Abstract
Purpose of review
Real-time 3-dimensional (3-D) imaging of cardiovascular injury and regeneration remains challenging. We introduced a multi-scale imaging strategy that uses light-sheet illumination to enable applications of cardiovascular injury and repair in models ranging from zebrafish to rodent hearts.
Recent findings
Light-sheet imaging enables rapid data acquisition with high spatiotemporal resolution and with minimal photo-bleaching or photo-toxicity. We demonstrated the capacity of this novel light-sheet approach for scanning a region of interest with specific fluorescence contrast, thereby providing axial and temporal resolution at the cellular level without stitching image columns or pivoting illumination beams during one-time imaging. This cutting-edge imaging technique allows for elucidating the differentiation of stem cells in cardiac regeneration, providing an entry point to discover novel micro-circulation phenomenon with clinical significance for injury and repair.
Summary
These findings demonstrate the multi-scale applications of this novel light-sheet imaging strategy to advance research in cardiovascular development and regeneration.
Keywords: light-sheet imaging, cardiovascular injury, regeneration, doxorubicin
Introduction
Real-time 3-dimensional (3-D) imaging of cardiovascular injury and repair remains a significant challenge. Conventional microscopes are confined to low light penetration and a small working distance for optical sectioning and parallelization that limit long-term biological interrogation (1–3). In addition, mechanical slicing likely distorts the intrinsic tissue integrity, and under-sampling occurs following 3-D reconstruction (4). Despite the advent of 3-D live imaging techniques such as positron emission tomography (PET) (5, 6), computed tomography (CT) (7, 8), and magnetic resonance image (MRI) (9, 10), these techniques are limited by spatial resolution (11–15). In addition, they are unable to capture fluorescently-labelled molecules that are used frequently in biomedical research. For these reasons, we hereby introduce a multi-scale and 3-D approach to imaging organisms ranging from live zebrafish embryos (~ 0.4 × 0.5 × 0.6 mm3) to mouse hearts (~2 × 2 × 4 mm3). This technique, called light-sheet fluorescence microscopy (LSFM) (16–20), has demonstrated high spatiotemporal resolution and specific labeling without needing to stitch image columns (21–23) or pivot the illumination beams and has minimal photo-bleaching and rapid acquisition time (24).
While fluorescence microscopy technologies in general have become increasingly powerful in terms of resolution, speed, and penetration, they are most often performed in thin and transparent samples (25, 26). The size and opacity of entire embryos, which are often a few millimeters in size, render it challenging to achieve single-cell resolution of ~10 μm (several hundred microns deep inside the intact embryos). Current imaging techniques such as confocal microscopy are limited by their intrinsic depth penetration and z-axis resolution. Images acquired with the conventional microscopy techniques incur 1) significant background noise due to out-of-focus illumination, and 2) low axial resolution due to a large depth of field. The dynamic range and axial penetration depth are also reduced due to ~95% of fluorescent blockage (4). Unlike the confocal and wide-field microscopy, LSFM has the capacity to localize the 4-D (3-D spatial + 1-D time or spectra) cellular phenomena with multi-fluorescence channels (27–31). Due to the intrinsic properties of the current optical microscopy design, depth-of-focus and resolution offset each other. Furthermore, optical sectioning for dynamic sample remains a challenging issue. Initially developed to image Caenohabditis elegans (32, 33), zebrafish embryos (34, 35), and Drosophilas (36, 37), LSFM has now been adapted to image rodent organs such as the mouse hippocampus (38–40) and mouse cochlea (41) with the introduction of optical clearing techniques (42). Thus, LSFM system has allowed for uncovering both mechanical and structural phenotypes at the cellular level (16–18, 20). The Comparative advantages and disadvantages among different optical imaging modalities are listed in Table 1. In this review, we will introduce the basic optical principle of LSFM. Next, we will provide three areas of applications: 1) analysis of vascular injury and regeneration in zebrafish embryos, 2) quantification of doxorubicin chemotherapy-induced cardiac injury and repair, and 3) 3-D localization of cardiac progenitor lineage in neonatal mouse hearts.
Table 1.
Comparative advantages and disadvantages among different imaging modalities
| Method | Resolution (Lateral-Axial, nm) | Penetration (μm) | Acquisition time per image | Advantage | Disadvantage |
|---|---|---|---|---|---|
| LSFM | 200–500 | >1000 | ms-s |
|
|
| WFM | 250 | --- | ms-min |
|
|
| CFM | 200–400 | 150 | s-min |
|
|
| SD-CFM | 200–400 | 150 | ms-min |
|
|
| MPM | 300–500 | 1000 | s-min |
|
|
| STED | 80–400 | 50 | min |
|
|
| PALM/STORM | 50–100 | 0.1 | min-h |
|
|
| FRET | 4–10 | --- | ms-s |
|
|
Legend: LSFM: light-sheet fluorescent microscopy; WFM: wide-field microscopy; CFM: confocal microscopy; SD-CFM: spinning disk confocal microscopy; MPM: multi-photon microscopy; STED: stimulated emission depletion; PALM: photoactivated localization microscopy; STORM: stochastic optical reconstruction microscopy; FRET: Förster resonance energy transfer. Adapted from [2].
1. Operating principle of light-sheet system and sample preparation
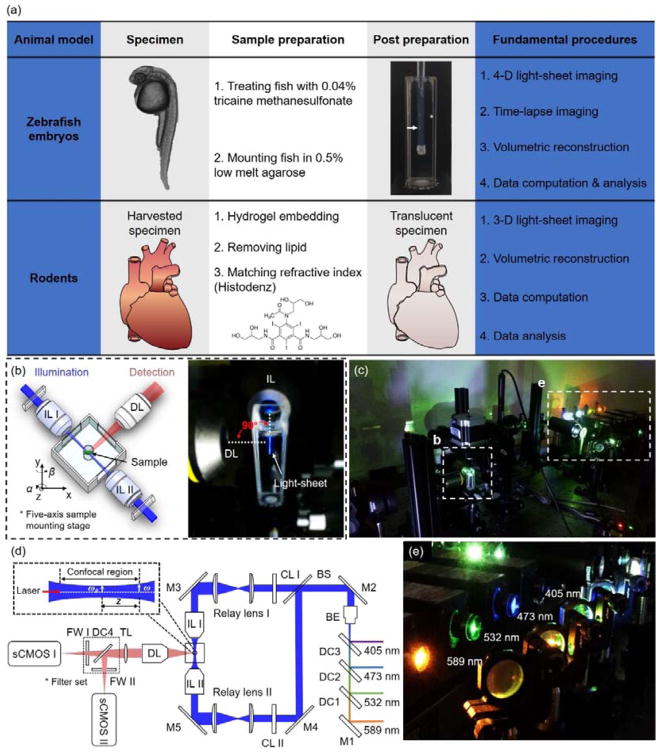
The custom-made light-sheet microscope (Figure 1a–c) utilized a continuous-wave laser (Laserglow Technologies, Canada) as the illumination source. The detection module was composed of the scientific CMOS (sCMOS, ORCA-Flash4.0, Hamamatsu, Japan) and a set of filters (Semrock, New York, USA). This detection module was perpendicularly installed to the illumination plane. The sample holder was oriented by a five-axis mounting stage for scanning the biological specimen. Both illumination and detection modules were controlled by a computer with a dedicated solid-state drive and the Redundant Array of Independent Disks level 0 (RAID 0) storage for fast data streaming. The lateral resolution of the LSFM is mainly governed by the numerical aperture (NA) of the objective lens, as denoted by dlateral=0.61λ/NA, where λ indicates the wavelength of excitation light. The axial resolution is determined by the waist of Gaussian beam and detection NA. The waist ω0 and Rayleigh range Z are used to define the light-sheet (Figure 1d): ω0=λf/πω, Z=λf2/πω2, where f is the focal length of the excitation objective, and ω denotes the half of the width of illumination beam before focusing. Generally, both waist ω0 and Rayleigh range Z drop as the ω increases.
Figure 1.
Fundamental concept of the light-sheet imaging strategy. (a) The critical procedures of the multi-scale imaging are indicated for both embryonic zebrafish and mouse studies. (b) The specimen is mounted at the intersection of the illumination lens (IL) with the detection lens (DL). The laser light-sheet is excited from the IL in a 2-D plane which is orthogonal to the detection axis. The LSFM system provides a long working distance with air objective lenses in comparison to water-dipping lenses in conventional light-sheet systems. (c–d) A photo and a schematic illustrate the layout of the light-sheeting imaging system. A cylindrical lens (CL) converts the laser beam to a sheet of laser light that can transversely illuminate a thin layer of the sample. The illuminated 2-D thin layer (fluorescent detection in red) is captured by the high-frame rate sCMOS camera. The filter wheels (FW I and II) in front of sCMOS cameras are used for multi-color acquisitions. (e) A photo depicts an array of laser beams aligned for multi-channel fluorescent detection. M: mirror; BS: beam splitter; BE: beam expander; TL: tube lens; DC: dichroic mirror; FW: filter wheel.
For the imaging data presented here, the detection objective was imaged through the liquid-air interface which introduced a spherical aberration-based point spread function (PSF) extension (43). Each image was acquired within a 10–50 ms exposure time. The stepping size of mechanical scanning was 1–5 μm, less than one half of the light sheet thickness in accordance with the Nyquist-Shannon sampling theorem. Thus, the resolving power of the LSFM in cross-section varied from 1 μm to 10 μm, while the waist ω0 ranged from 2 to 9 μm. All of the raw data were processed to remove stationary noise. The simplified tissue clearing method was used to process the mouse hearts as described in (44).
2. Light-sheet imaging of vascular injury and regeneration in zebrafish embryos
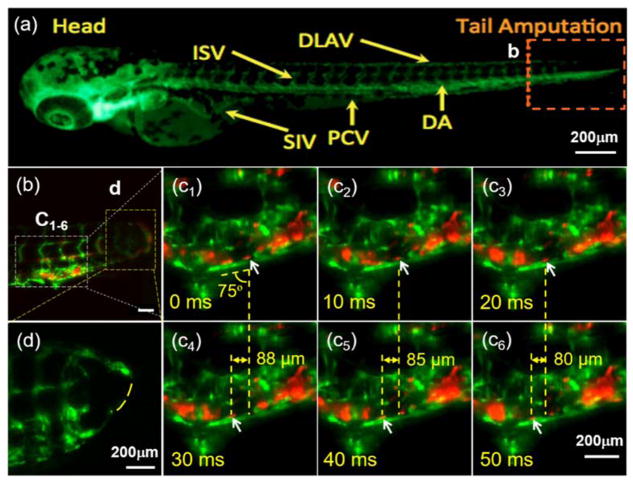
We studied zebrafish blood cell migration in response to vascular injury and regeneration by using the transgenic Tg(fli1:GFP; gata1:DsRed) and Tg(fli1:GFP; cmlc2:mCherry) lines, in which we were able to track blood cells in the vasculature in response to tail amputation at 3 days post fertilization (dpf). Using the transgenic Tg(fli1:GFP;gata1:DsRed) line in which the fli1 promoter drives expression of GFP in all vasculature throughout embryogenesis and gata1 promoter drives the expression of DsRed in blood cells (Figure 2a), we applied dual-channel detection to simultaneously track 1) the vascular loop connection between the dorsal aorta (DA) and the dorsal longitudinal anastomotic vessel (DLAV) at 3 dpf, and 2) the blood cells circulating to the injured site (Figure 2b). At 100 fps, the measured angle and displacement allowed computation of the velocity of the targeted cells as 2.9 ± 0.1 μm/ms (Figure 2c). This method may allow studies of non-linear shear rates in a low Reynolds number system (Re: 100 ~ 1000). Genetic manipulation of the ADAM10 inhibitor (which inhibits proteolytic cleavage of the Notch extracellular domain) also enabled us to elucidate Notch-mediated vascular regeneration to connect the loop between DLAV with DA (Figure 2d vs. 2b). Thus, LSFM enables the discovery of novel micro-circulation phenomenon with clinical significance for injury and repair.
Figure 2.
Light-sheet microscopic illumination of vascular regeneration and circulating blood cells in response to tail amputation. (a) An inverted fluorescence image of a transgenic Tg(fli1:GFP;gata1:DsRed) zebrafish embryo showing the vasculature (green) at 3 dpf. ISV: intersegmental vessel; DLAV: dorsal longitudinal anastomotic vessel; SIV: subintestinal vessel; PCV: posterior cardinal vein; DA: dorsal aorta. Box b indicates the site of tail amputation. (b) LSFM captures blood cells (red) proximal to the site of amputation and regeneration. Dashed yellow boxes indicate locations of higher power images in the subsequent panels (c1–6). Arrows indicate the position of an individual RBC (red) in relation to the vascular endothelial layer (green) acquired by LSFM at 100 fps. The average angle between the vein (green PCV) and vertical axis of the frame is 75°, and the relative displacement along the horizontal axis of the frame in each 30 ms period is 88 μm (c4-c1), 85 μm (c5-c2) and 80 μm (c6-c3), respectively. These measures correspond to a net velocity of nearly 2.9 ± 0.1 μm/ms for that blood cell of interest. (d) The dashed line indicates the incomplete vascular regeneration between DLAV and DA in a separate zebrafish embryo treated with an inhibitor of ADAM10 (GI254023X, Sigma) which blocks Notch signaling. Scale bars: 200 μm.
3. Light-sheet imaging to analyze doxorubicin chemotherapy-induced cardiac injury and repair
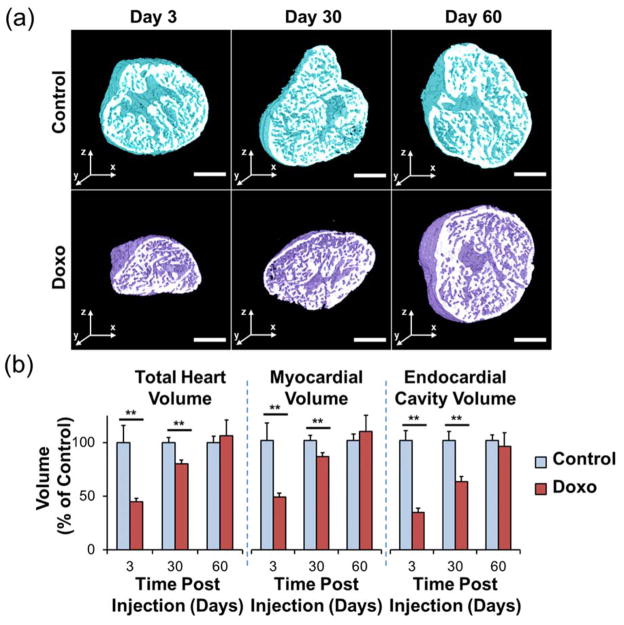
In adult zebrafish, the regenerating myocardium electrically couples with uninjured myocardium (45) and represents an evolutionarily-conserved model of cardiomyopathy (46). However, the small size and the two-chambered zebrafish heart limits precise morphologic assessment. In the setting of chemical clearing to achieve tissue transparency and laser light transmission, advances of light-sheet imaging coupled with automated image segmentation based on histogram analysis led to rapid and robust 3-D cardiac reconstruction, thereby unraveling the architecture of doxorubicin chemotherapy-induced cardiac injury and regeneration in adult zebrafish (47). By combining light-sheet imaging with automated segmentation, we observed dynamic changes in cardiac volumes in response to doxorubicin-induced cardiac toxicity. Precise 3-D reconstruction further enabled quantitation of cardiac volumes at days 3, 30, and 60 following chemotherapy treatment (Figure 3a). Compared to control fish, doxorubicin treatment led to an acute decrease in myocardial and endocardial volumes at day 3 (P < 0.01), demonstrating global cardiac injury (Figure 3b). This was followed by ventricular remodeling at day 30 (P < 0.01) and complete regeneration and restoration of normal architecture at day 60. Our results demonstrate the suitability of light-sheet imaging combined with automated segmentation as a high-throughput method to monitor 3-D cardiac ultrastructural changes in adult zebrafish, with translational implications to drug discovery and modifiers of chemotherapy-induced cardiomyopathy.
Figure 3.
Cardiac architecture following doxorubicin treatment. Following intraperitoneal treatment with doxorubicin or control vehicle, adult zebrafish hearts were harvested at days 3, 30, and 60. (a) Throughout the duration of the study, control hearts exhibited a preserved architecture. In contrast, doxorubicin-treated hearts demonstrated a profound cardiac remodeling leading to acute decrease in size at day 3, followed by gradual increase at day 30, and normalization at day 60. (b) Cardiac architecture characterization by quantitative analysis of the total heart, myocardial, and endocardial volumes compared to control values demonstrating the cardiac repair process following response to chemotherapy-induced injury. Legend. ** P < 0.01. Doxo: doxorubicin. Scale bar: 200 μm. (Reproduced with permission from: Packard et al. Sci Rep. 2017;7) (47).
4. Light-sheet to localize cardiac progenitor lineage in neonatal mouse hearts
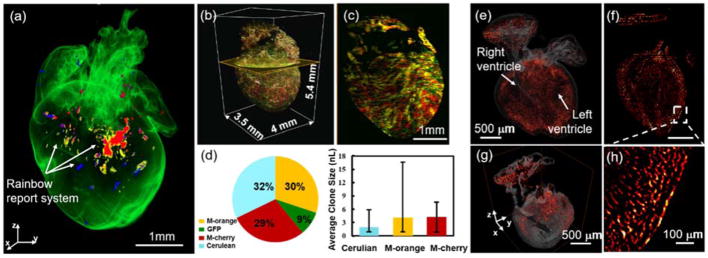
Accumulating evidence supports the ability of the mammalian heart to generate new cardiomyocytes during development and in response to injury (48, 49). However, 3-D distribution of cardiac progenitor cells to cardiac tissue formation remain poorly understood for both cardiac morphogenesis and regeneration (48, 49). We applied two reporter systems to demonstrate LSFM-imaged cardiac progenitor lineage. A rainbow multi-color reporter system (50) was used to retrospectively identify the source of new cardiomyocytes in a mouse heart at postnatal day 1 (P1) (Figure4). Cre-mediated recombination of paired lox P sites in the αMHCCre;R26VT2/GK mouse model resulted in expression of all four fluorescent proteins (Cerulean, GFP, mOrange and mCherry, Figure 4b–c). We applied a sub-voxel light-sheet imaging method to precisely localize and subsequently quantify the numbers of cells and size of the individual clones (Figure 4d). Another fetal mouse model (embryonic day 16.5, E16.5) was Mesp1Cre/+ crossed with Rosa26tdT/+ reporter system to study the contribution of tdT+ cell (red hot) in an intact fetal mouse heart (Figure 4e–h). 3-D LSFM imaging showed the majority of the cardiac cells that were labeled with tdT arose from a mesoderm posterior1 homolog (Mesp1) origin and allowed tracing of their 3-D distribution, proliferation, and tissue formation. Thus, light-sheet imaging allows for clonal analysis of cardiomyocyte generation during development and injury.
Figure 4.
3-D tracking of the cardiac progenitor lineage in the fetal and neonatal mouse hearts. (a) Expression of Cre in the neonatal mouse heart induces random recombination between mutated paired lox P sites leading to expression of Cerulean, GFP, mOrange and mCherry respectively. (b–c) Sub-voxel imaging of the rainbow heart was captured in four channels. Multi-view reconstruction was performed to enhance the spatial resolution for tracking and localizing the differentiation of cardiomyocytes in the intact neonatal heart. (d) Quantification of the number of labeled cells is illustrated in the pie chart, and the sizes of individual clones in a heart by the bar graphs. (e–f) Spatial distribution of tdT+ cells (red hot) in a fetal mouse heart which was Mesp1Cre/+ crossed with Rosa26tdT/+ reporter. (g) A 3-D orthogonal slice and (h) 2-D inset reveal the contribution of tdT+ cells in the heart. Scale bars: (a & c) 1 mm; (e–g) 500 μm; (h) 100 μm.
Conclusions and Discussion
We have established a holistic strategy for multi-scale applications of LSFM to elucidate cardiovascular injury and repair in both zebrafish and mouse models. This method provides an efficient and robust platform for detailed analysis of cardiovascular phenomena ranging from tissue clearing ex vivo to 4-D imaging in vivo. This framework builds on the high axial and temporal resolution provided by LSFM, which allows for long-term, 3-D and 4-D visualization of in vivo cellular events and organogenesis with minimal photo-bleaching or photo-toxicity.
Light-sheet imaging allows one to image the entire vasculature and cardiac architecture without the need for mechanical slicing or stitching of image columns. By this means, LSFM bypasses the need to move the tissue volume or light-sheet along the propagation of the illumination, thereby allowing us to carry out multi-scale imaging in a wide range of specimens, from embryos to adults, with the light-sheet thickness ranging from 1 μm to 18 μm (18, 20). Our methodology expands the field of view from hundreds of micrometers to tens of millimeters to cover the entire mouse heart with sufficient spatiotemporal resolution to track cardiovascular development in rodents. Our imaging strategy simplifies the operation during image acquisition, reduces the complexity of pre- and post-processing of cardiac images, and enhances the robustness of the imaging system to provide dual-channel in vivo imaging at over 100 fps. This strategy brings advanced imaging to studies of tissue injury and regeneration with multi-scale applications to fundamental studies of cardiovascular development and translational work in congenital cardiac anomalies and disease.
Acknowledgments
The authors would like to express gratitude to all lab members for discussion and insights. This study was supported by the National Institutes of Health HL118650 (T.K.H.), HL083015 (T.K.H.), HL111437 (T.K.H.), HL129727 (T.K.H.), AHA Scientist Development Grant 16SDG30910007 (R.R.S.P) and AHA Pre-Doctoral Fellowship 15PRE21400019 (J.L.).
Footnotes
Compliance with Ethical Standards
Conflict of Interest
Yichen Ding, Juhyun Lee, Jeffrey J. Hsu, Chih-Chiang Chang, Kyung In Baek, Sara Ranjbarvazirj, Reza Ardehali, René R. Sevag Packard, and Tzung K. Hsiai declare no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human subjects performed by any of the authors. All animal rights have been approved by AAALAC and USDA.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Huisken J, Stainier DY. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136(12):1963–75. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vos WH, Beghuin D, Schwarz CJ, Jones DB, van Loon JJ, Bereiter-Hahn J, et al. Invited Review Article: Advanced light microscopy for biological space research. Rev Sci Instr. 2014;85(10):101101. doi: 10.1063/1.4898123. [DOI] [PubMed] [Google Scholar]
- 3.Power RM, Huisken J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat Meth. 2017;14(4):360–73. doi: 10.1038/nmeth.4224. [DOI] [PubMed] [Google Scholar]
- 4.Bacallao R, Kiai K, Jesaitis L. Guiding principles of specimen preservation for confocal fluorescence microscopy. In: Pawley J, editor. Handbook of biological confocal microscopy. Springer; 2006. pp. 368–80. [Google Scholar]
- 5.Miyaoka R, Lewellen T, Yu H, McDaniel D. Design of a depth of interaction (DOI) PET detector module. IEEE T Nucl Sci. 1998;45(3):1069–73. [Google Scholar]
- 6.Tai Y-C, Chatziioannou AF, Yang Y, Silverman RW, Meadors K, Siegel S, et al. MicroPET II: design, development and initial performance of an improved microPET scanner for small-animal imaging. Phys Med Biol. 2003;48(11):1519. doi: 10.1088/0031-9155/48/11/303. [DOI] [PubMed] [Google Scholar]
- 7.Paxton R, Ambrose J. The EMI scanner. A brief review of the first 650 patients. Brit J Radiol. 1974;47(561):530–65. doi: 10.1259/0007-1285-47-561-530. [DOI] [PubMed] [Google Scholar]
- 8.Prokop M. General principles of MDCT. Eur J Radiol. 2003;45:S4–S10. doi: 10.1016/s0720-048x(02)00358-3. [DOI] [PubMed] [Google Scholar]
- 9.Kelly KA, Allport JR, Tsourkas A, Shinde-Patil VR, Josephson L, Weissleder R. Detection of vascular adhesion molecule-1 expression using a novel multimodal nanoparticle. Circ Res. 2005;96(3):327–36. doi: 10.1161/01.RES.0000155722.17881.dd. [DOI] [PubMed] [Google Scholar]
- 10.van der Graaf M. In vivo magnetic resonance spectroscopy: basic methodology and clinical applications. Eur Biophys J. 2010;39(4):527–40. doi: 10.1007/s00249-009-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel MR, Spertus JA, Brindis RG, Hendel RC, Douglas PS, Peterson ED, et al. ACCF proposed method for evaluating the appropriateness of cardiovascular imaging. J Am Coll Cardiol. 2005;46(8):1606–13. doi: 10.1016/j.jacc.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 12.Slavin GS, Bluemke DA. Spatial and temporal resolution in cardiovascular MR imaging: review and recommendations. Radiology. 2005;234(2):330–8. doi: 10.1148/radiol.2342031990. [DOI] [PubMed] [Google Scholar]
- 13.Sanz J, Fayad ZA. Imaging of atherosclerotic cardiovascular disease. Nature. 2008;451(7181):953. doi: 10.1038/nature06803. [DOI] [PubMed] [Google Scholar]
- 14.James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev. 2012;92(2):897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- 15.Plana JC, Galderisi M, Barac A, Ewer MS, Ky B, Scherrer-Crosbie M, et al. Expert consensus for multimodality imaging evaluation of adult patients during and after cancer therapy: a report from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging. 2014;15(10):1063–93. doi: 10.1093/ehjci/jeu192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Fei P, Lee J, Packard RRS, Sereti K-I, Xu H, Ma J, et al. Cardiac Light-Sheet Fluorescent Microscopy for Multi-Scale and Rapid Imaging of Architecture and Function. Sci Rep. 2016;6:22489. doi: 10.1038/srep22489. This study provides multi-scale visualization of architecture and function to drive cardiovascular research with translational implication in congenital heart diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan Z, Lee J, Jiang H, Dong S, Jen N, Hsiai T, et al. Compact plane illumination plugin device to enable light sheet fluorescence imaging of multi-cellular organisms on an inverted wide-field microscope. Biomed Opt Express. 2016;7(1):194–208. doi: 10.1364/BOE.7.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Lee J, Fei P, Packard RRS, Kang H, Xu H, Baek KI, et al. 4-Dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J Clin Invest. 2016;126(5):1679–90. doi: 10.1172/JCI83496. This study indicates that interfacing light-sheet imaging with the zebrafish system opens a fundamental direction for demonstrating shear stress modulation of trabeculation to influence contractile function via Notch signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Ding Y, Abiri A, Abiri P, Li S, Chang C-C, Baek KI, et al. Integrating light-sheet imaging with virtual reality to recapitulate developmental cardiac mechanics. JCI Insight. 2017;2(22):e97180. doi: 10.1172/jci.insight.97180. This study demonstrates an efficient and robust framework for creating a user-directed microenvironment in which we uncovered developmental cardiac mechanics and physiology with high spatiotemporal resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Y, Lee J, Ma J, Sung K, Yokota T, Singh N, et al. Light-sheet fluorescence imaging to localize cardiac lineage and protein distribution. Sci Rep. 2017;7:42209. doi: 10.1038/srep42209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buytaert JA, Dirckx JJ. Tomographic imaging of macroscopic biomedical objects in high resolution and three dimensions using orthogonal-plane fluorescence optical sectioning. Appl Opt. 2009;48(5):941–8. doi: 10.1364/ao.48.000941. [DOI] [PubMed] [Google Scholar]
- 22.Santi PA, Johnson SB, Hillenbrand M, GrandPre PZ, Glass TJ, Leger JR. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. Biotechniques. 2009;46(4):287. doi: 10.2144/000113087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buytaert JA, Descamps E, Adriaens D, Dirckx JJ. The OPFOS microscopy family: High-resolution optical sectioning of biomedical specimens. Anat Res Int. 2011;2012:1–8. doi: 10.1155/2012/206238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huisken J, Stainier DY. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM) Opt Lett. 2007;32(17):2608–10. doi: 10.1364/ol.32.002608. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y, Xie H, Peng T, Lu Y, Jin D, Teng J, et al. Laser oblique scanning optical microscopy (LOSOM) for phase relief imaging. Opt Express. 2012;20(13):14100–8. doi: 10.1364/OE.20.014100. [DOI] [PubMed] [Google Scholar]
- 26.Peng T, Xie H, Ding Y, Wang W, Li Z, Jin D, et al. CRAFT: Multimodality confocal skin imaging for early cancer diagnosis. J Biophoton. 2012;5(5–6):469–76. doi: 10.1002/jbio.201100124. [DOI] [PubMed] [Google Scholar]
- 27.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305(5686):1007–9. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 28.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135(6):1179–87. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 29.Mickoleit M, Schmid B, Weber M, Fahrbach FO, Hombach S, Reischauer S, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Meth. 2014;11(9):919–22. doi: 10.1038/nmeth.3037. [DOI] [PubMed] [Google Scholar]
- 30.Chhetri RK, Amat F, Wan Y, Höckendorf B, Lemon WC, Keller PJ. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat Meth. 2015;12:1171–8. doi: 10.1038/nmeth.3632. [DOI] [PubMed] [Google Scholar]
- 31.Dodt H-U, Saghafi S, Becker K, Jährling N, Hahn C, Pende M, et al. Ultramicroscopy: development and outlook. Neurophotonics. 2015;2(4):041407. doi: 10.1117/1.NPh.2.4.041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A, et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol. 2013;31(11):1032–8. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346(6208):1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322(5904):1065–9. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 35.Alvers AL, Ryan S, Scherz PJ, Huisken J, Bagnat M. Single continuous lumen formation in the zebrafish gut is mediated by smoothened-dependent tissue remodeling. Development. 2014;141(5):1110–9. doi: 10.1242/dev.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Meth. 2011;8(9):757–60. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 37.Krzic U, Gunther S, Saunders TE, Streichan SJ, Hufnagel L. Multiview light-sheet microscope for rapid in toto imaging. Nat Meth. 2012;9(7):730–3. doi: 10.1038/nmeth.2064. [DOI] [PubMed] [Google Scholar]
- 38.Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Meth. 2007;4(4):331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 39.Saghafi S, Becker K, Jährling N, Richter M, Kramer ER, Dodt HU. Image enhancement in ultramicroscopy by improved laser light sheets. J Biophot. 2010;3(10–11):686–95. doi: 10.1002/jbio.201000047. [DOI] [PubMed] [Google Scholar]
- 40.Saghafi S, Becker K, Hahn C, Dodt HU. 3D-ultramicroscopy utilizing aspheric optics. J Biophot. 2014;7(1–2):117–25. doi: 10.1002/jbio.201300048. [DOI] [PubMed] [Google Scholar]
- 41.Buytaert JA, Dirckx JJ. Design and quantitative resolution measurements of an optical virtual sectioning three-dimensional imaging technique for biomedical specimens, featuring two-micrometer slicing resolution. J Biomed Opt. 2007;12(1):014039–13. doi: 10.1117/1.2671712. [DOI] [PubMed] [Google Scholar]
- 42.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162(2):246–57. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, Sankaran S, et al. SPED light sheet microscopy: Fast mapping of biological system structure and function. Cell. 2015;163(7):1796–806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung K, Ding Y, Ma J, Chen H, Huang V, Cheng M, et al. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci Rep. 2016;6:30736. doi: 10.1038/srep30736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, Cao H, Kang BJ, Jen N, Yu F, Lee C-A, et al. Hemodynamics and ventricular function in a zebrafish model of injury and repair. Zebrafish. 2014;11(5):447–54. doi: 10.1089/zeb.2014.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, et al. Haploinsufficiency of Target of Rapamycin Attenuates Cardiomyopathies in Adult ZebrafishNovelty and Significance. Circ Res. 2011;109(6):658–69. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Packard RRS, Baek KI, Beebe T, Jen N, Ding Y, Shi F, et al. Automated Segmentation of Light-Sheet Fluorescent Imaging to Characterize Experimental Doxorubicin-Induced Cardiac Injury and Repair. Sci Rep. 2017;7:8603. doi: 10.1038/s41598-017-09152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13(8):970. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ and lineage restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476(7361):409–13. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]