Abstract
Allergen-specific type 2 helper T (TH2) cells play a central role in initiating and orchestrating the allergic and asthmatic inflammatory response pathways. One major factor limiting the use of such atopic disease–causing T cells as both therapeutic targets and clinically useful biomarkers is the lack of an accepted methodology to identify and differentiate these cells from overall nonpathogenic TH2 cell types. We have described a subset of human memory TH2 cells confined to atopic individuals that includes all allergen-specific TH2 cells. These cells are terminally differentiated CD4+ T cells (CD27− and CD45RB−) characterized by coexpression of CRTH2, CD49d, and CD161 and exhibit numerous functional attributes distinct from conventional TH2 cells. Hence, we have denoted these cells with this stable allergic disease–related phenotype as the TH2A cell subset. Transcriptome analysis further revealed a distinct pathway in the initiation of pathogenic responses to allergen, and elimination of these cells is indicative of clinical responses induced by immunotherapy. Together, these findings identify a human TH2 cell signature in allergic diseases that could be used for response-monitoring and designing appropriate immunomodulatory strategies.
INTRODUCTION
As part of their specialization, CD4+ effector T cells acquire functional and phenotypic characteristics to specifically respond against pathogens. Within different T helper (TH) cell subsets, the TH2 cell subset is characterized by the production of interleukin-4 (IL-4), IL-5, IL-9, and IL-13 cytokines, which promote both immunoglobulin E (IgE)–and eosinophil-mediated immune responses (1). Although TH2 cells were initially considered to be a homogeneous subset, their functional heterogeneity is now appreciated, as is the fact that additional TH2 sub-populations may determine TH2-driven pathology (2–4). For example, a recent study revealed a subpopulation of human memory TH2 cells that produces IL-17 along with cardinal TH2 cytokines (5). Remarkably, the proportion of these circulating TH17/TH2 cells was extremely low in nonatopic individuals compared to patients with chronic severe asthma, suggesting a possible role in the pathogenesis and severity of the disease. Another source of heterogeneity among CD4+ T cell subsets is at the level of T cell surface marker expression that determines their differentiation states, effector functions, and migratory capacity. With respect to the TH2 cell subset, our group recently demonstrated that pathogenic allergen-specific T cells are highly matured effector TH2 cells characterized by the lack of expression of CD27, a tumor necrosis factor receptor superfamily member of costimulatory molecules (6, 7). Similarly, distinct subpopulations of TH2 cells with enhanced function have been described in a murine model of allergic inflammation based on differential expression of CXCR3 and CD62L (8) or CCR8 (9) and in human allergic eosinophilic inflammatory diseases, according to the expression of the hematopoietic prostaglandin D synthase (hPGDS) (10) or IL-17RB (11). In these studies, the authors suggested that heterogeneity within TH2-mediated immune responses plays differential roles in immunopathology. Hence, we surmise that allergic individuals have specific subpopulations of TH2 cells associated with global atopic inflammatory disorders.
Until now, there has been no biological measurement to accurately reflect and quantify an underlying allergic disease process and ideally provide accurate surrogate end points to assess immunotherapy efficacy. A major impediment to the use of allergic disease–causing T cells as a therapeutic target and clinically useful biomarker is the lack of an accepted method to both identify these cells and differentiate them from the overall TH2 cell types. Recent progress in peptide–major histocompatibility complex (MHC) class II (pMHCII) tetramer staining has allowed direct ex vivo visualization of allergen-specific CD4+ T cells and enabled quantification and characterization of these cells in a setting closer to their natural physiological state (7, 12). Description of a set of T cell surface markers that are differentially expressed in allergen-specific TH2 cells as compared to classical TH2 cells would allow this issue to be addressed.
Here, we describe an allergic T cell signature characterized by the coexpression of the chemoattractant receptor CRTH2, the natural killer cell marker CD161, and the homing receptor CD49d in human terminally differentiated (CD45RBlow CD27−) CD4+ T cells. The vast majority of allergen-specific T cells in allergic individuals with either food, pollen, pet’s dander, mold, or house dust mite allergy fall into this subset and were preferentially deleted during allergen-specific immunotherapy (AIT). Hence, we have denoted this proallergic sub-population of TH2 cells, confined to atopic individuals, as the TH2A cell subset. Transcript analysis further highlights key functional differences between TH2A cells and conventional TH2 cells, providing molecular signatures that suggest specific contribution of the TH2A cell subset to allergic disease. Together, these findings identify a pathogenic TH2 cell signature unique to allergic individuals that could potentially be used as a clinically relevant biomarker and therapeutic target in atopic disorders.
RESULTS
Allergic disease–related phenotypic differences exist in the TH2 cell subset
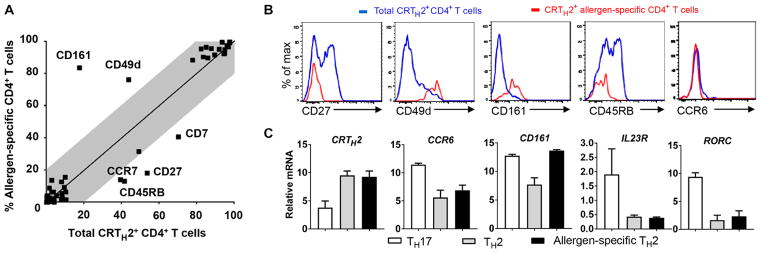
For many years, chemokine receptors and surface markers have been instrumental in the characterization of memory T cell subsets with distinct migratory capacity and effector functions. To determine whether a set of T cell surface markers can be differentially expressed in allergen-specific TH2 cells, we undertook a detailed ex vivo phenotypic profiling of total CD4+ T cells, conventional TH2 cells, and allergen-specific CD4+ T cells. Using alder pollen allergy as a model, freshly isolated peripheral blood mononuclear cells (PBMCs) from DR07:01- or DR15:01-restricted allergic individuals were stained with fluorescently labeled pMHCII tetramers, followed by magnetic column enrichment process to directly examine allergen-specific CD4+ T cell phenotypic profiles. Among TH2-associated surface markers, CRTH2, the prostaglandin D2 receptor chemoattractant receptor–homologous molecule expressed on TH2 cells, is reported as the most reliable marker to identify human TH2 cells (13). As a control, we examined the ex vivo phenotypic profile of total CRTH2+ CD4+ memory T cells to compare with the ex vivo enriched allergen-specific CD4+ T cells. During these flow cytometric screen analyses, fluorochrome-conjugated antibodies directed against cell surface marker antigens were selected to elucidate the differentiation, maturation, activation, and homing properties of each group (fig. S1 and table S1). Variation in surface marker expression between groups is shown in fig. S2 (A and B). As expected, ex vivo enriched allergen-specific CD4+ T cells from allergic individuals share numerous memory TH2 cell features with the conventional TH2 cell group featuring the expression of CD45RO, CCR4, CD200R, CD58, CD29, and CRTH2. However, we identified an allergic T cell signature that includes two up-regulated (CD161 and CD49d) and four down-regulated (CD27, CD45RB, CCR7, and CD7) T cell surface markers with significant differential expression (greater than 20% change; P < 0.001) between groups (Fig. 1A). The CD27low, CCR7low, CD7low, and CD45RBlow phenotypes, which are associated with terminally differentiated memory CD4+ T cells, likely reflect recurrent natural allergen exposure (14, 15). This is consistent with previous findings by our group demonstrating a strong relationship between pathogenicity of allergen-specific CD4+ T cells and the maturation stage of the cells (7, 16). Although loss of CD27 expression within CD4+ memory T cells is consistently associated with cells lacking CCR7 and CD7, we observed that CD27low CD4+ T cell subset can be subdivided into two groups by CD45RB expression (fig. S3). Thus, to define a smaller set of surface markers, we chose CD27 and CD45RB as convenient down-regulated markers reflecting allergic features.
Fig. 1. Allergic disease–related phenotypic differences emerged in the TH2 cell subset.
(A) Fluorescence-activated cell sorting (FACS)–based T cell surface expression screening revealed up-regulated and down-regulated T cell surface markers in ex vivo magnetically enriched allergen-specific CD4+ T cells compared to total CRTH2+ CD4+ T cells. Average expression levels for each T cell surface marker in the allergen-specific CD4+ T cell group and in total CRTH2+ CD4+ T cell group are plotted against each other. Data are means from four allergic subjects per group. The gray field depicted less than 20% expression variation between groups. Differences between groups were analyzed using the Mann-Whitney U test. (B) Examples of intensity distributions of total CRTH2+ CD4+ T cells (blue) and ex vivo magnetically enriched CRTH2+ allergen-specific CD4+ T cells tracked by pMHCII tetramer (red) stained with candidate cell surface markers. Data are representative of at least three allergic donors. (C) Real-time PCR analysis confirms that allergen-specific TH2 cells express CD161 but are not related to a type 17 phenotype. Data are means ± SEM from at least three subjects per group.
Another striking finding from this T cell profiling was the over-expression of the C-type lectin-like receptor CD161 (4.2-fold difference, P < 0.001) as part of the signature characterizing allergen-specific TH2 cells. Expression of CD161 on CD4+ T cells is typically associated with TH17 responses (17, 18), and like the conventional TH2 cell subset (CRTH2+ CD4+), allergen-specific TH2 cells do not express the TH17-associated chemokine receptor CCR6 (Fig. 1B). We next performed quantitative polymerase chain reaction (PCR) analysis on sorted cells from allergic donors and confirmed the higher expression of CD161 mRNA in CRTH2-expressing allergen-specific T cells compared to conventional TH2 cells (Fig. 1C). However, although allergen-specific TH2 cells express similar levels of CD161 as the TH17 cell subset (CCR6+ CXCR3− CD4+), these cells did not exhibit mRNA expression of TH17 phenotypic markers such as CCR6, IL23R, and the transcription factor RORC. Together, these data indicate that allergic disease–related phenotypic differences (not related to a type 17 phenotype) occur in the TH2 cell subset.
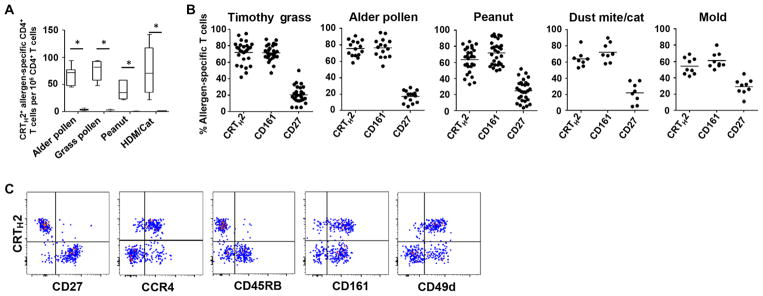
To demonstrate that our data were not restricted to tree pollen allergy, we next performed our ex vivo pMHCII tetramer approach to characterize allergen-specific CD4+ T cells in patients with either food allergy (peanut), perennial allergy (cat and house dust mite), mold allergy (Aspergillus and Alternaria), or seasonal pollen allergy (alder and timothy grass). We also used nonallergic individuals as controls. Whatever the allergen tested in this study, IgE-mediated allergic diseases were characterized by high frequencies of allergen-specific CRTH2+ T cells, which were strictly absent in nonallergic subjects, suggesting that the presence of these CD4+ effector T cells is necessary for allergy pathogenesis (Fig. 2A). In all allergic individuals tested, the vast majority of pMHCII tetramer–positive T cells were also characterized by the lack of CD27 expression along with expression of CD161 (Fig. 2B). Remarkably, CRTH2+ expression on allergen-specific CD4+ T cells was concomitant with a lack of CD45RB and CD27 expression as well as coexpression of CD161 and CD49d (Fig. 2C and fig. S4). Collectively, these data identify the pathogenic allergen-specific TH2 cell subset in atopic individuals as highly mature (CD27−CD45RBlow) TH2 cells coexpressing CD161 and CD49d.
Fig. 2. A unique allergic disease footprint across allergen-specific TH cells.
(A) Average frequencies of CRTH2+ allergen-specific T cells in allergic (white box) and nonallergic subjects (black box) are indicated for each allergen tested. Data are means ± SEM from at least six individuals per group. *P < 0.001. Differences between groups were analyzed by using the Mann-Whitney U test. (B) Percentage of CRTH2+, CD161+, and CD27+ cells among ex vivo magnetically enriched allergen-specific CD4+ T cells from allergic individuals is indicated for each allergen tested. Each dot represents a single donor. (C) Plots show representative ex vivo profile of alder pollen–specific CD4+ T cells in alder-allergic patient according to CD27, CCR4, CD45RB, CD161, CD49d, and CRTH2 expression. Data are representative of at least three donors.
A distinct TH2 cell subset is associated with type 1 allergic diseases
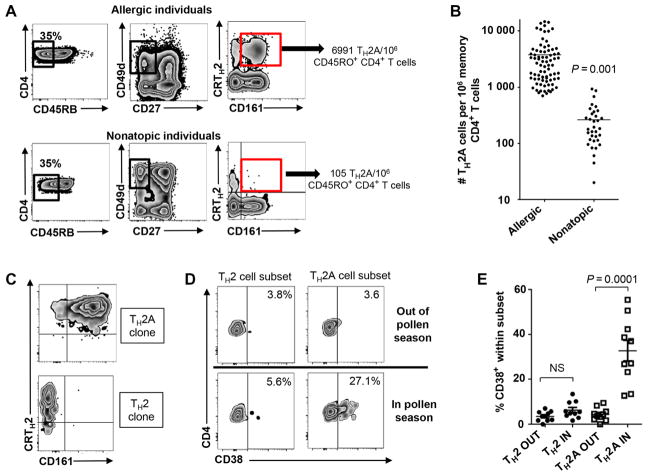
We next sought to determine whether the pathogenic T cell signature identified on allergen-specific TH2 cells could be used to define a subset of the TH2 cells that would reflect an underlying allergic disease process. Although it has been argued that CRTH2+ CD4+ T cells are present at higher frequency in allergic subjects, we observed that this difference is marginal (fig. S5A). Despite a substantially lower proportion of CD161-expressing CRTH2+ T cells in nonatopic individuals, this subset was not restricted to allergic subjects. However, we observed that at least two markers (that is, CD161 and CR45RB or CD27) were needed to subset the CRTH2+ CD4+ T cells to identify an allergy-prone TH2 subset virtually absent in the nonatopic group, which includes the vast majority of allergen-specific T cells from allergic individuals (fig. S5, B and C). Using the gating strategy depicted in Fig. 3A, we observed that all allergic individuals tested exhibited a significantly higher number (n = 80; mean ± SEM, 3766 ± 413 cells per 106 memory CD4+ T cells) of CD45lowCD49d+CD27− CRTH2+CD161+ cells relative to nonatopic individuals (n = 34; mean ± SEM, 259 ± 37 cells per 106 memory CD4+ T cells; P < 0.0001) (Fig. 3, A and B). Hence, we have named these proallergic TH2 cells (which are unique to allergic individuals) the TH2A cell subset.
Fig. 3. A distinct subset of TH2 cells include pathogenic allergen-specific CD4+ T cells.
(A) Gating strategy for defining proallergic TH2 cells (TH2A cells). PBMCs were first gated according to their size, expression of CD4 and CD45RO, and after the exclusion of dead cells. Gates then identify CD45RBlow cells among live memory (CD45RO+) CD4+ T cells, CD27−CD49d+ cell subset, and then CRTH2+CD161+ T cell subset. Representative staining in allergic individual and nonatopic subject is shown. (B) Frequency of CD45RBlowCD27−CRTH2+CD161+CD49d+ CD4+ T cells (TH2A) between allergic subjects (n = 80) and nonatopic individuals (n = 34). Each dot represents a single donor, and differences between groups were analyzed by using the Mann-Whitney U test. (C) TH2 and TH2A phenotype observed over a culture time of 6 weeks with subsequent T cell receptor (TCR) stimulations. (D and E) Percentage of TH2A and TH2 cells expressing CD38 in and out grass pollen season in grass-allergic individuals. Data are representative of at least three donors (A, C, and D). Differences between groups were analyzed by using the Wilcoxon matched pairs test. NS, not significant.
Remarkably, both conventional TH2 and TH2A cell subsets retain their respective phenotype after long-term clonal expansion, suggesting that they did not differ in activation or maturation status and can thus be used as a stable and relevant surrogate marker (Fig. 3C). To confirm that the TH2A cell subset is specifically involved in type I allergic diseases, we next followed 10 grass pollen–allergic individuals before and during the grass pollen season (May to August), a window of time that correlates with increased allergy symptoms and with up-regulation of the activation marker CD38 within grass pollen–reactive CD4+ T cells (7, 16). Consistent with direct access to allergy-prone TH2 cells according to CRTH2, CD27, CD45RB, CD49d, and CD161 differential expression, we observed that CD38 expression was specifically up-regulated within the TH2A subset during grass pollen season but not within the conventional TH2 cell subset or outside pollen season (Fig. 3, D and E). Collectively, our data demonstrate that the TH2A cell subset represents a phenotypically distinct TH2 sub-population, which may encompass the vast majority of pathogenic TH2 cells involved in type I allergic diseases.
The TH2A cell subset represents a suitable therapeutic target
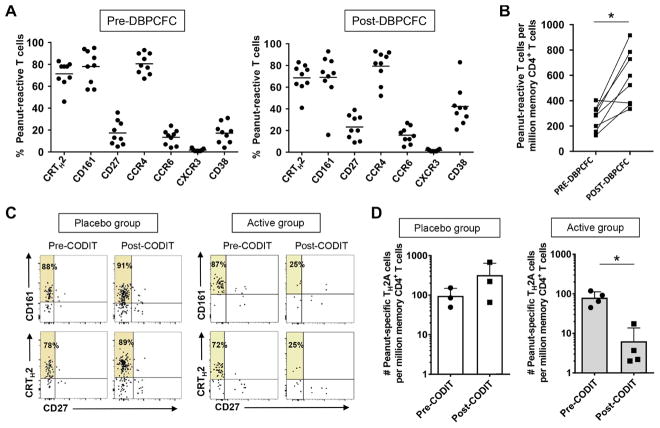
To determine whether the TH2A cell subset constitutes a clinically relevant therapeutic target in the allergy context, we next performed a longitudinal study in a subset of peanut-allergic patients completing characterized oral desensitization immunotherapy (CODIT) with AR101, an experimental orally administered biological drug containing the antigenic profile found in peanuts. During this randomized, double-blinded, placebo-controlled trial (ARC001), coded samples from subjects were provided to the operator at baseline both before and after double-blind, placebo-controlled food challenges (DBPCFC) with peanut flour, as well as at the end of the maintenance visit before DBPCFC. The magnitude and quality of peanut-specific T cell responses were determined ex vivo using the CD154 up-regulation assay (19) after short restimulation of PBMCs with a pool of peanut peptides library derived from Ara h 1, Ara h 2, Ara h 3, Ara h 6, and Ara h 8 peanut-allergic components. As expected, the vast majority of peanut-reactive CD4+ T cells were bona fide TH2A cells at baseline, and the DBPCFC protocol led to significant increased expression of the cell surface activation marker CD38 (Fig. 4A and fig. S6A), concomitant with an increased average frequency of these cells (Fig. 4B). Accordingly, only TH2A cells, and not conventional TH2 cells, were specifically activated after peanut oral food challenge (OFC) (fig. S6B).
Fig. 4. Peanut-specific TH2A cells are specifically targeted during immunotherapy.
(A) Ex vivo phenotype of peanut-reactive CD4+ T cells before and after DBPCFC with peanut flour. Each dot represents a single donor. (B) Ex vivo frequency of peanut-reactive CD4+ T cells before and after DBPCFC. (C) Plots show representative ex vivo profile of peanut-reactive CD4+ T cells according to CD27, CD161, and CRTH2 expression before and after CODIT both in placebo and active groups. Data are representative of at least three donors per group. Percentages of CD27− allergen-specific T cells expressing the given marker are indicated in the upper left quadrant. (D) Ex vivo peanut-specific TH2A cell frequencies before and after CODIT both in placebo (n = 3) and active (n = 4) groups. Differences between groups were analyzed by using the Wilcoxon matched pairs test (A and B) and unpaired t test (D). *P < 0.05.
As reported elsewhere (20), 100 and 78% of patients who completed the active treatment regimen (n = 23) tolerated a cumulative amount of peanut protein of 443 and 1043 mg, respectively, compared to 19 and 0% in the placebo group (n = 26). In such a setting, we observed a direct correlation between decrease in peanut-specific TH2A cell frequency and achievement of peanut desensitization in the active group compare to placebo (Fig. 4, C and D, and fig. S6C). Together, our data demonstrate that TH2A cells play a critical role in allergic disease pathogenesis and reinforce previous data by our group that the allergen-specific TH2 cell subset may represent a suitable therapeutic target and surrogate marker of clinical efficacy during AIT (7, 16, 21).
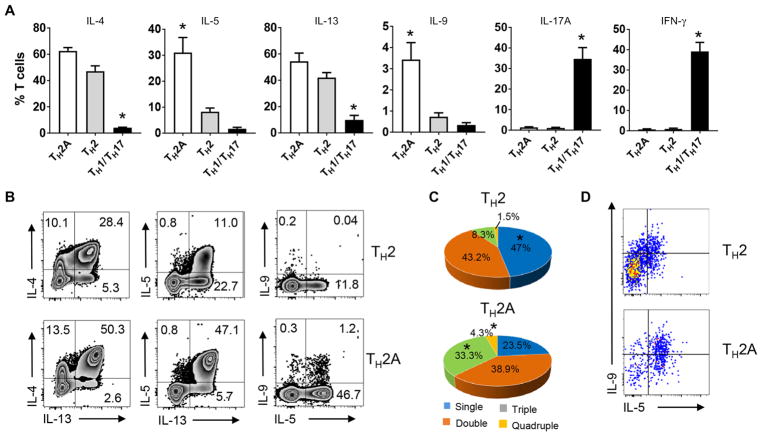
TH2A cells differentially contribute to TH2-driven pathology
To determine whether allergic disease–related functional differences could be identified in the TH2A cell subset, freshly isolated TH2A, TH2 (CD161−CRTH2+CD27−), and TH1/TH17 (CD161+CRTH2−CD27−) cell subsets from allergic individuals were subjected to polychromatic intracellular cytokine profile analysis. After polyclonal activation with phorbol 12-myristate 13-acetate(PMA)/ionomycin, a significantly higher proportion of TH2A cells expressed IL-5 and IL-9 compared to conventional TH2 cells (Fig. 5A). Conversely, interferon-γ (IFN-γ) and IL-17, the respective cytokines for TH1 and TH17 cell subsets, were restricted to the CD161+CRTH2−CD27+ TH cell population. The TH2A cell subset was also more polyfunctional, with a significantly greater proportion of cells producing simultaneously multiple TH2 effector cytokines compared to conventional TH2 cells (Fig. 5, B and C). As a comparison, expression of cardinal TH2 cytokine was also investigated within ex vivo enriched allergen-specific CD4+ T cells in allergic individuals and found to be restricted to the CD27−CRTH2+CD161+ allergen-specific CD4+ T cell subset (fig. S7). Remarkably, the unique secretion pattern of TH2A cell lines was quite stable over time, even after multiple rounds of stimulations over sequential 6-week cultures (Fig. 5D). Thus, human circulating TH2A cells may contribute differently to TH2-driven pathology than conventional TH2 cells by simultaneously producing multiple cardinal TH2 cytokines.
Fig. 5. TH2A cell subset may differentially contribute to TH2-driven pathology.
(A) Cytokine production by TH2A (white bar), conventional TH2 (gray bar), and TH1/TH17 (black bar) cell subset. T effector cell subset from allergic individuals was sorted by FACS and stimulated for 5 hours with PMA/ionomycin in the presence of a protein transport inhibitor. Data are means ± SEM of four subjects per group. Differences between groups were analyzed by using the Mann-Whitney U test. *P < 0.01. (B) Plots show representative ex vivo intracellular cytokine staining for IL-4, IL-13, IL-5, and IL-9 in FACS-sorted TH2 and TH2A subset. Numbers indicate relative percentages in each quadrant. (C) Pie charts show the proportion of cells producing simultaneously one, two, three, or four cardinal TH2 cytokines (IL-4, IL-5, IL-9, and IL-13) after polyclonal activation. Data are mean percentage of cytokine-producing cells from four allergic donors. Comparisons between groups were performed using Kruskal-Wallis one-way analysis of variance (ANOVA) on ranks. *P < 0.01. (D) Plots show representative intracellular cytokine staining for IL-5 and IL-9 in TH2 and TH2A cell clone from the same allergic individuals. Data are representative of at least three allergic donors (B and D).
Transcriptome analysis reveals unique pathway in TH2A cells
To further investigate the pathophysiologic meaning of the allergic T cell signature, we performed microarray analysis (Gene Expression Omnibus accession GSE93219) on freshly isolated TH2A cells compared to known T cell subsets (that is, TH1, TH17, and TH2) from different donor pools, which contained blood from two to three donors. This was necessary to obtain sufficient numbers of cells for microarray experiments. From the data sets comparing TH2A with TH2 cells, epithelium-derived cytokines receptors, such as the IL-25 receptor (IL-17RB), the IL-33 receptor (IL1RL1), and the thymic stromal lymphopoietin–receptor (CLRF2), which are well-known molecules involved in the allergic/asthmatic immune response (22–24), were more highly expressed in TH2A cells relative to conventional TH2 cells (Fig. 6, A and B). In addition, we confirmed that TH2A cells produced more IL-5 and IL-9 relative to conventional TH2 cells, whereas TH1-and TH17-related genes (IFN-γ, IL-17, RORC, IL23-R, and CCL20) were absent in TH2 and TH2A cell subset (Fig. 6B). TH2A cells also highly expressed genes involved in arachidonic acid signaling that have previously been linked to allergic disease such as hPGDS (10), the prostaglandin synthase PTGS2 (25, 26), the short-chain free fatty acid receptor GPR42 (27), and the peroxisome proliferator–activated receptor PPARγ (table S2) (21). Because of limitations of currently available anti-human ST2 and IL17RB reagents, we were unable to observe the differential expression of these two markers on the surface of peripheral CD4+ T cells by using flow cytometry. Thus, we wished to determine whether up-regulation of IL-17RB and IL1RL1 transcript identified in the TH2A cell subset was specifically observed on allergen-specific T cells from allergic individuals. To this aim, we performed a real-time PCR expression analysis on sorted pMHCII tetramer–positive T cells tracking peanut-specific CD4+ T cells in peanut-allergic subjects and in nonatopic individuals. Sorted conventional TH2 cells from the same allergic subjects were also used as control. As expected, we confirmed that gene transcripts, such as CD161, IL1RL1, and IL17RB, were expressed in allergen-specific CD4+ T cells from allergic individuals but were absent both in conventional TH2 cells and in allergen-specific T cells from nonallergic individuals (Fig. 6C). Although not causal, these data imply that pathological differences between TH2A and conventional TH2 cells in allergic individuals are fundamental to disease development (fig. S8).
Fig. 6. TH2A cell subset shows distinct gene expression patterns.
(A) Scatterplot of the average signal of TH2A versus conventional TH2 cell gene expression microarray data. Shown are genes whose transcription has been up-regulated (red) or down-regulated (blue) by a factor of 2. Genes that have previously been linked to allergic diseases are listed. (B) Hierarchical clustering heat map of all genes with expression fold changes of eight in one cell subset relative to the other three subsets. Data are mean normalized raw gene expression values from two independent microarray experiments on cells sorted from different donor pools (each pool containing blood from two to three donors). (C) Real-time PCR analysis showing mRNA expression profile of the most relevant genes up-regulated in TH2A cell subset in total CRTH2+ T cells (gray) and in allergen-specific T cells from nonallergic individuals (white) or allergic subjects (black). Data are means ± SEM from at least three subjects per group.
DISCUSSION
Although antigen-specific TH2 cells are at the core of the allergic process in atopic individuals, tracking and targeting these allergic disease–causing T cells without affecting other nonpathogenic TH2 processes have been a challenge. Using an ex vivo pMHCII tetramer–based T cell profiling, we have shown that in all type 1 allergic individuals, the differential expression of at least three markers (that is, CRTH2, CD161, and a differentiation stage marker such as CR45RB or CD27) is needed to define a pathogenic TH2 cell subset that is allergen-specific and virtually absent in nonatopic individuals (denoted here as TH2A subset).
Multiples lines of evidence suggested the pathogenic potential of TH2A cell subset in settings of allergic inflammatory disease. First, we observed that allergen-specific TH2 cells from allergic patients with either seasonal, perennial, fungus, or food allergy were virtually all contained in the terminally differentiated (CD27−) memory TH cell subset that coexpresses CRTH2 and CD161. Second, the overall number of cells from this subset was markedly higher in all allergic individuals as compared to nonatopic individuals. This particular proallergic TH cell subset is remarkable in that it can easily be detected directly ex vivo in every allergic individual due to its ability to include a broad array of allergen-specific TH2 cells. Hence, our data demonstrate that during a natural allergen challenge, such as pollen season or a peanut challenge test, the TH2A cell subset was distinctively activated (16, 28, 29). Finally, our data highlight key functional and molecular differences between pathogenic and conventional TH2 cells, recapitulating previous observation in their murine counterpart (8) and highlighting specific therapeutic targets.
CD161 expression has been described as a hallmark of human TH17 cells (17, 18). Therefore, its expression on a TH2 cell subset that does not express CCR6, RORC, or IL-17 cytokine is of great interest. Given that lectin-like transcript 1, the CD161 ligand, is expressed on respiratory epithelial cells during respiratory virus infection (30), it likely indicates the specialized role of allergen-specific TH2 cells and thus may be implicated in allergic pulmonary inflammation and asthma exacerbation. CD161 expression also provides gut-specific homing properties to T cells (31), and a higher proportion of CD161+ circulating CD4+ T cells have been previously described in allergic patients compared to nonatopic individuals (10, 32). Expression of CD161 on TH2 cells was also associated with IL-5–producing T effector cells associated with eosinophilic gastrointestinal disease (3). In support of these findings, our results show that IL-5 and IL-9 cytokines have some of the greatest fold changes of all up-regulated transcripts in the TH2A subpopulation compared with conventional TH2 cells. Our functional analysis also confirmed that TH2A cells exhibited profoundly superior functional activity compared to conventional TH2 cells, with individual cells capable of producing a larger amount of a broad spectrum of TH2 cytokines upon TCR activation. Because each TH2 cytokine has a well-defined and relatively specific function, it is likely that TH2A cells have greater adverse activity relative to conventional TH2 cells, which might reflect the wide array of clinical symptoms associated with allergic disorders (10, 33–35).
Understanding why some individuals elicit a pathogenic TH2 response to allergen might facilitate the development of improved vaccination strategies. It therefore raises the question of the origin of TH2A cells in atopic individuals. There is now growing evidence for a role of epithelium-derived cytokines in the differentiation of TH2 cells and in the establishment of airway inflammation (36). IL-33 and IL-25 pathways have been also associated with the induction of both IL-9 and IL-5 production in human TH2 cells that drive a cascade of downstream events (37–40). One possible mechanism to explain and integrate all these results into a cohesive schema is that upon allergen recognition, epithelial cells release cytokines that not only stimulate innate cell networks but may also act directly on CD4+ T cells to confer memory TH2 cell pathogenicity in atopic individuals, as recently suggested by Endo et al. (39). Whether local epithelial cytokines influence allergen-specific TH2 cell response requires further study, but our finding that TH2A cells specifically express IL-17RB and IL1RL1 supports the notion of a local checkpoint that restricts the optimal pathogenic TH2 responses to sites of tissue distress (10, 41). By establishing a clear link between the elimination of the allergen-specific TH2A cell subset in peanut-allergic patients and the clinical benefit induced by oral immunotherapy, our data reinforce previous reports by our group that the current immunotherapy approach, using crude preparation of intact allergens, restores a desensitization state in the allergic patients by means of preferential exhaustion/deletion of allergen-specific TH2 cells (7, 16, 42). TH2A cell subset shares multiple functional features with CCR8+ (9), hPGDS+ (10), and IL-17RB+ (11) pathogenic TH2 cell subsets that have been recently described in chronic atopic dermatitis, eosinophilic gastrointestinal diseases, and eosinophilic chronic rhinosinusitis, respectively. Therefore, it seems likely that TH2A cell subset described in this study may encompass various types of pathogenic TH2 cell populations involved in atopic diseases. Together, it supports the “disease induction model” proposed by Nakayama and colleagues (43–45), wherein the presence of a pathogenic CD4+ T cell subset with distinct phenotypic and functional properties might be sufficient for the pathogenesis of an immune-mediated disease, regardless of the balance of other TH subsets.
In summary, we have identified a proinflammatory human TH2 cell subpopulation unique to atopic individuals that is defined by stable coexpression of CRTH2, CD161, and CD49d and low expression of CD45RB and CD27. We suggest that TH2A cells are important in the pathogenesis of allergic diseases and should facilitate the detailed analysis of allergen-specific TH2 cell subset in allergic individuals. Therefore, further detailed studies focusing on the TH2A cell subset may prove useful in the diagnosis, molecular characterization, or the discovery of novel therapeutic targets to enhance the power of allergen vaccines.
MATERIALS AND METHODS
Study design
The main research objective of this study was to determine whether allergic individuals have specific subpopulations of TH2 cells associated with global atopic inflammatory disorders. To investigate allergic-related differences in peripheral T cells from allergic individuals, the profile of allergen-specific TH2 cell subset ex vivo using direct pMHCII tetramer staining was determined and compared to the profile of total TH2 cell subset. Candidate signature-associated markers were then tested in allergic patients and in nonatopic individuals. To evaluate this signature in the context of clinical intervention, a longitudinal study was conducted in patients receiving oral immunotherapy. Sample size was determined on the basis of the availability of fresh blood samples and with the intention to include samples before and after OFC and before and after therapy, where possible. All data generated were included in the analysis. Researchers performing the measurements were blinded to the treatment group and sample identity. To further explore the pathophysiologic meaning of this allergic T cell signature, we used real-time PCR, intracellular cytokine analysis and microarray analysis. Replication numbers for experiments are listed in the figure legends. Primary data for experiments where n < 20 are shown in table S3.
Subjects
Subjects were recruited at the Allergy Clinic at Virginia Mason Medical Center. All subjects were recruited with informed consent, and the study was approved by the Institutional Review Board of Benaroya Research Institute. Allergic subjects (n = 80) were selected on the basis of their clinical history, a positive prick test, and positive IgE reactivity to extract (test score, ≥0.35 kU/liter) using the ImmunoCAP test (Phadia AB). For subjects with no history of allergy (n = 34), the nonatopic status was confirmed by a lack of IgE reactivity and a negative in vitro basophil activation assay after stimulation with a pool of allergen extracts. All subjects were human leukocyte antigen (HLA)–typed by using sequence-specific oligonucleotide primers with UniTray SSP kits (Invitrogen).
CODIT study design and participants
In ARC001 (46), a multicenter, randomized, double-blind, placebo-controlled study of efficacy and safety of CODIT (Aimmune Therapeutics Inc.), peanut-allergic subjects aged 4 to 26 years were enrolled on the basis of clinical history of allergy to peanut, a serum IgE to peanut of ≥0.35 kU/liter (UniCAP) or positive skin prick test to peanut of >3 mm compared to control, and an allergic reaction at or before 100 mg of peanut protein during a screening DBPCFC, conducted in accordance with PRACTALL (Practical Issues in Allergology, Joint United States/European Union Initiative) guidelines. Participants were randomly assigned (1:1) to active treatment with AR101 or matched placebo. Subjects initiated the study with a single dose of 0.5 mg of study product and escalated biweekly over the course of about 20 weeks to the target maintenance dose of 300 mg/day. The primary clinical efficacy end point was the proportion of subjects in each group who tolerated at least 300 mg (443 mg cumulative) of peanut protein with no more than mild symptoms at the exit DBPCFC. Of 55 subjects enrolled in the ARC001 study, 10 participants were consented for additional volume of blood (10 to 15 ml) to be collected before and after the screening DBPCFC, and 7 participants (3 placebo and 4 active) were consented for additional volume of blood to be collected before and after CODIT.
Tetramer reagents
Biotinylated HLA-DR molecules were generated and purified as described (47). T cell epitopes were identified by tetramer-guided epitope mapping (table S4) (48). Epitope-specific pMHCII tetramer reagents were generated by loading specific peptides onto biotinylated soluble DR monomers and subsequently conjugated with phycoerythrin (PE)–streptavidin (47).
Ex vivo analysis of allergen-specific CD4+ T cells
Twenty million PBMCs in culture medium at a concentration of 150 million cells/ml were treated with dasatinib (49) for 10 min at 37°C, followed by staining with of PE-labeled pMHCII tetramers (20 μg/ml) at room temperature for 100 min. After tetramer staining, cells were then washed twice and incubated with anti-PE magnetic beads (Miltenyi Biotec) at 4°C for another 20 min. The cells were washed again and enriched using a magnetic column according to the manufacturer’s instructions (Miltenyi Biotec). Frequency was calculated as previously described (50). For unbiased FACS screen analysis, CRTH2-labeled PBMCs and cells in the tetramer-bound fractions were both stained with antibodies against markers of interest (table S1) or corresponding isotype-matched monoclonal antibodies. A combination of the vital dye Via-Probe (BD Pharmingen) as a viability marker, CD19 (eBioscience), and CD14 (eBioscience) was used to exclude dead cells, B cells, and monocytes from the analysis, respectively. A FACSAria II was used for multiparameter analysis, and data were analyzed with FlowJo software (Tree Star, Inc.).
TH2A cell subset analysis
TH2A cells were defined as CD4+CD45RO+CD27−CD45RBlowCRTH2+ CD161+CD49d+ T cell subset. The following antibodies were used in flow cytometric analysis: fluorescein isothiocyanate (FITC)–conjugated anti-CD45RB (clone MEM-55, AbD Serotec), phycoerythrin-Texas Red (ECD)–conjugated anti-CD45RO (clone UCHL1, Beckman Coulter), Alexa Fluor 647–conjugated anti-CRTH2 (clone BM16, BD Biosciences), antigen-presenting cell (APC)–H7–conjugated anti-CD27 (clone M-T271, BD Biosciences), V450-conjugated anti-CD38 (clone HIT2, eBioscience), eFluor 650–conjugated anti-CD3 (clone OKT3, eBioscience), PE-conjugated anti-CD161 (clone HP-3G10, eBioscience), PE-Cy7–conjugated anti-CD49d (clone 9F10, BioLegend), and BV605-conjugated anti-CD4 (clone OKT4, BioLegend). CD45RBlow cells were identified using a cutoff of 35% among live memory CD4+ T cells.
TH cell subset isolation
Freshly isolated PBMCs were labeled with V500-conjugated anti-CD4 (clone RPA-T4, BD Biosciences), Alexa Fluor 647–conjugated anti- CRTH2 (clone BM16, BD Biosciences), PE-Cy7–conjugated anti-CCR6 (clone R6H1, BD Biosciences), AF488-conjugated anti-CXCR3 (clone 1C6/CXCR3, BD Biosciences), APC-H7–conjugated CD27 (clone M-T271, BD Biosciences), ECD-conjugated anti-CD45RO (clone UCHL1, Beckman Coulter), PE-conjugated anti-CD161 (clone HP-3G10, eBioscience), and eFluor 650–conjugated anti-CD3 (clone OKT3, eBioscience). A combination of the vital dye Via-Probe (BD Pharmingen) as a viability marker, CD19 (eBioscience), and CD14 (eBioscience) was used to exclude dead cells, B cells, and monocytes from the analysis, respectively. TH2A cells (CD4+CD45RO+CD27−CRTH2+CD161+), conventional TH2 cells (CD4+CD45RO+CD27−CRTH2+CD161−), TH17 cell subset (CD4+CD45RO+CRTH2−CCR6+ CXCR3−), and TH1 cells (CD4+CD45RO+CRTH2−CCR6−CXCR3+) were isolated to a purity over 96% using FACSAria II (BD Biosciences) (fig. S9).
Intracellular cytokine staining
Intracellular staining was performed by using the Cytofix/Cytoperm buffer set (BD Biosciences) according to the manufacturer’s instructions. Briefly, cells were incubated for 5 hours at 37°C with 5% CO2 with PMA (50 ng/ml), ionomycin (500 ng/ml), and GolgiPlug (BD Biosciences), permeabilized with Cytofix/Cytoperm buffer, and stained with APC-conjugated anti-IL-5 (JES1-39D10, Miltenyi Biotec), FITC-conjugated anti–IL-4 (clone 8D4-8, eBioscience), PE-conjugated anti–IL-9 (clone MH9A4, BioLegend), PerCP/ Cy5.5-conjugated anti–IL-13 (clone JES10-5A2, BioLegend), BV510-conjugated anti–IFN-γ (clone 4S.B3, BioLegend), and APC/ Cy7-conjugated anti–IL-17 (clone BL168, BioLegend). After 30 min at 4°C, cells were washed and immediately analyzed by flow cytometry.
Real-time PCR expression analysis
The Fluidigm BioMark 96.96 Dynamic Array (51) was used to measure the gene expression in small cell populations. Ten cells per well were sorted by FACS in quadruplicate into 96-well plates containing a reaction mix for reverse transcription (CellsDirect One-Step qRT-PCR kit, Invitrogen) and preamplification with 96 selected gene primer pairs (Delta Gene assays, Fluidigm Corp.). After sorting, samples were reverse-transcribed and preamplified for 18 cycles. Primers and deoxynucleotide triphosphates were removed by incubation with Exonuclease I (New England Biolabs), and samples were diluted (five times) with TE buffer and stored at −20°C. Samples and assays (primer pairs) were prepared for loading onto 96.96 Fluidigm Dynamic Arrays according to the manufacturer’s recommendations. Briefly, the sample was mixed with 20× DNA binding dye sample loading reagent (Fluidigm Corp.) and 2× SsoFast EvaGreen Supermix with Low ROX (Bio-Rad). Assays were mixed with 2× assay loading reagent (Fluidigm Corp.) and TE buffer to a final concentration of 5 μM. The 96.96 Fluidigm Dynamic Arrays (Fluidigm Corp.) were primed and loaded on an IFC Controller HX (Fluidigm Corp.), and real-time PCR was run on a BioMark HD (Fluidigm Corp.). Data were collected and analyzed using Fluidigm Real-Time PCR analysis software (v4.1.2).
Microarray analysis and data analysis
Conventional TH1 cells, conventional TH17 cells, TH2A cells, and conventional TH2 cells were sorted from PBMCs of allergic subjects, as described above. Use of donor pools (each pool containing blood from two to three donors) was necessary to obtain sufficient numbers of cells for microarray experiments. Sorted TH subsets were stimulated for 6 hours with anti-CD3/CD28 beads (Life Technologies) or left unstimulated before extraction of RNA (RNeasy Mini kit, Qiagen). Replicates of RNA were obtained from each sample that passed quality control. Complementary RNA was prepared by amplification and labeling using the Illumina TotalPrep RNA Amplification kit (Life Technologies) and hybridized to human HT-12 Beadarray chips (Illumina). Beadchips were scanned on a HiScanSQ (Illumina). Background-subtracted data were generated using GenomeStudio software (Illumina). Data were processed by customized R/Bioconductor pipeline, including quantile normalization (52), flooring, log2 transformation, and PALO filtering (Present At Least Once; at least one sample must have had detection P < 0.01). Analyses were performed using R.
Statistical analysis
Prism software (GraphPad) was used for statistical analysis of flow cytometry data. No randomization or exclusion of data points was used. The nonparametric Mann-Whitney U test was used for unpaired comparisons between groups, whereas the nonparametric Wilcoxon matched pairs test was used for paired comparison.
Supplementary Material
Fig. S1. Flow cytometric plots showing phenotyping of ex vivo enriched allergen-specific CD4+ T cells in allergic subjects.
Fig. S2. Characteristics of allergic disease causing CD4+ T cells.
Fig. S3. Allergen-specific TH2 cells are highly mature cells.
Fig. S4. Allergen-specific TH2 cells fall into the CD27−CD161+CD45RB−CD49d+ CD4+ T cell subset.
Fig. S5. Discrimination between proallergic TH2A and conventional TH2 cell subset.
Fig. S6. Influence of OFC and oral immunotherapy on peanut-specific CD4+ T cells.
Fig. S7. Expression of TH2 cytokines is restricted to the allergen-specific TH2A cell subset.
Fig. S8. Overview of TH2A phenotype.
Fig. S9. Gating strategy for TH cell subset isolation.
Table S1. List of antibodies used in this study for the allergen-specific CD4+ T cell profiling.
Table S2. List of all up-regulated genes in the TH2A cell subset relative to conventional TH2 cells.
Table S3. Primary data.
Table S4. List of pMHCII tetramer reagents used in this study.
Acknowledgments
We thank K. Gilroy, S. Posso, G. Marchesini, and T.-S. Nguyen for the help with subject recruitment. We thank K. Arumuganathan for the expert advice on flow cytometry. We thank C. Cousens-Jacobs for the excellent secretarial assistance. We thank G. T. Nepom and J. Buckner for the comments on the manuscript. pMHCII reagents are available from the Tetramer Core under a material transfer agreement with the Benaroya Research Institute.
Funding: This work was supported by the NIH (grant R01AI108839 to E. Wambre, grant DP3DK110867 to P.S.L., and grant HHSN272200700046C to W.W.K.). Food Allergy Research and Education (FARE) contributed supplemental support to the Wambre Laboratory. Immune Tolerance Networks (ITN) and Aimmune Therapeutics provided support to the Benaroya Research Institute for the pilot experiments on samples from Aimmune’s clinical trial.
Footnotes
Competing interests: E. Wambre and W.W.K. are inventors on patent/patent application (EP 2681555 A1) held/submitted by the Benaroya Research Institute that covers “TH2A analysis: Detection of an immune response.” All other authors declare that they have no competing interests.
Author contributions: E. Wambre designed the study, planned and performed the experiments, analyzed the data, and wrote the manuscript. V.B., J.H.D., K.O., Q-A.N., and C.S. performed the experiments, analyzed the data, and reviewed the manuscript. M.F., D.J., D.R., and B.P.V. participated in patient recruitment, clinical data, and biological sample collection. B.P.V. directed the ARC001 study and provided coded biological sample from ARC001 study. E. Whalen, V.H.G., C.N., H.A.D., and P.S.L. statistically analyzed the microarray and PCR data and reviewed the manuscript. W.W.K. provided advice and technical support, analyzed the data, and reviewed the manuscript. E. Wambre, P.S.L., and W.W.K. raised the funding and supervised the project.
REFERENCES AND NOTES
- 1.Romagnani S. T-cell responses in allergy and asthma. Curr Opin Allergy Clin Immunol. 2001;1:73–78. doi: 10.1097/01.all.0000010988.60715.c8. [DOI] [PubMed] [Google Scholar]
- 2.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol. 2011;187:3111–3120. doi: 10.4049/jimmunol.1101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prussin C, Yin Y, Upadhyaya B. TH2 heterogeneity: Does function follow form? J Allergy Clin Immunol. 2010;126:1094–1098. doi: 10.1016/j.jaci.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wensky A, Marcondes MCG, Lafaille JJ. The role of IFN-γ in the production of Th2 subpopulations: Implications for variable Th2-mediated pathologies in autoimmunity. J Immunol. 2001;167:3074–3081. doi: 10.4049/jimmunol.167.6.3074. [DOI] [PubMed] [Google Scholar]
- 5.Cosmi L, Maggi L, Santarlasci V, Capone M, Cardilicchia E, Frosali F, Querci V, Angeli R, Matucci A, Fambrini M, Liotta F, Parronchi P, Maggi E, Romagnani S, Annunziato F. Identification of a novel subset of human circulating memory CD4+ T cells that produce both IL-17A and IL-4. J Allergy Clin Immunol. 2010;125:222–230. doi: 10.1016/j.jaci.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Gilles S, Traidl-Hoffmann C. CD27 expression on allergen-specific T cells: A new surrogate for successful allergen-specific immunotherapy? J Allergy Clin Immunol. 2012;129:552–554. doi: 10.1016/j.jaci.2011.12.967. [DOI] [PubMed] [Google Scholar]
- 7.Wambre E, DeLong JH, James EA, LaFond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J Allergy Clin Immunol. 2012;129:544–551. doi: 10.1016/j.jaci.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endo Y, Iwamura C, Kuwahara M, Suzuki A, Sugaya K, Tumes DJ, Tokoyoda K, Hosokawa H, Yamashita M, Nakayama T. Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity. 2011;35:733–745. doi: 10.1016/j.immuni.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, Lira SA, Charo IF, Luster AD. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ TH2 cells. Nat Immunol. 2011;12:167–177. doi: 10.1038/ni.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitson-Salazar A, Yin Y, Wansley DL, Young M, Bolan H, Arceo S, Ho N, Koh C, Milner JD, Stone KD, Wank SA, Prussin C. Hematopoietic prostaglandin D synthase defines a proeosinophilic pathogenic effector human TH2 cell subpopulation with enhanced function. J Allergy Clin Immunol. 2016;137:907–918. doi: 10.1016/j.jaci.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Lam EPS, Kariyawasam HH, Rana BMJ, Durham SR, McKenzie AN, Powell N, Orban N, Lennartz-Walker M, Hopkins C, Ying S, Rimmer J, Lund VJ, Cousins DJ, Till SJ. IL-25/IL-33–responsive TH2 cells characterize nasal polyps with a default TH17 signature in nasal mucosa. J Allergy Clin Immunol. 2016;137:1514–1524. doi: 10.1016/j.jaci.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wambre E, James EA, Kwok WW. Characterization of CD4+ T cell subsets in allergy. Curr Opin Immunol. 2012;24:700–706. doi: 10.1016/j.coi.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosmi L, Annunziato F, Galli MIG, Maggi RME, Nagata K, Romagnani S. CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur J Immunol. 2000;30:2972–2979. doi: 10.1002/1521-4141(200010)30:10<2972::AID-IMMU2972>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Tortorella C, Schulze-Koops H, Thomas R, Splawski JB, Davis LS, Picker LJ, Lipsky PE. Expression of CD45RB and CD27 identifies subsets of CD4+ memory T cells with different capacities to induce B cell differentiation. J Immunol. 1995;155:149–162. [PubMed] [Google Scholar]
- 15.Fritsch RD, Shen X, Sims GP, Hathcock KS, Hodes RJ, Lipsky PE. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J Immunol. 2005;175:6489–6497. doi: 10.4049/jimmunol.175.10.6489. [DOI] [PubMed] [Google Scholar]
- 16.Wambre E, DeLong JH, James EA, Torres-Chinn N, Pfutzner W, Möbs C, Durham SR, Till SJ, Robinson D, Kwok WW. Specific immunotherapy modifies allergen-specific CD4+ T-cell responses in an epitope-dependent manner. J Allergy Clin Immunol. 2014;133:872–879. doi: 10.1016/j.jaci.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, Rodolico G, Querci V, Abbate G, Angeli R, Berrino L, Fambrini M, Caproni M, Tonelli F, Lazzeri E, Parronchi P, Liotta F, Maggi E, Romagnani S, Annunziato F. Human interleukin 17–producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, Querci V, Fambrini M, Liotta F, Levings MK, Maggi E, Cosmi L, Romagnani S, Annunziato F. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol. 2010;40:2174–2181. doi: 10.1002/eji.200940257. [DOI] [PubMed] [Google Scholar]
- 19.Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, Scheffold A, Thiel A. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat Med. 2005;11:1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 20.Bird JA, Spergel JM, Jones SM, Rachid R, Assa’ad AH, Wang J, Leonard SA, Laubach SS, Kim EH, Vickery BP, Davis B, Heimall J, Cianferoni A, MacGinnitie AJ, Crestani E, Elfont RM, Sampson HA, Burks AW. In: EAACI Congress. Burks AW, editor. Vol. 70. EAACI; 2015. p. 110. [Google Scholar]
- 21.Bonvalet M, Moussu H, Wambre E, Ricarte C, Horiot S, Rimaniol AC, Kwok WW, Horak F, de Beaumont O, Baron-Bodo V, Moingeon P. Allergen-specific CD4+ T cell responses in peripheral blood do not predict the early onset of clinical efficacy during grass pollen sublingual immunotherapy. Clin Exp Allergy. 2012;42:1745–1755. doi: 10.1111/cea.12015. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler SF. The role of thymic stromal lymphopoietin (TSLP) in allergic disorders. Curr Opin Immunol. 2010;22:795–799. doi: 10.1016/j.coi.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portelli MA, Hodge E, Sayers I. Genetic risk factors for the development of allergic disease identified by genome-wide association. Clin Exp Allergy. 2015;45:21–31. doi: 10.1111/cea.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Noguchi E, Imoto Y, Nanatsue K, Takeshita K, Shibasaki M, Arinami T, Fujieda S. Upregulation of IL17RB during natural allergen exposure in patients with seasonal allergic rhinitis. Allergol Int. 2011;60:87–92. doi: 10.2332/allergolint.10-OA-0230. [DOI] [PubMed] [Google Scholar]
- 25.Chan IHS, Tang NLS, Leung TF, Ma SL, Zhang YP, Wong GWF, Wong CK, Lam CWK. Association of prostaglandin-endoperoxide synthase 2 gene polymorphisms with asthma and atopy in Chinese children. Allergy. 2007;62:802–809. doi: 10.1111/j.1398-9995.2007.01400.x. [DOI] [PubMed] [Google Scholar]
- 26.Fajt ML, Gelhaus SL, Freeman B, Uvalle CE, Trudeau JB, Holguin F, Wenzel SE. Prostaglandin D2 pathway upregulation: Relation to asthma severity, control, and TH2 inflammation. J Allergy Clin Immunol. 2013;131:1504–1512. doi: 10.1016/j.jaci.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puhl HL, III, Won Y-J, Lu VB, Ikeda SR. Human GPR42 is a transcribed multisite variant that exhibits copy number polymorphism and is functional when heterologously expressed. Sci Rep. 2015;5:12880. doi: 10.1038/srep12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pepper M, Linehan JL, Pagán AJ, Zell T, Dileepan T, Cleary PP, Jenkins MK. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol. 2010;11:83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendriks J, Xiao Y, Borst J. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J Exp Med. 2003;198:1369–1380. doi: 10.1084/jem.20030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satkunanathan S, Kumar N, Bajorek M, Purbhoo MA, Culley FJ. Respiratory syncytial virus infection, TLR3 ligands, and proinflammatory cytokines induce CD161 ligand LLT1 expression on the respiratory epithelium. J Virol. 2014;88:2366–2373. doi: 10.1128/JVI.02789-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinschek MA, Boniface K, Sadekova S, Grein J, Murphy EE, Turner SP, Raskin L, Desai B, Faubion WA, de Waal Malefyt R, Pierce RH, McClanahan T, Kastelein RA. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206:525–534. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gernez Y, Tirouvanziam R, Nguyen KD, Herzenberg LA, Krensky AM, Nadeau KC. Altered phosphorylated signal transducer and activator of transcription profile of CD4+CD161+ T cells in asthma: Modulation by allergic status and oral corticosteroids. J Allergy Clin Immunol. 2007;120:1441–1448. doi: 10.1016/j.jaci.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, Bernstein JA, Huston DP, Liu YJ. A novel subset of CD4+ TH2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207:2479–2491. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Angkasekwinai P, Spoede W, Huston D, Liu Y. Proinflammatory stimuli induce the generation of IL-17-producing TH2 memory cells that is associated with exacerbated allergic diseases. J Allergy Clin Immunol. 2009;123:S143. [Google Scholar]
- 35.Brough HA, Cousins DJ, Munteanu A, Wong YF, Sudra A, Makinson K, Stephens AC, Arno M, Ciortuz L, Lack G, Turcanu V. IL-9 is a key component of memory TH cell peanut-specific responses from children with peanut allergy. J Allergy Clin Immunol. 2014;134:1329–1338. doi: 10.1016/j.jaci.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 36.Bartemes KR, Kita H. Dynamic role of epithelium-derived cytokines in asthma. Clin Immunol. 2012;143:222–235. doi: 10.1016/j.clim.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blom L, Poulsen BC, Jensen BM, Hansen A, Poulsen LK. IL-33 induces IL-9 production in human CD4+ T cells and basophils. PLOS ONE. 2011;6:e21695. doi: 10.1371/journal.pone.0021695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurowska-Stolarska M, Kewin P, Murphy G, Russo RC, Stolarski B, Garcia CC, Komai-Koma M, Pitman N, Li Y, Niedbala W, McKenzie ANJ, Teixeira MM, Liew FY, Xu D. IL-33 induces antigen-specific IL-5+ T cells and promotes allergic-induced airway inflammation independent of IL-4. J Immunol. 2008;181:4780–4790. doi: 10.4049/jimmunol.181.7.4780. [DOI] [PubMed] [Google Scholar]
- 39.Endo Y, Hirahara K, Iinuma T, Shinoda K, Tumes DJ, Asou HK, Matsugae N, Obata-Ninomiya K, Yamamoto H, Motohashi S, Oboki K, Nakae S, Saito H, Okamoto Y, Nakayama T. The interleukin-33-p38 kinase axis confers memory T helper 2 cell pathogenicity in the airway. Immunity. 2015;42:294–308. doi: 10.1016/j.immuni.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Lee J-B, Chen C-Y, Liu B, Mugge L, Angkasekwinai P, Facchinetti V, Dong C, Liu Y-J, Rothenberg ME, Hogan SP, Finkelman FD, Wang Y-H. IL-25 and CD4+ TH2 cells enhance type 2 innate lymphoid cell–derived IL-13 production, which promotes IgE-mediated experimental food allergy. J Allergy Clin Immunol. 2016;137:1216–1225. doi: 10.1016/j.jaci.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Dyken SJ, Nussbaum JC, Lee J, Molofsky AB, Liang HE, Pollack JL, Gate RE, Haliburton GE, Ye CJ, Marson A, Erle DJ, Locksley RM. A tissue checkpoint regulates type 2 immunity. Nat Immunol. 2016;17:1381–1387. doi: 10.1038/ni.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wambre E. Effect of allergen-specific immunotherapy on CD4+ T cells. Curr Opin Allergy Clin Immunol. 2015;15:581–587. doi: 10.1097/ACI.0000000000000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama T, Hirahara K, Onodera A, Endo Y, Hosokawa H, Shinoda K, Tumes DJ, Okamoto Y. Th2 cells in health and disease. Annu Rev Immunol. 2016;35:53–84. doi: 10.1146/annurev-immunol-051116-052350. [DOI] [PubMed] [Google Scholar]
- 44.Endo Y, Hirahara K, Yagi R, Tumes DJ, Nakayama T. Pathogenic memory type Th2 cells in allergic inflammation. Trends Immunol. 2014;35:69–78. doi: 10.1016/j.it.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Hirahara K, Nakayama T. CD4+ T-cell subsets in inflammatory diseases: Beyond the Th1/ Th2 paradigm. Int Immunol. 2016;28:163–171. doi: 10.1093/intimm/dxw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aimmune Therapeutics. Developing an orally administered biologic immunotherapy for the treatment of peanut allergy. Aimmune Therapeutics; 2017. www.aimmune.com/clinical-trials/ar101-for-peanut-allergy/ [Google Scholar]
- 47.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4+ T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, Koelle DM, Kwok WW. Tetramer-guided epitope mapping: Rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J Immunol. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 49.Lissina A, Ladell K, Skowera A, Clement M, Edwards E, Seggewiss R, van den Berg HA, Gostick E, Gallagher K, Jones E, Melenhorst JJ, Godkin AJ, Peakman M, Price DA, Sewell AK, Wooldridge L. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods. 2009;340:11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, Robinson D. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol. 2010;125:1407–1409. doi: 10.1016/j.jaci.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Flow cytometric plots showing phenotyping of ex vivo enriched allergen-specific CD4+ T cells in allergic subjects.
Fig. S2. Characteristics of allergic disease causing CD4+ T cells.
Fig. S3. Allergen-specific TH2 cells are highly mature cells.
Fig. S4. Allergen-specific TH2 cells fall into the CD27−CD161+CD45RB−CD49d+ CD4+ T cell subset.
Fig. S5. Discrimination between proallergic TH2A and conventional TH2 cell subset.
Fig. S6. Influence of OFC and oral immunotherapy on peanut-specific CD4+ T cells.
Fig. S7. Expression of TH2 cytokines is restricted to the allergen-specific TH2A cell subset.
Fig. S8. Overview of TH2A phenotype.
Fig. S9. Gating strategy for TH cell subset isolation.
Table S1. List of antibodies used in this study for the allergen-specific CD4+ T cell profiling.
Table S2. List of all up-regulated genes in the TH2A cell subset relative to conventional TH2 cells.
Table S3. Primary data.
Table S4. List of pMHCII tetramer reagents used in this study.