript levels are unchanged by the presence of the ABI3 transgene and/or ABA exposure. These results suggested that ABI5 might be an important mediator of these transgenes' effects. However, comparison of ectopic ABI3-dependent marker gene expression in wild-type versus abi5-1 backgrounds showed that ABI5 function was not required for strong induction of the marker genes, although in some cases it was needed for full induction.
Attempts to correlate the degree of marker gene expression with any of the ABI genes indicated that different markers showed differential dependence on the ABI genes, but that strong expression of one ABI gene could reduce reliance on the others. To date, all of our analyses have focused on correlations among transcript levels and it is possible that these may not reflect the level of active protein for each of the ABI genes. It is also possible that the relatively subtle phenotypic effects of severe biochemical lesions in ABI4 and ABI5 reflect genetic redundancy. ABI5 is a member of a bZIP subfamily that includes at least four genes involved in ABA response that are expressed in young seedlings (Choi et al., 2000); these other family members may maintain ABI3-dependent vegetative expression of the “seed-specific” genes in an abi5-1 background. No close ABI4 homologs have been reported and we do not know whether any of the weakly related homologs play a role in ABA signaling.
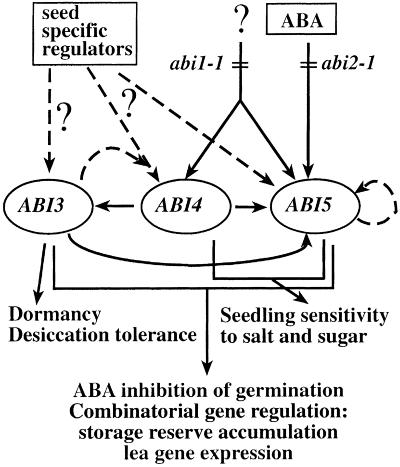
In summary, initial studies of genetic interactions indicated that ABI3, ABI4, and ABI5 were likely to act in a common seed-specific signaling pathway (Finkelstein, 1994) that could be introduced into vegetative tissue by ectopic expression of ABI3 (Parcy et al., 1994). It was subsequently shown that none of these three loci exhibited truly seed-specific expression (Finkelstein et al., 1998; Rohde et al., 1999; Finkelstein and Lynch, 2000), consistent with the possibility that the effects of ectopic ABI3 expression were dependent on either endogenous or induced expression of ABI4 and/or ABI5. We have now shown that ABI3 and ABI5 cross-regulate one another and that ectopic ABI4 expression confers ABA-inducible expression of ABI3 and ABI5. ABI3, ABI4, and ABI5 are all expressed throughout seed development and it is quite possible that expression of all three is regulated by as yet unidentified seed-specific regulators. In addition to the cross-regulation of ABI3, ABI4, and ABI5 loci described above, ABI1 and ABI2 appear to auto- and inter-regulate each other (Leung et al., 1997). Furthermore, the effects of ectopic ABI3 expression have been found to depend partially on ABI1 function (Parcy and Giraudat, 1997). A hypothetical model depicting the observed cross-regulation among the ABI loci is shown in Figure 11.
Figure 11.
Proposed model of regulatory interactions among the ABI loci, ABA, and unidentified seed-specific regulators. Solid arrows represent cross-regulation among ABI loci demonstrated by reduced expression in mutants and/or increased expression in ectopic expression lines. Dashed arrows indicate speculative autoregulation (e.g. ABI5), regulation by unknown seed-specific factors, or stage-specific cross-regulation (e.g. full ABI4 expression requires ABI3 function in seeds, but is not induced by ectopic ABI3 expression in vegetative tissues). Subsets of responses regulated by one or more ABI loci are listed at the bottom.
Although the ectopic expression studies show that abnormally high expression of ABI3 or ABI4 is sufficient for ABA hypersensitivity, this may not accurately reflect their regulatory roles in wild-type plants. These results are consistent with the hypothesis that ABI3, ABI4, and ABI5 act in a combinatorial network, each making differential contributions to regulation of genes expressed in seeds, and in response to ABA or assorted stresses. Direct physical interactions between rice homologs of ABI3 and ABI5 have recently been shown to enhance ABA-regulated gene expression in protoplasts derived from rice suspension-cultured cells (Hobo et al., 1999). This network is also likely to include additional genes already demonstrated to interact with members of this network, e.g. LEC1 and FUS3 (Parcy et al., 1997), and possibly functionally redundant genes, such as other members of the ABI5-related bZIP subfamily. Various subsets of this network could participate in “enhancesome” formation regulating expression of individual genes.
MATERIALS AND METHODS
Plant Material
The abi4-1 and abi5-1 mutant lines were isolated from the Columbia (Col) and Ws backgrounds, respectively, as described in Finkelstein (1994). The abi4-101, abi4-102, and abi4-103 mutants were isolated as sugar insensitive (sis) mutants in the Col background, as described in Laby et al. (2000). The aba1-1, abi1-1, abi2-1, and abi3-1 mutant lines were isolated from the Ler background, as described in Koornneef et al. (1982, 1984). The fus3-3 and era1-2 mutants were isolated from the Col background, as described in Keith et al. (1994) and Cutler et al. (1996). The lec1-1 mutant was isolated from the Ws background, as described in West et al. (1994).
The 35S::ABI4 transgenic lines were constructed in the Col and Ws backgrounds by vacuum infiltration (Bechtold et al., 1993) with Agrobacterium tumefaciens carrying the plasmids described below. Lines 114, 118, and 138 contain a transgene with a 53-bp 5′-UTR, whereas lines 130 and 131 contain 0.5-kb of 5′-UTR between the CaMV35S promoter and the initiating ATG. ABI4::GUS lines were constructed in the Col background.
The EN35S::ABI3 (isolate C7A19) and ABI3::GUS (isolate CAG3–11) transgenic lines were constructed in the C24 background as described in Parcy et al. (1994). To obtain lines with a EN35S::ABI3 or ABI3::GUS transgene in the mutant backgrounds, recombinant lines of abi4-1 and abi5-1 carrying the closely linked pyrimidine-requiring (py) mutation were crossed to the transgenic lines. Kanamycin-resistant F2 individuals were selected and screened for thiamine auxotrophy resulting from the py mutation. Families homozygous for the transgene were identified as 100% kanamycin resistant in the F3 generation.
For RNA isolation from siliques, plants were grown in soil in continuous light at 22°C. Siliques were harvested in pools corresponding to four developmental stages: maturation (8–11 dpa), post-abscission (12–16 dpa), late embryogenesis (17–21 dpa), and dry seed (>21 dpa). Seedlings were grown aseptically on Murashige-Skoog medium (1962) with 1% (w/v) Suc and 0.55% (w/v) agar for 11 d at 22°C in continuous light (50–70 μE m−2 s−1) then transferred to fresh Murashige-Skoog medium with 1% (w/v) Suc, 0.7% (w/v) agar, and 0 or 50 μm ABA for an additional 2 d before harvest. All tissues for RNA isolation were weighed, flash frozen in liquid nitrogen, and stored at −70°C until extraction.
For histochemical staining of GUS activity in 4-week-old plants, flowers, and developing seeds, plants were grown in soil in continuous light at 22°C. To test for induction of ABI4 promoter activity by ABA, 4-week-old plants were sprayed to run-off with 0.02% (w/v) Triton X-100, with or without 100 μm ABA, then tented with plastic wrap. Plants were sprayed once per day over 2 d, then harvested 24 h after the last application. Saline stress was imposed by watering plants with 100 mm NaCl on the 1st d, followed by 150 mm NaCl on the 2nd d. Tissues were harvested for GUS staining 24 h after the last treatment. Cold stress was imposed by incubation for 2 d in a dimly illuminated cold room at 6°C. Drought stress was imposed by not watering the plants for 13 d. For GUS staining of younger plants, seedlings were grown aseptically on Murashige-Skoog medium with 1% (w/v) Suc for 5 or 11 d at 22°C in continuous light (50–70 μE m−2 s−1), then transferred to fresh Murashige-Skoog medium with 1% (w/v) Suc and 0 or 100 μm ABA, 150 mm NaCl, or 500 mm sorbitol for an additional 2 d before harvest. To test for induction of ABI4 promoter activity by cold, one set of seedlings on control media was placed at 6°C for 2 d. To test for Glc-inducibility of ABI4 promoter activity, seedlings were grown on minimal medium (Haughn and Somerville, 1986) or 0.5× Murashige-Skoog medium with 0 or 1% (w/v) Suc (Murashige and Skoog, 1962) for 1 or 2 weeks, then transferred to fresh media with or without 7% (w/v) Glc for an additional 2 d before harvest.
Transgene Constructs
Ectopic ABI4 expression transgenes were constructed in pGA643 (An et al., 1988) with varying lengths of 5′-UTR between the CaMV35S promoter and the initiating codon of ABI4. One construct fused an intact SspI genomic fragment into the BglII site of pGA643, blunted by filling in the ends with the Klenow fragment of DNA polymerase I. The resulting construct contained 0.5 kb 5′ to the initiating codon of ABI4, as well as the coding region and 420 bp 3′ to the stop codon, including several possible polyadenylation sites. The other construct was a PCR-generated derivative of this SspI fragment with the same 3′ end, but only 53 bp of 5′-UTR.
Antisense ABI4 transgenes were constructed by subcloning a BglII-BamHI fragment spanning 1 kb of 5′ sequence and 0.9 kb of coding sequence in reverse orientation into the BglII site of pGA643, blunted as described above.
ABI4::GUS transgenes were constructed by subcloning a series of ABI4 genomic fragments into pBI101.3. All of the fragments terminated at an AluI site near the start of the ABI4 coding sequence, thereby creating translational fusions including the first four codons of ABI4. Although there was substantial quantitative variability among the transgenic lines, the qualitative expression patterns were very similar. The data presented displays results with the lines containing the largest promoter fragment, a 3-kb EcoRI-AluI fragment.
Binary plasmids carrying the transgenes were introduced into A. tumefaciens line GV3101 by direct transformation, followed by selection for growth on appropriate antibiotics (kanamycin for pBI101 derivatives, tetracycline and kanamycin for pGA643 derivatives).
One-Hybrid Assays of Transcription Activation Function
Fusions between varying portions of ABI4 and the GAL4-BD were constructed in the pGBD-C1 vector (James et al., 1996). All used an EcoRI site present at the 5′ end of a cDNA clone to make translational fusions starting at codon 3 of ABI4. The “full length” ABI4 fusion terminated at an EcoRI site that truncates the final product by one amino acid. Restriction sites used for the additional truncation products and the extent of each clone are described in Table I. All gene fusions were transformed into the yeast cell line PJ69–4A using the Alkali-Cation yeast transformation kit, according to the manufacturers instructions (Bio101, Vista, CA). Quantitative assays of GAL4-BD driven β-galactosidase gene expression were performed as described at http://www.fhcrc.org/∼gottschling/Bgal.html.
Germination Assays
For germination assays, 60 to 100 seeds per treatment were surface sterilized in 5% (w/v) hypochlorite and 0.02% (w/v) Triton X-100, then rinsed three to four times with sterile water before plating on minimal medium (Haughn and Somerville, 1986) containing 0.7% (w/v) agar and ABA (mixed isomers, Sigma, St. Louis) at 0, 3, 10, or 30 μm in 15- × 100-mm Petri dishes. The dishes were incubated 3 d at 4°C to break any residual dormancy, then transferred to 22°C in continuous light; germination was scored daily.
Root Growth Assays
For root growth assays, seeds were surface sterilized as described above before plating on germination medium (Valvekens et al., 1988) containing 0.7% (w/v) agar. Petri plates were incubated 3 d at 4°C, then transferred to 22°C in continuous light (50–70 μE m−2 s−1). After 2 d, germinated seedlings were transferred to germination medium supplemented with 0 or 3 μm ABA. Plates were incubated vertically, with seedlings placed with their root tips pointing up such that new root growth would occur along the surface of the plates in the opposite direction from the original growth. New root growth was measured after 4 d, then average growth was calculated for each genotype and treatment and expressed as a percentage of the growth on hormone-free media.
Tests of Saline or Osmotic Stress Tolerance
Seeds were surface sterilized as described above before plating on germination medium (Valvekens et al., 1988) containing 0.7% (w/v) agar. Petri plates were incubated 3 d at 4°C, then transferred to 22°C in continuous light (50–70 μE m−2 s−1). After 2 d, germinated seedlings were transferred to germination medium supplemented with nothing, 50, or 100 mm NaCl, or 150 or 300 mm sorbitol. After 2 weeks additional growth, samples of 10 to 15 plants were harvested, blotted dry of surface moisture, and weighed. Dry weights were determined after an additional 2 to 3 d incubation in a drying oven at 65°C. Average weights per plant were calculated and expressed as a percentage of those grown on control medium.
RNA Gel-Blot Analysis
RNA was isolated from seeds and vegetative tissue by hot phenol extraction as described previously (Finkelstein, 1993). Additional seed RNA preps were based on the procedure of Vicient and Delseny (1999), modified by grinding directly in sintered glass homogenizers with extraction buffer rather than an initial grinding in liquid nitrogen with sterile quartz powder. RNA from siliques was isolated by extraction in 0.2 m Tris [tris(hydroxymethyl)-aminomethane], pH 9, 0.4 m NaCl, 25 mm EDTA, 1% (w/v) SDS, 5 mg mL−1 polyvinylpolypyrrolidone, and 0.5 mg mL−1 proteinase K, followed by precipitation of polysaccharides, proteins, and other contaminants by incubation on ice with 18.3 mg mL−1 BaCl2 and 150 mm KCl, as described in Finkelstein et al. (1998). RNA concentrations were estimated based on absorbance at 260 and 280 nm.
Total RNA (1–20 μg per lane) was size fractionated on MOPS [3-(N-morpholino)-propanesulfonic acid]-formaldehyde gels (Sambrook et al., 1989), then transferred to Nytran (Schleicher & Schuell, Keene, NH) membranes using 20 × sodium chloride/sodium phosphate/EDTA as blotting buffer. RNA was bound to the filters by UV-crosslinking (120 mJ cm−2 at 254 nm). Uniformity of loading and transfer was assayed qualitatively by methylene blue staining of the filters (Herrin and Schmidt, 1988) and eventually hybridization to an rDNA probe. Transcripts from At2S3, CRC, AtEm1, ABI3, and the RAB18 homolog were detected by hybridization to cDNA clones as described in Finkelstein (1994), labeled by random-priming to a specific activity of 108 cpm μg−1. The AtEm6 mRNA was detected by hybridization to a genomic clone encompassing the entire transcribed region and 0.8-kb 5′ flanking sequences. The ABI5 probe was a PCR-amplified genomic fragment excluding most of the conserved bZIP domain. The ABI4 probe was an EcoRI fragment from a cDNA clone encompassing all but the first two and final codons of the coding sequence. Hybridization conditions were (50% [w/v] formamide, 5× sodium chloride/sodium phosphate/EDTA, 5× Denhardts, 0.1% [w/v] SDS, and 200 μg mL−1 DNA) at 43°C or (7% [w/v] SDS, 0.5 m sodium-phosphate, pH 7.2, 1 mm EDTA, and 1% [w/v] bovine serum albumin) at 65°C for 16 to 24 h (Church and Gilbert, 1984) in a Hyb-Aid rotisserie oven. Filters were washed twice at 60°C in 2× SSC, 0.1% (w/v) SDS and once at 60°C in 0.2× SSC, 0.1% (w/v) SDS for 30 to 60 min. All hybridization reagents were prepared as described in Sambrook et al. (1989). Hybridization intensity was quantified by either phosphorimage analysis (Storm 640, Molecular Dynamics, Sunnyvale, CA) or densitometric analysis of autoradiograms scanned on a flatbed scanner (Vista S-8, Umax, Fremont, CA) using Photoshop to capture the images and NIH Image to calculate intensities, then normalized relative to hybridization to a radish rDNA probe (Delseny et al., 1983).
Measurement of GUS Activity
Soluble extracts of seeds were assayed fluorometrically for GUS activity using 4-methylumbelliferyl glucuronide (Rose Scientific, Canada) as substrate, as described in Jefferson et al. (1987).
GUS activity in intact plants or organs was detected histochemically by infiltration with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, as described in Jefferson et al. (1987). Plant material was cut and incubated in GUS staining solution containing 50 mm sodium phosphate, pH 7.0, 0.1% (w/v) Triton X-100, K3/K4 FeCN 0.5 mm, and 1 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid at 37°C for 2 to 72 h depending on staining intensity. Tissues were cleared of chlorophyll in ethanol. Photographs of whole-mounted tissues were taken using a stereomicroscope.
ACKNOWLEDGMENTS
We thank Dr. Douglas Bush for critical review of the manuscript, Drs. Jerome Giraudat and Francois Parcy for the EN35S::ABI3 transgenic lines, Dr. Michel Delseny for the EST cDNAs used for gene expression analyses, Dr. Peter McCourt for the fus3-3 and era1-2 mutants, and Dr. John Harada for the lec1-1 mutant.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9728297 to R.R.F.) and by the United States Department of Agriculture (grant no. 95–37304–2217 to R.R.F.). E.M.S. was supported by a postdoctoral fellowship from The Swedish Foundation for International Cooperation in Research and Higher Education.
LITERATURE CITED
- An G, Ebert P, Mitra A, Ha S. Binary vectors. In: Gelvin S, Schilperoort R, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1988. pp. 1–19. [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Bonetta D, McCourt P. Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 1998;3:231–235. [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim S. ABFs, a family of ABA-responsive element binding factors. J Biol Chem. 2000;275:1723–1730. doi: 10.1074/jbc.275.3.1723. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Delseny M, Cooke R, Penon P. Sequence heterogeneity in radish nuclear ribosomal RNA genes. Plant Sci. 1983;30:107–119. [Google Scholar]
- Finkelstein RR. Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant gene. Mol Gen Genet. 1993;238:401–408. doi: 10.1007/BF00291999. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR. Three classes of abscisic acid-insensitive mutations of Arabidopsis define genes that control overlapping subsets of abscisic acid responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Zeevaart JAD. C Somerville and E Meyerowitz, eds, Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. Gibberellin and abscisic acid biosynthesis and response. [Google Scholar]
- Gibson SI, Laby RJ, Kim D (1999) Sugar-insensitive mutants of Arabidopsis with defects in phytohormone metabolism and/or response. Presented at 10th International Conference on Arabidopsis Research, July 4–8, 1999, Melbourne, Australia
- Giraudat J, Hauge B, Valon C, Smalle J, Parcy F, Goodman H. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Herrin D, Schmidt G. Rapid, reversible staining of northern blots prior to hybridization. BioTechniques. 1988;6:196–200. [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA. 1999;96:15348–15353. doi: 10.1073/pnas.96.26.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser JC, Pego J, Kortstee A, Smeekens S (1999) The sugar sensing mutant sun6 is insensitive to abscisic acid: involvement of abscisic acid and the ABI genes in sugar sensing. Presented at 10th International Conference on Arabidopsis Research, July 4–8, 1999, Melbourne, Australia
- James P, Halladay J, Craig E. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannette E, Rona J-P, Bardat F, Cornel D, Sotta B, Miginiac E. Induction of RAB18 gene expression and activation of K+ outward rectifying channels depend on extracellular perception of ABA in Arabidopsis thaliana suspension cells. Plant J. 1999;18:13–22. doi: 10.1046/j.1365-313x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. Fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna M, Brinkhorst-van der Swan D, Karssen C. The isolation of abscisic acid (ABA)-deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen C. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- Laby R, Kincaid M, Kim D, Gibson S. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nambara E, Naito S, McCourt P. A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new ABI3 allele. Plant J. 1992;2:435–441. [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J, Caster B, Villarroel R, Van Montagu M, Jofuku K. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Giraudat J. Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 1997;11:693–702. doi: 10.1046/j.1365-313x.1997.11040693.x. [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science. 1998;282:287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce M, Micol J. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;154:421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynal M, Guilleminot J, Gueguen C, Cooke R, Delseny M, Gruber V. Structure, organization and expression of two closely related novel Lea (late-embryogenesis-abundant) genes in Arabidopsis thaliana. Plant Mol Biol. 1999;40:153–165. doi: 10.1023/a:1026403215270. [DOI] [PubMed] [Google Scholar]
- Robertson D. The genetics of vivipary in maize. Genetics. 1955;40:745–760. doi: 10.1093/genetics/40.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W. The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ. 1999;22:261–270. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat-DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient C, Delseny M. Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem. 1999;268:412–413. doi: 10.1006/abio.1998.3045. [DOI] [PubMed] [Google Scholar]
- Wang ML, Belmonte S, Kim U, Dolan M, Morris JW, Goodman HM. A cluster of ABA-regulated genes on Arabidopsis thaliana BAC T07M07. Genome Res. 1999;9:325–333. [PubMed] [Google Scholar]
- West MAL, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]