Abstract
The members of the Order Nidovirales share a similar genome organization with two overlapping nonstructural polyproteins encoded in the 5′ two-thirds and the structural proteins encoded in the 3′ third. They also express their 3′ region proteins from a nested set of 3′ co-terminal subgenomic messenger RNAs (sg mRNAs). Some but not all of the Nidovirus sg mRNAs also have a common 5′ leader sequence that is acquired by a discontinuous RNA synthesis mechanism regulated by multiple 3′ body transcription regulating sequences (TRSs) and the 5′ leader TRS. Initial studies detected a single major body TRS for each 3′ sg mRNA with a few alternative functional TRSs reported. The recent application of advanced techniques, such as next generation sequencing and ribosomal profiling, in studies of arteriviruses and coronaviruses has revealed an expanded sg mRNA transcriptome and coding capacity.
Keywords: Nidovirus, Coronavirus, Arterivirus, Transcription regulatory sequences, Subgenomic mRNAs, Next generation sequencing, Discontinuous RNA synthesis, Leader-body junction sequences
Highlights
-
•
Nidoviruses express some of their genes from a nested set of 3′ co-terminal sg mRNAs.
-
•
The sg mRNAs of some but not all Nidoviruses have a common 5′ leader sequence.
-
•
Transcription regulating sequences in the genome are sites of premature minus stand termination.
-
•
Multiple alternative transcriptional regulating sequences were identified for some 3′ORFs.
-
•
Next generation sequencing and ribosomal profiling identified additional coding capacity.
1. Subgenomic mRNA production by members of the Order Nidovirales
The order Nidovirales consists of enveloped, single-stranded positive-sense RNA viruses that are currently classified into four virus families, Coronaviridae, Arteriviridae, Mesoniviridae, and Roniviridae (Gorbalenya et al., 2006, Zirkel et al., 2013). Due to the large number of new Nidoviruses recently identified by next generation sequencing (NGS) efforts, additional revisions to the Nidovirales taxonomy classification are currently being discussed. Among the four families, the Coronaviridae and Arteriviridae mainly infect mammalian hosts while the Mesoniviridae infect insects and the Roniviridae infect crustaceans. Although nidovirus genomes range in size from ~12 to ~30 kb, they share similarities in gene organization (two large, overlapping, nonstructural protein ORFs encoded in the 5′ two thirds and multiple structural protein ORFs encoded in the 3′ third) and produce a nested set of 3′ co-terminal subgenomic messenger RNAs (sg mRNA) in infected cells (Cong et al., 2017). Among the arteriviruses, simian hemorrhagic fever virus (SHFV) has the largest genome (15.7 kb) compared to those of equine arteritis virus (EAV), porcine reproductive and respiratory syndrome virus (PRRSV) and lactate dehydrogenase-elevating virus (LDV) (Snijder et al., 2013).
An early sucrose gradient sedimentation analysis of coronavirus murine hepatitis virus (MHV) viral RNA extracted from late harvest, sucrose gradient-purified virus detected several smaller, polyadenylated RNA fragments in addition to the full length genomic RNA (Lai and Stohlman, 1978). These RNAs were initially thought to be genomic degradation products but subsequent studies identified them as members of a nested set of six 3′ co-terminal, subgenomic mRNAs that also possessed a 5′ leader sequence, identical to that of the 5′ region of the genome (Lai et al., 1981). Equine arteritis virus (EAV), the prototype of the family Arteriviridae, was later found to also produce six sg mRNAs that were 3′ co-terminal and contained a common 5′ leader sequence (de Vries et al., 1990b, van Berlo et al., 1982, van Berlo et al., 1986). A subsequent study showed that although most coronavirions contained a single copy of genome RNA, a few virions also contained a sg mRNA, suggesting that sg mRNAs can be packaged but inefficiently (Zhao et al., 1993).
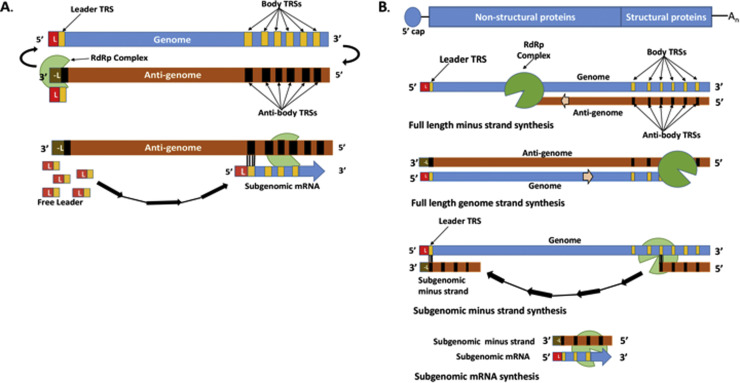
The synthesis of sg mRNAs during an infection is not exclusive to the Nidovirales. However, the use of a discontinuous transcription mechanism for their generation and the resulting 5–3′ co-terminal nested sg mRNAs produced are unique features of the coronaviruses and arteriviruses (Miller and Koev, 2000). Two models were proposed to explain the acquisition of the 5′ leader sequence by coronavirus sg mRNAs. The initial model, a leader-primed transcription model (Lai, 1986) suggested that discontinuous transcription occurred during the synthesis of plus strand RNA with a single, full-length, minus strand anti-genome RNA serving as the template for synthesis of nascent genomic and sg mRNAs ( Fig. 1A). It was hypothesized that short leader sequences synthesized from the 3′ region of the anti-genome by the viral RNA dependent RNA polymerase served as primers to initiate RNA synthesis at multiple complementary intergenic sequences (internal promoters later called anti-transcription regulating sequences) located within the 5′ region of the anti-genomic template. Elongation from these primers produced a set of 5–3′ co-terminal sg mRNAs (Lai, 1986).
Fig. 1.
Diagrams of models of nidovirus sg RNA generation. A. Leader-primed transcription model. Short leader fragments synthesized from the 3′ region of the anti-genomic RNA function as primers for the synthesis of the sg mRNAs at multiple complementary anti-TRSs in the 5′ region of the anti-genome. B. Discontinuous minus strand synthesis model. Minus strand synthesis from the genomic RNA template terminates at various genomic body TRSs and after the nascent minus strand anneals to the complementary genomic leader TRS, minus strand sg RNA synthesis is completed. The minus sg RNAs are then used as templates for sg mRNA synthesis.
An alternate model later proposed suggested that the discontinuous step occurred during the synthesis of minus strand RNA from the genome (Sawicki and Sawicki, 1990, Sethna et al., 1989). The discovery of multiple subgenomic length minus strand RNAs as well as of multiple sg RNA replicative intermediates in cells infected with the coronaviruses, porcine transmissible gastroenteritis virus (TGEV) or MHV, provided evidence supporting a discontinuous minus strand synthesis model (Sawicki and Sawicki, 1990, Sethna et al., 1989) (Fig. 1B). According to this model, full-length genomic minus strand RNAs as well as a 5′ nested set of subgenomic length minus strand RNAs that have a common 3′ anti-leader sequence are first synthesized from the genomic RNA and then serve as templates for genomic and sg mRNA synthesis, respectively. Subgenomic replicative intermediate and replicative form viral RNAs were subsequently identified in cells infected with the arterivirus EAV indicating that the same mechanism of sg RNA production is also used by a member of the family Arteriviridae (den Boon et al., 1996). Body transcription regulating sequences (TRSs) in the 3′ region of the genome RNA are located at each of the premature termination sites for sg RNAs during minus strand RNA synthesis and were identified by amplifying and sequencing the leader-body TRS junction regions in the sg mRNAs produced. Each junction region consists of a 3′ sequence fused to the 5′ leader (van Marle et al., 1999a ). In the EAV genome, the single 5' leader TRS 6 nt core sequence was shown to be located on the loop of a stem loop structure with two 10–15 nt flanking sequences forming the stem (Van Den Born et al., 2004, van den Born et al., 2005). Whether the leader regions of other arteriviruses form similar structures is not known. However, the leader TRS sequence of multiple coronavirus genomes has been shown to be located in the loop of a short RNA stem loop structure (Masters and Perlman, 2013) (Masters, 2013) and a cis-acting 5′ proximal enhancer element was reported to be located in the leader region of the MHV genome (Wang and Zhang, 2000). The results of a study using a transfected in vitro synthesized sg mRNA indicated that long sg mRNAs which contain multiple TRSs can also function as templates for the synthesis of prematurely terminated minus strand RNAs (Wu and Brian, 2010).
The subsequent identification of multiple 5′-3′ co-terminal sg mRNAs for additional coronavirus and arteriviruses led to the adoption of the view that all Nidoviruses utilize the same discontinuous minus strand synthesis strategy to produce a nested set of 3′ co-terminal sg mRNAs with a common 5′ leader in infected cells. However, data from studies on equine torovirus (EToV) (previously called Berne virus) and gill-associated virus (GAV), showed that not all nidoviruses produce 3′ co-terminal sg mRNAs with a common 5′ leader (Pasternak et al., 2006). EToV, produces a 3′ nested set of four sg mRNAs, three of which (RNA 3, 4, and 5) do not possess a 5′ leader sequence while RNA 2, the longest EToV sg mRNA, has a short 5′ leader sequence (Smits et al., 2005, van Vliet et al., 2002). These findings suggest that EToV uses both discontinuous and non-discontinuous RNA synthesis mechanisms to produce its sg mRNAs. A study on GAV, a crustacean virus in the family Roniviridae, identified two 3′ co-terminal sg mRNAs of different lengths that are initiated from the same intergenic sequence site. Neither of these sg mRNAs possesses a 5′ leader sequence (Cowley et al., 2002).
2. Identification of functional alternative TRSs for known structural proteins
The TRSs for individual coronavirus and arterivirus sg mRNAs were initially identified by RT-PCR amplification and sequencing of leader-body junction regions of the sg mRNAs produced in infected cells. Typically a single major TRS was identified for each sg mRNA and it was assumed that the majority of the sg mRNA generated for each structural gene was regulated by the major TRS. However, two alternative body TRSs (also known as non-canonical TRSs) for sg mRNA3 were identified for EAV by amplification, cloning and sequencing of sg mRNA leader-body junctions (den Boon et al., 1996). A later EAV study not only confirmed the use of alternative TRSs for sg mRNA3, but also identified one alternative TRS for sg mRNA4 and one for sg mRNA5 (Pasternak et al., 2000). One alternative TRS was also identified for sg mRNA4 and one for sg mRNA7 of the PRRSV strain VR2332 (Nelsen et al., 1999). A different alternative TRS for sg mRNA7 was later identified in the PRRSV strain tw91 (Lin et al., 2002). Mutation of the major TRS for sg mRNA4 in the EAV genome showed that the sg mRNAs produced from alternative TRSs although less abundant were still sufficient to generate infectious progeny virus but at a reduced titer, suggesting that alternative body TRSs can function as “back-ups” when the major TRS is not functional (Pasternak et al., 2000). Studies on coronaviruses have also shown that alternative body TRSs can function to produce sg mRNAs when the major body TRS of a 3′ region ORF is mutated (Ozdarendeli et al., 2001, Schelle et al., 2005, Zuniga et al., 2004).
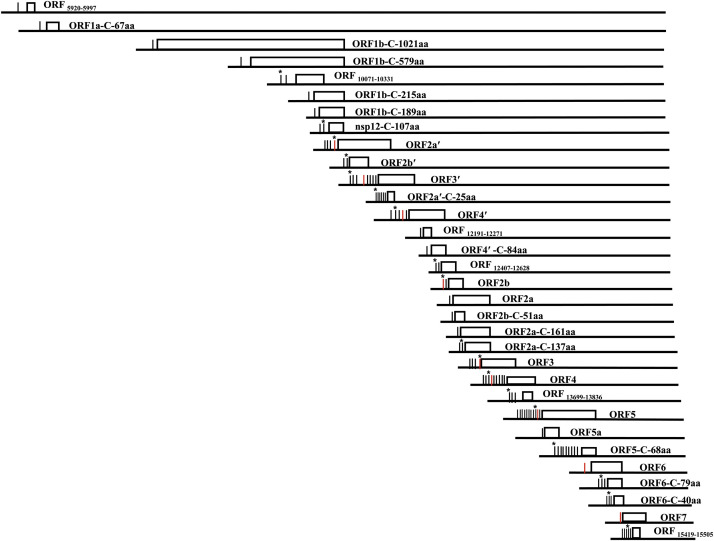
Although amplification, cloning and sequencing of sg mRNA leader-body junctions is an effective means of identifying TRSs that produce abundant sg mRNAs, it was less effective in identifying the TRSs that produce low abundance sg mRNAs due to the difficulty of detecting the RT-PCR product bands after electrophoresis on gels or to the limited number of clones screened. Although only one alternative EAV TRS was identified for sg mRNA4 and one for sg mRNA5, when both the known major and alternative TRSs for sg mRNA4 or sg mRNA5 were mutated, the mutant virus produced was still infectious and able to spread to neighboring cells, suggesting the possible existence of additional unidentified functional alternative TRSs producing sg mRNAs for each of these genes (Pasternak et al., 2000). Because NGS detects all sg mRNAs produced in RNA samples harvested from infected cells, including those at very low abundance, it can be used to identify all of the functional TRSs in a nidovirus genome and assess the relative abundance of each sg mRNA produced. The first NGS analysis of an arterivirus transcriptome identified a total of 96 functional body TRSs in the SHFV genome. Most of these were TRSs for alternative sg mRNAs of the SHFV structural proteins, with 8 identified for GP4 and 10 for GP5. (Di et al., 2017) ( Fig. 2). A mass spectrometry analysis showed that for the majority of the SHFV structural proteins, the relative protein levels produced for each ORF correlated well with the combined sg mRNA abundance produced from the major TRS and all of the alternative TRSs. These results suggested that instead of just being “back-ups” for the major TRS, the alternative body TRSs are an integral part of the viral expression strategy and essential for the production of the optimal amount of each viral protein (Di et al., 2017). Because similar NGS analyses have not yet been performed for other arteriviruses it is currently not known whether the SHFV genome is unique in having a high number of alternative TRSs for some of its ORFs or whether this is also a characteristic feature of other arteriviruses.
Fig. 2.
Diagram of the SHFV transcriptome. The ORFs are represented by white boxes. Identified functional TRSs are indicated by vertical lines to the left each ORF. Red vertical lines represent the nine major SHFV body TRSs published prior to the recent SHFV NGS study. For ORFs with multiple functional TRSs, an asterisk indicates the TRS that produces the highest abundance of sg mRNA for that ORF. (Published in Di et al., 2017. Proc Natl Acad Sci USA 114(42): E8895-E8904).
3. Identification of sg mRNAs for in-frame, C-terminal fragments of larger proteins
In addition to the alternative TRSs for PRRSV sg mRNAs for GP4 and N, a TRS located downstream of the AUG start codon of GP5 was identified in the PRRSV, strain VR2332, genome (Nelsen et al., 1999) Compared to sg mRNA5, this TRS generated a shorter sg mRNA (sg mRNA5-1) that is predicted to encode an in-frame, C-terminal peptide of GP5. A sg mRNA band migrating between sg mRNA5 and sg mRNA6 was also detected on SHFV Northern blots (Di et al., 2017, Vatter et al., 2014). Amplification, cloning and sequencing of the leader-body junctions revealed that this band was composed of seven different sg mRNAs of similar sizes that were generated from seven unique body TRSs located between TRS5 and TRS6. All seven of these sg mRNAs encode the same in-frame, C-terminal peptide of GP5 (Di et al., 2017) (Fig. 2). Sg mRNAs encoding an in-frame, C-terminal peptide from each of the SHFV structural proteins, GP2′, GP4′, GP2 and M, were also identified in infected cells (Di et al., 2017) (Fig. 2). Two previous studies on the coronaviruses, severe acute respiratory syndrome virus and infectious bronchitis virus, identified a novel sg mRNA encoding an in-frame, C-terminal peptide of the virion spike protein (Bentley et al., 2013, Hussain et al., 2005). Similarly, both a previous study and a recent NGS and ribosome profiling study of the coronavirus MHV detected translation initiation at an internal AUG within ORF5 (Irigoyen et al., 2016, Thiel and Siddell, 1994). Although both coronaviruses and arteriviruses appear to produce C-terminal fragments of larger viral proteins, the functions of these peptides are currently not known. Mutagenesis of the AUGs of five of the SHFV C-terminal peptides in an infectious clone provided preliminary evidence for the functional relevance of the GP5 and M C-terminal peptides. Both of these mutant viruses displayed a reduced viral yield and plaque size in cell culture (Di et al., 2017).
4. Identification of sg mRNAs for alternative frame proteins
Knowledge of the coding capacity of arteriviruses and coronaviruses has expanded over the years with the discovery of additional ORFs. Arteriviruses were initially thought to encode three minor structural proteins, GP2, GP3 and GP4 and three major structural proteins GP5, membrane protein (M) and N (Snijder et al., 1999). An early Northern blot analysis of SHFV sg mRNA production in infected MA104 cells identified six strong sg mRNA bands. Based on their sizes, these sg mRNAs were predicted to encode the six known arterivirus minor and major structural proteins (Zeng et al., 1995). An extra set of minor structural protein ORFs for GP2′, GP3′ and GP4′ was later shown to be encoded in the 3′ region of the SHFV genome (Smith et al., 1997). A subsequent sg mRNA leader-body junction region amplification, cloning and sequencing study identified body TRSs for GP2′ and GP4′ but not for GP3′ and it was proposed that GP3′ was also expressed from the sg mRNA encoding GP2′ (Godeny et al., 1998). A recent reanalysis of the SHFV sg mRNAs by Northern blotting using a more sensitive 5′ leader probe detected ten sg mRNA bands including a sg mRNA3′ for GP3′ (Vatter et al., 2014). Additional structural protein ORFs, E and ORF5a, were later discovered in the genomes of EAV and PRRSV that were predicted to be expressed bicistronically from the sg mRNAs for GP2 and GP5, respectively (Firth et al., 2011, Johnson et al., 2011, Snijder et al., 1999). Bicistronic expression for the GP2′ and E′ proteins and for the E and GP2 proteins of SHFV was also initially proposed. However, separate sg mRNAs for both E′ and GP2 were later identified in the NGS study of SHFV transcriptome (Di et al., 2017). Comparison of the relative sg mRNA and protein abundances suggested that the majority of E′ is expressed from the 5′ ORF of multiple E′ sg mRNAs. In contrast, the abundance of the sg mRNAs with GP2 as the 5′ ORF was much lower than the relative abundance of this protein suggesting that the majority of GP2 is translated as a second ORF from the E sg mRNAs. Some coronavirus proteins initially thought to be expressed only bicistronically from the second 5′ ORF of a longer sg mRNA were later shown to also be translated from the 5′ ORF of a different sg mRNA, for example, the mouse hepatitis virus (MHV) E protein (O'Connor and Brian, 2000, Zhang and Liu, 2000).
An additional alternative frame ORF overlapping ORF5 (GP5) was identified in a PRRSV genome and shown to express a protein designated ORF5a (Firth et al., 2011, Johnson et al., 2011). Although a separate sg mRNA encoding ORF5a as the 5′ proximal ORF was not identified in that study, the NGS study of the SHFV transcriptome identified a sg mRNA for ORF5a at very low abundance (Di et al., 2017). An early study of PRRSV found that four USA PRRSV isolates, including VR2385 and ISU79, produced an additional sg mRNA3-1 that encodes an alternative frame ORF (ORF3-1) that overlaps ORF3 (GP3) (Meng et al., 1996). The production of this protein has not yet been confirmed. A recent bioinformatic study suggested the existence of a small ORF7a protein encoded in the +2 reading frame inside ORF7 (N) of type I and type II PRRSV genomes (Olasz et al., 2016). The recent SHFV NGS study identified five functional body TRSs that each generated sg mRNAs encoding ORF15419–15505 in the +2 reading frame located inside SHFV ORF7 (N) (Fig. 1) (Di et al., 2017). The SHFV NGS study also identified sg mRNAs encoding five additional alternative frame ORFs (Fig. 1). The predicted sizes of the proteins that would be translated from these alternative frame ORFs are 86aa, 25aa, 28aa, 73aa, 26aa, and 45aa. The 86 aa protein expressed from the largest of these alternative frame ORFs (ORF10071–10331) was detected by mass spectrometry in infected MA104 extracts (Di et al., 2017). The low abundance of the sg mRNAs of each these proteins and the technical challenges of detecting small proteins from gels by MassSpec may be the reasons the rest of these small proteins were not detected. Studies on coronaviruses have also identified alternative frame proteins such as an alternative ORF located inside the N gene that encodes an accessary protein (Fischer et al., 1997, Liu et al., 2014, Meier et al., 2006). A recent analysis of coronavirus MHV protein ORF translation by ribosome profiling and NGS detected the translation of multiple previously unidentified small alternative frame ORFs (Irigoyen et al., 2016). Many of these MHV alternative frame ORFs were initiated from non-canonical start codons such as CUG, suggesting an even larger coding potential for Nidovirus genomes. The authors proposed that some of these small alternative frame ORFs may function as regulators of the translation of downstream ORFs in the same sg mRNA (Irigoyen et al., 2016). Alternatively, increasing evidence from multiple host species suggests that small, functional proteins, some as short as 11 aa, translated from in frame or alternative frame ORFs in cellular mRNAs are involved in regulating a variety of cellular processes (Landry et al., 2015, Pueyo et al., 2016).
A coronavirus was recently found to also produce short non-coding RNAs. Three 18–22 nt small noncoding viral RNAs (svRNAs) were identified in SARS-CoV infected cells that were derived from the nsp3 (svRNA-nsp3.1 and -nsp3.2) and N (svRNA-N) genomic regions (Morales et al., 2017). Although similar in size to miRNAs, the generation of these RNAs was found to be RNase III, cell type, and host species independent. The functional relevance of one of these RNAs was indicated by the observed reduction in lung pathology and pro-inflammatory cytokine expression when svRNA-N was inhibited with an antagomir.
5. Identification of sg mRNA production from the genomic 5′ nonstructural region
Although functional body TRSs were thought to be located only in the 3′ structural protein region of Nidovirus genomes, a previous study on the arterivirus LDV identified a functional body TRS in the nonstructural protein ORF1b region by amplifying, cloning and sequencing the leader-body junction of a long sg mRNA (Chen et al., 1993). This sg mRNA encoded an in-frame, C-terminal portion of ORF1b (200 aa) but translation of this truncated ORF1b polyprotein was not confirmed. A study on the coronavirus MHV identified three functional body TRSs in the ORF1a region by amplifying, cloning and sequencing the leader-body junctions (La Monica et al., 1992). The recent SHFV NGS study identified two functional body TRSs in ORF1a and eight in ORF1b that generated ten long sg mRNAs. Among these sg mRNAs, six encode different lengths of in-frame, C-terminal portions of ORF1b with the shortest one encoding only the C-terminus of nsp12 and the longest one encoding the C-terminus of nsp9 plus full-length nsp10, nsp11 and nsp12 (Fig. 2) (Di et al., 2017). ORF1b has been reported to be translated at a much lower efficiency (15–20%) than ORF1a due to the requirement of a -1 frameshift near the 3′ end of ORF1a for ORF1a/1b translation to occur (den Boon et al., 1991, Snijder et al., 2013). Analysis of SHFV protein production by mass spectrometry detected 3 times more nsp12 and 1.2 times more nsp11 than nsp8, which is encoded at the 3′ end of the ORF1a, suggesting that the long sg mRNAs produced from TRSs in the ORF1b region provide additional sources of nsp10, nsp11 and especially nsp12. However, the relative half-lives of the individual SHFV nonstructural proteins are not known and could also affect the relative abundances of these proteins. Previous studies on PRRSV detected the production of atypical sg mRNAs (designated heteroclite sg mRNAs) in the ORF1a region. These sg mRNAs differed from typical sg mRNAs in having their 3′ genomic sequences fused to various sites within ORF1a, rather than to a common 5′ leader (Yuan et al., 2000, Yuan et al., 2004). Production of heteroclite sg mRNAs has not been reported for other arteriviruses.
6. Relative abundance of individual sg mRNAs and junction sequence heterogeneity
Northern blotting studies of virus infected cell extracts suggested that although the relative abundances of the individual viral sg mRNAs detected differed, the abundance of each sg mRNA was consistent at different times after infection (Di et al., 2017). The recent SHFV NGS analysis allowed calculation of the relative abundance of each viral sg mRNA. The data indicated that the relative sg mRNA abundance produced from each identified body TRS, regardless of whether it is a major or alternative TRS, was consistent at early and later times after infection in both primary macrophages and MA104 cells, suggesting that sg mRNA production is primarily regulated by viral not cellular factors (Di et al., 2017).
Two previous PRRSV leader-body junction sequence analysis studies identified heterogeneous junction sequences (Lin et al., 2002, Meulenberg et al., 1993). The junction sequence of a sg mRNA is determined by the location within the particular body TRS where the polymerase disassociates and the location within the leader TRS where the nascent minus strand anneals. The SHFV NGS data indicated that about one-third of the total body TRSs identified, including both major and alternative TRSs, produced sg mRNAs with two or three heterogeneous junction sequences (Di et al., 2017). Heterogeneity in sg mRNA junction sequences produced from the same body TRS has also been detected for coronaviruses by amplification, cloning and sequencing the junction sequences or by NGS (Irigoyen et al., 2016, Makino et al., 1988, Schaad and Baric, 1993). The heterogeneity of sg mRNA junction sequences suggests that the fusion position between the leader and body TRSs can vary but the observation that the abundances of the sg mRNAs with each of the alternative junction sequences are consistent suggests that this variation is controlled by the characteristics of the viral elements regulating discontinuous minus strand synthesis at these sites.
7. Possible modes of sg mRNA regulation
The means by which discontinuous RNA synthesis is regulated by Nidoviruses is not well understood. Previous studies on both coronaviruses and arteriviruses have suggested that shorter sg mRNAs generated from TRSs located closer to the 3′ end of the genome are usually in higher abundance (de Vries et al., 1990a, Krishnan et al., 1996, Pasternak et al., 2004). Studies of two closely located body TRSs showed that the downstream TRS could suppress the function of the upstream one (Joo and Makino, 1995, Pasternak et al., 2004). However, data from the SHFV NGS study clearly showed that across the genome there is not a linear correlation between the distance of a TRS from the 3′ end of the genome and the corresponding sg mRNA abundance produced from that TRS. Instead, multiple genomic regions containing TRSs regulating high levels of discontinuous minus strand synthesis were flanked by regions containing TRSs regulating low levels of discontinuous synthesis (Di et al., 2017). Previous studies also observed that when mutations in a body TRS increased the duplex stability between the leader and body TRS sequences, the corresponding sg mRNA abundance increased with a few exceptions (Pasternak et al., 2001, Pasternak et al., 2003, Sola et al., 2005, Zuniga et al., 2004). However, some sequence motifs were found in the viral genomes with 100% similarity to the leader TRS core sequence that were not utilized as TRSs, suggesting that duplex stability is necessary but not sufficient for regulating discontinuous RNA synthesis. The duplex stability between the leader TRS and each of the seven closely located functional body TRSs for SHFV ORF5C was calculated and found not to show a linear correlation with the corresponding sg mRNA abundance (Di et al., 2017). These findings support previous suggestions that the local RNA secondary structure of each body TRS, which may be dynamic, as well as long distance genomic RNA-RNA interactions play important roles in regulating discontinuous RNA synthesis (Mateos-Gomez et al., 2013, Moreno et al., 2008, Pasternak et al., 2000, Sola et al., 2005).
EAV nsp1 plays an essential role in sg RNA generation but little is known about how protein-protein and protein-RNA interactions regulate sg RNA synthesis (Tijms et al., 2007). It was suggested that the interaction of nsp1 with a particular body TRS could stall a viral polymerase complex copying minus RNA from the genome at that site and that nsp1 may also facilitate targeting and base pairing between the complementary body TRS and the leader TRS. The EAV nsp1 consists of the regions equivalent to nsp1α and nsp1β of other arteriviruses due to mutation of the nsp1α cleavage site. The zinc finger located in the nsp1α region was shown to be critical for sg RNA production. However, a subsequent study found that viral sg mRNA abundance was regulated by an intricate interaction network involving all of the EAV nsp1 subdomains (Nedialkova et al., 2010). EAV nsp10 was also shown to play an important role in sg RNA synthesis (van Marle et al., 1999b).
8. Conclusions
Following the initial discovery that coronaviruses produce a nested set of 3′ co-terminal sg mRNAs with a common 5′ leader sequence, the main emphasis of research was focused on determining the mechanism by which these sg mRNAs were generated. Minus strand sg RNAs were shown to be generated from the genomic template by a discontinuous minus strand synthesis mechanism. These sg RNAs then function as templates for sg mRNA synthesis. The amplification, cloning and sequencing of the leader-body junction sequences of the sg mRNAs produced for each viral gene typically identified a single major TRS that had sequence homology to the leader TRS. However, the detection of alternative regulatory sequences mediating discontinuous RNA synthesis (heteroclite sg mRNAs) and of 3′ co-terminal sg mRNAs without a 5′ leader for some nidoviruses indicates the use of additional mechanisms for the generation of sg mRNAs among viruses in this order. Recent studies employing NGS and ribosome profiling have provided a greatly expanded view of the transcriptomes and coding capacities of both an arterivirus and a coronavirus. The finding that an arterivirus and a coronavirus may encode a number of small previously unidentified proteins opens new avenues for investigation of the cell-virus interaction.
Acknowledgements
Support for the work described was provided by Public Health Service research grant AI073824 to M. Brinton and by a Georgia State University Molecular Basis of Disease seed grant to H. Di. H. Di was also supported by a Georgia State University Molecular Basis of Disease fellowship.
References
- Bentley K., Keep S.M., Armesto M., Britton P. Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J. Virol. 2013;87:2128–2136. doi: 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Kuo L., Rowland R.R., Even C., Faaberg K.S., Plagemann P.G. Sequences of 3' end of genome and of 5' end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5' leader and bodies of seven subgenomic mRNAs. J. General. Virol. 1993;74(Pt 4):643–659. doi: 10.1099/0022-1317-74-4-643. [DOI] [PubMed] [Google Scholar]
- Cong Y., Verlhac P., Reggiori F. The Interaction between Nidovirales and autophagy components. Viruses. 2017:9. doi: 10.3390/v9070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley J.A., Dimmock C.M., Walker P.J. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3'-coterminal subgenomic mRNAs that do not possess 5'-leader sequences. J. Gen. Virol. 2002;83:927–935. doi: 10.1099/0022-1317-83-4-927. [DOI] [PubMed] [Google Scholar]
- de Vries A.A., Chirnside E.D., Bredenbeek P.J., Gravestein L.A., Horzinek M.C., Spaan W.J. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries A.A., Chirnside E.D., Bredenbeek P.J., Gravestein L.A., Horzinek M.C., Spaan W.J. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Kleijnen M.F., Spaan W.J., Snijder E.J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J. Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Snijder E.J., Chirnside E.D., de Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronavirus like superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di H., Madden J.C., Jr., Morantz E.K., Tang H.Y., Graham R.L., Baric R.S., Brinton M.A. Expanded subgenomic mRNA transcriptome and coding capacity of a nidovirus. Proc. Natl. Acad. Sci. USA. 2017;114 doi: 10.1073/pnas.1706696114. (E8895-e8904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Zevenhoven-Dobbe J.C., Wills N.M., Go Y.Y., Balasuriya U.B.R., Atkins J.F., Snijder E.J., Posthuma C.C. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J. General. Virol. 2011;92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Peng D., Hingley S.T., Weiss S.R., Masters P.S. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 1997;71:996–1003. doi: 10.1128/jvi.71.2.996-1003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeny E.K., de Vries A.A., Wang X.C., Smith S.L., de Groot R.J. Identification of the leader-body junctions for the viral subgenomic mRNAs and organization of the simian hemorrhagic fever virus genome: evidence for gene duplication during arterivirus evolution. J. Virol. 1998;72:862–867. doi: 10.1128/jvi.72.1.862-867.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., Wu Y., Li Z., Zhu Y., Tien P., Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N., Firth A.E., Jones J.D., Chung B.Y., Siddell S.G., Brierley I. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12:e1005473. doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.R., Griggs T.F., Gnanandarajah J., Murtaugh M.P. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J. Gen. Virol. 2011;92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., Makino S. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 1995;69:272–280. doi: 10.1128/jvi.69.1.272-280.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R., Chang R.Y., Brian D.A. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology. 1996;218:400–405. doi: 10.1006/viro.1996.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Monica N., Yokomori K., Lai M.M. Coronavirus mRNA synthesis: identification of novel transcription initiation signals which are differentially regulated by different leader sequences. Virology. 1992;188:402–407. doi: 10.1016/0042-6822(92)90774-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M. Coronavirus leader-RNA-primed transcription: an alternative mechanism to RNA splicing. BioEssays : News Rev. Mol., Cell. Dev. Biol. 1986;5:257–260. doi: 10.1002/bies.950050606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Brayton P.R., Armen R.C., Patton C.D., Pugh C., Stohlman S.A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J. Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Stohlman S.A. RNA of mouse hepatitis virus. J. Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C.R., Zhong X., Nielly-Thibault L., Roucou X. Found in translation: functions and evolution of a recently discovered alternative proteome. Curr. Opin. Struct. Biol. 2015;32:74–80. doi: 10.1016/j.sbi.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Chang R.Y., Chueh L.L. Leader-body junction sequence of the viral subgenomic mRNAs of porcine reproductive and respiratory syndrome virus isolated in Taiwan. J. Vet. Med Sci. 2002;64:961–965. doi: 10.1292/jvms.64.961. [DOI] [PubMed] [Google Scholar]
- Liu D.X., Fung T.S., Chong K.K., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Soe L.H., Shieh C.K., Lai M.M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J. Virol. 1988;62:3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology Sixth Edition. 2013. pp. 825–858. [Google Scholar]
- Mateos-Gomez P.A., Morales L., Zuniga S., Enjuanes L., Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013;87:177–186. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.J., Paul P.S., Morozov I., Halbur P.G. A nested set of six or seven subgenomic mRNAs is formed in cells infected with different isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1996;77(Pt 6):1265–1270. doi: 10.1099/0022-1317-77-6-1265. [DOI] [PubMed] [Google Scholar]
- Meulenberg J.J., de Meijer E.J., Moormann R.J. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J. Gen. Virol. 1993;74(Pt 8):1697–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- Miller W.A., Koev G. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology. 2000;273:1–8. doi: 10.1006/viro.2000.0421. [DOI] [PubMed] [Google Scholar]
- Morales L., Oliveros J.C., Fernandez-Delgado R., tenOever B.R., Enjuanes L., Sola I. SARS-CoV-encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe. 2017;21:344–355. doi: 10.1016/j.chom.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.L., Zuniga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., Gorbalenya A.E., Snijder E. Aterivirus nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLOS Pathog. 2010;6:31000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen C.J., Murtaugh M.P., Faaberg K.S. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 1999;73:270–280. doi: 10.1128/jvi.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J.B., Brian D.A. Downstream ribosomal entry for translation of coronavirus TGEV gene 3b. Virology. 2000;269:172–182. doi: 10.1006/viro.2000.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olasz F., Denes B., Balint A., Magyar T., Belak S., Zadori Z. Immunological and biochemical characterisation of 7ap, a short protein translated from an alternative frame of ORF7 of PRRSV. Acta Vet. Hung. 2016;64:273–287. doi: 10.1556/004.2016.027. [DOI] [PubMed] [Google Scholar]
- Ozdarendeli A., Ku S., Rochat S., Williams G.D., Senanayake S.D., Brian D.A. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 2001;75:7362–7374. doi: 10.1128/JVI.75.16.7362-7374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Gultyaev A.P., Spaan W.J., Snijder E.J. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 2000;74:11642–11653. doi: 10.1128/jvi.74.24.11642-11653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol. 2004;78:8102–8113. doi: 10.1128/JVI.78.15.8102-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., Spaan W.J., Snijder E.J. Nidovirus transcription: how to make sense...? J. Gen. Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. The stability of the duplex between sense and antisense transcription-reguating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 2003;77:1175–1183. doi: 10.1128/JVI.77.2.1175-1183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueyo J.I., Magny E.G., Couso J.P. New peptides under the s(ORF)ace of the genome. Trends Biochem. Sci. 2016;41:665–678. doi: 10.1016/j.tibs.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad M.C., Baric R.S. Evidence for new transcriptional units encoded at the 3' end of the mouse hepatitis virus genome. Virology. 1993;196:190–198. doi: 10.1006/viro.1993.1467. [DOI] [PubMed] [Google Scholar]
- Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P.B., Hung S.L., Brian D.A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.L., Wang X., Godeny E.K. Sequence of the 3' end of the simian hemorrhagic fever virus genome. Gene. 1997;191:205–210. doi: 10.1016/S0378-1119(97)00061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., van Vliet A.L., Segeren K., el Azzouzi H., van Essen M., de Groot R.J. Torovirus non-discontinuous transcription: mutational analysis of a subgenomic mRNA promoter. J. Virol. 2005;79:8275–8281. doi: 10.1128/JVI.79.13.8275-8281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., van Tol H., Pedersen K.W., Raamsman M.J., de Vries A.A. Identification of a novel structural protein of arteriviruses. J. Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Moreno J.L., Zuniga S., Alonso S., Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Internal ribosome entry in the coding region of murine hepatitis virus mRNA 5. J. Gen. Virol. 1994;75(Pt 11):3041–3046. doi: 10.1099/0022-1317-75-11-3041. [DOI] [PubMed] [Google Scholar]
- Tijms M.A., Nedialkova D.D., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Snijder E.J. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 2007;81:10496–10505. doi: 10.1128/JVI.00683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo M.F., Horzinek M.C., van der Zeijst B.A. Equine arteritis virus-infected cells contain six polyadenylated virus-specific RNAs. Virology. 1982;118:345–352. doi: 10.1016/0042-6822(82)90354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo M.F., Rottier P.J., Horzinek M.C., van der Zeijst B.A. Intracellular equine arteritis virus (EAV)-specific RNAs contain common sequences. Virology. 1986;152:492–496. doi: 10.1016/0042-6822(86)90154-6. [DOI] [PubMed] [Google Scholar]
- Van Den Born E., Gultyaev A.P., Snijder E.J. Secondary structure and function of the 5'-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. (New York, N.Y.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E., Posthuma C.C., Gultyaev A.P., Snijder E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., van Dinten L.C., Spaan W.J., Luytjes W., Snijder E.J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A.L., Smits S.L., Rottier P.J., de Groot R.J. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatter H.A., Di H., Donaldson E.F., Baric R.S., Brinton M.A. Each of the eight simian hemorrhagic fever virus minor structural proteins is functionally important. Virology. 2014;462–463C:351–362. doi: 10.1016/j.virol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang X. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 2000;74:10571–10580. doi: 10.1128/jvi.74.22.10571-10580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Brian D.A. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl. Acad. Sci. USA. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Murtaugh M.P., Faaberg K.S. Heteroclite subgenomic RNAs are produced in porcine reproductive and respiratory syndrome virus infection. Virology. 2000;275:158–169. doi: 10.1006/viro.2000.0639. [DOI] [PubMed] [Google Scholar]
- Yuan S., Murtaugh M.P., Schumann F.A., Mickelson D., Faaberg K.S. Characterization of heteroclite subgenomic RNAs associated with PRRSV infection. Virus Res. 2004;105:75–87. doi: 10.1016/j.virusres.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Zeng L., Godeny E.K., Methven S.L., Brinton M.A. Analysis of simian hemorrhagic fever virus (SHFV) subgenomic RNAs, junction sequences, and 5' leader. Virology. 1995;207:543–548. doi: 10.1006/viro.1995.1114. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu R. Identification of a noncanonical signal for transcription of a novel subgenomic mRNA of mouse hepatitis virus: implication for the mechanism of coronavirus RNA transcription. Virology. 2000;278:75–85. doi: 10.1006/viro.2000.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Shaw K., Cavanagh D. Presence of subgenomic mRNAs in virions of coronavirus IBV. Virology. 1993;196:172–178. doi: 10.1006/viro.1993.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkel F., Roth H., Kurth A., Drosten C., Ziebuhr J., Junglen S. Identification and characterization of genetically divergent members of the newly established family Mesoniviridae. J. Virol. 2013;87:6346–6358. doi: 10.1128/JVI.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Alonso S., Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]